Open Access Article
Open Access ArticleSurface properties modulate protein corona formation and determine cellular uptake and cytotoxicity of silver nanoparticles†
Marianna
Barbalinardo
 a,
Jessika
Bertacchini
b,
Linda
Bergamini
c,
Maria Sara
Magarò
a,
Jessika
Bertacchini
b,
Linda
Bergamini
c,
Maria Sara
Magarò
 b,
Luca
Ortolani
d,
Alessandra
Sanson
c,
Carla
Palumbo
b,
Massimiliano
Cavallini
b,
Luca
Ortolani
d,
Alessandra
Sanson
c,
Carla
Palumbo
b,
Massimiliano
Cavallini
 a and
Denis
Gentili
a and
Denis
Gentili
 *a
*a
aConsiglio Nazionale delle Ricerche, Istituto per lo Studio dei Materiali Nanostrutturati (CNR-ISMN), via P. Gobetti 101, 40129 Bologna, Italy. E-mail: denis.gentili@cnr.it
bDepartment of Biomedical, Metabolic and Neural Sciences, Section of Human Morphology, University of Modena and Reggio Emilia, Via del Pozzo 71, 41124 Modena, Italy
cConsiglio Nazionale delle Ricerche, Istituto di Scienza e Tecnologia dei Materiali Ceramici (CNR-ISTEC), via Granarolo 64, 48018 Faenza, Italy
dConsiglio Nazionale delle Ricerche, Istituto per la Microelettronica e Microsistemi (CNR-IMM), via P. Gobetti 101, 40129 Bologna, Italy
First published on 19th July 2021
Abstract
Nanoparticles (NPs) have been studied for biomedical applications, ranging from prevention, diagnosis and treatment of diseases. However, the lack of the basic understanding of how NPs interact with the biological environment has severely limited their delivery efficiency to the target tissue and clinical translation. Here, we show the effective regulation of the surface properties of NPs, by controlling the surface ligand density, and their effect on serum protein adsorption, cellular uptake and cytotoxicity. The surface properties of NPs are tuned through the controlled replacement of native ligands, which favor protein adsorption, with ligands capable of increasing protein adsorption resistance. The extent and composition of the protein layer adsorbed on NPs are strongly correlated to the degree of ligands replaced on their surface and, while BSA is the most abundant protein detected, ApoE is the one whose amount is most affected by surface properties. On increasing the protein resistance, cellular uptake and cytotoxicity in mouse embryonic fibroblasts of NPs are drastically reduced, but the surface coating has no effect on the process by which NPs mainly induce cell death. Overall, this study reveals that the tuning of the surface properties of NPs allows us to regulate their biological outcomes by controlling their ability to adsorb serum proteins.
Introduction
Over the past decades, nanoparticles (NPs) have been extensively studied for biomedical applications, ranging from prevention, early diagnosis and treatment of diseases as their unique and highly tunable physical, chemical, and biological properties promise to overcome some critical shortcomings of conventional medicine, such as poor bioavailability and low target specificity, as well as facing the challenges related to improving the quality of life of patients during treatments. However, the lack of the basic understanding of how NPs interact with the biological environment and, in turn, with living systems has severely limited their potential in medicine.1–6 Although significant progress has been made to overcome some critical limits, to date, independently of material (organic or inorganic) and targeting strategy (active or passive), the delivery of NPs to the target tissue is less than 1% of the injected dose and their clinical translation has been limited. Although NPs can be engineered with precise functional properties for specific and nonspecific cellular targeting, they have to face physical and biological barriers before reaching the target tissue, resulting in a very low nanoparticle delivery efficiency.7,8 There is no simple correlation between biological responses and synthetic identity of NPs (i.e., size, shape and surface properties post-synthesis) as, in biological fluids, their physicochemical properties can drastically change following the interactions and adsorption onto the surface of extracellular proteins and other biological molecules, creating the so-called protein corona. The structure and composition of the protein corona, which depend on the combination of synthetic identity of NPs with the nature of the biological fluid and exposure time, can be sufficiently long lived to determine the biological identity of NPs and, therefore, can critically regulate their physiological responses.5,9–11 Thus, elucidating the role of each physicochemical parameter of NPs is needed to control their identity in biological media and, in turn, to develop effective strategies to design long-circulating and target-specific NPs with increased delivery efficiency and improved therapeutic response.12–16 Among all physicochemical parameters that affect the nature and amount of protein adsorption, the surface properties of NPs have been reported to play the most significant role in the formation of the protein corona.17,18 Silver nanoparticles (AgNPs) have been extensively studied for biomedical applications and, because of their antibacterial and antifungal properties, they are one of the most diffuse nanomaterials in customer products.1,19 We have recently demonstrated that surface coating of AgNPs strongly affects the adsorption of serum proteins and, in turn, their cellular uptake and cytotoxicity in mouse embryonic fibroblasts. In particular, citrate-coated AgNPs are promptly coated and stabilized by a protein corona and show a time- and dose-dependent toxicity, while the passivation of the NP surface with an oligo(ethylene glycol) (OEG)-terminated alkanethiol drastically reduces both the adsorption of proteins and the cytotoxicity.20,21 However, the engineering of nanomaterials capable of overcoming biological barriers needs the basic understanding of how key design parameters of NPs affect the serum protein adsorption and the resulting biological outcomes.In the present work, we report the effective gradual modulation of the surface properties of NPs, by controlling the surface ligand density, and their effect on serum protein adsorption, cellular uptake and cytotoxicity, aiming to provide further insight for the engineering of nanomaterials exploiting the manipulation of the protein corona. As schematically shown in Scheme 1, we use citrate-coated AgNPs and gradually modify their surface properties through the controlled displacement of citrate ions with an OEG-terminated alkanethiol, namely (11-mercaptoundecyl)hexa(ethylene glycol) (hereafter indiscriminately referred to as “EG6OH” or “ligand”, see Fig S1 in the ESI† for molecular structures). The gradual modification of the surface properties as well as the consequent modulation of the resistance to the protein adsorption of the AgNPs are studied by UV-vis spectroscopy, anion-exchange chromatography, and electrolyte-induced aggregation tests and by assessing the hydrodynamic diameter and zeta potential.
 | ||
| Scheme 1 Schematic illustration of the effect of ligand (EG6OH) density on protein adsorption (left), cellular uptake and cytotoxicity (right) of AgNPs. | ||
The increase of ligand density gradually increases the resistance to protein adsorption of the AgNPs, thus drastically affecting both the composition and amount of proteins that constitute their protein corona. These changes in the protein corona have, in turn, a significant impact on the biological identity of nanoparticles and, therefore, on their interactions with cells. We show that the uptake and cytotoxic activity of AgNPs on mouse embryonic fibroblasts strongly correlate with the surface properties, in fact, both proportionally decrease with the increasing ligand density, but the process by which AgNPs mainly induce cell death remains the same. These results reveal that the surface properties of NPs regulate their interactions with cells and, in turn, dictate their biological outcomes. The synergism between the surface coverage and biological identity of NPs can be exploited for their rational design and engineering, thus thoroughly tailoring their interactions with living systems to specific applications and minimizing the side effects.
