Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The conjugation strategy affects antibody orientation and targeting properties of nanocarriers†
Maximilian
Brückner‡
ab,
Johanna
Simon‡
ab,
Katharina
Landfester
 b and
Volker
Mailänder
b and
Volker
Mailänder
 *ab
*ab
aDermatology Clinic, University Medical Center of the Johannes Gutenberg-University Mainz, Langenbeckstr. 1, 55131 Mainz, Germany. E-mail: mailaend@mpip-mainz.mpg.de
bMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany
First published on 11th May 2021
Abstract
Antibody-modified drug delivery systems in the nano-range have the ability to overcome current challenges for treating diseases due to their high specificity towards the targeted body region. However, no antibody-bound nanocarrier has been clinically approved to date. This missing clinical approval may be a result of the conjugation strategy that influences the spatial orientation of the attached antibody on the nanocarriers’ surface. What is not missing, however, is a diverse selection of antibody to nanocarrier conjugation strategies that determine the success of an antibody functionalized drug delivery system. In this paper, two antibody conjugation strategies were compared by conjugating the surface of cross-linked starch iron oxide nanocarriers with specifically modified CD11c monoclonal antibodies. The antibody nanocarrier conjugates, synthesized either by the chemistry of thiol–maleimide coupling or copper-free click chemistry, were analyzed by flow cytometry to determine their binding affinity towards a murine dendritic cell line (DC2.4). In the cell uptake, different antibody amounts on the nanocarrier could induce a dendritic cell uptake for both conjugation strategies. However, blocking experiments further highlighted the importance of the orientation of the antibody on to the nanocarriers’ surface. While the antibodies which were attached via the copper-free click chemistry were oriented, maleimide synthesized conjugates presented their antibodies randomly on the surface. Lastly, to evaluate the in vivo properties of the antibody modified nanocarriers, targeting experiments with mouse plasma were performed, and it was proven that the biomolecular corona does not diminish the targeting efficiency.
Introduction
The attachment of an antibody to the surface of a nanocarrier is the transfer of the specific and selective recognition ability of the antibody towards a nanocarrier. In return, nanocarriers offer a stable drug encapsulation, reduce toxicity, and control drug release.1 Therefore, it is of great interest to maintain the antibody's targeting ability to recognize its antigen for a site- and cell-directed delivery of a nanocarrier. Also, the non-binding parts like the constant region (Fc) of the antibody should not be exposed because Fc parts can enhance the mistargeting of nanocarriers to Fc-receptor containing cells like macrophages.2 Therefore, it is essential that an appropriate conjugation and orientation of the antibody on the nanocarriers’ surface is ensured for a targeted drug delivery via nanocarriers.3Over the past decades, numerous strategies have been developed to attach antibodies to the surface of nanocarriers. These strategies include the physical adsorption, covalent bonding, adapter molecules, or the combination of these concepts.4 Despite the simplicity or straightforwardness of these conjugation concepts in theory, the implementation is challenging.5 For instance, the Fab regions of the antibody should remain unaffected during the modification and immobilization process, because this region mediates the antigen recognition.6
The major goal of the synthesized antibody nanocarrier conjugates is the targeted transport of the cargo. As an example, in the field of immunotherapy, the nanocarrier can allow a co-delivery of different immunomodulators (e.g. adjuvant and antigen). The targeted transport of both substances to specific cells (e.g. antigen presenting cells, APCs) will further innate all down-stream processes, e.g. an antigen-specific cytotoxic T-lymphocyte (CTL), which can eventually enable a tumor regression7,8. In this process, dendritic cells, which are APCs, play a crucial role because they initiate and regulate these immune responses.9
The aim of this study was to compare two bioconjugation methods for the attachment of an antibody on a nanocarriers’ surface. For this purpose, we used red-fluorescent nanocarriers that are composed of a magnetite core because of their ease in detection and magnetic separation. These nanocarriers are further equipped with a shell of cross-linked hydroxyethyl starch with amino groups on the surface. Anti-CD11c antibodies were either attached via the chemistry of thiol–maleimide coupling or via copper-free click chemistry on the surface of magnetic nanocarriers to specifically target dendritic cells. In our study, we found that both, the amount of functionalized antibody on the nanocarriers’ surface and the conjugation chemistry, had a significant influence on the spatial orientation of the covalently bound antibody and thus on the targeting efficiency.
