Open Access Article
Open Access ArticleNano-bio interface between human plasma and niosomes with different formulations indicates protein corona patterns for nanoparticle cell targeting and uptake†
Esther
Imperlini‡
 a,
Christian
Celia‡
a,
Christian
Celia‡
 b,
Armando
Cevenini‡
b,
Armando
Cevenini‡
 cd,
Annalisa
Mandola
cd,
Annalisa
Mandola
 de,
Maddalena
Raia
de,
Maddalena
Raia
 d,
Massimo
Fresta
d,
Massimo
Fresta
 f,
Stefania
Orrù
f,
Stefania
Orrù
 *de,
Luisa
Di Marzio
*de,
Luisa
Di Marzio
 *b and
Francesco
Salvatore
*b and
Francesco
Salvatore
 *cd
*cd
aIRCCS SDN, Napoli, Italy
bDipartimento di Farmacia, Università di Chieti-Pescara “G. d'Annunzio”, Chieti, Italy. E-mail: luisa.dimarzio@unich.it
cDipartimento di Medicina Molecolare e Biotecnologie Mediche, Università degli Studi di Napoli “Federico II”, Napoli, Italy. E-mail: salvator@unina.it
dCEINGE-Biotecnologie Avanzate S.c.a r.l., Napoli, Italy. E-mail: orru@uniparthenope.it
eDipartimento di Scienze Motorie e del Benessere, Università “Parthenope”, Napoli, Italy
fDipartimento di Scienze della Salute, Università “Magna Graecia” di Catanzaro, Campus Universitario “S. Venuta”, Catanzaro, Italy
First published on 5th March 2021
Abstract
Unraveling the proteins interacting with nanoparticles (NPs) in biological fluids, such as blood, is pivotal to rationally design NPs for drug delivery. The protein corona (PrC), formed on the NP surface, represents an interface between biological components and NPs, dictating their pharmacokinetics and biodistribution. PrC composition depends on biological environments around NPs and on their intrinsic physicochemical properties. We generated different formulations of non-ionic surfactant/non-phospholipid vesicles, called niosomes (NIOs), using polysorbates which are biologically safe, cheap, non-toxic and scarcely immunogenic. PrC composition and relative protein abundance for all designed NIOs were evaluated ex vivo in human plasma (HP) by quantitative label-free proteomics. We studied the correlation of the relative protein abundance in the corona with cellular uptake of the PrC-NIOs in healthy and cancer human cell lines. Our results highlight the effects of polysorbates on nano-bio interactions to identify a protein pattern most properly aimed to drive the NIO targeting in vivo, and assess the best conditions of PrC-NIO NP uptake into the cells. This study dissected the biological identity in HP of polysorbate-NIOs, thus contributing to shorten their passage from preclinical to clinical studies and to lay the foundations for a personalized PrC.
1. Introduction
Passive and active targeting are the two major mechanisms involved for the most proper delivery of payloads in selected cancer cells and tissues. Passive targeting takes advantage of the “enhanced permeability and retention effect” (EPR), which steers intravenously injected nanoparticles (NPs) to accumulate more in the tumor than in normal organs and tissues, due to the fenestrated endothelium of tumor blood vessels and to the lack of lymphatic drainage in the tumor.1 On the other hand, active targeting exploits the capability of molecules conjugated/adsorbed onto the surface of nanocarriers, such as antibodies or their fragments, to bind specific receptors/antigens over-expressed by cancer cells and tissues.2In the blood stream, systemically injected NPs interact with biological fluids and tissues; here the high free energy of NP surface is responsible for the binding of biomolecules, mainly proteins, able to form a layer, called protein corona (PrC);3 this latter represents a sort of bio-nanointerface, between NPs and biological systems mediating NP cytotoxicity, their cellular internalization and specific targeting to cancer cells.4,5
The qualitative and quantitative composition of PrC depends on the chemical nature, size, shape and surface charge of the NPs, besides the composition of the biological fluid encountered by the NPs.6 Protein coating may increase the size of bare synthetic and organic NPs (e.g. NPs made of metals, polystyrene, silica, lipids),7 whereas polyethylene glycol modification (PEGylation) partially modifies the adsorption of biomolecules on the NP surface8 and these changes depend on PEG configuration and properties, i.e. molecular weight, grafting density, random coil shape and chain entropy.9,10 In fact, proteins forming PrC can penetrate less dense PEG and interact with the NP surface; while dense grafted and planar PEG hampers the adsorption of proteins on the NP surface.11 These events depend on the curvature radius of the NPs; in fact, largest particles have a small curvature radius allowing proteins to interact deeply with the NP surface because PEG are unstretchable random coiled chains showing less grafting density.8 Furthermore, PrCs adsorbed on PEGylated NPs have been investigated, both in human and rodents, mainly ex vivo (static condition) rather than in vivo (dynamic condition): however further studies are needed as the two conditions determined different results in terms of protein composition and relative abundance.12,13
PrC confers additional biological properties to NPs14 by replacing the synthetic derivative with a new biological identity, i.e. the circulating PrC-NP complex, which effectively controls their biodistribution and targeting towards specific cells and tissues.15 As a matter of fact, proteins adsorbed on NPs form a dynamic layer that is continuously adsorbed and desorbed from NP surface. This phenomenon, named Vroman effect, depends on the affinity interactions between NP surface and PrC components but is independent from the protein abundance in the biological environment.16–18 This model has been widely debated and implies the existence of a highly dynamic PrC formed over time, where the total amount of adsorbed molecules remains relatively constant.18–20
Vroman effect also suggests the presence of two different corona layers adsorbed on the NP surface: hard and soft corona. The former is considered the first tightly bound layer of proteins that has a longer exchange time (many hours), whereas the latter is represented by the outer layer of proteins (not directly bound to NPs) that undergoes faster exchanges over time (seconds or minutes).21 In fact, soft corona, once formed after systemic injection, is completely removed in patients where NPs are recovered 10 minutes post injection13 and changes significantly in terms of abundance and composition for organic13,22 and synthetic PEGylated NPs.8,23 Given the high dynamicity of soft corona, optimized strategies for its isolation are challenging.13 On the other side, the internal hard corona, representing the real NP-biomolecule interface, plays a fundamental role in the biological interactions of NPs since it is usually recognized by the target tissue.24 Hence, PrC affects the biodistribution, the trafficking and the interaction of NPs with cell receptors,22 and may dictate, by manipulating the surface properties, their delivery,25 as well as the pharmacokinetic of payloads,14 thus representing an example of active natural targeting and offering new targeting opportunities.26
Alternative co-polymers to PEG, showing the same long-circulating properties and providing a similar stealth effect, are polysorbates; Tweens® 20 and 80 are used to synthesize non-ionic surfactant/non-phospholipid vesicles, also called non-ionic liposomes or niosomes (NIOs)27–29 that display physicochemical and biopharmaceutical properties similar to liposomes,30 such as delivering payloads with different physicochemical properties. NIOs are more stable than liposomes, and are low-cost, easy handling carriers31 that can be injected in vivo and used for anticancer therapies.32 Based on these features, NIOs can represent a suitable alternative to liposomes in pharmaceutical science of drug delivery with the relative advantage to avoid drawbacks, like aggregation, fusion, hydrolysis and oxidation, related to the use of phospholipids, as in liposomes.33
Hence, ex vivo and in vitro evaluation of PrC effect on the surface properties of NIOs may be instrumental for future in vivo applications. For this reason, the aim of this work is to study ex vivo the protein composition and the relative protein abundance of the PrC adsorbed on differently designed NIO nanoparticles, after incubation in human plasma (HP), and to evaluate the in vitro intracellular uptake and internalization of PrC-NIOs in healthy and cancer human cell lines. Present data can provide a further opportunity to simulate and modulate PrC properties on NIOs and to understand the effect of PEG-mimetic polysorbate moieties on nano-biomolecules interactions in order to identify a protein pattern devoted to the best effective targeting of NIOs in vivo.
2. Methods
2.1 Materials
Tween 20 (TW20), 9-(diethylamino)-5H-benzo [R] fenossazin-5H-one (Nile-Red, NR) and cholesterol (Chol) were obtained from Acros Organics (Acros Organics BVBA, Geel, Belgium). Isopropyl alcohol, Tween 21 (TW21), Tween 80 (TW80), 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (Hepes), human plasma (HP) and Nucleopore® track-etched polycarbonate membranes (cut-off 100 and 200 nm, 25 mm) were purchased from Sigma Aldrich (Milano, Italy). Oregon Green™ 488 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Oregon Green™ 488 DHPE) was obtained by Thermo Fisher Scientific (Waltham, MA, US). All other products and reagents were of analytical grade. HDF (human primary normal dermic fibroblasts), HeLa (human cervix adenocarcinoma) and HCT-116 (human colon carcinoma) cell lines were obtained from Cell Culture Facility of CEINGE – Biotecnologie Avanzate (Naples, Italy). DMEM (Dulbecco's modified Eagle's medium) RPMI (Roswell Park Memorial Institute) and McCoy's medium, fetal bovine serum (FBS), penicillin, streptomicyn and L-glutamin, trypsin/EDTA, MTT. Ammonium bicarbonate, dithiothreitol iodoacetamide were obtained from Sigma Aldrich (Milano, Italy).Colloid Blue Stain Reagent (Thermo Fisher Scientific, US). Dulbecco's phosphate buffered saline (DPBS), BDTMP100 blood collection system was obtained by Becton Dickinson (BD, NJ, USA).
2.2 Niosome preparation
Niosomes (NIOs) were made up from surfactants (TW20, TW21 or TW80) and Chol at different molar ratio (Table 1). TW20, TW21, and TW80 were used at a molar concentration below their critical micellar concentration (CMC) as previously reported.34,35 NIOs were synthesized using the thin layer evaporation method with some modifications as previously reported.34 Briefly, surfactants and Chol were dissolved in the organic solvent (chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol, 3
methanol, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The organic solvent was removed using a rotary evaporator (Heidolph, model 4000 type Laborota, Delchimica, Naples, Italy) at room temperature. The dried film was hydrated with Hepes buffer (10 mM, pH 7.4) and subsequently extruded at room temperature using a Lipex Extruder® device (Vancouver, Canada) through polycarbonate-membrane filters having pore sizes of 200 nm (NIO-200) and 100 nm (NIO-100). Fluorescent NIOs for cellular uptake studies were obtained using the same procedure herein reported. The fluorescent dye Oregon Green (OG) was added to the mixture of surfactant/Chol at the final molar concentration of 0.1% (w/v). OG-NIOs were thus obtained and used for intracellular uptake experiments.
1, v/v). The organic solvent was removed using a rotary evaporator (Heidolph, model 4000 type Laborota, Delchimica, Naples, Italy) at room temperature. The dried film was hydrated with Hepes buffer (10 mM, pH 7.4) and subsequently extruded at room temperature using a Lipex Extruder® device (Vancouver, Canada) through polycarbonate-membrane filters having pore sizes of 200 nm (NIO-200) and 100 nm (NIO-100). Fluorescent NIOs for cellular uptake studies were obtained using the same procedure herein reported. The fluorescent dye Oregon Green (OG) was added to the mixture of surfactant/Chol at the final molar concentration of 0.1% (w/v). OG-NIOs were thus obtained and used for intracellular uptake experiments.
| Niosome components | Acylic chains | Ethylene oxide units (PEG)e | NIO1 | Niosome NIO2 (molar) | Formulations NIO3 (ratio) | NIO4 |
|---|---|---|---|---|---|---|
| a TW20: Tween 20. b TW21: Tween 21. c TW80: Tween 80. d Chol: cholesterol. e PEG: polyethylene glycol chains. | ||||||
| TW20a | Lauric acid (C12) | 20 | 1 | — | 0.5 | — |
| TW21b | Lauric acid (C12) | 4 | — | — | 0.5 | 0.5 |
| TW80c | Oleic acid (C18) | 20 | — | 1 | — | 0.5 |
| Chold | — | — | 1 | 1 | 1 | 1 |
2.3 Physicochemical characterization of niosomes
Average sizes, polydispersity index (PDI) of NIOs were evaluated by using dynamic light scattering (DLS) analysis, whereas zeta potential was measured by laser doppler electrophoresis (LDE). Samples were diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 v/v) in Hepes buffer (10 mM, pH 7.4), to avoid a multi-scattering phenomenon, and then analyzed by Malvern Zetasizer Nano ZS (Malvern Instruments Ltd, UK). Buffer was previously filtered through 0.45 μm cellulose filters (Millex® Syringe Filters, Millipore) to remove dust particles. Measurements were replicates of three different batches (5 replications for each batch). Results showed the hydrodynamic radius (nm), PDI and zeta potential (mV) as a function of electrophoretic mobility. The amount of surfactants self-assembled into the NIOs (w/w%) were quantified spectrophotometrically as previously reported.34
30 v/v) in Hepes buffer (10 mM, pH 7.4), to avoid a multi-scattering phenomenon, and then analyzed by Malvern Zetasizer Nano ZS (Malvern Instruments Ltd, UK). Buffer was previously filtered through 0.45 μm cellulose filters (Millex® Syringe Filters, Millipore) to remove dust particles. Measurements were replicates of three different batches (5 replications for each batch). Results showed the hydrodynamic radius (nm), PDI and zeta potential (mV) as a function of electrophoretic mobility. The amount of surfactants self-assembled into the NIOs (w/w%) were quantified spectrophotometrically as previously reported.34
2.4 Serum stability
The serum stability of NIOs was evaluated as previously reported.36 Briefly, NIOs (400 μl) were incubated with a solution (1 ml, final volume) containing HP and Hepes buffer (10 mM, pH 7.4) 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, v/v. A time course study was performed up to 24 h at room temperature under continuous shaking (400 rpm). At different time of incubation, 50 μl of samples was collected and replaced with fresh solution. Samples were collected at different times of incubation and changes of physicochemical parameters (average size and PDI) due to the interaction between plasma proteins and NIOs were analyzed by DLS as herein reported in the previous section. Sterile conditions were maintained during the experiments in order to avoid any potential bacterial contaminations that can affect sample analysis.