Results and discussion
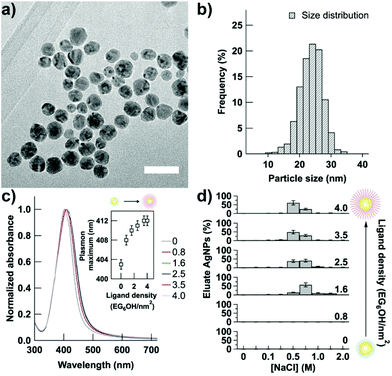
Alkanethiols terminated with an OEG unit have been extensively studied for their ability to form protein resistant self-assembled monolayers (SAMs) on gold and silver surfaces,22–25 even when used in mixed SAMs.26–28 The resistance to protein adsorption is determined by the internal and terminal hydrophilicity, the number of ethylene glycol units and lateral packing density of OEG-terminated alkanethiol used. The lateral packing density, which in turn determines the molecular conformation of the OEG moieties, can be affected both by the assembly solvent and the substrate material.23,24 Among all, EG6OH is an OEG-terminated alkanethiol capable of forming robust protein resistant SAMs on planar silver surfaces,22,23 as well as on AgNPs,20,21 regardless of the assembly solvent used.24 Therefore, we have tuned the protein resistance properties of AgNPs through the controlled replacement of citrate ions, which favor surface protein adsorption,20 with EG6OH. Citrate-coated AgNPs having an average diameter of 22 ± 4 nm, as determined from transmission electron microscopy (TEM) analysis (Fig. 1a and b), were prepared as previously reported29 and functionalized with increasing density of EG6OH. Because thiol containing molecules bind strongly to the gold and silver surfaces and can displace other weakly bound ligands, such as citrate ions,30 the density of the EG6OH on the nanoparticle surface was controlled by varying the EG6OH-to-nanoparticle ratio and quantified by a thiol depletion assay (see the Experimental section). The studied ligand densities range from AgNPs coated exclusively with citrate ions, i.e. NPs with EG6OH density = 0 (hereafter indiscriminately referred to such NPs as “0” or “citrate-coated AgNPs”), to saturating density of EG6OH (4.0 ± 0.1 EG6OH per nm2 – see the ESI†). Noteworthily, the saturation ligand density is in good agreement with values reported on planar surfaces22 and gold nanoparticles.31,32 Citrate-coated AgNPs show the characteristic localized surface plasmon resonance (LSPR) band centered at ∼400 nm (S1a in the ESI†) that is red-shifted, without shape variation, upon surface functionalization with the ligand (Fig. 1c and a zoomed-in view of it in the ESI – Fig S1b†). We noticed the absence of the broadening of the LSPR band also for very low density (i.e. 0.6–0.8 EG6OH per nm2), thus revealing that there is no aggregation of AgNPs, contrary to what we previously reported for gold nanoparticles.33Fig. 1c inset shows the ligand density dependence of the plasmon band. Below the saturation density, the plasmon band maximum is found to red shift rapidly increasing the EG6OH density, as a result of the gradual change of the refractive index of the surrounding environment.34 On approaching saturation, the plasmon band maximum of AgNPs increases more slowly to reach its maximum value. Noteworthily, this trend is reproducible on all batches of citrate-coated AgNPs we have prepared and functionalized with EG6OH (see for a different batch, ESI – Fig. S1c†).The gradual replacement of citrate ions adsorbed on the nanoparticle surface with EG6OH molecules was further confirmed by anion-exchange chromatography on diethylaminoethyl (DEAE) sepharose. Although AgNPs bind to the positively charged resin regardless of EG6OH density, because they show a negative zeta potential (see below), their elution profile is drastically different as a function of surface coverage. As shown in Fig. 1d, AgNPs with low densities (≤0.8 EG6OH per nm2) bind irreversibly to the DEAE resin as they were not eluted even with a high concentration of NaCl (2 M). However, on increasing the ligand density, the elution of AgNPs takes place at lower concentrations of NaCl and almost complete elution of NPs was obtained with 0.5 M NaCl approaching the saturation density. The gradual increase of AgNP stability against irreversible adsorption onto resin is due to the replacement of weakly associated citrate ions on their surface, which can be easily displaced by the functional groups of the resin, with EG6OH, in contrast, binding strongly to the nanoparticle surface. In addition, as we already reported for gold nanoparticles, the replacement of citrate ions with EG6OH molecules leads to a decrease of electrostatic repulsions and a simultaneous increase of steric ones between the nanoparticles proportional to the extent of surface ligand substitution.33 In agreement, the electrolyte-induced aggregation test showed that citrate-coated AgNPs are unstable at a salt concentration of 0.05 M, but the stability of NPs in saline solution increases following the increase of EG6OH density to show good stability also in 1 M saline solution (the highest value tested) when the density reaches the saturation value (see ESI – Fig. S2†).
We have recently reported the key role of the adsorption of serum proteins onto the surface of AgNPs in mediating their cellular uptake and cytotoxic activity.20 Therefore, to investigate the effect of EG6OH density on serum protein adsorption, we measured the change of hydrodynamic diameter and zeta potential of the NPs after incubation in a growth medium containing fetal bovine serum (FBS) and subsequent extensive purification to remove unbound or weakly adsorbed proteins (see the Experimental section). As we already reported, the mean hydrodynamic diameter of citrate-coated AgNPs is strongly increased by the incubation in serum-containing media. However, as shown in Fig. 2a, the discrepancy between the hydrodynamic diameter before and after incubation in the cellular medium becomes less significant increasing the density of EG6OH and disappears, within the experimental error, when the saturation value is reached. As shown in Fig. 2b, the variation of zeta potential values before and after exposure to serum proteins shows a trend that is completely in agreement with what was observed with hydrodynamic diameters. Before incubation with serum proteins, the zeta potential of AgNPs becomes less negative with the increasing ligand density, as expected by the replacement of negatively charged citrate ions with EG6OH molecules. Following the incubation with serum proteins and purification, the zeta potential values are lowered; however, this variation decreases as the EG6OH density increases and it becomes insignificant when approaching the saturation density. Interestingly, the final value of zeta potential of AgNPs after incubation in the growth medium is almost the same regardless of the ligand density.
 | ||
| Fig. 2 (a) Hydrodynamic diameter and (b) zeta potential of AgNPs with different EG6OH densities before (blue circle) and after (red square) incubation in growth media (exposure to serum proteins), and the values are summarized in Table S1 in the ESI.† (c) SDS-PAGE gel of adsorbed serum proteins on AgNPs with different EG6OH densities. The molecular weight ladder is shown in lane 1. SDS-PAGE of products extracted from citrate-coated AgNPs not incubated in the growth medium was included as a control (ctrl, lane 2). (d) Densitometric analysis as a function of EG6OH density of PAGE scan shown in Fig. 2c. Relative band intensity was calculated by measuring for each lane identical area corresponding to those marked by blue dashed squares only in lane 3 for convenience of clarity; (blue circle) sum of all (1–7) areas for each lane and (red square) only area 6. (e) Heat map of protein molar fractions identified and determined by proteomic mass spectrometry in the areas marked in lane 3 of Fig. 2c by blue dashed squares: sum of (A) 1–7 and (B) 2, 3, 4, 6 and 7 areas. Values were calculated from the emPAI of each protein identified by LC-MS/MS according to the equation: Protein content (mol %) = emPAI/∑(emPAI).35 Abbreviated protein names are explained in the ESI – Table S2.† | ||
DLS and zeta potential data clearly show that the protein resistance of AgNPs is strictly correlated to their surface coverage, in fact the extent of protein adsorption on their surface increases with the increasing ligand density.
To further investigate the role of surface coverage, after incubation in a growth medium containing FBS and subsequent purification, serum proteins adsorbed on the surface of AgNPs were isolated and separated using polyacrylamide gel electrophoresis (PAGE) coupled with a protein stain (see the Experimental section). As shown in Fig. 2c, proteins separated from citrate-coated AgNPs (lane 3) showed several bands, marked by blue dashed squares and labeled 1–7, that were not present in the control (ctrl, lane 2), confirming their ability to adsorb a broad range of serum proteins. The proteins isolated from AgNPs functionalized with higher ligand density showed similar band patterns (Fig. 2c, lanes 4–9). However, the intensity of some bands decreases down to disappear with increasing ligand density, revealing that the composition of the serum protein layer adsorbed on the AgNPs is strongly dependent on the surface coverage. In agreement, semiquantitative densitometry analysis of PAGE lanes shows that the total amount of adsorbed proteins contained in the areas corresponding to bands 1–7 drastically decreases with the increasing EG6OH density (blue circle, Fig. 2d); however, the adsorption of proteins was not completely eliminated even at the saturation density. Noteworthily, our results are in agreement with what was observed for the adsorption of a single protein on a planar gold surface functionalized either with a mixed layer of EG6OH and n-alkanethiols26,27 or with hydroxyl-terminated poly(ethylene glycol) (PEG)36 and for the adsorption of serum proteins on gold nanoparticles functionalized with methoxy-terminated PEG.37
A more detailed investigation of the protein corona composition was performed using label-free liquid chromatography mass spectrometry (LC-MS/MS). Proteins separated in the seven bands from citrate-coated AgNPs (lane 3, Fig. 2c) were excised, digested with trypsin and analyzed by LC-MS/MS. Protein molar fractions of serum proteins that constitute the corona of citrate-coated AgNPs are reported in Fig. 2e (column A) and summarized in Table S2 in the ESI.† A total of 29 serum proteins were identified in seven bands and their relative abundances were estimated quantitatively by calculating their exponentially modified protein abundance index (emPAI).35 Overall, the most abundant protein detected in the corona is bovine serum albumin (BSA, 21% mol) followed by apolipoprotein E (ApoE, 16% mol), which is a protein involved in lipid transport and uptake.38,39 Both the proteins were identified on different types of nanoparticles but, while the adsorption of albumin has been reported to protect nanomaterials from opsonization (dysopsonin effect), thus leading to an increase in their blood circulation times,40–42 the adsorption of ApoE is reported to mediate the interaction of nanomaterials with low-density lipoprotein receptors (LDLR), promoting their uptake into endothelial cells38,39 and hepatocytes.43
As mentioned above, the protein pattern changes with surface coverage, however, not all the bands (1–7 in Fig. 2c) show a significant decrease of intensity with increasing ligand density. As shown in Fig. 2c, bands 2, 3, 4, 6, and 7 show a marked decrease of the intensity upon the increase of ligand density, while bands 1 and 5 are not affected significantly, suggesting that not all proteins that constitute the corona of citrate-coated AgNPs are affected by replacement of citrate ions with EG6OH molecules.
Therefore, for a deeper insight, we compared the relative protein abundance calculated considering the sum of all (1–7) bands versus the relative protein abundance calculated considering only bands whose intensity decreases more drastically with the increase of ligand density (i.e. bands 2, 3, 4, 6, and 7 in Fig. 2c). As schematically reported in Fig. 2e and summarized in Table S3 in the ESI,† while BSA is the most abundant protein that constitutes the corona (Fig. 2e, column A), ApoE is the most abundant protein when only PAGE bands whose intensity is clearly affected by the surface coverage are considered (Fig. 2e, column B), revealing that the content of ApoE in the corona is more affected by the ligand density than that of BSA. In addition, since ApoE was identified only in band 6, we compared the trend of the intensity of this band with the sum of all bands (1–7); as revealed by densitometric analysis, the amount of proteins in band 6 (red square, Fig. 2d) decreases more drastically with the increasing EG6OH density than that of the sum of all bands (blue circle, Fig. 2d). Taking into account that ApoE represents almost the 60% mol of proteins contained in band 6 (see ESI – Table S4†), these results further confirm that the adsorption of ApoE, more than those of BSA, is influenced by the EG6OH density.
To evaluate the impact of EG6OH density on the interactions between the NPs and living systems, we performed studies of toxicity, based on 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay, and cellular uptake using inductively coupled plasma optical emission spectroscopy (ICP-OES). Mouse embryonic fibroblasts (NIH-3T3) were selected as a model cell line, due to their susceptibility to the coating-dependent toxicity of AgNPs, and exposed to 40 μg mL−1 AgNPs because at this concentration citrate-coated AgNPs are toxic, while AgNPs completely coated with EG6OH do not affect significantly the cell viability.20,21 After 24 h of exposure to NPs, as shown in Fig. 3a, the MTT viability assay revealed that the surface coverage significantly determines the toxic activity of the AgNPs; cell viability drastically increases with the increasing EG6OH density on the surface of AgNPs and, in agreement with what was previously observed,20 becomes not significantly affected compared to control (ctrl), i.e. cells treated with vehicle solution (2 mM sodium citrate), when the saturation density of the ligand on the NP surface is reached. In agreement with the viability results, double fluorescence labeling of actin (red) and nucleus (blue) showed surface coating-depending toxicity of AgNPs (see Fig. 3b and Fig S3 in the ESI† for non-merged images). At low EG6OH density values, cytoskeleton size and density of fibroblasts treated with AgNPs are reduced, thus revealing an impaired ability to replicate and less adhesion to the substrate. The increase of ligand density clearly decreases the toxic effects of AgNPs, in fact, approaching the saturation density; the morphology and density of cells treated with AgNPs are similar to those of cells treated with vehicle solution (control, see the ESI – Fig. S4†). Note that the quantitative analysis of NIH-3T3 nuclei per area showed a trend that is completely in agreement with what was observed above (see Table S5 in the ESI†).
For a deeper understanding of the role of surface coating on the mode of cell death, NIH-3T3 cells were double-stained with acridine orange/ethidium bromide (AO/EB) following 48 h of exposure to AgNPs. Data were extracted from the fluorescence microscopy images based on the percentages of green (viable cells), red (necrotic cells), yellow (early apoptosis), and orange (late apoptosis) fluorescence cells (see Fig. S5 in the ESI† for representative images),44 and the results are shown in Fig. 4a and summarized in Table S6 in the ESI.† Consistently with the MTT viability assay, the inhibition of cell proliferation after 48 h of exposure to AgNPs shows a trend that is completely in agreement with what was observed at 24 h (Fig. 3a), in fact, the increase of EG6OH density on the nanoparticle surface drastically reduces their cytotoxic activity, further confirming its key role in the interaction between NPs and cells. Moreover, double-staining assay clearly showed that, independently of the ligand density, AgNPs induce cell death leading mainly to their apoptosis and, to a lesser extent, necrosis. These results are in agreement with what was previously observed.45–48 Noteworthily, while the percentage of total apoptotic cells is inversely proportional to EG6OH density, there is no correlation with the percentage of necrotic cells; however, in all cases, necrosis is the least relevant cell death process. From these results, we can infer that, although the surface coverage plays a key role in the extent of cytotoxic activity, the process by which AgNPs mainly induce cell death remains the same independently of the surface coverage of the nanoparticles. Considering that AgNPs carry out their toxic effects after cell internalization,20,49 these results suggested that EG6OH density determines the toxic activity of AgNPs by regulating their cellular uptake. In order to probe whether the variation of surface coverage was correlated with the variation of NP internalization, we have assessed the relationship between ligand density and uptake of AgNPs in NIH-3T3 cells by ICP-OES analysis. The cellular uptake was measured following a 6 h incubation with the same concentration of AgNPs functionalized with different EG6OH densities and extensive washing of NIH-3T3 cells, therefore silver per cell ratios represent AgNPs that were either tightly bound to the cell membrane or actually internalized (see the Experimental section). A shorter incubation time than that used for previous studies was chosen to ensure the nanoparticle uptake, but preventing cell detachment that follows the toxic activity of the nanoparticles. This was confirmed by assessing that the cell number after 6 h of exposure to AgNPs was found to be similar to that of cells treated with vehicle solution (2 mM sodium citrate), regardless of the ligand density. As shown in Fig. 4b, silver uptake drastically decreases with the increasing EG6OH density, in fact, cells treated with citrate-coated AgNPs (ligand density = 0) show a silver uptake 200 times higher than those treated with nanoparticles completely coated with EG6OH (ligand density = 4). Noteworthily, this large uptake difference was also qualitatively revealed by the presence of dark spots, due to fluorescence quenching by metal NPs, in the actin cytoskeleton image of cells treated with citrate-coated AgNPs (see white arrows in Fig. 4c and Fig S6 in the ESI† for non-merged images), but they are not present in those of cells treated with AgNPs completely coated with EG6OH (Fig. 4d). Therefore, cellular uptake of NPs, as well as the extent and composition of the protein layer adsorbed on their surface and their cytotoxic activity, is strongly correlated to the degree of ligands replaced on their surface, while they induce cell death mainly by apoptosis, regardless of their surface coverage.
Overall, considering that citrate-coated AgNPs are taken up into the NIH-3T3 cells via receptor-mediated endocytosis,20 our results suggest that the increase of EG6OH density does not affect the NP uptake pathway, but rather the endocytosis process is gradually inhibited by the following decrease of protein adsorption, regulating their cellular uptake and, in turn, their cytotoxic effect.
Experimental
Materials
Silver nitrate (AgNO3), sodium citrate (C6H5O7Na3), tannic acid (C76H52O46), (11-mercaptoundecyl)hexa(ethylene glycol) (EG6OH), Ellman's reagent, dimethyl sulfoxide (DMSO), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT), MEM Non Essential Amino Acids (NEAA), Dulbecco's phosphate buffered saline (DPBS) and Dulbecco's modified Eagle's medium (DMEM) were purchased from Sigma-Aldrich and used without further purification. All aqueous solutions were prepared with deionized water obtained using an ultrafiltration system (Milli-Q, Millipore) with a measured resistivity of above 18 MΩ. Fetal bovine serum (FBS) was purchased from Gibco and heat-inactivated before use.Synthesis of citrate-coated AgNPs
Citrate-stabilized AgNPs were prepared following the method reported elsewhere.29,50 In brief, 100 mL of an aqueous solution of sodium citrate (5 mM) and tannic acid (0.025 mM) was refluxed and an aqueous solution of silver nitrate (1 mL, 25 mM) was added quickly. Then, the reaction mixture was refluxed for 30 min, resulting in a bright yellow colloidal silver solution, and was then allowed to cool down to room temperature. The aqueous suspension of AgNPs was purified by two rounds of centrifugation (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 2 h) and resuspension in 10 mL of an aqueous solution of sodium citrate (2 mM).
000g for 2 h) and resuspension in 10 mL of an aqueous solution of sodium citrate (2 mM).
Characterization of AgNPs
UV-Vis-NIR spectra were recorded on a Jasco V-560 UV-Vis spectrophotometer and a Thermo Scientific Varioskan Flash Multimode Reader. Dynamic light scattering (DLS) and zeta potential measurement were performed in phosphate buffer (1 mM, pH = 7) and KCl (1 mM) on a NanoBrook Omni Particle Size Analyser (Brookhaven Instruments Corporation, USA) equipped with a 35 mW red diode laser (nominal 640 nm wavelength). AgNPs were characterized using a Transmission Electron Microscopy (TEM) FEI Tecnai F20 ST, equipped with a CCD Camera Gatan MSC704 and an X-Ray EDS spectrometer EDAX EDS PV9761. TEM samples were prepared by drop casting 5 microliters of nanoparticle solutions onto a carbon-coated copper grid, then blotted with filter paper to remove excess solution and dried in air for 20 min. The average size and size distribution of citrate-stabilized AgNPs were measured using ImageJ software by counting 1000 particles. Silver concentrations were measured using an Inductively Coupled Plasma Optical Emission Spectrometer (ICP-OES) Varian Liberty 200. The concentration of silver was measured by ICP-OES titration at λAg = 328.068.Saturation ligand density
The concentration of AgNPs was measured by combining ICP-OES titration and TEM. The as-prepared water dispersed AgNPs were thoroughly mixed at room temperature overnight with a freshly prepared aqueous solution of EG6OH (∼2 mM) adjusting the added volume to achieve a stoichiometry of ∼10 EG6OH per nm2 that ensures to exceed the maximum ligand density. Nanoparticle-free control samples containing an equal concentration of EG6OH were prepared. After incubation, AgNPs were pelleted by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min) and the supernatant containing the excess of thiol molecules was transferred to a new tube. The thiol concentrations both in the supernatant and in a nanoparticle-free control sample were measured with Ellman's reagent51 and used to calculate the saturation ligand density.
000g for 30 min) and the supernatant containing the excess of thiol molecules was transferred to a new tube. The thiol concentrations both in the supernatant and in a nanoparticle-free control sample were measured with Ellman's reagent51 and used to calculate the saturation ligand density.
Functionalization of AgNPs
The as-prepared water dispersed AgNPs were thoroughly mixed at room temperature overnight with a freshly prepared aqueous solution of EG6OH adjusting the thiol concentration to achieve several samples with stoichiometry in the range between 0 EG6OH per nm2 and saturation. Nanoparticle-free control samples containing an equal concentration of EG6OH were prepared. After incubation, AgNPs were purified by four rounds of centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min) and resuspension in an aqueous solution of sodium citrate (2 mM). The thiol concentrations of nanoparticle-free control samples were measured with Ellman's reagent51 and used to calculate the ligand density.
000g for 30 min) and resuspension in an aqueous solution of sodium citrate (2 mM). The thiol concentrations of nanoparticle-free control samples were measured with Ellman's reagent51 and used to calculate the ligand density.
Salt stability of AgNPs
AgNPs were mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 volume ratio with aqueous solutions of NaCl in sodium citrate (2 mM).
19 volume ratio with aqueous solutions of NaCl in sodium citrate (2 mM).
Ion exchange chromatography
Diethylaminoethyl (DEAE) Sepharose was prepared as specified by the manufacturer and equilibrated in sodium citrate (2 mM). 30 μL of nanoparticles were incubated with 50 mg of resin in a 1.5 mL tube for 1 h on a rotating wheel. The unbound nanoparticles were recovered and successive washes of the resin were then performed using phosphate buffer (10 mM, pH = 7) with an increasing amount of NaCl. The amount of the recovered nanoparticles was quantified by the measurement of absorbance at 400 nm.Serum protein adsorption
Silver nanoparticles (100 μL) were mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 volume ratio with complete medium (DMEM supplemented with 10% (v/v) heat-inactivated FBS, 2 mM L-glutamine, 0.1 mM MEM Non Essential Amino Acids (NEAA), 100 U mL−1 of penicillin and 100 U mL−1 of streptomycin) and incubated overnight at 37 °C. The nanoparticles were purified by three rounds of centrifugation (22
5 volume ratio with complete medium (DMEM supplemented with 10% (v/v) heat-inactivated FBS, 2 mM L-glutamine, 0.1 mM MEM Non Essential Amino Acids (NEAA), 100 U mL−1 of penicillin and 100 U mL−1 of streptomycin) and incubated overnight at 37 °C. The nanoparticles were purified by three rounds of centrifugation (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C) and resuspended in 100 μL of an aqueous solution of phosphate buffer (1 mM, pH = 7) and KCl (1 mM).
000g for 30 min at 4 °C) and resuspended in 100 μL of an aqueous solution of phosphate buffer (1 mM, pH = 7) and KCl (1 mM).
Polyacrylamide gel electrophoresis (PAGE)
AgNPs (100 μL) were washed by two rounds of centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C) and resuspended in 40 μL of Tris-Cl (10 mM, pH 7.4). 8 μl of 6× Laemmli SDS sample reducing buffer (Bio Rad) was added to the nanoparticles and incubated at 100 °C for 5 min to reduce and denature proteins on the surface of the particles. Nanoparticles were removed by centrifugation (13
000g for 20 min at 4 °C) and resuspended in 40 μL of Tris-Cl (10 mM, pH 7.4). 8 μl of 6× Laemmli SDS sample reducing buffer (Bio Rad) was added to the nanoparticles and incubated at 100 °C for 5 min to reduce and denature proteins on the surface of the particles. Nanoparticles were removed by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C) and the supernatants containing the free proteins, along with a molecular weight ladder (Bio Rad), were loaded on 10% SDS-PAGE and resolved at 100 V for 60 min. The gel was fixed with a solution of 25% isopropyl alcohol, 10% glacial acetic acid for 60 min and stained with colloidal Coomassie Blue G-250 for 2 h. Gel densitometry was performed using image J software.
000g for 20 min at 4 °C) and the supernatants containing the free proteins, along with a molecular weight ladder (Bio Rad), were loaded on 10% SDS-PAGE and resolved at 100 V for 60 min. The gel was fixed with a solution of 25% isopropyl alcohol, 10% glacial acetic acid for 60 min and stained with colloidal Coomassie Blue G-250 for 2 h. Gel densitometry was performed using image J software.
Mass spectrometry analysis and protein identification
As reported above, protein separation was performed by PAGE using a 4–20% precast polyacrylamide gel (Bio Rad). The resulting gel was stained overnight with colloidal Coomassie Blue G-250 and seven bands, labeled A–G, were excised for mass spectrometry analysis. Protein digestion was performed following a protocol reported in the literature.52 Coomassie-stained bands were diced into 1 mm3 cubes, destained for 30 min with 50% acetonitrile in 100 mM ammonium bicarbonate buffer, dehydrated with 100% acetonitrile for 10 min, reduced with 10 mM DTT (1,4-dithiothreitol, Sigma Aldrich) for 30 min at 56 °C and alkylated with 55 mM iodoacetamide (IAA, Sigma Aldrich) for 30 min in the dark. After alkylation, gel pieces were dehydrated with 100% acetonitrile and digested overnight with MS-grade trypsin (Thermo Fisher, 13 ng μl−1 in ammonium bicarbonate buffer 10 nM with 10% acetonitrile) at 37 °C. The digestion products were extracted with a solution of 5% formic acid and acetonitrile 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 for 15 min at 37 °C in a shaker and were then lyophilized using a SpeedVac Concentrator (Savant). Dry peptides from bands were resuspended in 40 μL of a mixture of water
2 for 15 min at 37 °C in a shaker and were then lyophilized using a SpeedVac Concentrator (Savant). Dry peptides from bands were resuspended in 40 μL of a mixture of water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile
acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) formic acid 97
formic acid 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, sonicated for 10 min at room temperature and centrifuged (12
2, sonicated for 10 min at room temperature and centrifuged (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100g for 10 min). Analyses were performed on an ESI† Q Exactive Mass spectrometer (Thermo Scientific) controlled using Xcalibur (v. 29 build 2926) and interfaced with an Ultimate 3000 UHPLC pump. The column (Zorbax SB-C18 RRHT, 2.1 × 50 mm, 1.8 μ particle size, Agilent Technologies) was equilibrated with 0.3 ml min−1 of water 0.1% formic acid (A) with 2% acetonitrile (B); after sample injection (18 μl), B% was raised from 2 to3%, then linearly increased from 3 to 21% in 19 min; B% was then brought to 90% in 4 min and kept at 90% B for 3 min before the reconditioning step. The total runtime was 35 min. The ESI source was operated in positive mode; probe was heated at 290 °C, the capillary temperature was set at 270 °C; the following nitrogen flows (arbitrary units) were used to assist the ionization: Sheath Gas 40, Aux Gas 30, Sweep Gas 3; capillary voltage was set to 3.8 kV, the S-Lens RF level was set at 45 (arbitrary units). Profile MS and MS2 spectra were recorded from 200 to 2000 m/z in FULL MS/dd-MS2 (TOP5) mode, at a resolution of 70
100g for 10 min). Analyses were performed on an ESI† Q Exactive Mass spectrometer (Thermo Scientific) controlled using Xcalibur (v. 29 build 2926) and interfaced with an Ultimate 3000 UHPLC pump. The column (Zorbax SB-C18 RRHT, 2.1 × 50 mm, 1.8 μ particle size, Agilent Technologies) was equilibrated with 0.3 ml min−1 of water 0.1% formic acid (A) with 2% acetonitrile (B); after sample injection (18 μl), B% was raised from 2 to3%, then linearly increased from 3 to 21% in 19 min; B% was then brought to 90% in 4 min and kept at 90% B for 3 min before the reconditioning step. The total runtime was 35 min. The ESI source was operated in positive mode; probe was heated at 290 °C, the capillary temperature was set at 270 °C; the following nitrogen flows (arbitrary units) were used to assist the ionization: Sheath Gas 40, Aux Gas 30, Sweep Gas 3; capillary voltage was set to 3.8 kV, the S-Lens RF level was set at 45 (arbitrary units). Profile MS and MS2 spectra were recorded from 200 to 2000 m/z in FULL MS/dd-MS2 (TOP5) mode, at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 17
000 and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, respectively. The five most intense multi-charged ions were selected for MS2 nitrogen-promoted collision-induced dissociation (NCE = 28). A precursor active exclusion of 20 seconds was set; peptide-like isotope pattern ions were preferred. The mass spectrometer was calibrated before the start of the analyses. Raw data, converted into mascot generic format using MsConvert (v. 3.0.10730, ProteoWizard tools), were searched against Swiss-Prot (accessed Oct 2018; 6002 sequences for Bos Taurus) for peptide sequences and C-RAP for contaminants with MASCOT (Version 2.4, Matrix Science, London, UK). Trypsin as proteolytic enzyme and carbamidomethyl cysteine as fixed modification were set in search parameters. Deamidated (NQ) and oxidated (M) were set as variable modifications. One missed cleavage was allowed. Mass tolerances were set at 10 ppm for the precursor ions and 0.05 Da for the product ions. Automatic decoy database search was used to estimate the false discovery rate, which was adjusted to ≤1%.
500, respectively. The five most intense multi-charged ions were selected for MS2 nitrogen-promoted collision-induced dissociation (NCE = 28). A precursor active exclusion of 20 seconds was set; peptide-like isotope pattern ions were preferred. The mass spectrometer was calibrated before the start of the analyses. Raw data, converted into mascot generic format using MsConvert (v. 3.0.10730, ProteoWizard tools), were searched against Swiss-Prot (accessed Oct 2018; 6002 sequences for Bos Taurus) for peptide sequences and C-RAP for contaminants with MASCOT (Version 2.4, Matrix Science, London, UK). Trypsin as proteolytic enzyme and carbamidomethyl cysteine as fixed modification were set in search parameters. Deamidated (NQ) and oxidated (M) were set as variable modifications. One missed cleavage was allowed. Mass tolerances were set at 10 ppm for the precursor ions and 0.05 Da for the product ions. Automatic decoy database search was used to estimate the false discovery rate, which was adjusted to ≤1%.
Cell cultures
Mouse embryonic fibroblast (NIH-3T3) cells were cultured under standard conditions in the DMEM medium supplemented with 10% (v/v) heat-inactivated FBS, 2 mM L-glutamine, 0.1 mM MEM Non Essential Amino Acids (NEAA), 100 U mL−1 of penicillin and 100 U mL−1 of streptomycin in a humidified incubator set at 37 °C with 5% CO2. The cells were seeded in 96-well plates at a density 1 × 104 cells per well and grown for 24 h before exposure to nanoparticles.53 For the control, the cell culture medium was diluted with sodium citrate (2 mM) to ensure that dilution of the medium by the solution of nanoparticles has no impact on the cell performance.Actin and nucleus staining
The cells were fixed with 4% paraformaldehyde in DPBS, washed with DPBS. They were then permeabilized with 0.001% Triton-X 100. The cells were labelled with TRITC-conjugated phalloidin (FAK100, Merck Millipore) for 1 h, followed by rinses with DPBS. Actin staining was critical to map the local orientation of actin filaments within the cells. Nuclear counterstaining was performed by incubation with DAPI (FAK100, Merck Millipore) for 3 min, followed by rinses with DPBS. The samples were examined using a Nikon Eclipse 80i microscope equipped for fluorescence analysis.Cell viability (cytotoxicity assay)
Cell viability was determined by 3-(4,5-di-methyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay, measuring the intracellular reduction of tetrazolium salts into purple formazan by viable cells.54 Briefly, the cells were seeded in 96-well plates with complete medium with or without nanoparticles for 24 h and 48 h under standard conditions. After incubation times, the medium with or without AgNPs was discarded, and the cells were washed with 100 μL of DPBS and then 100 μL of complete medium was added. Afterwards, 10 μL of sterile MTT solution (5 mg mL−1 in DPBS) was added to each well and incubated for 2 h at 37 °C with 5% CO2. Subsequently, the medium was discarded and 200 μL of DMSO was added to each sample to solubilize formazan crystals. Optical density (OD) was read on a microplate reader (Thermo Scientific Varioskan Flash Multimode Reader) at 550 nm as a working wavelength and 640 nm as a reference. Cell viability was calculated as the proportion of the mean OD of the replicated wells relative to that of the control.Fluorescence microscopic analysis of cell death
A combination of fluorescent DNA-binding dyes acridine orange (AO) and ethidium bromide (EB) was used to detect the cells’ death morphology, according to the standard procedure.55 The cells were seeded in 24-well plates at a density 4 × 104 cells per well and grown for 24 h before exposure to nanoparticles. After 48 h of exposure, the cells were treated with trypsine-EDTA 1×, neutralized with DPBS and collected by centrifugation. Then, fluorescent dyes AO (1 g L−1) and EB (1 g L−1) were added to the cellular pellet. The cell suspension was dropped on a glass slide and covered by a coverslip. Slides were observed with a Nikon Eclipse 80i fluorescence microscope equipped with a UV-mercury lamp. Percentages of viable, apoptotic and necrotic cells were measured by analyzing 100 cells.In vitro uptake of AgNPs
The uptake of AgNPs by NIH-3T3 cells was quantified using Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES). AgNPs were incubated for 6 h with pre-seeded NIH-3T3 cells in 12-well plates at a density of 1 × 105 cells per well. After exposure, the medium was discarded and the cells were washed three times with DPBS to remove residual NPs. Then, the cells were treated with trypsin-EDTA 1×, neutralized with DPBS, counted and collected by centrifugation. The recovered pellets were treated with 200 μL of lysis buffer (RIPA buffer), digested overnight with freshly prepared aqua regia (HNO3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCl = 1
HCl = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and then diluted with 2% HNO3. The concentration of silver was measured by ICP-OES at 328.068 nm. Operating conditions of the ICP-OES are listed below: RF power, 1200 W; plasma Ar flow rate, 12 L min−1; nebulizer Ar flow rate, 0.70 L min−1; uptake time, 25 s; stabilization time, 15 s. A series of silver standard solutions (20, 10, 5, 2.5, 1.5, 0.5, and 0 ppm) in HNO3 2% were prepared to obtain a calibration curve used to determine the silver amount taken up by the cells in each sample, and the concentration reported for each sample is the mean value of 5 different measures.
