Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interfacial interactions between protective, surface-engineered shells and encapsulated bacteria with different cell surface composition†
Hao
Wei
a,
Xiao-Yu
Yang
 bc,
Wei
Geng
d,
Henny C.
van der Mei
bc,
Wei
Geng
d,
Henny C.
van der Mei
 *a and
Henk J.
Busscher
*a
*a and
Henk J.
Busscher
*a
aUniversity of Groningen and University Medical Center Groningen, Department of Biomedical Engineering, Antonius Deusinglaan 1, 9713 AV Groningen, The Netherlands. E-mail: h.c.van.der.mei@umcg.nl; h.j.busscher@umcg.nl
bWuhan University of Technology, State Key Laboratory of Advanced Technology for Materials Synthesis and Processing, Luoshi Road 122, 430070, Wuhan, China
cSchool of Engineering and Applied Sciences, Harvard University, Cambridge, Massachusetts 02138, USA
dSouthern Marine Science and Engineering Guangdong Laboratory (Zhuhai) & School of Chemical Engineering and Technology & School of Materials, Sun Yat-Sen University, Guangdong, 510275, China
First published on 31st March 2021
Abstract
Surface-engineered encapsulation is a non-genetic method to protect living organisms against harsh environmental conditions. Different cell encapsulation methods exist, yielding shells with different interfacial-interactions with encapsulated, bacterial surfaces. However, the impact of interfacial-interactions on the protection offered by different shells is unclear and can vary for bacteria with different surface composition. Probiotic bacteria require protection against gastro-intestinal fluids and antibiotics. Here, we encapsulated two probiotic strains using ZIF-8 (zeolitic imidazolate framework) biomineralization (strong-interaction by coordinate–covalent bonding), alginate gelation (intermediate-interaction by hydrogen bonding) or protamine-assisted packing of SiO2 nanoparticles yielding a yolk–shell (weak-interaction across a void between shells and bacterial surfaces). The surface of probiotic Lactobacillus acidophilus was rich in protein, yielding a hydrophilic, positively-charged surface below and a negatively-charged one above pH 4.0. Probiotic Bifidobacterium infantis had a hydrophilic, uncharged surface, rich in polysaccharides with little proteins. Although amino groups are required for coordinate–covalent bonding of zinc and hydrogen bonding of alginate, both L. acidophilus and B. infantis could be encapsulated using ZIF-8 biomineralization and alginate gelation. Weakly, intermediately and strongly interacting shells all yielded porous shells. The strongly interacting ZIF-8 biomineralized shell made encapsulated bacteria more susceptible to antibiotics, presumably due to the cell wall damage already inflicted during Zif-8 biomineralization. Overall, weakly interacting yolk–shells and intermediately interacting alginate gels protected best and maintained probiotic activity of encapsulated bacteria. The impact of interfacial-interactions between shells and encapsulated bacteria on different aspect of protection described here, contributes to the further development of effective surface-engineered shells and its application for protecting bacteria.
Introduction
Maintaining bacterial viability under harsh environmental conditions using surface-engineered encapsulation is a rapidly emerging field.1 Surface-engineered encapsulation can significantly extend the efficacy of living organisms in biosensors,2 bioreactors,3 food industry4 and probiotic prevention of gastro-intestinal infection.5 For instance, use of antibiotics may cause a disbalance in the gastro-intestinal microflora resulting in diarrhea in around 30% of all patients after antibiotic treatment.6–8 A balanced gastro-intestinal microflora during antibiotic treatment can be better preserved and more rapidly restored through the use of probiotic bacteria. However, without protective encapsulation either antibiotics, bile-salts or low gastro-intestinal pH may kill the majority of probiotic bacteria on their way through the gastro-intestinal tract.9 In general, an effective surface-engineered shell should allow exchange of nutrients and metabolites, and at the same time protect a bacterium against the host immune system, antimicrobials, pH, (self-produced) toxins, elevated temperature, UV light and other environmental challenges.10–14 Surface-engineered shells do not alter the genetic code of an organism and are therefore generally preferred above genetic manipulation of bacteria.15 Surface-engineered shells are temporary and break upon bacterial replication16 or chemical removal of the shells,17 rendering bacteria with an unaltered genetic code and environmental susceptibility as prior to encapsulation.Numerous methods have been proposed to encapsulate bacteria. Nanoparticles such as nanorods, nanospheres, nanotubes or nanosheets, can be deposited to a cell surface after cationic modification of the nanoparticle surface to allow in situ deposition mediated by electrostatic double-layer attraction to generally negatively-charged bacterial cell surfaces.18 Alternatively, deposition of negatively-charged nanoparticles to cell surfaces can be mediated by poly-electrolyte adsorption to the cell surface to create a positively-charged surface. Unfortunately, viability may suffer from direct deposition methods and approaches leaving a small gap between the cell surface and a surface-engineered shell (“yolk–shells”) are preferred.19 Moreover, many cell encapsulation methods based on surface-engineering are published without distinguishing between tissue cells, yeasts and bacteria.17,20–22 Yet, their surface compositions and physico-chemical properties can be very different. Even within a particular microbial strain or species, cell surface hydrophobicity23 and charge24,25 can hugely differ, depending on the surface chemical composition. For bacteria, the outermost cell surface to which encapsulation chemistry is applied, can either be rich in proteins or polysaccharides, combined with phosphate groups, originating from surface exposed lipoteichoic acids. The interfacial interactions occurring between the encapsulating shells and the encapsulated bacterial cell surface are of critical importance in maintaining bacterial viability and enhancing their functionality. Yet, there are no systematic studies that relate interfacial interactions with bacterial cell surface composition, resulting viability and enhanced functionality of encapsulated bacteria.
Therefore, the aim of this paper is to determine the role of interfacial interaction between surface-engineered shells and two different probiotic bacterial strains with widely differing cell surface compositions. Three different encapsulation methods have been applied (see Scheme 1) that include (1) biomineralization of a 2-methylimidazole zinc–salt based, metal–organic-framework (ZIF-8)17 interacting with bacterial cell surfaces through strong coordinate covalent bonding, (2) alginate gelation11 through hydrogen bonding yielding intermediate interaction with the bacterial cell surface and (3) protein-assisted yolk–shell packing of SiO2 nanoparticles19 with the aid of a cell penetrating protein (protamine). Yolk–shells typically leave a void between the shell and the encapsulated bacteria, and interfacial interactions are absent. Since the presence of bacterial cell surface proteins is required to establish bonding in ZIF-8 biomineralization and alginate gelation (see Scheme 1), two probiotic bacterial strains (Lactobacillus acidophilus and Bifidobacterium infantis) were employed in this study that differ in their amounts of cell surface proteins. Both strains are important in the production of dairy products and restoration of a healthy microflora in different sites of the human body, most notable restoration of a balanced intestinal microflora after or during antibiotic use.10 Concentrations of key-chemicals to be applied in the different encapsulation methods and in direct contact with the bacterial cell surface were determined by optimizing the viability of bacteria under different conditions. Physico-chemical bacterial cell surface properties were determined using particulate micro-electrophoresis (zeta potentials), two-phase partitioning, water contact angle measurements (cell surface hydrophobicity) and X-ray photoelectron spectroscopy (surface chemical composition). Encapsulation efficacy was evaluated on the basis of the protection offered by the different encapsulation methods against simulated gastro-intestinal fluids and exposure to different antibiotics. In addition, it was evaluated whether encapsulation affects release of biosurfactive metabolites (“biosurfactants”) that are characteristic to probiotic bacteria and crucial for their probiotic action.26
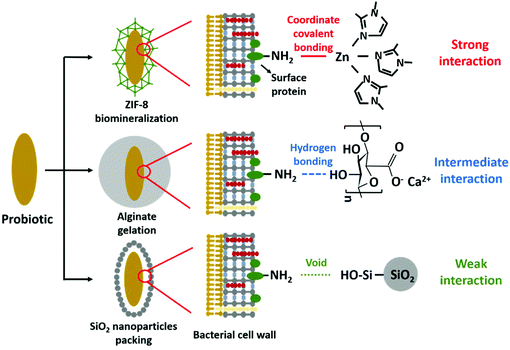 | ||
| Scheme 1 Overview of the different encapsulation methods applied and their interaction with bacterial cell surface proteins. | ||
Results
Effects of encapsulation on bacterial viability
First, the different encapsulation methods were applied at different concentrations of the key-chemicals that directly contact the bacterial cell surfaces in order to optimize the viability of encapsulated bacteria. Viability was determined using agar plating and enumeration of the number of colonies forming units (CFUs) prior to and after different encapsulations. Agar plating is the gold-standard in microbiology to demonstrate viability and other types of measurements do not reflect viability. Fluorescence-based metabolic activity assays for instance, negate the fact that bacteria can survive long periods of time without showing metabolic activity,27 while many so-called live–dead stains measure cell wall damage that does not necessarily reflect lack of viability.28 Decreases in viability (Fig. 1) were analyzed for statistical and microbiological significance, considering that microbiological differences in viability less than 2 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log units are generally considered meaningless.29L. acidophilus was able to remain viable upon applications of higher concentrations of dimethylimidazole and zinc acetate than B. infantis, while sodium alginate, protamine and SiO2 nanoparticles did not affect their viabilities. Within these considerations, concentrations of different key-chemicals in direct contact with the bacterial cell surfaces were derived yielding maximal viability for each strain, as summarized in Table S1.† These concentrations were further applied in the remainder of this study.
