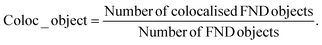Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Not all cells are created equal – endosomal escape in fluorescent nanodiamonds in different cells†
Yue
Zhang
a,
Rokshana
Sharmin
a,
Alina
Sigaeva
 a,
Carline W. M.
Klijn
a,
Aldona
Mzyk
ab and
Romana
Schirhagl
a,
Carline W. M.
Klijn
a,
Aldona
Mzyk
ab and
Romana
Schirhagl
 *a
*a
aUniversity of Groningen, University Medical Center Groningen, Antonius Deusinglaan 1, 9713 AW Groningen, Netherlands. E-mail: romana.schirhagl@gmail.com
bInstitute of Metallurgy and Materials Science, Polish Academy of Sciences, Reymonta St. 25, 30-059 Krakow, Poland
First published on 7th July 2021
Abstract
Successful delivery of fluorescent nanodiamonds (FNDs) into the cytoplasm is essential to many biological applications. Other applications require FNDs to stay within the endosomes. The diversity of cellular uptake of FNDs and following endosomal escape are less explored. In this article, we quantify particle uptake at a single cell level. We report that FNDs enter into the cells gradually. The number of internalized FNDs per cell differs significantly for the cell lines we investigated at the same incubation time. In HeLa cells we do not see any significant endosomal escape. We also found a wide distribution of FND endosomal escape efficiency within the same cell type. However, compared with HeLa cells, FNDs in HUVECs can easily escape from the endosomes and less than 25% FNDs remained in the vesicles after 4 h incubation time. We believe this work can bring more attention to the diversity of the cells and provide potential guidelines for future studies.
Introduction
Fluorescent nanodiamonds have gained popularity in the biomedical fields over the past decades. They have for instance been proven useful in the drug delivery field.1,2 Here their rich surface chemistry offers possibilities for chemists to attach drugs and engineer their properties.3,4 They are infinitely photostable and do not bleach.5–7 Additionally, their excellent biocompatibility was demonstrated in many different cell types and even in vivo.8–11 Thus they are promising fluorescent labels, especially when long term labeling is required.12,13 Nanodiamonds are also well visible with several different imaging techniques, which makes them interesting for correlative microscopy.14More recently also their unusual magnetic sensing abilities have proven to be useful.15 FNDs contain fluorescent defects, which change their optical properties based on their magnetic surrounding.16 Since optical signals are easier to detect than magnetic signals, this technique is unprecedentedly sensitive and allows nanoscale magnetic resonance measurements.17,18 This effect was first utilised by physicists for nanoscale sensing of magnetic proteins,19 magnetic structures like hard drives,20 domain walls21 or nanoparticles.22 Recently, this method was applied in cell biology. There nanodiamonds have been utilised for nanoscale temperature measurements23,24 or to measure free radical generation during stress responses.25
For all these applications it is crucial to determine where exactly the nanodiamonds end up. There are a lot of articles, which report on nanodiamond uptake in many different types of cells.26–29 Unless nanodiamonds are injected or otherwise forced into the cells,14,30 the most common way to enter the cells is endocytosis.31 More specifically, Liu et al. have shown that they enter via so-called clathrin mediated endocytosis.32 This pathway is a complex energy dependent endocytic portal into cells through which cargo is packaged into vesicles with the aid of a clathrin coat (and other compounds).33 If particles are following this uptake route they usually end up in early endosomes multivesicular bodies (MVBs) and finally in lysosomes. Vaijayanthimala et al. have also shown clathrin mediated endocytosis in HeLa cells. This was achieved by blocking different pathways specifically while monitoring changes in uptake.26 Sigaeva et al. have observed evidence for macropinocytosis in HT29 cells (a colon cancer cell line).34 However, we know much less about what happens after they have entered. Chu et al. have shown that particles escape the endosomes in HepG2 cells (a liver carcinoma cell line).35,36 We show here that there is a large variability between different cell types. As illustrated in Fig. 1, we systematically compare uptake in different cell types and determine dynamics and variability in the uptake process.