40, v/v. A time course study was performed up to 24 h at room temperature under continuous shaking (400 rpm). At different time of incubation, 50 μl of samples was collected and replaced with fresh solution. Samples were collected at different times of incubation and changes of physicochemical parameters (average size and PDI) due to the interaction between plasma proteins and NIOs were analyzed by DLS as herein reported in the previous section. Sterile conditions were maintained during the experiments in order to avoid any potential bacterial contaminations that can affect sample analysis.
2.5 Turbiscan Lab Expert® analysis of niosomes
NIOs were filled in a cylinder glass holder and analyzed using Turbiscan Lab Expert® as previously reported.37 The stability of NIOs was carried out by measuring the variation of backscattering (ΔBS) profiles after 1 h of analysis at room temperature. ΔBS of NIOs was calculated by applying equations reported in the software's manufacture and in a previous work.37 A pulsed near infrared LED at a wavelength of 880 nm; two different synchronous optical sensors received the light transmitted through and backscattered by samples at an angle of 180° and 45° with respect to the incident radiation were used for the analysis of samples. NIOs (10 ml) were scanned for 1 h for the full height of samples by the sensors herein reported, and the results were correlated as percentage to the light flux of two reference standards of polystyrene latex suspension (absence of transmission and maximum backscattering) and silicon oil (maximum transmission and absence of backscattering) used to set-up the instrument.2.6 HP collection and niosome incubation with HP
Human whole blood was obtained at the laboratories of CEINGE-Biotecnologie Avanzate of Naples, Italy, in accordance with the relevant laws and guidelines existing in Italy and in accordance with Helsinki tenets, after having obtained informed consent from blood plasma donors, and following the guidelines of the Ethics Committees of the University of Naples Federico II, of which CEINGE is part, and that of the Istituto Nazionale Tumori IRCCS “Fondazione G. Pascale”, Naples, Italy.Blood samples were collected using BDTMP100 blood collection system with K2EDTA-containing vials and protease inhibitors cocktail. After clot formation, plasma was isolated by centrifugation at 1000g for 5 min. After checking the absence of hemolysis, plasma samples were pooled to reduce the overall subject-to-subject variation and then aliquoted and stored at −80 °C, until further use. For analysis, the aliquots were thawed at 4 °C and then allowed to warm at room temperature.
Pooled plasma samples (450 μl) were incubated with 450 μl of NIO suspension (1.67 mg ml−1) at 37 °C for 1 h. Afterwards, PrC-NIO complexes were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. Each pellet was washed with PBS twice. After each washing, tubes were changed to reduce contamination from plasma proteins. Then, the pellets were resuspended in Laemmli buffer.
000g for 10 min. Each pellet was washed with PBS twice. After each washing, tubes were changed to reduce contamination from plasma proteins. Then, the pellets were resuspended in Laemmli buffer.
2.7 Electrophoretic separation and in gel digestion
Proteins resuspended in Laemmli buffer were incubated at 95 °C for 5 min, and then separated by 12% (v/v) SDS-PAGE. Protein bands were stained with colloid blue stain reagent (Thermo Fisher Scientific, USA). Gel image was acquired using the scanner GS-800 Calibrated Densitometer (Bio-Rad, USA) supported by PDquest 7.1 software (Bio-Rad Laboratories Srl, Segrate (MI), Italy).The whole gel lanes were manually cut to create 2 mm gel slices and treated as previously reported.38 Briefly, each slice was washed with acetonitrile and 50 mM ammonium bicarbonate. The protein bands were reduced in 10 mM dithiothreitol for 45 min at 56 °C and then alkylated in 55 mM iodoacetamide for 30 min in the dark at room temperature. Protein bands were submitted to enzymatic digestion using modified trypsin (Sigma-Aldrich, Germany) (10 ng μl−1) at 4 °C as previously reported.39 Peptide mixtures were extracted and resuspended for the following mass spectrometry analysis.39
2.8 LC-MS/MS analysis and protein identification/quantitation
Mass spectrometry analyses were performed using the LC/MSD Trap XCT Ultra (Agilent Technologies, Palo Alto, CA) equipped with an 1100 HPLC system and a chip cube (Agilent Technologies) as previously described.40Raw MS/MS data files were converted into a Mascot format text to identify proteins using Mascot software (Matrix Science, UK). The protein search was carried out against NCBI database out as previously reported.41 Briefly, the following standard parameters were used: Homo Sapiens; one missed cleavage; carboxyamidomethylation of Cys; partial Met oxidation and putative modification of Gln to pyro-Glu; mass tolerance of 300 ppm on precursor ions and 0.6 Da on the product ions. In particular, individual ion scores >43 indicate identity or extensive homology (p < 0.05).
To estimate plasma protein abundance in the corona of all NIO formulations, MS/MS outputs were analyzed by the Protein Discoverer platform (Thermo Scientific), interfaced with the in-house Mascot server, and submitted to label-free quantitation method based on spectral counting (SpC).42 For each identified protein, in fact, SpC was divided by protein length obtaining the spectral abundance factor (SAF) that, in turn, was normalized to the total sum of SpC, in a given lane, defining the normalized spectral abundance factor (NSAF). Moreover, SpC log ratio (RSC) was calculated to compare the expression of the same protein between two different NIO formulations.
2.9 Bioinformatics analysis
The identified proteins were classified according to the Database for Annotation, Visualization and Integrated Discovery (DAVID; version v6.7 (http://david.abcc.ncifcrf.gov/) as previously reported.43 This tool adopts the Fisher's exact test to measure the protein enrichment in annotation terms. If the p value is ≤0.05, a protein is considered significantly enriched in the annotation categories.2.10 Cell cultures
HDF (human primary normal dermic fibroblasts), HeLa (human cervix adenocarcinoma) and HCT-116 (human colon carcinoma) cell lines were obtained from Cell Culture Facility of CEINGE – Biotecnologie Avanzate (Naples, Italy).The three cell lines were cultured in 10 cm tissue culture plates containing specific culture media (DMEM for HDF, RPMI medium for HeLa, McCoy's medium for HCT-116), supplemented with 10% (v/v) of fetal bovine serum (FBS), 1% (v/v) penicillin, 1% (v/v) streptomycin and 1% (v/v) L-glutamine. Cells were grown in an incubator at 37 °C in a humidified atmosphere of 5% CO2 until they reached about 70–80% confluence. The culture media were then withdrawn and cells were washed twice with 1× DPBS without calcium and magnesium, detached by trypsin/EDTA, collected with the complete medium and centrifuged at 250g for 5 min at room temperature. The supernatant was discarded and the cells were resuspended in fresh medium, diluted and appropriately seeded as following reported. All operations were performed under laminar flow hood, in sterile conditions.
2.11 Cell culture treatments
For cell viability analyses at different concentration of NIOs, HCT-116 was seeded at a density of 9 × 104 cells per cm2 in 96-well plates. After 24 hours of incubation, culture medium was replaced with fresh medium (supplemented with 10% (v/v) FBS) containing NIOs. Cells were then incubated in standard culture conditions and assayed at 3 h and 24 h.For cell viability and uptake analyses with PrC-NIOs and bare NIOs, cells were seeded at a density of 3 × 104 cells per cm2 (HDF), 5 × 104 cells per cm2 (HeLa) or 9 × 104 cells per cm2 (HCT-116) in six-well plates. On the following day and 2 hours before NIO administration, culture medium was replaced with serum-free medium after washing twice with PBS. In the meanwhile, fluorescent NIOs were incubated with HP to allow the formation of PrC. In particular, as described above, a solution with 1.67 mg ml−1 of NIOs in Hepes (10 mM, pH 7.4) was added 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) to HP and incubated for 1 h at 37 °C under gently shaken at 250g. After incubation, in order to avoid any type of physical changes, NIOs were not centrifuged to eliminate the HP. The entire solution was diluted in serum-free medium and added to the cell culture at a final concentration of 20 μg ml−1 of NIOs and 1% (v/v) HP. Cells treated with bare NIOs were used as a control. To this aim, a solution with 1.67 mg ml−1 of NIOs in Hepes (10 mM, pH 7.4) was diluted in serum-free medium and added to cell culture at a final concentration of 20 μg ml−1. The HP was also added at 1% (v/v) final concentration in cell culture medium before the analysis. Another control was represented by untreated cells in serum-free medium with HP 1% (v/v). Cells were assayed after 3 h (viability and uptake) and 24 h (viability).
1 (v/v) to HP and incubated for 1 h at 37 °C under gently shaken at 250g. After incubation, in order to avoid any type of physical changes, NIOs were not centrifuged to eliminate the HP. The entire solution was diluted in serum-free medium and added to the cell culture at a final concentration of 20 μg ml−1 of NIOs and 1% (v/v) HP. Cells treated with bare NIOs were used as a control. To this aim, a solution with 1.67 mg ml−1 of NIOs in Hepes (10 mM, pH 7.4) was diluted in serum-free medium and added to cell culture at a final concentration of 20 μg ml−1. The HP was also added at 1% (v/v) final concentration in cell culture medium before the analysis. Another control was represented by untreated cells in serum-free medium with HP 1% (v/v). Cells were assayed after 3 h (viability and uptake) and 24 h (viability).
2.12 MTT assay
For cell viability analyses cells were seeded in 96-well plates. The cells were assayed by MTT as previously described.28,44 Briefly in each well the culture medium was replaced with 100 μl serum-free medium containing 0.5 mg ml−1 MTT dye and cells were incubated for 4 h at 37 °C. The medium was then removed, cells were washed with PBS and 100 μl of isopropanol were added to each well. After 10 min of gentle shaken, plates were put in a plate-reader and absorbance at 570 nm of wavelength was measured. Results obtained were normalized with respect to the negative control (untreated cells).2.13 Cellular uptake of niosomes
For the NIO uptake experiments, cells were seeded in six-well plates. After 3 h of incubation with different OG-NIOs, adherent cells were washed twice with magnesium and calcium-free PBS, detached from the plate with trypsin/EDTA, collected in PBS and centrifuged at 250g for 10 min at 4 °C to obtain cell pellets. The supernatant was removed and the cells were resuspended in 500 μl of PBS containing 1 mg ml−1 trypan blue (to quench extracellular fluorescence)45,46 and were analyzed by flow cytometry on a Beckman Coulter FC500. Cellular uptake was determined by measuring the mean fluorescence (MFI) of cells, that was measured in three independent replicate experiments, and opportunely elaborated. In particular the average of the untreated cell MFI was considered as background and was subtracted. Moreover, the MFI values were normalized with the respective fluorescence (excitation/emission (nm): 501/526) of the various formulations. The fluorescence curves at various concentrations (diluted in Hepes 10 mM, pH 7.4) were measured with an Enspire (PerkinElmer) spectrofluorimeter and the MFI were normalized with the mean value obtained with NIOs. In order to have normalized MFI related to the number of NIOs instead of μg of surfactants, the values were normalized with the NIO surface area (assuming that a niosome with a wider surface contains more molecules of fluorescent lipid). The surface areas were calculated from the respective mean Dh. Results are reported as mean values ± SD from three independent experiments. Statistical significance was determined by one-way two-tail paired t-test.3. Results and discussion
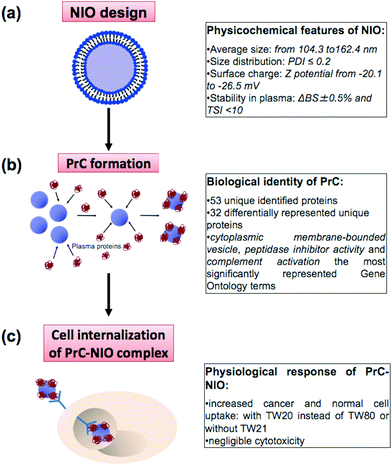
Nanotechnology and proteomics integrated with bioinformatics tools and cell biology allowed to set-up a multi-dimensional workflow able to combine: (i) niosome (NIO) design using different surfactants/cholesterol formulations, (ii) proteomic skills to define protein corona (PrC) composition and relative protein abundance in human plasma (HP) and (iii) the evaluation of the uptake and cytotoxicity in healthy and tumor human cell lines (Fig. 1).3.1 Physicochemical characterization of niosomes
Four NIO formulations, at 100 nm- and 200 nm-extrusion size, were synthesized using different Tween® derivatives (TW20, TW21 and TW80), and cholesterol (Chol) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Table 1). TW20 and TW80 were used alone or in combination with TW21 (Table 1). TW20, TW21 and TW80 differ both for the acylic chains of fatty acids (FAs) conjugated to the sorbitan backbone structure and for the number of polyethylene glycol (PEG) units (Table 1). In particular, TW20 and TW80 have the same units of PEG but the former contains saturated lauric acid (C12:0) and the latter unsaturated oleic acid (C18:1) conjugated to sorbitan; conversely, TW21 and TW20 have the same acylic chains (C12:0) but a different number of PEG chains (4 versus 20, respectively) (Table 1). Hence, the formulations adopted here allowed to cluster NIOs in two further types: TW21-lacking NIOs (NIO1 and NIO2) and TW21-containing NIOs (NIO3 and NIO4). We hypothesized that these differences could affect the PrC composition and the relative abundance of the proteins adsorbed on the surface of NIOs, as well as, obviously, the nano-bio interface of PrC-NIO complexes.
1) (Table 1). TW20 and TW80 were used alone or in combination with TW21 (Table 1). TW20, TW21 and TW80 differ both for the acylic chains of fatty acids (FAs) conjugated to the sorbitan backbone structure and for the number of polyethylene glycol (PEG) units (Table 1). In particular, TW20 and TW80 have the same units of PEG but the former contains saturated lauric acid (C12:0) and the latter unsaturated oleic acid (C18:1) conjugated to sorbitan; conversely, TW21 and TW20 have the same acylic chains (C12:0) but a different number of PEG chains (4 versus 20, respectively) (Table 1). Hence, the formulations adopted here allowed to cluster NIOs in two further types: TW21-lacking NIOs (NIO1 and NIO2) and TW21-containing NIOs (NIO3 and NIO4). We hypothesized that these differences could affect the PrC composition and the relative abundance of the proteins adsorbed on the surface of NIOs, as well as, obviously, the nano-bio interface of PrC-NIO complexes.
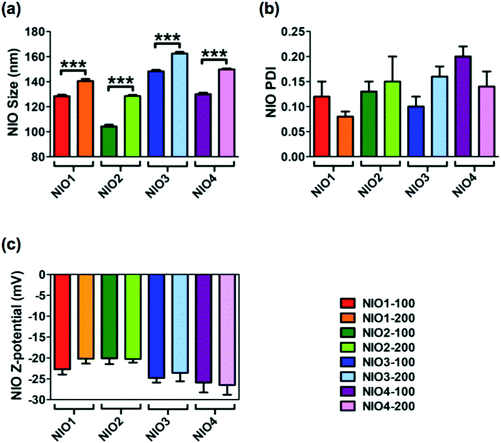
Average size, polydispersity index (PDI) and zeta potential of bare NIOs were measured by dynamic light scattering (DLS) and used as reference standard (Fig. 2). All bare NIO formulations, extruded both at 100 and at 200 nm, displayed an average size below 200 nm (from 104.3 ± 1.4 nm to 162.4 ± 1.4 nm; Fig. 2). Such a finding could be due to the presence of Chol, that mimics the second lipophilic chain of surfactants, able to stabilize the NIO bilayer;47 Chol causes a condensation effect thus likely forcing the acylic chains of surfactants to assume a trans-configuration in the hydrophobic compartment of the different NIOs, making their bilayers as rigid as those made of distearoyl phosphatidylcholine.48 Average size of NIO2 was smaller than that of NIO1, suggesting an acylic chain-dependent effect (p value <0.0001, Fig. 2a). In fact, TW80, bearing an unsaturated fatty acid (FA) has a wider area per molecule in its bilayer than TW20, bearing a saturated FA. As a matter of fact, TW80 bilayer could host Chol better than TW20 one, thus increasing the packaging of its unsaturated hydrocarbon chains.31
TW21-containing NIOs (NIO3 and NIO4) showed slightly increased average sizes compared to NIO1 and NIO2 (p value <0.0001), but still smaller than 200 nm (Fig. 2). These results could depend on the number of ethylene oxide units forming the PEG-mimetic chains of NIO3 and NIO4. A greater number of ethylene oxide units (such as in TW20 and TW80) determines a higher PEG molecular weight and affects the packing of the corresponding NIO bilayer; as a consequence, TW21-containing NIOs are more fluid than NIO1/2, and have a liquid crystal configuration.
DLS analysis showed that all the bare NIOs had a PDI ≤ 0.2 (Fig. 2a) and a negative zeta potential (from −20.10 ± 1.39 to −26.50 ± 2.33 mV; Fig. 2b), thus showing that all formulations were stable and homogeneously distributed. Furthermore, the percentage of self-assembled TWs in each NIO formulation was 40 ± 5%, in agreement with our previous data.34,49
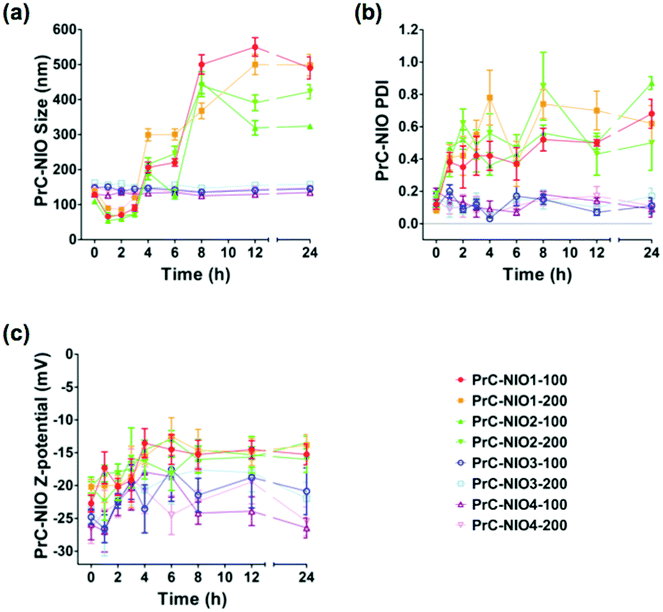
To study the effects of PrC on physicochemical properties of NIO formulations, DLS analysis was performed again after the incubation in HP, at 37 °C for 24 h (Fig. 3). The average sizes of PrC-NIO1 and PrC-NIO2, at both extrusion size, increased after 24 h (up to ≈500 nm for PrC-NIO1, up to 300–400 nm for PrC-NIO2; Fig. 3a), even if in the first 4 h they decreased below 100 nm. Conversely, PrC formation did not change NIO3 and NIO4 average size, suggesting that the phenomenon was driven by PEG features. Such a behavior suggests that the more stable and smaller size of TW21-containing PrC-NIO complexes (PrC-NIO3 and PrC-NIO4) makes them more suitable for drug delivery applications if compared with the two TW21-lacking counterparts (PrC-NIO1 and PrC-NIO2).
Likewise, after HP incubation, PDI increased (>0.3) for NIO1 and NIO2, regardless of extrusion size, negatively affecting their stability and size distribution (Fig. 3b); the opposite behavior was observed for TW21-containing formulations (NIO3 and NIO4), which hence have a superior biological stability and narrow size distribution. Indeed, we performed these stability analyses only at one concentration of HP; however, it is conceivable that small variations of HP concentration could not change significantly the stable behavior of TW21-containing formulations. These interesting findings regarding the effect of TW21 addition in the NIO formulations are not completely surprising, since they are in agreement with some previously reported data,12,13 indicating that the PrC formation can generate two divergent effects: (i) the shrinkage of nanocarriers due to the osmotic pressure generated by the biological environment, thus determining a reduction of their diameter,36 (ii) the increase of NP size due to the greater amount of bound proteins.50
For all NIOs, zeta potential of different formulations remained negative after incubation with HP, regardless of their composition and size (Fig. 3c). These conserved charge properties are not surprising, since they partially agreed with some previously reported data stating that the electrostatic interaction between negatively charged NPs and positively charged proteins is not the only force affecting the nano-bio interface50 but it also depends on (i) the modification of charged groups on the surface of PEG, (ii) the chelation of cations by the PEG chains51 and (iii) the resulting net negative charge generated by some plasma proteins at physiological pH.52
The stability of the dispersion of NIOs in plasma was tested by using a Turbiscan Lab® apparatus in order to evaluate the putative in situ aggregation of plasma proteins (such as fibrinogen) in ex vivo PrC-NIO complexes (see ESI† for experimental details). This type of analysis is able to predict the long-term stability of emulsions, solutions and suspensions37 and has been widely used to study the stability of different nanocarriers and to correlate the migration of particles with destabilization phenomena, like sedimentation.53–55 As regards to the backscattering (ΔBS) signals measured by Turbiscan, they were considered significant only when relative to the zone comprised between 2 mm and 8 mm (ESI Fig. S1†). In this length interval the ΔBS peaks were within a ±2% range for all the tested NIO formulations (ESI Fig. S1†). The presence of positive or negative ΔBS peaks below 2 mm and over 8 mm did not depend on migration of NIO particles (sedimentation) during the analysis, but they depended on the air at the bottom and at the top of the cylindrical glass tube holding samples (ESI Fig. S1†).37 On the other hand, the transmission stability index (TSI) was below 6, for all the tested formulations except for NIO4-100 nm, which had a TSI slightly over 10 after 1 h of incubation (ESI Fig. S2†). These mentioned ranges of ΔBS and TSI values were suggestive of stable NP dispersions as previously reported.37 Hence, we may state that all the tested NIOs showed stable dispersion in HP, and this stability was independent of the extrusion procedure and of chemical composition (ESI Fig. S1 and S2†).
3.2 Identification of plasma proteins in the corona
To characterize the PrC in all NIOs, each formulation was separately incubated in HP and, after the PrC elution, the adsorbed proteins were separated by SDS-PAGE (ESI Fig. S3†). To identify the PrC components associated with each NIO formulation, the protein bands were in-gel digested with trypsin and analyzed by LC-MS/MS. We identified a total of 53 unique species, listed in ESI Tables S1–S4,† which include all the details of the mass spectrometry (MS) analysis. In particular, we found 33 proteins for NIO1-100, 27 for NIO1-200, 25 for NIO2-100, 37 for NIO2-200, 24 for NIO3-100, 35 for NIO3-200, 9 for NIO4-100 and 7 for NIO4-200. Among the identified proteins, we found that the composition of PrC, eluted from all the analyzed NIO formulations, was characterized by the presence of several immunoglobulin (Ig) chains. Among them, the immunoglobulin kappa light chain VLJ region (IGK) and Ig mu chain C region (IGHM) were common to the PrC of all tested NIO formulations. In general, plasma proteins like immunoglobulins are the typical components of PrC for many types of NPs, such as polystyrene, copolymer, TiO2, ZnO, SiO2, carbon nanotubes and quarts NPs.56,57 This finding probably depends on the opsonization phenomenon, a process occurring in plasma and mediated by immunoglobulins and complement proteins (namely opsonins), that neutralize the non-self antigens. Opsonins promote phagocytosis by labeling specific antigens, and recruit the mononuclear phagocytic system (MPS) modulating the human immune response, thus eventually affecting NP clearance.58By comparing, within each NIO formulation, the number of identified immunoglobulins against the total number of identified species, we also observed an immunoglobulin enrichment in the TW21-containing NIOs, particularly NIO4 formulation. Such an absorption of opsonins on the NIO4 surface could be strictly related to the less dense and compact structure of PEG.59,60
We functionally analyzed proteins identified in the PrC-NIO using bioinformatics tools, such as DAVID software. The enriched gene ontology (GO) terms, belonging either to the cellular component (CC) category, or to the molecular function (MF) category, or to the biological process (BP) category, are listed in Table 2. Bioinformatics analysis showed that all the analyzed proteins were clustered in different functional categories except for the PrC of NIO4 formulation, due to the low number of identified species for this formulation.
| GOsa | Proteins | p-Value |
|---|---|---|
| a GOs: gene ontologies. | ||
| NIO1-100 | ||
| Complement activation | C1QA, C1QB, C3, FCN3, CFI, C4BPA, C1QC | 1.4 × 10−10 |
| Cytoplasmic membrane-bounded vesicle | STOM, TF, FGG, A2M, APOA1, F5, ALB, HRG, SERPINA1, THBS1 | 7.7 × 10−7 |
| Heparin binding | APOE, HRG, VTN, THBS1 | 7.6 × 10−4 |
| Plasma lipoprotein particle | APOA1, APOE, HPR | 2.3 × 10−3 |
| Peptidase inhibitor activity | A2M, C3, HRG, SERPINA1 | 2.4 × 10−3 |
| Regulation of lipid transport | APOA1, APOE, THBS1 | 1.5 × 10−3 |
| Response to endogenous stimulus | C1QB, A2M, SERPINA1, THBS1 | 4.2 × 10−2 |
| Cellular chemical homeostasis | TF, APOE, SLC4A1, HPR | 3.5 × 10−2 |
| NIO1-200 | ||
| Complement activation | C1QA, C1QB, FCN3, C4BPA, C1QC | 4.6 × 10−7 |
| Cytoplasmic membrane-bounded vesicle | STOM, TF, APOA1, F5, ALB, HRG, SERPINA1, THBS1 | 2.5 × 10−5 |
| Heparin binding | APOE, HRG, VTN, THBS1 | 3.6 × 10−4 |
| High-density lipoprotein particle | APOA1, APOE, HPR | 8.3 × 10−4 |
| Cellular homeostasis | TF, APOE, HPX, SLC4A1, HPR | 5.2 × 10−3 |
| Regulation of blood coagulation | APOE, HRG, THBS1 | 1.4 × 10−3 |
| Cortical cytoskeleton | ACTB, SLC4A1, SPTB | 3.2 × 10−3 |
| Regulation of phosphate metabolic process | APOA1, APOE, HPX, THBS1 | 3.8 × 10−2 |
| NIO2-100 | ||
| Cytoplasmic membrane-bounded vesicle | STOM, TF, APOA1, ALB, FGB, HRG, SERPINA1 | 7.4 × 10−5 |
| Plasma lipoprotein particle | APOA1, APOE, HPR | 1.1 × 10−3 |
| Complement activation | C1QB, C3, FCN3 | 1.4 × 10−3 |
| Cellular homeostasis | TF, APOE, HPR, SLC4A1 | 2.3 × 10−2 |
| Peptidase inhibitor activity | C3, HRG, SERPINA1 | 1.7 × 10−2 |
| NIO2-200 | ||
| Cytoplasmic membrane-bounded vesicle | STOM, TF, FGG, A2M, APOA1, F5, ALB, FGB, HRG, SERPINA1, THBS1 | 1.9 × 10−7 |
| Peptidase inhibitor activity | A2M, C3, SERPINA3, HRG, SERPINA1 | 3.5 × 10−4 |
| Complement activation | C1QB, C3, FCN3 | 9.6 × 10−5 |
| Plasma lipoprotein particle | APOA1, APOE, HPR | 2.8 × 10−3 |
| Hemostasis | FGG, F5, FGB, SERPINA1 | 1.6 × 10−3 |
| Heparin binding | APOE, HRG, VTN, THBS1 | 1.5 × 10−3 |
| Regulation of lipid transport | APOA1, APOE, THBS1 | 1.9 × 10−3 |
| Cholesterol metabolic process | APOA1, APOE, CAT | 1.6 × 10−2 |
| NIO3-100 | ||
| Complement activation | C1QA, C1QB, FCN3, C1QC | 2.2 × 10−5 |
| Plasma lipoprotein particle | APOA1, APOE, HPR | 1.1 × 10−3 |
| Cytoplasmic membrane-bounded vesicle | STOM, APOA1, ALB, HRG, SERPINA1 | 7.8 × 10−4 |
| Heparin binding | APOE, HRG, VTN | 6.1 × 10−3 |
| Peptidase inhibitor activity | SERPINA3, HRG, SERPINA1 | 1.3 × 10−2 |
| NIO3-200 | ||
| Cytoplasmic membrane-bounded vesicle | STOM, TF, FGG, A2M, APOA1, ALB, FGB, SERPINA1 | 1.8 × 10−4 |
| Peptidase inhibitor activity | A2M, C3, C4B, SERPINA3, SERPINA1 | 4.0 × 10−4 |
| Positive regulation of immune response | C1QB, C3, FCN3, C4B, HPX | 2.8 × 10−4 |
| Cellular ion homeostasis | TF, APOE, HPX, SLC4A1, CP, HPR | 1.3 × 10−3 |
| Plasma lipoprotein particle | APOA1, APOE, HPR | 2.8 × 10−3 |
| Negative regulation of apoptosis | ALB, APOE, PRDX2, CAT | 4.3 × 10−2 |
As for CC terms, the most represented term was cytoplasmic membrane-bounded vesicle; whereas “peptidase inhibitor activity” was the significantly represented MF term among proteins adsorbed onto NIO formulations except for NIO1-200. Likewise, “complement activation” was one of the most relevant BP terms involving all analyzed PrCs except for NIO3-200 (Table 2). It is well known that the activation of the complement system via the classical pathway requires the close proximity of Fc fragments of immunoglobulin proteins;61 interestingly, in our system, PrC were immunoglobulin-rich mixtures, that might be able to trigger in vivo the complement activation.
The GO analysis also showed that other identified proteins, such as apolipoprotein E (APOE) and apolipoprotein A-I (APOA1), were significantly represented in all the PrCs (Table 2). This apolipoprotein enrichment in the PrC of NIOs indicates that, under physiological conditions, lipoproteins are typical corona components able to bind specific cell-surface receptors.12,13 The adsorption of these proteins can modify the physicochemical properties of NIOs, such as hydrophobicity, thus affecting their biopharmaceutic and pharmacokinetic features. In fact, apolipoproteins, but also albumin (ALB), act as desoponins by prolonging the blood circulation time.56 Among the identified proteins in the corona, also ALB was found to bind all the designed NIOs, thus allowing to speculate about ALB effect on NIO biodistribution. It's noteworthy that ALB-coated gold NPs showed more accumulation in the lungs and brain than APOE-coated ones.57 On the other hand, the adsorption of APOB and APOE onto lipid NPs coated with TW80 enhances drug delivery across the blood brain barrier.56 Moreover, apolipoproteins such as APOA1, identified in the hard corona of other PrC-NP complexes, are used to evaluate the rate of PrC coating for specific NPs.14,62 Interestingly, APOA1 and APOA2 adsorption on mesoporous silica NPs showed enhanced hepatic biodistribution of drug loaded in these NPs.57
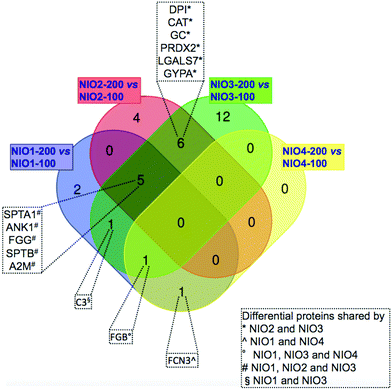
We compared the identified proteins between NIOs having the same composition but different sizes. For each protein, Tables 3–6 report the gene name and the RSC value representing the log2 ratio between its relative abundance in NIO-200 versus NIO-100. For all the compared NIO formulations, 32 differentially represented unique proteins were found with RSC ≥ 1.40 or ≤−1.40: particularly 10 species for NIO1, 15 for NIO2, 23 for NIO3 and 2 for NIO4 (Tables 3–6). As reported in Fig. 4, each pairwise comparison, except for NIO4, showed formulation-specific differentially represented proteins: 2 species for NIO1, 4 for NIO2 and 12 for NIO3. Conversely, we also found common species in at least two pairwise comparisons (Fig. 4). As reported in Fig. 4, NIO2 and NIO3 (200 nm versus 100 nm) shared the following 6 differentially represented proteins: desmoplakin I (DPI), catalase (CAT), serum vitamin D-binding protein precursor (GC), peroxiredoxin-2 (PRDX2), galectin-7 (LGALS7), glycophorin alpha GYPA. Among these 6 proteins, CAT, GC, PRDX2 and GYPA are all over-represented in both NIO2 and NIO3, while DPI and LGALS7 are under-represented (Tables 5 and 6). Moreover, ficolin-3 isoform 1 precursor (FCN3), the only differential protein exclusively shared by NIO1 and NIO4, is over-represented in the former formulation (200 nm versus 100 nm) and under-represented in the latter (Tables 3 and 6). Furthermore, human fibrinogen (FGB) is differentially represented in all NIO formulations extruded at 200 nm compared to those extruded at 100 nm (Tables 3, 5 and 6), with the exception of the formulation NIO2 (Table 5). In all formulations extruded at 200 nm versus 100 nm, except for NIO4, the following 5 proteins were differentially represented (Fig. 4): spectrin alpha, erythrocytic 1 (SPTA1), ankyrin-1 isoform 4 (ANK1), fibrinogen gamma chain isoform gamma-A precursor (FGG), spectrin beta chain (SPTB), alpha-2-macroglobulin precursor (A2M). Interestingly, similar proteins were identified in vivo in the PrC of PEGylated liposomes (Caelyx) collected 10 min after injection in human13 and mice.12 In our experimental setting, all these proteins are under-represented in NIO1 and over-represented in both NIO2 and NIO3 (Tables 3–5). Similarly, the chain A, human complement component C3 (C3), the only differential protein exclusively shared by NIO1 and NIO3, is under-represented in the former formulation and over-represented in the latter (Tables 3 and 5). In particular, these two last comparisons, between formulations extruded at 200 nm versus 100 nm, show that the same trend in protein profiling (from under-representation to over-representation of plasma proteins common to NIO1 and NIO3) could be related to the presence of TW21 in combination with TW20. Interestingly, those proteins identified as over-represented in the PrC-NIO3 (200 nm versus 100 nm) and with opposite trend in PrC-NIO1, such as A2M and FGG, are plasma proteins whose levels are higher in pathological conditions (Alzheimer's disease and diabetes, respectively);63 this observation leads to hypothesize that a NIO formulation, such as NIO3 extruded at 200 nm, could be potentially applied for drug delivery in specific disorders characterized by alteration of plasma proteins known to be preferentially adsorbed to the NIO of interest. Therefore, the obtained results demonstrate that the specific formulations, together with the extrusions sizes, can quantitatively rather than qualitatively affect the composition of PrC.
| Protein | Gene | R SC |
|---|---|---|
| a R SC: log2 ratio between protein levels of PrC-NIO1-200 versus PrC-NIO1-100. Proteins with RSC ≥ 1.40 or ≤−1.40 were considered differentially represented. | ||
| Ficolin-3 isoform 1 precursor | FCN3 | 1.95 |
| Band 3 anion transport protein | SLC4A1 | −2.09 |
| Ankyrin-1 isoform 4 | ANK1 | −2.28 |
| Spectrin alpha, erythrocytic 1 | SPTA1 | −2.33 |
| Spectrin beta chain, erythrocytic isoform b | SPTB | −2.61 |
| Chain B, structure of complement C3b: insights into complement activation and regulation | CFI | −2.63 |
| Fibrinogen gamma chain isoform gamma-A precursor | FGG | −2.63 |
| Alpha-2-macroglobulin precursor | A2M | −3.74 |
| Chain B, crystal structure of human fibrinogen | FGB | −3.74 |
| Chain A, human complement component C3 | C3 | −4.48 |
| Coagulation factor V precursor | F5 | 1.35 |
| CD5 antigen-like precursor | CD5L | 1.02 |
| Complement C1q subcomponent subunit C precursor | C1QC | 0.184 |
| Haptoglobin-like protein | HPR | 0.521 |
| Apolipoprotein E | APOE | 0.297 |
| Albumin | ALB | 0.496 |
| Stomatin peptide | STOM | −0.353 |
| Actin, cytoplasmic 1 | ACTB | −0.127 |
| Apolipoprotein A-I preproprotein | APOA1 | −0.210 |
| C1q B-chain precursor | C1QB | −0.335 |
| Vitronectin | VTN | −0.270 |
| Histidine-rich glycoprotein precursor | HRG | −1.18 |
| Collectin 10 | COLEC10 | −0.111 |
| Alpha-1-antitrypsin | SERPINA1 | 0.389 |
| Serotransferrin precursor | TF | −0.332 |
| Thrombospondin-1 precursor | THBS1 | −0.314 |
| C4b-binding protein alpha chain precursor | C4BPA | −0.452 |
| Ig mu chain C region | IGHM | 0.538 |
| Hemopexin precursor | HPX | 0.180 |
| Ig G1 H Nie | IGHG1 | 0.358 |
| Complement C1q subcomponent subunit A precursor | C1QA | −0.133 |
| Immunoglobulin kappa light chain VLJ region | IGK | 0.370 |
| Immunoglobulin lambda-chain | IGL | 0.814 |
| Peroxiredoxin-2 | PRDX2 | 1.29 |
| Protein | Gene | R SC |
|---|---|---|
| a R SC: log2 ratio between protein levels of PrC-NIO2-200 versus PrC-NIO2-100. Proteins with RSC ≥ 1.40 or ≤−1.40 were considered differentially represented. | ||
| Spectrin alpha, erythrocytic 1 | SPTA1 | 4.75 |
| Ankyrin-1 isoform 4 | ANK1 | 4.49 |
| Coagulation factor V precursor | F5 | 4.17 |
| Vitronectin | VTN | 3.62 |
| Protein 4.1 isoform 6 | EPB41 | 3.43 |
| Alpha-2-macroglobulin precursor | A2M | 2.98 |
| Glycophorin alpha | GYPA | 2.69 |
| Thrombospondin-1 precursor | THBS1 | 2.69 |
| Catalase | CAT | 2.69 |
| Spectrin beta chain, erythrocytic isoform b | SPTB | 2.40 |
| Peroxiredoxin-2 | PRDX2 | 2.32 |
| Serum vitamin D-binding protein precursor | GC | 2.32 |
| Fibrinogen gamma chain isoform gamma-A precursor | FGG | 1.73 |
| Desmoplakin I | DPI | −3.96 |
| Galectin-7 | LGALS7 | −3.96 |
| C1q B-chain precursor | C1QB | 0.0704 |
| Complement C1q subcomponent subunit C precursor | C1QC | −0.698 |
| CD5 antigen-like precursor | CD5L | 0.424 |
| Chain A, human complement component C3 | C3 | 1.12 |
| Serotransferrin precursor | TF | 0.294 |
| Chain B, crystal structure of human fibrinogen | FGB | −0.125 |
| Stomatin peptide | STOM | 0.434 |
| Albumin | ALB | 0.00641 |
| Band 3 anion transport protein | SLC4A1 | 0.513 |
| Apolipoprotein A-I preproprotein | APOA1 | −0.101 |
| Apolipoprotein E | APOE | −0.163 |
| Alpha-1-antitrypsin | SERPINA1 | 0.0612 |
| Actin, cytoplasmic 1 | ACTB | −0.616 |
| Haptoglobin-like protein | HPR | 0.645 |
| Ficolin-3 isoform 1 precursor | FCN3 | −0.803 |
| Ig mu chain C region | IGHM | −1.26 |
| Histidine-rich glycoprotein precursor | HRG | −0.489 |
| Alpha1-antichymotrypsin, partial | SERPINA3 | 1.29 |
| Ig G1 H Nie | IGG | −0.0716 |
| Ig alpha-1 chain C region, partial | IGHA1 | −0.960 |
| Delta-aminolevulinate dehydratase | ALAD | −0.489 |
| Ig gamma-2 chain C region | IGHG2 | −0.382 |
| Immunoglobulin kappa light chain VLJ region | IGK | 0.126 |
| Ig J-chain, partial | IGJ | −0.491 |
| Protein | Gene | R SC |
|---|---|---|
| a R SC: log2 ratio between protein levels of PrC-NIO3-200 versus PrC-NIO3-100. Proteins with RSC ≥ 1.40 or ≤−1.40 were considered differentially represented. | ||
| Spectrin beta chain, erythrocytic isoform b | SPTB | 6.19 |
| Spectrin alpha, erythrocytic 1 | SPTA1 | 5.14 |
| Alpha-2-macroglobulin precursor | A2M | 3.59 |
| Catalase | CAT | 3.38 |
| Chain B, human complement C4 | C4B | 3.38 |
| Chain B, crystal structure of human fibrinogen | FGB | 3.38 |
| Serum vitamin D-binding protein precursor | GC | 3.38 |
| Fibrinogen gamma chain isoform gamma-A precursor | FGG | 3.14 |
| Ankyrin-1 isoform 4 | ANK1 | 2.87 |
| Glycophorin alpha | GYPA | 2.49 |
| Hemopexin precursor | HPX | 2.49 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 2.49 |
| Caspase-14 precursor | CASP14 | 2.49 |
| Delta-aminolevulinate dehydratase | ALAD | 2.00 |
| Ceruloplasmin, partial | CP | 2.00 |
| Peroxiredoxin-2 | PRDX2 | 1.74 |
| Chain A, human complement component C3 | C3 | 1.74 |
| Haptoglobin-like protein | HPR | 1.61 |
| Galectin-7 | LGALS7 | −2.65 |
| Desmoplakin I | DPI | −2.65 |
| Plakoglobin | JUP | −2.65 |
| Complement C1q subcomponent subunit A precursor | C1QA | −4.25 |
| Histidine-rich glycoprotein precursor | HRG | −4.59 |
| Band 3 anion transport protein | SLC4A1 | 1.16 |
| Alpha1-antichymotrypsin, partial | SERPINA3 | 0.962 |
| CD5 antigen-like precursor | CD5L | −0.0120 |
| Serotransferrin precursor | TF | 0.747 |
| Albumin | ALB | 0.494 |
| Alpha-1-antitrypsin | SERPINA1 | 0.306 |
| Stomatin peptide | STOM | 0.415 |
| Complement C1q subcomponent subunit C precursor | C1QC | −1.02 |
| C1q B-chain precursor | C1QB | −0.994 |
| Vitronectin | VTN | 0.124 |
| Apolipoprotein A-I preproprotein | APOA1 | 0.747 |
| Apolipoprotein E | APOE | −0.0199 |
| Ficolin-3 isoform 1 precursor | FCN3 | −0.232 |
| Ig mu chain C region | IGHM | −1.39 |
| Ig gamma-2 chain C region | IGHG2 | −1.17 |
| Immunoglobulin lambda light chain VLJ region | IGL | −0.0845 |
| Ig J-chain, partial | IGJ | −0.570 |
| Immunoglobulin kappa light chain VLJ region | IGK | 0.754 |
| Protein | Gene | R SC |
|---|---|---|
| a R SC: log2 ratio between protein levels of PrC-NIO4-200 versus PrC-NIO4-100. Proteins with RSC ≥ 1.40 or ≤−1.40 were considered differentially represented. | ||
| Chain B, crystal structure of human fibrinogen | FGB | −2.20 |
| Ficolin-3 isoform 1 precursor | FCN3 | −2.20 |
| Serum albumin | ALB | 1.18 |
| Ig mu chain C region | IGHM | −0.451 |
| CD5 antigen-like precursor | CD5L | 0.0865 |
| Immunoglobulin alpha-2 heavy chain | IGHA2 | −1.10 |
| Apolipoprotein E | APOE | −0.342 |
| Immunoglobulin heavy chain variable region | IGHV3 | −0.368 |
| Immunoglobulin kappa light chain VLJ region | IGK | 1.13 |
We evaluated and compared the relative abundance of identified PrC components, based on their MS spectral counts,64 in all NIO formulations (Table 7). Among them, a set of 5 species were found to bind all the NIOs, but with different abundance: ALB, APOE, CD5 molecule like (CD5L), IGK and IGHM; ALB, for example, binds different NIOs varying from 10.3% in NIO3-100 to 25.8% in NIO4-200 (Table 7). These results strengthen the concept that the PrC depends on physicochemical properties of NPs, particularly on their sizes and composition.65–68
| Gene | NSAF | NSAF | NSAF | NSAF | NSAF | NSAF | NSAF | NSAF |
|---|---|---|---|---|---|---|---|---|
| NIO1-100 | NIO1-200 | NIO2-100 | NIO2-200 | NIO3-100 | NIO3-200 | NIO4-100 | NIO4-200 | |
| ALB | 11.0% | 12.3% | 16.4% | 17.9% | 10.3% | 14.8% | 13.8% | 25.8% |
| APOE | 8.07% | 8.30% | 8.74% | 8.62% | 7.34% | 7.93% | 13.5% | 11.6% |
| CD5L | 1.81% | 3.11% | 1.66% | 2.47% | 2.33% | 2.54% | 6.36% | 6.37% |
| IGK | 20.3% | 22.0% | 15.8% | 18.8% | 11.0% | 20.0% | 2.78% | 6.95% |
| IGHM | 19.0% | 21.5% | 14.2% | 7.21% | 18.0% | 8.78% | 28.0% | 24.4% |
| FCN3 | 0.750% | 2.60% | 3.67% | 2.32% | 4.57% | 4.28% | 1.48% | — |
| TF | 0.835% | 0.556% | 1.12% | 1.52% | 0.363% | 0.695% | — | — |
| STOM | 0.934% | 0.599% | 0.726% | 1.13% | 0.703% | 1.07% | — | — |
| SLC4A1 | 3.64% | 0.758% | 2.00% | 3.09% | 1.11% | 2.66% | — | — |
| APOA1 | 1.34% | 0.969% | 1.17% | 1.22% | 0.948% | 1.82% | — | — |
| HPR | 1.42% | 1.73% | 1.03% | 1.81% | 0.291% | 1.14% | — | — |
| SERPINA1 | 0.644% | 0.722% | 0.751% | 0.878% | 0.605% | 0.844% | — | — |
| C1QB | 5.32% | 3.58% | 1.65% | 1.93% | 3.20% | 1.74% | — | — |
| C1QC | 4.21% | 4.05% | 3.20% | 2.16% | 6.61% | 3.60% | — | — |
In addition to NP characteristics, however, we cannot exclude that PrC composition also depends on qualitative/quantitative alterations of plasma proteome in different physiological and pathological conditions.63,69 In fact, ALB is the most abundant protein in plasma as well as the first protein to be adsorbed on different type of NPs: changes of ALB levels in the plasma indicate specific pathological conditions, such as infections or an inflammatory state.70 Importantly, HP is a heterogeneous fluid and several proteins, whose levels change in the plasma, are used as diagnostic biomarkers for different diseases. In addition to ALB, in fact, also the modulation of other plasma proteins, such as immunoglobulin IgM and complement proteins, have been reported in patients with neurological disorders or cancer. In addition to disease type and progress, plasma composition in terms of protein abundance/concentration also depends on other factors such as age, gender and ethnicity. As a consequence, both health and disease conditions affect the PrC composition: plasma from healthy donors of the same gender and age induced the formation of not always consistent PrC on the same NPs; whereas plasma from patients with the same disorders generated PrC with slight differences in composition.69,70 Hence, such dependence on physicochemical properties of NPs and on plasma proteome affects the interactions at the nano-bio interface and the PrC composition, and consequently cytotoxicity, intracellular uptake and potentially the biological fate of PrC-NP complexes.
3.3 Cellular uptake of PrC-NIO
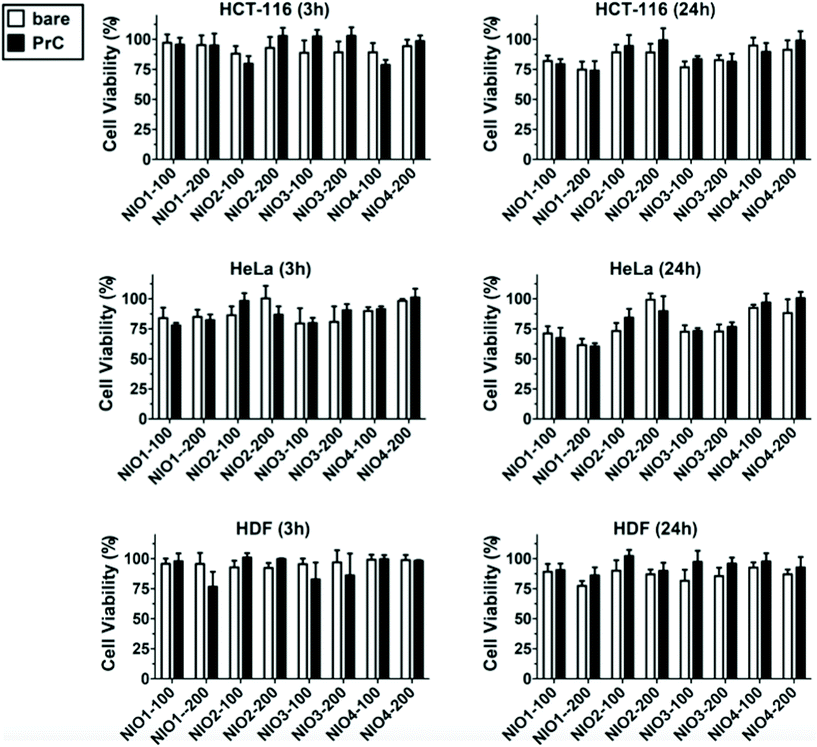
To evaluate NIO cytotoxicity and cellular uptake, we tested their biological effect onto two cancer cell lines (HCT-116 cells for colorectal cancer and HeLa cells for cervix cancer) and onto healthy primary cells (HDF, human dermic fibroblasts) that were used as a control. Colorectal and cervix cancer represent two forms of cancer with high incidence in human,71 and can have some benefits using NPs as potential therapeutic approach for cure and treatment.In order to test dose-dependent cytotoxicity of NIO formulations, we performed MTT assays on HCT-116 cells treated for 3 h and 24 h with different concentrations of NIOs (from 0.8 to 100 μg ml−1; ESI Fig. S4†). The resulting data clearly showed that no NIO formulation, at a concentration of 20 μg ml−1, after 3 h of incubation, caused a cell viability reduction below the 80% when compared with untreated cells, while higher concentrations or longer incubation times caused a more evident cytotoxicity of some formulation. Thus, we considered a concentration of 20 μg ml−1 and an incubation time of 3 h as ideal conditions for further cell culture experiments.
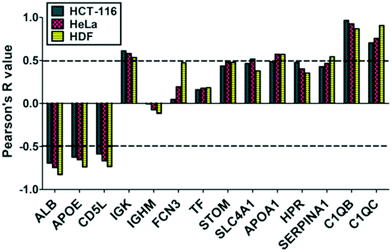
To analyze the cellular uptake of PrC-NIO, we used fluorescent NIO formulations (OG-NIOs) and measured the mean fluorescence intensity (MFI) of cells by cytofluorimetric analysis after 3 h of incubation (ESI Fig. S5†). For this analysis, the PrC formation was obtained at the same HP concentration used for proteomic analysis. As control, we also evaluated the cellular uptake of bare fluorescent NIOs in the same experimental condition of PrC-NIOs (in the cell culture with 1% HP) (Fig. 5) and in protein-free condition (ESI Fig. S6†), thus demonstrating a similar trend when the cellular uptake of PrC-NIOs was compared to the bare NIOs added into cell culture either without HP or with low HP. The uptake profile of bare and PrC-NIOs are similar among the three considered cell lines; the uptake level pairwise comparison between bare and the corresponding PrC-NIO showed some significant differences. In particular, in HeLa cells we found significantly different uptake between bare NIOs and the relative PrC-NIOs only for NIO3-200 and NIO4-200. Conversely, in HCT-116 and HDF these differences were evidenced in most NIO formulations. Moreover, in some cases PrC-NIOs showed higher uptake than bare NIOs, while in other cases the vice versa occurred. Not surprisingly, smaller NIOs were internalized better than bigger ones; in fact, all NIO-100 showed a higher uptake than the corresponding NIO-200. The various formulations showed statistically relevant differences in their uptake levels. In particular, the fluorescent PrC-NIO1 and PrC-NIO3 (TW20-containing NIOs) were internalized better than fluorescent PrC-NIO2 and PrC-NIO4; moreover, TW21-containing NIOs showed a reduced uptake compared with TW21-lacking formulations. In other words the uptake levels are enhanced by the presence of TW20, instead of TW80 and they are impaired by the addition of TW21. Moreover, this trend was similar in the three cell lines as shown by Pearson's correlation (Table 8).
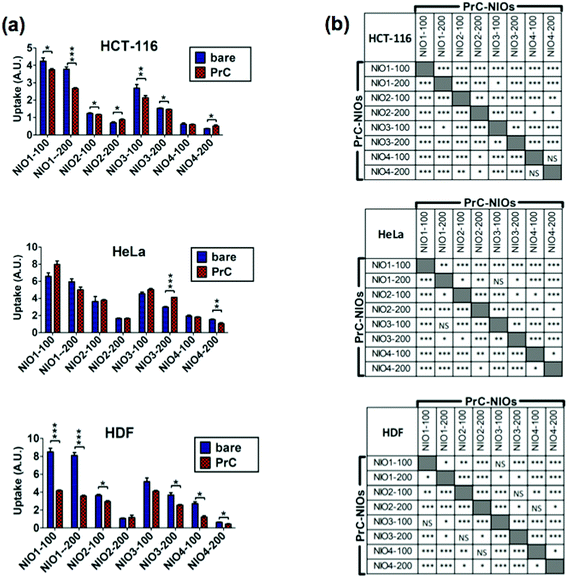 | ||
| Fig. 5 Cellular uptake of bare NIOs and PrC-NIOs. HCT-116, HeLa and HDF cells were incubated in serum-free medium containing 1% human plasma (HP) enriched in fluorescent bare NIOs and fluorescent PrC-NIOs (100 nm and 200 nm). Their uptake was quantified by flow cytometry measuring the mean fluorescence (MFI) of cells. The MFI raw data (Fig. S5†), measured in three independent replicate experiments, were properly normalized. (a) Uptake levels are reported in graphs as the mean values ± SD and the statistical significance of pairwise comparison between bare and the corresponding coated NIO is showed. (b) Statistical significance of pairwise comparison among PrC-NIOs. * = p < 0.05; ** = p < 0.01; *** = p < 0.005; NS = not significant; A. U., arbitrary units. | ||
| R | ||
|---|---|---|
| a R: the Pearson's correlation coefficient. | ||
| PrC-NIOs | HCT-116 versus HeLa | 0.96 |
| HCT-116 versus HDF | 0.87 | |
| HeLa versus HDF | 0.93 | |
| PrC-NIOs versus bare NIOs | HCT-116 | 0.98 |
| HeLa | 0.94 | |
| HDF | 0.89 | |
We also evaluated if PrC-NIO uptake can depend on the amount of the most represented proteins in the PrC. To this aim we selected the proteins, which were identified by proteomics in at least six out of eight formulations and we calculated the Pearson's coefficients between the spectral counts and the uptake levels for each protein and cell line (Fig. 6). Indeed, five proteins showed a significant Pearson's R in all the three cell lines: ALB, APOE, CD5L, complement C1q B chain (C1QB) and C chain (C1QC). ALB, APOE and CD5L showed a strong negative correlation indicating that a higher content of these proteins in the PrC was associated to a lower NIO uptake; while C1QB and C1QC showed a strong positive correlation, which imply that a higher content of these complement proteins matched a higher NIO uptake. In other terms, based on these results, NP endocytosis might be enhanced through a modulation of the content of ALB, APOE, CD5L, C1QB and C1QC in the PrC-NP. It should be noticed that the levels of the five considered proteins show a significant correlation in all the three tested cell lines, including the HDF cells (Fig. 6). A NP with a generalized high propensity to be internalized by different types of cells, including the healthy ones, has obvious drawbacks in terms of drug delivery specificity in vivo. Moreover, contrarily to what happens for C1QB, the levels of the other four mentioned proteins (ALB, APOE, CD5L and C1QC) show a stronger correlation (positive for C1QC and negative for ALB, APOE, CD5L) with the uptake in HDF cells than in both cancer cell lines (HCT-116 and HeLa) (Fig. 6). The ideal formulation might be one with a middle tier of generalized cellular uptake and possibly a higher tendency to be internalized by cancer cells rather than by healthy cells. In this scenario, among the tested formulations, NIO3 ones fit better these characteristics. In fact, NIO3 formulations absorb relatively low amount of ALB, APOE and CD5L and relatively high amount of C1QC, in the corona. At the same time, the presence of PrC gives to NIO3 formulations (particularly to NIO3-200) a higher propensity to be internalized by HeLa cells and lower tendency to be internalized by HDF cells, if compared with correspondent bare NIOs (Fig. 5a).
Indeed, ALB has been widely and extensively used and studied both as NP constituent and NP coating.72,73 Undoubtedly, many evidences support the idea that ALB occurrence on NP surface is able to modify the biodistribution of such NPs, improving their circulation in the bloodstream, decreasing their clearance by MPS and enhancing their accumulation in the tumor.72,73 Surprisingly, we found that an increase of ALB content in the PrC corresponds to a decreased endocytosis of the PrC-NIO complexes. This finding is in apparent contrast with the usefulness of ALB coatings as a mean to increase the delivery properties of NPs. However, accordingly with our results, some authors reported that the presence of ALB on the surface of NPs can impair their uptake in different cell lines.74,75 We may hypothesize that the propensity to internalize NPs, as consequence of ALB presence onto NP surface, can depend on NP type and dimensions, and on cell type. Therefore, the effect of an ALB coating is not easily predictable and should be ad hoc evaluated. We may not exclude that, in certain contexts, an ALB coating onto NPs impairs their uptake levels in tumor cells; while a more favorable biodistribution counterbalances such a negative effect promoting their accumulation in the tumor environment, resulting in a net anti-cancer effect. If we hypothesize that the PEG-mimetic effect of the Tween derivatives might guarantee reasonable circulation times in the bloodstream, the ideal choice could be NIO formulations like NIO3 which, among the tested NIOs, absorb relatively low amount of ALB and which are efficiently internalized by cancer cells.
As mentioned before, another protein whose abundance in the PrC negatively correlates with the uptake levels is APOE. This is a component of the plasma lipoproteins, has a major role in the transport of lipids through the plasma and interstitial fluids and is involved in the production, conversion and clearance of plasma lipoproteins themselves.74,76 APOE is also a ligand for low density lipoprotein (LDL) receptors and is involved in cholesterol metabolism.76,77 APOE content is higher in TW80-containing NIOs (NIO2 and NIO4) accordingly to what is elsewhere reported about TW80-NPs.78 Currently, there are several studies testing APOE-enriched coronas of different NPs to investigate drug delivery to cells of central nervous system78 and to hepatocytes:79 in fact, thanks to the binding of APOE to LDL receptors, APOE-enriched coronas can facilitate the NP trafficking into the brain, across the blood–brain barrier,78 and into the liver.79 Intriguingly, APOE enrichment on PEGylated lipid NP surface can either enhance or hamper the uptake in hepatocarcinoma cells, depending on PEG chain length and NP size.79
CD5L is a circulating protein also known as “apoptosis inhibitor of macrophage” (AIM) that acts as a marker for phagocytes so that they can efficiently recognize and engulf cell debris as their targets.80 CD5L is secreted mainly by tissue macrophages and can act with an autocrine mechanisms on the macrophages themselves to modulate inflammatory responses and to enhance the mycobactericidal activity of these cells.81,82 CD5L has been also identified in the PrC of different types of NPs,83,84 however the role of this protein onto the surface of NPs is not completely understood. In agreement with our results showing that CD5L-enriched coronas correlate negatively with uptake levels of NIOs, we may hypothesize that CD5L could enhance in vivo the clearance of NPs by MPS. From this point of view, once again NIO3 formulations seem to be the ideal choice, since they showed relatively low abundance of CD5L in their PrC, if compared with the other tested NIOs.
Among the most interesting proteins, identified in the corona, there are complement factors: in fact, we found C1QB and C1QC, two subunits of C1q protein, the first activator of complement cascade via classical pathway, produced by macrophages and immature dendritic cells and involved in the clearance of immunocomplexes.85 In this context, immunoglobulins, such as IgM and IgG, binding self- and non-self-antigens, modulate the complement activation. In our ex vivo experimental setting, we identified and quantified C1QB and C1QC in all formulations except for NIO4, whose PrC is mostly composed by immunoglobulins. We cannot exclude that the efficiency of complement activation depends on the differences in the PrC composition, particularly on the different immunoglobulin content.86 On the other hand, the major C1QB and C1QC deposition on the NIO1/3 might favor the cellular uptake, whereas C1QB/C-lacking coronas eventually do not promote internalization.
We also evaluated if PrC-NIO uptake could depend by factors other than relative protein abundance, such as the size of naked NPs: but this parameter did not show significant correlation with the uptake in any of the three tested cell lines (Table S5†).
In addition to the cellular uptake analysis, we studied the cytotoxicity of bare and PrC-NIO in serum-free medium containing 1% (v/v) HP to investigate the relationship between internalization of NIOs and their effect on the cell viability. MTT assays were performed at 3 h and 24 h (Fig. 7) on HDF, HeLa and HCT-116 cells after the treatment with NIO formulations. The presence of pre-formed PrC showed no significant variation in cell viability, even for NIO1-100, NIO1-200 that are better taken up from all cell lines than the other NIO formulations, possibly thanks to the safety and low toxicity of TWs.27,49 A comparable cell viability among all treatments enforces cellular uptake data, excluding a contribution of cell death to the observed differences in NIO internalization (Fig. 5).87,88 Indeed, only a slight decrease in cell viability was noticed in the cancer cells (particularly in HeLa) and particularly at 24 h in the case of incubation with NIO1 and NIO3 which had a higher propensity to be internalized by cells. Moreover, the general impact of both PrC-NIO complexes and bare NIOs on cell viability was lower for HDF than for the two tested cancer cell lines. This observation further strengthens the usefulness of NIOs as means for cancer therapy.
4. Conclusions
Up to now, the effects of PrC on NPs have been already studied in human using PEGylated liposomes, that are nanomedicine approved by food and drug administration (FDA) and European Medicine Agency (EMA).13,22 Nevertheless, very few NPs are used as drug delivery system in clinical practice. The gap between pre-clinical and clinical studies probably lies in overlooking some aspects that contribute to the nano-bio interactions. In this context, we studied the nano-bio interface between human plasma (HP, from healthy subjects) and different types of NIOs to define candidate formulations to be potentially applied for drug delivery. Currently, there are no NIO formulations approved by FDA and EMA for intravenous injection and systemic treatment of cancer, despite their potential advantages, herein presented, as drug delivery systems.31 Hence, we investigated the effects of polysorbates (TW20, TW21, TW80), as PEG-mimetic species, on NIO physicochemical features, PrC composition and cellular uptake of PrC-NIO complexes (Fig. 1). The present study demonstrates that the key factors to be considered for an appropriate design of NIOs are: (i) physicochemical properties of NPs and (ii) interactions between NPs and their biological environment. From the physicochemical point of view, the presence of TW21, particularly in combination with TW20 (as observed for NIO3 formulation), guarantees the maintenance of biological stability and size distribution after PrC formation. On the other hand, we observed that PrC composition depends on particle sizes as well as on their net surface charge and formulation. Indeed, size and Z-potential of PrC-NIOs changed over time (Fig. 3) and therefore we cannot exclude that the composition and the relative abundance of PrC might change too.13,18,20,50,52 Although in our experimental protocol the relative abundance of PrC was not analyzed over time, it is highly conceivable that no great change may occur to the hard corona. In addition, we suggest and support that also PrC of polysorbate-NPs, such as NIOs, shows a molecular complexity related to the plasma proteome.In the light of the recent concept of personalized PrC, plasma proteins of patients with different diseases are now believed to generate PrC with different compositions.63,69 In addition, also healthy subjects may display a distinct plasma proteome leading to the formation of individual PrC around the NPs. Characterizing health- and disease-related PrC is fundamental to design therapeutic NPs and to predict their biological response and clinical efficacy. In view of that, this study, testing eight different NIO formulations with the same HP, certainly points out the dynamic nature of PrC whose variations are both qualitative and quantitative. Despite this, we identified differentially represented proteins common to two or three pairwise comparisons (between the same formulations at the two extrusion sizes). In our case, the identification in the corona of disease-related plasma proteins, preferentially and quantitatively adsorbed on the different designed NIOs, strengthens the concept of a personalized PrC.
Moreover, we demonstrated that the type of plasma proteins and their relative abundance affect the intracellular uptake modulating the relative internalization of NIOs in human healthy and cancer cells. In particular, TW21-containing NIOs slightly impaired cellular uptake, suggesting that an alternate pattern of high/low-MW PEG chains seems to be not favorable for NP internalization. Despite this, TW21-containing NIOs display a better biological stability and a narrower size distribution; such a finding should be taken into consideration, if not considered a precious advantage in nanomedicine, provided that TW21 has PEG similar biopharmaceutical features30 at a lower cost and it is able to increase the final steric hindrance of colloidal NPs with a slight modification of the surface properties.
As for PrC composition, it's noteworthy that the relative abundance of five plasma proteins, identified in the corona of all designed NIOs, shows a significant correlation (positive for C1QB/C1QC and negative for ALB, APOE, CD5L) with the uptake in all the three tested cell lines. This could enable to enhance NIO internalization by appropriately modulating the content of the five specific plasma proteins in the PrC-NIO.
Summing up the objective evidence, in all tested cells, PrC-NIOs show a very negligible cytotoxicity confirming the safe nature of polysorbates, thus encouraging their use in nanomedicine applications above all as drug delivery systems; moreover, NIO3 formulations (containing both TW21 and TW20) show the better balance among the key factors quoted above. In fact, these specific NPs guarantee proper physicochemical features and biological stability, absorbs relatively low amount of ALB, APOE and CD5L and high amount of C1QC in the corona and is efficiently internalized by cancer cells.
Given the importance of proteins in the NP corona, our data are highly promising in foreseeing the biological performance of NIOs in different tissues. Further studies need to be extended on the corona of NIOs and we also expect to explore the novel field for all nanomaterials, concerning the so called biomolecular corona, mainly composed of proteins, but also of other biomolecules, such as lipids and metabolites. Characterizing these biomolecules by lipidomics and other metabolomics approaches is still challenging, due to experimental limits; however, they will be instrumental for a comprehensive overview of the phenomena occurring at the nano-bio interface needed to predict the physiological and therapeutic outcome of NPs.
Author contributions
AC, CC, EI, LDM, SO: conceptualization, methodology, investigation, writing – original draft. FS and MF: overall supervision and project planning and administration, writing – review & editing, visualization and revision. In particular, CC and LDM performed the experiments for the synthesis and physicochemical characterization and stability of niosomes, analyzed the resulting data and discussed the relative results. AM and EI performed proteomic analyses. EI analyzed the resulting data, performed bioinformatic experiments and discussed the relative results. AC performed cell culture experiments, analyzed the resulting data and discussed the relative results. AC and MR performed cytofluorimetric analyses and AC analyzed the resulting data and discussed the relative results. SO supervised proteomic, bioinformatic and cell culture experiments. FS and MF drafted and supervised the editing of manuscript and critically revised the results and the whole content of the paper.Conflicts of interest
All authors declare no conflict of interest.Acknowledgements
This manuscript was partially supported by: Ministero dell’Istruzione, dell’Università e della Ricerca [FAR 2017, FAR 2018 (D56C18000780005), FAR 2019 (D54I19002790005)] to C. C. and L. D. M.; Ministero della Salute [RF-2010-23183729] to F. S., and Regione Campania [CIRO project: infrastructures and scientific instrumentation; SATIN "Neoplasia studies" POR Campania FESR 2014/2020; “Predictive Medicine in neoplasia”, L. Regione Campania 752/2019 and 38/2020] all to F. S.References
- A. Cevenini, C. Celia, S. Orrù, D. Sarnataro, M. Raia, V. Mollo, M. Locatelli, E. Imperlini, N. Peluso and R. Peltrini, Pharmaceutics, 2020, 12, 559 CrossRef CAS.
- M. A. Subhan and V. P. Torchilin, Transl. Res., 2019, 214, 62–91 CrossRef CAS PubMed.
- S. T. Yang, Y. Liu, Y. W. Wang and A. Cao, Small, 2013, 9, 1635–1653 CrossRef CAS PubMed.
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Aberg, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Aberg, E. Mahon and K. A. Dawson, Nat. Nanotechnol., 2013, 8, 137–143 CrossRef CAS.
- M. T. Zhu, G. J. Nie, H. Meng, T. Xia, A. Nel and Y. L. Zhao, Acc. Chem. Res., 2013, 46, 622–631 CrossRef CAS.
- T. Cedervall, I. Lynch, M. Foy, T. Berggad, S. C. Donnelly, G. Cagney, S. Linse and K. A. Dawson, Angew. Chem., Int. Ed., 2007, 46, 5754–5756 CrossRef CAS PubMed.
- B. Pelaz, P. del Pino, P. Maffre, R. Hartmann, M. Gallego, S. Rivera-Fernandez, J. M. de la Fuente, G. U. Nienhaus and W. J. Parak, ACS Nano, 2015, 9, 6996–7008 CrossRef CAS PubMed.
- D. Y. Joh, Z. Zimmers, M. Avlani, J. T. Heggestad, H. B. Aydin, N. Ganson, S. Kumar, C. M. Fontes, R. K. Achar and M. S. Hershfield, Adv. Healthcare Mater., 2019, 8, 1801177 CrossRef PubMed.
- J. Zarzar, W. Shatz, N. Peer, R. Taing, B. McGarry, Y. Liu, D. G. Greene and I. E. Zarraga, Biophys. Chem., 2018, 236, 22–30 CrossRef CAS PubMed.
- P. Harder, M. Grunze, R. Dahint, G. M. Whitesides and P. E. Laibinis, J. Phys. Chem. B, 1998, 102, 426–436 CrossRef CAS.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Mazza, R. F. Collins, K. Dawson and K. Kostarelos, ACS Nano, 2015, 9, 8142–8156 CrossRef CAS PubMed.
- M. Hadjidemetriou, S. McAdam, G. Garner, C. Thackeray, D. Knight, D. Smith, Z. Al-Ahmady, M. Mazza, J. Rogan, A. Clamp and K. Kostarelos, Adv. Mater., 2019, 31, e1803335 CrossRef PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. T. Furman, F. Salvatore and E. Tasciotti, Nanomedicine, 2016, 11, 81–100 CrossRef CAS.
- G. Caracciolo, O. C. Farokhzad and M. Mahmoudi, Trends Biotechnol., 2017, 35, 257–264 CrossRef CAS PubMed.
- L. Vroman and A. L. Adams, J. Biomed. Mater. Res., 1969, 3, 43–67 CrossRef CAS PubMed.
- L. Vroman, Nature, 1962, 196, 476–477 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- M. P. Monopoli, F. B. Bombelli and K. A. Dawson, Nat. Nanotechnol., 2011, 6, 11–12 CrossRef CAS.
- M. Hadjidemetriou, Z. Al-Ahmady and K. Kostarelos, Nanoscale, 2016, 8, 6948–6957 RSC.
- S. Milani, F. B. Bombelli, A. S. Pitek, K. A. Dawson and J. Radler, ACS Nano, 2012, 6, 2532–2541 CrossRef CAS PubMed.
- M. Papi, D. Caputo, V. Palmieri, R. Coppola, S. Palchetti, F. Bugli, C. Martini, L. Digiacomo, D. Pozzi and G. Caracciolo, Nanoscale, 2017, 9, 10327–10334 RSC.
- S. Schrittwieser, B. Pelaz, W. J. Parak, S. Lentijo-Mozo, K. Soulantica, J. Dieckhoff, F. Ludwig and J. Schotter, Sci. Rep., 2017, 7, 4752 CrossRef PubMed.
- E. Mahon, A. Salvati, F. B. Bombelli, I. Lynch and K. A. Dawson, J. Controlled Release, 2012, 161, 164–174 CrossRef CAS.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS.
- S. Palchetti, D. Caputo, L. Digiacomo, A. L. Capriotti, R. Coppola, D. Pozzi and G. Caracciolo, Pharmaceutics, 2019, 11, 31 CrossRef CAS PubMed.
- R. Primavera, P. Palumbo, C. Celia, F. Cilurzo, B. Cinque, E. Carata, M. Carafa, D. Paolino, M. G. Cifone and L. Di Marzio, Eur. J. Pharm. Biopharm., 2018, 128, 259–259 CrossRef.
- M. Di Francesco, C. Celia, R. Primavera, N. D'Avanzo, M. Locatelli, M. Fresta, F. Cilurzo, C. A. Ventura, D. Paolino and L. Di Marzio, Int. J. Pharm., 2017, 528, 18–32 CrossRef CAS PubMed.
- C. Marianecci, L. Di Marzio, E. Del Favero, L. Cantu, P. Brocca, V. Rondelli, F. Rinaldi, L. Dini, A. Serra, P. Decuzzi, C. Celia, D. Paolino, M. Fresta and M. Carafa, Langmuir, 2016, 32, 1241–1249 CrossRef CAS PubMed.
- C. Marianecci, L. Di Marzio, F. Rinaldi, C. Celia, D. Paolino, F. Alhaique, S. Esposito and M. Carafa, Adv. Colloid Interface Sci., 2014, 205, 187–206 CrossRef CAS PubMed.
- S. Moghassemi and A. Hadjizadeh, J. Controlled Release, 2014, 185, 22–36 CrossRef CAS PubMed.
- D. Cosco, D. Paolino, R. Muzzalupo, C. Celia, R. Citraro, D. Caponio, N. Picci and M. Fresta, Biomed. Microdevices, 2009, 11, 1115–1125 CrossRef CAS.
- X. M. Ge, M. Y. Wei, S. N. He and W. E. Yuan, Pharmaceutics, 2019, 11, 55 CrossRef CAS PubMed.
- L. Di Marzio, C. Marianecci, M. Petrone, F. Rinaldi and M. Carafa, Colloids Surf., B, 2011, 82, 18–24 CrossRef CAS PubMed.
- Z. M. Zheng and J. P. Obbard, Water Res., 2002, 36, 2667–2672 CrossRef CAS.
- J. Wolfram, K. Suri, Y. Yang, J. L. Shen, C. Celia, M. Fresta, Y. L. Zhao, H. F. Shen and M. Ferrari, Colloids Surf., B, 2014, 114, 294–300 CrossRef CAS PubMed.
- C. Celia, E. Trapasso, D. Cosco, D. Paolino and M. Fresta, Colloids Surf., B, 2009, 72, 155–160 CrossRef CAS PubMed.
- M. Caterino, A. Aspesi, E. Pavesi, E. Imperlini, D. Pagnozzi, L. Ingenito, C. Santoro, I. Dianzani and M. Ruoppolo, Proteomics, 2014, 14, 2286–2296 CrossRef CAS.
- M. Caterino, R. J. Chandler, J. L. Sloan, K. Dorko, K. Cusmano-Ozog, L. Ingenito, S. C. Strom, E. Imperlini, E. Scolamiero, C. P. Venditti and M. Ruoppolo, Mol. BioSyst., 2016, 12, 566–574 RSC.
- E. Imperlini, M. Gnecchi, P. Rognoni, E. SabidÃ, M. C. Ciuffreda, G. Palladini, G. Espadas, F. M. Mancuso, M. Bozzola and G. Malpasso, Sci. Rep., 2017, 7, 1–19 CrossRef CAS.
- E. Imperlini, S. Spaziani, A. Mancini, M. Caterino, P. Buono and S. Orru, Proteomics, 2015, 15, 1813–1818 CrossRef CAS.
- E. Nigro, E. Imperlini, O. Scudiero, M. L. Monaco, R. Polito, G. Mazzarella, S. Orru, A. Bianco and A. Daniele, Respir. Res., 2015, 16, 74 CrossRef CAS PubMed.
- V. Capobianco, M. Caterino, L. Iaffaldano, C. Nardelli, A. Sirico, L. Del Vecchio, P. Martinelli, L. Pastore, P. Pucci and L. Sacchetti, Sci. Rep., 2016, 6, 25270 CrossRef CAS PubMed.
- M. Costanzo, A. Cevenini, E. Marchese, E. Imperlini, M. Raia, L. Del Vecchio, M. Caterino and M. Ruoppolo, Int. J. Mol. Sci., 2018, 19, 3580 CrossRef PubMed.
- A. Parodi, S. G. Haddix, N. Taghipour, S. Scaria, F. Taraballi, A. Cevenini, I. K. Yazdi, C. Corbo, R. Palomba, S. Z. Khaled, J. O. Martinez, B. S. Brown, L. Isenhart and E. Tasciotti, ACS Nano, 2014, 8, 9874–9883 CrossRef CAS PubMed.
- S. Vranic, N. Boggetto, V. Contremoulins, S. Mornet, N. Reinhardt, F. Marano, A. Baeza-Squiban and S. Boland, Part. Fibre Toxicol., 2013, 10, 2 CrossRef CAS.
- M. Manconi, D. Valenti, C. Sinico, F. Lai, G. Loy and A. M. Fadda, Int. J. Pharm., 2003, 260, 261–272 CrossRef CAS.
- D. Paolino, M. L. Accolla, F. Cilurzo, M. C. Cristiano, D. Cosco, F. Castelli, M. G. Sarpietro, M. Fresta and C. Celia, Colloids Surf., B, 2017, 155, 266–275 CrossRef CAS.
- R. Primavera, M. Di Francesco, A. De Cola, V. De Laurenzi, D. Paolino, M. Ciancaioni, M. Carafa, C. Celia, C. Di Ilio, A. Di Stefano, M. Fresta, M. Locatelli and L. Di Marzio, Colloids Surf., B, 2015, 135, 575–580 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- R. A. Sperling, T. Pellegrino, J. K. Li, W. H. Chang and W. J. Parak, Adv. Funct. Mater., 2006, 16, 943–948 CrossRef CAS.
- A. L. Barran-Berdon, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Lagana, Langmuir, 2013, 29, 6485–6494 CrossRef CAS PubMed.
- M. Di Francesco, R. Primavera, S. Fiorito, M. C. Cristiano, V. A. Taddeo, F. Epifano, L. Di Marzio, S. Genovese and C. Celia, Planta Med., 2017, 83, 482–491 CAS.
- A. Gagliardi, S. Bonacci, D. Paolino, C. Celia, A. Procopio, M. Fresta and D. Cosco, Heliyon, 2019, 5, e02422 CrossRef PubMed.
- M. C. Cristiano, F. Froiio, R. Spaccapelo, A. Mancuso, S. P. Nistico, B. P. Udongo, M. Fresta and D. Paolino, Pharmaceutics, 2019, 12, 6 CrossRef PubMed.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, J. Nanobiotechnol., 2013, 11, 26 CrossRef CAS PubMed.
- R. Cai and C. Chen, Adv. Mater., 2019, 31, 1805740 CrossRef CAS PubMed.
- Y. K. Lee, E. J. Choi, T. J. Webster, S. H. Kim and D. Khang, Int. J. Nanomed., 2015, 10, 97–112 Search PubMed.
- E. V. Amirgoulova, J. Groll, C. D. Heyes, T. Ameringer, C. Rocker, M. Moller and G. U. Nienhaus, ChemPhysChem, 2004, 5, 552–555 CrossRef CAS PubMed.
- J. Groll, E. V. Amirgoulova, T. Ameringer, C. D. Heyes, C. Rocker, G. U. Nienhaus and M. Moller, J. Am. Chem. Soc., 2004, 126, 4234–4239 CrossRef CAS PubMed.
- K. Prapainop and P. Wentworth, Eur. J. Pharm. Biopharm., 2011, 77, 353–359 CrossRef CAS.
- S. Winzen, S. Schoettler, G. Baier, C. Rosenauer, V. Mailaender, K. Landfester and K. Mohr, Nanoscale, 2015, 7, 2992–3001 RSC.
- C. Corbo, R. Molinaro, M. Tabatabaei, O. C. Farokhzad and M. Mahmoudi, Biomater. Sci., 2017, 5, 378–387 RSC.
- E. Imperlini, I. Colavita, M. Caterino, P. Mirabelli, D. Pagnozzi, L. D. Vecchio, R. D. Noto, M. Ruoppolo and S. Orrù, J. Cell. Biochem., 2013, 114, 2577–2587 CrossRef CAS.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS.
- D. E. Owens and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef CAS PubMed.
- J. Kreuter, D. Shamenkov, V. Petrov, P. Ramge, K. Cychutek, C. Koch-Brandt and R. Alyautdin, J. Drug Targeting, 2002, 10, 317–325 CrossRef CAS PubMed.
- K. Ogawara, K. Furumoto, S. Nagayama, K. Minato, K. Higaki, T. Kai and T. Kimura, J. Controlled Release, 2004, 100, 451–455 CrossRef CAS PubMed.
- M. G. Vence, M. D. Chantada-Vazquez, S. Vazquez-Estevez, J. M. Cameselle-Teijeiro, S. B. Bravo and C. Nunez, Clin. Chim. Acta, 2020, 501, 102–111 CrossRef PubMed.
- C. Corbo, A. Cevenini and F. Salvatore, Proteomics: Clin. Appl., 2017, 11, 5–6 Search PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, CA-Cancer J. Clin., 2019, 69, 7–34 CrossRef PubMed.
- J. Mariam, S. Sivakami and P. M. Dongre, Drug Delivery, 2016, 23, 2668–2676 CrossRef CAS PubMed.
- K. Bolanos, M. J. Kogan and E. Araya, Int. J. Nanomed., 2019, 14, 6387–6406 CrossRef CAS PubMed.
- H. A. X. Cai and P. Yao, Colloids Surf., B, 2014, 123, 900–906 CrossRef CAS PubMed.
- D. C. Kennedy, H. Qian, V. Gies and L. Yang, Environ. Sci.: Nano, 2018, 5, 863–867 RSC.
- R. W. Mahley, Science, 1988, 240, 622–630 CrossRef CAS PubMed.
- R. W. Mahley, J. Mol. Med., 2016, 94, 739–746 CrossRef CAS PubMed.
- Z. Li, Y. Wang, J. Zhu, Y. Zhang, W. Zhang, M. Zhou, C. Luo, B. Cai, S. Gui, Z. He and J. Sun, J. Controlled Release, 2020, 320, 1–18 CrossRef CAS PubMed.
- D. Y. Chen, S. Ganesh, W. M. Wang and M. Amiji, Nanoscale, 2019, 11, 8760–8775 RSC.
- S. Arai and T. Miyazaki, Semin. Immunopathol., 2018, 40, 567–575 CrossRef CAS PubMed.
- L. Sanjurjo, N. Amezaga, G. Aran, M. Naranjo-Gomez, L. Arias, C. Armengol, F. E. Borras and M. R. Sarrias, Autophagy, 2015, 11, 487–502 CrossRef PubMed.
- L. Sanjurjo, G. Aran, N. Roher, A. F. Valledor and M. R. Sarrias, J. Leukocyte Biol., 2015, 98, 173–184 CrossRef CAS PubMed.
- F. Pederzoli, G. Tosi, F. Genovese, D. Belletti, M. A. Vandelli, A. Ballestrazzi, F. Forni and B. Ruozi, Nanomedicine, 2018, 13, 407–422 CrossRef CAS PubMed.
- M. D. Chantada-Vazquez, A. C. Lopez, M. G. Vence, S. Vazquez-Estevez, B. Acea-Nebril, D. G. Calatayud, T. Jardiel, S. B. Bravo and C. Nunez, J. Proteomics, 2020, 212, 103581 CrossRef PubMed.
- R. A. van Schaarenburg, J. Suurmond, K. L. L. Habets, M. C. Brouwer, D. Wouters, F. A. S. Kurreeman, T. W. J. Huizinga, R. E. M. Toes and L. A. Trouw, Mol. Immunol., 2016, 78, 164–170 CrossRef CAS PubMed.
- V. P. Vu, G. B. Gifford, F. F. Chen, H. Benasutti, G. K. Wang, E. V. Groman, R. Scheinman, L. Saba, S. M. Moghimi and D. Simberg, Nat. Nanotechnol., 2019, 14, 298–298 CrossRef CAS PubMed.
- G. E. Lombardo, V. Maggisano, M. Celano, D. Cosco, C. Mignogna, F. Baldan, S. M. Lepore, L. Allegri, S. Moretti, C. Durante, G. Damante, M. Fresta, D. Russo, S. Bulotta and E. Puxeddu, Mol. Cancer Ther., 2018, 17, 1187–1195 CrossRef CAS PubMed.
- D. Cosco, F. Cilurzo, J. Maiuolo, C. Federico, M. Di Martino, M. C. Cristiano, P. Tassone, M. Fresta and D. Paolino, Sci. Rep., 2015, 5, 17579 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr07229j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |