Open Access Article
Open Access ArticleA multiplexed phospholipid membrane platform for curvature sensitive protein screening†
Eider
Berganza
 *a,
Mirsana P.
Ebrahimkutty
b,
Srivatsan K.
Vasantham
a,
Chunting
Zhong
a,
Alexander
Wunsch
c,
Alexander
Navarrete
c,
Milos
Galic
*a,
Mirsana P.
Ebrahimkutty
b,
Srivatsan K.
Vasantham
a,
Chunting
Zhong
a,
Alexander
Wunsch
c,
Alexander
Navarrete
c,
Milos
Galic
 *b and
Michael
Hirtz
*b and
Michael
Hirtz
 *a
*a
aInstitute of Nanotechnology (INT) & Karlsruhe Nano Micro Facility (KNMF), Karlsruhe Institute of Technology (KIT), Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany. E-mail: eider.berganza.eguiarte@gmail.com
bUniversity of Münster, Robert-Koch-Straße 31, 48149, Münster, Germany
cInstitut für Mikroverfahrenstechnik (IMVT), Karlsruher Institut für Technologie (KIT), Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
First published on 16th June 2021
Abstract
The curvature of lipid membranes plays a key role in many relevant biological processes such as membrane trafficking, vesicular budding and host–virus interactions. In vitro studies on the membrane curvature of simplified biomimetic models in the nanometer range are challenging, due to their complicated nanofabrication processes. In this work, we propose a simple and low-cost platform for curvature sensitive protein screening, prepared through scanning probe lithography (SPL) methods, where lipid bilayer patches of different compositions can be multiplexed onto substrate areas with tailored local curvature. The curvature is imposed by anchoring nanoparticles of the desired size to the substrate prior to lithography. As a proof of principle, we demonstrate that a positive curvature membrane sensitive protein derived from the BAR domain of Nadrin2 binds selectively to lipid patches patterned on substrate areas coated with 100 nm nanoparticles. The platform opens up a path for screening curvature-dependent protein–membrane interaction studies by providing a flexible and easy to prepare substrate with control over lipid composition and membrane curvature.
Introduction
Lipid bilayers constitute 5–7 nanometer thick selectively permeable barriers of self-assembled amphiphilic molecules that separate intracellular components from the external environment. Cell membranes play an important role in cell signaling1 although this functionality was long believed to be solely endowed by the membrane proteins embedded into them.2 However, many recent studies point out the relevance of the chemical lipid composition and their curvature, which are claimed to be synergized parameters,3 mediating many relevant biological processes such as membrane trafficking, vesicular budding, viral interactions, mitosis, and membrane fusion.4,5The BAR domain proteins are a good example of a family of proteins extensively involved in these processes.6 They feature a three helix coiled coil core that results in a positively charged banana-shaped surface, with high binding affinity towards negatively charged lipids. Based on their nanometer scale curvature preferences, proteins of this family are selectively enriched at either highly curved positive membrane deformations (i.e. N-BAR), shallow positive deformations (i.e. F-BAR), or negative membrane deformations (i.e. I-BAR). However, although the curvature-selectivity of individual proteins is well established, their lipid-selectivity remains elusive.4 Hence, despite their high biomedical relevance,7 the molecular mechanisms driving selectivity during sensing, generation and stabilization of curved structures in living cells are not fully understood.
A ubiquitous approach to study cell membranes with reduced complexity is to build bottom-up in vitro models that mimic naturally occurring mechanisms. In this regard, tubulation assays carried out by different groups have provided valuable insights into membrane deformation mechanisms.8,9 However, these methods are usually quite time consuming, as the generation of tubules is performed individually at a speed range of a few microns per second.10 Amongst other biomimetic membrane types, supported lipid bilayers (SLB) offer a solid platform where phospholipid vesicles form substrate stabilized membranes by vesicle fusion and permit incubation of the targeted proteins or conducting thorough characterization via surface sensitive techniques.11,12
Despite SLB being originally conceived to be built on planar substrates, some studies have already recreated curvature by patterning lipids or directly depositing cells onto previously lithographed substrates, though mainly in the micron range.13–16 The creation of local curvature effects at the nanoscale is particularly challenging due to the needed resolution and high cost of the available lithography techniques. Thus, the experimental works addressing this topic remain scarce. The use of nanoparticles (NPs) as a source of local curvature has proven to be a cost effective way to overcome this problem,17 as phospholipid molecules can form a layer around silica NPs.18 Nonetheless, the number of works leveraging the use of nanoparticles for this purpose are still scarce and these studies rely on vesicle fusion-based techniques,19 which do not yield lipid patches in the desired shape or precise control over spatial resolution as is desirable for screening applications.
Hence, the vesicle fusion approach does not allow multiplexing or patterning lipid patches of different compositions close to each other. In contrast, scanning probe lithography (SPL) derived techniques enable phospholipids to be deposited with a very high spatial resolution by transferring them from a sharp probe to the desired region of the substrate.20
There is an increasing trend in biophysics to add complexity to artificially created biomimetic models that, for instance, recreate a more realistic cell microenvironment,21 or reliably replicate the behaviour of natural membranes.22 Establishing such complex synthetic systems, however, critically relies on a profound understanding of the fundamental interaction properties of their components. These, however, are to date still missing for curvature-dependent protein–membrane interactions. In this work, we present a convenient approach for the fabrication of multiplexed phospholipid patches onto surfaces with tuned specific nanoscale curvature, on the way to building more realistic membrane models. We conceive an in vitro platform for the screening of curvature sensing proteins, using dip-pen nanolithography with phospholipids (L-DPN), similar to the one depicted in Fig. 1.23,24 In this manuscript, however, different configurations of the same idea are presented, with the aim to optimize conditions for each particular experiment.
 | ||
| Fig. 1 Schematic representation of a platform for the multiplexing of lipid patches via dip-pen nanolithography onto locally curved surfaces. The inset shows a schematic of a curvature sensitive protein binding to a tailored curved phospholipid bilayer (adapted with permission from ref. 25). | ||
Results and discussion
The generation of a curvature-dependent screening platform requires addressing several critical aspects that arise during the different steps of this bottom-up fabrication process, as well as testing its functionality. Most experiments were carried out using 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) admixed with 1 mol% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) for visualization, unless otherwise stated. This is a commonly used fluid phase phospholipid ink (at room temperature) for DPN patterning.Tuning curvature by nanoparticle coating
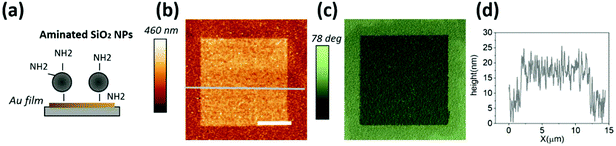
As discussed above, the use of nanoparticle coated substrates as a source of local curvature represents a cost effective alternative to conventional nanolithography approaches. Due to the many different applications that rely on gold nanoparticle coating ranging from sensing of biomolecules,26 to plasmonics27 or the enhancement of the Raman signals,28 different methods seeking to immobilize the NPs on a surface have been extensively reported in the literature.29,30 A widely used protocol for NP coating typically starts with the functionalization of the substrate with (3-aminopropyl)triethoxysilane (APTMS) and subsequent exposure of the substrate to a gold NP suspension.31 As patterning phospholipid patches via L-DPN relies on dragging a low force constant cantilever (typically about 0.1 N m−1) AFM probe covered with the desired phospholipid mixture ink on the tip edge over the substrate (Fig. 1), the forces affecting the bound NPs must be considered. For regular L-DPN, the process is usually carried out on planar substrates, where the combination of the high lateral friction forces exerted by the tip and the ink viscosity do not constitute a problem for the integrity of the substrate. In the initial experiments, it turned out that the widely adopted method of NP immobilization30 (gold NPs on an amine functionalized surface, see section 1 of the ESI†) was not stable enough for use in L-DPN. After coating the whole substrates with NPs, square lipid patches were patterned via L-DPN as detailed in the Experimental section. Subsequent atomic force microscopy (AFM) imaging showed that DPN on gold NPs led to NP detachment during the patterning process.Conversely, the application of an inverted approach (amine carrying silica NPs on a gold coated substrate, see Fig. 2a) provided sufficient binding strength to allow successful patterning of the phospholipid patches without affecting the NP coating (Fig. 2b and c). The topographic image and profile (Fig. 2d) show a square lipid patch of approximately 9 nm thickness on average (one and a half bilayers), successfully printed over the nanoparticle coated substrate.
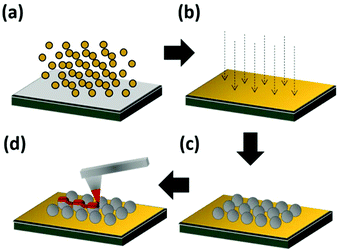
These results show that in processes with higher lateral forces exerted on NPs such as L-DPN, the binding method can be critical. Fig. 3 describes the four steps followed to obtain strong NP–substrate binding: (1) a 5 nm chromium layer is sputtered onto a silicon substrate to ensure good adhesion, followed by an 80 nm thick gold layer (Fig. 3a). (2) The gold layer is then rinsed and treated with O2 plasma, which leaves the gold clean of any organic residue remaining (Fig. 3b). As shown in section 2 of the ESI,† this step is crucial for the successful binding of NPs. (3) The amine functionalized silica NPs are incubated on the substrate in a deionized (DI) water colloidal suspension, allowing them to bind to the gold film (Fig. 3c).32,33 (4) Phospholipid patches with the desired mixture composition and feature shape are patterned with L-DPN, under controlled humidity (Fig. 3d). Further details can be found in the Experimental section.
This simple yet critical change of strategy (using amine group functionalized NPs rather than amine functionalized substrates) makes it possible to successfully write lipid patches using a DOPC phospholipid ink on the NP coated surface without nanoparticle detachment. Furthermore, by lowering the scanning rate to 0.1 microns per second, we optimized writing conditions to minimize damage caused by friction.
Quality assessment of lipid membranes by AFM
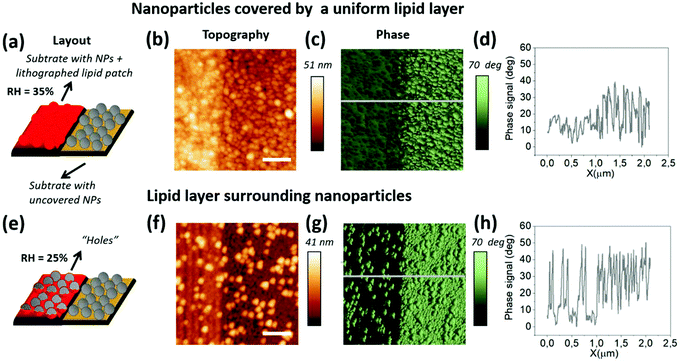
AFM imaging is a well-established nanoscale characterization technique, particularly capable of simultaneously providing topographic and compositional information even for soft matter (such as lipid membranes).34,35 In the amplitude modulation mode (AM-AFM), the amplitude is kept constant through force-feedback and the topographic and compositional information is therefore decoupled into amplitude and phase shift channels.Thus, the oscillation phase image is commonly used as an indicator of compositional differences in multicomponent systems. In our system, the highly different mechanical and compositional properties between silica NPs and the phospholipid membrane make it a particularly good system to observe notable material contrast in the phase channel. For the proposed platform, the assessment of the sample quality through AFM is of great importance to ensure that the phospholipid membrane is successfully curved, or in other words, to assess whether the NPs are well covered by one or more phospholipid bilayers. Fig. 4 shows AFM images of two samples with the same layout prepared under different lipid writing conditions on substrates fully covered with NPs leading to either a fully formed membrane or single lipid lines (the sample shown in Fig. 4a–d was patterned at 35% RH and the sample in Fig. 4e–h was patterned under 25% RH, leading to less lipid deposition36). Interestingly, due to the elastic nature of the phospholipids, the patterned lipid patches display a similar roughness to that of the bare NP coated substrate (see section 3 of the ESI† for further details). Fig. 4b shows a fully lipid covered part on the area on the left. Its corresponding phase image (Fig. 4c), depicted in the green scale, shows a rather homogeneous dark contrast in this same area, as compared to the bare NP part on the right.
Fig. 4f shows the thinner printed lines that have not successfully merged into a continuous membrane (as they were written under L-DPN conditions depositing less lipid material.36 Here, the non-covered NPs can be distinguished in all areas of the phase image (Fig. 4g) clearly sticking out of the surrounding lipid membrane by their phase contrast.
Another notable difference between the examples shown in Fig. 4b and f is the nanoparticle density. It has been reported that in lipids patterned on NP decorated surfaces, beyond 22 nm NP diameter, holes can form37 which can become critical if we try to cover bigger nanoparticles.
A reliable way to avoid NPs puncturing through the membrane was thus (I) the deposition of a sufficient amount of lipid material during L-DPN, and (II) maintaining a high NP density, leaving no room for gaps. See section 4 of the ESI† for more details.
Stability of lipid patches in aqueous solution
As the L-DPN process takes place in the air under ambient conditions, for protein binding experiments, the substrates need to be transferred into liquid. Upon immersion into liquid, reorganization of the lipid membranes takes place to accommodate the energetically most favourable configuration.38,39 Thus, new samples were prepared (similarly to the one shown in Fig. 2f or 4b) to trial the stability of the system proposed in this work during the immersion process. For this purpose, a sample with a mixture of DOPC with Rhod-PE patches was fabricated onto a surface homogeneously coated with 25 nm diameter NPs and later imaged before and during immersion into deionized water. Fig. 5 shows that the patches remain in place, although the exposure time was slightly elevated for the image in liquid, due to the expected loss of excess bilayers that are washed away in the presence of water and dampening of the light intensity in the liquid.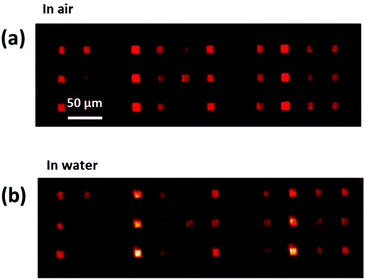 | ||
| Fig. 5 Fluorescently labelled lipid patches were imaged (a) first in the air (as prepared) and then (b) during immersion into water, showing the stability of the written structures. | ||
To further characterize the system, the ability of the lipid membranes to protect the sample surface from unspecific protein adhesion was trialed. Here, the integrity of membrane patches in buffer (phosphate buffered saline, PBS) was demonstrated by incubating the sample with fluorescently labelled BSA to block the sample areas where no lipids were previously deposited (see section 5 of the ESI†). The green fluorescence is only observed on the bare substrate around the lipid patches, while the patches themselves remain dark. These results unequivocally demonstrate the stability of the membrane structures on the NPs for experimentation in liquid.
Towards a multiplexed platform for curvature sensing
As pointed out in the Introduction, one aim of this highly versatile platform is to screen for curvature-dependent protein–membrane interactions. In order to create such systems, it is highly desirable not only to harness the multiplexing capability of DPN itself but also to be able to create regions with a specific nanoscale curvature in different areas of the same substrate (see Fig. 6a). This was possible to achieve using microchannel cantilever spotting (μCS)40 (Fig. 6b) to deposit the NP suspension (adding 20% glycerol to prevent evaporation in the spotting tip). Subsequent AFM imaging (Fig. 6c) shows the obtained result after solvent evaporation, using 100 nm nanoparticles. Despite the high boiling point of glycerol, when the sample is taken out from the controlled humidity chamber, the water and glycerol mixture evaporates within a few minutes with no need for further heating because of their small (femtoliter) volume. The same experiment was repeated spotting 25 nm NPs (see section 6 of the ESI†) and should be extensible to other particle sizes in the nanoscale range.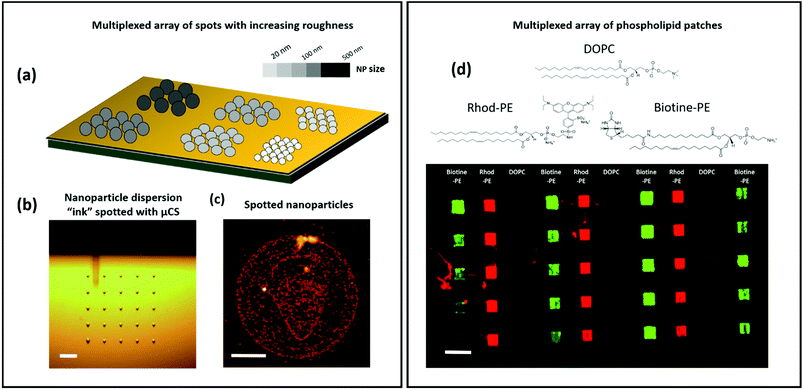 | ||
| Fig. 6 (a) Sketch showing multiplexed substrate areas displaying NPs of increasing size. (b) Microscopy image of an array of water–glycerol based 100 nm nanoparticle dispersion ink droplets, produced by microchannel cantilever spotting (μCS). The μCS tip is hovering above the substrate (therefore slightly out of focus) in the position to deposit the next type of NP ink (cf. ESI Fig. S6† for the completed 100 nm/25 nm multiplexed array). Scale bar is 50 μm. (c) AFM image of 100 nm nanoparticles spotted with μCS, after evaporation of the solvent. Scale bar is 5 μm. (d) Example of a multiplexed array of phospholipid patches (three different compositions in columns), patterned onto a substrate fully covered with nanoparticles. Scale bar is 50 μm. | ||
Additionally, functional lipids of varying compositions can be patterned with very high spatial control on the locally curved substrates. Leveraging the multiplexing capability of DPN as shown in Fig. 6d, where we combine 3 different lipid mixtures, namely pure DOPC, biotinylated DOPC (DOPC admixed with 5 mol% 1-oleoyl-2-(12-biotinyl(aminododecanoyl))-sn-glycero-3-phosphoethanolamine) and fluorescently labelled DOPC (with 1 mol% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt)), multiplexed lipid patch arrays can be created, which demonstrates the possibilities that DPN offers to build a protein screening platform, with lipid patches of varying compositions in close proximity. On incubation with fluorescently labelled streptavidin, which binds selectively to the biotinylated DOPC patches, a clear distinctive fluorescence pattern occurs (a green signal on biotinylated patches and a red signal on Rhod-PE containing patches) proving the functionality of the lipids on the nanoparticle coated substrate. In principle, the combination of multiplexed L-DPN with μCS can generate arrays of NPs of different sizes enabling a very flexible choice of curvature and membrane composition within one microarray for protein screening applications.
Binding of curvature sensitive proteins
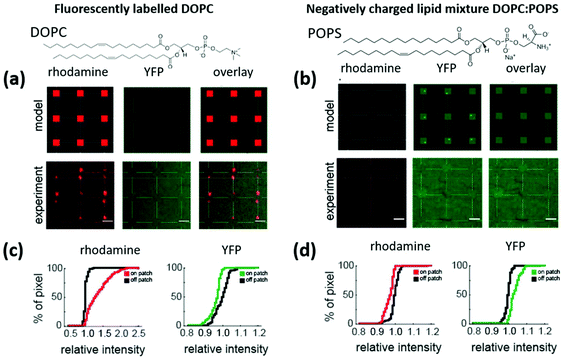
Finally, to showcase the functionality of the locally curved lipid patches for curvature sensitive protein binding, experiments were conducted using the BAR domain of Nadrin2 labelled with the yellow fluorescent protein (YFP). For this, lipid patches deposited onto substrates covered with 100 nm NPs were incubated with the protein extract. This particular protein (Nadrin 2) is expected to bind to the curved membranes on NPs of approximately 100 nm diameter.6 The experiments shown in this part were carried out on 3 × 3 grids of lipid patches patterned on substrates fully covered with 100 nm NPs.As previously explained, in spite of the existing evidence of the interplay between membrane curvature and composition, whether curvature alone is sufficient to promote BAR domain binding is still unknown. Our results, however, suggest that pure DOPC (electrically neutral lipids) patches are not sufficient to assist protein enrichment at curved membrane sections. Fig. 7 shows the results of binding experiments on lipid patches of neutral DOPC with the minor addition of Rho-PE as the fluorescent label (Fig. 7a). If curvature alone is not sufficient for BAR domain binding, no binding events would be detected on the neutral DOPC/Rho-PE curved lipid patches (as indicated in the “model” row). The quantification of the corresponding binding events is shown in Fig. 7c. Here, cumulative intensity distributions are shown for the rhodamine signal (red, indicating the Rho-PE to unambiguously identify the patch positions) and the YFP signal (green, indicating the presence of the curvature sensitive protein receptor), respectively. Compared to the area surrounding the patches (black curve), the rhodamine signal from the lipid patches (red curve) displays a shift of the intensity distribution curve towards higher values (indicated by a +35.4% increase in the intensity on the patches relative to the off-patch positions). In contrast, there is no intensity increase in the protein signal on the patch positions (green curve) compared to off-patch positions (black curve). Intriguingly, there is even a slight decrease in the signal of the curvature sensitive protein on the patches (−3.6%), indicating reduced fluorescence transmission through the lipid bilayer or a better blocking of unspecific adhesion on the lipid membranes compared to the surrounding substrate area (n = 84 lipid dots from N = 2 glass plates; rhodamine: +35.4%, P = 3.0 × 10−29; Nadrin2: −3.6%, P = 2.3 × 10−11). P values refer to the measure of significance by the rank-sum test in MATLAB.
Considering that binding for the BAR protein relies not only on membrane curvature but also on the strength of electrostatic interactions with individual lipids in the membrane, we performed additional experiments with negatively charged lipids added to the mixture, namely 30 mol% (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt)) POPS. The corresponding experiments are shown in section 7 of the ESI,† where a shift in the intensity profiles of the fluorescence images suggests that there is indeed protein receptor binding as a result of the combined effect of membrane curvature and the negative charge. These results are, however, obscured by the crosstalk of the rhodamine added to the lipids in the green fluorescent channel used to screen protein binding. Therefore, the experiments were repeated using the same DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS mixture (70
POPS mixture (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) on 100 nm NPs, but in the absence of Rho-PE (Fig. 7b). Here, no signal in the rhodamine channel is expected (as no Rho-PE is present in the patches), while specific protein binding should be indicated by a signal in the YFP channel, as exemplified in the “model” row. The quantification of the experimental results with the cumulative intensity distributions (Fig. 7d) reveals the expected results: no increase in intensity is observed for the rhodamine signal between on and off patch positions (red curve vs. black curve). However, a clear shift towards higher fluorescence intensity (+3.8%) on the lipid patches relative to the off-patch positions can be seen on the charged lipid patches with curvature (n = 90 dots from N = 2 glass plates; rhodamine = −2.9%; P = 4.5 × 10−16 and protein = +3.8%, p = 8.2 × 10−22). To ensure that negative charge alone does not promote binding, a negative control with charged lipid patches on a flat gold surface was conducted. As expected, no protein receptor enrichment was found in this case (see section 8 of the ESI†). Summarizing the above results, the curved membranes along with the negatively charged lipids are crucial for facilitating the recruitment of curvature sensing protein receptors at these sites. Besides showing the functionality of the proposed method to screen curvature sensing proteins, we can conclude from these results that the binding of curvature-sensing proteins requires both, appropriate lipid bilayer curvature combined with specific lipid composition.
30) on 100 nm NPs, but in the absence of Rho-PE (Fig. 7b). Here, no signal in the rhodamine channel is expected (as no Rho-PE is present in the patches), while specific protein binding should be indicated by a signal in the YFP channel, as exemplified in the “model” row. The quantification of the experimental results with the cumulative intensity distributions (Fig. 7d) reveals the expected results: no increase in intensity is observed for the rhodamine signal between on and off patch positions (red curve vs. black curve). However, a clear shift towards higher fluorescence intensity (+3.8%) on the lipid patches relative to the off-patch positions can be seen on the charged lipid patches with curvature (n = 90 dots from N = 2 glass plates; rhodamine = −2.9%; P = 4.5 × 10−16 and protein = +3.8%, p = 8.2 × 10−22). To ensure that negative charge alone does not promote binding, a negative control with charged lipid patches on a flat gold surface was conducted. As expected, no protein receptor enrichment was found in this case (see section 8 of the ESI†). Summarizing the above results, the curved membranes along with the negatively charged lipids are crucial for facilitating the recruitment of curvature sensing protein receptors at these sites. Besides showing the functionality of the proposed method to screen curvature sensing proteins, we can conclude from these results that the binding of curvature-sensing proteins requires both, appropriate lipid bilayer curvature combined with specific lipid composition.
Conclusions
In this work, we present a facile and reliable method for the fabrication of biomimetic membrane models with tailored curvature. The use of NPs for the creation of membrane curvature implies a highly desirable degree of freedom to select the required curvature for specific experiments eluding time consuming and expensive cleanroom nanolithography processes. Our results demonstrate that the patterning of phospholipid patches on NPs can be achieved without damaging the NP coating, by choosing a suitable surface functionalization that ensures their anchoring to the substrate. Importantly, the patterned bilayer remains stable upon immersion into water or buffer.This versatile platform is well suited to further grow our understanding of membrane curvature mediated processes and curvature sensitive proteins, such as the BAR family proteins. The versatility of the proposed method will, for instance, enable detailed studies with different membrane curvatures and variations in the composition of the lipids.
In summary, the use of SPL methods for the fabrication of the curvature sensitive protein screening platform allows combining the multiplexing of both NPs of different sizes, creating substrate areas with different membrane curvatures, as well as phospholipid patches with different compositions, with very high spatial control which in turn can lead to in-depth studies of the interplay between membrane composition and curvature in cellular processes.
Experimental section
Sample fabrication
Preparation of the substrates: Route 1: Glass substrates were cleaned by immersion in 20 vol% H2O2/80 vol% H2SO4 for 30 min and then by rinsing 3 times with DI water. They were dipped in APTMS solution for 30 minutes and rinsed subsequently with ethanol. The substrates were left in a gold nanoparticle suspension for 24 hours and then rinsed with DI water. Route 2: Glass substrates were cleaned in a sonication bath using acetone, ethanol and DI water sequentially. An 80 nm gold layer was sputtered on the glass substrates. Subsequently, the substrates were activated with O2 plasma for 2 minutes (after storage time exceeding a week, cleaning time was extended to 5 minutes). Silica nanoparticles functionalized with amine head-groups in aqueous solution were purchased from Creative Diagnosis (NY, USA). The substrates were covered with the nanoparticle suspension for 3 hours (25 nm NPs) and 20 hours (100 nm NPs) and then rinsed with DI water and dried under a nitrogen flow.
Preparation of phospholipid mixtures: The phospholipids, dissolved in a chloroform solution, were purchased from Avanti Polar Lipids (USA). Three different phospholipid mixtures were prepared with DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) as a carrier solution in all cases. (1) Fluorescently labelled Liss Rhod PE was admixed to obtain 1 mol% concentration. (2) The biotinylated Biotinyl Cap PE was added to obtain 5 mol%. (3) For the Nadrin2 protein binding experiments, DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS (70
POPS (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) mol% mixtures were prepared, with and without Liss Rhod PE contents. All the mixtures were sonicated for a few minutes to improve homogeneity. Dip-pen nanolithography writing: The lipid writing was performed using an NLP2000 system (NanoInk, USA) with F-type one-dimensional cantilever arrays (NanoInk). Matching inkwells were loaded with 1 μL of each of the prepared mixtures and then allowed to dry in air for a few minutes. To ink the types, the cantilever ends were left in contact with the loaded inkwells for 15 minutes at high humidity (70% RH). Before writing on the substrates, the excess ink was removed from the tips by writing on a sacrificial area. The lipid patches lithographed on the prepared substrates were typically written keeping the relative humidity at 30%, with a line pitch of 100 nm and a writing speed of 0.1 μm s−1 to avoid nanoparticle detachment due to high lateral force. Nanoparticle multiplexing via μCS: In order to create different nanoscale curvatures on the same surface, the commercially purchased nanoparticle aqueous solutions were admixed with 25% volume glycerol to prevent evaporation during spotting. The spotting process was performed on a nanolithography platform (NLP 2000 system, NanoInk, USA) using surface patterning cantilevers (SPT-S-C10S, Bioforce Nanosciences). 2 μL of NP ink was deposited on the pen. A probe dwell time of 10 s was chosen, to ensure that the amount of nanoparticles transferred to the surface is large. Spotting was performed at a relative humidity of 80%.
30) mol% mixtures were prepared, with and without Liss Rhod PE contents. All the mixtures were sonicated for a few minutes to improve homogeneity. Dip-pen nanolithography writing: The lipid writing was performed using an NLP2000 system (NanoInk, USA) with F-type one-dimensional cantilever arrays (NanoInk). Matching inkwells were loaded with 1 μL of each of the prepared mixtures and then allowed to dry in air for a few minutes. To ink the types, the cantilever ends were left in contact with the loaded inkwells for 15 minutes at high humidity (70% RH). Before writing on the substrates, the excess ink was removed from the tips by writing on a sacrificial area. The lipid patches lithographed on the prepared substrates were typically written keeping the relative humidity at 30%, with a line pitch of 100 nm and a writing speed of 0.1 μm s−1 to avoid nanoparticle detachment due to high lateral force. Nanoparticle multiplexing via μCS: In order to create different nanoscale curvatures on the same surface, the commercially purchased nanoparticle aqueous solutions were admixed with 25% volume glycerol to prevent evaporation during spotting. The spotting process was performed on a nanolithography platform (NLP 2000 system, NanoInk, USA) using surface patterning cantilevers (SPT-S-C10S, Bioforce Nanosciences). 2 μL of NP ink was deposited on the pen. A probe dwell time of 10 s was chosen, to ensure that the amount of nanoparticles transferred to the surface is large. Spotting was performed at a relative humidity of 80%.
Protein binding experiments
Preparing cell lysates: 70% confluent HEK cells were transfected with the Nadrin 2 BAR domain using Lipofectamine 2000 (Invitrogen, 11668-019) according to the manufacturer's instructions. 24 hours post transfection, the cells were lysed with IP Lysis buffer (Pierce, 87787) supplemented with Protease inhibitors (Cytoskeleton, PIC02), followed by sedimentation on a table-top centrifuge (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm). Only the supernatant was used for the experiment. Nadrin2 protein binding: The coverslips were activated with 1× PBS (Gibco, 1891-2014), followed by short incubation with 1 mg ml−1 BSA (Sigma, A90885) and biotinylated BSA (Pierce, 29130) (1
000 rpm). Only the supernatant was used for the experiment. Nadrin2 protein binding: The coverslips were activated with 1× PBS (Gibco, 1891-2014), followed by short incubation with 1 mg ml−1 BSA (Sigma, A90885) and biotinylated BSA (Pierce, 29130) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Excess BSA was removed by rinsing with 1× PBS, and the coverslips were exposed to cell lysates containing the curvature-sensitive BAR domain of Nadrin2 (AA1-256).41Streptavidin binding to biotinylated lipid patches: To prevent non-specific binding, the samples were incubated in a solution of 1% Bovine Serum Albumin (BSA) for 15 minutes. Subsequently, the BSA solution was removed and the sample was washed several times. The sample was then incubated for 15 minutes with a solution of 1
1). Excess BSA was removed by rinsing with 1× PBS, and the coverslips were exposed to cell lysates containing the curvature-sensitive BAR domain of Nadrin2 (AA1-256).41Streptavidin binding to biotinylated lipid patches: To prevent non-specific binding, the samples were incubated in a solution of 1% Bovine Serum Albumin (BSA) for 15 minutes. Subsequently, the BSA solution was removed and the sample was washed several times. The sample was then incubated for 15 minutes with a solution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 streptavidin-cy3 (Sigma-Aldrich, Germany) in PBS and washed again.
200 streptavidin-cy3 (Sigma-Aldrich, Germany) in PBS and washed again.
Sample characterization
Atomic force microscopy imaging: AFM images were recorded on a Dimension Icon system (Bruker, Germany). Measurements in the air were done in amplitude modulation with a 40 N m−1 probe from Nanosensors, with a typical oscillation amplitude of 10 nm. Fluorescence imaging: Fluorescence microscopy images were obtained on a Zeiss set-up equipped with SARFUS Software (Nanolane) or a Zeiss upright Axio Imager confocal microscope or an inverted Nikon Eclipse Ti, equipped with a confocal spinning disk unit and EMCCD camera using 10×, 20× or 60× objectives. Line scans of lipid patches were acquired using ImageJ.42 Relative intensities of the peak vs. adjacent shoulders were plotted and statistically probed (rank-sum test) in Matlab.All procedures were done at room temperature.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was carried out with the support of the Karlsruhe Nano Micro Facility (KNMF, http://www.knmf.kit.edu), a Helmholtz Research Infrastructure at the Karlsruhe Institute of Technology (KIT, http://www.kit.edu). E. B. acknowledges the Alexander von Humboldt Foundation for a postdoctoral fellowship. M. H. acknowledges the additional support by the Helmholtz Association in the form of a Helmholtz ERC Recognition Award.References
- M.-J. Park, R. Sheng, A. Silkov, D.-J. Jung, Z.-G. Wang, Y. Xin, H. Kim, P. Thiagarajan-Rosenkranz, S. Song, Y. Yoon, W. Nam, I. Kim, E. Kim, D.-G. Lee, Y. Chen, I. Singaram, L. Wang, M. H. Jang, C.-S. Hwang, B. Honig, S. Ryu, J. Lorieau, Y.-M. Kim and W. Cho, Mol. Cell, 2016, 62, 7–20 CrossRef CAS PubMed.
- C. M. Revankar, D. F. Cimino, L. A. Sklar, J. B. Arterburn and E. R. Prossnitz, Science, 2005, 307, 1625–1630 CrossRef CAS PubMed.
- S. Vanni, H. Hirose, H. Barelli, B. Antonny and R. Gautier, Nat. Commun., 2014, 5, 4916 CrossRef CAS PubMed.
- M. P. Ebrahimkutty and M. Galic, BioEssays, 2019, 41, 1900068 CrossRef PubMed.
- K. R. Rosholm, N. Leijnse, A. Mantsiou, V. Tkach, S. L. Pedersen, V. F. Wirth, L. B. Oddershede, K. J. Jensen, K. L. Martinez, N. S. Hatzakis, P. M. Bendix, A. Callan-Jones and Di. Stamou, Nat. Chem. Biol., 2017, 13, 724–729 CrossRef CAS PubMed.
- B. J. Peter, H. M. Kent, I. G. Mills, Y. Vallis, P. J. G. Butler, P. R. Evans and H. T. McMahon, Science, 2004, 303, 495–499 CrossRef CAS PubMed.
- J. S. Park, D. H. Kim, H. N. Kim, C. J. Wang, M. K. Kwak, E. Hur, K. Y. Suh, S. S. An and A. Levchenko, Nat. Mater., 2016, 15, 792–801 CrossRef CAS PubMed.
- J. Varkey, J. M. Isas, N. Mizuno, M. B. Jensen, V. K. Bhatia, C. C. Jao, J. Petrlova, J. C. Voss, D. G. Stamou, A. C. Steven and R. Langen, J. Biol. Chem., 2010, 285, 32486–32493 CrossRef CAS PubMed.
- E. Evans, H. Bowman, A. Leung, D. Needham and D. Tirrell, Science, 1996, 273, 933–935 CrossRef CAS PubMed.
- A. Roux, G. Cappello, J. Cartaud, J. Prost, B. Goud and P. Bassereau, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5394–5399 CrossRef CAS PubMed.
- R. P. Richter and A. R. Brisson, Biophys. J., 2005, 88, 3422–3433 CrossRef CAS PubMed.
- M. P. Mingeot-Leclercq, M. Deleu, R. Brasseur and Y. F. Dufrêne, Nat. Protoc., 2008, 3, 1654–1659 CrossRef PubMed.
- S. Chand, P. Beales, F. Claeyssens and B. Ciani, Exp. Biol. Med., 2019, 244, 294–303 CrossRef CAS PubMed.
- B. Sanii, A. M. Smith, R. Butti, A. M. Brozell and A. N. Parikh, Nano Lett., 2008, 8, 866–871 CrossRef CAS PubMed.
- R. Parthasarathy, C. Yu and J. T. Groves, Langmuir, 2006, 22, 5095–5099 CrossRef CAS PubMed.
- X. Li, L. Matino, W. Zhang, L. Klausen, A. F. McGuire, C. Lubrano, W. Zhao, F. Santoro and B. Cuip, A nanostructure platform for live-cell manipulation of membrane curvature, Springer US, 2019, vol. 14 Search PubMed.
- S.-R. Lee, Y. Park and J.-W. Park, J. Phys. Chem. B, 2020, 124(41), 8984–8988 CrossRef CAS PubMed.