Materials and methods
Reagents and materials
Oxygen terminated red fluorescent nitrogen-vacancy nanodiamonds (FNDs, Adamas Nanotechnology) were used in this study. The average particle size of FNDs in the stock solutions (1 mg mL−1 in deionised water) is 70 nm. According to the vendor, these FNDs were produced by a high-pressure, high temperature (HPHT) process followed by size separation and acid cleaning. Calcein, paraformaldehyde, gelatin, Triton X-100, Bovine Serum Albumin (BSA), FITC-phalloidin, 4′,6-diamidino-2-phenylindole (DAPI) and HEPES were purchased from Sigma Aldrich, Dulbecco's Modified Eagle's Medium supplemented with 4.5 mg mL−1 glucose (DMEM-HG), fetal bovine serum (FBS), penicillin/streptomycin (P/S) and trypsin/EDTA from Gibco Life Technologies. Microvascular Endothelial Cell Growth Medium (EGM™-2MV) is commercially available from Lonza, (CC-3202, Switzerland) and glutaraldehyde from Merck. Phosphate-buffered saline (PBS) was purchased from Invitrogen.Preparation of FNDs (FBS coated)
Following a previous report, FND stock solutions were diluted in pure FBS to prevent the formation of large FND clusters in the cell culture medium.37 FBS-coated FNDs were further mixed with serum-free medium to get the desired FND concentration and 10% FBS. FND in the following text refers to FBS-coated FNDs unless specified differently.FNDs were prepared similarly for endosomal escape experiments in complete cell culture medium, supplemented with 0.25 mM calcein.
Characterization of FNDs
The average hydrodynamic diameter of FNDs was determined by dynamic light scattering (DLS) at 25 °C. The tests were performed by Zetasizer Nano ZS ZEN3500 (Malvern) equipped with a 633 nm He–Ne laser using back-scattering detection. The material is widely used in literature and its size, shape and surface chemistry are well known from literature.38,39Cellular toxicity of FNDs
HeLa cells (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) or HUVECs (60
000 cells per well) or HUVECs (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2) were seeded in 96-well plates and incubated for 24 h until the confluency reached 70–90%. Then cells were incubated with 10 or 2.5 μg mL−1 FNDs for 24 h. Then, 20 μL MTT solution (0.75 μg mL−1 final concentration) was added to each well. The plates were further incubated at 37 °C for 3 h. Finally, the media were replaced with 200 μL dimethyl sulfoxide (DMSO). The absorbance at OD = 590 nm was determined with a synergy H1 microplate reader (BioTek). Untreated cells were used as the control and DMSO was used as blank. The concentrations used in this article had no effects on metabolic activity (see ESI†).
000 cells per cm2) were seeded in 96-well plates and incubated for 24 h until the confluency reached 70–90%. Then cells were incubated with 10 or 2.5 μg mL−1 FNDs for 24 h. Then, 20 μL MTT solution (0.75 μg mL−1 final concentration) was added to each well. The plates were further incubated at 37 °C for 3 h. Finally, the media were replaced with 200 μL dimethyl sulfoxide (DMSO). The absorbance at OD = 590 nm was determined with a synergy H1 microplate reader (BioTek). Untreated cells were used as the control and DMSO was used as blank. The concentrations used in this article had no effects on metabolic activity (see ESI†).
Quantification of cellular uptake of FNDs
The quantification procedure was done in the same way as reported previously.34 In this study we chose to investigate HeLa cells as well as HUVECs because of their relevance in nanoparticle uptake studies. In addition they are also readily available and relatively easy to work with. HeLa cells were cultured in DMEM-HG medium, supplemented with 10% FBS and 1% P/S. 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded in each quarter of 35 mm four compartment glass-bottom Petri dishes (Greiner) and cultured in an incubator (37 °C and 5% CO2) for 24 h. When the confluency reached 70–90%, the cells were incubated with 10 μg mL−1 FNDs for 1 h, 2 h, 4 h, 8 h or 10 h in the incubator. After co-incubation, HeLa cells were rinsed twice with PBS, fixed with 3.7% PFA and stored at 4 °C. Cells cultured in complete DMEM-HG medium without FNDs were used as the control group.
