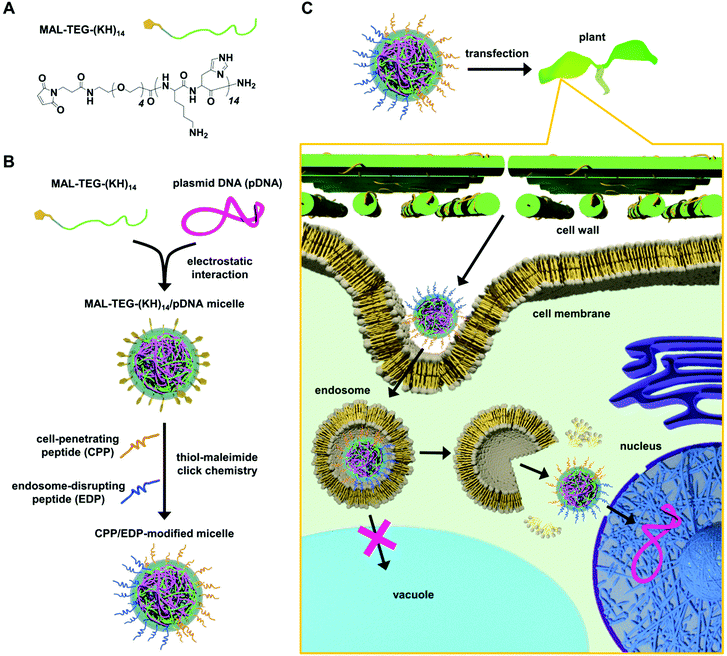
Endosome-escaping micelle complexes dually equipped with cell-penetrating and endosome-disrupting peptides for efficient DNA delivery into intact plants†
Takaaki
Miyamoto
 a,
Kousuke
Tsuchiya
*b and
Keiji
Numata
a,
Kousuke
Tsuchiya
*b and
Keiji
Numata
 *ab
*ab
aBiomacromolecules Research Team, RIKEN Center for Sustainable Resource Science, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan. E-mail: numata.keiji.3n@kyoto-u.ac.jp
bDepartment of Material Chemistry, Graduate School of Engineering, Kyoto University, Kyoto-Daigaku-Katsura, Nishikyo-ku, Kyoto 615-8510, Japan
First published on 12th February 2021
Abstract
The delivery of DNA to plants is crucial for enhancing their ability to produce valuable compounds and adapt to climate change. Peptides can provide a versatile tool for delivering DNA to a specific target organelle in various plant species without the use of specialized equipment. However, peptide-mediated DNA delivery suffers from endosomal entrapment and subsequent vacuolar degradation of the DNA cargo, which leads to poor transfection efficiency. To overcome the lack of a reliable approach for bypassing vacuolar degradation in plants, we herein present an endosome-escaping micelle. The micelle surface is dually modified with cell-penetrating (CPP) and endosome-disrupting peptides (EDP) and the core is composed of plasmid DNA condensed with cationic peptides. Due to the functions of CPP and EDP, the dual peptide-modified micelles efficiently undergo endocytic internalization and escape from endosomes to the cytosol, thereby achieving significantly enhanced transfection of intact plants with negligible cytotoxicity. The present study offers a robust strategy for efficient intracellular DNA delivery to plants without vacuolar degradation, and can facilitate plant bioengineering for diverse biotechnological applications.
Introduction
Rationally designed synthetic carriers enable the effective delivery of genetic cargoes not only into mammalian cells for therapeutic purposes but also into plant cells for conferring biotic and abiotic stress tolerance and for enhancing the production of valuable compounds.1,2 Recent studies have shown that synthetic carriers based on peptides and nanomaterials can be superior to existing delivery tools available for plants,3–24 such as Agrobacterium or particle bombardment,25,26 due to their potential to deliver various types of genetic cargoes into a wide range of plant species without the use of specialized instruments. Among synthetic carriers, peptides present attractive features, including their diverse functions and ease of preparation. Various peptide-based carriers with DNA-binding, cell-penetrating, and organelle-targeting functions have been designed and utilized for selective DNA delivery to the nucleus,11,14,18,22,24,27 chloroplast,10,13 and mitochondrion in intact plants.12,13,16 Peptide carriers have remarkable organelle selectivity, which can provide a distinct advantage over other tools, but they generally exhibit low transfection efficiency, which limits their utility in plant gene delivery. Peptide carriers and their ionic complexes with DNA cargo can be internalized into plant cells through endocytosis,10,27 but most of them are entrapped in endosomes and subsequently transferred to the vacuole, which is equivalent to the lysosome in animal cells, and eventually degraded by vacuolar enzymes.14,27 This vacuolar degradation severely reduces the transfection efficiency of the peptide carrier. Accordingly, endosomal escape ability is a key requirement for efficient gene delivery to plants. However, a robust approach for enhancing endosomal escape in plant systems is still missing.Similar to the situation in plant cells, endosomal escape is regarded as a bottleneck for intracellular DNA delivery to mammalian cells.28 Much effort has been devoted to developing materials for overcoming endosomal entrapment.29 One of the most commonly used materials is a histidine-containing peptide that can destabilize endosomes through the proton sponge effect,30–32 where the protonation of the histidine imidazole ring (pKa = ∼6.0) at weakly acidic endosomal pH (below 6.0) is considered to induce an extensive inflow of ions and water into endosomes, resulting in the rupture of the endosomal membrane.33 In anticipation of the proton sponge effect, we previously tested a peptide carrier containing multiple histidine residues for DNA delivery to plants, but the carrier/DNA complex failed to escape from the endosome and accumulated in the vacuole lumen of plant cells.14 These findings suggest that the proton sponge effect may not be reliable in plant cells even though the pH in the plant endosomes is sufficiently acidic (pH 5.5–6.3) for histidine protonation.34,35 An alternative strategy is thus needed to facilitate endosomal escape in plant cells.
We focused on an endosome-disrupting peptide (hereafter referred to as EDP) that directly interacts with the endosomal membrane to promote endosomal escape through different mechanisms from the proton sponge effect. LAH4-L1 and GALA peptides have been developed as pH-sensitive EDPs.36–39 The LAH4-L1 peptide with an α-helical conformation aligns parallel to the membrane surface and induces strong membrane disruption when the four histidine residues in the peptide are positively charged at an endosomal pH (5.0), whereas it exhibits less membrane disruption ability when the histidines are uncharged at a neutral pH (7.4).40 The GALA peptide turns from a random coil to an amphipathic α-helix with membrane-disrupting ability when the negative charge of the glutamic acid residues is neutralized by reducing the pH from neutral (∼7.0) to acidic (∼5.0).38 In another example, Dowdy and coworkers showed that the inclusion of a hydrophobic peptide motif, an EED4 peptide, in a cationic peptide dramatically promotes endosomal escape through a pH-independent mechanism.41 Although EDPs provide a promising strategy to overcome endosomal entrapment in plant cells, their implementation is still challenging. For example, our previous studies showed that a synthetic peptide containing the EED4 peptide sequence induced cytotoxicity in plants,42 leading to limited improvement in transfection efficiency.27 The pH-sensitive LAH4-L1 and GALA peptides seem to be less toxic, but their potentials for plant systems have yet to be explored. In addition to being nontoxic, EDPs need to be efficiently internalized into plant cells together with a DNA cargo. Thus, EDPs must be used concomitantly with a cell-penetrating peptide (CPP) that induces efficient endocytosis-mediated cellular uptake and a DNA-binding polycation peptide that facilitates ionic complex formation with the DNA cargo.
Herein, we present an “all-in-one” peptide-based gene delivery system that can bypass endosomal entrapment and subsequent vacuolar degradation in plants without causing cytotoxicity. The system relies on a surface modifiable micelle complex composed of a maleimide-containing polycation peptide (MAL-TEG-(KH)14, Fig. 1A) and plasmid DNA (pDNA). According to our previous study,43 the micelle was intended to display maleimide groups on the surface and enabled simultaneous surface modification with multiple types of peptides via thiol-maleimide click chemistry. We used this surface modification strategy to prepare dual CPP/EDP-modified micelles (Fig. 1B) and explored their ability to deliver pDNA into a model plant system (Fig. 1C). The CPP/EDP-modified micelles enabled pDNA delivery into intact plants with high transfection efficiency and low cytotoxicity owing to their enhanced cellular uptake and cytosolic translocation. Notably, we demonstrated that EDP-mediated endosomal escape operated through different mechanisms from the proton sponge effect, which was considered to be unreliable in plant cells. Our approach utilizing CPP/EDP-modified micelles could offer a promising solution to overcome endosomal entrapment and facilitate intracellular DNA delivery in plant systems.