- S. Mornet, O. Lambert, E. Duguet and A. Brisson, Nano Lett., 2005, 5, 281–285 CrossRef CAS PubMed.
- Y. Roiter, M. Ornatska, A. R. Rammohan, J. Balakrishnan, D. R. Heine and S. Minko, Nano Lett., 2008, 8, 941–944 CrossRef CAS PubMed.
- S. Sekula, J. Fuchs, S. Weg-Remers, P. Nagel, S. Schuppler, J. Fragala, N. Theilacker, M. Franzreb, C. Wingren, P. Ellmark, C. A. K. Borrebaeck, C. A. Mirkin, H. Fuchs and S. Lenhert, Small, 2008, 4, 1785–1793 CrossRef CAS PubMed.
- M. Hippler, K. Weißenbruch, K. Richler, E. D. Lemma, M. Nakahata, B. Richter, C. Barner-Kowollik, Y. Takashima, A. Harada, E. Blasco, M. Wegener, M. Tanaka and M. Bastmeyer, Sci. Adv., 2020, 6, eabc2648 CrossRef CAS PubMed.
- T. Ren, M. Erbakan, Y. Shen, E. Barbieri, P. Saboe, H. Feroz, H. Yan, S. McCuskey, J. F. Hall, A. B. Schantz, G. C. Bazan, P. J. Butler, M. Grzelakowski and M. Kumar, Adv. Biosyst., 2017, 1, 1–10 Search PubMed.
- G. Liu, M. Hirtz, H. Fuchs and Z. Zheng, Small, 2019, 15, 1900564 CrossRef PubMed.
- S. Lenhert, P. Sun, Y. Wang, H. Fuchs and C. A. Mirkin, Small, 2007, 3, 71–75 CrossRef CAS PubMed.
- T. Wu, Z. Shi and T. Baumgart, PLoS One, 2014, 9, e93060 CrossRef PubMed.
- Q. Zhang, C. Xue, Y. Yuan, J. Lee, D. Sun and J. Xiong, Sensors, 2012, 12, 2729–2741 CrossRef CAS PubMed.
- P. Rooney, A. Rezaee, S. Xu, T. Manifar, A. Hassanzadeh, G. Podoprygorina, V. Böhmer, C. Rangan and S. Mittler, Phys. Rev. B: Condens. Matter Mater. Phys., 2008, 77, 235446 CrossRef.
- K. Kneipp, A. S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. E. Shafer-Peltier, J. T. Motz, R. R. Dasari and M. S. Feld, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- K. M. Woeppel, X. S. Zheng and X. T. Cui, J. Mater. Chem. B, 2018, 6, 3058–3067 RSC.
- X. Rao, C. Guyon, S. Ognier, B. Da Silva, C. Chu, M. Tatoulian and A. A. Hassan, Appl. Surf. Sci., 2018, 439, 272–281 CrossRef CAS.
- M. Ben Haddada, J. Blanchard, S. Casale, J. Krafft, A. Vallée, C. Méthivier and S. Boujday, Gold Bull., 2013, 46, 335–341 CrossRef.
- T. Sainsbury, T. Ikuno, D. Okawa, D. Pacile, J. M. J. Frechet, A. Zettl, D. Pacilé, J. M. J. Fréchet and A. Zettl, J. Phys. Chem. C, 2007, 111, 12992–12999 CrossRef CAS.
- G. Lee, H. Lee, K. Nam, J.-H. Han, J. Yang, S. Lee, D. Yoon, K. Eom and T. Kwon, Nanoscale Res. Lett., 2012, 7, 608 CrossRef PubMed.
- R. García, R. Magerle and R. Perez, Nat. Mater., 2007, 6, 405–411 CrossRef PubMed.
- S. N. Magonov, V. Elings and M.-H. Whangbo, Surf. Sci., 1997, 375, L385–L391 CrossRef CAS.
- A. Urtizberea and M. Hirtz, Nanoscale, 2015, 7, 15618–15634 RSC.
- Y. Roiter, M. Ornatska, A. R. Rammohan, J. Balakrishnan, D. R. Heine and S. Minko, Langmuir, 2009, 25, 6287–6299 CrossRef CAS PubMed.
- M. Hirtz, A. Oikonomou, T. Georgiou, H. Fuchs and A. Vijayaraghavan, Nat. Commun., 2013, 4, 2591 CrossRef PubMed.
- N. Willems, A. Urtizberea, A. F. Verre, M. Iliut, M. Lelimousin, M. Hirtz, A. Vijayaraghavan and M. S. P. Sansom, ACS Nano, 2017, 11, 1613–1625 CrossRef CAS PubMed.
- J. Atwater, D. S. Mattes, B. Streit, C. von Bojničić-Kninski, F. F. Loeffler, F. Breitling, H. Fuchs and M. Hirtz, Adv. Mater., 2018, 30, 1801632 CrossRef PubMed.
- I. Begemann, T. Saha, L. Lamparter, I. Rathmann, D. Grill, L. Golbach, C. Rasch, U. Keller, B. Trappmann, M. Matis, V. Gerke, J. Klingauf and M. Galic, Nat. Phys., 2019, 15, 848–857 Search PubMed.
- M. D. Abràmoff, P. J. Magalhães and S. J. Ram, Biophotonics Int., 2004, 11, 36–42 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr01133b |
| This journal is © The Royal Society of Chemistry 2021 |