000 cells were seeded in each quarter of 35 mm four compartment glass-bottom Petri dishes (Greiner) and cultured in an incubator (37 °C and 5% CO2) for 24 h. When the confluency reached 70–90%, the cells were incubated with 10 μg mL−1 FNDs for 1 h, 2 h, 4 h, 8 h or 10 h in the incubator. After co-incubation, HeLa cells were rinsed twice with PBS, fixed with 3.7% PFA and stored at 4 °C. Cells cultured in complete DMEM-HG medium without FNDs were used as the control group.
Human umbilical vein endothelial cells (HUVECs), obtained from Lonza (CC-2519, Switzerland), were cultured in EGM™-2 medium supplemented with EGM™-2 MV Single Quot Kit Supplements and Growth factors (Lonza). Cells were maintained at 37 °C with 5% CO2/95% air until passage 5 or 6. Meanwhile, a Petri dish was pretreated with 1% gelatin solution for 30 minutes. After that, the gelatin solution was replaced with 0.5% glutaraldehyde solution for 15 min to fix the coating. The Petri dish was washed three times with Ca2+ or Mg2+ free PBS and EGM™-2MV medium and kept in an incubator with 300 μL EGM™-2MV medium for 30 minutes before use. 35 × 103 cells per cm2 were seeded in the coated Petri dish and cultured in an incubator (37 °C and 5% CO2) for 24 hours. The following steps were the same as for HeLa cells with 2.5 μg mL−1 FNDs. Cells cultured in EGM™-2MV medium without FNDs were used as the control group.
After fixation, the actin cytoskeleton and nuclei of cells were stained with FITC-phalloidin and DAPI. The z-stack images were acquired using a Zeiss LSM780 confocal microscope with a 63× objective. The signals of FNDs were obtained using 561 nm excitation laser, and emission was collected between 670 nm and 740 nm (NV centers emit above 600 nm; however, collecting above 670 nm gives a better signal to noise ratio in presence of strong backgrounds). The signals of FITC–phalloidin were obtained using a 488 nm excitation laser and the emission was collected between 500 nm and 550 nm. The signal of DAPI was obtained using 405 nm excitation laser and the emission was collected between 420 nm and 480 nm. Z-Stack images were obtained with voxel sizes of 190 × 190 × 200 nm. To count diamond particles inside cells the FITC–phalloidin labelled volume was used to identify each cell. For each FND it was then checked whether it is within or outside the cell border.
Assessment of FNDs endosomal escape
To evaluate the endosomal escape efficiency of FNDs, 30.000 HeLa cells were seeded in each quarter of a glass-bottom Petri dish and incubated at 37 °C and 5% CO2 for 24 h. Then cells were incubated with 10 μg mL−1 FNDs in cell culture medium (containing 0.25 mM calcein) for 4 h in an incubator. After that, the cells were rinsed twice with PBS to remove free FNDs. For the 4 + 0 h groups, the cells were fixed immediately. For 4 + 4 h groups, the cells were further incubated another 4 h in the standard complete medium before the fixation. The cells were fixed with warm 3.7% PFA for 3 min and stored at 4 °C in the dark. The images were taken within 2 days after the cell fixation to keep high quality. The cells cultured in the complete medium (containing 0.25 mM calcein) were used as the control group. The cells cultured in normal complete medium were used as control group for image analysis. The same experimental design was also applied to HUVECs with 2.5 μg mL−1 FNDs. The FND concentrations for two different cell lines were chosen based on the previous experiences in our group, with the intention to have enough internalized distinguishable FND objects per cell for statistical analysis.The z-stack images were acquired using a Zeiss LSM780 confocal microscope with a 63× objective. The signals of FNDs were obtained using 561 nm excitation laser and emission was collected between 670 nm and 740 nm; the signals of calcein were obtained using 488 nm excitation laser and emission was collected between 510 nm and 540 nm. Z-Stack images were obtained with voxel sizes of 190 × 190 × 200 nm.
