 Open Access Article
Open Access ArticleFolding DNA into origami nanostructures enhances resistance to ionizing radiation†
Leo
Sala
 a,
Agnes
Zerolová
ab,
Alvaro
Rodriguez
a,
Agnes
Zerolová
ab,
Alvaro
Rodriguez
 a,
Dan
Reimitz
a,
Marie
Davídková
a,
Dan
Reimitz
a,
Marie
Davídková
 c,
Kenny
Ebel
c,
Kenny
Ebel
 d,
Ilko
Bald
d,
Ilko
Bald
 d and
Jaroslav
Kočišek
d and
Jaroslav
Kočišek
 *a
*a
aJ. Heyrovský Institute of Physical Chemistry of the CAS, Dolejškova 3, 18223 Prague, Czech Republic. E-mail: jaroslav.kocisek@jh-inst.cas.cz; Fax: +420 26605 3910; Tel: +420 26605 2011
bDepartment of Chemistry, Technical University of Liberec, 46117, Liberec, Czech Republic
cDepartment of Radiation Dosimetry, Nuclear Physics Institute of the CAS, Na Truhlářce 39/64, 180 00 Prague, Czech Republic
dInstitute of Chemistry-Physical Chemistry, Universität Potsdam, Karl-Liebknecht-Straße 24-25, D-14476 Potsdam, Germany
First published on 18th June 2021
Abstract
We report experimental results on damage induced by ionizing radiation to DNA origami triangles which are commonly used prototypes for scaffolded DNA origami nanostructures. We demonstrate extreme stability of DNA origami upon irradiation, which is caused by (i) the multi-row design holding the shape of the origami even after severe damage to the scaffold DNA and (ii) the reduction of damage to the scaffold DNA due to the protective effect of the folded structure. With respect to damage induced by ionizing radiation, the protective effect of the structure is superior to that of a naturally paired DNA double helix. Present results allow estimating the stability of scaffolded DNA origami nanostructures in applications such as nanotechnology, pharmacy or in singulo molecular studies where they are exposed to ionizing radiation from natural and artificial sources. Additionally, possibilities are opened for scaffolded DNA use in the design of radiation-resistant and radio-sensitive materials.
Introduction
DNA damage plays a pivotal role in IR effects on living organisms including mutations and cell death.1 Targeted damage to DNA is now a basis for radiation therapy of cancer. Sensitization of DNA to radiation by chemotherapeutics can result in synergistic effects, minimizing the dose delivered and subsequently lessening the damage to surrounding tissues.2 DNA origami nanostructures prove useful in the development of such techniques.3–6 On the one hand, they can be used as robust substrates to fundamentally study the damage and mechanisms caused by radiation to sensitized DNA strands4,7,8 while on the other, they can be used directly as drug delivery vehicles to transport therapeutic or theranostic agents into target cells.5,6,9–14Besides radiation therapy of cancer and related laboratory experiments where DNA origami structures are directly exposed to high levels of IR, there are other applications where they can be exposed to natural IR sources. These include in singulo experiments in chemistry, biology or physics15–18 or scaffolded DNA use as building blocks in nanotechnology and pharmacy.19,20 Information about how scaffolded DNA origami respond to IR are therefore essential to rationally design them for the envisaged applications and open novel ways of their processing by IR.21
DNA strands are known to undergo single, clustered, or double-stranded breakage when exposed to IR through direct and indirect mechanisms.22–25 Direct effects result from the ionization of DNA molecules from their immediate interaction with IR. In the environment, DNA is surrounded by water molecules and other chemical species either from the stabilizing buffer in laboratory conditions or proteins and other biomolecules in the cell nuclei. In this case, indirect effects take part when reactive species produced from the radiolysis of surrounding molecules, e.g. radicals and low-energy electrons (E ≈ 10 eV), further react with DNA.3,26 Does the high sensitivity of DNA strands to ionizing radiation automatically imply that novel scaffolded DNA nanomaterials are also easily damaged by ionizing radiation?