3) and then diluted with 2% HNO3. The concentration of silver was measured by ICP-OES at 328.068 nm. Operating conditions of the ICP-OES are listed below: RF power, 1200 W; plasma Ar flow rate, 12 L min−1; nebulizer Ar flow rate, 0.70 L min−1; uptake time, 25 s; stabilization time, 15 s. A series of silver standard solutions (20, 10, 5, 2.5, 1.5, 0.5, and 0 ppm) in HNO3 2% were prepared to obtain a calibration curve used to determine the silver amount taken up by the cells in each sample, and the concentration reported for each sample is the mean value of 5 different measures.
Statistical analysis
All data represented the mean ± standard deviation (SD) of at least 4 independent culture experiments. Statistical significance was determined using a one-way analysis of variance with Tukey's test for multiple comparisons using Origin 8 software (OriginLab Corporation). Differences were considered significant when p < 0.005 and p < 0.001.Conclusions
In the present study, we have shown the role of surface coverage of nanoparticles in serum protein adsorption, cellular uptake and cytotoxicity. The surface properties of AgNPs were modulated through the controlled displacement of citrate ions with an oligo(ethylene glycol)-based ligand; the ligand density has been varied between no ligand (only citrate ions) and saturation. The exposure to serum-containing media revealed that the extent and composition of the serum proteins layer adsorbed on the AgNPs, i.e. protein corona, is strongly dependent on the EG6OH density. The protein resistance of AgNPs increases following the increase of EG6OH density; however, the adsorption of proteins was not completely eliminated even at the saturation density. Although BSA was the most abundant protein detected in the corona, our results suggest that ApoE is the one whose amount is most affected by the ligand density. The different extent and composition of protein corona on the AgNPs significantly determine their cellular uptake and cytotoxicity, in fact, both silver uptake and toxic activity drastically decrease with the increasing EG6OH density. However, the surface coating has no effect on the process by which AgNPs mainly induce cell death that, in fact, occurs mainly through apoptosis regardless of ligand density. We can infer that the drastically different biological outcomes of AgNPs as a function of their surface coverage can be ascribed to the different ability to adsorb serum proteins which, in turn, regulate the toxic activity of AgNPs regulating their cellular uptake via receptor-mediated endocytosis.In summary, our results reveal that the synergism between the surface coverage and biological identity of NPs can be exploited for their rational design and engineering, thus thoroughly tailoring the interaction of NPs with living systems to specific applications and minimizing the side effects.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Fondazione Cassa di Risparmio di Modena for funding the UHPLC-ESI-Q Exactive system at the Centro Interdipartimentale Grandi Strumenti (CIGS). M. B. was supported by the project MIUR-PRIN prot. 2017YH9MRK.References
- G. Y. Chen, I. Roy, C. H. Yang and P. N. Prasad, Chem. Rev., 2016, 116, 2826–2885 CrossRef CAS PubMed.
- L. Cheng, C. Wang, L. Feng, K. Yang and Z. Liu, Chem. Rev., 2014, 114, 10869–10939 CrossRef CAS PubMed.
- J. A. Barreto, W. O'Malley, M. Kubeil, B. Graham, H. Stephan and L. Spiccia, Adv. Mater., 2011, 23, H18–H40 CrossRef CAS.
- E.-K. Lim, T. Kim, S. Paik, S. Haam, Y.-M. Huh and K. Lee, Chem. Rev., 2015, 115, 327–394 CrossRef CAS PubMed.
- M. Rahman, S. Laurent, N. Tawil, L. H. Yahia and M. Mahmoudi, Protein-Nanoparticle Interactions: The Bio-Nano Interface, Springer Berlin Heidelberg, 2013 Search PubMed.
- Editorial, Nat. Nanotechnol., 2019, 14, 1083–1083 CrossRef PubMed.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Nat. Rev. Mater., 2016, 1, 16014 CrossRef CAS.
- Q. Dai, S. Wilhelm, D. Ding, A. M. Syed, S. Sindhwani, Y. Zhang, Y. Y. Chen, P. MacMillan and W. C. W. Chan, ACS Nano, 2018, 12, 8423–8435 CrossRef CAS PubMed.
- C. D. Walkey and W. C. W. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- M. P. Monopoli, F. B. Bombelli and K. A. Dawson, Nat. Nanotechnol., 2011, 6, 11–12 CrossRef CAS.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050 CrossRef CAS PubMed.
- R. Lima, A. B. Seabra and N. Durán, J. Appl. Toxicol., 2012, 32, 867–879 CrossRef CAS PubMed.
- J. Skalska and L. Strużyńska, Folia Neuropathol., 2015, 53, 281–300 CrossRef PubMed.
- R. B. Foldbjerg and H. Autrup, Arch. Basic Appl. Med., 2013, 1, 5–15 Search PubMed.
- M. E. Samberg and N. A. Monteiro-Riviere, in Encyclopedia of Nanotechnology, ed. B. Bhushan, Springer Netherlands, Dordrecht, 2012, pp. 1069–1077 Search PubMed.
- N. Durán, G. Nakazato and A. B. Seabra, Appl. Microbiol. Biotechnol., 2016, 100, 6555–6570 CrossRef PubMed.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Delivery Rev., 2009, 61, 428–437 CrossRef CAS PubMed.
- N. Liu, M. Tang and J. Ding, Chemosphere, 2020, 245, 125624 CrossRef CAS PubMed.
- M. E. Vance, T. Kuiken, E. P. Vejerano, S. P. McGinnis, M. F. Hochella Jr., D. Rejeski and M. S. Hull, Beilstein J. Nanotechnol., 2015, 6, 1769–1780 CrossRef CAS PubMed.
- M. Barbalinardo, F. Caicci, M. Cavallini and D. Gentili, Small, 2018, 14, 1801219 CrossRef PubMed.
- F. Decataldo, M. Barbalinardo, D. Gentili, M. Tessarolo, M. Calienni, M. Cavallini and B. Fraboni, Adv. Biosyst., 2020, 4, 1900204 CrossRef CAS PubMed.
- P. Harder, M. Grunze, R. Dahint, G. M. Whitesides and P. E. Laibinis, J. Phys. Chem. B, 1998, 102, 426–436 CrossRef CAS.