Image processing and data analysis
All confocal z-stack images were processed and analysed by the free FIJI software. The z-stack images were deconvolved by FIJI plugins “Diffraction PSF 3D” and “Iterative Deconvolve 3D” before the following process to achieve better signal to noise ratio.Similarly as reported before,34 the FND objects were identified by the FIJI plugin “3D Object Counter”.40 It's worth noting that these measurements only reveal an estimate rather than the precise amount of objects due to limitations of the conventional confocal resolution and FND brightness. However, it's still possible to compare numbers to reveal trends. The threshold is determined by the brightness of the control group (a threshold was selected that gives zero detected objects in absence of particles). In cellular uptake experiments, we used the FITC signals to define the cells’ area and the FND objects outside the cells or on the image edges were removed. More than 30 complete cells (Table S1†) from three independent experiments were randomly selected for the counting.
The colocalization analysis was performed on a single cell level. Every single cell was manually selected and separated as in Fig. S1.† At least 15 complete cells with more than 3 FND objects from three independent experiments were considered.
To assess the fraction of FNDs colocalized with endosomes at different timepoints, two methods were used. Manders’ Colocalization Coefficients (MCC) were calculated based on a pixel-level analysis. For two probes, denoted as R (Red, FNDs) and G (Green, Calcein), two different MCC values are derived, M1, the fraction of R in compartments containing G and M2, the fraction of G in compartments containing R.41 We are interested in the proportion of colocalized FNDs, which is described by M1 in this case. The M1 coefficient is simply calculated by JACoP as:
Another method is object-based colocalization. It is calculated as:
The number of total and colocalized FND objects were calculated based on centres of mass-particles coincidence by Fiji plugin “JACoP”.
Statistical analysis
Nonparametric one-way analysis of variance (Kruskal–Wallis test, one-way ANOVA) was performed to evaluate the statistical significance among the different groups in FND cellular uptake experiments. The multiple comparisons between each pair of groups were done by Dunn's test. The unpaired t-test was used to compare the colocalization results from the two timepoints. All tests were two sided with a significance level of 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). The statistical analysis was performed in GraphPad Prism 7.Results and discussion
Characterization of FNDs
Bare FNDs are relatively colloidally stable in H2O and easily aggregate in high salt solution or cell culture medium.42 Coating with FBS is a simple and effective way to prevent the formation of large FND clusters. The size distribution of FNDs in different media were measured by DLS. Fig. 2 shows that, compared to bare FNDs in H2O, the size of FNDs suspended in cell culture medium increased to around 150–200 nm. The slight increase might be from protein coating or small aggregation, but an increase in this size range is still acceptable for further experiments. No significant difference was found between the two different media.Quantifications of cellular uptake of FNDs
In a limited number of cases, it has been shown that FNDs enter the cells via energy-dependent, clathrin-mediated and receptor-mediated endocytosis.33 These processes can differ per cell type.43,44 Some immune cells like macrophages are more likely to ingest the particles while HT-29 cells are not.34 Besides, the shape and size of the cells vary even within the same cell line which is less explored.45 To have a certain amount of FNDs for the endosomal escape study and know more about the cells’ diversity, we investigated the relationship between the number of FNDs in the cells and the incubation time. Overall, compared with HeLa cells, HUVECs ingest more FNDs. As shown in Fig. 3, in both HeLa cells and HUVECs, the number of FND objects keeps increasing within 4 h incubation while a very low number of FNDs were found in the first 2 h. Therefore, we chose 4 h incubation for the following endosomal escape experiments. The number of FNDs inside the cells reaches a maximum after 8 h and 10 h. A possible reason is that the confluency of cells is much higher after long time incubation and might influence the cellular uptake of FNDs. Specifically, as shown in Fig. 3, particle numbers per cell vary significantly among different cells even after 10 h incubation. Even cells from the same type differ in their ability to engulf FNDs. Interestingly particle uptake differs for other nanoparticles and Torrano et al. for instance found that silica particles are taken up more by HeLa cells rather than HUVECs.46Assessment of FNDs endosomal escape