Results and discussion
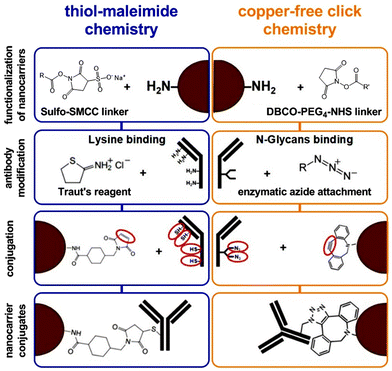
Two conjugation strategies were compared to covalently attach a targeting antibody onto a nanocarriers’ surface (Fig. 1). First, the amine groups on the nanocarriers’ surface were functionalized with two different linkers via NHS chemistry (size and charge of the nanocarrier and synthesized nanocarrier conjugates are listed in Table S1†). Each linker provides a specific reactive moiety, which is necessary for the antibody attachment. The modification of the antibodies was carried out by either the random thiolation through Traut's reagent (2-Iminothiolan) or by a site-directed enzymatic modification at the Fc region. In a second step, the linker-functionalized nanocarriers were conjugated to the modified antibodies by using either the thiol–maleimide or the copper-free click chemistry. The targeting efficiency of the resulting nanocarrier conjugates was analyzed in a cell-based assay via flow cytometry and confocal laser scanning microscopy (cLSM).The modification of monoclonal CD11c antibodies
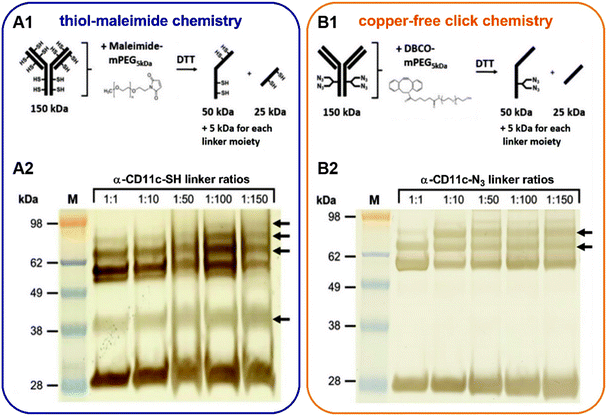
In the case of the thiol–maleimide chemistry, antibodies were chemically modified by reacting the primary amines of lysine residues with Traut's reagent in the absence of a regiochemical control, generating antibodies with free sulfhydryl groups. For the copper-free click chemistry, antibodies were enzymatically modified at their N-glycans, which are positioned at the Fc region to generate antibodies with free azido groups.The validation of the antibody modification was investigated by SDS-PAGE and visualized by silver staining (Fig. 2). For this concept, modified antibodies were reacted with a functional linker (PEG5kDa-Malemide or PEG5ka-DBCO) and further applied to SDS-PAGE under reducing conditions (A1 and B1). This enabled a mass-based separation of the different antibody linker species and a distinguishable observation between the heavy and the light chain modification of the antibody. The thiol–maleimide chemistry approach showed next to the unconjugated heavy chain at 50 kDa up to three additional heavy chain linker species ranging between 55, 60, and 65 kDa (A2, black arrows). This modification was achieved at equal molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), carrying up to two linker moieties. At the lower molecular weight region, the light chain was observed at around 30 kDa. An additional band at around 40 kDa was detected, representing a conjugation of two linker units to the light chain (A2 black arrow). In contrast, via the use of the copper-free click chemistry the antibody was only modified at the heavy chain (B2 black arrows) and not the light chain. The addition of the PEG5ka-DBCO linker molecule generated two additional heavy chain linker species at 55 and 60 kDa for all tested linker ratios. The linker attachment efficacy was also demonstrated for the whole antibody molecules (150 kDa) and for the isotype control antibodies under non-reducing and reducing conditions (S1–S5).
1), carrying up to two linker moieties. At the lower molecular weight region, the light chain was observed at around 30 kDa. An additional band at around 40 kDa was detected, representing a conjugation of two linker units to the light chain (A2 black arrow). In contrast, via the use of the copper-free click chemistry the antibody was only modified at the heavy chain (B2 black arrows) and not the light chain. The addition of the PEG5ka-DBCO linker molecule generated two additional heavy chain linker species at 55 and 60 kDa for all tested linker ratios. The linker attachment efficacy was also demonstrated for the whole antibody molecules (150 kDa) and for the isotype control antibodies under non-reducing and reducing conditions (S1–S5).
The results therefore show that the antibody modification via Traut's reaction for the thiol–maleimide conjugation was mainly unspecific and modified the heavy and light chain. This broad modification can be explained by the high number of lysine residues (∼80) on an antibody with around 10 of them being chemically accessible.10 This availability of lysine residues indicates that a precise control of the conjugation site cannot be guaranteed with the lysine amide coupling approach. For instance, some of the lysine residues are positioned in the antigen recognition, and therefore, a modification could potentially reduce the binding affinity of the antibody. Moreover, the abundance of lysine residues increases the number of conjugated antibody-nanocarrier configurations, which might have an impact on pharmacokinetics, pharmacodynamics, and cytotoxicity, as it has been observed for antibody–drug conjugates.11 Especially, antibodies attached with the two Fab regions onto the surface of the nanocarrier will exhibit the Fc part of the antibody and therefore, can promote a mistargeting.
On the contrary, the enzymatic antibody modification for the copper-free click reaction was highly site-specific because only the heavy chain was modified (B2, black arrows). In this case, antibodies were selectively modified on the sugar residues present at the Fc-region. All immunoglobulin G (IgG) subclasses are N-glycosylated at a single, conserved region at the Fc portion. This glycosylation site is positioned within the heavy chains CH2 domain at the amino acid asparagine 297 (N297).12 The attached carbohydrate chain has a predominantly biantennary complex structure, which is usually composed of N-acetylglucosamine (GlcNAc) and mannose, followed by a variable addition of galactose (Gal), N-acetylneuraminic acid (NeuNAc), fucose (Fuc), as well as bisecting GlcNAc residues.13 In our case, using this site for the conjugation involved the unmasking of the GlcNac residues by removing the terminal Gal residues with β-galactosidase, followed by the enzymatic β-galactosyl transferase reaction. The mutant bovine enzyme (Y289L-Gal-T1) transfers N-azidoacetylglucosamine (GalNAz) selectively to the unmasked GlcNAc residues of the N-linked IgG glycans.14,15 The resulting azide-modified tags can be used for the strain-promoted copper-free click reaction between the alkyne-functionalized (DBCO) nanocarrier and the azide-modified antibody. This strategy is of great advantage because it can be applied for any IgG isotypes with various N-glycosylation profiles.11
In summary, the enzymatic modification provides the ability to attach a linker molecule to the antibody in a site-specific manner under mild reaction conditions without altering the antigen binding sites. In contrast, conventional attachment strategies, like the lysine approach, result in a non-specific antibody functionalization.5,11,16
The amount of antibodies on the nanocarrier is crucial for a specific dendritic cell uptake
In a next step, different antibody amounts were attached onto the nanocarriers’ surface in order to investigate the lowest quantity that still evokes a specific cell response towards the murine dendritic cell line DC2.4. For this purpose, the integrin αX or CD11c was chosen as a target surface receptor because of its wide use as a defining marker for dendritic cells17 and its ability to mediate phagocytosis of, for example, antibody-bound nanocarriers.18 The CD11c antibody-based targeting was performed with an additional isotype control antibody, which exhibits a similar composition to that of the CD11c antibody, however, it lacks in a specific target binding. This allows to determine the contribution of non-specific binding via the Fc region of the antibody contrary to a specific binding via the Fab region.For this investigation, five different antibody quantities of 0.05 μg, 0.5 μg, 2.5 μg, 7.5 μg, and 12.5 μg were chosen to be conjugated to 500 μg of nanocarriers (Fig. 3). In a first step, all nanocarrier conjugates were tested for the presence of the antibody on the surface using a labeled secondary antibody and analyzed by flow cytometry (Fig. 3A1 and B1). For the nanocarrier conjugates using the thiol–maleimide or the copper-free click chemistry, the CD11c and isotype antibodies were detected in a concentration dependent manner (Fig. 3A1 and B1).
The DC2.4 dendritic cell uptake was conducted with 1.5 × 105 cells per mL in a culture medium containing fetal bovine serum (FBS, ThermoFisher, Germany) (Fig. 3A2 and B2). Additional serum proteins in the medium lead to the formation of a biomolecular corona around the surface of the nanocarriers representing a more physiological environment.19 The lowest antibody quantity on the thiol–maleimide nanocarrier conjugates showed no significant uptake by the dendritic cells. Starting at an antibody amount of 0.5 μg, a significant cell uptake of the CD11c conjugate compared to the isotype conjugate was observed (P < 0.001***). This specificity increased for the 2.5 μg group to an even higher median fluorescence intensity (MFI). However, for higher antibody quantities of 7.5 and 12.5 μg, the unspecific cell uptake of the isotype conjugates was also increased.
In case of the copper-free click chemistry, the lowest antibody amount of 0.05 μg showed no significant cell uptake (Fig. 3B2). Starting from the 0.5 μg CD11c conjugate group, a slightly increased intensity was detected, compared to the corresponding isotype conjugate (P < 0.05*). For the higher antibody amounts, the cellular uptake was strongly increased with an amount of 2.5 μg and reached a maximum at 7.5 μg (P < 0.001***).
In a nutshell, different amounts of antibodies on the nanocarrier's surface showed that there is a certain antibody amount acting as a threshold that leads to a specific cell targeting. In fact, the actual amount of antibody molecules on a nanocarrier, which are believed to initiate an intended cell-based response, is an important value. However, reports about the introduction of a precise and controllable number of antibodies per nanocarrier are largely missing. Nevertheless, it is possible to generate nanocarriers with exactly one or two conjugated antibodies. In this attempt, gold nanocarriers with only one attached antibody suggested the best targeting efficiency in mice bearing MCF-7 xenografts.20 Furthermore, bioanalytical systems and biosensors have also demonstrated a better antigen-capturing activity for lower antibody densities on nanocarriers.21,22
Blocking dendritic targeting sides reveals a spatial orientation of the antibody on the nanocarrier
To further investigate the specific targeting ability of the nanocarrier conjugates additional blocking experiments were conducted. This included the blocking of the targeted CD11c ligands on dendritic cells.For the blocking experiment, a CD11c antibody concentration of 7.5 μg mL−1 with an incubation time of 30 min at 4 °C prior to the cell uptake has been proven to be sufficient (S6). All tested conjugates were synthesized with an antibody amount of 7.5 μg (Fig. 4 and Fig. S7†).
Blocking CD11c had virtually no effect on the thiol–maleimide generated nanocarriers, because there was no difference between the unblocked condition (Medium−Serum, white bar) and the blocked condition (grey bar). According to our data, which showed that the CD11c thiol–maleimide conjugates were taken up to a significant higher amount compared to the isotype conjugate, it can be assumed that an alternative uptake mechanism is addressed. This difference highlights that CD11c thiol–maleimide conjugates have a heterogeneous antibody arrangement on their surface. As soon as the CD11c integrin is blocked and targeting through the Fab region of the antibody is no longer possible, the misplaced antibodies pointing their Fc region to the outside will cause the cell uptake by Fc receptors. Therefore, thiol–maleimide generated antibody nanocarriers compensate the uptake mechanism by a different arrangement of the antibody molecules on their surface. Moreover, copper-free click generated CD11c conjugates are effectively blocked to a comparable level of the unspecific isotype conjugate (4B). In addition, the visualization of the blocking experiment by confocal laser scanning microscopy revealed the same outcome (Fig. S7†). Blocking had no influence on the thiol–maleimide generated conjugates (high uptake for both CD11c conjugate and IgG conjugate), whereas the copper-free click generated conjugates showed almost no uptake (low uptake for both CD11c conjugate and IgG conjugate).
Further, we investigated the detection of unbound antibodies in the supernatant and washes (Fig. S8†) to attribute the Fab-mediated targeting efficiency of the copper-free click generated CD11c conjugates to their spatial orientation. This investigation should highlight that there is no difference in the number of conjugated antibodies between the two conjugation chemistries. As a result, we demonstrated that in each magnetically separated wash fraction, the level of unbound antibodies were on a comparable detection level for each conjugation chemistry (Fig. S8A†) and that a certain amount of nanocarriers was lost during the washing process (Fig. S8B†).
Therefore, the conjugation strategy of linking an antibody on to the nanocarrier had a significant influence on the spatial orientation of the antibody molecule. In case of the copper-free click attached antibodies, this spatial orientation is referring to the conjugation of the antibody via its azido-modified N-glycans of the Fc region with the DBCO-functionalized nanocarrier surface. Given this spatial orientation, the unspecific Fc-region is hidden from any cell surface interactions, while the specific Fab region is pointing away from the surface being accessible to target the CD11c surface receptors.
Mouse plasma incubation influences the targeting ability of CD11c nanocarriers
To investigate the targeting efficiency of the antibody nanocarrier conjugates in a more physiological context, cellular experiments using mouse plasma (GeneTex, USA) were performed. In the biological environment, e.g. in blood, proteins are in constant interactions with the surface of the nanocarrier, forming the biomolecular corona.19 Therefore, it is of great importance to examine the binding affinity of the antibody nanocarrier conjugates in the presence of the biomolecular corona.In Fig. 5, the cellular uptake experiments were performed using three different cell culture conditions. There was no significant difference for the conditions with and without FBS. The pre-coating of the nanocarrier conjugates with mouse plasma slightly reduced the cellular uptake, however, it did not diminish the targeting efficiency.
In literature, it is shown that different protein sources alter the physicochemical properties of nanocarriers,23,24 which can affect the antigen–antibody interaction, cellular targeting, and uptake behavior.25–27 Furthermore, it is important to consider a possible loss of antigen binding ability or a complete change of antibody nanocarrier functionality due to the corona. Indeed, our group already showed this effect for coupled and adsorbed antibodies on polystyrene nanocarriers. We demonstrated that pre-adsorbed CD63 antibodies keep their targeting ability and are not covered by the biomolecular corona compared to conjugated antibodies.28
To additionally verify the intracellular localization of the nanocarrier-conjugates, confocal laser scanning microscopy images were taken (Fig. 6). Two cell culture conditions, one being a medium with 5% FBS and the other being pre-incubated with mouse plasma were analyzed. Because there were no visual differences between these conditions, only mouse plasma incubated samples are representatively shown in the Fig. 6. Additional images are summarized in Fig. S9.† The images clearly indicate the difference between the targeting specificity of the CD11c conjugates in contrast to the isotype control or the DBCO linker-attached nanocarrier. No uptake was detected in the case of linker-functionalized nanocarriers (first row). The CD11c conjugates are mostly internalized in dendritic cells, with some of them being randomly attached to the surface of the cells (second row, enlarged image). The isotype conjugates are only taken up by a few dendritic cells or form clusters outside of the cells (third row, enlarged image). We additionally visualized the nanocarriers by TEM to confirm their colloidal stability in cell culture media under the same sample concentration (Fig. S10†). According to our data, we observed that the pristine and antibody-modified nanocarriers formed small clusters that approx. matched in size to the values obtained by multi angle DLS (Table S1†).
Studies have shown that the cell uptake significantly increases during the first minutes and gradually slows down until reaching a plateau within two to three hours.29–31 Therefore, in the first phase, antibody nanocarrier conjugates might directly adsorb onto the cell membrane, while at a later state, internalization through an endocytic pathway is initiated. CD11c conjugates which have been incubated for two hours show this behavior as some are still attached to the cell membrane, while others are already internalized (Fig. 6).
For a quantitative analysis of the murine biomolecular corona, the label-free mass spectrometry was applied to identify and characterize the biological identity of the antibody targeted nanocarriers. In Fig. 7, all identified proteins were divided into nine groups, and the respective values were plotted as a percentage. The comparison of the individual groups illustrated almost no difference between the identified proteins of the nanocarrier and the antibody conjugated nanocarrier groups. Only for the DBCO linker-functionalized nanocarrier conjugate without the antibody, lower amounts of serum albumin and higher amounts of coagulation proteins were observed. Serum albumin is not only the most abundant murine corona protein found on our antibody nanocarrier conjugates, but moreover, it is the most abundant serum protein for many species, including mouse and human.32,33 Dai et al. have examined the targeting ability of antibody–mimetic ligand-functionalized nanocarriers under different conditions using human serum or human serum albumin alone. They showed that the mixture of different proteins in the human serum corona inhibited cell association, while human serum albumin alone enhanced the targeting ability of the nanocarriers.34 In comparison to their findings, we detected a general enrichment of murine serum albumin on our targeted nanocarriers, which could potentially support an uptake by dendritic cells. However, having a complex mixture of murine plasma proteins acting as a corona, the balance between the targeting ability of the conjugated antibody in cooperation with serum albumin or other candidates, either facilitates or impedes a precise cell targeting.
For a more detailed analysis, the top 20 most abundant proteins are highlighted in a heat map (Fig. 8). As already shown in the identified protein groups, serum albumin is the most abundant protein detected in the corona of the antibody nanocarrier conjugates. Linker-functionalized nanocarrier conjugates (DBCO conjugate) highly adsorbed platelet factor 4. Additional abundant mouse plasma proteins included ceruloplasmin, complement C3 and complement C4-B, histidine-rich glycoprotein, kininogen-1, serine protease inhibitor A3K, as well as vitronectin. Other murine biomolecular corona studies on differently composed nanocarriers have also identified these proteins to be commonly adsorbed.33,35
In general, the findings of the murine biomolecular corona suggest that the targeting ability of CD11c antibody conjugated nanocarriers is not induced by corona proteins, although they influence the targeting process. Therefore, antibody targeted nanocarrier conjugates must be examined in the in vivo mouse model.
Conclusion
In this study, it was demonstrated that copper-free click chemistry generated nanocarrier conjugates surpass the efficacy of thiol–maleimide chemistry produced conjugates. We proved that the enzymatic modification of CD11c antibodies is specific with modified antibodies carrying up to four azide groups. In contrast, the Traut's modification is highly unspecific and modifies unwanted antibody regions, such as the light chain or N-terminus. Evaluating the targeting specificity of nanocarriers with different amounts of conjugated CD11c antibodies in vitro have highlighted a limit up to which a specific uptake by dendritic cells is evoked. Especially, blocking experiments have emphasized the importance of a site-directed conjugation chemistry, which considers the spatial orientation of the attached antibody on the nanocarriers’ surface. Additionally, targeting experiments with mouse plasma proved that the biomolecular corona does not impair the binding efficiency of antibody modified nanocarriers towards dendritic cells. This underlines the great potential of the here presented copper-free click generated antibody nanocarriers, which can be used in immunotherapy and nanomedicine.Author contributions
Maximilian Brückner: Conceptualization, data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing – original draft and writing – review & editing; Johanna Simon: Conceptualization, data curation, formal analysis, investigation, methodology, project administration, supervision, validation, visualization, writing – original draft and writing – review & editing; Katharina Landfester: Conceptualization, funding acquisition, resources, supervision and validation; Volker Mailänder: Conceptualization, funding acquisition, project administration, resources, supervision, validation and writing – review & editing.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
For financial support, the authors gratefully acknowledge the Deutsche Forschungsgemeinschaft (DFG Sonderforschungsbereich 1066/CRC1066). In addition, the authors would like to express their sincere thanks to Francesca Mazzotta and Ingo Lieberwirth for performing the TEM images as well as Christine Rosenauer for measuring the multi angle DLS and Elke Muth for performing the ζ-potential. For the design of the table of contents, we highly thank Stefan Schuhmacher. Last but not least, we gratefully thank Karolin Eberle for her help in revising the scientific language in the final readings of the manuscript. Open Access funding provided by the Max Planck SocietyReferences
- Z. Cheng, A. Al Zaki, J. Z. Hui, V. R. Muzykantov and A. Tsourkas, Science, 2012, 338, 903–910 CrossRef CAS PubMed.
- M. Daëron, in Fc Receptors, Springer International Publishing, 2014, pp. 131–164, DOI:10.1007/978-3-319-07911-0_7.
- J. M. Montenegro, V. Grazu, A. Sukhanova, S. Agarwal, J. M. de la Fuente, I. Nabiev, A. Greiner and W. J. Parak, Adv. Drug Delivery Rev., 2013, 65, 677–688 CrossRef CAS PubMed.
- E. Polo, S. Puertas, M. Moros, P. Batalla, J. M. Guisan, J. M. de la Fuente and V. Grazu, Methods Mol. Biol., 2013, 1051, 149–163 CrossRef CAS PubMed.
- A. J. Sivaram, A. Wardiana, C. B. Howard, S. M. Mahler and K. J. Thurecht, Adv. Healthcare Mater., 2018, 7 DOI:10.1002/adhm.201700607.
- S. Puertas, M. Moros, R. Pacheco, M. R. Ibarra, V. Grazu and J. Fuente, J. Phys. D: Appl. Phys., 2010, 474012 CrossRef.
- J. M. Silva, M. Videira, R. Gaspar, V. Preat and H. F. Florindo, J. Controlled Release, 2013, 168, 179–199 CrossRef CAS PubMed.
- M. S. Singh and S. Bhaskar, ImmunoTargets Ther., 2014, 3, 121–134 Search PubMed.
- M. Diwan, P. Elamanchili, H. Lane, A. Gainer and J. Samuel, J. Drug Targeting, 2003, 11, 495–507 CrossRef CAS PubMed.
- R. V. Chari, Acc. Chem. Res., 2008, 41, 98–107 CrossRef CAS PubMed.
- K. Tsuchikama and Z. An, Protein Cell, 2018, 9, 33–46 CrossRef CAS PubMed.
- J. Xue, L. P. Zhu and Q. Wei, Glycoconjugate J., 2013, 30, 735–745 CrossRef CAS PubMed.
- F. Higel, A. Seidl, F. Sorgel and W. Friess, Eur. J. Pharm. Biopharm., 2016, 100, 94–100 CrossRef CAS PubMed.
- E. Boeggeman, B. Ramakrishnan, M. Pasek, M. Manzoni, A. Puri, K. H. Loomis, T. J. Waybright and P. K. Qasba, Bioconjugate Chem., 2009, 20, 1228–1236 CrossRef CAS PubMed.
- B. Ramakrishnan and P. K. Qasba, J. Biol. Chem., 2002, 277, 20833–20839 CrossRef CAS PubMed.
- G. Chaubet, F. Thoreau and A. Wagner, Drug Discovery Today: Technol., 2018, 30, 21–26 CrossRef PubMed.
- J. Wu, H. Wu, J. An, C. M. Ballantyne and J. G. Cyster, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 6786–6791 CrossRef CAS PubMed.
- H. Matsuo, M. Somiya, M. Iijima, T. Arakawa and S. I. Kuroda, J. Nanobiotechnol., 2018, 16, 59 CrossRef PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- M. Colombo, L. Fiandra, G. Alessio, S. Mazzucchelli, M. Nebuloni, C. De Palma, K. Kantner, B. Pelaz, R. Rotem, F. Corsi, W. J. Parak and D. Prosperi, Nat. Commun., 2016, 7, 13818 CrossRef CAS PubMed.
- B. Saha, T. H. Evers and M. W. Prins, Anal. Chem., 2014, 86, 8158–8166 CrossRef CAS PubMed.
- N. A. Byzova, I. V. Safenkova, E. S. Slutskaya, A. V. Zherdev and B. B. Dzantiev, Bioconjugate Chem., 2017, 28, 2737–2746 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772 CrossRef CAS PubMed.
- S. Schöttler, K. Klein, K. Landfester and V. Mailänder, Nanoscale, 2016, 8, 5526–5536 RSC.
- D. Pozzi, G. Caracciolo, A. L. Capriotti, C. Cavaliere, G. La Barbera, T. J. Anchordoquy and A. Lagana, J. Proteomics, 2015, 119, 209–217 CrossRef CAS PubMed.
- H. de Puig, I. Bosch, M. Carré-Camps and K. Hamad-Schifferli, Bioconjugate Chem., 2017, 28, 230–238 CrossRef CAS PubMed.
- M. Tonigold, J. Simon, D. Estupiñán, M. Kokkinopoulou, J. Reinholz, U. Kintzel, A. Kaltbeitzel, P. Renz, M. P. Domogalla, K. Steinbrink, I. Lieberwirth, D. Crespy, K. Landfester and V. Mailänder, Nat. Nanotechnol., 2018, 13, 862–869 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- M. P. Desai, V. Labhasetwar, E. Walter, R. J. Levy and G. L. Amidon, Pharm. Res., 1997, 14, 1568–1573 CrossRef CAS PubMed.
- D. Guarnieri, A. Guaccio, S. Fusco and P. A. Netti, J. Nanopart. Res., 2011, 13, 4295 CrossRef CAS.
- M. Leeman, J. Choi, S. Hansson, M. U. Storm and L. Nilsson, Anal. Bioanal. Chem., 2018, 410, 4867–4873 CrossRef CAS PubMed.
- A. Solorio-Rodriguez, V. Escamilla-Rivera, M. Uribe-Ramirez, A. Chagolla, R. Winkler, C. M. Garcia-Cuellar and A. De Vizcaya-Ruiz, Nanoscale, 2017, 9, 13651–13660 RSC.
- Q. Dai, Y. Yan, J. Guo, M. Björnmalm, J. Cui, H. Sun and F. Caruso, ACS Macro Lett., 2015, 4, 1259–1263 CrossRef CAS.
- L. Shen, S. Tenzer, W. Storck, D. Hobernik, V. K. Raker, K. Fischer, S. Decker, A. Dzionek, S. Krauthauser, M. Diken, A. Nikolaev, J. Maxeiner, P. Schuster, C. Kappel, A. Verschoor, H. Schild, S. Grabbe and M. Bros, J. Allergy Clin. Immunol., 2018, 142, 1558–1570 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional figures are summarized in the supporting information. In a separate Excel File all identified proteins (LC-MS) are summarized. See DOI: 10.1039/d0nr08191d |
| ‡ Shared first author. |
| This journal is © The Royal Society of Chemistry 2021 |