- S. Herrwerth, W. Eck, S. Reinhardt and M. Grunze, J. Am. Chem. Soc., 2003, 125, 9359–9366 CrossRef CAS PubMed.
- L. Y. Li, S. F. Chen, J. Zheng, B. D. Ratner and S. Y. Jiang, J. Phys. Chem. B, 2005, 109, 2934–2941 CrossRef CAS PubMed.
- D. Gentili, P. D'Angelo, F. Militano, R. Mazzei, T. Poerio, M. Brucale, G. Tarabella, S. Bonetti, S. L. Marasso, M. Cocuzza, L. Giorno, S. Iannotta and M. Cavallini, J. Mater. Chem. B, 2018, 6, 5400–5406 RSC.
- K. L. Prime and G. M. Whitesides, Science, 1991, 252, 1164–1167 CrossRef CAS PubMed.
- K. L. Prime and G. M. Whitesides, J. Am. Chem. Soc., 1993, 115, 10714–10721 CrossRef CAS.
- L. Duchesne, D. Gentili, M. Comes-Franchini and D. G. Fernig, Langmuir, 2008, 24, 13572–13580 CrossRef CAS PubMed.
- N. G. Bastus, F. Merkoci, J. Piella and V. Puntes, Chem. Mater., 2014, 26, 2836–2846 CrossRef CAS.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103–1170 CrossRef CAS PubMed.
- H. Hinterwirth, S. Kappel, T. Waitz, T. Prohaska, W. Lindner and M. Lämmerhofer, ACS Nano, 2013, 7, 1129–1136 CrossRef CAS PubMed.
- M. D. Torelli, R. A. Putans, Y. Tan, S. E. Lohse, C. J. Murphy and R. J. Hamers, ACS Appl. Mater. Interfaces, 2015, 7, 1720–1725 CrossRef CAS PubMed.
- D. Gentili, G. Ori, L. Ortolani, V. Morandi and M. Cavallini, ChemNanoMat, 2017, 3, 874–878 CrossRef CAS.
- H. Chen, X. Kou, Z. Yang, W. Ni and J. Wang, Langmuir, 2008, 24, 5233–5237 CrossRef CAS.
- Y. Ishihama, Y. Oda, T. Tabata, T. Sato, T. Nagasu, J. Rappsilber and M. Mann, Mol. Cell. Proteomics, 2005, 4, 1265–1272 CrossRef CAS PubMed.
- L. D. Unsworth, H. Sheardown and J. L. Brash, Langmuir, 2008, 24, 1924–1929 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- S. Wagner, A. Zensi, S. L. Wien, S. E. Tschickardt, W. Maier, T. Vogel, F. Worek, C. U. Pietrzik, J. Kreuter and H. von Briesen, PLoS One, 2012, 7, e32568 CrossRef CAS PubMed.
- A. L. Barran-Berdon, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Lagana, Langmuir, 2013, 29, 6485–6494 CrossRef CAS PubMed.
- H. Gao and Q. He, Expert Opin. Drug Delivery, 2014, 11, 409–420 CrossRef CAS.
- V. H. Nguyen, N. M. Meghani, H. H. Amin, T. T. D. Tran, P. H. L. Tran, C. Park and B. J. Lee, Colloids Surf., B, 2018, 170, 179–186 CrossRef CAS PubMed.
- Y. Yan, K. T. Gause, M. M. Kamphuis, C. S. Ang, N. M. O'Brien-Simpson, J. C. Lenzo, E. C. Reynolds, E. C. Nice and F. Caruso, ACS Nano, 2013, 7, 10960–10970 CrossRef CAS PubMed.
- A. Akinc, W. Querbes, S. De, J. Qin, M. Frank-Kamenetsky, K. N. Jayaprakash, M. Jayaraman, K. G. Rajeev, W. L. Cantley, J. R. Dorkin, J. S. Butler, L. Qin, T. Racie, A. Sprague, E. Fava, A. Zeigerer, M. J. Hope, M. Zerial, D. W. Sah, K. Fitzgerald, M. A. Tracy, M. Manoharan, V. Koteliansky, A. Fougerolles and M. A. Maier, Mol. Ther., 2010, 18, 1357–1364 CrossRef CAS PubMed.
- K. Liu, P.-c. Liu, R. Liu and X. Wu, Med. Sci. Monit. Basic Res., 2015, 21, 15–20 CrossRef PubMed.
- P. V. AshaRani, G. Low Kah Mun, M. P. Hande and S. Valiyaveettil, ACS Nano, 2009, 3, 279–290 CrossRef CAS.
- N. Miura and Y. Shinohara, Biochem. Biophys. Res. Commun., 2009, 390, 733–737 CrossRef CAS PubMed.
- Y.-H. Lee, F.-Y. Cheng, H.-W. Chiu, J.-C. Tsai, C.-Y. Fang, C.-W. Chen and Y.-J. Wang, Biomaterials, 2014, 35, 4706–4715 CrossRef CAS PubMed.
- Y.-H. Hsin, C.-F. Chen, S. Huang, T.-S. Shih, P.-S. Lai and P. J. Chueh, Toxicol. Lett., 2008, 179, 130–139 CrossRef CAS PubMed.
- E. Caballero-Díaz, C. Pfeiffer, L. Kastl, P. Rivera-Gil, B. Simonet, M. Valcárcel, J. Jiménez-Lamana, F. Laborda and W. J. Parak, Part. Part. Syst. Charact., 2013, 30, 1079–1085 CrossRef.
- J. Hühn, C. Carrillo-Carrion, M. G. Soliman, C. Pfeiffer, D. Valdeperez, A. Masood, I. Chakraborty, L. Zhu, M. Gallego, Z. Yue, M. Carril, N. Feliu, A. Escudero, A. M. Alkilany, B. Pelaz, P. del Pino and W. J. Parak, Chem. Mater., 2016, 29, 399–461 CrossRef.
- C. K. Riener, G. Kada and H. J. Gruber, Anal. Bioanal. Chem., 2002, 373, 266–276 CrossRef CAS PubMed.
- A. Shevchenko, H. Tomas, J. Havli, J. V. Olsen and M. Mann, Nat. Protoc., 2007, 1, 2856 CrossRef PubMed.
- M. Barbalinardo, D. Gentili, F. Lazzarotto, F. Valle, M. Brucale, M. Melucci, L. Favaretto, M. Zambianchi, A. I. Borrachero-Conejo, E. Saracino, V. Benfenati, D. Natalini, P. Greco, M. G. Di Carlo, G. Foschi and M. Cavallini, Small Methods, 2018, 2, 1700377 CrossRef.
- P. R. Twentyman and M. Luscombe, Br. J. Cancer, 1987, 56, 279–285 CrossRef CAS PubMed.
- M. K. Squier and J. J. Cohen, Mol. Biotechnol., 2001, 19, 305–312 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr08259g |
| This journal is © The Royal Society of Chemistry 2021 |