The calcein assay is a method to evaluate whether particles enter the cells and whether they can escape the endosome. Calcein is a cell membrane impermeant fluorescent dye and emits green light in endosomes or lysosomes. If a particle escapes the endosome, it no longer colocalizes with the green vesicles. First of all, from Fig. 4, we can easily find separated FND signals and green vesicles in the cells (representative 3D reconstruction in Fig. S2†). Then, we performed pixel and object based colocalization analysis. Both methods showed similar results. Fig. 5 confirms that also here there are large differences between individual cells. It's not surprising that the percentage of FNDs colocalized with endosomes in single cells has a wide range. There are several possible reasons for this, including cells being in different stages of their cell cycle or slight variations in the microenvironment. This is quite different from the whole image analysis which leads to relatively narrow distributed numbers in some papers.35,47 Normally, the result from one image is the average of several cells and these differences are averaged out. In contrast, the single cell analysis provides more detailed information, but also brings more uncertainty. Nevertheless, we observed considerable differences in our two cell types. FNDs in the HUVECs can escape from the endosomes more easily within the extra 4 h incubation. At 4 + 4 h, the FND colocalization in HUVECs is below 25% and significantly different from the previous timepoint. The situation in HeLa cells is different. Although the FND colocalization decreased slightly after 4 + 4 h incubation, there are no statistically significant differences between the two timepoints. FNDs endosomal escape is complex and difficult to evaluate precisely. Han et al. reported that FND clusters appeared outside the HeLa's membrane as well as in endosomal vesicles as detected by correlative light-electron microscopy (CLEM).48–50 Chu et al. found that prickly FNDs were able to rupture the membranes in HepG2 cells and easily escape from the endosomes.35 Overall, we have shown that the cell type influences the efficiency of the endosomal escape. Here, we show that calcein is useful to evaluate this process on a single cell level. A possible reason might be in the biological function of the two cell types. HUVECS are endothelial cells from veins of the umbilical cord while HeLa cells originated in the cervix. Thus, the two cell lines are specialised to receive their nutrients from a very different environment which might alter their uptake behaviour. A reason for these discrepancies in endosomal escape between different cells could also be caused by the formation of a protein corona. Unless it is specifically prevented,51 this corona formation always happens and has also been reported for nanodiamonds.36,52–55 Depending on the cell type different proteins might be treated in a different way. Another explanation could be that the two cell types use a different uptake mechanism. Besides clathrin mediated endocytosis there are many other mechanisms which can be differentiated by specific blocking.26 Yet another explanation could be that their endosomes are just more or less permeable due to differences in the lipid composition of their endosomes.56Conclusions
It has been reported already earlier that there are large differences between different cell types in their ability to ingest nanodiamond particles. We confirm this also for the cells we have investigated. We also performed a single cell analysis which reveals large differences between cells from the same cell type. Similarly, the endosomal escape ratio differs greatly between the two investigated cell types. While in HUVECs less than 25% remain in the endosomes within 4 h, in HeLa cells up to 50% remain in the endosomes within the same time period. Being aware of the differences between cells is essential for labelling and sensing applications. Furthermore, our data can be used as a guideline to select the best cell types to investigate processes in specific organelles or to adjust the timing and conditions of FND internalization.Author contributions
A. Mzyk and Y. Zhang designed the study under the supervision of R. Shirhagl. Y. Zhang, R. Sharmin and A. Mzyk conceived the study and acquired data. Y. Zhang, A. Sigaeva and C. W. M. Klijn analysed the data. Y. Zhang, R. Sharmin and R. Shirhagl wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Y. Zhang is thankful for the financial support via her CSC scholarship (No. 201908320456). R. Sharmin acknowledges financial support from the Bangabandhu Fellowship Trust. R. Schirhagl acknowledges the financial support from the European Commission via the ERC starting grant 714289 - Stress Imaging.Notes and references
- Y. Li, X. Zhou, D. Wang, B. Yang and P. Yang, J. Mater. Chem., 2011, 21(41), 16406–16412 RSC.
- X. Wang, X. C. Low, W. Hou, L. N. Abdullah, T. B. Toh, M. Mohd Abdul Rashid, D. Ho and E. K. H. Chow, ACS Nano, 2014, 8(12), 12151–12166 CrossRef CAS PubMed.
- X. Q. Zhang, R. Lam, X. Xu, E. K. Chow, H. J. Kim and D. Ho, Adv. Mater., 2011, 23(41), 4770–4775 CrossRef CAS PubMed.
- X. Q. Zhang, M. Chen, R. Lam, X. Xu, E. Osawa and D. Ho, ACS Nano, 2009, 3(9), 2609–2616 CrossRef CAS PubMed.