Results and discussion
Formation and characterization of MAL-TEG-KH14/pDNA micelle complexes
We prepared MAL-TEG-(KH)14/pDNA micelles at N/P ratios (defined as the molar ratio of cationic peptide nitrogen to anionic pDNA phosphate) of 1–5 from pDNA encoding luciferase as a reporter (p35S-Nluc-tNOS).11 The micelles were characterized by electrophoretic mobility shift assays (EMSAs), dynamic light scattering (DLS), and zeta potential measurements. In the EMSA, two distinct bands corresponding to supercoiled and open circular forms of pDNA were observed at N/P 0 (in the absence of the peptide), whereas pDNA migration was retarded at N/P ratios of 1–5 (Fig. S1A†), indicating electrostatic interaction between MAL-TEG-(KH)14 and pDNA. According to the DLS and zeta potential measurements, the mean hydrodynamic diameter of the micelle decreased from 229 to 81 nm as the N/P ratio was increased from 1 to 4 (Fig. S1B†), while the zeta potential increased from −34 to 30 mV (Fig. S1D†). These changes in hydrodynamic diameter and zeta potential reached a plateau above N/P 4. The polydispersity index (PDI) value was less than 0.2 at N/P 2 and greater than 0.3 at the other N/P ratios (Fig. S1C†). Owing to the lowest PDI value (0.141), the micelle prepared at N/P 2 was selected for surface modification with functional peptides.Selection of an optimal CPP for micelle surface modification with respect to transfection efficiency in a model plant system
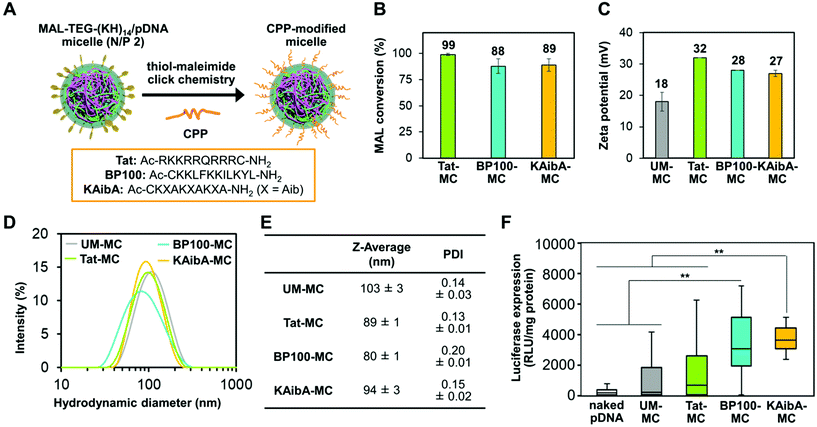
As the choice of CPP considerably affected the efficacy of peptide-mediated transfection in plants,18,44 we aimed to optimize CPP for micelle surface modification prior to investigating CPP/EDP-modified micelles. To this end, using thiol-maleimide click chemistry, we modified the MAL-TEG-KH14/pDNA micelle (prepared at N/P 2) with three different CPPs (Fig. 2A): a Tat peptide,45 an arginine-rich cationic CPP that has been intensively studied in mammalian cells; a BP100 peptide,46 a lysine-rich amphipathic CPP that efficiently permeated the plasma membrane of plant cells; or a KAibA peptide,47 a synthetic CPP with a lysine (K)-α-aminoisobutyric acid (Aib)-alanine (A) repeat that was applicable to plant cells with long-term stability against enzymatic degradation. The successful modification of the micelle with each CPP was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Fig. S2B†). We determined the conversion rate of maleimide groups (MAL conversion) for each CPP-modified micelle through reversed-phase high-performance liquid chromatography (RP-HPLC) (Fig. S2A†). Almost complete MAL conversion (>99%) was detected for the Tat-modified micelle complex (referred to as Tat-MC), while the MAL conversion was determined to be approximately 90% for the BP100- and KAibA-modified micelles (referred to as BP100-MC and KAibA-MC, respectively) (Fig. 2B). We compared the physicochemical properties of the CPP-modified micelles with those of unmodified micelles (referred to as UM-MC). The zeta potentials and hydrodynamic diameters of Tat-MC (32 mV, 89 nm), BP100-MC (28 mV, 80 nm), and KAibA-MC (27 mV, 94 nm) were more positive and smaller than those of UM-MC (18 mV, 103 nm) (Fig. 2C–E). The PDI values of Tat-MC (0.13) and KAibA-MC (0.15) were similar to that of UM-MC (0.14), while BP100-MC showed a slightly higher PDI (0.20) than the other micelles (Fig. 2D and E).Next, we compared the transfection efficiency of the micelles in seedlings of Arabidopsis thaliana, which serves as a model for dicot plants. The micelles containing the luciferase-coded pDNA (p35S-Nluc-tNOS) were introduced into the seedlings via the vacuum/compression method.11 The transfection efficiency was quantified on the basis of the luciferase expression level in the seedlings transfected by each micelle system or naked pDNA (control). BP100-MC and KAibA-MC exhibited significantly higher transfection efficiency than UM-MC, whereas Tat-MC and UM-MC did not significantly differ in efficiency (Fig. 2F). BP100-MC appeared to have higher efficacy than Tat-MC, but no significant difference was observed between them (Fig. 2F). In contrast, KAibA-MC was significantly more effective than Tat-MC in transfection (Fig. 2F). From these results, although the difference in transfection efficiency between KAibA-MC and BP100-MC was not significant (Fig. 2F), we selected the KAibA peptide as the CPP to prepare CPP/EDP-modified micelles.
Since the MAL conversion and physicochemical properties were similar among the CPP-modified micelles, the differences in their transfection efficiency would be derived from the sequence of the CPP. The higher transfection efficiency of KAibA-MC compared to that of Tat-MC is consistent with our previous findings that the KAibA peptide (not conjugated to the micelle) was internalized into plant cells more efficiently than the Tat peptide due to the greater resistance of the former peptide to enzymatic degradation.47 In mammalian cells, arginine-rich CPPs, such as the Tat peptide, are considered to undergo cellular uptake more effectively than lysine-rich CPPs. This could be because a guanidino group of arginine residues can strongly interact with the sulfate group of heparan sulfate proteoglycans, which are often found at the cell surface in animal species, through bidentate hydrogen bonds, while an ε-amino group of lysine residues can form a monodentate hydrogen bond with the sulfate group.48 However, heparan sulfate proteoglycans are generally not observed at the surface of plant cells, negating the benefits of arginine-rich CPPs found in mammalian cells. Moreover, arginine-rich CPPs could be entrapped in the cell wall of plant cells more persistently than lysine-rich CPPs, possibly due to the strong interaction between the arginine side chain and the carboxylate moieties of pectin, one of the major components in plant cell walls. These factors might cause the differences in transfection efficiency among the CPP-modified micelles.
Preparation and characterization of CPP/EDP-modified micelles
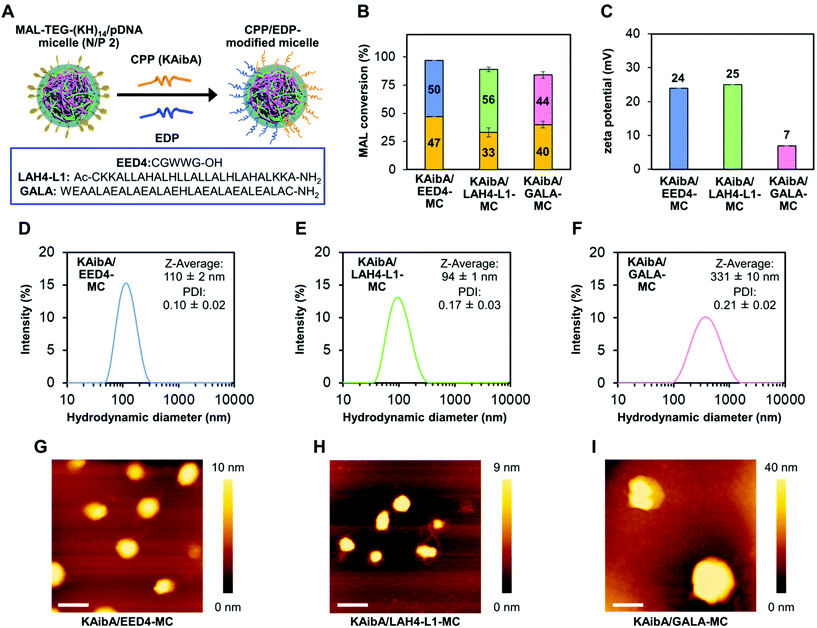
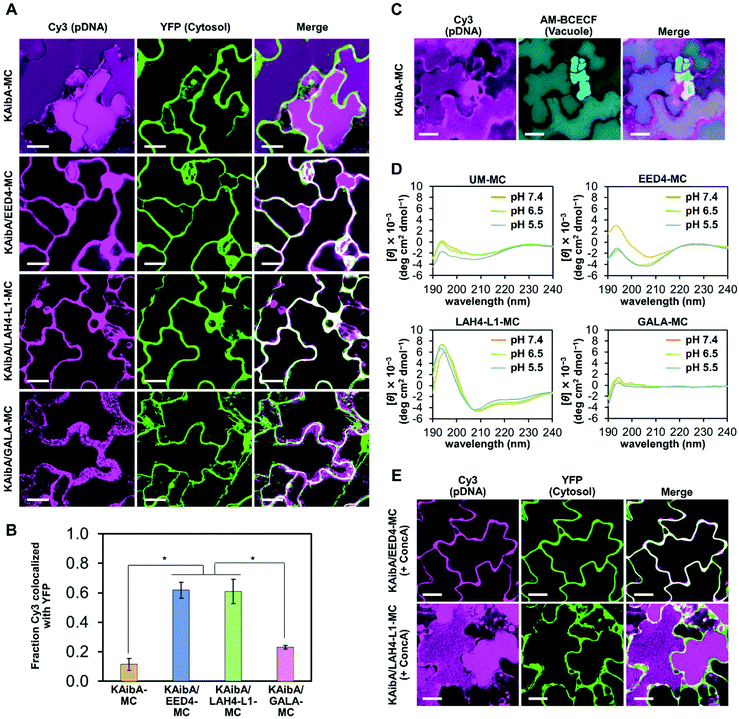
For the simultaneous surface modification of the MAL-TEG-KH14/pDNA micelle (prepared at N/P 2), the KAibA peptide was used in combination with three different EDPs (Fig. 3A): the EED4 peptide,41 a tryptophan-containing hydrophobic EDP that functions independently of its secondary structure or endosomal pH; the LAH4-L1 peptide,37 a histidine-rich amphipathic EDP that destabilizes lipid membranes with positively charged histidine side chains located on one side of its α-helix at the weakly acidic pH in endosomes; or the GALA peptide,39 a glutamic acid-rich anionic EDP that disrupts lipid membranes in response to its structural conversion from a random coil to an α-helix at the endosomal pH. Accordingly, we obtained three CPP/EDP-modified micelles, referred to as KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC, via thiol-maleimide conjugation (Fig. 3A). From RP-HPLC and MALDI-TOF MS analyses (Fig. S3†), the MAL conversions of KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC were estimated to be 47/50, 33/56, and 40/44%, respectively (Fig. 3B). The zeta potential measurements indicated that the surface charges of KAibA/EED4-MC and KAibA/LAH4-L1-MC were positive (24 and 25 mV), while that of KAibA/GALA-MC was neutral (7 mV) due to the negative charge of the GALA peptide (Fig. 3C). According to DLS measurements, KAibA/EED4-MC and KAibA/LAH4-L1-MC were similar in hydrodynamic diameter (110 and 94 nm) and PDI (0.10 and 0.17), whereas KAibA/GALA-MC exhibited a larger diameter (331 nm) and higher PDI (0.21) than the other two micelles (Fig. 3D–F).In the atomic force microscopy (AFM) height images, all CPP/EDP-modified micelles showed a similar globular shape, although KAibA/GALA-MC was apparently larger in diameter than the others (Fig. 3G–I). Similar globular morphologies were also observed for KAibA-MC and UM-MC (Fig. S4†).
Cellular uptake of the CPP/EDP-modified micelles via endocytosis in the model plant
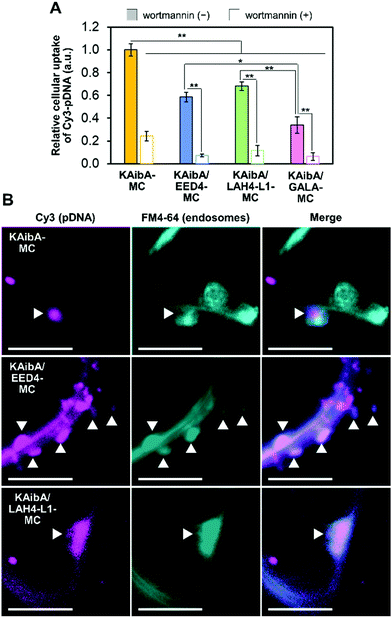
We prepared KAibA-MC, KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC from fluorescently labeled pDNA (Cy3-pDNA) to evaluate their cellular uptake. The fluorescent labeling did not greatly affect the particle size and size distribution of the micelles (Fig. S5†). As a plant tissue used for cellular uptake experiments, A. thaliana cotyledon (leaf of seedlings) was selected because the cellular uptake of a peptide/pDNA complex was mainly observed in the cotyledon rather than in the root for A. thaliana seedlings.11 We introduced fluorescently labeled micelles into the cotyledons via the vacuum/compression method. After incubation for 12 h, the cotyledons were treated with cell wall-degrading enzymes to remove the micelles entrapped in the cell walls. Accordingly, protoplasts, plant cells lacking cell walls, were isolated. The protoplast was intensively washed to remove the micelles that were nonspecifically absorbed on the cell surface and then used for confocal laser scanning microscopy (CLSM) to confirm the intracellular Cy3-pDNA. According to the CLSM images of the protoplast, KAibA-MC was observed in the vacuole, while KAibA/EED4-MC and KAibA/LAH4-L1-MC appeared to be located in the cytosol (Fig. S6A†). KAibA/GALA-MC was rarely observed in the protoplast, implying poor cellular uptake (Fig. S6A†). The Cy3-pDNA was extracted from the protoplast, and its fluorescence intensity was measured to quantify the cellular uptake efficiency of each micelle. All CPP/EDP-modified micelles (KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC) exhibited lower efficiency than the CPP-modified micelle (KAibA-MC) (Fig. 4A), likely because less CPP (the KAibA peptide, responsible for cellular uptake) was conjugated to the maleimide groups in the former micelles than in the latter (Fig. 2B and 3B). Comparing the CPP/EDP-modified micelles showed that KAibA/EED4-MC and KAibA/LAH4-L1-MC had higher uptake efficiency than KAibA/GALA-MC (Fig. 4A). This difference could be related to the size-exclusion limit of the cell wall, which was reported to be ∼50 nm.49,50 Specifically, fluorescently labeled KAibA/EED4-MC and KAibA/LAH4-L1-MC with diameters of 40–300 nm were still allowed to permeate through the cell wall, but KAibA/GALA-MC with diameters of 100–1000 nm was not (Fig. S5A†).To explore the cellular internalization pathways of the micelles, we assessed the uptake efficiency in the presence of wortmannin, a phosphatidylinositol 3-kinase inhibitor. In plant cells, wortmannin inhibits endocytosis, likely by inducing the aggregation of clathrin-coated pits at the plasma membrane.51 Prior to the cellular uptake experiments, we confirmed that the presence of wortmannin did not affect the particle size and size distribution of the fluorescently labeled micelles (Fig. S5B†). The cellular uptake of all micelles was significantly suppressed by wortmannin (Fig. 4A and S6B†), suggesting that endocytosis is the major internalization pathway for micelles in A. thaliana cotyledons. This finding is consistent with our previous data on the cellular uptake of a peptide/pDNA complex into A. thaliana protoplasts.27 To further probe the cellular internalization of the micelles, we employed FM4-64, a plasma membrane-staining fluorescent dye that has often been used as a marker for endocytosis in plants. The cotyledon was infiltrated with fluorescently labeled KAibA-MC, KAibA/EED4-MC, or KAibA/LAH4-L1-MC and subsequently incubated for 3 h in the presence of FM4-64. In the CLSM images, we observed FM4-64-stained particles of various sizes (Fig. 4B), which can be attributed to endosomal compartments including the trans-Golgi network (which is equivalent to the early endosome and thus referred to as TGN/EE) and the prevacuolar compartment (which is equivalent to the late endosome and thus referred to as PVC/LE).52 Some FM4-64-stained particles colocalized with Cy3 fluorescence signals (Fig. 4B), suggesting that the micelles were internalized into the cell through the endocytic pathway. However, the colocalization of KAibA/GALA-MC with endosomal compartments could not be detected, likely due to insufficient cellular uptake (Fig. 4A).
Subcellular localization of the CPP/EDP-modified micelles in the model plant
To evaluate the cytosolic translocation of the micelles following endocytic uptake, we introduced fluorescently labeled micelles into the cotyledons of transgenic A. thaliana seedlings expressing YFP in the cytosol.17 After 12 h of incubation, the colocalization of the micelle and YFP was quantitatively assessed through CLSM observation. KAibA-MC was distributed throughout the vacuole and did not colocalize with cytosolic YFP (Fig. 5A and B). To ensure the localization of KAibA-MC in the vacuole, we infiltrated wild-type A. thaliana cotyledons with KAibA-MC and then stained them with AM-BCECF, a vacuole-staining dye. At 12 h post infiltration, the colocalization of KAibA-MC with vacuoles was confirmed in the CLSM image (Fig. 5C). In contrast, KAibA/EED4-MC and KAibA/LAH4-L1-MC were mainly located in the cytosol (Fig. 5A and B), indicating that the EED4 and LAH4-L1 peptides can destabilize endosomes to promote the cytosolic translocation of micelles in plant cells. However, the CLSM images for KAibA/GALA-MC showed punctate Cy3 fluorescence signals that were not localized in the cytosol (Fig. 5A), which might be ascribed to the poor cellular uptake of KAibA/GALA-MC (Fig. 4A) and/or the dysfunction of GALA peptide. We further confirmed the translocation of KAibA/EED4-MC and KAibA/LAH4-L1-MC to the nucleus at 24 h post infiltration (Fig. S7†).The α-helical conformation of LAH4-L1 and GALA peptides at weakly acidic endosomal pH is essential for their endosome-disrupting ability,38,40 while the EED4 peptide is considered to destabilize lipid membranes regardless of its confromation.41 To investigate the secondary structure of EDPs, we prepared EDP-modified micelles (EED4-MC, LAH4-L1-MC, and GALA-MC) (Fig. S8†) and compared their circular dichroism (CD) spectra with those of unmodified micelles (UM-MC). The CD measurements were performed at pH values ranging from 7.4 to 5.5, which corresponded to the pH of the transfection solution and the plant endosome (TGN/EE),34 respectively. The CD spectra of EED4-MC showed that the EED4 peptide did not adopt specific secondary structures at pH 7.4–5.5 (Fig. 5D). The LAH4-L1-MC exhibited distinctive CD spectra with negative peaks at 208 and 222 nm, which are characteristic of an α-helical conformation, at pH 7.4–5.5 (Fig. 5D), similar to the free (i.e., nonconjugated) LAH4-L1 peptide (Fig. S9†). This result indicates that the LAH4-L1 peptide maintained the α-helical conformation at neutral and weakly acidic pH even when it was conjugated to the micelle. In contrast, the GALA peptide did not form an α-helical structure at pH 7.4–5.5 when it was conjugated to the micelle (Fig. 5D), although it was observed to adopt an α-helical below pH 6.5 in a free state (Fig. S9†). This loss of the α-helical conformation was possibly due to the electrostatic interaction between the anionic GALA peptide and the cationic MAL-TEG-(KH)14 in the micelle. The presence of the α-helical conformation of EDPs may reflect the different subcellular localization of the micelles. KAibA/LAH4-L1-MC, in which the LAH4-L1 peptide was able to maintain the α-helical conformation necessary for its function, was successfully translocated to the cytosol, whereas KAibA/GALA-MC, in which the GALA peptide was unable to adopt the α-helical conformation to exert its function, was not. These findings highlight the importance of an appropriate choice of EDP in micelle-mediated gene delivery to plants.
We next investigated the effect of endosomal pH on the cytosolic translocation of KAibA/EED4-MC and KAibA/LAH4-L1-MC. The EED4 peptide was expected to exert its endosome-disrupting ability regardless of endosomal pH, while the LAH4-L1 peptide was expected to destabilize endosomal membranes below pH 6.0 in response to the protonation of its histidine sidechains (their pKa was reported to be 5.7–6.2).40 We utilized concanamycin A (ConcA), a specific inhibitor of vacuolar-type H+-ATPase (V-ATPase). Notably, a low concentration of ConcA (100–500 nM) has been shown to cause a significant alkalization of TGN/EE from pH 5.5–6.3 to ∼6.9 without blocking vesicle transport to the vacuole.35 The A. thaliana cotyledon expressing YFP was treated with ConcA (250 nM) followed by infiltration with the micelles and subsequent incubation for 12 h in the presence of ConcA (250 nM). The particle size and size distribution of fluorescently labeled KAibA/EED4-MC and KAibA/LAH4-L1-MC were not considerably affected by ConcA (Fig. S5B†). The CLSM images clearly indicated the cytosolic location of KAibA/EED4-MC and the vacuolar accumulation of KAibA/LAH4-L1-MC (Fig. 5E), suggesting that the ConcA-induced alkalization of TGN/EE did not affect the endosomal escape ability of KAibA/EED4-MC but counteracted that of KAibA/LAH4-L1-MC. These observations validated that the EED4 and LAH4-L1 peptides on the micelle surface can disrupt endosomes in pH-independent and pH-dependent manners, respectively.
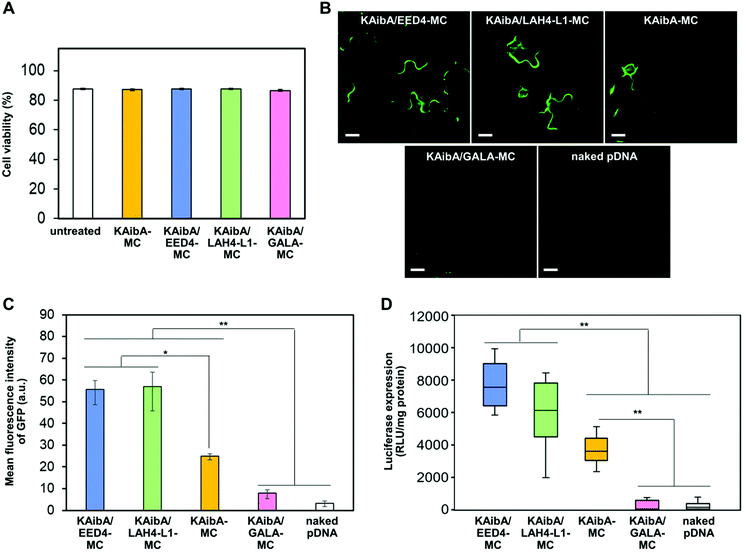
Cytotoxicity and transfection efficiency of the CPP/EDP-modified micelles in the model plant
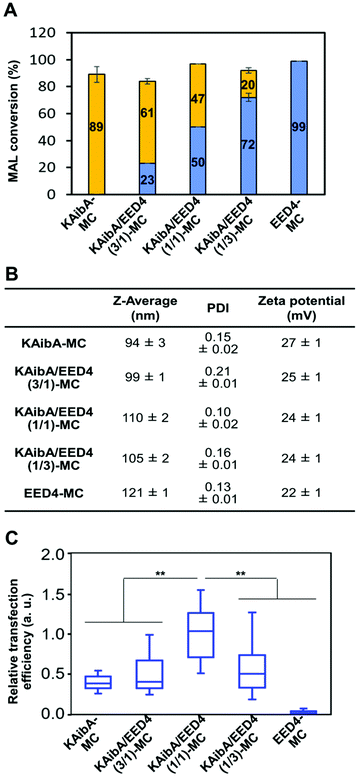
We tested the cytotoxicity of KAibA-MC, KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC to A. thaliana seedlings through the Evans blue assay. The seedlings were treated with each micelle under the same conditions as the transfection experiments described below. The micelle-treated seedlings exhibited similar viability to the untreated seedlings (Fig. 6A), indicating that not all micelles caused cytotoxicity. Next, we compared the transfection efficiency of the micelles in A. thaliana seedlings using GFP- or luciferase-coding pDNA. Each micelle containing the GFP-coding pDNA exhibited similar physicochemical properties (zeta potential, hydrodynamic diameter, and PDI) to the corresponding micelle containing the luciferase-coding pDNA (Table S1†). The seedlings were transfected with micelles containing GFP- or luciferase-coding pDNA and incubated for 24 h prior to CLSM observations to confirm GFP expression or luciferase assays to quantify luciferase expression. According to the GFP expression level estimated from the CLSM images of the transfected plants (Fig. 6B), KAibA/EED4-MC and KAibA/LAH4-L1-MC exhibited significantly higher transfection efficiency than KAibA-MC, while KAibA/GALA-MC demonstrated comparable efficiency to a control (naked pDNA) (Fig. 6C). The efficiency based on luciferase expression showed a similar trend to that based on GFP expression (Fig. 6D). Although KAibA-MC exhibited the highest cellular uptake efficiency among the micelles (Fig. 4A), it mainly accumulated in the vacuole lumen (Fig. 5A, C, and S6A†) and could be degraded by vacuolar enzymes, resulting in lower transfection compared to KAibA/EED4-MC and KAibA/LAH4-L1-MC (Fig. 6B and C). In contrast, KAibA/EED4-MC and KAibA/LAH4-L1-MC were successfully translocated to the cytosol (Fig. 5A and S6A†) due to the endosomal escape ability of the EED4 and LAH4-L1 peptides, leading to their significantly enhanced transfection (Fig. 6B and C). The lowest efficiency of KAibA/GALA-MC among the micelles, which was equal to that of the control, can be explained by its insufficient cellular internalization, probably due to its large hydrodynamic diameter (Fig. 4A), and/or by the dysfunction of the GALA peptide resulting from the loss of its α-helical conformation (Fig. 5D).To verify the contribution of the KAibA peptide, we evaluated the transfection efficiency of the EDP-modified micelles (EED4-MC, LAH4-L1-MC, and GALA-MC), which did not contain the KAibA peptide. All EDP-modified micelles were significantly less effective than KAibA-MC for transfection (Fig. S10†), even though KAibA-MC showed lower efficiency than KAibA/EED4-MC and KAibA/LAH4-L1-MC owing to the lack of endosomal escape ability (Fig. 6C and D). These results suggest that, as well as the endosomal escape ability of the EDPs, the function of the KAibA peptide enabling efficient cellular internalization is essential for transfection. This finding led us to further examine the optimal ratio of CPP/EDP for micelle-mediated transfection. We focused on the combination of the KAibA and EED4 peptides and prepared a series of micelles with different KAibA/EED4 conversion ratios (Fig. 7A): “KAibA-MC”, in which 89% of MAL was conjugated to the KAibA peptide; “KAibA/EED4 (3/1)-MC”, in which 61% and 23% of MAL was conjugated to the KAibA and EED4 peptides, respectively; “KAibA/EED4 (1/1)-MC”, in which 47% and 50% of MAL was conjugated to the KAibA and EED4 peptides, respectively; “KAibA/EED4 (1/3)-MC”, in which 20% and 72% of MAL was conjugated to the KAibA and EED4 peptides; “EED4-MC”, in which 99% of MAL was conjugated to the EED4 peptide. The DLS and zeta potential measurements of these micelles indicated that the KAibA/EED4 conversion ratio did not severely affect their physicochemical properties (Fig. 7B). According to the comparative transfection study using the seedlings, KAibA/EED4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)-MC exhibited the highest efficiency among the micelles (Fig. 7C), suggesting that both KAibA and EED4 peptides would synergistically function and contribute to the transfection.
1)-MC exhibited the highest efficiency among the micelles (Fig. 7C), suggesting that both KAibA and EED4 peptides would synergistically function and contribute to the transfection.
Conclusions
A robust approach to overcome endosomal entrapment has yet to be established for DNA delivery into plants, even though various strategies have been reported for mammalian cells. In this study, we have described a dual CPP/EDP-modified micelle system that can escape from the endosomal/vacuolar degradation pathway and enable efficient DNA delivery into intact plants without causing severe cytotoxicity. Our data highlight the synergistic contribution of CPP and EDP and the importance of appropriate selection for the successful micelle-mediated transfection of plants. Notably, the CPP/EDP-modified micelles achieved efficient endosomal escape in plant cells, where the proton sponge effect commonly used for mammalian cells was not apparent. The present study implies that micelle-mediated transfection may be hindered by other steps, including cell wall penetration and DNA release, as well as by endosomal entrapment. Addressing these issues could be future directions for further enhancement of transfection. We previously reported a zwitterionic polypeptide that disrupted the cellulose network in the cell walls of living plant cells and a reducible DNA-binding peptide that enabled efficient DNA release in response to intracellular glutathione.14,53 The integration of these peptides into the present micelle system would be useful to promote cell wall penetration and DNA release, leading to more efficient transfection of plants. While we have utilized the micelle system for DNA delivery to the nucleus, it may also be applicable to chloroplast- or mitochondrion-targeting DNA delivery by modifying its surface with organelle-targeting peptides. We envision that the combination of endosome-escaping micelles and organelle-targeting peptides represents a promising strategy for achieving highly selective and efficient DNA delivery to specific target organelles in intact plants.Experimental
Peptides, pDNA, and plants
Peptides including MAL-TEG-(KH)14 (sequence: MAL-TEG-KHKHKHKHKHKHKHKHKHKHKHKHKHKH-NH2; Mw: 4130), BP100 (sequence: Ac-CKKLFKKILKYL-NH2; Mw: 1566), KAibA (sequence: Ac-CKXAKXAKXA-NH2, X = α-aminoisobutyric acid (Aib); Mw: 1015), Tat (sequence: Ac-RKKRRQRRRC-NH2; Mw: 1483), EED4 (sequence: CGWWG-OH; Mw: 608), LAH4-L1 (sequence: Ac-CKKALLAHALHLLALLALHLAHALKKA-NH2; Mw: 2923), and GALA (sequence: WEAALAEALAEALAEHLAEALAEALEALAC-NH2; Mw: 3062), were obtained from the Research Resources Division of RIKEN Center for Brain Science (Wako, Japan). The purity of each peptide, determined by RP-HPLC analysis, was over 95%. The plasmids used in this study encoded an engineered Oplophorus luciferase (Nluc) or a green fluorescent protein (GFP) gene with the cauliflower mosaic virus 35S promoter and the Agrobactrium tumefaciens NOS terminator (p35S-Nluc-tNOS or p35S-GFP-tNOS).11,54 Detailed information on the plasmid vector is available on the Addgene Repository website (http://n2t.net/addgene:80127; Addgene plasmid #80127; RRID: Addgene_80127). Wild-type (Col-0) or transgenic (YFP-expressing) Arabidopsis thaliana (a model dicot plant species) seedlings were grown under previously described conditions.11,17 Seedlings 7–10 days after germination were used for the experiments.Preparation and characterization of MAL-TEG-(KH)14/pDNA micelles
MAL-TEG-(KH)14 aqueous solutions (2 mg mL−1) were added to Milli-Q water containing pDNA (20 μg) at various N/P ratios. The final volume and pDNA concentration were fixed to 800 μL and 25 μg mL−1, respectively. The solution was vortexed and incubated for 30 min at 25 °C to stabilize the MAL-TEG-(KH)14/pDNA complexes. After incubation, MAL-TEG-(KH)14/pDNA complex solutions (800 μL), prepared at various N/P ratios, were used for characterization by EMSA, DLS and zeta potential measurements as previously reported.27Preparation and characterization of peptide-modified micelles
The MAL-TEG-(KH)14/pDNA micelle, prepared at N/P 2, was modified with CPP in HEPES buffer (5 mM, pH 7.6) to obtain CPP-modified micelles. The final concentration of MAL-TEG-(KH)14 was fixed to 5 μM, whereas that of CPP was adjusted to 6 μM. The mixture (final volume, 820 μL) was stirred at 25 °C and 1200 rpm for 1 h with a mixer (Thermomixer comfort, Eppendorf, Hamburg, Germany). To obtain CPP/EDP-modified micelles, MAL-TEG-(KH)14/pDNA micelles (prepared at N/P 2) were simultaneously modified with KAibA and EDP in HEPES buffer (5 mM, pH 7.4). The final concentration of MAL-TEG-(KH)14 was 5 μM, whereas the concentrations of the KAibA peptide and EDP were adjusted to 3 μM. The mixture (820 μL) was stirred at 25 °C and 1200 rpm for 1 h with the mixer. The reaction mixture containing each peptide-modified micelle was directly used for characterization and transfection experiments. The micelles were characterized through MALDI-TOF MS, RP-HPLC, DLS, zeta potential, and AFM analyses as previously reported.27,43Cellular uptake of peptide-modified micelles in plants
Peptide-modified micelles (KAibA-MC, KAibA/EED4-MC, KAibA/LAH4-L1-MC, and KAibA/GALA-MC) were prepared from Cy3-labeled pDNA, which was obtained using a Label IT Nucleic Acid Labeling Kit (Mirus Bio, LLC, Madison, WI, USA) by the method described above. The fluorescently labeled micelles were analyzed by DLS as described above. Approximately 300 cotyledons were obtained from A. thaliana seedlings (7–10 days after germination) and transferred into a 1.5 mL microtube. Then, the cotyledons were infiltrated with a solution (400 μL) containing each fluorescently labeled micelle (Cy3-pDNA = 10 μg) alone or in combination with wortmannin (30 μM) (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) via the previously described vacuum/compression method, followed by incubation at 22 °C for 12 h in the presence or absence of wortmannin (30 μM). To remove the micelles aggregated in intracellular spaces or entrapped in the cell wall, the cotyledons were treated with an enzyme solution consisting of cellulase R10 (1.5% w/v), macerozyme R10 (0.4% w/v), mannitol (0.4 M), KCl (20 mM), CaCl2 (10 mM), bovine serum albumin (0.1% w/v), and MES (20 mM, pH 5.7) for 2 h with rotation at 20 rpm. The enzyme solution was filtered through nylon mesh (95 μm) and centrifuged at 1000 rpm for 5 min to obtain protoplasts in pellets. The protoplast pellets were intensively washed with “solution A” containing mannitol (0.4 M), CaCl2 (70 mM), and MES (20 mM, pH 5.7) to remove the micelles remaining on the cell surface prior to CLSM observation with LSM 880 (Carl Zeiss, Oberkochen, Germany) at excitation/emission (Ex/Em) wavelengths of 561/570–620 nm (for Cy3) and 561/650–735 nm (for chlorophyll). To extract the intracellular Cy3-pDNA, the protoplasts were lysed by sonication in Milli-Q and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm for 5 min. The lysate (5 μL) was used to measure the fluorescence intensity of Cy3-pDNA with an FP-8500 spectrophotometer (JASCO, Tokyo, Japan) at Ex/Em wavelengths of 550/570 nm. The lysate (2 μL) was mixed with Bradford reagent (100 μL) (APRO SCIENCE, Tokushima, Japan) to quantify the protein amount based on the absorbance at 595 nm. The fluorescence intensity (RFU) was divided by the protein amount (mg protein) to obtain the “RFU/mg protein” value. Each RFU per mg protein value was normalized to the average value obtained from 6 biologically independent samples transfected with KAibA (one sample was obtained from ∼300 cotyledons). The normalized value was considered to be the relative cellular uptake of Cy3-pDNA. To trace the endocytosis-mediated uptake of the micelles, a solution containing the fluorescently labeled micelle (200 μL, Cy3-pDNA = 5 μg) was infiltrated into A. thaliana cotyledons (obtained 7–10 days after germination) as described above. The infiltrated cotyledons were incubated with FM4-64 (20 μM) (Thermo Fisher Scientific. Waltham, MA, USA) at 22 °C for 3 h. After washing with Milli-Q water, the cotyledon was subjected to CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 561/560–595 nm (for Cy3) and 488/660–695 nm (for FM4-64).
500 rpm for 5 min. The lysate (5 μL) was used to measure the fluorescence intensity of Cy3-pDNA with an FP-8500 spectrophotometer (JASCO, Tokyo, Japan) at Ex/Em wavelengths of 550/570 nm. The lysate (2 μL) was mixed with Bradford reagent (100 μL) (APRO SCIENCE, Tokushima, Japan) to quantify the protein amount based on the absorbance at 595 nm. The fluorescence intensity (RFU) was divided by the protein amount (mg protein) to obtain the “RFU/mg protein” value. Each RFU per mg protein value was normalized to the average value obtained from 6 biologically independent samples transfected with KAibA (one sample was obtained from ∼300 cotyledons). The normalized value was considered to be the relative cellular uptake of Cy3-pDNA. To trace the endocytosis-mediated uptake of the micelles, a solution containing the fluorescently labeled micelle (200 μL, Cy3-pDNA = 5 μg) was infiltrated into A. thaliana cotyledons (obtained 7–10 days after germination) as described above. The infiltrated cotyledons were incubated with FM4-64 (20 μM) (Thermo Fisher Scientific. Waltham, MA, USA) at 22 °C for 3 h. After washing with Milli-Q water, the cotyledon was subjected to CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 561/560–595 nm (for Cy3) and 488/660–695 nm (for FM4-64).
Subcellular localization of peptide-modified micelles in plants
Using the abovementioned method, a solution containing the fluorescently labeled micelles (200 μL, Cy3-pDNA = 5 μg) was infiltrated into the cotyledon of transgenic A. thaliana seedlings (7–10 days after germination), which expressed YFP in the cytosol. After incubation at 22 °C for 12 h, the cotyledon was washed with Milli-Q and subjected to CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 561/570–625 nm (for Cy3) and 488/500–540 nm (for YFP). The confocal imaging data obtained from 5 biologically independent samples for each micelle were used to quantify the Cy3 fraction colocalized with YFP. Specifically, using Fiji ImageJ software, the colocalization fraction in each confocal image was calculated by dividing the “colocalized area”, where the Cy3 and YFP fluorescence overlapped, by the “Cy3 area”, where the Cy3 fluorescence was detected. To examine the effect of endosomal pH on subcellular localization, transgenic cotyledons were infiltrated with a solution (200 μL) containing fluorescently labeled micelles (KAibA/EED4-MC or KAibA/LAH4-L1-MC, Cy3-pDNA = 5 μg) and concanamycin A (200 nM) (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). After incubation at 22 °C for 12 h, the cotyledon was washed with Milli-Q and used for CLSM observations under the abovementioned conditions. For fluorescence imaging of the localization of KAibA-MC to the vacuole, cotyledons obtained from wild-type A. thaliana seedlings (7–10 days after germination) were infiltrated with a solution (200 μL) containing the fluorescently labeled micelle (KAibA-MC, Cy3-pDNA = 5 μg) as described above. After incubation at 22 °C for 12 h, the cotyledon was treated with BCECF-AM (20 μM) (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for 10 min, washed with Milli-Q, and used for CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 488/500–545 nm (for BCECF-AM) and 561/570–610 nm (for Cy3). For fluorescence imaging of the localization of KAibA/EED4-MC or KAibA/LAH4-L1-MC to the nucleus, cotyledons obtained from wild-type A. thaliana seedlings (7–10 days after germination) were infiltrated with a solution (200 μL) containing the fluorescently labeled micelle (KAibA/EED4-MC or KAibA/LAH4-L1-MC, Cy3-pDNA = 5 μg) as described above. After incubation at 22 °C for 12 h, the cotyledon was treated with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (20 μg mL−1) (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for 10 min, washed with Milli-Q, and used for CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 405/450–530 nm (for Hoechst) and 561/570–625 nm (for Cy3).
342 (20 μg mL−1) (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for 10 min, washed with Milli-Q, and used for CLSM observations with an LSM 880 microscope at Ex/Em wavelengths of 405/450–530 nm (for Hoechst) and 561/570–625 nm (for Cy3).
Secondary structure of EDPs on the micelle surface
EDP-modified micelles were prepared by stirring a mixture (820 μL) composed of MAL-TEG-(KH)14/pDNA micelles (prepared at N/P 2), EDP (6 μM), and HEPES (5 mM, pH 7.4) at 25 °C and 1200 rpm for 1 h. The MAL conversion and physicochemical properties of each EDP-modified micelle were determined by MALDI-TOF MS, RP-HPLC, DLS, and zeta potential analyses in the abovementioned method. The pH of the solution containing the EDP-modified micelle (400 μL) was adjusted to 7.4, 6.5, and 5.5 by the addition of sodium hydroxide. The pH-adjusted solution was transferred into a quartz cuvette with a 0.1 cm path length to measure CD spectra using a J-820 spectropolarimeter (Jasco, Tokyo, Japan). The spectra were recorded from 190 to 240 nm at 25 °C under a nitrogen atmosphere. Data are reported as the mean residual mass ellipticity (deg cm2 dmol−1). As controls, the spectra of free EDPs, which were not conjugated to the micelle, were measured under the abovementioned conditions.Cytotoxicity of peptide-modified micelles to plants
Four seedlings of wild-type A. thaliana (at 7–8 days after germination) were infiltrated with a solution (130 μL) containing each peptide-modified micelle (KAibA-MC, KAibA/EED4-MC, KAibA/LAH4-L1-MC, or KAibA/GALA-MC) in a 1.5 mL microtube as described above. After incubation at 22 °C for 24 h on a 1/2 MS medium plate, the seedlings were treated with Evans blue solution (400 μL, 150 μg mL−1) at 25 °C for 30 min and then intensively washed with Milli-Q. The seedlings were then incubated with a water/methanol (50/50) mixture (200 μL) containing SDS (1%, w/v) at 25 °C for 30 min to extract Evans blue, followed by spectrophotometric quantification at 600 nm. Seedlings not treated with micelles (considered “untreated” samples) or boiled at 100 °C for 30 min (considered as “100% dead” samples) were used as controls. Data were obtained from 6 biologically independent samples (each sample consisted of 4 infiltrated seedlings).Transfection efficiency of micelles in plants
For transfection experiments based on GFP expression, peptide-modified micelles (KAibA-MC, KAibA/EED4-MC, KAibA/LAH4-L1-MC, or KAibA/GALA-MC) were prepared from GFP-coding pDNA (p35S-GFP-tNOS) according to the abovementioned method. A solution (130 μL) containing each peptide-modified micelle or naked pDNA (25 μg mL−1) was infiltrated into seedlings of wild-type A. thaliana (at 7–10 days after germination) as described above. The seedlings were incubated on a 1/2 MS medium plate at 22 °C for 24 h. Cotyledons from the transfected seedlings were used for CLSM observation with an LSM 880 microscope at Ex/Em wavelengths of 488/490–540 nm (for GFP). The GFP fluorescent signal in the transfected cells was quantified using Fiji ImageJ. Specifically, fluorescence intensities were determined from at least five different regions of interest in confocal images obtained from four independent experiments. The background signal from adjacent cells was subtracted from these fluorescence intensities to obtain the GFP expression levels in the transfected cells. For transfection experiments based on luciferase expression, peptide-modified micelles were prepared from luciferase-coding pDNA (p35S-Nluc-tNOS). A solution (130 μL) containing each peptide-modified micelle or naked pDNA (25 μg mL−1) was infiltrated into 10 seedlings of wild-type A. thaliana (7–10 days after germination) as described above. After 24 h of incubation on a 1/2 MS medium plate (at 22 °C), the luciferase expression level in the transfected seedlings was quantified by the previously reported luciferase assay.27 Data were obtained from 20 biologically independent samples (one sample consisted of 10 transfected seedlings) for each system.Optimization of KAibA/EED4 modification ratios for micelle-mediated transfection
The micelles, including KAibA-MC, KAibA/EED4-MC, and EED4-MC, were prepared from luciferase-coding pDNA (p35S-Nluc-tNOS) as described above. In addition to these micelles, KAibA/EED4 (3/1)-MC and KAibA/EED4 (1/3)-MC were prepared by mixing the MAL-TEG-(KH)14/pDNA micelles (prepared at N/P 2 using p35S-Nluc-tNOS) and the KAibA (4.5 or 1.5 μM) and EED4 (4.5 or 1.5 μM) peptides in HEPES buffer (820 μL, 5 mM, pH 7.4) at 25 °C and 1200 rpm for 1 h. The MAL conversion and physicochemical properties of the micelles were determined by MALDI-TOF MS, RP-HPLC, DLS, and zeta potential analyses in the abovementioned method. The transfection efficiency of the micelles was quantified by the previously described luciferase assay.Statistical analysis
The statistical significance of differences in the cellular uptake, Cy3/YFP colocalization, and GFP-based transfection experiments were evaluated by Welch's t-tests, while those in the luciferase-based transfection experiments were assessed by Mann–Whitney U-tests. The level of significance was set at P < 0.05 (*, two-tailed) and P < 0.01 (**, two-tailed) in Welch's t-test, whereas it was set at P < 0.05 (*, two-tailed) and P < 0.01 (**, two-tailed) in the Mann–Whitney U-test.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Grants-in-Aid from the Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; Grant No. JPMJER1602 to K. N.) and from the Japan Society for the Promotion of Science for Scientific Research (JSPS-KAKENHI; Grant No. JP19K15411 to T. M.). We acknowledge the Support Unit for Bio-Material Analysis, RIKEN Center for Brain Science Research Resources Division, for performing the peptide syntheses.References
- D. Nandy, A. Maity and A. K. Mitra, J. Biosci., 2020, 45, 30 CrossRef CAS.
- F. J. Cunningham, N. S. Goh, G. S. Demirer, J. L. Matos and M. P. Landry, Trends Biotechnol., 2018, 36, 882–897 CrossRef CAS.
- G. S. Demirer, H. Zhang, N. S. Goh, R. L. Pinals, R. Chang and M. P. Landry, Sci. Adv., 2020, 6, eaaz0495 CrossRef CAS.
- H. Zhang, G. S. Demirer, H. Zhang, T. Ye, N. S. Goh, A. J. Aditham, F. J. Cunningham, C. Fan and M. P. Landry, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7543–7548 CrossRef CAS.
- S.-Y. Kwak, T. T. S. Lew, C. J. Sweeney, V. B. Koman, M. H. Wong, K. Bohmert-Tatarev, K. D. Snell, J. S. Seo, N.-H. Chua and M. S. Strano, Nat. Nanotechnol., 2019, 14, 447–455 CrossRef CAS.
- G. S. Demirer, H. Zhang, J. L. Matos, N. S. Goh, F. J. Cunningham, Y. Sung, R. Chang, A. J. Aditham, L. Chio, M.-J. Cho, B. Staskawicz and M. P. Landry, Nat. Nanotechnol., 2019, 14, 456–464 CrossRef CAS.
- G. S. Demirer, H. Zhang, N. S. Goh, E. González-Grandío and M. P. Landry, Nat. Protoc., 2019, 14, 2954–2971 CrossRef CAS.
- X. Zhao, Z. Meng, Y. Wang, W. Chen, C. Sun, B. Cui, J. Cui, M. Yu, Z. Zeng, S. Guo, D. Luo, J. Q. Cheng, R. Zhang and H. Cui, Nat. Plants, 2017, 3, 956–964 CrossRef CAS.
- N. Mitter, E. A. Worrall, K. E. Robinson, P. Li, R. G. Jain, C. Taochy, S. J. Fletcher, B. J. Carroll, G. Q. Lu and Z. P. Xu, Nat. Plants, 2017, 3, 16207 CrossRef CAS.
- C. Thagun, J.-A. Chuah and K. Numata, Adv. Sci., 2019, 6, 1902064 CrossRef CAS.
- K. Midorikawa, Y. Kodama and K. Numata, Sci. Rep., 2019, 9, 271 CrossRef.
- T. Macmillan, A. Ziemienowicz, F. Jiang, F. Eudes and I. Kovalchuk, Plant Biotechnol. Rep., 2019, 13, 11–23 CrossRef.
- T. Yoshizumi, K. Oikawa, J.-A. Chuah, Y. Kodama and K. Numata, Biomacromolecules, 2018, 19, 1582–1591 CrossRef CAS.
- J.-A. Chuah and K. Numata, Biomacromolecules, 2018, 19, 1154–1163 CrossRef CAS.
- M. Lakshmanan, T. Yoshizumi, K. Sudesh, Y. Kodama and K. Numata, Plant Biotechnol., 2015, 32, 39–45 CrossRef CAS.
- J.-A. Chuah, T. Yoshizumi, Y. Kodama and K. Numata, Sci. Rep., 2015, 5, 7751 CrossRef CAS.
- K. Numata, M. Ohtani, T. Yoshizumi, T. Demura and Y. Kodama, Plant Biotechnol. J., 2014, 12, 1027–1034 CrossRef CAS.
- M. Lakshmanan, Y. Kodama, T. Yoshizumi, K. Sudesh and K. Numata, Biomacromolecules, 2013, 14, 10–16 CrossRef CAS.
- T. Mizuno, M. Miyashita and H. Miyagawa, J. Pept. Sci., 2009, 15, 259–263 CrossRef CAS.
- A. Chugh, E. Amundsen and F. Eudes, Plant Cell Rep., 2009, 28, 801–810 CrossRef CAS.
- A. Chugh and F. Eudes, FEBS J., 2008, 275, 2403–2414 CrossRef CAS.
- C.-P. Chen, J.-C. Chou, B. R. Liu, M. Chang and H.-J. Lee, FEBS Lett., 2007, 581, 1891–1897 CrossRef CAS.
- N. Unnamalai, B. G. Kang and W. S. Lee, FEBS Lett., 2004, 566, 307–310 CrossRef CAS.
- N. Zheng, Z. Song, Y. Liu, L. Yin and J. Cheng, Front. Chem. Sci. Eng., 2017, 11, 521–528 CrossRef CAS.
- S. B. Gelvin, Microbiol. Mol. Biol. Rev., 2003, 67, 16–37 CrossRef CAS.
- J. C. Sanford, Physiol. Plant., 1990, 79, 206–209 CrossRef CAS.
- T. Miyamoto, K. Tsuchiya and K. Numata, Biomacromolecules, 2020, 21, 2735–2744 CrossRef CAS.
- A. K. Varkouhi, M. Scholte, G. Storm and H. J. Haisma, J. Controlled Release, 2011, 151, 220–228 CrossRef CAS.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS.
- Z. Meng, L. Luan, Z. Kang, S. Feng, Q. Meng and K. Liu, J. Mater. Chem. B, 2017, 5, 74–84 RSC.
- A. Mann, V. Shukla, R. Khanduri, S. Dabral, H. Singh and M. Ganguli, Mol. Pharm., 2014, 11, 683–696 CrossRef CAS.
- S. L. Lo and S. Wang, Biomaterials, 2008, 29, 2408–2414 CrossRef CAS.
- P. Midoux, C. Pichon, J.-J. Yaouanc and P.-A. Jaffrès, Br. J. Pharmacol., 2009, 157, 166–178 CrossRef CAS.
- J. M. Dragwidge, S. Scholl, K. Schumacher and A. R. Gendall, J. Cell Sci., 2019, 132, jcs226472 CrossRef CAS.
- J. Shen, Y. Zeng, X. Zhuang, L. Sun, X. Yao, P. Pimpl and L. Jiang, Mol. Plant, 2013, 6, 1419–1437 CrossRef CAS.
- A. J. Mason, C. Leborgne, G. Moulay, A. Martinez, O. Danos, B. Bechinger and A. Kichler, J. Controlled Release, 2007, 118, 95–104 CrossRef CAS.
- A. J. Mason, A. Martinez, C. Glaubitz, O. Danos, A. Kichler and B. Bechinger, FASEB J., 2006, 20, 320–322 CrossRef CAS.
- W. Li, F. Nicol and F. C. Szoka, Adv. Drug Delivery Rev., 2004, 56, 967–985 CrossRef CAS.
- N. K. Subbarao, R. A. Parente, F. C. Szoka, L. Nadasdi and K. Pongracz, Biochemistry, 1987, 26, 2964–2972 CrossRef CAS.
- J. Wolf, C. Aisenbrey, N. Harmouche, J. Raya, P. Bertani, N. Voievoda, R. Süss and B. Bechinger, Biophys. J., 2017, 113, 1290–1300 CrossRef CAS.
- P. Lönn, A. D. Kacsinta, X.-S. Cui, A. S. Hamil, M. Kaulich, K. Gogoi and S. F. Dowdy, Sci. Rep., 2016, 6, 32301 CrossRef.
- J.-A. Chuah, M. Odahara, Y. Kodama, T. Miyamoto, K. Tsuchiya, Y. Motoda, T. Kigawa and K. Numata, bioRxiv, 2019, 630301, DOI:10.1101/630301..
- T. Miyamoto, K. Tsuchiya and K. Numata, Biomacromolecules, 2019, 20, 653–661 CrossRef CAS.
- K. Numata, Y. Horii, K. Oikawa, Y. Miyagi, T. Demura and M. Ohtani, Sci. Rep., 2018, 8, 10966 CrossRef.
- E. Vivès, P. Brodin and B. Lebleu, J. Biol. Chem., 1997, 272, 16010–16017 CrossRef.
- K. Eggenberger, C. Mink, P. Wadhwani, A. S. Ulrich and P. Nick, ChemBioChem, 2011, 12, 132–137 CrossRef CAS.
- K. Terada, J. Gimenez-Dejoz, Y. Miyagi, K. Oikawa, K. Tsuchiya and K. Numata, ACS Biomater. Sci. Eng., 2020, 6, 3287–3298 CrossRef CAS.
- S. Futaki and I. Nakase, Acc. Chem. Res., 2017, 50, 2449–2456 CrossRef CAS.
- E. Corredor, P. S. Testillano, M.-J. Coronado, P. González-Melendi, R. Fernández-Pacheco, C. Marquina, M. R. Ibarra, J. M. de la Fuente, D. Rubiales, A. Pérez-de-Luque and M.-C. Risueño, BMC Plant Biol., 2009, 9, 45 CrossRef.
- T. Eichert, A. Kurtz, U. Steiner and H. E. Goldbach, Physiol. Plant., 2008, 134, 151–160 CrossRef CAS.
- E. Ito, M. Fujimoto, K. Ebine, T. Uemura, T. Ueda and A. Nakano, Plant J., 2012, 69, 204–216 CrossRef CAS.
- A. L. Contento and D. C. Bassham, J. Cell Sci., 2012, 125, 3511–3518 CrossRef CAS.
- K. Tsuchiya, N. Yilmaz, T. Miyamoto, H. Masunaga and K. Numata, Biomacromolecules, 2020, 21, 1785–1794 CrossRef CAS.
- Y. Fujii and Y. Kodama, Plant Biotechnol., 2015, 32, 81–87 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr08183c |
| This journal is © The Royal Society of Chemistry 2021 |