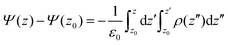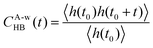Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
How stereochemistry of lipid components can affect lipid organization and the route of liposome internalization into cells†
Stefano
Borocci
 *ab,
Giuseppina
Bozzuto
*ab,
Giuseppina
Bozzuto
 c,
Cecilia
Bombelli
c,
Cecilia
Bombelli
 d,
Francesca
Ceccacci
d,
Francesca
Ceccacci
 d,
Giuseppe
Formisano
c,
Annarita
Stringaro
d,
Giuseppe
Formisano
c,
Annarita
Stringaro
 c,
Agnese
Molinari
c,
Agnese
Molinari
 *c and
Giovanna
Mancini
*c and
Giovanna
Mancini
 *b
*b
aDipartimento per la Innovazione nei sistemi Biologici, Agroalimentari e Forestali (DIBAF), Università degli Studi della Tuscia, L.go dell'Università, s.n.c., 01100 Viterbo, Italy. E-mail: borocci@unitus.it
bCNR, Istituto per i Sistemi Biologici, Area della Ricerca di Roma 1, SP35d 9, 00010 Montelibretti, Roma, Italy. E-mail: giovanna.mancini@cnr.it
cCentro Nazionale per la Ricerca e la Valutazione preclinica e clinica dei Farmaci, Istituto Superiore di Sanità, Viale Regina Elena 299, 00161 Roma, Italy. E-mail: agnese.molinari@iss.it
dCNR, Istituto per i Sistemi Biologici, Sede Secondaria di Roma-Meccanismi di Reazione c/o Dipartimento di Chimica Università degli Studi di Roma “Sapienza”, P.le A. Moro 5, 00185 Roma, Italy
First published on 2nd July 2021
Abstract
Though liposome-based drugs are in clinical use, the mechanism of cell internalization of liposomes is yet an object of controversy. The present experimental investigation, carried out on human glioblastoma cells, indicated different internalization routes for two diastereomeric liposomes. Molecular dynamics simulations of the lipid bilayers of the two formulations indicated that the different stereochemistry of a lipid component controls some parameters such as area per lipid molecule and fluidity of lipid membranes, surface potential and water organization at the lipid/water interface, all of which affect the interaction with biomolecules and cell components.
Introduction
The development of liposome technology has grown fast in the last 20–25 years, and a number of liposome-based drugs were approved for clinical use and more are in various phases of clinical trials.1–5 However, the parameters that control the interaction of these lipid vesicles with biological molecules and their biological targets, the uptake of their payloads and, hence, their efficacy, are not fully elucidated. It was reported that the lipid composition, vesicle size, surface electrical features and fluidity/rigidity of lipid membranes control the interaction with cells and the pathway of internalization.6,7 Once in the biological environment, liposomes interact with surrounding biomolecules, which adsorb on their surface and form a biomolecular corona that might affect targeting.8 Furthermore, the water bound to the surface of biomembranes typically called “glassy” or “biological water” plays a critical role in the interaction between cells and in the fusogenic properties of lipid membranes.9–11We previously reported that liposomes composed of dimyristoyl-sn-glycero-phosphocholine, DMPC, and one of the two diastereomeric cationic gemini amphiphiles, 1a or 1b (Fig. 1), showed different efficacy in the delivery of the photosensitizer meta-tetrahydroxyphenylchlorin (m-THPC) to malignant glioma cells, DMPC/1b liposomes being more efficient than DMPC/1a liposomes.12 Furthermore, it was found that the different stereochemistry of the gemini component also controls the intracellular distribution of m-THPC.12 These findings strongly suggest that the ‘diastereomeric’ liposomes might follow different pathways of internalization.
Herein we report on experimental and theoretical investigations aimed at elucidating, on the one hand, the pathways of internalization and the intracellular trafficking of the two liposomes (DMPC/1a, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and DMPC/1b, 6
4, and DMPC/1b, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and, on the other hand, the parameters that might control cell internalization. Appropriate inhibitors along with flow cytometry, antibody labelling along with laser scanning confocal microscopy (LSCM), and transmission electron microscopy (TEM) observations were used to investigate the potential differences in internalization pathways and intracellular trafficking. Molecular dynamics (MD) simulations of the lipid bilayers of the two formulations allowed us to shed light on the control of gemini component stereochemistry on the organization of lipids and on parameters such as lipid fluidity, surface electrical features and distribution of water bound to the surface of liposomes.
4) and, on the other hand, the parameters that might control cell internalization. Appropriate inhibitors along with flow cytometry, antibody labelling along with laser scanning confocal microscopy (LSCM), and transmission electron microscopy (TEM) observations were used to investigate the potential differences in internalization pathways and intracellular trafficking. Molecular dynamics (MD) simulations of the lipid bilayers of the two formulations allowed us to shed light on the control of gemini component stereochemistry on the organization of lipids and on parameters such as lipid fluidity, surface electrical features and distribution of water bound to the surface of liposomes.
Results and discussion
Inhibition of specific internalization routes
Multiple pathways of internalization have been described for liposomes such as fusion with the cell membrane, exchange of lipids with the plasma membrane and endocytosis,6,13–15 all of which were affected by liposome physicochemical features. Endocytic internalization can engage different cell components, whose involvement defines different cellular entry such as clathrin- or caveolae-mediated endocytosis, phagocytosis, macropinocytosis and non clathrin-, non caveolae-dependent endocytosis, each one affecting liposome intracellular fate.6,13,14In this study we investigated by flow cytometry analysis, in human glioblastoma LN229 cells, the effect of different endocytosis and trafficking inhibitors on the uptake of fluorescent m-THPC loaded in DMPC/1a and/or DMPC/1b liposomes. In particular, we used (i) chloropromazine, a cationic amphiphilic drug which interferes with clathrin-mediated endocytosis at multiple levels, by inhibiting the function of AP2, one of the key adaptor proteins in clathrin-mediated endocytosis, and by trapping receptors inside the endosomes, thus blocking their recycling; (ii) filipin that inhibits caveolae-mediated endocytosis by forming complexes with 3-β-hydroxysterols in the plasma membrane, thus inducing the disassembly of filamentous caveolin-1-coating; (iii) bafilomycin A1, a macrolide antibiotic that affects the clathrin pathway by specifically inhibiting vacuolar-type H(+)-ATPase, thus preventing the acidification of endosomes and lysosomes; and (iv) LY294002 that inhibits macropinocytosis by interacting with phosphatidylinositol 3-kinase whose activity controls the arrangement of actin filaments.16
The results of uptake experiments of liposome-included m-THPC, carried out after pre-treatment of cells with the different inhibitors, are reported in Table 1 and show that the uptake of m-THPC mediated by the two formulations was influenced to a different extent by the various endocytic inhibitors (Table 1), though a total inhibition never occurred because different processes can be functioning in parallel for cell internalization. The uptake of m-THPC delivered by DMPC/1a was mainly inhibited by filipin (∼75%) and, to a lesser extent, by chloropromazine (∼50%). On the other hand the uptake of m-THPC delivered by DMPC/1b liposomes was strongly inhibited by chlorpromazine (∼80%) and, to a lesser extent, by filipin (∼40%). Therefore, both formulations enter cells via endocytosis and cell internalization involves more than a single specific pathway. The extent of inhibition exerted by filipin and chlorpromazine indicates that caveolae are mainly involved in the internalization of DMPC/1a whereas the clathrin-mediated pathway is mainly involved in the internalization of DMPC/1b. Bafilomycin A1 and LY294002 inhibited to some extent (∼40% and ∼47%, respectively) the uptake of m-THPC loaded in DMPC/1b liposomes while they did not affect that mediated by DMPC/1a liposomes, showing on the one hand that the acidic compartments are involved only in the final destination of the DMPC/1b formulation and, on the other hand, that macropinocytosis is a concomitant route of its internalization.
| Inhibitor | Percentage of inhibition | Inhibition route | |
|---|---|---|---|
| DMPC/1a | DMPC/1b | ||
| Chloropromazine | 53 ± 5 | 80 ± 3 | Clathrin-mediated endocytosis |
| Bafilomycin A | 0 | 40 ± 8 | Acidification of endosomes |
| Filipin | 75 ± 4 | 38 ± 5 | Caveolae-mediated endocytosis |
| LY294002 | 0 | 47 ± 4 | Macropinocytosis |
Tracing cell entry and intracellular trafficking of DMPC/1 liposomes
In order to deeply investigate the mechanism of DMPC/1a and DMPC/1b internalization, we carried out immunofluorescence experiments by LSCM. In these experiments we used DMPC/1a and DMPC/1b liposomes fluorescently labelled with a NBD (4-nitrobenzo-2-oxa-1,3-diazole) tagged lipid. To trace cell entry and intracellular trafficking of liposomes we performed co-localization analysis by labeling the subcellular compartments involved in the endocytic pathway. In particular, we used antibodies against caveolin (CV), clathrin (CT), early endosomes (anti-Rab5), late endosomes (anti-Rab7), and lysosomes (anti-Lamp-1). Cells were first incubated with fluorescent DMPC/1a and DMPC/1b liposomes and then labeled with antibodies. LSCM optical sections relative to double-labeling experiments are reported in Fig. 2A (DMPC/1a) and 2B (DMPC/1b); merging of color-coded channels indicates colocalization and traces the pathway of internalization.Fluorescent DMPC/1a liposomes (green) co-localize with caveolin-rich regions (red), as shown by the regions of yellow pixels in panel VI of Fig. 2A, and with organelles labeled with Rab5-antibody (blue signal), as shown by light blue regions (early endosomes, panel IX, Fig. 2A). Localization of the same liposomes is hardly observed in clathrin (panel III of Fig. 2A), Rab7-positive or Lamp-1-positive organelles (late endosomes and lysosomes, respectively; panels XII and XV of Fig. 2A). Conversely, merging of color-coded channels shows that DMPC/1b liposomes localize preferentially in cytoplasmic regions rich in clathrin (yellow regions in panel III of Fig. 2B), in early and late endosomes and in the lysosomes (light blue regions in panels IX, XII and XV of Fig. 2B), whereas colocalization of DMPC/1b liposomes with caveolin (panel VI of Fig. 2B) is sporadic.