Unlike typical linear and plasmid DNA strands, DNA origami nanostructures are observed to mostly remain intact, especially in dry state, upon exposure to low doses of low-energy electrons and UV.4,27 At more intense exposures to UVB and UVC (200–300 nm), however, significant damage and eventual degradation of the nanostructures occur.28,29 Irradiation of DNA origami nanostructures by high-energy radiation are yet to be explored. We therefore present here the results on damage induced to DNA origami nanostructures after exposure to gamma rays (representing low linear energy transfer LET radiation) and protons (representing high LET radiation).
An overview of the experimental procedure is illustrated in Fig. 1. A 2D triangular DNA origami according to Rothemund's design30 was selected as a model scaffolded DNA nanostructure and irradiated by various doses of IR. We selected the design due to its known stability and ubiquitous use in various applications.5,31–33 The structures were irradiated in dry state on silicon substrates and in aqueous solution to observe damage contributions from direct and indirect causes. Atomic force microscopy (AFM) was used to assess the damage to the overall structure of the DNA origami. To evaluate the damage to the scaffold strand of the DNA origami, we performed agarose gel electrophoresis (AGE) in low magnesium and TAE buffer solutions, which separated the DNA origami into their components (scaffold strand and staples). This conveniently allows visualization and estimation of strand breaks that are otherwise difficult to resolve in AFM. Furthermore, by varying the radical scavenger concentration, we observed that most of the damage in the nanostructures in aqueous medium is caused by radicals produced from water radiolysis.
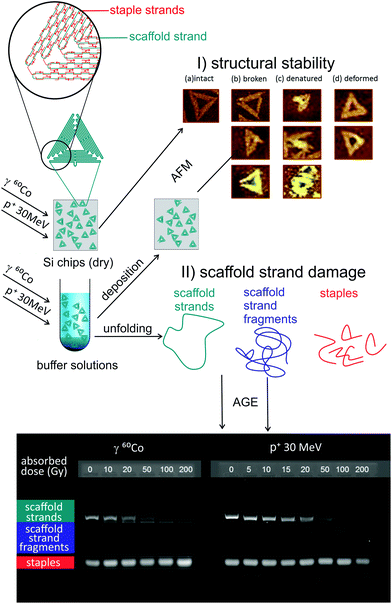 | ||
| Fig. 1 Overview of the performed experiments. Triangular 2D DNA origami nanostructures according to Rothemund's design30 each consisting of a scaffold strand (ss-m13mp18) and 208 staples (short ss-DNA segments) were deposited on the surface of silicon chips or placed in buffer solutions with varying scavenger concentrations. After irradiation with 60Co gamma rays (1.17, 1.33 MeV) or protons (30 MeV), we (I) probed the stability of the nanostructures using atomic force microscopy (AFM) or (II) unfolded the structures and evaluated the damage to the scaffold strand using agarose gel electrophoresis (AGE). In section I of the figure, one can see various types of damage to DNA origami that can be detected using AFM. In section II of the figure, one can see examples of AGE results (100 V, 35 min, 1% agarose) for DNA origami triangles in H2O exposed to various doses of gamma or proton radiation. | ||
Results and discussion
Structural damage
First, we will discuss IR damage to DNA origami triangles on the basis of AFM analysis, which allows the observation of changes in their overall shape. AFM images of the non-irradiated control and samples irradiated with high doses of gamma photons from three different irradiation conditions are shown in Fig. 2.On the leftmost panel, we can see the case corresponding to a surface type experiment, where DNA origami structures are deposited on Si wafer. Under this dry condition, the irradiated sample (b) appears mostly similar to the control (a). Overall triangular shape is preserved even at the highest absorbed dose of 300 Gy. There is an increase in the number of damage at the triangle corners which are more vulnerable as they are only held additionally by three single strand bridging extensions with 1–3 thymine bases, as opposed to the sides with 9 rows of crosslinked dsDNA. Features of the triangles counted as damaged are shown in section I of Fig. 1 and are consistent with the damage classification employed by Kielar et al.34 The percentage of remaining intact triangles in the AFM images is shown in Fig. 3 for several absorbed doses, which enables a linear fit to the data with a slope of −0.014% Gy−1. Therefore, approximately 4% of the triangles change their structure at the absorbed dose of 300 Gy.
The central panels (c) and (d) of Fig. 2 show the case corresponding to a typical biochemical experiment where the triangles are kept in their folding buffer solution (1× TAE, 12.5 mM MgCl2). The situation is similar to the previous case where the shape persists even at the highest absorbed dose of 300 Gy. The enhancement of the damage can be seen only on the overall decrease of the number of intact triangles as shown in Fig. 3. The percentage decrease is now 0.03% Gy−1 in comparison to 0.014% Gy−1 for the dry case. Both these numbers are extremely low due to absence of secondary radiation effects. In the case of the dry sample, there are only few secondary radicals formed from the water tightly bound to DNA origami. In the buffer solution, most of the radicals are scavenged by high amounts of tris [tris(hydroxymethyl)aminomethane]. When tris is removed from the solution the damage rises as shown in Fig. 2(e) and (f). In H2O (∼0 tris) the triangles are less stable as already observed in the DNA origami solution stability study of Kielar et al.35 Therefore, we can see a significant amount of damaged triangles (40%) already in the non-irradiated control (Fig. 2(e) and first point of the H2O (∼0 tris) data plot in Fig. 3). This reduction in the number of intact triangles in the solution together with higher damage rate of ∼0.08% Gy−1 (see Fig. 3) enabled a reproducible study only up to an absorbed dose of 200 Gy.
In Fig. 3, we can also compare the damage induced by gamma irradiation to that induced by a 30 MeV proton beam. We can see that the observed enhancement in the damage induced by protons is about two times more for dry solutions and around 1.3× more in the hydrated ones.
The overall structural damage as estimated by AFM imaging is very low under both dry and solution conditions. The damage enhancement from dry to solution conditions does not correlate with enhancements known from plasmid DNA studies which falls within the range of 100×–1000×.25,36 In DNA origami, the enhancement is only ∼6× (0.08/0.014). This means that the damage via individual strand breaks, nicks, and base modifications correlate with the rise in the number of radical species in solution, but the shape of the DNA origami remains intact; or the damage does not correlate with the increase in the number of radical species, which implies that the DNA origami prevents radical damage to its scaffold DNA strand. These effects cannot be resolved by AFM analysis alone; therefore, we resorted to AGE.
Scaffold strand damage
As we have shown in the previous part, the structure of DNA origami triangles is extremely resistant to damage induced by IR. The question remains as to whether it is caused by the multi-row design of the folded DNA which prevents disintegration of the triangular structure, or the structure provides a protective environment to the scaffold DNA strand. To address this question, we performed AGE analysis of the damage to the DNA origami nanostructures and the single-stranded scaffold (ss-m13mp18).In high tris and high Mg2+ concentrations, the AGE analysis of DNA origami triangles exhibits a single strong band corresponding to intact triangles in agreement with AFM results (Fig. S2 & S3†). At low Mg2+ and tris concentrations, however, electrophoresis in pure TAE buffer can separate the scaffold from the staples as two distinct bands, which coincide with the mobilities of the pure scaffold strand and pure staples (Fig. S2†). Section II of Fig. 1 shows a strong dose-dependent change in the intensity of the band corresponding to the scaffold DNA strand, which further broadens and shifts downwards at high absorbed doses. This broadening and shift can only be attributed to a non-specific fragmentation of the scaffold strand. Estimates on the dose-dependent decay of the scaffold strand are shown in Fig. 4. From the exponential character of the graph, we can already see that the amount of damage to the scaffold under gamma-irradiation is much higher than the DNA origami damage evaluated using AFM. DNA origami in solutions fragment by 0.026 scaffold per Gy or about an average of 5 strand breaks can be estimated for each DNA origami after an absorbed dose of 200 Gy, while only a few percent of the DNA origami change their overall shape as shown in the previous section. The shape of DNA origami remains stable even after significant damage to the scaffold DNA strand by IR.
 | ||
| Fig. 4 Shown on panel (a) is the dose-dependent decay of the DNA origami scaffold strand with exponential fits to estimate the number of strand breaks in gamma-irradiated and proton-irradiated DNA origami solutions with different tris concentrations. At the highest tris concentration shown (10 mM tris, hollow triangles), the line represents the decay of the intact triangle band as the scaffold band does not separate in these conditions (see ESI†). Panel (b) plots the decay rates of DNA origami and ss-m13mp18 as a function of the OH scavenging capacity based on the concentrations of scavengers (tris or glycerol) in the solution. | ||
AGE shows a more amplified damage compared to AFM as the strand breaks responsible for fragmenting the scaffold will not manifest in AFM unless they are clustered in a specific point in the triangle. This indicates that DNA origami triangles may be more sensitive to high LET, which is known to induce more direct damage resulting in more clustered DNA lesions.37,38 Indeed, our results for protons support this hypothesis when approximately 30% enhancement for both the triangle damage analyzed by AFM (Fig. 3) and scaffold damage analyzed by gel electrophoresis (Fig. 4) is observed for protons in comparison to photons. The value is higher than the expected increase of around 10% known from relative biological effectiveness of IR39 or from experiments irradiating plasmid DNA.25
Does the structural stability of DNA origami explain the overall stability of DNA origami templates under IR? The answer is no. Panel (b) of Fig. 4 compares the IR induced damage to the scaffold strand of DNA origami and the single strand (ss-m13mp18) DNA of the same base sequence at different concentrations of radical scavenger tris and glycerol (OH radical reaction rate constants ktris = 1.5 × 109 M−1 s−1 and kglycerol = 1.9 × 109 M−1 s−1).40 We can see that damage to ss-m13mp18 is several times higher than the damage to the scaffold DNA strand. The DNA origami folding clearly has a protective effect on the scaffold strand. The effect is most probably caused by the limited access of reactive secondary radicals to the scaffold strand as indicated by already discussed low difference in the DNA origami damage at dry and hydrated conditions. Also, the dependence of the damage on the scavenging capacity (Fig. 4b) is weaker than that of ss-m13mp18.
Finally, as the scaffold strand of the origami is practically fully paired, it will be interesting to compare the IR induced damage to the scaffold strand of the origami to the damage of one of the strands of naturally paired double stranded m13mp18 DNA. Evaluation of single strand breaks in double-stranded DNA is however not as straightforward as the evaluation of the single strand damage reported in the previous text and the data cannot be compared directly. On the basis of the data evaluated using the model of Cowan et al.41 (Fig. S1 and Table S1, ESI†), we can get the lowest estimate for comparison. Single strand breaks in ds-m13mp18 DNA in water are induced by IR at a rate of 0.38 single strand breaks per molecule and Gy of radiation. This value is in a good agreement with previous studies of IR induced damage in various types of double stranded DNA.24,25 At the same time, the value is more than 10 times higher than the value of DNA strand breaks incurred into the DNA origami (Fig. S1 and Table S1, ESI†). We can therefore see that Rothemund's triangle design is more resistant to IR than a naturally paired double helix.
Conclusions
We measured the damage induced to DNA origami triangles by ionizing radiation. Denaturation of DNA origami during AGE allowed us to disentangle the overall triangle disintegration from scaffold DNA strand breaks. We show that even though the scaffold strand is damaged by IR, the DNA origami keeps its shape due to its multi-row design. Thus, DNA origami can be utilized as robust substrates with low sensitivity to IR for studies of IR effects on molecules immobilized on their surfaces.The triangle corners are, however, more sensitive to IR as the multi-row structure is broken in these areas. Experiments with 30 MeV protons indicate that DNA origami nanostructures will be more sensitive to high LET radiation, which can induce more direct and clustered damage.
Finally, the folding also has a protective effect on the scaffold DNA strand. Sensitivity of DNA origami to damage induced by reactive oxygen species is much lower than that of plasmid DNA, even though this remains the primary source of IR induced damage. Comparing the damage to naturally paired supercoiled double stranded m13mp18 DNA, the damage to the DNA strands of the origami scaffold is (∼10 times lower under no radical scavenger conditions. The protective effect of folding may be further used in nanotechnology, e.g. for the rational design of radiation resistive materials, design of targeted radiosensitizers, or radiation processing of scaffolded DNA nanostructures.
Materials and methods
Preparation of DNA origami and plasmid samples
DNA origami triangles were prepared according to Rothemund's protocol.30 A closed-loop single-stranded m13mp18 viral DNA scaffold (5 nM, Tilibit Nanosystems) composed of 7249 bases was mixed with an excess concentration of 208 oligonucleotide staples (Metabion International AG) in 1× TAE buffer with 12.5 mM MgCl2. The resulting solution was then heated to 90 °C and slowly cooled down to 10 °C at a rate of 0.7 °C min−1 on a BioSan CH-100 heating block customized using arduino platform to precisely regulate and program temperature ramping. The DNA origami solutions were then purified to remove excess staples by spin filtering them three times through 100 kDa Amicon filters at 6000 rpm for 6 min each cycle.Solutions of single-stranded m13mp18 scaffold DNA and double-stranded m13mp18 plasmid DNA (New England Biolabs) come in 10 mM tris-HCl buffer. This was first adjusted to meet the same concentration of tris (40 mM) and MgCl2 (12.5 mM) as the DNA origami folding buffer by addition of appropriate volumes of 0.1 M tris-HCl (pH = 8) and 0.04 M MgCl2 stock solutions. This buffer solution is then exchanged with the desired solution for irradiation.
Buffer exchange
Buffers with different tris concentrations were prepared and exchanged with the DNA folding buffer through spin filtration.35 Sixty microliters of DNA solution was introduced inside a 100 kDa Amicon filter with additional 440 μL of the desired buffer. The solution was then spun at 7000 rpm for 4 min. About 50–60 μL were left inside the filter after filtration. A second and third filtration cycle was done with additional 440 μL of the desired buffer added after each cycle. The volume of the resulting solution was then measured and subsequently adjusted to meet the desired DNA concentration (20–30 ng μL−1). The approximate concentrations of tris and Mg2+ were estimated from the remaining volume after the filtration cycles and the additional buffer solution added during the concentration adjustments. After exchange with pure water, the average DNA concentration is 5.5 ± 0.9 nM with estimated average Mg2+ and tris concentrations of 10 ± 3 μM and 32 ± 8 μM, respectively.Atomic force microscopy (AFM)
Irradiated dry samples were readily imaged in AFM without any additional treatment. DNA origami structures from irradiated solutions needed to be first immobilized on plasma-treated Si substrates. For the preparation of dry samples and deposition of irradiated DNA solution, one microliter of the origami solution (20–30 ng μL−1 as measured using the NanoDrop™ One Microvolume UV-Vis Spectrophotometer, Thermo Scientific™) and 15 μL 10× TAE buffer (125 mM MgCl2) were dropped onto a plasma-cleaned Si chip (∼0.6 cm × 0.6 cm, MicroChemicals) and left to incubate for 60 min over an ethanol bath. The samples were then rinsed with 1 mL of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethanol–H2O solution and gently blown with air to dry completely. AFM was done in air using the Dimension Icon AFM (Bruker) operating with PeakForce Tapping Technology and ScanAsyst probes (40 kHz, 0.4 N m−1). Areas of 4 × 4 μm2 and 2 × 2 μm2 were scanned for each sample. Four to seven images were taken for each absorbed dose from 2–3 trials, each result reflecting total counts between 200–1000 triangles per absorbed dose. Surface coverages may vary from 5–50 triangles per μm2. Intact and damaged triangles were identified and counted following the classification adopted by Kielar et al.34 All images were flattened with the same criteria using the Gwyddion software.
1 ethanol–H2O solution and gently blown with air to dry completely. AFM was done in air using the Dimension Icon AFM (Bruker) operating with PeakForce Tapping Technology and ScanAsyst probes (40 kHz, 0.4 N m−1). Areas of 4 × 4 μm2 and 2 × 2 μm2 were scanned for each sample. Four to seven images were taken for each absorbed dose from 2–3 trials, each result reflecting total counts between 200–1000 triangles per absorbed dose. Surface coverages may vary from 5–50 triangles per μm2. Intact and damaged triangles were identified and counted following the classification adopted by Kielar et al.34 All images were flattened with the same criteria using the Gwyddion software.
Agarose gel electrophoresis (AGE)
Samples were run in 1% agarose gel (Serva Electrophoresis) for DNA origami and ss-m13mp18 samples and in 0.8% agarose gel for ds-m13mp18. The gels were stained with fluorescent dye SYBR Green I (dilution 1/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Sigma-Aldrich) and run under 100 V in 0.5× TAE buffer (20 mM tris, 10 mM acetic acid, 0.5 mM EDTA, pH 8.0) for 35–90 min for DNA origami, ss-m13mp18 and 120 min for ds-m13mp18 samples. The DNA solutions were mixed with 6× loading dye (EZ Vision® Two) in a 5
000, Sigma-Aldrich) and run under 100 V in 0.5× TAE buffer (20 mM tris, 10 mM acetic acid, 0.5 mM EDTA, pH 8.0) for 35–90 min for DNA origami, ss-m13mp18 and 120 min for ds-m13mp18 samples. The DNA solutions were mixed with 6× loading dye (EZ Vision® Two) in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DNA–dye solution volume ratio prior to loading. Gels were observed using the Vilber Quantum CX5 Edge Gel Documentation System which can readily capture an image of the gel illuminated with 312 nm UV transilluminator stage. The profile of each well was extracted using the ImageJ software and the bands were fitted with Gaussian functions using the FityK software to estimate the areas corresponding to their intensities.
1 DNA–dye solution volume ratio prior to loading. Gels were observed using the Vilber Quantum CX5 Edge Gel Documentation System which can readily capture an image of the gel illuminated with 312 nm UV transilluminator stage. The profile of each well was extracted using the ImageJ software and the bands were fitted with Gaussian functions using the FityK software to estimate the areas corresponding to their intensities.
Gamma irradiation
Dry DNA origami on Si chips as well as DNA origami solutions were irradiated at the Accredited Metrology Center of the Nuclear Physics Institute of the CAS using a 60Co source Chisostat (Chirana) emitting gamma rays with energies of 1.17 and 1.33 MeV. The irradiation was carried out at room temperature and in a water phantom to allow precise delivery of the absorbed dose. Solutions were placed inside 1.5 mL Eppendorf tubes while the dry samples were placed inside 2 mL tubes, orienting the DNA-deposited surface vertically, facing the source. The tubes were then immersed in water, the centers of which were fixed at exactly 3.35 cm from the front wall of the phantom placed 26.8 cm from the source, corresponding to a dose rate of 3 Gy min−1.Proton irradiation
DNA origami and plasmid DNA samples were irradiated with 30 MeV protons produced by the U-120M isochronous cyclotron at the Center of Accelerators and Nuclear Analytical Methods (CANAM) of the Nuclear Physics Institute of the Czech Academy of Sciences, the details of which are described elsewhere.25 Liquid samples (14 μL, 20–30 ng μL−1 DNA) were irradiated in polypropylene microtubes with the conical tip placed within the width of the beam. Dry samples of DNA origami on Si were fixed on plastic petridishes and oriented facing the beam using a dedicated holder. In the low-dose regime (5–20 Gy), a dose rate of 4 Gy min−1 was used and at higher absorbed doses (50–200 Gy), the rate was increased to 50 Gy min−1. At these higher absorbed doses, the liquid samples were immediately transferred to nonirradiated tubes to avoid potential sample contamination from activated tubes. NE 2581 ionization chamber (Nuclear Enterprises Ltd, UK) was positioned at the sample holder to monitor the doses and dose rates delivered. Before the irradiation, the homogeneity of the beam at the sample position was verified using Gafchromic films (XR-RV3, Ashland, USA) irradiated by doses of ∼10 Gy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Czech Science Foundation, project no. 19-01159S. The mobility of researchers was supported by bilateral project of the Czech Academy of Sciences and the German Academic Exchange service project no. 57448878 DAAD 19-02/ (K. E. and L. S.) I. B. and K. E. acknowledge support by the European Research Council (ERC; consolidator grant no. 772752). A. R. acknowledges the support from European Regional Development Fund; OP RDE; project “CARAT” no. CZ.02.1.01/0.0/0.0/16\_026/0008382.Notes and references
- T. J. Jorgensen, Strange Glow: The Story of Radiation, Princeton University Press, Princeton, New Jersey, 2016 Search PubMed.
- T. Y. Seiwert, J. K. Salama and E. E. Vokes, Nat. Rev. Clin. Oncol., 2007, 4, 86–100 CrossRef CAS PubMed.
- R. Schürmann, S. Vogel, K. Ebel and I. Bald, Chem. – Eur. J., 2018, 24, 10271–10279 CrossRef PubMed.
- J. Rackwitz, J. Kopyra, I. Dabkowska, K. Ebel, M. L. Ranković, A. R. Milosavljević and I. Bald, Angew. Chem., Int. Ed., 2016, 55, 10248–10252 CrossRef CAS PubMed.
- Q. Zhang, Q. Jiang, N. Li, L. Dai, Q. Liu, L. Song, J. Wang, Y. Li, J. Tian, B. Ding and Y. Du, ACS Nano, 2014, 8, 6633–6643 CrossRef CAS PubMed.
- X. Zhuang, X. Ma, X. Xue, Q. Jiang, L. Song, L. Dai, C. Zhang, S. Jin, K. Yang, B. Ding, P. C. Wang and X.-J. Liang, ACS Nano, 2016, 10, 3486–3495 CrossRef CAS PubMed.
- I. Bald and A. Keller, Molecules, 2014, 19, 13803–13823 CrossRef PubMed.
- R. Schürmann, T. Tsering, K. Tanzer, S. Denifl, S. V. K. Kumar and I. Bald, Angew. Chem., Int. Ed., 2017, 56, 10952–10955 CrossRef PubMed.
- A. Udomprasert and T. Kangsamaksin, Cancer Sci., 2017, 108, 1535–1543 CrossRef CAS PubMed.
- L. Song, Q. Jiang, J. Liu, N. Li, Q. Liu, L. Dai, Y. Gao, W. Liu, D. Liu and B. Ding, Nanoscale, 2017, 9, 7750–7754 RSC.
- R. Huang, N. He and Z. Li, Biosens. Bioelectron., 2018, 109, 27–34 CrossRef CAS PubMed.
- A. Keller and V. Linko, Angew. Chem., Int. Ed., 2020, 59, 15818–15833 CrossRef CAS PubMed.
- N. Li, X.-Y. Wang, M.-H. Xiang, J.-W. Liu, R.-Q. Yu and J.-H. Jiang, Anal. Chem., 2019, 91, 2610–2614 CrossRef CAS PubMed.
- X. Huang, N. T. Blum, J. Lin, J. Shi, C. Zhang and P. Huang, Mater. Horiz., 2021, 8, 78–101 RSC.
- N. V. Voigt, T. Tørring, A. Rotaru, M. F. Jacobsen, J. B. Ravnsbæk, R. Subramani, W. Mamdouh, J. Kjems, A. Mokhir, F. Besenbacher and K. V. Gothelf, Nat. Nanotechnol., 2010, 5, 200–203 CrossRef CAS PubMed.
- A. Keller, I. Bald, A. Rotaru, E. Cauët, K. V. Gothelf and F. Besenbacher, ACS Nano, 2012, 6, 4392–4399 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 874–890 CrossRef CAS PubMed.
- D. Huang, K. Patel, S. Perez-Garrido, J. F. Marshall and M. Palma, ACS Nano, 2019, 13, 728–736 CrossRef CAS PubMed.
- N. C. Seeman, Nat. Rev., 2017, 3, 17068 Search PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- M. Cleland, L. Parks and S. Cheng, Nucl. Instrum. Methods Phys. Res., Sect. B, 2003, 208, 66–73 CrossRef CAS.
- Advances in Radiation Biology, ed. J. T. Lett and W. K. Sinclair, Elsevier, 1993, vol. 17, pp. 121–180 Search PubMed.
- C. Leloup, G. Garty, G. Assaf, A. Cristovão, A. Breskin, R. Chechik, S. Shchemelinin, T. Paz-Elizur, Z. Livneh, R. Schulte, V. Bashkirov, J. Milligan and B. Grosswendt, Int. J. Radiat. Biol., 2005, 81, 41–54 CrossRef CAS PubMed.
- H. M. Dang, M. J. van Goethem, E. R. van der Graaf, S. Brandenburg, R. Hoekstra and T. Schlathölter, Eur. Phys. J. D, 2011, 63, 359 CrossRef CAS.
- L. Vyšín, K. Pachnerová Brabcová, V. Štěpán, P. Moretto-Capelle, B. Bugler, G. Legube, P. Cafarelli, R. Casta, J. P. Champeaux, M. Sence, M. Vlk, R. Wagner, J. Štursa, V. Zach, S. Incerti, L. Juha and M. Davídková, Radiat. Environ. Biophys., 2015, 54, 343–352 CrossRef PubMed.
- S. K. Sahbani, S. Girouard, P. Cloutier, L. Sanche and D. J. Hunting, Radiat. Res., 2014, 181, 99–110 CrossRef PubMed.
- S. Vogel, K. Ebel, R. M. Schürmann, C. Heck, T. Meiling, A. R. Milosavljevic, A. Giuliani and I. Bald, ChemPhysChem, 2019, 20, 823–830 CrossRef CAS PubMed.
- W. Fang, M. Xie, X. Hou, X. Liu, X. Zuo, J. Chao, L. Wang, C. Fan, H. Liu and L. Wang, J. Am. Chem. Soc., 2020, 142, 8782–8789 CrossRef PubMed.
- H. Chen, R. Li, S. Li, J. Andréasson and J. H. Choi, J. Am. Chem. Soc., 2017, 139, 1380–1383 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- A. M. Hung, C. M. Micheel, L. D. Bozano, L. W. Osterbur, G. M. Wallraff and J. N. Cha, Nat. Nanotechnol., 2010, 5, 121–126 CrossRef CAS PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z.-G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- J. Prinz, C. Heck, L. Ellerik, V. Merk and I. Bald, Nanoscale, 2016, 8, 5612–5620 RSC.
- C. Kielar, Y. Xin, X. Xu, S. Zhu, N. Gorin, G. Grundmeier, C. Möser, D. M. Smith and A. Keller, Molecules, 2019, 24, 2577–2588 CrossRef CAS PubMed.
- C. Kielar, Y. Xin, B. Shen, M. A. Kostiainen, G. Grundmeier, V. Linko and A. Keller, Angew. Chem., Int. Ed., 2018, 57, 9470–9474 CrossRef CAS PubMed.
- C. von Sonntag, The Chemical Basis of Radiation Biology, Taylor & Francis, London, 1987 Search PubMed.
- D. Goodhead, Int. J. Radiat. Biol., 1994, 65, 7–17 CrossRef CAS PubMed.
- A. Yokoya, S. M. T. Cunniffe and P. O'Neill, J. Am. Chem. Soc., 2002, 124, 8859–8866 CrossRef CAS PubMed.
- H. Paganetti, Phys. Med. Biol., 2014, 59, R419–R472 CrossRef PubMed.
- G. V. Buxton, C. L. Greenstock, W. P. Helman and A. B. Ross, J. Phys. Chem. Ref. Data, 1988, 513, 8859–8866 Search PubMed.
- R. Cowan, C. M. Collis and G. W. Grigg, J. Theor. Biol., 1987, 127, 229–245 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Specific details of data analysis by AGE and representative AFM images of DNA origami in various irradiation conditions. See DOI: 10.1039/d1nr02013g |
| This journal is © The Royal Society of Chemistry 2021 |