log units are generally considered meaningless.29L. acidophilus was able to remain viable upon applications of higher concentrations of dimethylimidazole and zinc acetate than B. infantis, while sodium alginate, protamine and SiO2 nanoparticles did not affect their viabilities. Within these considerations, concentrations of different key-chemicals in direct contact with the bacterial cell surfaces were derived yielding maximal viability for each strain, as summarized in Table S1.† These concentrations were further applied in the remainder of this study.
Biosurfactant release and physico-chemical surface properties of unencapsulated bacteria
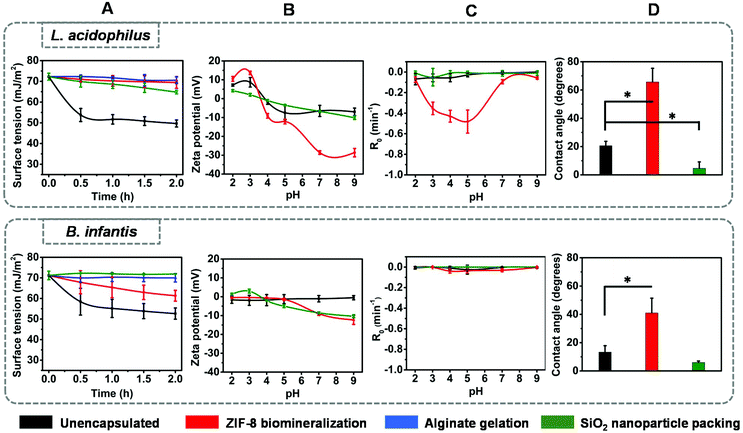
One of the most important characteristics of probiotic bacteria that can be demonstrated using physico-chemical methods, is biosurfactant production. Both strains, when suspended in buffer, decreased the buffer surface tension within 2 h from 71 mJ m−2 to 50 mJ m−2 and 52 mJ m−2 for L. acidophilus and B. infantis, respectively (Fig. 2A). Considering bacteria that decrease the surface tension of a bacterial suspension droplet by more than 8 mJ m−2 after 2 h as biosurfactant releasing,30 this attests to the ability of both strains to produce and release biosurfactants, as one of their probiotic mechanisms of action.Zeta potentials of L. acidophilus were pH-dependent, ranging from +10 mV at acidic pH to −10 mV above their iso-electric point (IEP) at pH 4.0. Zeta potentials of B. infantis on the other hand, hovered around zero over the entire pH-range from 2 to 9 (Fig. 2B). Both strains showed little partitioning from an aqueous phase into a hydrophobic phase (Fig. 2C), classifying both strains as hydrophilic. Hydrophilicity was confirmed by water contact angle measurements on macroscopic, hydrated bacterial lawns prepared on membrane filters23 and amounted 20 ± 3 degrees and 13 ± 4 degrees for L. acidophilus and B. infantis, respectively. Differences in surface chemical composition of the bacterial cell surfaces underlying the differences in bacterial zeta potential and hydrophobicity were measured on lyophilized bacteria using XPS. Elemental surface compositions of bacteria and yeast determined by XPS have previously been shown to relate with water contact angles, two-phase partitioning and zeta potentials.23,31 The main differences between both strains involved the amounts of nitrogen and oxygen at their outermost surface Fig. 2D–I. L. acidophilus had a nitrogen-rich surface (Fig. 2D) compared with B. infantis (Fig. 2G). Decomposition of the N1s photo-electron binding energy peak demonstrated that surface nitrogen in L. acidophilus is due to amino-groups and amide groups (Fig. 2E), confirming that nitrogen reflects protein presence.32 For B. infantis (Fig. 2G) absence of major amounts of nitrogen and phosphorus were concurrent with the lack of charged groups over the entire pH range, as observed from their zeta potentials (Fig. 2B). This, combined with the high amount of oxygen at the B. infantis surface with a major O1s component located at 532.6 eV (Fig. 2I), suggests that its surface is mainly composed of polysaccharides.
Use of these elemental surface compositions in a generalized model describing the bacterial cell surface as being composed of proteins, polysaccharides and lipoteichoic acids (see also ESI†),33 confirmed the above suggestions and yielded the conclusion that the L. acidophilus surface was composed for 62% of proteins, 33% of teichoic acid and 5% of hydrocarbon-like components. The B. infantis surface had an entirely different chemical composition containing only 20% of proteins but 40% of polysaccharides. Lipoteichoic acids occurred in minor amounts (17%), while 23% of the surface was composed of hydrocarbon-like components.
In summary, L. acidophilus and B. infantis both have a similarly hydrophilic surface and the ability to produce biosurfactants. The main difference between both strains involves their zeta potentials, which hover around 0 mV over the pH range 2 to 9 for B. infantis, while showing an iso-electric point at pH 4 for L. acidophilus. XPS confirmed that these differences are due to the protein-rich surface of L. acidophilus as compared with B. infantis surface, that is rich in polysaccharides with little proteins.
Biosurfactant release and physico-chemical surface properties of surface-engineered shells around L. acidophilus and B. infantis
Biosurfactant release by both strains was severely hampered by all surface-engineered encapsulating shells (Fig. 3A). However, biosurfactant release through the shells was different for shells formed on the protein-rich L. acidophilus surface than on the carbohydrate-rich B. infantis surface. Encapsulated L. acidophilus demonstrated slow decreases of the suspension surface tension upon protein-assisted packing of SiO2 nanoparticles. For B. infantis, a slow decrease in surface tension indicative of biosurfactant release was only observed for biomineralized ZIF-8 shells.Different shells around the two bacterial strains had different physico-chemical properties. Protamine-assisted yolk–shell packing of SiO2 nanoparticles requires adsorption and subsequent internalization of protamine after packing of SiO2 nanoparticles. Internalization of pre-adsorbed protamine was confirmed by time-dependent zeta potential measurements, XPS and FTIR of L. acidophilus and B. infantis after protamine adsorption. Immediately after protamine adsorption, both bacterial strains acquired a more positively-charged zeta potential between pH 4 to pH 9 and using XPS, S was detected due to protamine sulphate at the bacterial cell surfaces (Fig. S1 and Table S2,† respectively). Zeta potentials returned to their pre-adsorption value within 60 min (Fig. S1†), while S became below detection by XPS (Table S2†). FTIR spectra of both strains showed an increased AmI/CH ratio immediately after protamine adsorption (Fig. S1C–1E†). However, amide absorption band ratios of both strains remained invariably high after 60 min. Since FTIR represents the composition of both the bacterial interior and the surface, collectively these results demonstrate that both bacterial cell surfaces allow internalization of protamine to leave a void between a nanoparticle shell and the bacterial cell surface. Due to this void, interaction is weak (Scheme 1). The presence of adsorbed protamine immediately after adsorption, accommodated packing of SiO2 nanoparticles into a shell with more negative zeta potentials above pH 4 for B. infantis but not for L. acidophilus. Presence of SiO2 nanoparticles was further evidenced by the detection of around 30% Si at an Si to O ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (see Table 1). The pronounced presence of Si and O in a 1
2 (see Table 1). The pronounced presence of Si and O in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio as in SiO2, combined with the near disappearance of N1s and P2p peaks, attest to a thick shell composed of SiO2 nanoparticles around both strains. SEM micrographs showed the shells as a clear coat around the bacteria (Fig. S2A†). EDX line scans demonstrated similar elements as observed using XPS, but due to the larger depth of information of EDX as compared with XPS, the elemental composition from EDX relates not only to the surface but also to the interior of the bacteria. From Fig. S2A,† the presence of a SiO2 nanoparticle shell can be clearly observed from the Si signal intensity when the line-scan passes over the bacterium.
2 ratio as in SiO2, combined with the near disappearance of N1s and P2p peaks, attest to a thick shell composed of SiO2 nanoparticles around both strains. SEM micrographs showed the shells as a clear coat around the bacteria (Fig. S2A†). EDX line scans demonstrated similar elements as observed using XPS, but due to the larger depth of information of EDX as compared with XPS, the elemental composition from EDX relates not only to the surface but also to the interior of the bacteria. From Fig. S2A,† the presence of a SiO2 nanoparticle shell can be clearly observed from the Si signal intensity when the line-scan passes over the bacterium.
| Encapsulation method | Elemental surface composition (at%) | |||||||
|---|---|---|---|---|---|---|---|---|
| C | N | O | P | Si | Zn | Ca | Na | |
| a Denotes below detection. b Alginate gelation yielded shells with a high amount of Cl for L. acidophilus and B. infantis surfaces (14.6 and 13.4%, respectively). | ||||||||
| L. acidophilus ATCC 4356 | ||||||||
| Unencapsulated | 66.2 | 12.2 | 19.2 | 2.5 | —a | — | — | — |
| ZIF-8 biomineralization | 61.5 | 19.5 | 10.4 | — | — | 7.8 | — | — |
| Alginate gelationb | 47.2 | 2.1 | 21.5 | 1.5 | — | — | 3.7 | 9.4 |
| Protein assisted SiO2 nanoparticles packing | 8.3 | 1.6 | 59.4 | — | 30.8 | — | — | — |
| B. infantis ATCC 15697 | ||||||||
| Unencapsulated | 65.2 | 3.3 | 30.4 | 1.1 | — | — | — | — |
| ZIF-8 biomineralization | 58.0 | 15.9 | 18.0 | — | — | 7.1 | — | — |
| Alginate gelationb | 45.5 | 2.9 | 24.5 | 0.9 | — | — | 2.8 | 10.0 |
| Protein assisted SiO2 nanoparticles packing | 9.6 | 1.0 | 59.5 | — | 29.8 | — | — | — |
Alginate gelation yielded a shell that interacted with intermediate hydrogen bonding (Scheme 1) and with an elemental surface composition that was nearly identical for both strains (Table 1). After alginate encapsulation, percentages of carbon, nitrogen and phosphorus present in the L. acidophilus cell surface and carbon and oxygen percentages for the B. infantis decreased, indicating encapsulation with a relatively thick shell. In addition, Ca was observed after alginate gelation, serving to bridge between alginate molecules upon exchange with Na. However, not all Na was exchanged upon bridging (see also Table 1). Neither zeta potentials nor two-phase partitioning could be reliably measured due to the large diameter of hydrated alginate-hydrogel encapsulated bacteria. SEM micrographs were also impossible to take, while the alginate hydrogel did not contain unique elements to demonstrate its presence in an EDX line-scan.
ZIF-8 encapsulation through strong coordinate interaction (Scheme 1) caused more negative zeta potentials above pH 4 for both strains (Fig. 3B). Removal of ZIF-8 encapsulated bacteria from an aqueous phase by hexadecane was only increased for lactobacilli, while water contact angles of both strains were strongly increased after ZIF-8 encapsulation (Fig. 3C and D). XPS confirmed ZIF-8 encapsulation and detected 7%–8% Zn and increased amounts of nitrogen as compared with unencapsulated strains in the absence of a detectable phosphorus signal (Table 1). The ratio of Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N was 0.40 for ZIF-8 encapsulated L. acidophilus and 0.45 for ZIF-8 encapsulated B. infantis. SEM micrographs and EDX line-scans confirmed the presence of a Zn shell around the bacteria (Fig. S2B†). The experimental Zn
N was 0.40 for ZIF-8 encapsulated L. acidophilus and 0.45 for ZIF-8 encapsulated B. infantis. SEM micrographs and EDX line-scans confirmed the presence of a Zn shell around the bacteria (Fig. S2B†). The experimental Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratio is higher than the theoretical ratio expected based on the molecular structure of ZIF-8 (0.25). Likely, the high Zn/N ratio is caused by the nanoscale depth of information of XPS. On the outer surface of the ZIF-8 layer, there will be undercoordinated Zn-ions34 with less than tetrahedral coordination which will likely cause higher, experimental Zn/N ratio. Collectively this indicates that ZIF-8 had formed a thick shell around both bacterial strains.
N ratio is higher than the theoretical ratio expected based on the molecular structure of ZIF-8 (0.25). Likely, the high Zn/N ratio is caused by the nanoscale depth of information of XPS. On the outer surface of the ZIF-8 layer, there will be undercoordinated Zn-ions34 with less than tetrahedral coordination which will likely cause higher, experimental Zn/N ratio. Collectively this indicates that ZIF-8 had formed a thick shell around both bacterial strains.
Finally, porosities of the different shells were determined using nitrogen adsorption/desorption isotherms. BET surface areas and pore diameters were derived from the adsorption/desorption isotherms presented in Fig. S3,† as summarized in Table 2. Note that in case of unencapsulated bacteria, nitrogen adsorption/desorption isotherms were too flat to derive BET surface areas and pore diameters. All encapsulation methods yielded pores with a diameter between 9 and 17 nm, regardless of the strength of interaction with the bacterial cell surfaces. Alginate gelation yielded a low porosity compared with ZIF-8 biomineralization and protamine assisted SiO2 nanoparticle packing, which is probably a result of the collapse of alginate-hydrogel shell during measurement. ZIF-8 biomineralization and protamine assisted SiO2 nanoparticle packing yielded large increases in porosity that were similar for L. acidophilus and B. infantis in case of weakly interacting yolk–shells. However, strongly interacting ZIF-8 biomineralized shells had a two-fold lower porosity when encapsulating protein-rich L. acidophilus surfaces than observed for B. infantis with a polysaccharide-rich surface.
