DNA modification and visualization on an origami-based enzyme nano-factory†
Abstract
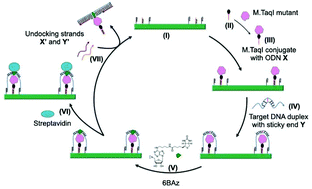
The past decade has seen enormous progress in DNA nanotechnology through the advent of DNA origami. Functionalizing the DNA origami for multiple applications is the recent focus of this field. Here we have constructed a novel DNA enzyme nano-factory, which modifies target DNA embedded on a DNA origami platform. The enzyme is programmed to reside in close proximity to the target DNA which enhances significantly the local concentration compared to solution-based DNA modification. To demonstrate this we have immobilized DNA methyltransferase M·TaqI next to the target DNA on the DNA origami and used this enzyme to sequence-specifically modify the target DNA with biotin using a cofactor analogue. Streptavidin binding to biotin is applied as a topographic marker to follow the machine cycle of this enzyme nano-factory using atomic force microscopy imaging. The nano-factory is demonstrated to be recyclable and holds the potential to be expanded to a multi-enzyme, multi-substrate operating system controlled by simple to complex molecules made of DNA, RNA or proteins.
- This article is part of the themed collection: Editor’s Choice: Recent breakthroughs in nanobiotechnology


 Please wait while we load your content...
Please wait while we load your content...