These results nicely confirmed those obtained in the inhibition experiments, indicating that caveolae and clathrin-coated vesicles serve as preferential “gates of entry” for DMPC/1a and DMPC/1b liposomes, respectively. In fact, DMPC/1a liposomes were mainly internalized in caveolae and in early endosomes (Rab5+ organelles) whereas DMPC/1b liposomes in clathrin-positive vesicles, late endosomes (Rab7+ organelles) and lysosomes (Lamp-1+ organelles). These findings are in agreement with previous studies reporting that the mechanism of caveolae uptake does not transport the internalized material to late endosomes and lysosomes, whereas clathrin-mediated endocytosis causes the endocytosed material to end up in degradative lysosomes.6,12
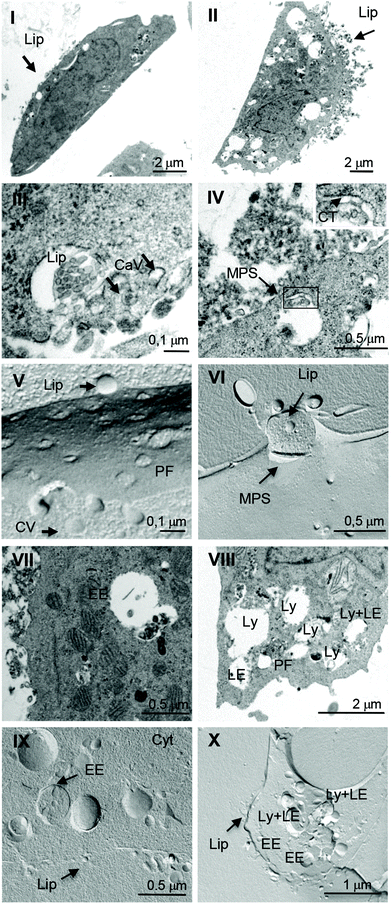
Ultrastructural investigation
The interaction of liposomes with the plasma membrane and intracytoplasmic organelles of glioblastoma cells was investigated at high resolution by transmission electron microscopy (TEM). After the treatment with liposomes, cells were processed by both ultrathin sectioning of resin embedded samples and freeze fracturing of frozen samples. Fig. 3 shows the results of the experiments on ultrathin sectioned (panels I–IV, VII, VIII) and freeze-fractured samples (panels V, VI, IX, X) relative to the experiments on DMPC/1a (left panels) and DMPC/1b (right panels) liposomes. A comparison of panel I and II shows that DMPC/1b liposomes (Lip) interacting with the plasma membrane on the apical surface of LN229 cells (panel II) are more in number with respect to DMPC/1a liposomes (panel I). Moreover, the intracytoplasmic vacuoles (endosomes and lysosomes) are more evident in cells interacting with DMPC/1b liposomes with respect to cells interacting with DMPC/1a liposomes. Panel III shows DMPC/1a liposomes interacting with the plasma membrane of a LN229 cell. Liposomes are visible in spherical invaginations of cell membranes that are caveolae surrounded by a hardly detectable fine meshwork (see arrows), likely representing the integral membrane protein caveolin located on the cytoplasmic side of the cell membrane. The observation of the same freeze fractured sample (panel V) confirms this evidence; in fact DMPC/1a liposomes strictly adhere to the cell membrane and interact with caveolae (arrow heads), which appear clustered on an area of the protoplasmic fracture face (PF). On the other hand, DMPC/1b liposomes interact with the cell surface preferentially with clathrin-coated areas (CT) that appear as spike-like arrays lining the cytoplasmic side of the plasma membrane (panel IV, inset, arrows). DMPC/1a liposomes are internalized in early endosomes (panels VII and IX), whereas DMPC/1b liposomes in early endosomes, late endosomes and lysosomes (panels VIII and X). The pictures reported in panels IV and VI suggest an internalization of DMPC/1b liposomes by macropynocitosis (MPS), thus confirming the results of inhibition experiments.Therefore, TEM observations confirm different endocytic routes, caveolae- and clathrin-mediated, for DMPC/1a and DMPC/1b, respectively, as traced by inhibition and immunofluorescence experiments. Actually, the different endocytic routes of the two formulations could explain why DMPC/1a liposomes were less efficient than DMPC/1b in delivering m-THPC in the same range of time.12 In fact, it is known that the uptake of caveolae-mediated endocytosis occurs at a much slower rate than that of clathrin-mediated endocytosis.6
We know from a previous investigation17 that DMPC/1a liposomes are characterized by a higher transition temperature and by a minor extent of lipid miscibility with respect to DMPC/1b liposomes. It was also found that DMPC/1a liposomes feature a surface potential, Ψs, higher than DMPC/1b liposomes.12 At 37 °C DMPC/1a and DMPC/1b liposomes feature a similar diameter (150–200 nm) after extrusion, and the ultrastructural investigation reported above did not suggest different sizes of the liposomes processed for internalization, which is/are the parameters that control the route of cell entry and how?
Molecular dynamics simulations
Molecular dynamic simulations (MD) can be a powerful tool to investigate at the atomic level how the stereochemistry of the gemini component dictates the differences in lipid organization that might account for the different physicochemical features and, hence, for the different biological behavior. Two main limitations characterize the use of MD to study lipid membranes: the size of the system and the accessible time scales; furthermore, simulations are based on a number of simplifying approximations and assumptions and hence cannot fully match the experimental conditions. Nevertheless, with the appropriate choice of parameters it is possible to gather information on relatively fast and localized molecular and intermolecular interactions responsible for the macroscopic properties of lipid membranes and hence their interaction with the biological environment.We carried out MD simulations of DMPC/1a, DMPC/1b (at 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 molar ratio) and DMPC bilayers, the last one to both investigate the effect of the gemini components on the physicochemical features of the lipid bilayer and validate our calculations, since the DMPC bilayer has been largely investigated.18–31 In particular, we took into consideration molecular interactions and organization that might control parameters such as fluidity, electrical features, and surface interaction with biomolecules.
4 molar ratio) and DMPC bilayers, the last one to both investigate the effect of the gemini components on the physicochemical features of the lipid bilayer and validate our calculations, since the DMPC bilayer has been largely investigated.18–31 In particular, we took into consideration molecular interactions and organization that might control parameters such as fluidity, electrical features, and surface interaction with biomolecules.
Area per lipid
First, we determined the average area per lipid, 〈AL〉, because it is related to the nature of lipid interaction and is an important property often used to validate a lipid force field32 and assess whether a lipid bilayer system has reached equilibrium through simulation.〈AL〉 was calculated by dividing the lateral (XY) dimensions of the simulated box by the number of lipid molecules in each leaflet. In the case of DMPC, 〈AL〉 was 0.638 ± 0.002 nm2 at 310 K, in agreement with experimental data and previous molecular dynamics simulation,18 whereas it was 0.716 ± 0.002 nm2 and 0.721 ± 0.003 nm2 for DMPC/1a and DMPC/1b, respectively. The increase of the area per lipid molecule in DMPC/1 bilayers is in agreement with literature data relative to other cationic mixed lipid bilayers. In particular, it is reported that while a low content of the cationic component causes a condensing effect and hence a decrease of area per lipid molecule,33,34 a high concentration of the cationic component, as in the case of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 DMPC/1 systems, involves electrostatic repulsion between charged headgroups and hence leads to an increase of the area per lipid with respect to pure phosphocholine.
4 DMPC/1 systems, involves electrostatic repulsion between charged headgroups and hence leads to an increase of the area per lipid with respect to pure phosphocholine.
To investigate the area per lipid of each component in the mixed lipid bilayer we applied the Voronoi tessellation using a set of selected key atoms as implemented in APL@voro,35 in particular, the phosphorus atom of DMPC and the mass center of the two stereogenic centers of the gemini headgroup. In the DMPC/1a system, the values of the area per lipid were 0.696 ± 0.003 nm2 and 0.719 ± 0.002 nm2 for DMPC and 1a, respectively, while in the DMPC/1b system the values were 0.706 ± 0.003 nm2 and 0.743 ± 0.006 nm2 for DMPC and 1b, respectively. Thus the two formulations feature a different averaged area per lipid (slightly larger in the case of DMPC/1b), and this difference is mostly due to the larger area per lipid of 1b with respect to the 1a component; the headgroup interactions involve also a slightly larger area of DMPC in DMPC/1b with respect to the DMPC/1a formulation (Fig. S1 in the ESI†). The larger area per lipid of 1b and DMPC, due to a higher extent of charge repulsion, suggests a higher charge exposure in gemini 1b with respect to 1a.
The area per lipid and its variation over simulation time were used to calculate the area compressibility modulus, KA, that provides a measure of the elastic properties of lipid bilayers, higher values of KA corresponding to higher rigidity of lipid bilayers36,37 (see the Material and methods section). The average KA calculated for the DMPC bilayer was 334 ± 22 mN m−1, whereas in the case of DMPC/1a and DMPC/1b it was 283 ± 19 mN m−1 and 253 ± 18 mN m−1, respectively. Therefore, the larger area per lipid, that characterizes the DMPC/1b bilayer with respect to the DMPC/1a one, involves an increase of the elasticity of the DMPC/1b membrane with respect to the DMPC/1a membrane.
Headgroup position and organization
The deepness and orientation of headgroups in lipid bilayers affect lipid compaction, surface electrical features and the capability of the system to bind counterions and water molecules. Therefore, we investigated by various means the position and organization of lipid headgroups.Fig. 4 shows the density distribution of the most relevant atoms and functional groups of lipid components at the lipid/water interface, namely nitrogen (NDMPC), phosphorus (PDMPC) and carbonyl groups (COsn-1 and COsn-2) of DMPC, both nitrogen N1a and N1b and both oxygen, O1a and O1b, of 1a and 1b, respectively. The analysis of mass density shows that in DMPC/1a and DMPC/1b bilayers nitrogen atoms of the gemini component are located close to the phosphorus atom of DMPC and replace the nitrogen atom of choline in the formation of charge-pairs. The electrostatic repulsion between DMPC choline groups and gemini ammonium groups induces a reorientation of phosphocholine headgroups and a consequent exposure of their nitrogen atom (NDMPC) toward the water phase. These findings are in agreement with results obtained by molecular dynamics simulation of cationic lipid membranes reported previously.33,38 The presence of the gemini component also induces a decrease of the thickness of the lipid bilayer, calculated as the distance between the peaks of the phosphate groups from the density profile, which was 3.57 nm in the case of DMPC (Fig. S2†), 3.22 nm for DMPC/1a and 3.21 nm for DMPC/1b, this being in agreement with the increase of surface area per lipid and fluidity, described above.
On the other hand, a difference between DMPC/1a and DMPC/1b bilayers concerns the time-averaged position of methoxy group oxygen atoms of 1a and 1b (O1a and O1b) that suggests a different conformation of gemini headgroups and a different orientation of 1a and 1b methoxy groups.
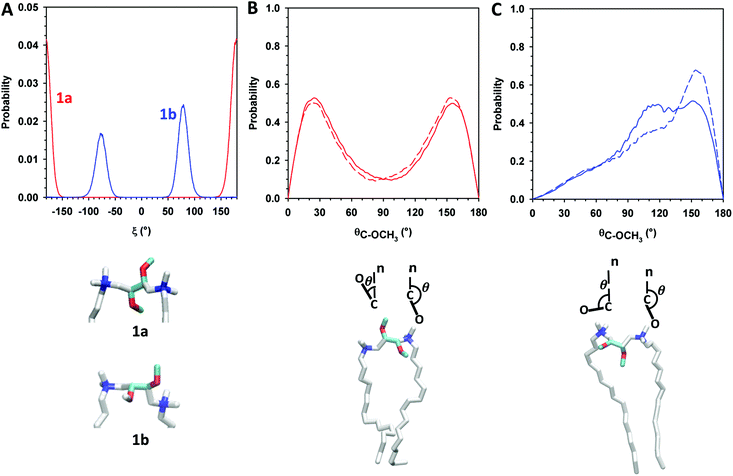
Next, we evaluated O–C–C–O dihedral angle distribution of gemini headgroups and calculated the angular distribution of C → OCH3 vectors of gemini and P–N vectors with respect to the normal to the lipid bilayer. The evaluation of the first parameter gave us detailed information on the effect of the different configuration of stereogenic centers of 1a and 1b on the conformation of the O–C–C–O headgroup portion. In the case of 1a, the O–C–C–O segment adopts exclusively an anti conformation whereas in 1b it adopts exclusively a gauche conformation (g+ = 57.9%, g− = 42.1%) as shown in Fig. 5A.
The orientation of C → OCH3 vectors with respect to the normal to the lipid bilayer confirms the different conformation adopted by the O–C–C–O portion in the gemini components. In 1a, methoxy groups are oriented in opposite directions, one towards the lipid bilayer hydrophobic region and the other one toward the water phase ( – Fig. 5B), whereas in 1b, both methoxy groups are mainly oriented towards the hydrophobic region (
– Fig. 5B), whereas in 1b, both methoxy groups are mainly oriented towards the hydrophobic region ( ), as shown in Fig. 5C and in the snapshots of DMPC/1a and DMPC/1b bilayers reported in Fig. 6.
), as shown in Fig. 5C and in the snapshots of DMPC/1a and DMPC/1b bilayers reported in Fig. 6.
The orientation of the P–N vector is strongly affected by the presence of the gemini component as it switches from 84.5° in DMPC to ∼36.0° in the case of both mixed lipid bilayers (Fig. S3 in the ESI†); however, it is not affected by the gemini stereochemistry.
A detailed picture of the short-range order of headgroups at the lipid bilayer surface was obtained by calculating the 2D radial distribution function (2D-RDF) between the phosphate group of DMPC and significant atoms of the gemini component. The 2D-RDF (g2D(r)) between the phosphate group and nitrogen atoms of gemini, reported in Fig. 7A, shows an intense peak at a lateral distance of 0.47 nm due to the first shell of neighbor nitrogen atoms around the phosphate group, and two less intense peaks at 0.77–1.0 nm range distance, for both DMPC/1a and DMPC/1b bilayers. This analysis does not give information on the orientation of the whole gemini molecule with respect to phosphorus atoms, namely it does not indicate if both nitrogen atoms of the gemini face the phosphate group. Insight into this point was given by the analysis of 2D-RDF between the DMPC phosphate group and the center of mass of gemini stereogenic centers (Fig. 7B). In both systems the 2D-RDF shows a first peak at a lateral distance of r = 0.49 nm and a more intense peak at 0.76 nm. However, in the case of DMPC/1b the peak at 0.49 is less intense with respect to DMPC/1a indicating that this configuration is less abundant. Representative snapshots of DMPC/1 pairs selected from MD trajectories contributing to the peaks in the radial distribution (Fig. 7C and D) illustrate the different organization of headgroups in the two lipid bilayers. The first peak at a lateral distance of r = 0.49 nm in the RDF can be ascribed to interactions between DMPC and gemini molecules that orient both nitrogen atoms toward the phosphate group (Fig. 7C). In the case of 1a the head-group conformation of gemini allows a tighter interaction between the head groups of DMPC and gemini, with the phosphate group coordinating two nitrogen atoms of the same gemini molecule. The second and more intense peak at 0.76 nm is due to the interaction between the phosphate and only one nitrogen atom of the neighbor gemini, with the second nitrogen atom far from the phosphate group of DMPC (Fig. 7D). These findings are in agreement with the results relative to the area per lipids.
Alkyl chains
Chain ordering and orientational mobility of methylene groups of alkyl hydrophobic tails affect lipid membrane fluidity, and are affected in turn by surface organization. Therefore, we investigated these parameters as a function of lipid composition by calculating the deuterium order parameter for sn-1 and sn-2 chains of DMPC in DMPC/1 bilayers.The order parameter, SCD, is defined as
Because we used a united atom force field, the order parameter of each methylene group was calculated based on the position of neighboring methylene and assuming a tetrahedral geometry.18,39Fig. 8A and B show the averaged order parameter, |SCD|, of methylene groups of sn-1 and sn-2 alkyl chains in the DMPC bilayer in the presence and in the absence of the gemini component. In all simulated systems, the value of |SCD| for the methylenes of DMPC tails is lower than 0.25, which indicates the occurrence of a fluid lipid bilayer with disordered alkyl chains.40 Actually the three bilayers were investigated at 310 K, i.e. above their transition temperature and are all in the liquid crystal phase. The presence of gemini involves a decrease of the order parameter of DMPC tails, indicating less ordered tails in the mixed bilayers with respect to the pure phospholipid, with very small differences due to the nature of the gemini. On the other hand, a relevant difference between the mixed systems concerns the averaged order parameter of methylene groups of gemini alkyl chains (Fig. 8C), the first four methylenes of 1a alkyl chains showing a higher order parameter (lower orientational mobility) with respect to 1b. The anti orientation of 1a methoxy groups involves a steric hindrance on the first methylenes of alkyl chains that reduces their orientational mobility. Conversely, in 1b the gauche conformation of the O–C–C–O torsional angle and the consequent syn orientation of methoxy groups allow a higher flexibility of the first methylenes of alkyl chains.
It is worth noting that these results are in good agreement with those concerning the surface area per lipid, as it is known that the SCD of alkyl chains is related to the surface area per lipid, a larger area per lipid corresponding to a lower order parameter, and, hence, to more disordered alkyl chains.41,42
Electrostatic potential
The organization of lipids at the lipid/water interface affects surface charge of lipid bilayers that in turn affects the interaction of liposomes with cells.43–46 Hence, we calculated the electrostatic potential as a function of the distance from the center of the lipid bilayer to characterize the electrostatic properties of DMPC/1 bilayers. Fig. 9A shows the profiles of electrostatic potential of DMPC, DMPC/1a and DMPC/1b with respect to the potential of the lipid bilayer center set to zero.Both mixed lipid bilayers feature a positive electrostatic potential with respect to bulk water; however, they feature a different boundary potential, Ψb, (defined as the difference between the electrostatic potential found at the center of the lipid bilayer and the electrostatic potential of the water phase), the Ψb of DMPC/1b being higher (1.175 V) than that of DMPC/1a (1.062 V). We analyzed also the surface charge density, σ(z), of the cationic lipid bilayers as a function of the distance from the center of the lipid bilayer, and the relative profiles are reported in Fig. 9B. In DMPC/1b the value of σ(z) close to the surface of the lipid bilayer, in the region of DMPC phosphate groups and of the headgroups of gemini (1.0 < z < 1.9 nm), is positive and slightly higher with respect to DMPC/1a. For z > 2.0 nm, in the region of nitrogen atoms of DMPC, surface charge density of DMPC/1a switches to a higher value with respect to DMPC/1b.
These results are not in contrast with those reported previously12 and obtained by experimental data that, as mentioned above, gave a higher surface potential in the case of DMPC/1a. In fact, the experimental determination of surface potential was carried out by an indirect method based on the effect of the microenvironment at the water/lipid interface of liposomes on the pKa of the umbelliferone fluorescent probe. The different headgroup organization and lipid packing of DMPC/1a and DMPC/1b bilayers could determine a different position of the probe at the lipid/water interface providing the value of the detected potential surface of slightly different zones in the case of DMPC/1a and DMPC/1b.
Water organization and dynamics
Some features of water molecules at the lipid/water interface influence electrostatic and other surface properties of lipid bilayers.47 In addition, water bound at the lipid membrane surface might control the interaction of liposomes with biomolecules.9–11 Therefore, we analyzed the penetration, the orientation and the mobility of water molecules as a function of their position with respect to the lipid bilayer surface, the last being defined as the region containing phosphorus (DMPC) and nitrogen (gemini) atoms (as described in the Material and methods section).The analysis of water properties with respect to the membrane surface took into account the roughness of the surface. Fig. 10A shows the water density profiles relative to DMPC, DMPC/1a and DMPC/1b bilayers calculated as a function of the distance d from the surface of the bilayer (d = 0). All the systems do not show a smooth density profile and water density defines four regions: region I (−1.0 < d < −0.14 nm for DMPC and −1.0 < d < 0.05 nm for DMPC/1) includes water molecules close to carbonyl groups; region II (−0.14 < d < 0.40 nm for DMPC and 0.05 < d < 0.52 nm for DMPC/1) includes the first shell of water molecules close to phosphate and ammonium ions of DMPC and gemini; region III (0.40 < d < 0.80 nm for DMPC and 0.52 < d < 1.0 nm for DMPC/1) the second shell of water molecules close to ammonium ions of DMPC; region IV (d > 0.80 nm for DMPC and d > 1.0 nm for DMPC/1) bulk water.
Fig. 10 shows that the hydrophobic region close to carbonyl groups of phosphocholine (region I) in the case of the DMPC bilayer is characterized by a deeper penetration of water molecules with respect to the DMPC/1 systems. Some differences between the cationic bilayers were observed in water density close to the lipid surface. In fact, DMPC/1a shows a higher density of water molecules in region I, whereas DMPC/1b shows it higher in the region of the second solvation shell of DMPC ammonium groups (region II).
Water molecule orientation with respect to the lipid bilayer was then evaluated by analyzing the average cosine of the angle formed by the dipole vector and the normal to the lipid bilayer (n). Note that for water randomly oriented the value of the averaged cosine is zero. On the other hand, positive and negative values of the averaged cosine correspond to an averaged orientation of water molecules with oxygen atoms pointing inward and outward the lipid bilayer, respectively. Hence, higher positive or negative values of averaged cosine correspond to a larger extent of orientation of water molecules. Fig. 10B shows water dipole orientation as a function of the distance d from the surface of DMPC/1 bilayers. In both systems, the oxygens of water molecules are oriented toward the lipid bilayer; however water molecules of the second hydration shell of DMPC ammonium ions (region III) bound to the DMPC/1a bilayer feature a higher grade of orientation with respect to those bound to DMPC/1b; only negligible differences were found in the other regions of hydration.
Lateral diffusion of water (Dlat) and the relaxation time (τ1) of dipolar moment of water molecules as a function of the distance d from the surface of the lipid bilayer were also calculated, because these parameters allow evaluating the translational mobility and the rotational diffusion of water close to the lipid bilayer.
For the model of water we used (SPC), at 310 K we obtained for the bulk D = (5.01 ± 0.02) × 10−5 cm2 s−1 and τ1 = 2.5 ± 0.2 ps. In all simulated systems we observed (Table 2) a slowdown of lateral mobility and rotational relaxation of water close to the lipid surface with respect to bulk water. In particular, in the region I the mobility and rotational relaxation of water are reduced, with respect to bulk water, by ∼10 and ∼80 times, respectively, due to deep penetration into the lipid bilayer. In the region II, the mobility and rotational relaxation of water are reduced, with respect to bulk water, by ∼4 and ∼16 times, respectively. Finally, in the region III and IV both lateral diffusion and rotational relaxation of water approach the value of bulk water. However, while in the case of the DMPC bilayer the values of both parameters relative to region IV are similar to those of bulk water, in the cases of DMPC/1 bilayers they are reduced with respect to bulk water, thus indicating that the perturbation effect of the lipid bilayer and chloride ions on the organization and dynamics of water extends beyond 1.5 nm from the surface of the lipid membrane. Therefore, this analysis indicates that the presence of the gemini component affects the mobility of water molecules at the surface of the lipid bilayer in regions II–IV; furthermore, it shows that its stereochemistry does not affect lateral mobility of water molecules (see also the ESI†), while slightly affecting water rotational relaxation in regions II and III. In particular water molecules bound to the DMPC/1b bilayer show a faster relaxation with respect to those bound to the DMPC/1a bilayer.
| Region | DMPC | DMPC/1a | DMPC/1b | |||
|---|---|---|---|---|---|---|
| D lat | τ 1 | D lat | τ 1 | D lat | τ 1 | |
| I | 0.47 ± 0.02 | 154.9 ± 6.0 | 0.46 ± 0.04 | 188.9 ± 7.0 | 0.48 ± 0.03 | 194.1 ± 7.0 |
| II | 1.94 ± 0.03 | 31.3 ± 2.0 | 1.46 ± 0.03 | 42.3 ± 3.0 | 1.47 ± 0.02 | 38.9 ± 3.0 |
| III | 3.98 ± 0.02 | 4.4 ± 0.2 | 3.31 ± 0.02 | 7.3 ± 0.2 | 3.32 ± 0.03 | 6.5 ± 0.3 |
| IV | 4.90 ± 0.02 | 2.6 ± 0.2 | 4.47 ± 0.02 | 2.9 ± 0.2 | 4.45 ± 0.02 | 3.0 ± 0.2 |
Lateral and rotational diffusion of water are influenced by the capacity of forming and breaking hydrogen bonds with other molecules.48 Therefore, in order to better characterise the structure and dynamics of water molecules close to the surface of DMPC/1 bilayers we analysed hydrogen bonds between water molecules close to the lipid surface and those between water molecules and oxygen atoms of lipids.
Calculations indicated that in DMPC/1a and DMPC/1b bilayers each molecule of DMPC forms 5.8 and 5.7 hydrogen bonds with neighbour water molecules, respectively, a number slightly lower with respect to a mere DMPC bilayer where each molecule of DMPC forms 6.2 hydrogen bonds.
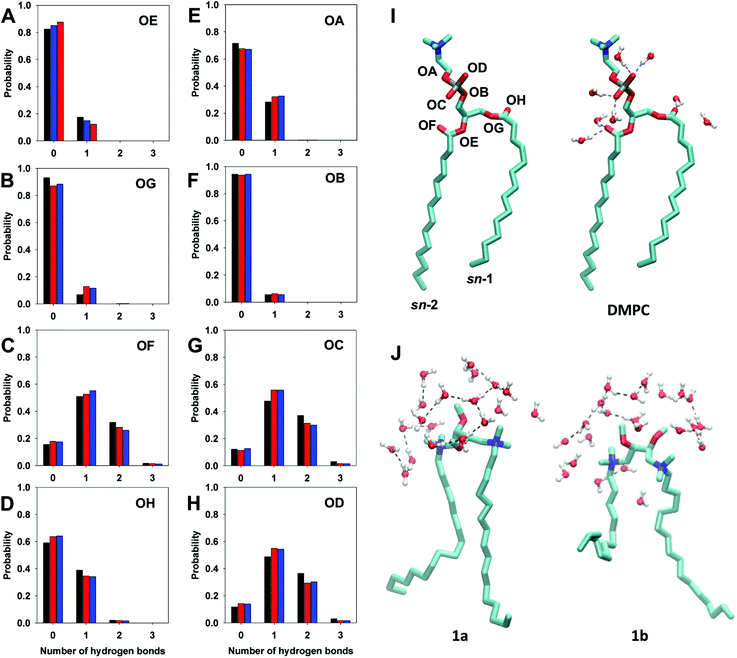
Fig. 11 shows the average probability of hydrogen bond formation between water molecules and oxygen atoms of DMPC. The different probability to form hydrogen bonds reflects the different exposure of DMPC oxygen atoms to the solvent and hence their different environment.21,23
The analysis, shown in Fig. 11, indicates that ester oxygens OA, OB, OE, and OG (OA and OB belonging to C–O–R bonds and OE and OG to P–O–R bonds) have a lower tendency to form hydrogen bonds with respect to carbonylic oxygen atoms, OF and OH (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and non-ester oxygen of the phosphate group OC and OD. In fact, in the DMPC bilayer the ester oxygens OA, OB, OE, and OG give rise exclusively to a single hydrogen bond with a probability of 30% for OA and of ∼15% for OB, OG and OE. This tendency slightly increases in the presence of the gemini component, except for OE, which shows a modest reduction with respect to the bilayer of pure DMPC.
O) and non-ester oxygen of the phosphate group OC and OD. In fact, in the DMPC bilayer the ester oxygens OA, OB, OE, and OG give rise exclusively to a single hydrogen bond with a probability of 30% for OA and of ∼15% for OB, OG and OE. This tendency slightly increases in the presence of the gemini component, except for OE, which shows a modest reduction with respect to the bilayer of pure DMPC.
In the case of OC and OD oxygen atoms the probability of forming one hydrogen bond increases in the presence of the gemini (56% in DMPC/1 bilayers versus 48% in DMPC, Fig. 11G and H), whereas the probability to form two hydrogen bonds for each oxygen is reduced (30% in DMPC/1 bilayers versus 37% in DMPC). The same trend was observed also in the case of OF oxygen atoms (sn-2 carbonyl oxygen), the probability of forming one hydrogen bond being 54% in DMPC/1 bilayers versus 50% in DMPC bilayer, and for the formation of two hydrogen bonds 25% in DMPC/1versus 32% in DMPC. The other carbonylic oxygen atom, sn-1 OH, shows a lower tendency to bind water molecules with respect to sn-2 OF, (Fig. 11C and D), this difference reflecting the different orientation of the sn-1 and sn-2 carbonyl groups with respect to the lipid surface and a different hydration of the two ester groups, as evidenced by the analysis of the radial distribution function (Fig. S4 in the ESI†).
The stereochemistry of the gemini component does not affect the capability of DMPC to bind water molecules by hydrogen bonds.
Differently from what observed in the case of DMPC oxygen atoms, the oxygen atoms of gemini headgroups form hydrogen bonds with water molecules very rarely. In fact, an averaged value of 1.2 hydrogen bonds was calculated over all 52 molecules of 1. The absence in the gemini components of functional groups able to form stable intermolecular hydrogen bonds with water and the hydrophobic nature of the charged ammonium groups lead to the formation of “clathrate-like” structures of water around the headgroups of cationic gemini molecules (Fig. 11J) that play a key role in the structure and dynamics of water close to the lipid bilayer.20,49,50
The dynamical properties of water molecules close to the lipid bilayer surface were then investigated by analysing their ability to break and reform hydrogen bonds between themselves and with DMPC. The analysis of lifetime of hydrogen bonds gave in the case of relaxation time (τ) of water–water hydrogen bonds τw–wHB = 3.2 ps in the case of DMPC, τw–wHB = 4.7 ps in DMPC/1a and τw–wHB = 4.4 ps in DMPC/1b.
The hydrogen bonds between water and DMPC have a longer lifetime with respect to water–water hydrogen bonds, namely τhg-wHB = 41.5 ps, τhg-wHB = 50.0 ps and τhg-wHB = 49.4 ps in DMPC, DMPC/1a and DMPC/1b bilayers, respectively.
Therefore, in the interfacial region of DMPC/1 bilayers we observed a stronger interaction between water molecules and between water and lipid molecules with respect to the DMPC bilayer due to longer lasting hydrogen bonds. This could be due to lower density of water in the interfacial region of DMPC/1 with respect to DMPC (Fig. 10) that reduces hydrogen bond switching between water molecules.51 In addition, the orientation of water molecules around gemini headgroups and the formation of “clathrate-like” structures might play a key role in the increase of the lifetime of water–water hydrogen bonds. In fact, in “clathrate-like” structures formed around hydrophobic surfaces, water molecules are ordered and translational and rotational diffusion is reduced.52–57
Conclusion
This study highlights how the stereochemical molecular difference can have a cascade effect on liposome features, by deeply affecting lipid organization and, in turn, liposome physic-chemical properties, finally translating into a different biological fate of the aggregates.The two liposomes, DMPC/1a and DMPC/1b, differ exclusively for the stereochemistry of the gemini component, 1a being chiral because of the S configuration of both its stereogenic centres and 1b being a meso form, i.e. achiral. This difference, that might seem subtle, was found to control some of the physicochemical features12,17 of the considered lipid membranes such as the transition temperatures of the two liposomes and the lipid miscibility. In this work it was shown that the stereochemical information also controls the biological fate of DMPC/1a and DMPC/1b liposomes, the first formulation entering cells mainly by caveolae mediated endocytosis and the second one by clathrin mediated endocytosis and partly by macropinicytosis. Hence the stereochemistry of the gemini component controls the intracellular target of the DMPC/1 carriers and of their payloads. In the case of DMPC/1a the target is early endosomes whereas in the case of DMPC/1b it is lysosomes; this means that, if a plasma membrane were the target of a given drug, DMPC/1a would be the formulation of choice, whereas if the nucleus were the target, as in the case of DNA, then DMPC/1b would be the carrier for reaching it. This is confirmed by our previous results that indicated DMPC/1b as a better formulation than DMPC/1a for the delivery of genetic materials such as a DNA plasmid58 or siRNA.59,60
Finding a connection between the stereochemical structure, the physicochemical properties and finally the biological fate of liposomes is a complicated task, and one should keep in mind that physicochemical properties are interrelated and contribute altogether in defining liposome biological features.
The MD simulation allowed understanding what happens at the molecular level, providing information that gathered together give us a clear picture of the two systems, DMPC/1a and DMPC/1b. Its results are summarized in Table 3 together with those obtained in the biological evaluation. The stereochemistry dictates the conformation of the head group of the gemini component embedded in the lipid bilayer. The different conformation assumed by the two stereoisomers results in a different exposure of charges. From this difference derive the differences between DMPC/1a and DMPC/1b in terms of extent of charge repulsion between gemini molecules and interaction with the phosphate group of DMPC, and hence the difference in area per lipid (higher in the case of DMPC/1b and 1b itself with respect to DMPC/1a and 1a, respectively) with a consequent effect on the elasticity of lipid bilayers. The different conformation assumed by 1a and 1b also implies a different order parameter of the respective alkyl tails and hence a different fluidity of DMPC/1a and DMPC/1b, DMPC/1b being less ordered and more fluid than DMPC/1a. The different reciprocal organization of lipids components due to the different stereochemistry of gemini components is also reflected in the surface electrical features and in the penetration and organization of water at the lipid/water interface. In fact, the DMPC/1b bilayer features a higher boundary potential than DMPC/1a. Penetration of water is different in the regions defined with respect to the surface of the lipid bilayer, in particular, in the case of DMPC/1b water density is higher than in DMPC/1a, in the region that includes the first shell of water molecules close to phosphate groups and ammonium ions of DMPC and gemini (region II), at the boundary with the region of the second shell of water molecules close to ammonium ions of DMPC (region III). This implies a higher probability of hydrogen bond switching between water molecules and a shorter hydrogen bond lifetime with respect to water at the surface of the DMPC/1a bilayer. Consistently, water molecules in regions II and III of the DMPC/1a bilayer were more oriented and characterized by a longer rotational dipolar relaxation time with respect to those in the DMPC/1b bilayer.
| Liposome | Biological features | Lipid organization and physicochemical features | Water at surface | H-Bond | |||
|---|---|---|---|---|---|---|---|
| Cell internalization | Intracellular trafficking | 〈AL〉 (nm2) | K A (mN m−1) | Ψ b (V) | τ 1 region III (ps) | τ w-wHB (ps) | |
| DMPC/1a | Caveolae | Early endosomes | 0.716 ± 0.002 | 287 ± 20 | 1.062 | 7.3 ± 0.2 | 4.7 ± 0.2 |
| DMPC/1b | Clathrin and macropinocytosis | Late endosomes and lysosomes | 0.722 ± 0.002 | 257 ± 18 | 1.175 | 6.5 ± 0.3 | 4.4 ± 0.2 |
All these differences, some marked and other subtle, account for the different biological behaviour of the formulations, because they all affect the interactions with biomolecules and the cell membrane. In particular, the organization of water and the surface potential control the interaction with protein in serum and thus the nature of biomolecular corona that in turn control the interaction with cell components and the intracellular fate.6,7
The message is that ascribing the control of biological features exclusively to membrane fluidity or particle charge or size or biological water might be too generic and restrictive since, as shown here, physicochemical parameters are interrelated and it is their whole to control the biological outcome.
Experimental materials and methods
Materials
Dimyristoyl-sn-glycero-phosphatidylcholine (DMPC, purity >99%) was purchased from Avanti Polar Lipids (Alabaster, AL); m-tetrahydroxyphenylchlorin (m-THPC) was a kind gift by Biolitec (Jena, Germany); RPE and HPLC grade solvents (chloroform, ethanol, isopropanol, bidistilled water) were purchased from Carlo Erba Reagenti (Milano, Italy); polycarbonate membranes were purchased from Whatman Nuclepore (Toronto, ON, Canada); chlorpromazine, Filipin III from Streptomyces filipinensis, Bafilomycin A1 from Streptomyces griseus, LY294002, phosphate buffered saline (PBS) and Hanks’ balanced salt solution (HBSS) were purchased from Sigma-Aldrich (St Louis, MO, USA); Dulbecco's modified Eagle's medium (DMEM) and streptomycin were purchased from Gibco Life Technologies (Paisley, UK); fetal bovine serum (FBS) was purchased from Hyclone (Carmlington, UK); Rab-5 and Rab-7 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); Lamp-1 was purchased from BD Transduction Laboratories™; Alexa Fluor® Dye was purchased from Molecular Probes (Poortgebouw, Netherlands); Epon 812 resin was purchased from Electron Microscopy Science (Fort Washington, PA).Gemini surfactants (S,S)-2,3-dimethoxy-1,4-bis(N-hexadecyl-N,N-dimethylammonium)butane bromide, 1a, and (S,R)-2,3-dimethoxy-1,4-bis(N-hexadecyl-N,N-dimethylammonium)butane bromide, 1b, were synthesized as previously described.61,62
Preparation of liposomes
Aqueous dispersions of DMPC/1 liposomes were prepared according to a reported procedure.63 Briefly, a film of lipids (total 25.0 μmol) and m-THPC was prepared on the inside wall of a round-bottom flask by evaporation of a CHCl3 solution containing appropriate amounts of DMPC and 1 to obtain the 60/40 molar percentage mixture. The lipid films were kept overnight under reduced pressure (0.4 mbar) and 2.0 mL of PBS buffer solution (10−2, M pH 7.4) was added in order to obtain 12.5 mM lipid dispersions. The aqueous suspensions were vortex-mixed and freeze-thawed six times from liquid nitrogen to 313 K. Dispersions were then extruded (10 times) through a 100 nm polycarbonate membrane. The extrusions were carried out at 307 K, well above the transition temperature of DMPC (297.2 K), using a 2.5 mL extruder (Lipex Biomembranes, Vancouver, Canada).m-THPC containing liposomes were prepared by adding an appropriate volume of a m-THPC(5 × 10−4 M, EtOH abs) stock solution to the chloroform solution of the lipids to obtain, after hydration, a 50 μM m-THPC final concentration.
Cell cultures
A human (LN229) glioblastoma cell line was grown as the monolayer in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 1% penicillin (50 IU mL−1) and streptomycin (50 IU mL−1) under a humidified atmosphere of 5% CO2 in a water jacketed incubator at 37 °C.Flow cytometry
Cells were pre-treated with inhibitors and, subsequently, with DMPC/1a or DMPC/1b liposomes loaded with m-THPC, for 1 h. The inhibition of clathrin function was achieved by pre-treating LN229 cells with 28 μM chlorpromazine for 60 min at 37 °C. To investigate the caveolae-mediated uptake, cells were treated with 9 μg mL−1 of Filipin III from Streptomyces filipinensis for 60 min at 37 °C. In order to investigate the dependence of liposome uptake on the acidification of endosomes, cells were treated with 100 nM Bafilomycin A1 from Streptomyces griseus (Sigma-Aldrich) for 60 min at 37 °C. The role of the actin cytoskeleton in endocytosis was investigated by pre-treating cells with 30 μM LY294002 for 60 min at 37 °C.At the end of each treatment, cells were washed with ice-cold Hank's balanced salt solution (HBSS), detached with EDTA and 0.25% trypsin, resuspended in ice-cold PBS and immediately analyzed for the photosensitizer content. Fluorescence signals were analyzed with a FACScan™ flow cytometer (Becton Dickinson, Mountain View, CA) equipped with a 15 mW, 488 nm and air-cooled argon ion laser. The fluorescence emission was collected through a 670 nm band-pass filter and acquired in “log” mode. At least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were analyzed. The m-THPC content was evaluated as fluorescence intensity, expressed as the mean fluorescence channel (MFC). The analysis was performed using CellQuest™ software (Becton Dickinson). Results analyzed were reported as mean percent of inhibition obtained from 3 independent experiments.
000 events were analyzed. The m-THPC content was evaluated as fluorescence intensity, expressed as the mean fluorescence channel (MFC). The analysis was performed using CellQuest™ software (Becton Dickinson). Results analyzed were reported as mean percent of inhibition obtained from 3 independent experiments.
Laser scanning confocal microscopy
For the study of the internalization pathway and localization of the liposomes within LN229 cells, liposomes fluorescently tagged with a NDB (4-nitrobenzo-2-oxa-1,3-diazole) and antibodies against clathrin, caveolin, early endosomes (Rab-5), late endosomes (Rab-7) and lysosomes (Lamp-1) were used.Cells grown for 24 h on glass coverslips were incubated with NBD tagged liposomes for 18 h at 37 °C. At the end of the treatment LN229 cells were fixed with 3.7% paraformaldehyde in PBS for 15 min at room temperature, washed twice in PBS, and permeabilized with 0.04% Triton in PBS + 1% BSA. Cells were washed twice, incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 primary antibodies in PBS + 1% BSA for 45 min at 37 °C, washed twice and incubated with 1
100 primary antibodies in PBS + 1% BSA for 45 min at 37 °C, washed twice and incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 secondary antibody 633 nm Alexa Fluor® Dyes in PBS + 1% BSA for 30 min at 37 °C. Finally, samples were washed and observed under a Leica TCS SP2 confocal microscope (Leica Microsystems, Mannheim, Germany).
50 secondary antibody 633 nm Alexa Fluor® Dyes in PBS + 1% BSA for 30 min at 37 °C. Finally, samples were washed and observed under a Leica TCS SP2 confocal microscope (Leica Microsystems, Mannheim, Germany).
Transmission electron microscopy
For transmission electron microscopy observations, LN229 cells were treated with DMPC/1a or DMPC/1b liposomes for 18 h at 37 °C. At the end of the treatment, cells were fixed with glutaraldehyde 2.5% solution in 0.2 M cacodylate buffer (pH 7.3) for 1 h at room temperature. For ultrathin sectioning, after washing in 0.2 M cacodylate buffer (pH 7.3), cells were post-fixed in 1% osmium tetroxide (OsO4) solution in 0.2 M cacodylate buffer (pH 7.3) for 2 h at room temperature. After post-fixation with OsO4, cells were dispersed in liquid agar and then let to solidify on ice. Samples were then dehydrated through graded ethanol concentrations, with a final propylene oxide dehydration. Samples were then embedded in Epon 812 resin. Ultrathin sections of embedded samples were stained with lead citrate and 2% uranyl acetate solutions and examined with a Philips EM 208S electron microscope (FEI Company, Eindhoven, The Netherlands).For freeze-fracturing cells were washed twice in 0.1 M cacodylate buffer, resuspended in the same buffer containing 30% glycerol, and incubated overnight at 4 °C. Samples were then put on carriers and quickly frozen in Freon 22, partially solidified at the liquid nitrogen temperature. The mounted carriers were then transferred into a Bal-Tec BAF 060 freeze-etch unit (BAL-TEC, Balzers, Liechtenstein), cleaved at −100 °C at a pressure of 2–4 × 10−7 mbar, shadowed with 2 nm of platinum–carbon and replicated with a 20 nm carbon film. Cells were digested for 2 h from the replicas by chlorox. The replicas were mounted on grids, and examined with a Philips 208S transmission electron microscope (FEI Company, Eindhoven, The Netherlands).
Computational methods
Molecular dynamics simulations were performed on mixed lipid bilayers consisting of 76 molecules of DMPC and 52 molecules of gemini (1a or 1b) embedded in 5123 SPC (Single Point Charge)64 water molecules and 104 chloride ions to neutralize the electric charges of gemini components. The number of lipid molecules (128 total) was chosen based on recent reports.65–67Starting coordinates of the lipid bilayer, composed of 128 molecules of DMPC, were taken from the website of Biocomputing laboratory at the University of Calgary (http://wcm.ucalgary.ca/tieleman/downloads) and solvated with 5227 water molecules in a rectangular box of 6.5 × 6.5 × 7.0 nm3. The mixed bilayers of DMPC and gemini components were obtained by a random selection of 52 molecules of DMPC replaced by the same number of gemini molecules. The positive charges of the mixture lipid bilayers were neutralised by replacing 104 water molecules with chloride ions.
Simulation parameters
All molecular dynamics simulations were performed with the version 5.1.5 of GROMACS software package.68 The lipids and the gemini molecules were described by using the Berger force field69 with the GROMOS G53a6 bonding parameters.70 The chloride ions were described by using the OPLS parameters.71The partial atomic charges of the headgroups of gemini molecules were evaluated using the RESP fit method72 of the electrostatic potential obtained from the HF/6-31G(d) wave function using the Merz–Singh–Kollman scheme. The electrostatic potential was calculated on the B3LYP/6-31 G(d,p) optimized geometry of the gemini headgroups by using a high point density around the molecule (17 points per Å2 and 10 layers around the van der Waals molecular surface). The RESP fit of the electrostatic potential was performed by using the antechamber module of AmberTools16.73 The quantum mechanical calculations were performed by using the GAUSSIAN03 program package.74 The partial atomic charges and the atom type of the headgroups of gemini molecules are reported in the ESI.†
The torsional parameters for O-CH2-CH2-O and CH3-O-CH2-CH2 dihedrals of the gemini molecules were parameterized as described in the ESI.†
Lennard–Jones and electrostatic interactions were calculated using a cut-off of 1.0 nm and the long-range electrostatic interactions were accounted by using the particle mesh Ewald method (PME).75
All bonds were constrained using the P-LINCS algorithm76,77 whereas the geometry of water molecules was fixed with the SETTLE algorithm.78 The simulations were carried out with a time step of 2 fs in the NPT ensemble. After the initial energy minimization, the simulated systems were warmed up by five consecutive unrestrained MD from 100 K to 323 K in 500 ps. After 120 ns of equilibration at 323 K the temperature of each system was cooled down from 323 K to 310 K in 1.5 ns by four consecutive MD and finally simulated for 500 ns at 310 K.
The lipids, gemini surfactants, water and ions were coupled separately to a temperature bath using the velocity rescale method79 with a time constant of 0.1 ps. The pressure was kept at 1 bar by weakly coupling to a pressure bath,80 using a coupling constant of 1.0 ps and an isothermal compressibility of 4.5 × 10−5 bar−1 and in the last 400 ns the Parrinello–Rahman barostat81 (P = 1 bar, τP = 4.0 ps) was used. Pressure coupling was applied semi-isotropically: the z and the x–y (isotropic) box dimensions were allowed to vary independently. Periodic boundary conditions were applied in all three dimensions.
For each simulated system three simulations with different random initial velocity were performed.
Analysis
The analyses of MD trajectories were performed by using the GROMACS analysis tools, VMD 1.9.3 program82 and in-house scripts. The initial 200 ns of the production phase at 310 K were excluded for the analysis of the equilibrium properties. The remaining 300 ns at 310 K were used for the analysis. All figures of molecular structures have been produced by using VMD.Area per lipid
The average area per lipid of the lipid bilayer was calculated from the lateral (XY) dimension of the simulated box divided by the number of the lipids in each leaflet. The area per lipid of each component in the mixed lipid bilayer was calculated by using the Voronoi tessellation as implemented in APL@voro.35 We used, as selected key atoms for tessellation, the phosphorus atom of DMPC and the center of mass of the two stereogenic carbon atoms of gemini headgroups.Isothermal area compressibility modulus
The isothermal area compressibility modulus was calculated fromwhere kB is the Boltzmann constant, T is the simulation temperature, 〈AL〉 is the average area per lipid and σ(AL) is the variance of AL.
Electrostatic potential
The electrostatic potential of the lipid bilayer was calculated by double integration of the averaged charge density ρ(z), across the bilayerwhere the electrostatic potential of the water phase was set to zero.
The surface charge density as a function of z was calculated using the relationship:
Water density and lateral diffusion
The analysis of the water density, water orientation and the lateral diffusion of water molecules with respect to the membrane surface was performed taking into account the roughness of the surface considering, as a bilayer surface, the surface containing the phosphorus (DMPC) and nitrogen (gemini) atoms. Each water molecule was classified as a function of its distance from the surface of the bilayer. First, at any frame of trajectory, we projected the coordinates of phosphorus and nitrogen (gemini) atoms on the plane z = 0. Next, the coordinates of each water molecule were projected on the plane z = 0 and we identified the closest P or N atom with the least value of the distance in the XY-plane. Finally the distance from the surface of the bilayer, d, (vertical distance) was defined as the distance between the z coordinate of water oxygen and the z coordinates of the closest P or N atom identified previously. The classification of water molecules with our algorithm is consistent with other methods used in the literature.9,84The lateral diffusion coefficient Dlat of water molecules as a function of the distance d from the surface of the lipid bilayer was obtained by dividing the d dimension into 0.2 nm slabs.85 The lateral diffusion coefficient Dlat was calculated from the mean square displacement (MSD) of water molecules using the Einstein equation:
The MSD of water in each zone was calculated over 20 ps intervals by considering only the water molecules located in the same zone at time t0 and t.
The rotational dynamics of water molecules in the different regions of lipid bilayers was investigated by the analysis of the dipole autocorrelation function
 and
and  are the unit electric dipole moment of the water molecules at time 0 and t, respectively.
are the unit electric dipole moment of the water molecules at time 0 and t, respectively.
The relaxation time of dipole autocorrelation function was determined according to
The mean and standard error of diffusion coefficients were obtained by splitting the trajectories into pieces of 20 ns and using the block averaging method.86
Hydrogen bond dynamics
The structure and dynamics of the water molecules at the lipid/water interface were investigated by the analysis of hydrogen bonds between water–water and water–lipid molecules.Hydrogen bonds formed by water with other water molecules or with oxygen atoms of lipid molecules were identified by using the geometric criteria; we considered two water molecules or a water molecule and an oxygen atom linked by hydrogen bonds when the oxygen–oxygen distance was less than 0.35 nm and the angle formed by the O–H bond of the donor molecule with the O–O direction was smaller than 30°.87
The number of hydrogen bonds formed by a DMPC lipid or gemini molecule was obtained by counting the number of water molecules bound to oxygen atoms of the bilayer component (according the geometric criteria) and by averaging over time (300 ns) and over all DMPC or gemini molecules.
The time correlation functions of hydrogen bonds were calculated by using the following equation88
where the correlation function CA-wHB(t) was fitted with a three-exponential function to obtain a more accurate value of lifetime τA-wHB.
Author contributions
C.B., S.B., G.M. G.B and A.M. conceived and designed the experiments, G.B., G.F., A. M. and A.S. performed inhibition experiments by flow cytometry, co-localization experiments by LSCM and ultrastructural characterization by TEM. C.B. prepared liposome formulations for the experiments on cells. S.B. designed and performed the molecular dynamics simulations. S.B., F.C., G.B., G.M. and A.M. analyzed all data and wrote the paper.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by Regione Lazio (LIPOBARR project), by CNCCS (Rare Diseases project) and by the “Departments of Excellence-2018” Program (Dipartimenti di Eccellenza) of the Italian Ministry of Education, University and Research, DIBAF – Department of University of Tuscia, Project “Landscape 4.0 – food, wellbeing and environment”.Notes and references
- H.-I. Chang and M.-K. Yeh, Int. J. Nanomed., 2012, 7, 49 CAS.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Nat. Rev. Cancer, 2017, 17, 20 CrossRef CAS PubMed.
- Y. Fan and Q. Zhang, Asian J. Pharm. Sci., 2013, 8, 81 CrossRef CAS.
- L. Belfiore, D. N. Saunders, M. Ranson, K. J. Thurecht, G. Storm and K. L. Vine, J. Controlled Release, 2018, 277, 1 CrossRef CAS PubMed.
- N. Lamichhane, T. S. Udayakumar, W. D. D'Souza, C. B. Simone II, S. R. Raghavan, J. Polf and J. Mahmood, Molecules, 2018, 23, 288 CrossRef PubMed.
- H. Hillaireau and P. Couvreur, Cell. Mol. Life Sci., 2009, 66, 2873 CrossRef CAS PubMed.
- Z. Dai, M. Yu, X. Yi, Z. Wu, F. Tian, Y. Miao, W. Song, S. He, E. Ahmad, S. Guo, C. Zhu, X. Zhang, Y. Li, X. Shi, R. Wang and Y. Gan, ACS Nano, 2019, 13, 7676 CrossRef CAS PubMed.
- V. Francia, K. Yang, S. Deville, C. Reker-Smit, I. Nelissen and A. Salvati, ACS Nano, 2019, 13, 11107 CrossRef CAS PubMed.
- S. A. Pandit, D. Bostick and M. L. Berkowitz, J. Chem. Phys., 2003, 119, 2199 CrossRef CAS.
- S. Aeffner, T. Reusch, B. Weinhausen and T. Salditt, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9678 CrossRef CAS PubMed.
- P. M. Kasson, E. Lindahl and V. S. Pande, J. Am. Chem. Soc., 2011, 133, 3812 CrossRef CAS PubMed.
- C. Bombelli, A. Stringaro, S. Borocci, G. Bozzuto, M. Colone, L. Giansanti, R. Sgambato, L. Toccaceli, G. Mancini and A. Molinari, Mol. Pharmaceutics, 2010, 7, 130 CrossRef CAS PubMed.
- R. Duncan and S. C. W. Richardson, Mol. Pharmaceutics, 2012, 9, 2380 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182 CrossRef CAS PubMed.
- T.-G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176 CrossRef CAS.
- U. S. Huth, R. Schubert and R. Peschka-Süss, J. Controlled Release, 2006, 110, 490 CrossRef CAS PubMed.
- S. Aleandri, M. G. Bonicelli, F. Bordi, S. Casciardi, M. Diociaiuti, L. Giansanti, F. Leonelli, G. Mancini, G. Perrone and S. Sennato, Soft Matter, 2012, 8, 5904 RSC.
- D. Poger and A. E. Mark, J. Chem. Theory Comput., 2010, 6, 325 CrossRef CAS PubMed.
- J. E. Davies, O. Rahaman and S. Patel, Biophys. J., 2009, 96, 385 CrossRef PubMed.
- K. V. Damodaran and K. M. Merz Jr., Biophys. J., 1994, 66, 1076 CrossRef CAS PubMed.
- M. Pasenkiewicz-Gierula, Y. Takaaoka, H. Miyagawa, K. Kitamura and A. Kusumi, Biophys. J., 1999, 76, 1228 CrossRef CAS PubMed.
- P. B. Moore, C. F. Lopez and M. L. Klein, Biophys. J., 2001, 81, 2484 CrossRef CAS PubMed.
- C. F. Lopez, S. O. Nielsen, M. L. Klein and P. B. Moore, J. Phys. Chem. B, 2004, 108, 6603 CrossRef CAS.
- C.-J. Högberg and A. Lyubartsev, J. Phys. Chem. B, 2006, 110, 14326 CrossRef PubMed.
- J. Wohlert and O. Edholm, J. Chem. Phys., 2006, 125, 204703 CrossRef PubMed.
- J. S. Hub, T. Salditt, M. C. Rheinstädter and B. L. de Groot, Biophys. J., 2007, 93, 3156 CrossRef CAS PubMed.
- F. H. Hansen, G. H. Peters, H. Taub and A. Miskowiec, J. Chem. Phys., 2012, 137, 204910 CrossRef CAS PubMed.
- J. Das, E. Flenner and I. Kosztin, J. Chem. Phys., 2013, 139, 065102 CrossRef PubMed.
- J. Yang, C. Calero and J. Marti, J. Chem. Phys., 2014, 140, 104901 CrossRef CAS PubMed.
- E. Lee, A. Kundu, J. Jeon and M. Cho, J. Chem. Phys., 2019, 151, 114705 CrossRef PubMed.
- C. Calero, H. E. Stanley and G. Franzese, Materials, 2019, 9, 319 CrossRef PubMed.
- D. Poger and A. E. Mark, J. Chem. Theory Comput., 2012, 8, 4807 CrossRef CAS PubMed.
- A. A. Gurtovenko, M. Patra, M. Karttunen and I. Vattulainen, Biophys. J., 2004, 86, 3461 CrossRef CAS PubMed.
- W. Zhao, A. A. Gurtovenko, I. Vattulainen and M. Karttunen, J. Phys. Chem. B, 2012, 116(1), 269 CrossRef CAS PubMed.
- G. Lukat, J. Krüger and B. Sommer, J. Chem. Inf. Model., 2013, 53, 2908 CrossRef CAS PubMed.
- R. M. Venable, F. L. H. Brown and R. W. Pastor, Chem. Phys. Lipids, 2015, 192, 60 CrossRef CAS PubMed.
- J. B. Klauda, J. Chem. Phys., 2018, 149, 220901 CrossRef PubMed.
- J. A. S. Almeida, E. F. Marques, A. S. Jurado and A. A. C. C. Pais, Phys. Chem. Chem. Phys., 2010, 12, 14462 RSC.
- T. J. Piggot, J. R. Allison, R. B. Sessions and J. W. Essex, J. Chem. Theory Comput., 2017, 13, 5683 CrossRef CAS PubMed.
- L. S. Vermeer, B. L. de Groot, V. Réat, A. Milon and J. Czaplicki, Eur. Biophys. J., 2007, 36, 919 CrossRef CAS PubMed.
- S. E. Feller and R. W. Pastor, J. Chem. Phys., 1999, 111, 1281 CrossRef CAS.
- P. Khakbaz and J. B. Klauda, Chem. Phys. Lipids, 2015, 192, 12 CrossRef CAS PubMed.
- C. R. Miller, B. Bondurant, S. D. McLean, K. A. McGovern and D. O'Brien, Biochemistry, 1998, 37, 12875 CrossRef CAS PubMed.
- Y. Li, J. Wang, Y. Gao, J. Zhu, M. G. Wientjes and J. L.-S. Au, AAPS J., 2011, 13, 585 CrossRef CAS PubMed.
- S. Patil, A. Sandberg, E. Heckert, W. Self and S. Seal, Biomaterials, 2007, 28, 4600 CrossRef CAS PubMed.
- E. Fröhlich, Int. J. Nanomed., 2012, 7, 5577 CrossRef PubMed.
- M. L. Berkowitz, D. L. Bostick and S. Pandit, Chem. Rev., 2006, 106, 1527 CrossRef CAS PubMed.
- C. Calero and G. Franzese, J. Mol. Liq., 2019, 273, 488 CrossRef CAS.
- M. Pasenkiewicz-Gierula, Y. Takaoka, H. Miyagawa, K. Kitamura and A. Kusumi, J. Phys. Chem. A, 1997, 101, 3677 CrossRef CAS.
- H. Alper, D. Bassolino and T. R. Stouch, J. Chem. Phys., 1993, 98, 9798 CrossRef CAS.
- Z. Zhang and M. L. Berkowitz, J. Phys. Chem. B, 2009, 113, 7676 CrossRef CAS PubMed.
- J. T. Titantah and M. Karttunen, J. Am. Chem. Soc., 2012, 134, 9362 CrossRef CAS PubMed.
- A. A. Bakulin, M. S. Pshenichnikov, H. J. Bakker and C. Petersen, J. Phys. Chem. A, 2011, 115, 1821 CrossRef CAS PubMed.
- Y. Rezus and H. J. Bakker, Phys. Rev. Lett., 2007, 99, 148301 CrossRef CAS PubMed.
- J. Qvist and B. Halle, J. Am. Chem. Soc., 2008, 130, 10345 CrossRef CAS PubMed.
- K.-J. Tielrooij, J. Hunger, R. Buchner, M. Bonn and H. J. Bakker, J. Am. Chem. Soc., 2010, 132, 15671 CrossRef CAS PubMed.
- D. Kraemer, M. L. Cowan, A. Paarmann, N. Huse, E. T. J. Nibbering, T. Elsaesser and R. J. Dwayne Miller, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 437 CrossRef CAS PubMed.
- C. Bombelli, F. Faggioli, P. Luciani, G. Mancini and M. G. Sacco, J. Med. Chem., 2005, 48, 5378 CrossRef CAS PubMed.
- V. Perri, M. Pellegrino, F. Ceccacci, A. Scipioni, S. Petrini, E. Gianchecchi, A. Lo Russo, S. De Santis, G. Mancini and A. Fierabracci, PLoS One, 2017, 12, e0175784 CrossRef PubMed.
- M. Pellegrino, F. Ceccacci, S. Petrini, A. Scipioni, S. De Santis, M. Cappa, G. Mancini and A. Fierabracci, Nanomedicine, 2019, 18, 371 CrossRef CAS PubMed.
- C. Bello, C. Bombelli, S. Borocci, P. di Profio and G. Mancini, Langmuir, 2006, 22, 9333 CrossRef CAS PubMed.
- D. Seebach, H. O. Kalinowski, B. Bastani, G. Crass, H. Daum, H. Doerr, N. P. DuPreez, V. Ehrig and W. Langer, Helv. Chim. Acta, 1977, 60, 301 CrossRef CAS.
- M. J. Hope, R. Nayar, L. D. Mayer and P. R. Cullis, in Liposome Technology, ed. G. Gregoriadis, CRC Press, Boca Raton FL, 2nd edn, 1992, vol. 2, pp. 123–139 Search PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren and J. Hermans, in Intermolecular Forces, ed. B. Pullman, D. Reidel Publishing Company, Dordrecht, 1981, pp. 331–342 Search PubMed.
- A. West, V. Zoni, W. E. Teague Jr., A. N. Leonard, S. Vanni, K. Gawrisch, S. Tristram-Nagle, J. N. Sachs and J. B. Klauda, J. Phys. Chem. B, 2020, 124, 828 CrossRef CAS PubMed.
- M. Blumer, S. Harris, M. Li, L. Martinez, M. Untereiner, P. N. Saeta, T. S. Carpenter, H. I. Ingólfsson and W. F. Drew Bennett, Front. Cell Dev. Biol., 2020, 8, 575 CrossRef PubMed.
- A. Olzẏnśka, W. Kulig, H. Mikkolainen, T. Czerniak, P. Jurkiewicz, L. Cwiklik, T. Rog, M. Hof, P. Jungwirth and I. Vattulainen, Langmuir, 2020, 36, 10438 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19 CrossRef.
- O. Berger, O. Edholm and F. Jähnig, Biophys. J., 1997, 72, 2002 CrossRef CAS PubMed.
- C. Oostenbrink, A. Villa, A. E. Mark and W. F. van Gunsteren, J. Comput. Chem., 2004, 13, 1656 CrossRef PubMed.
- T. P. Lybrand, I. Ghosh and J. A. McCammon, J. Am. Chem. Soc., 1985, 107, 7793 CrossRef CAS.
- C. I. Bayly, P. Cieplak, W. D. Cornel and P. A. A. Kollman, J. Phys. Chem., 1993, 97, 10269 CrossRef CAS.
- D. A. Case, R. M. Betz, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, H. T. Nguyen, I. Omelyan, A. Onufriev, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, W. M. Botello-Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao and P. A. Kollman, AMBER 2016, University of California, San Francisco, 2016 Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery Jr., T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez and J. A. Pople, Gaussian 03, Revision C.02, Gaussian, Inc., Wallingford CT, 2004 Search PubMed.
- D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089 CrossRef.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463 CrossRef CAS.
- B. Hess, J. Chem. Theory Comput., 2007, 4, 116 CrossRef PubMed.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 14101 CrossRef PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. Di Nola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684 CrossRef CAS.
- M. Parinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182 CrossRef.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33 CrossRef CAS PubMed.
- A. A. Gurtovenko, M. Miettinen, M. Karttunen and I. Vattulainen, J. Phys. Chem. B, 2005, 109, 21126 CrossRef CAS PubMed.
- J. Das, E. Flenner and I. Kosztin, J. Chem. Phys., 2013, 139, 065102 CrossRef PubMed.
- S. W. I. Siu, R. Vácha, P. Jungwirth and R. A. Böckmann, J. Chem. Phys., 2008, 128, 125103 CrossRef PubMed.
- B. Hess, J. Chem. Phys., 2002, 116, 209 CrossRef CAS.
- A. Luzar and D. Chandler, Phys. Rev. Lett., 1996, 76, 928 CrossRef CAS PubMed.
- D. C. Rapaport, Mol. Phys., 1983, 50, 1151 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr02175c |
| This journal is © The Royal Society of Chemistry 2021 |