Open Access Article
Open Access ArticlePeptide foldamer-based self-assembled nanostructures containing cyclic beta-amino acids
Monika
Szefczyk

Department of Bioorganic Chemistry, Faculty of Chemistry, Wroclaw University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wrocław, Poland. E-mail: monika.szefczyk@pwr.edu.pl
First published on 14th June 2021
Abstract
Peptide soft materials belong to an emerging branch of materials sciences due to their growing importance as responsive materials in diagnostics, therapeutics, and biomedical applications. The diversity provided by easily modifiable peptide sequences can be further increased by introducing nonnatural amino acids such as cyclic β-amino acids, leading to the formation of foldamers. Moreover, it is possible to combine peptide chains with other polymers, aromatic compounds, etc. to create hybrids with completely new properties and applications. In this review, we focus on the cis/trans enantiomers of three cyclic β-amino acids: 2-aminocyclobutane-1-carboxylic acid (ACBC), 2-aminocyclopentane-1-carboxylic acid (ACPC) and 2-aminocyclohexane-1-carboxylic acid (ACHC). The peptides discussed here either contain exclusively β-amino acids or are α,β-peptides, and they undergo self-assembly by forming different interactions that lead to the creation of well-defined nanostructures.
Introduction
Peptide-based self-assembled structures possess many advantages over other organic and inorganic systems due to their chemical diversity, ability to adopt distinct secondary structures, biocompatibility, resemblance to proteins, large potential for modification, relatively easy and scalable synthesis, and ability to associate spontaneously.1,2 The type, number, and sequence of amino acids can be modified to design peptides that can self-assemble to form nanotubes,3 nanofibers,4 nanoribbons,5 nanospheres,6 nanotapes7 and nanorods8 – all with controllable properties and a broad spectrum of applications in biotechnology,9 bioengineering,10 biomedicine11,12 and more. Most of the nanostructures described in the literature consist of α-amino acid residues with different charges, hydrophobicities, polarities, and sizes that allow the creation of various complex nanostructures with different characteristics.13,14 Incorporation of unnatural amino acids such as β-amino acids into the sequence significantly increases the number of possible nanostructures.15Review
β-Amino acids as conformationally constrained building blocks
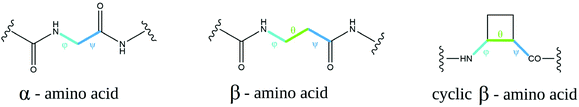
β-Amino acids are considered “nonnatural” amino acids; however, there are many more β-amino acids in nature than there are proteinogenic amino acids.16 The most common and most important is H-βhGly-OH, the so-called “β-alanine” (a component of the naturally occurring peptides anserine and carnosine17), as well as pantothenic acid (vitamin B5, a component of coenzyme A18). There are several advantages of using peptides with incorporated β-amino acids depending on their application. β-Amino acids are resistant to the enzymatic cleavage of peptides, and they seem to be extraordinarily stable in metabolic processes occurring in mammals, insects, and plants; additionally, they undergo microbial degradation very slowly. Many studies conducted thus far have demonstrated negligible biocidal or toxic properties of these peptides.16The conformation of α-amino acids can be represented by two torsion angles, denoted φ (phi) and ψ (psi). There is an extra carbon atom in the β-amino acid structure; hence, there is an additional torsion angle, θ (theta) (Fig. 1). In cyclic β-residues, this θ angle depends on φ and ψ, and its gauche conformation is inherently stable.19 Changing the conformation of the additional torsional angle in β-peptides can even profoundly affect the folding of these molecules. 2-Aminocyclopentane-1-carboxylic acid (ACPC)-based peptides with the cis conformation at the θ torsion angle have been shown to lead to an extended strand with a zigzag pattern rather than to the helix fold observed when θ is in the trans conformation.20 In amino acids with constrained conformations, C2 and C3 atoms are part of the ring, and cyclization at these atoms imposes an additional constraint on five- and six-membered rings because θ is now constrained to ±60 degrees due to sp3 hybridization.21 Consequently, β-amino acids have φ and ψ values that are arranged in a Ramanathan diagram similar to that of a glycine residue,22 and thus, β-amino acids tend to adopt the conformational space normally accessible only to glycine residues. This in turn opens a range of possible conformations available for foldamers.
The main advantage of β-peptides is their ability to adopt various types of helices, β-sheets, turns, and extended structures.23 Their ability to form well-defined helical structures stabilized by hydrogen bonds is particularly valuable. β-Peptides demonstrate a higher folding propensity than α-peptides of comparable length; for instance, properly designed β-peptides with only six residues adopt helical conformations in water,24 while α-peptides usually need to be two or three times longer to exhibit significant helicity.25 Moreover, among a few types of hydrogen-bonded helices known for α-peptides, the most common are only three: α-, 310- and π-helices, whereas β-peptides have been shown to form a variety of different helical H-bonded structures,26e.g., 10-helix,27 12-helix28 and 14-helix.29 If both α- and β-residues are introduced into the sequence, the number of possible helical types is even higher.30,31 Importantly, the type of secondary structure adopted can be predicted using a method called stereochemical patterning, which is based on the known tendency of individual units to prefer dihedral angles of a certain magnitude and sign.32 Moreover, β-amino acids introduced into the α-amino acid chain may induce changes in the peptide conformation, mainly by stabilizing the secondary structure,33 which translates into biological functions34 and a propensity for supramolecular self-assembly.35 Additionally, it is possible to control the α,β-helix handedness through small changes in the peptide termini.36 It was also shown that changing the relative configurations of the β-peptide backbone may lead to the control of the secondary structure and the self-assembly pattern and, thus, the morphology of the obtained nanostructure.37
The basic and most versatile structures of cyclic β-amino acids are shown in Fig. 2.
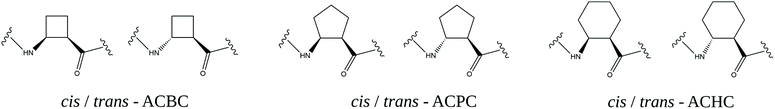 | ||
| Fig. 2 Cis and trans enantiomers of 2-aminocyclobutane-1-carboxylic acid (ACBC), 2-aminocyclopentane-1-carboxylic acid (ACPC) and 2-aminocyclohexane-1-carboxylic acid (ACHC) residues. | ||
Self-assembly process in nanostructure formation
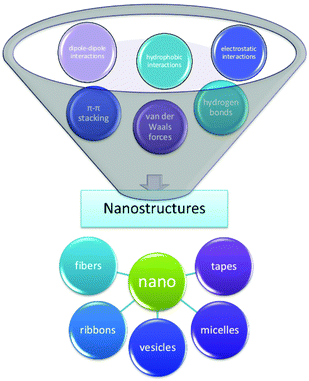
The term self-assembly is used to describe a wide variety of processes in which molecules associate into larger species.38 This spontaneous organization of small units into higher-order structures is possible due to the collective balance of noncovalent interactions, mainly electrostatic interactions, hydrogen bonding, and hydrophobic interactions, with a contribution from π–π stacking and other dispersive forces.39,40 While these interactions are rather weak compared to covalent bonds, they account for relatively stable higher-order self-assembled structures, thanks to the additive effect of these secondary forces; i.e., self-assembled structures are thermodynamically more stable since their Gibbs free energy values are lower than the energy of the individual components.41 The formation of peptide aggregates may or may not be reversible; they may be amorphous or highly structured aggregates, and they may assemble both in solution and on surfaces after adsorption.42 The precise mechanisms of self-aggregation of foldameric peptides remain insufficiently understood, despite many efforts already made in different fields of peptide research, e.g., research on beta-amyloid aggregation.43 The elucidation of the self-assembly mechanism is not trivial, mainly due to the complexity of the interactions between molecules building nanostructures. Numerous hydrogen bond patterns play an important role in supporting various elements of the secondary structure, among which α-helices and β-strands have been mainly observed in peptide materials.44 For example, peptides forming β-sheets exhibit hydrogen bond interactions along the backbone but can also be found in arrays with ionic interactions, hydrophobic interactions, van der Waals interactions, and water-mediated hydrogen bonds.45 In particular, the “bottom up” approach mechanisms require a deep understanding; therefore, rational methods of nanostructure formation should be developed considering the structure and the ability of the individual molecular building blocks to self-aggregate (Fig. 3).46Peptide foldamer-based self-assembled nanostructures containing cyclic beta-amino acids
Peptide foldamers are oligomers containing nonnatural amino acids that fold into well-defined three-dimensional structures.47 If they are correctly designed, they can undergo a self-assembly process within minutes and without external stimuli, forming distinct nanostructures.46 Examples of the different nanostructures formed by particular building blocks containing cyclic β-amino acids along with reported interactions/mechanisms leading to their self-association are described in Table 1.| β-Amino acid | Building blocks | Type of nanostructure | Interactions/mechanism | Ref. |
|---|---|---|---|---|
| cis-ACBC | (ACBC)4 (β-strands) | Ribbon-like fibrils; gels | Interactions between amphiphilic molecules; hydrogen bonds | 48 |
| (ACBC)2–8 (β-strands) | Fibers; gels | Hydrogen bonds; hydrophobic interactions | 49 | |
| TTF-functionalized (ACBC)2 | Tape-like fibers | Head-to-head interactions; complex twisted bilayers | 50 | |
| hcptpy-functionalized (ACBC)4 | Helical aggregates; fibers; xerogels | π–π stacking; hydrogen bonds | 51 | |
| Lipophilic chain anchored to ACBC | Fibers; spherical micelles; liquid crystals | Hydrogen bonds; charge-dipole interactions | 52 | |
| (trans-ACBC)(cis-ACBC) | Helical aggregate; solid-like networks; xerogels | Head-to-tail molecular arrangement; hydrogen bonds | 53 | |
| 1,2-difunctionalized ACBC | Vesicles; fibers (bilayer tubes) | Hydrogen bonds | 54 | |
| trans-ACBC | hcptpy-functionalized (ACBC)2 | Helical aggregates; fibers; xerogels | π–π stacking; hydrogen bonds | 51 |
| Lipophilic chain anchored to ACBC | Fibers; spherical micelles; liquid crystals | Hydrogen bonds | 52 | |
| (ACBC)2 | Helical aggregates; solid-like networks; xerogels | Head-to-head molecular arrangement; hydrogen bonds | 53 | |
| 1,2-difunctionalized ACBC | Irregular aggregates; fibers (bilayer tubes) | Hydrogen bonds | 54 | |
| cis-ACPC | (ACPC)4–6 | Nonpolar strand; ribbonlike fibrils; fibrils of pleated-sheet sandwiches | Hydrophobic forces; hydrogen bonds | 55 |
| trans-ACPC | α,β-Peptides (helices) | Helical bundles | Hydrophobic forces; polarization/ionic interactions | 56 |
| N-Boc-(trans-(S,S)-ACPC)7-OBn (12-helix) | Windmill-shaped supramolecular architectures | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 57 | |
| BocNH-(trans-(R,R)-ACPC)6-OBn (12-helix) | Square plate; molar tooth | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 58 | |
| BocNH-(ACPC)6-OH (12-helix) | Rhombic plate; rhombic rod | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 59 | |
| BocNH-(Aib-ACPC)3-Aib-OBn (11-helix) | Tripod shape; trigonal bipyramids | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 60 | |
| BocNH-(Aib-ACPC)3-OBn (11-helix) | Microrods; parallelogram plates | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 61 | |
| Boc-(ACPC)8-OBn (12-helix) | Rectangular plate | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 62 | |
| Boc-(ACPC)4-OBn (12-helix) | Tubular foldectures | Hydrophobic forces; head-to-tail intermolecular hydrogen bonds | 63 | |
| BocNH-(ACPC)6-Leu2-OBn (12-helix) | Hexagonal plate | Hydrophobic forces; hydrogen bonds | 64 | |
| α,β-Peptides (helices) | Fibrils | Cyclopentyl zipper | 46 | |
| cis-ACHC | H2N-(ACHC)4 or 6 (10/12-helix) | Vesicles | Hydrophobic forces | 65 |
| Boc-ACHC-Aib-Phe-OMe (turn-type β-sheet) | Fibers; organogels | Hydrogen bonds; π–π stacking interactions | 66 | |
| trans-ACHC | (ACHC-ACHC-β3-Lys)n (14-helix) | Globular aggregates; nanofibers; LC phase | Cyclohexyl zipper; interactions between amphiphilic molecules | 67 |
| β-Peptides (14-helix) | Helix bundles; vesicle-forming membranes; lyotropic liquid crystals | Cyclohexyl zipper; interactions between amphiphilic helix units | 68 | |
| (ACHC)3–4 (helix) | Spherical objects; helix–bundle membranes | Interactions between amphiphilic helix units; head-to-head and head-to-tail contacts; hydrophobic forces | 55 | |
| (ACHC)4 (10-helix) (ACHC)5–6 (14-helix) | Helical bundles | Hydrophobic forces | 69 |
β-Peptides containing only a few residues of cyclic β-amino acids are known to form well-defined secondary structures that are stable in solution and possess a high propensity to form different nanostructures. Hetenyi et al. showed that trans-2-aminocyclohexane-1-carboxylic acid (trans-ACHC) homooligomers can form different types of helices depending on the number of residues in the sequence.69 For instance, (trans-ACHC)4 adopts a 10-helix motif, whereas (trans-ACHC)5 and (trans-ACHC)6 form 14-helices. All these peptides can form helical bundles in solution. Rua and coworkers combined the rigidity and amphiphilic properties of another tetrapeptide, namely, β-strands mimicking cis-2-aminocyclobutane-1-carboxylic acid (cis-ACBC)4, and obtained ribbon-like fibrils that formed a gel in some media.49 Vertically amphiphilic self-stabilizing helix units containing 3–4 residues of trans-ACHC were also reported, in which the hydrophobic surface of the helix was large enough to facilitate solvent-driven association into a continuous helix-bundle motif.55 The cog-wheel-like mutual fit of the helix side chains further enhances the stabilizing hydrophobic surface, and head-to-head and head-to-tail contacts of the helices enable the formation of a membrane. More information about the interaction forces between amphiphilic molecules in aggregates and how they are affected by solution conditions can be found elsewhere.70 Other interesting examples of β-peptides were described by Torres et al.49 They reported the synthesis of cis-ACBC derivatives and their incorporation into β-peptides consisting of 2–8 residues bearing different N-protecting groups. The obtained oligomers adopted a β-strand-type conformation induced by the formation of intraresidue six-membered hydrogen-bonded rings to form cis-fused [4.2.0]octane structural units that show the high rigidity of these β-peptides. The synthesized peptides self-associated to form nanofibers and, under certain conditions, gels. The authors elucidated a model in which a parallel molecular arrangement is preferred and the conformation is similar to that observed in a solution in which both hydrogen bonding and hydrophobic interactions are responsible for assembly. As demonstrated by Martinek and coworkers, 4–6 residues of cis-ACPC can self-assemble to form ribbons 2–3 nm high, which corresponds to the distance between the N and C termini of a single (cis-ACPC)n strand.55 It was concluded that since the width of the ribbon increased with the size of the oligomer and the polarity of the solvent, the ribbons must be organized by hydrophobic forces between the side chains in the corresponding dimension, thus leading to a continuous pleated-sheet sandwich tertiary structural motif. The supramolecular structure was organized by interstrand hydrogen bonds formed by peptide moieties parallel to the long fibril axis. Mandity and coworkers showed that not only the α-helix but also the β-peptidic H10/12 helix can tolerate side chains containing alicyclic rings.65 Conformational polymorphism was observed for the alternating cis-ACHC hexamer, in which chemical exchange occurred between the major left-handed H10/12 helix and a minor folded conformation. For this peptide, vesicles with diameters of ∼100 nm formed as a result of hydrophobic self-assembly.
Most of the published research concerns nanostructures in which the building element is the trans enantiomer rather than the cis enantiomer (Table 1). This is most likely related to the difficulties in the synthesis and the purchase cost of the ci enantiomer s. Several authors have accepted the challenge of comparing the conformations and properties of peptides, including those of each enantiomer. For example, Gorrea et al. described studies on two β-dipeptides, one with two trans-ACBC fragments and another with one trans residue and one cis residue.53 Studies on noncovalent interactions responsible for the self-assembly, morphology and size of the obtained nanostructure showed head-to-head (a dipeptide with two trans residues) or head-to-tail (a dipeptide with trans and cis ACBC residues) molecular arrangements present in the studied peptides. Helical structures corresponding to hydrogen-bonded single chains that interact with each other in an antiparallel way to form bundles were obtained. Hydrogen bonds formed between the two NH groups of one molecule and the CO3 and CO9 of the other molecule. These hydrogen bonds propagate along the axis of the aggregates, inducing a helical aggregation mode. The work of Pi-Boleda and coworkers aimed to investigate the influence of the stereochemistry and regiochemistry of peptides on their ability to serve as surfactants and undergo self-organization.54 The authors showed that the interplay between stereoisomerism (cis- versus trans-1,2 disubstituted cyclobutane) and regiochemistry (N-centered versus C-centered amide bonds), enhanced by the rigidity of the cyclobutane ring, had a significant impact on the physicochemical properties of the obtained peptides. Regiochemistry has been found to govern the morphology of supramolecular aggregates (i.e., long fibers versus plates or spherical assemblies), while the critical micelle concentration (cmc value) is governed mainly by stereochemistry. A similar issue was investigated by Sorrenti and coworkers, who studied the effect of cis/trans stereochemistry on the molecular organization and recognition properties of small diastereomeric anionic amphiphiles based on a rigid cyclobutane β-amino acid scaffold.52 Peptides containing lipophilic chains anchored to the amino group of ACBC moieties self-assembled to form fibers, spherical micelles, and liquid crystals with a morphology that depends on the stereochemistry (Fig. 4). The self-association process was mediated by the formation of hydrogen bonds and cooperative hydrogen bond networks between secondary amide groups in the heads of neighboring molecules. The authors concluded that the relative stereochemistry mainly affects the solvation of the headgroup and anionic charge stabilization in a dilute solution, resulting in better stabilization of the cis diastereoisomer due to intramolecular hydrogen bonds and/or charge–dipole interactions. Furthermore, the relative configuration of ACBC residues affects the chiral recognition ability of spherical micelles for bilirubin enantiomers.