- V. Vaijayanthimala, P. Y. Cheng, S. H. Yeh, K. K. Liu, C. H. Hsiao, J. I. Chao and H. C. Chang, Biomaterials, 2012, 33(31), 7794–7802 CrossRef CAS PubMed.
- O. Faklaris, V. Joshi, T. Irinopoulou, P. Tauc, M. Sennour, H. Girard, C. Gesset, J. C. Arnault, A. Thorel, J. P. Boudou and P. A. Curmi, ACS Nano, 2009, 3(12), 3955–3962 CrossRef CAS PubMed.
- C. Y. Fang, V. Vaijayanthimala, C. A. Cheng, S. H. Yeh, C. F. Chang, C. L. Li and H. C. Chang, Small, 2011, 7(23), 3363–3370 CrossRef CAS PubMed.
- X. Zhang, W. Hu, J. Li, L. Tao and Y. Wei, Toxicol. Res., 2012, 1(1), 62–68 CrossRef CAS.
- S. R. Hemelaar, B. Saspaanithy, S. R. L'Hommelet, F. P. Perona Martinez, K. J. Van der Laan and R. Schirhagl, Sensors, 2018, 18(2), 355 CrossRef PubMed.
- T. C. Hsu, K. K. Liu, H. C. Chang, E. Hwang and J. I. Chao, Sci. Rep., 2014, 4, 5004 CrossRef CAS PubMed.
- K. van der Laan, M. Hasani, T. Zheng and R. Schirhagl, Small, 2018, 14(19), 1703838 CrossRef PubMed.
- S. Haziza, N. Mohan, Y. Loe-Mie, A. M. Lepagnol-Bestel, S. Massou, M. P. Adam, X. L. Le, J. Viard, C. Plancon, R. Daudin and P. Koebel, Nat. Nanotechnol., 2017, 12(4), 322–328 CrossRef CAS PubMed.
- B. M. Chang, H. H. Lin, L. J. Su, W. D. Lin, R. J. Lin, Y. K. Tzeng, R. T. Lee, Y. C. Lee, A. L. Yu and H. C. Chang, Adv. Funct. Mater., 2013, 23(46), 5737–5745 CrossRef CAS.
- S. R. Hemelaar, P. De Boer, M. Chipaux, W. Zuidema, T. Hamoh, F. P. Martinez, A. Nagl, J. P. Hoogenboom, B. N. G. Giepmans and R. Schirhagl, Sci. Rep., 2017, 7(1), 1–9 CrossRef PubMed.
- G. Balasubramanian, I. Y. Chan, R. Kolesov, M. Al-Hmoud, J. Tisler, C. Shin, C. Kim, A. Wojcik, P. R. Hemmer, A. Krueger and T. Hanke, Nature, 2008, 455(7213), 648–651 CrossRef CAS PubMed.
- R. Schirhagl, K. Chang, M. Loretz and C. L. Degen, Annu. Rev. Phys. Chem., 2014, 65, 83–105 CrossRef CAS PubMed.
- M. S. Grinolds, S. Hong, P. Maletinsky, L. Luan, M. D. Lukin, R. L. Walsworth and A. Yacoby, Nat. Phys., 2013, 9(4), 215–219 Search PubMed.
- H. J. Mamin, M. Kim, M. H. Sherwood, C. T. Rettner, K. Ohno, D. D. Awschalom and D. Rugar, Science, 2013, 339(6119), 557–560 CrossRef CAS PubMed.
- I. Lovchinsky, A. O. Sushkov, E. Urbach, N. P. de Leon, S. Choi, K. De Greve, R. Evans, R. Gertner, E. Bersin, C. Müller and L. McGuinness, Science, 2016, 351(6275), 836–841 CrossRef CAS PubMed.
- P. Maletinsky, S. Hong, M. S. Grinolds, B. Hausmann, M. D. Lukin, R. L. Walsworth, M. Loncar and A. Yacoby, Nat. Nanotechnol., 2012, 7(5), 320–324 CrossRef CAS PubMed.
- D. M. Juraschek, Q. N. Meier, M. Trassin, S. E. Trolier-McKinstry, C. L. Degen and N. A. Spaldin, Phys. Rev. Lett., 2019, 123(12), 127601 CrossRef CAS PubMed.
- D. Le Sage, K. Arai, D. R. Glenn, S. J. DeVience, L. M. Pham, L. Rahn-Lee, M. D. Lukin, A. Yacoby, A. Komeili and R. L. Walsworth, Nature, 2013, 496(7446), 486–489 CrossRef CAS PubMed.
- G. Kucsko, P. C. Maurer, N. Y. Yao, M. Kubo, H. J. Noh, P. K. Lo, H. Park and M. D. Lukin, Nature, 2013, 500(7460), 54–58 CrossRef CAS PubMed.
- M. Fujiwara, S. Sun, A. Dohms, Y. Nishimura, K. Suto, Y. Takezawa, K. Oshimi, L. Zhao, N. Sadzak, Y. Umehara and Y. Teki, Sci. Adv., 2020, 6(37), p.eaba9636 CrossRef PubMed.
- A. Morita, A. C. Nusantara, F. P. P. Martinez, T. Hamoh, V. G. Damle, K. J. van der Laan, A. Sigaeva, T. Vedelaar, M. Chang, M. Chipaux and R. Schirhagl, arXiv, 2020, preprint arXiv:2007.16130.
- V. Vaijayanthimala, Y. K. Tzeng, H. C. Chang and C. L. Li, Nanotechnology, 2009, 20(42), 425103 CrossRef PubMed.
- E. Perevedentseva, S. F. Hong, K. J. Huang, I. T. Chiang, C. Y. Lee, Y. T. Tseng and C. L. Cheng, J. Nanopart. Res., 2013, 15(8), 1834 CrossRef.
- K. K. Liu, C. L. Cheng, C. C. Chang and J. I. Chao, Nanotechnology, 2007, 18(32), 325102 CrossRef.
- L. P. McGuinness, Y. Yan, A. Stacey, D. A. Simpson, L. T. Hall, D. Maclaurin, S. Prawer, P. Mulvaney, J. Wrachtrup, F. Caruso and R. E. Scholten, Nat. Nanotechnol., 2011, 6(6), 358–363 CrossRef CAS PubMed.
- O. Loh, R. Lam, M. Chen, N. Moldovan, H. Huang, D. Ho and H. D. Espinosa, Small, 2009, 5(14), 1667–1674 CrossRef CAS PubMed.
- B. Zhang, X. Feng, H. Yin, Z. Ge, Y. Wang, Z. Chu, H. Raabova, J. Vavra, P. Cigler, R. Liu and Y. Wang, Sci. Rep., 2017, 7, 46462 CrossRef CAS PubMed.
- K. K. Liu, C. C. Wang, C. L. Cheng and J. I. Chao, Biomaterials, 2009, 30(26), 4249–4259 CrossRef CAS PubMed.
- H. T. McMahon and E. Boucrot, Nat. Rev. Mol. Cell Biol., 2011, 12(8), 517–533 CrossRef CAS PubMed.
- A. Sigaeva, A. Morita, S. R. Hemelaar and R. Schirhagl, Nanoscale, 2019, 11(37), 17357–17367 RSC.
- Z. Chu, S. Zhang, B. Zhang, C. Zhang, C. Y. Fang, I. Rehor, P. Cigler, H. C. Chang, G. Lin, R. Liu and Q. Li, Sci. Rep., 2014, 4, 4495 CrossRef PubMed.
- Z. Chu, K. Miu, P. Lung, S. Zhang, S. Zhao, H. C. Chang, G. Lin and Q. Li, Sci. Rep., 2015, 5, 11661 CrossRef PubMed.
- S. R. Hemelaar, A. Nagl, F. Bigot, M. M. Rodríguez-García, M. P. de Vries, M. Chipaux and R. Schirhagl, Microchim. Acta, 2017, 184(4), 1001–1009 CrossRef PubMed.
- S. Y. Ong, R. J. J. Van Harmelen, N. Norouzi, F. Offens, I. M. Venema, M. H. Najafi and R. Schirhagl, Nanoscale, 2018, 10(36), 17117–17124 RSC.
- A. Morita, F. P. P. Martinez, M. Chipaux, N. Jamot, S. R. Hemelaar, K. J. van der Laan and R. Schirhagl, Part. Part. Syst. Charact., 2019, 36(8), 1900116 CrossRef.
- S. Bolte and F. P. Cordelières, J. Microsc., 2006, 224(3), 213–232 CrossRef CAS PubMed.
- K. W. Dunn, M. M. Kamocka and J. H. McDonald, Am. J. Physiol.: Cell Physiol., 2011, 300(4), C723–C742 CrossRef CAS PubMed.
- T. Zheng, F. Perona Martínez, I. M. Storm, W. Rombouts, J. Sprakel, R. Schirhagl and R. De Vries, Anal. Chem., 2017, 89(23), 12812–12820 CrossRef CAS PubMed.
- E. Perevedentseva, S. F. Hong, K. J. Huang, I. T. Chiang, C. Y. Lee, Y. T. Tseng and C. L. Cheng, J. Nanopart. Res., 2013, 15(8), 1–12 CrossRef.
- O. Faklaris, V. Joshi, T. Irinopoulou, P. Tauc, M. Sennour, H. Girard, C. Gesset, J. C. Arnault, A. Thorel, J. P. Boudou and P. A. Curmi, ACS Nano, 2009, 3(12), 3955–3962 CrossRef CAS PubMed.
- A. Garz, M. Sandmann, M. Rading, S. Ramm, R. Menzel and M. Steup, Biophys. J., 2012, 103(5), 1078–1086 CrossRef CAS PubMed.
- A. A. Torrano and C. Bräuchle, Beilstein J. Nanotechnol., 2014, 5(1), 1616–1624 CrossRef PubMed.
- E. Hebisch, M. Hjort, D. Volpati and C. N. Prinz, Small, 2021, 2006421 CrossRef CAS PubMed.
- S. Han, M. Raabe, L. Hodgson, J. Mantell, P. Verkade, T. Lasser, K. Landfester, T. Weil and I. Lieberwirth, Nano Lett., 2019, 19(3), 2178–2185 CrossRef CAS PubMed.
- N. Prabhakar, M. H. Khan, M. Peurla, H. C. Chang, P. E. Hänninen and J. M. Rosenholm, ACS Omega, 2017, 2(6), 2689–2693 CrossRef CAS PubMed.
- W. Liu, B. Naydenov, S. Chakrabortty, B. Wuensch, K. Hübner, S. Ritz, H. Cölfen, H. Barth, K. Koynov, H. Qi and R. Leiter, Nano Lett., 2016, 16(10), 6236–6244 CrossRef CAS PubMed.
- V. Merz, J. Lenhart, Y. Vonhausen, M. E. Ortiz-Soto, J. Seibel and A. Krueger, Small, 2019, 15(48), 1901551 CrossRef CAS PubMed.
- A. E. Garcia-Bennett, A. Everest-Dass, I. Moroni, I. D. Rastogi, L. M. Parker, N. H. Packer and L. J. Brown, J. Mater. Chem. B, 2019, 7(21), 3383–3389 RSC.
- I. Machova, M. Hubalek, T. Belinova, A. Fucikova, S. Stehlik, B. Rezek and M. H. Kalbacova, Carbon, 2020, 162, 650–661 CrossRef CAS.
- D. Khanal, Q. Lei, G. Pinget, D. A. Cheong, A. Gautam, R. Yusoff, B. Su, S. Yamaguchi, A. Kondyurin, J. C. Knowles and G. Georgiou, Nanoscale Adv., 2020, 2(10), 4798–4812 RSC.
- Y. Zou and N. Komatsu, Carbon, 2020, 163, 395–401 CrossRef CAS.
- R. Wallbrecher, T. Ackels, R. A. Olea, M. J. Klein, L. Caillon, J. Schiller, P. H. Bovée-Geurts, T. H. van Kuppevelt, A. S. Ulrich, M. Spehr and M. J. Adjobo-Hermans, J. Controlled Release, 2017, 256, 68–78 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr02503a |
| This journal is © The Royal Society of Chemistry 2021 |