| Encapsulation method | Surface area (m2 g−1) | Pore diameter (nm) |
|---|---|---|
| a Could not be derived from the adsorption/desorption isotherms. | ||
| L. acidophilus ATCC 4356 | ||
| Unencapsulated | —a | — |
| ZIF-8 biomineralization | 34 | 12 |
| Alginate gelation | 1 | 12 |
| Protein assisted SiO2 nanoparticles packing | 134 | 9 |
| B. infantis ATCC 15697 | ||
| Unencapsulated | — | — |
| ZIF-8 biomineralization | 76 | 17 |
| Alginate gelation | 2 | 14 |
| Protein assisted SiO2 nanoparticles packing | 132 | 9 |
Protection offered by different shells against gastro-intestinal fluids and antibiotics
The protection of the different shells offered towards exposure to simulated gastric (SGF) and intestinal fluids (SIF) and three antibiotics carrying different charge, was evaluated using agar plating and CFU enumeration. In absence of encapsulation, neither of the two strains survived exposure to acidic simulated gastric fluid at pH 2 (Fig. 4A) and only L. acidophilus survived at pH 3 without encapsulation (Fig. 4B). Intermediately interacting alginate gelation and strongly interacting ZIF-8 biomineralization protected L. acidophilus against simulated gastric fluid at pH 2 (Fig. 4A). None of the methods achieved protection of B. infantis against simulated gastric fluid at pH 2 (Fig. 4A), but the strain was protected by all encapsulation methods at pH 3 (Fig. 4B). Both bacterial strains survived well in bile-salt rich, alkaline simulated intestinal fluid (pH 8). After encapsulation however, particularly by alginate gelation and ZIF-8 biomineralization, both strains became more susceptible to simulated intestinal fluid (Fig. 4C). | ||
| Fig. 4 Viability of L. acidophilus ATCC 4356 and B. infantis ATCC 15697 encapsulated by different surface-engineered shells after the exposure to simulated gastro-intestinal fluids and the antibiotics tobramycin, amoxicillin and tetracycline. Viability was assessed by agar plating after removal of the shells. (A) The number of CFU mL−1 as a function of exposure time to simulated gastric fluid (SGF) at pH 2 for L. acidophilus and B. infantis. (B) Same as panel (A), now for exposure to SGF at pH 3. (C) Same as panel (A), now for exposure to simulated, bile-salt rich intestinal fluid (SIF). (D) The number of CFU mL−1 after 24 h exposure to different antibiotics at 5× MIC (see Table S3†) for L. acidophilus and B. infantis. The horizontal band represents the number of CFU mL−1 for L. acidophilus and B. infantis not exposed to antibiotics. All error bars indicate standard deviations over triplicate experiments with separately cultured and encapsulated bacteria. Statistically significant differences (p < 0.05, Student t-test) within each group of antibiotics with respect to unexposed bacteria are indicated by hash-decks, while differences with respect to unencapsulated bacteria are indicated by spanning bars with asterisks. | ||
Tobramycin, amoxicillin and tetracycline represent positively charged, zwitter-ionic and negatively-charged antibiotics, respectively. Their Minimal Inhibitory Concentrations (MIC) were almost identical for L. acidophilus and B. infantis, but differed for each antibiotic (Table S3†). Tobramycin required the highest concentration for growth inhibition of both strains, followed by tetracycline and amoxicillin. Significant protection against positively-charged tobramycin was offered by alginate gelation and protein assisted SiO2 nanoparticle packing towards L. acidophilus, while only an alginate shell protected B. infantis against tobramycin (Fig. 4D). Neither of the three encapsulation methods protected B. infantis against zwitter-ionic amoxicillin, while SiO2 nanoparticle packing provided some protection against zwitter-ionic amoxicillin for L. acidophilus. None of the different surface-engineered shells yielded protection against negatively-charged tetracycline and ZIF-8 biomineralization even increased the susceptibility of L. acidophilus to tetracycline.
Pathogen killing by differently encapsulated lactobacilli and bifidobacteria
Maintenance of pathogen killing by encapsulated L. acidophilus and B. infantis was evaluated by co-culturing Escherichia coli in the presence of either probiotic strain and enumerating the number of surviving E. coli. The presence of unencapsulated L. acidophilus or B. infantis in a co-culture significantly reduced the number of viable E. coli (Fig. 5). Probiotic-mediated reductions in E. coli CFUs were maintained best by protein assisted SiO2 nanoparticle packing. Encapsulation by alginate gelation or ZIF-8 biomineralization of neither probiotic strain yielded any reduction in E. coli killing.Discussion
This paper aimed to determine the influence of interfacial interactions between bacterial cell surfaces with different composition and surface-engineered, encapsulating shells. Two bacterial strains were selected for this study that were demonstrated using XPS to possess either a protein-rich (L. acidophilus) or polysaccharide-rich surface with little proteins (B. infantis). The protein-rich bacterial strain had pH-dependent zeta potentials, while the polysaccharide-rich strain was nearly uncharged over the entire pH-range. Both strains classified as hydrophilic.Among the three encapsulation methods involved, ZIF-8 biomineralization mainly relies on the presence of amino-groups on the bacterial cell surfaces, attracting zinc to stimulate ZIF-8 mineralization through NH2–Zn2+ coordinate covalent bonding, yielding a strong interaction. Alginate gelation is achieved by cross-linking of alginate ligands caused by the exchange of calcium ions and sodium ions, while hydroxyl groups abundantly present in the alginate applied here (Scheme 1), form hydrogen bonds with amino groups on the bacterial cell surfaces. In a hydrogen bond, a shared electron pair is formed between a hydrogen atom and another atom,35 giving rise to weak interactions with typical binding energies up to 4 kcal mol−1.36 Opposite to hydrogen bonding, coordinate covalent bonding involves electrons from only one of the interacting atoms to form a shared electron pair with another atom, yielding strong interactions for zinc with amino acid side chains between 195 and 364 kcal mol−1, depending on the coordination geometry.37 Both the protein-rich surface of L. acidophilus (62% protein) as well as the protein-poor surface of B. infantis (20% protein) could be encapsulated by ZIF-8 and alginate, yielding the conclusion that for effective encapsulation only small amounts of cell surface proteins are needed.
SiO2 nanoparticle packing is assisted by protamine adsorption to the bacterial cell surface and its subsequent internalization through the bacterial cell wall which occurred for both strains (Fig. S1 and Table S2†) despite their widely different surface composition. This indicates that protamine internalization does not depend on the composition of the outer bacterial cell surfaces, but likely on the penetrability to protamine of the rigid peptidoglycan layer present in the cell walls of both strains. Previously, protamine was also demonstrated to penetrate cyanobacterium Synechocystis sp. PCC 7002,19 the blood brain barrier,38 the intestinal-epithelial cell wall,39 retina40 and neurons.41 After the internalization of protamine, a void interface develops which minimizes the interfacial interaction between the bacterial surface and the SiO2 nanoparticles constituting the encapsulating yolk–shell.
Below we present a comprehensive discussion of several aspects of the different encapsulation methods applied in relation with the interfacial interactions between encapsulating shells and composition of the cell surfaces of these two probiotic bacterial strains.
Impact of bacterial cell surface properties upon bacterial viability after encapsulation
The concentration of key-chemicals in direct contact with the bacterial cell surfaces during encapsulation had a major impact on the viability of the encapsulated bacteria. Alginate gelation (Fig. 1B) and protamine-assisted SiO2 yolk–shell packing (Fig. 1C) were well tolerated by both bacterial strains due to the mild interactions between key-chemicals and the bacterial cell surfaces, although minor reductions in viability were observed for B. infantis. Strongly interacting key-chemicals involved in ZIF-8 biomineralization strongly reduced the viability of encapsulated bacteria due to coordinate bonding, requiring optimizing the concentrations of key-chemicals in direct contact with the cell wall with respect to viability (see Fig. 1A and Table S1†).Possession of a protein-rich surface by L. acidophilus was of great influence on averting a negative impact on viability of key-chemicals involved in encapsulation by ZIF-8 biomineralization. Viabilities of B. infantis were significantly reduced by ZIF-8 biomineralization involving strong coordinate bonding to the small number of proteins present on the B. infantis cell surface. Proteins in lactobacilli are incorporated in a paracrystalline layer, called S-layer that contributes to the protection offered by its natural cell wall components against environmental conditions42 and making it less susceptible to ZIF-8 biomineralization than the protein-poor/polysaccharide-rich surface of B. infantis.
Hydrophobicity, zeta potentials and porosity of encapsulated bacteria
Encapsulation altered the physico-chemical properties of the bacteria, and most notably increased their hydrophobicity after ZIF-8 biomineralization, regardless of the bacterial cell surface composition. The ubiquitous increase in hydrophobicity after ZIF-8 biomineralization can be explained by the known hydrophobicity of ZIF-8.43Zeta potentials and their pH dependence however, largely reflected the charge properties of the unencapsulated bacterial cell surface (see Fig. 3A). Zeta potentials as measured using electrophoretic micro-electrophoresis, reflect the electrostatic potential at the plane of shear. For “hard” particles, the plane of shear is located several nanometers away from the surface in the fluid phase, with the exact distance depending on ionic strength conditions. In case of “soft” particles44 however, liquid is able to penetrate the surface and the plane of shear will be located inside the soft particle. This will also be the case for shells encapsulating bacteria as they are designed to possess porosity (see also Table 2) to allow exchange with their environment. This explains why zeta potentials and their pH-dependence of unencapsulated and encapsulated bacteria bear similarity and confirms the porosity of the shells as observed using nitrogen adsorption/desorption isotherms (Table 2). Importantly, it confirms that the low porosity observed for alginate gelation (Table 2) is an artefact due to collapse of the gel during nitrogen adsorption/desorption.
Protective effects of different encapsulation methods
Protection is the main reason why surface-engineered encapsulation methods are being designed. Porosity is crucial for maintaining metabolite exchange of an encapsulated bacterium with its environment but also in providing protection. Like metabolite exchange, protection often depends on adsorption and absorption in the shell and therewith is time- and concentration-dependent. In line with their thick, highly porous shells, alginate gelation and ZIF-8 biomineralization already provided protection to L. acidophilus against SGF at pH 2 (see Fig. 4A), while all encapsulation methods yielded protection of B. infantis against SGF at pH 3 (see Fig. 4B). Accordingly, it shows that the weak interfacial interaction between bacterial surfaces and an SiO2 nanoparticle layer does block acids less than alginate and ZIF-8 encapsulation. However, the difference in pH sensitivity of both strains makes a comparison of the protective properties of shells between strains based on acid attack difficult.Protection against antibiotic attack is easier for comparison of the protective properties of the shells, because the MIC of L. acidophilus and B. infantis against each of the three antibiotics evaluated are nearly identical (Table S3†). Moreover, also their molecular dimensions are equally small, based on their nearly identical molecular weight (see also Table S3†). This implies that none of the shells possesses pores that can physically block penetration of these antibiotics and instead, antibiotic protection must be caused by their physico-chemical adsorption inside the shells and affinity to the bacterial cell surface. None of the encapsulations evaluated provided protection towards negatively-charged tetracycline (see Fig. 4D), suggesting Lifshitz–van der Waals attraction of tetracycline to the bacterial cell surfaces and lack of its adsorption and absorption in the shells due to electrostatic repulsion. However, since L. acidophilus can create an acidic micro-environment, its zeta potential can be positive in the confinement of a shell (see Fig. 2B) and electrostatic attraction may also contribute to the tetracycline susceptibility of the lactobacilli after encapsulation. Also, none of the encapsulations protected uncharged B. infantis with its protein-poor/polysaccharide-rich surface against zwitter-ionic amoxicillin, while the protein-rich L. acidophilus was protected against amoxicillin by SiO2 nanoparticle yolk–shells (see also Fig. 4D). Amoxicillin is composed of cationic groups that are repelled by positive charges on the L. acidophilus cell surface in its acidic micro-environment. In addition, amoxicillin readily absorbs to negatively-charged SiO2 nanoparticles.19 The ZIF-8 shell is hydrophobic and therefore will not absorb and protect against hydrophilic amoxicillin dissolved in an aqueous phase. Both strains were protected against positively-charged tobramycin upon alginate gelation and SiO2 nanoparticle packing only protected L. acidophilus against tobramycin. Its positive charge makes tobramycin readily adsorb to negatively-charged shell components and causes repulsion towards positively-charged lactobacilli in their acidic micro-environment. Though without presenting it as an explicit reason, Li et al.10 successfully exploited the ease of providing protection to lactobacilli towards tobramycin by alginate gelation to demonstrate that alginate encapsulated lactobacilli eradicated methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa in co-culture when co-administered with the antibiotic.
Drawbacks of effective encapsulation
Encapsulating shells not only offer protection but can also backfire on the encapsulated bacteria. Strongly interacting ZIF-8 biomineralization increased susceptibility of L. acidophilus to tetracycline (Fig. 4D). Similarly, none of the two probiotic strains required protection against the high bile-salt concentration of SIF when unencapsulated (see Fig. 4C), but neither of the two strains survived exposure to SIF when encapsulated with intermediately interacting alginate or strongly interacting ZIF-8 shells. This first of all suggests adsorption and absorption of bile-salt in the shell to create a high bile-salt concentration in the immediate micro-environment of the bacteria in which they do not survive. However, this suggestion is not valid for weakly-interacting packed SiO2 nanoparticle yolk–shells despite their high porosity. This points to another reason why alginate gelation and ZIF-8 biomineralization may backfire on antibiotic protection. As a second suggestion to explain the “inverse protection” of encapsulation, it is offered that relatively strong interfacial interactions cause damage to the bacterial cell walls that assist antibiotic action opposite to the weak interactions across the void between a yolk–shell and the bacterial cell surface.Along the same line of thinking, release of biosurfactants, essential for probiotic activity, can be severely hampered by encapsulation. Biosurfactant release of both strains were inhibited by all three encapsulations, which is caused by the slow diffusion rate of biosurfactant through the shells. Since adsorption and absorption in a shell and also transport of biosurfactants through a shell are time-dependent phenomena, it is important to realize, that any conclusion about protection offered by any type of shell, should be placed in the context of a relevant timeframe of the challenged endured by encapsulated bacteria during their application.
Maintenance of pathogen killing by encapsulated bacteria
Interestingly, there is a 1 to 1 correspondence between the maintenance of biosurfactant release by L. acidophilus within the timeframe of our experiments and its ability to kill E. coli (compare Fig. 3A and 5A). This confirms the importance of biosurfactants in the probiotic activity of L. acidophilus. However, the lack of correspondence between maintenance of biosurfactant release and E. coli killing in B. infantis (compare Fig. 3A and 5B) demonstrates that maintenance of biosurfactant release is not a conditio sine qua non for an effective encapsulation that maintains probiotic activity. This is due to the temporary nature of surface-engineered shells. Due to growth, bacteria will eventually break through a shell and function as an unencapsulated bacterium. Therewith for probiotic bacteria, it is more important that the protection offered by encapsulation allows them to survive SGF, SIF and antibiotics encountered on their way to their intestinal target site than maintaining release of biosurfactants through a shell. Breaking through the shells once they have reached their target site will ensure biosurfactant release and suffices for effective probiotic action at the target site.Maintenance of pathogen killing by encapsulated bacteria is the combined effect of all available killing mechanisms that can be applied by a probiotic bacterium. Therewith it can be concluded from Fig. 5, that encapsulation methods like ZIF-8 biomineralization with a strong interfacial interaction with bacterial cell surface, are undesirable, leaving weakly interaction yolk–shells as a preferable method for encapsulating probiotic bacteria.
Conclusions
In summary, we have demonstrated that an L. acidophilus strain, possessing a protein-rich surface and a B. infantis strain with a protein-poor/polysaccharide-rich surface can both be encapsulated with methods that require the presence of cell surface proteins on the bacterial cells surfaces. Low numbers of cell surface proteins as on B. infantis surfaces suffice for strong coordinate covalent interaction between zinc and amino groups in ZIF-8 biomineralization and for the intermediate interaction of alginate gels with amino groups through hydrogen bonding.Weakly, intermediately and strongly interacting shells of different nature all yielded porous shells, that on the one hand protected against acids, bile salts and selected antibiotics, but on the other hand shells with a strong interaction with bacterial cell surfaces made bacteria more susceptible to antibiotics. Overall, weakly interacting protamine assisted SiO2 nanoparticle packing of yolk–shells best maintained probiotic activity of encapsulated lactobacilli and Bifidobacteria.
In short, surface-engineering of protective shells, must account for the impact of the strength of the interfacial interactions between shells and bacterial cell surfaces. The impact of interfacial interactions can differ amongst different bacterial strains, depending on their cell surface composition. Herewith, this study contributes to the further development of effective encapsulating shells and its application for protecting probiotic bacteria on their way in the gastro-intestinal tract in patients during antibiotic use.
Experimental section
Bacterial growth and harvesting
L. acidophilus ATCC 4356 was first grown on MRS (De Man, Rogosa and Sharpe, Merck KGaA, Germany) agar plates under 5% CO2 at 37 °C for 48 h. Then, a single colony was picked from the agar plate and re-suspended in 10 mL MRS broth. After culturing, also under 5% CO2 and at 37 °C for 24 h, the pre-culture was transferred into 200 mL of MRS broth and cultured under the same conditions for 18 h. After 18 h, bacteria were harvested by centrifugation (5000g, 5 min, 10 °C) centrifuged and washed with 10 mM potassium phosphate buffer three times and re-suspended in 5 mL of 10 mM phosphate buffer. B. infantis ATCC 15697 and E. coli ATCC 25922, were cultured essentially as described above for L. acidophilus, but using RCM (Reinforced Clostridial) agar or in RCM broth (Becton, Dickinson and Company, France) for B. infantis and BHI (brain heart infusion) agar or in BHI broth (Oxoid Ltd, UK) for E. coli. In additon, B. infantis was cultured under anaerobic conditions (85% N2, 10% H2, 5% CO2, at 37 °C,) and E. coli under aerobic conditions. Bacterial concentrations after suspending in phosphate buffer were determined in a Bürker-Türk counting chamber and fixed at 3 × 109 mL−1 for L. acidophilus and 3 × 1010 mL−1 for B. infantis.Alginate gelation
For alginate gelation,11L. acidophilus or B. infantis were added to 5 mL 10 mM phosphate buffer together with 2.5 wt% alginate to yield a final bacterial concentration of 3 × 108 mL−1. The mixture was subsequently vortexed for 1 min and gelation of alginate initiated by adding the mixture into 100 mL of 0.1 M CaCl2 at a controlled speed of 200 μL min−1, using a syringe pump. The bacterial suspension with the alginate in the CaCl2 solution were stirred for 30 min at room temperature to allow completely gelation. Finally, clumps of encapsulated bacteria were collected with a sterilized tweezer and stored in 10 mM phosphate buffer for further experiments.Protamine-assisted SiO2 nanoparticles packing
For protamine-assisted SiO2 yolk–shell nanoparticles packing,19L. acidophilus suspended in a 2 mg mL−1 protamine sulfate salt (Sigma-Aldrich, USA) solution at a concentration of 3 × 108 mL−1 were allowed to settle for 15 min after which bacteria were collected by centrifugation (5000g, 5 min). Subsequently, the bacteria were added to a 5 mg mL−1 SiO2 suspension (Ludox HS-40, Sigma-Aldrich, USA), and allowed to settle for 15 min to allow deposition of the nanoparticles. For B. infantis, lower concentrations of protamine (1 mg mL−1) and SiO2 (1 mg mL−1) were used to yield optimal viability after encapsulation (Fig. 1). Finally, encapsulated bacteria were collected by centrifugation (5000g, 5 min) and re-suspended by vortexing in 10 mM phosphate buffer.ZIF-8 biomineralization
For ZIF-8 biomineralization,17L. acidophilus or B. infantis were suspended in 5 mL 10 mM phosphate buffer containing 160 mM 2-methylimidazole (Sigma-Aldrich, USA) to concentration of 3 × 108 mL−1 and 3 × 109 mL−1, respectively. A higher concentration of B. infantis was used to yield optimal viability after encapsulation. Next, 5 mL 40 mM zinc acetate dihydrate (Sigma-Aldrich, USA) was added, the mixture was shaken at 37 °C at 150 rpm for 10 min after which the encapsulated bacteria were collected by centrifugation. The bacterial pellet was re-suspended by vortexing in 10 mM phosphate buffer.Viability of encapsulated probiotic bacteria
Agar plating and CFU enumeration were applied to determine the viability of unencapsulated and encapsulated bacteria. Encapsulating shells were removed prior to agar plating. ZIF-8 biomineralized shells were removed by exposure to 10 mM EDTA for 5 min, alginate-hydrogel shells were removed by 15 min exposure to 55 mM sodium citrate solution and SiO2 nanoparticle packed shell were removed by sonication at 130 W in an ice-water bath for 30 s. After removal of shells, bacteria were collected by centrifugation (5000g, 5 min, 10 °C) and re-suspended by vortexing in 10 mM phosphate buffer. Serial dilutions of the bacterial suspensions were made in 10 mM phosphate buffer and plated on corresponding agar plates (MRS for L. acidophilus, RCM for B. infantis). After 48 h culturing at 37 °C (L. acidophilus in 5% CO2 and B. infantis in an anaerobic chamber), the numbers of colony forming units (CFUs) were enumerated.E. coli killing by probiotic bacteria
A co-culture model was used to determine E. coli killing by unencapsulated and encapsulated probiotic bacteria. In the co-culture model, L. acidophilus, 3 × 108 mL−1 with or without a surface-engineered shell were co-cultured with 3 × 106 mL−1E. coli in MRS broth at 37 °C in 5% CO2 for 24 h. For B. infantis, concentrations of B. infantis and E. coli were 3 × 109 mL−1 and 3 × 104 mL−1, respectively. Both strains were co-cultured in RCM medium. After 24 h, the number of viable E. coli in the co-culture medium was determined by plate counting on BHI agar plates. BHI agar is selective for E. coli, and neither L. acidophilus nor B. infantis can grow on BHI agar.Protection against simulated gastric fluid (SGF)
SGF was prepared by dissolving 2.0 g NaCl and 6.0 g pepsin (porcine, Sigma-Aldrich, USA) in 7 mL 37% HCl. The solution was diluted with demineralized water to 1 L after which pH was adjusted to pH 2 or pH 3 with 1 M NaOH. For evaluating the protection offered by the shells, 3 × 108 mL−1 unencapsulated or encapsulated probiotic bacteria were exposed to SGF. After shaking at 37 °C (150 rpm) for different periods of time up to 2.5 h, bacteria were collected by centrifugation, washed with 10 mM phosphate buffer and their viability assessed by plate counting on MRS plates for L. acidophilus and RCM plates for B. infantis.Protection against simulated intestinal fluid (SIF)
SIF was prepared by dissolving 1 mg mL−1 pancreatin (porcine, Sigma-Aldrich, USA), 1.5 mg mL−1 bile salts (porcine, Sigma-Aldrich, USA), 22 mM NaCl, 3.2 mM KCl and 7.6 mM NaHCO3 in 1 L demineralized water (pH 8.0). Protection was subsequently evaluated as described for the evaluation of the protection offered by SGF.Protection against antibiotics
Antibiotic solutions were prepared by dissolving antibiotics in 10 mM phosphate buffer and added to MRS growth medium for L. acidophilus and RCM for B. infantis to an antibiotic concentration of 5 times their minimal inhibitory concentration (MIC). After culturing for 24 h, bacterial viability was evaluated as described for the evaluation of the protection offered by SGF. The MICs of tobramycin, amoxicillin and tetracycline against both strains were determined in 96-well plates. A stock solution of the different antibiotics with the concentration of 6.4 mg mL−1 were prepared in demineralized water. The stocks were serially diluted with the appropriate growth media to obtain concentrations ranging from 0.125 μg mL−1 to 128 μg mL−1. Then, bacteria were suspended in medium (MRS for L. acidophilus and RCM for B. infantis) to a concentration of 3 × 106 mL−1. Finally, 180 μL of bacterial suspension and 20 μL of diluted antibiotic solution were added to a 96-well plate. For the control, 20 μL of phosphate buffer was added instead of antibiotic solution. The 96-well plates were incubated at 37 °C for 24 h in the incubator (5% CO2 for L. acidophilus and anaerobic chamber for B. infantis). The MIC was the lowest concentration which showed no visible growth.Zeta potentials
Zeta potentials were measured on a Malvern ZetaSizer (model: ZEN3600, serial nr: MAL1037113) in 10 mM phosphate buffer with pH adjusted to between 2 and 9 using a PHM220 laboratory pH meter (Radiometer Analytical SAS, France), by adding 0.1 M HCl or KOH solution. Then, unencapsulated or encapsulated probiotic bacteria were suspended in phosphate buffer with different pH to yield a concentration of 3 × 107 mL−1 and subsequently the zeta potentials were measured.X-ray photoelectron spectroscopy
XPS was performed with an S-Probe (Surface Science Instrument, USA) with an Al-anode (1486 eV). To this end, unencapsulated or encapsulated probiotic bacteria suspended in demineralized water were freeze-dried (Leybold, Germany) for 48 h. The freeze-dried bacterial powders obtained were pressed into small stainless-steel cups, and put into the XPS chamber. X-ray production occurred by means of a magnesium anode (10 kV, 22 mA) using a spot size of 250 μm × 1000 μm. Scans were made of the overall spectrum in the binding energy range of 1–1200 eV at low resolution, then peaks over a 20 eV binding energy range were recorded for Nls and O1s. The C1s binding energy was set at 284.8 eV. The area under each peak, after background subtraction, was used for the calculation of peak intensities, yielding elemental surface concentration ratios for nitrogen, oxygen, and phosphorus to carbon.Two-phase partitioning
Unencapsulated or encapsulated bacteria were suspended in the phosphate buffer over the pH range 2–9 to an OD600 nm of 0.4–0.6 (A0), as measured using a visible spectrophotometer (GENESYS 30, Thermo Fisher Scientific). Next, 3 mL of the bacterial suspension was mixed with 150 μL of n-hexadecane (Sigma-Aldrich) in a glass tube, vortexed for 10 s, allowed to settle for 10 min and the OD600 nm measured again (At). The procedure above was repeated for 5 times. Finally, (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (At/A0) × 100) was plotted against the vortexing time and the initial removal rate R0 (min−1) of bacteria by the hexadecane phase was calculated from its slope using linear least squares fitting yields.
(At/A0) × 100) was plotted against the vortexing time and the initial removal rate R0 (min−1) of bacteria by the hexadecane phase was calculated from its slope using linear least squares fitting yields.
Water contact angles
Water contact angles were measured on lawns of unencapsulated or encapsulated bacteria deposited on a membrane filter (0.45 μm HA filter, Millipore, USA)23 using the sessile drop method (droplet volume 2.5 μL). Filters were dried till plateau water contact angles could be measured, representing bacteria in a hydrated state but without free water on their surfaces. Water contact angles were measured using a contour monitor and are all verified plateau values.Fourier transform infrared spectroscopy (FTIR)
FTIR spectra were measured on an Agilent Cary 600 series FTIR spectrometer (Agilent Technologies, USA). Freeze-dried bacteria were mixed with KBr powder and pressed into a tablet for measurements. FTIR spectra were recorded over the wavenumber range of 4000 and 400 cm−1 with a resolution of 4 cm−1. 32 scans were measured and averaged. A KBr pellet without bacteria was used as background. The areas of absorption bands were determined by integration after linear background subtraction and normalized with respect to the integrated intensity of the entire CH stretching region around 2930 cm−1.Scanning electron microscope and energy dispersive X-ray analysis
Unencapsulated and encapsulated probiotic bacteria were first suspended in deminralized water to a final concentration of 3 × 106 mL−1. Next, 20 μL suspension droplets were placed on an aluminum support and dried at 40 °C for 5 min. Micrographs were taken with a Tescan LYRA SEM-focused ion beam, dual beam microscope at 10 kV and 8.4 mm working distance. EDX line scans were taken along the width of a bacterium.Biosurfactants release
Biosurfactant release of unencapsulated or encapsulated bacteria was measured using Axisymmetric drop shape analysis by profile (ADSA-P).28 The shape of a liquid droplet depends on the liquid surface tension. The profile of a liquid droplet can be mathematically described by the Laplace equation | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46,47 to yield the liquid surface tension, the contact angle and the exact droplet volume as output parameters. Measurements on one bacterial suspension droplet was done as a function of time up to 2 h in an enclosed chamber at room temperature. Bacteria were considered to be biosurfactant releasing when the surface tension of the bacterial suspension droplet decreased by more than 8 mJ m−2 after 2 h.48
46,47 to yield the liquid surface tension, the contact angle and the exact droplet volume as output parameters. Measurements on one bacterial suspension droplet was done as a function of time up to 2 h in an enclosed chamber at room temperature. Bacteria were considered to be biosurfactant releasing when the surface tension of the bacterial suspension droplet decreased by more than 8 mJ m−2 after 2 h.48
Porosity measurements
Nitrogen adsorption/desorption isotherms of differently encapsulated L. acidophilus and B. infantis were determined to derive the BET (Brunauer–Emmett–Teller) surface area and pore diameters of the shells using a Tristar (Micromeritics, USA). To this end, unencapsulated or encapsulated probiotic bacteria suspended in demineralized water, were freeze-dried as described above (section: X-ray photoelectron spectroscopy). Lyophilized samples were further degassed at 40 °C for 12 h and measurements were subsequently performed in liquid nitrogen (77 K). BET surface areas were derived by fitting the adsorption/desorption isotherms to | (2) |
 | (3) |
 | (4) |
Statistical analysis
All experiments were done in triplicate with separately cultured and encapsulated bacteria, with the exception of XPS and the measurement of nitrogen adsorption/desorption isotherms. Results were shown as mean ± standard deviation and statistically significant differences (p < 0.05) were determined using a Student t-test.Conflicts of interest
HJB is also director of a consulting company, SASA BV (GN Schutterlaan 4, 9797 PC Thesinge, The Netherlands). The authors declare no potential conflicts of interest with respect to authorship and/or publication of this article. Opinions and assertions contained herein are those of the authors and are not construed as necessarily representing views of their respective employers.Acknowledgements
The authors thank Joop de Vries (Biomedical Engineering, UMCG) for XPS measurements and Anko C. Eissens (Pharmaceutical Technology and Biopharmacy, University of Groningen) for nitrogen adsorption measurements. The work was financially supported by the UMCG, Groningen, The Netherlands and FRFCU (19lgzd16, 19lgpy112), NSFC (52002414), GBABRF (2019A1515110590).References
- R. F. Fakhrullin, A. I. Zamaleeva, R. T. Minullina, S. A. Konnova and V. N. Paunov, Chem. Soc. Rev., 2012, 41, 4189–4206 RSC.
- P. Li, M. Muller, M. W. Chang, M. Frettloh and H. Schonherr, ACS Appl. Mater. Interfaces, 2017, 9, 22321–22331 CrossRef CAS PubMed.
- C. Desitti, M. Beliavski, S. Tarre, R. Avrahami, E. Zussman and M. Green, J. Water Process Eng., 2017, 19, 205–211 CrossRef.
- S. Khem, V. Bansal, D. M. Small and B. K. May, Food Hydrocolloids, 2016, 54, 162–169 CrossRef CAS.
- A. C. Anselmo, K. J. McHugh, J. Webster, R. Langer and A. Jaklenec, Adv. Mater., 2016, 28, 9486–9490 CrossRef CAS PubMed.
- J. G. Bartlett, Clin. Infect. Dis., 1992, 15, 573–581 CrossRef CAS PubMed.
- C. Hogenauer, H. F. Hammer, G. J. Krejs and E. C. Reisinger, Clin. Infect. Dis., 1998, 27, 702–710 CrossRef CAS PubMed.
- S. Hempel, S. J. Newberry, A. R. Maher, Z. Wang, J. N. V. Miles, R. Shanman, B. Johnsen and P. G. Shekelle, J. Am. Med. Dir. Assoc., 2012, 18, 1959–1969 Search PubMed.
- M. Hickson, A. L. D'Souza, N. Muthu, T. R. Rogers, S. Want, C. Rajkumar and C. J. Bulpitt, Br. Med. J., 2007, 335, 80–83 CrossRef PubMed.
- Z. Li, A. M. Behrens, N. Ginat, S. Y. Tzeng, X. Lu, S. Sivan, R. Langer and A. Jaklenec, Adv. Mater., 2018, 30, e1803925 CrossRef PubMed.
- M. T. Cook, G. Tzortzis, D. Charalampopoulos and V. V. Khutoryanskiy, J. Controlled Release, 2012, 162, 56–67 CrossRef CAS PubMed.
- J. K. Sakkos, L. P. Wackett and A. Aksan, Sci. Rep., 2019, 9, 3158 CrossRef PubMed.
- N. Jiang, X. Y. Yang, G. L. Ying, L. Shen, J. Liu, W. Geng, L. J. Dai, S. Y. Liu, J. Cao, G. Tian, T. L. Sun, S. P. Li and B. L. Su, Chem. Sci., 2015, 6, 486–491 RSC.
- L. H. Granicka, M. Wdowiak, A. Kosek, S. Swiezewski, D. Wasilewska, E. Jankowska, A. Werynski and J. Kawiak, Cell Transplant., 2005, 14, 323–330 CAS.
- S. B. Levy and B. Marshall, Nat. Med., 2004, 10, 122–129 CrossRef PubMed.
- M. H. Wahid, E. Eroglu, S. M. LaVars, K. Newton, C. T. Gibson, U. H. Stroeher, X. Chen, R. A. Boulos, C. L. Raston and S.-L. Harmer, RSC Adv., 2015, 5, 37424–37430 RSC.
- K. Liang, J. J. Richardson, J. Cui, F. Caruso, C. J. Doonan and P. Falcaro, Adv. Mater., 2016, 28, 7910–7914 CrossRef CAS PubMed.
- V. Berry, A. Gole, S. Kundu, C. J. Murphy and R. F. Saraf, J. Am. Chem. Soc., 2005, 127, 17600–17601 CrossRef CAS PubMed.
- L. Wang, Y. Li, X.-Y. Yang, B.-B. Zhang, N. Ninane, H. J. Busscher, Z.-Y. Hu, C. Delneuville, N. Jiang, H. Xie, G. V. Tendeloo, T. Hasan and B.-L. Su, Natl. Sci. Rev., 2020, 1–17 Search PubMed.
- W. Chen, S. Kong, M. Lu, F. Chen, W. Cai, L. Du, J. Wang and C. Wu, Soft Matter, 2020, 16, 270–275 RSC.
- W. Li, Z. Liu, C. Liu, Y. Guan, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2017, 56, 13661–13665 CrossRef CAS PubMed.
- P. E. Johnson, P. Muttil, D. MacKenzie, E. C. Carnes, J. Pelowitz, N. A. Mara, W. M. Mook, S. D. Jett, D. R. Dunphy, G. S. Timmins and C. J. Brinker, ACS Nano, 2015, 9, 6961–6977 CrossRef CAS PubMed.
- H. C. van der Mei, R. Bos and H. J. Busscher, Colloids Surf., B, 1998, 11, 213–221 CrossRef CAS.
- A. T. Poortinga, R. Bos, W. Norde and H. J. Busscher, Surf. Sci. Rep., 2002, 47, 1–32 CrossRef CAS.
- W. W. Wilson, M. M. Wade, S. C. Holman and F. R. Champlin, J. Microbiol. Methods, 2001, 43, 153–164 CrossRef CAS.
- G. Reid, J. A. Younes, H. C. van der Mei, G. B. Gloor, R. Knight and H. J. Busscher, Nat. Rev. Microbiol., 2011, 9, 27–38 CrossRef CAS PubMed.
- E. Grela, J. Kozłowska and A. Grabowiecka, Acta Histochem., 2018, 120, 303–311 CrossRef CAS PubMed.
- M. Rosenberg, N. F. Azevedo and A. Lvask, Sci. Rep., 2019, 9, 6483 CrossRef PubMed.
- Y. Liu, L. Shi, L. Su, H. C. van der Mei, P. C. Jutte, Y. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC.
- W. van der Vegt, H. C. van der Mei, J. Noordmans and H. J. Busscher, Appl. Microbiol. Biotechnol., 1991, 35, 766–770 CrossRef CAS.
- C. J. P. Boonaert and P. G. Rouxhet, Appl. Environ. Microbiol., 2000, 66, 2548–2554 CrossRef CAS PubMed.
- L. Leone, J. Loring, S. Sjöberg, P. Persson and A. Shchukarev, Surf. Interface Anal., 2006, 38, 202–205 CrossRef CAS.
- H. C. van der Mei, J. De Vries and H. J. Busscher, Surf. Sci. Rep., 2000, 39, 1–24 CrossRef CAS.
- F. Tian, A. M. Cerro, A. M. Mosier, H. K. Wayment-Steele, R. S. Shine, A. Park, E. R. Webster, L. E. Johnson, M. S. Johal and L. Benz, J. Phys. Chem. C, 2014, 118, 14449–14456 CrossRef CAS.
- F. Weinhold and R. A. Klein, Mol. Phys., 2012, 110, 565–579 CrossRef CAS.
- S. Scheiner, T. Kar and Y. Gu, J. Biol. Chem., 2001, 276, 9832–9837 CrossRef CAS PubMed.
- L. Rulíšek and Z. Havlas, J. Am. Chem. Soc., 2000, 122, 10428–10439 CrossRef.
- P. H. D. Gunnar and B. Mathias, Brain Res. Bull., 2005, 68, 103–114 CrossRef PubMed.
- K. Noriyasu, M. Mariko and T. Kozo, J. Controlled Release, 2009, 136, 179–186 CrossRef PubMed.
- E. M. Barnett, B. Elangovan, K. E. Bullok and D. Piwnica-Worms, Vis. Sci., 2006, 47, 2589–2595 Search PubMed.
- P. H. D. Gunnar, K. Ertugrul and B. Mathias, Mol. Cell. Neurosci., 2002, 21, 29–37 CrossRef PubMed.
- D. Jean, F. Thierry, D. Marie, P. Emmanuelle and H. Pascal, J. Gen. Appl. Microbiol., 1999, 76, 159–184 Search PubMed.
- Z. W. Li, Y. H. Deng and C. Y. Chao, J. Mater. Chem. A, 2018, 6, 3258–3263 RSC.
- H. Hiroyuki, J. Colloid Interface Sci., 1994, 163, 474–483 CrossRef.
- R. Aveyard and D. A. Haydon, Cambridge University Press, London, 1973 Search PubMed.
- Y. Rotenberg, L. Boruvka and A. W. Neumann, J. Colloid Interface Sci., 1983, 93, 169–183 CrossRef CAS.
- J. Noordmans and H. J. Busscher, Colloids Surf., 1991, 58, 239–249 CrossRef CAS.
- M. M. C. Velraeds, H. C. van der Mei, G. Reid and H. J. Busscher, Colloids Surf., B, 1996, 8, 51–61 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr09204e |
| This journal is © The Royal Society of Chemistry 2021 |