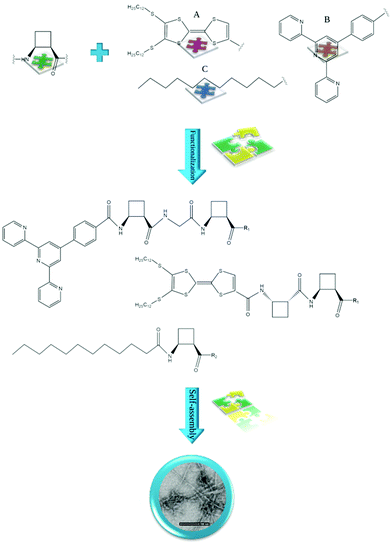 | ||
| Fig. 4 Examples of cis-ACBC residues functionalized with: (A) TTF,50 (B) hcptpy,51 and (C) lipophilic chain52 self-assembling to form fibrils/fibers. R1 = OMe and R2 = ONa. | ||
Introducing cyclic β-amino acids into peptide sequences can prompt hydrophobically induced aggregation or can help to direct association in the desired direction.71 The plausible advantages of the substitution of α-residues by β-residues depend on the application and whether a single modification or an entire β-peptide is considered. Szefczyk et al. proposed a hierarchical approach for the construction of nanofibrils based on α,β-peptide foldamers, in which the incorporation of a helix-promoting trans-ACPC residue at the outer positions of the model coiled-coil peptide led to its increased conformational stability and allowed for additional interactions between coiled-coil-like structures to direct the self-assembly process toward the formation of well-defined nanofibrils.46 In their publications, Pomerantz and coworkers described globally amphiphilic helices undergoing a hydrophobically driven association process by interdigitating ACHC residues at the interface between neighboring β-peptides, creating a “cyclohexyl zipper”.67,68 It has been revealed that hydrophobically driven assembly processes are promoted not only by the overall lipophilicity of a molecule but also by specific arrangements of lipophilic elements on the molecular surface and specific associations of lipophilic elements in intermolecular contacts. Unpaired ACHC residues that elongated into nanofibers were the predominant self-assembled structures leading to the lyotropic LC phase. Additionally, β3 → cyclic replacements that showed generally good accommodation in helix-bundle quaternary structures were also investigated.56 The authors investigated α/β-peptide foldamers with a regular α-residue/α-residue/β-residue (ααβ) backbone motif, which exhibited a four-helix bundle quaternary structure in the crystalline state. The folded structures were stabilized by the close packing of hydrophobic side chains in the core, with additional interactions between the polar residues. Kwon and coworkers described several peptides containing highly hydrophobic helices arising from the presence of trans-ACPC residues displayed on the lateral exterior of the helix and N- and C-termini protection by the Boc group and the benzyl group, respectively.57–60,62,63 The described peptides were able to self-assemble, creating nanostructures of various shapes, i.e., windmill-shaped structures, square plate (in water) and molar tooth (in surfactant) structures, microrods (in water), and parallelogram plates (in surfactant), depending on the sequence modifications and the presence of a surfactant in the solution. The authors concluded that the helices self-assembled in an aqueous solution via lateral hydrophobic interactions between the helical faces. Head-to-tail intermolecular hydrogen bonds and highly ordered anisotropic molecular packing motifs present in the foldamer building blocks were responsible for their unique shapes.
Peptides containing β-amino acids can also be functionalized with various molecules to obtain nanostructures with interesting properties. Torres et al. described homochiral dipeptides composed of cis-ACBC residues that were functionalized with a π-electron-rich tetrathiafulvalene (TTF) moiety (Fig. 4).50 Microscopic analysis revealed the formation of fibers in which the molecules were packed into dimeric tapes with the peptide head groups at the center. There are complex twisted bilayers whose deformation is caused partially by the chiral nature of the molecular components. The films of the obtained material showed electrical conductivity. Porcar-Tost and coworkers described dipeptides and tetrapeptides built with either cis- or trans-ACBC residues functionalized with a terpyridine derivative, namely, 4′-(4-carboxy)phenylterpyridine (hcptpy) (Fig. 4).51 The obtained conjugates self-associated as a result of the π–π interactions provided by the aromatic moieties that cooperate with intermolecular hydrogen bonds between NH and CO in the amide groups. The morphology of the created nanostructures, i.e., helical aggregates, fibers or xerogel, is strongly dependent on the solvent. Moreover, ruthenium(II) complexes derived from these ligands showed electrochemical behavior and activity as catalysts; as organogelators, these conjugates were able to gel various solvents. The mechanistic aspects of the supramolecular ordering of β-amino acid-based peptides in several organic solvents and fuel oils were investigated by Konda and coworkers.66 The studied peptide Boc-cisACHC-Aib-Phe-OMe was able to self-assemble into a bundle of nanofibers and entrap various organic solvents and oils, governing the formation of self-supporting gels. The self-assembly process was mediated by hydrogen-bonding and π–π stacking interactions.
In summary, both cis and trans enantiomers of 2-aminocyclobutane-1-carboxylic acid (ACBC), 2-aminocyclopentane-1-carboxylic acid (ACPC) and 2-aminocyclohexane-1-carboxylic acid (ACHC) proved to possess propensities for the formation of diverse self-assembled nanostructures, e.g., spherical micelles, fibers and gels. In the reported research, the self-assembly process was mediated mainly by hydrogen bond formation, hydrophobic interactions, head-to-head and head-to-tail molecular arrangement, and/or interactions between amphiphilic molecules. In some cases, the spontaneous arrangement of the building blocks was shown to be a hierarchical process leading to the formation of nanostructures with different organization levels, e.g., globular aggregates → nanofibers → liquid crystals. Moreover, it was proven that the morphology of the nanostructures can be controlled, for instance, by the appropriate modification of the peptide sequence or addition of a surfactant. The above findings, together with the continuous development of high-resolution methods (e.g., XRD, SAXS/WAXS, and cryo-TEM) for the characterization of foldameric nanostructures and intense research on another group of self-aggregating peptides – amyloids – may in the near future lead to a full mechanistic description of the self-association process of peptides and produce a breakthrough in the wide application of soft materials.
Conclusions
In conclusion, the presence of β-residues increases the diversity in peptide sequences, which is translated to the functions and applications of the obtained peptides. Cyclic β-amino acids help to form peptide foldamers with well-defined 3D structures. The self-assembly process, which is mediated through various types of interactions and enhanced by hydrophobic interactions, provides numerous nanostructures that can result in interesting properties and can be used in many applications. β-Peptide modularity for tailoring properties is indisputable, but there are still ongoing efforts to clarify the relationship among the β-peptide sequence, its 3D structure, and the self-assembly process leading to the nanostructure formation. In particular, more studies are essential to elucidate rational methods for peptide design that can explain the fundamental interactions that enhance their self-association and help to drive the self-assembly process in the desired direction.Abbreviations
| ACBC | 2-Aminocyclobutane-1-carboxylic acid |
| ACPC | 2-Aminocyclopentane-1-carboxylic acid |
| ACHC | 2-Aminocyclohexane-1-carboxylic acid |
| Aib | α-Amino isobutyric acid |
| Boc | tert-Butyloxycarbonyl |
| cmc | Critical micelle concentration |
| hcptpy | 4′-(4-Carboxy)phenylterpyridine |
| Phe | Phenylalanine |
| SAXS | Small-angle X-ray scattering |
| TEM | Transmission electron microscopy |
| TTF | Tetrathiafulvalene |
| WAXS | Wide-angle X-ray scattering |
| XRD | X-ray diffraction |
Conflicts of interest
The authors declare no potential conflicts of interest.Acknowledgements
The authors would like to gratefully acknowledge the National Science Centre, Poland, for their financial support through grant no. 2017/26/D/ST5/00341.References
- D. M. Raymond and B. L. Nilsson, Multicomponent Peptide Assemblies, Chem. Soc. Rev., 2018, 47, 3659–3720 RSC.
- E. Gazit, Self-Assembled Peptide Nanostructures: the Design of Molecular Building Blocks and their Technological Utilization, Chem. Soc. Rev., 2007, 36, 1263–1269 RSC.
- Y. Zhao, X. Hu, L. Zhang, D. Wang, S. M. King, S. E. Rogers, J. Wang, J. R. Lu and H. Xu, Monolayer Wall Nanotubes Self-Assembled from Short Peptide Bolaamphiphiles, J. Colloid Interface Sci., 2021, 583, 553–562 CrossRef CAS PubMed.
- L. Lu, D. Morrison and L. D. Unsworth, Controllable Self-Patterning Behaviours of Flexible Self-Assembling Peptide Nanofibers, Nanoscale, 2020, 12, 8133–8138 RSC.
- A. Rani, I. Kavianinia, L. M. De Leon-Rodriguez, D. J. McGillivray, D. E. Williams and M. A. Brimble, Nanoribbon Self-Assembly and Hydrogel Formation from an NOctanoyl Octapeptide Derived from the Antiparallel β-Interface of a Protein Homotetramer, Acta Biomater., 2020, 114, 233–243 CrossRef CAS PubMed.
- Q. Zhang, P. Zhang, S. Jian, J. Li, F. Li, X. Sun, H. Li, Y. Zeng, Y. Zeng, S. Liang, P. Chen and Z. Liu, Drug-Bearing Peptide-Based Nanospheres for the Inhibition of Metastasis and Growth of Cancer, Mol. Pharmaceutics, 2020, 17, 3165–3176 CrossRef CAS PubMed.
- A. Dehsorkhi and I. W. Hamley, Silica Templating of a Self-Assembling Peptide Amphiphile that Forms Nanotapes, Soft Matter, 2014, 10, 1660–1664 RSC.
- W. Wang and Y. Chau, Self-Assembled Peptidenanorods as Building Blocks of Fractal Patterns, Soft Matter, 2009, 5, 4893–4898 RSC.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Biomimetic Peptide Self-Assembly for Functional Materials, Nat. Rev. Chem., 2020, 4, 615–634 CrossRef CAS.
- Self-assembling peptides in biomedicine and bioengineering: Tissue engineering, regenerative medicine, drug delivery, and biotechnology in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, ed. S. Koutsopoulos, Woodhead Publishing, 2018, pp. 387–408 Search PubMed.
- L. Sun, C. Zheng and T. J. Webster, Self-Assembled Peptide Nanomaterials for Biomedical Applications: Promises and Pitfalls, Int. J. Nanomed., 2016, 12, 73–86 CrossRef PubMed.
- X. Zhang, C. Gong, O. U. Akakuru, Z. Su, A. Wu and G. Wei, The Design and Biomedical Applications of Self-Assembled Two-Dimensional Organic Biomaterials, Chem. Soc. Rev., 2019, 48, 5564–5595 RSC.
- S. H. Yoo and H.-S. Lee, Foldectures: 3D Molecular Architectures from Self-Assembly of Peptide Foldamers, Acc. Chem. Res., 2017, 50, 832–841 CrossRef CAS PubMed.
- E. De Santis and M. G. Ryadnov, Peptide Self-Assembly for Nanomaterials: the Old New Kid on the Block, Chem. Soc. Rev., 2015, 44, 8288–8300 RSC.
- S. Shankar, J. U. Rahim and R. Rai, Self-Assembly in Peptides Containing β-and γ-amino Acids, Curr. Protein Pept. Sci., 2020, 21, 584–597 CrossRef CAS PubMed.
- D. Seebach, A. K. Beck and D. J. Bierbaum, The World of Beta- and Gamma-Peptides Comprised of Homologated Proteinogenic Amino Acids and Other Components, Chem. Biodivers., 2004, 1, 1111–1239 CrossRef CAS PubMed.
- P. J. Quinn, A. A. Boldyrev and V. E. Formazuyk, Carnosine: Its Properties, Functions and Potential Therapeutic Applications, Mol. Aspects Med., 1992, 13, 379–444 CrossRef CAS PubMed.
- C. Spry, K. Kirk and K. J. Saliba, Coenzyme A Biosynthesis: an Antimicrobial Drug Target, FEMS Microbiol. Rev., 2008, 32, 56–106 CrossRef CAS.
- T. Beke, I. G. Csizmadia and A. Perczel, On the Flexibility of β-Peptides, J. Comput. Chem., 2004, 25, 285–307 CrossRef CAS PubMed.
- T. A. Martinek, G. K. Toth, E. Vass, M. Hollosi and F. Fulop, Cis-2-Aminocyclopentanecarboxylic Acid Oligomers Adopt a Sheetlike Structure: Switch from Helix to Nonpolar Strand, Angew. Chem., Int. Ed., 2002, 41, 1718–1721 CrossRef CAS.
- G. Ramanathan and A. Duley, Cyclic Beta-Amino Acids as Conformational Constraints in Biomolecular Forms and Functions, ed. M. B. N. Srinivasan, World Scientific Publishing Co. Pte. Ltd., Singapore, 2013, pp. 282–295 Search PubMed.
- G. N. Ramachandran, C. Ramakrishnan and V. Sasisekharan, Stereochemistry of Polypeptide Chain Configurations, J. Mol. Biol., 1963, 7, 95–99 CrossRef CAS PubMed.
- T. A. Martinek and F. Fulop, Peptidic Foldamers: Ramping up Diversity, Chem. Soc. Rev., 2012, 41, 687–702 RSC.
- D. H. Appella, J. J. J. Barchi, S. R. Durell and S. H. Gellman, Formation of Short, Stable Helices in Aqueous Solution by β-Amino Acid Hexamers, J. Am. Chem. Soc., 1999, 121, 2309–2310 CrossRef CAS.
- A. Chakrabarty and R. L. Baldwin, Stability of α-Helices, Adv. Protein Chem., 1995, 46, 141–176 CrossRef.
- R. P. Cheng, S. H. Gellman and W. F. DeGrado, β-Peptides: from Structure to Function, Chem. Rev., 2001, 101, 3219–3232 CrossRef CAS.
- T. D. W. Claridgea, J. M. Goodman, A. Moreno, D. Angus, S. F. Barker, C. Taillefumier, M. P. Watterson and G. W. J. Fleet, 10-Helical Conformations in Oxetane β-Amino Acid Hexamers, Tetrahedron Lett., 2001, 42, 4251–4255 CrossRef.
- C. Fernandes, S. Faure, E. Pereira, V. Thery, V. Declerck, R. Guillot and D. J. Aitken, 12-Helix Folding of Cyclobutane beta-Amino Acid Oligomers, Org. Lett., 2010, 12, 3606–3609 CrossRef CAS.
- D. H. Appella, L. A. Christianson, I. L. Karle, D. R. Powell and S. H. Gellman, β-Peptide Foldamers: Robust Helix Formation in a New Family of β-Amino Acid Oligomers, J. Am. Chem. Soc., 1996, 118, 13071–13072 CrossRef CAS.
- L. K. A. Pilsl and O. Reiser, α/β-Peptide Foldamers: State of the Art, Amino Acids, 2011, 41, 709–718 CrossRef CAS.
- W. S. Horne and S. H. Gellman, Foldamers with Heterogeneous Backbones, Acc. Chem. Res., 2008, 41, 1399–1408 CrossRef CAS PubMed.
- I. M. Mandity, E. Weber, T. A. Martinek, G. Olajos, G. K. Toth, E. Vass and F. Fulop, Design of Peptidic Foldamer Helices: a Stereochemical Patterning Approach, Angew. Chem., Int. Ed., 2009, 48, 2171–2175 CrossRef CAS.
- J. L. Price, W. S. Horne and S. H. Gellman, Structural Consequences of β-Amino Acid Preorganization in a Self-Assembling α/β-Peptide: Fundamental Studies of Foldameric Helix Bundles, J. Am. Chem. Soc., 2010, 132, 12378–12387 CrossRef CAS PubMed.
- C. Cabrele, T. A. Martinek, O. Reiser and L. Berlicki, Peptides Containing β-Amino Acid Patterns: Challenges and Successes in Medicinal Chemistry, J. Med. Chem., 2014, 57, 9718–9739 CrossRef CAS.
- K. Kulkarni, N. Habila, M. P. Del Borgo and M.-I. Aguilar, Novel Materials From the Supramolecular Self-Assembly of Short Helical β3-Peptide Foldamers, Front. Chem., 2019, 7, 70 CrossRef CAS PubMed.
- M. Szefczyk, E. Weglarz-Tomczak, P. Fortuna, A. Krzyszton, E. Rudzinska-Szostak and L. Berlicki, Controlling the Helix Handedness of ααβ-Peptide Foldamers through Sequence Shifting, Angew. Chem., Int. Ed., 2017, 56, 2087–2091 CrossRef CAS PubMed.
- T. A. Martinek, I. M. Mandity, L. Fulop, G. K. Toth, E. Vass, M. Hollosi, E. Forro and F. Fulop, Effects of the Alternating Backbone Configuration on the Secondary Structure and Self-Assembly of β-Peptides, J. Am. Chem. Soc., 2006, 128, 13539–13544 CrossRef CAS PubMed.
- Y. Lin and C. Mao, Bio-Inspired Supramolecular Self-Assembly towards Soft Nanomaterials, Front. Mater. Sci., 2011, 5, 247–265 CrossRef PubMed.
- L. Wang, C. Gong, X. Yuan and G. Wei, Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review, Nanomaterials, 2019, 9, 285 CrossRef CAS PubMed.
- K. L. Wooley, J. S. Moore, C. Wu and Y. Yang, Novel Polymers: Molecular to Nanoscale Order in Three-Dimensions, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 11147–11148 CrossRef CAS PubMed.
- P. Kumaraswamy, S. Sethuraman, J. V. Yakhmi and U. M. Krishnan, Hierarchical Self-Assembled Peptide Nano-ensembles, in Handbook of Nanomaterials Properties, ed. B. Bhushan, D. Luo, S. Schricker, W. Sigmund and S. Zauscher, Springer, Berlin, Heidelberg, 2014 Search PubMed.
- K. L. Zapadka, F. J. Becher, A. L. Gomes dos Santos and S. E. Jackson, Factors Affecting the Physical Stability (Aggregation) of Peptide Therapeutics, Interface Focus, 2017, 7, 20170030 CrossRef PubMed.
- F. Chiti and C. M. Dobson, Protein Misfolding, Amyloid Formation, and, Human Disease: A Summary of Progress Over the Last Decade, Annu. Rev. Biochem., 2017, 86, 27–68 CrossRef CAS.
- E. De Santis and M. G. Ryadnov, Peptide Self-Assembly for Nanomaterials: The Old New Kid on the Block, Chem. Soc. Rev., 2015, 44, 8288–8300 RSC.
- S. Zhang, Discovery and Design of Self-Assembling Peptides, Interface Focus, 2017, 7, 20170028 CrossRef PubMed.
- M. Szefczyk, N. Szulc, M. Gasior-Glogowska, A. Modrak-Wojcik, A. Bzowska, W. Majstrzyk, M. Taube, M. Kozak, T. Gotszalk, E. Rudzinska-Szostak and L. Berlicki, Hierarchical Approach for the Rational Construction of Helix-Containing Nanofibrils Using α,β-Peptides, Nanoscale, 2021, 13, 4000–4015 RSC.
- C. M. Goodman, S. Choi, S. Shandler and W. F. DeGrado, Foldamers as Versatile Frameworks for the Design and Evolution of Function, Nat. Chem. Biol., 2007, 3, 252–262 CrossRef CAS PubMed.
- F. Rua, S. Boussert, T. Parella, I. Diez-Perez, V. Branchadell, E. Giralt and R. M. Ortuno, Self-Assembly of a Cyclobutane Beta-Tetrapeptide to Form Nanosized Structures, Org. Lett., 2007, 9, 3643–3645 CrossRef CAS PubMed.
- E. Torres, E. Gorrea, K. K. Burusco, E. Da Silva, P. Nolis, F. Rua, S. Boussert, I. Diez-Perez, S. Dannenberg, S. Izquierdo, E. Giralt, C. Jaime, V. Branchadella and R. M. Ortuno, Folding and Self-Assembling with Beta-Oligomers Based on (1R,2S)-2-Aminocyclobutane-1-Carboxylic Acid, Org. Biomol. Chem., 2010, 8, 564–575 RSC.
- E. Torres, J. Puigmarti-Luis, A. Perez del Pino, R. M. Ortuno and D. B. Amabilino, Use of Unnatural β-Peptides as a Self-Assembling Component in Functional Organic Fibres, Org. Biomol. Chem., 2010, 8, 1661–1665 RSC.
- O. Porcar-Tost, B. Pi-Boleda, J. García-Anton, O. Illa and R. M. Ortuno, Cyclobutane-Based Peptides/Terpyridine Conjugates: Their Use in Metal Catalysis and as Functional Organogelators, Tetrahedron, 2018, 74, 7252–7260 CrossRef CAS.
- A. Sorrenti, O. Illa, R. Pons and R. M. Ortuno, Chiral Cyclobutane β-Amino Acid-Based Amphiphiles: Influence of Cis/Trans Stereochemistry on Solution Self-Aggregation and Recognition, Langmuir, 2015, 31, 9608–9618 CrossRef CAS.
- E. Gorrea, P. Nolis, E. Torres, E. Da Silva, D. B. Amabilino, V. Branchadell and R. M. Ortuno, Self–Assembly of Chiral trans–Cyclobutane–Containing β–Dipeptides into Ordered Aggregates, Chem. – Eur. J., 2011, 17, 4588–4597 CrossRef CAS PubMed.
- B. Pi-Boleda, A. Sorrenti, M. Sans, O. Illa, R. Pons, V. Branchadell and R. M. Ortunno, Cyclobutane Scaffold in Bolaamphiphiles: Effect of Diastereoisomerism and Regiochemistry on Their Surface Activity Aggregate Structure, Langmuir, 2018, 34, 11424–11432 CrossRef CAS PubMed.
- T. A. Martinek, A. Hetenyi, L. Fulop, I. M. Mandity, G. K. Toth, I. Dekany and F. Fulop, Secondary Structure Dependent Self–Assembly of β–Peptides into Nanosized Fibrils and Membranes, Angew. Chem., Int. Ed., 2006, 45, 2396–2400 CrossRef CAS PubMed.
- J. L. Price, W. S. Horne and S. H. Gellman, Structural Consequences of β-Amino Acid Preorganization in a Self-Assembling α/β-Peptide: Fundamental Studies of Foldameric Helix Bundles, J. Am. Chem. Soc., 2010, 132, 12378–12387 CrossRef CAS PubMed.
- S. Kwon, A. Jeon, S. H. Yoo, I. S. Chung and H. S. Lee, Unprecedented Molecular Architectures by the Controlled Self-Assembly of a β-Peptide Foldamer, Angew. Chem., Int. Ed., 2010, 49, 8232–8236 CrossRef CAS PubMed.
- S. Kwon, H. S. Shin, J. Gong, J.-H. Eom, A. Jeon, S. H. Yoo, I. S. Chung, S. J. Cho and H.-S. Lee, Self-Assembled Peptide Architecture with a Tooth Shape: Folding into Shape, J. Am. Chem. Soc., 2011, 133, 17618–17621 CrossRef CAS PubMed.
- S. H. Yoo, T. Eom, S. Kwon, J. Gong, J. Kim, S. J. Cho, R. W. Driver, Y. Lee, H. Kim and H.-S. Lee, Foldecture as a Core Material with Anisotropic Surface Characteristics, J. Am. Chem. Soc., 2015, 137, 2159–2162 CrossRef CAS PubMed.
- J.-H. Eom, J. Gong, S. Kwon, A. Jeon, R. Jeong, R. W. Driver and H.-S. Lee, A Hollow Foldecture with Truncated Trigonal Bipyramid Shape from the Self-Assembly of an 11-Helical Foldamer, Angew. Chem., Int. Ed., 2015, 54, 13204–13207 CrossRef CAS PubMed.
- J.-H. Eom, J. Gong, R. Jeong, R. W. Driver and H.-S. Lee, Parallelogram Plate Shaped Foldecture from the Controlled Self-Assembly of α/β-peptide Foldamer, Solid State Sci., 2015, 48, 39–43 CrossRef CAS.
- S. Kwon, B. J. Kim, H.-K. Lim, K. Kang, S. H. Yoo, J. Gong, E. Yoon, J. Lee, I. S. Choi, H. Kim and H.-S. Lee, Magnetotactic Molecular Architectures from Self-Assembly of β-Peptide Foldamers, Nat. Commun., 2015, 6, 8747 CrossRef CAS.
- J. Kim, S. Kwon, S. H. Kim, C. K. Lee, J. H. Lee, S. J. Cho, H.-S. Lee and H. Ihee, Microtubes with Rectangular Cross-Section by Self-Assembly of a Short β-Peptide Foldamer, J. Am. Chem. Soc., 2012, 134, 20573–20576 CrossRef CAS PubMed.
- E. Yoon, J. Gong, Y. Jung, W. Lee, R. W. Driver and H.-S. Lee, Unambiguous Characterization of Anisotropic Foldamer Packing in a Foldecture with an Elongated Hexagonal Plate Shape, Chem. Commun., 2016, 52, 5250–5253 RSC.
- I. M. Mandity, L. Fulop, E. Vass, G. K. Toth, T. A. Martinek and F. Fulop, Building β-Peptide H10/12 Foldamer Helices with Six-Membered Cyclic Side-Chains: Fine-Tuning of Folding and Self-Assembly, Org. Lett., 2010, 12, 5584–5587 CrossRef CAS PubMed.
- M. Konda, I. Maity, D. B. Rasale and A. K. Das, A New Class of Phase-Selective Synthetic β-Amino Acid Based Peptide Gelator: From Mechanistic Aspects to Oil Spill Recovery, ChemPlusChem, 2014, 79, 1482–1488 CrossRef CAS.
- W. C. Pomerantz, N. L. Abbott and S. H. Gellman, Lyotropic Liquid Crystals from Designed Helical β-Peptides, J. Am. Chem. Soc., 2006, 128, 8730–8731 CrossRef CAS PubMed.
- W. C. Pomerantz, V. M. Yuwono, C. L. Pizzey, J. D. Hartgerink, N. L. Abbott and S. H. Gellman, Nanofibers and Lyotropic Liquid Crystals from a Class of Self-Assembling β-Peptides, Angew. Chem., Int. Ed., 2008, 47, 1241–1244 CrossRef CAS PubMed.
- A. Hetenyi, I. M. Mandity, T. A. Martinek, G. K. Toth and F. Fulop, Chain-Length-Dependent Helical Motifs and Self-Association of β-Peptides with Constrained Side Chains, J. Am. Chem. Soc., 2005, 127, 547–553 CrossRef CAS PubMed.
- J. N. Israelachvili, 20 – Soft and Biological Structures, in Intermolecular and Surface Forces, ed. J. N. Israelachvili, Academic Press, 3rd edn, 2011, pp. 535–576 Search PubMed.
- R. D. Gopalan, M. P. Del Borgo, A. I. Mechler, P. Perlmutter and M. I. Aguila, Geometrically Precise Building Blocks: the Self-Assembly of β-Peptides, Chem. Biol., 2015, 22, 1417–1423 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |