Towards a point-of-care SERS sensor for biomedical and agri-food analysis applications: a review of recent advancements
Jayakumar
Perumal
,
Yusong
Wang
 ,
Amalina Binte Ebrahim
Attia
,
Amalina Binte Ebrahim
Attia
 ,
U. S.
Dinish
,
U. S.
Dinish
 * and
Malini
Olivo
*
* and
Malini
Olivo
*
Laboratory of Bio-Optical Imaging, Singapore Bioimaging Consortium (SBIC), Agency for Science Technology and Research (A*STAR), Singapore. E-mail: Dinish@sbic.a-star.edu.sg; Malini_Olivo@sbic.a-star.edu.sg
First published on 8th December 2020
Abstract
The growing demand for reliable and robust methodology in bio-chemical sensing calls for the continuous advancement of sensor technologies. Over the last two decades, surface-enhanced Raman spectroscopy (SERS) has emerged as one of the most promising analytical techniques for sensitive and trace analysis or detection in biomedical and agri-food applications. SERS overcomes the inherent sensitivity limitation associated with Raman spectroscopy, which provides vibrational “fingerprint” spectra of molecules that makes it unique and versatile among other spectroscopy techniques. This paper comprehensively reviews the recent advancements of SERS for biomedical, food and agricultural applications over the last 6 years, and we envision that, in the near future, some of these platforms have the potential to be translated as a point-of-care and rapid sensor for real-life end-user applications. The merits and limitations of various SERS sensor designs are analysed and discussed based on critical features such as sensitivity, specificity, usability, repeatability and reproducibility. We conclude by highlighting the opportunities and challenges in the field while stressing the technological gaps to be addressed in realizing commercially viable point-of-care SERS sensors for practical biomedical and agri-food technological applications.
1. Introduction
Molecular-level detection of chemicals, biological analytes and metabolites can be the gateway to elucidating biological processes, gaining insight to cellular functions and evaluating therapeutic efficacy. Detection and the unique identification of target analytes have become integral in the fields of medical diagnostics,1 disease monitoring,1 forensic science,2 water or environmental monitoring,3 food nutrient content4 and food safety4 testing. This warrants a sensing method that is rapid, molecular specific and sensitive enough to detect trace amounts in very small volumes and in complex environments. Established chemical sensing methods include mass spectrometry (MS), liquid chromatography (LC) or the combination of these two approaches.5 While the LC/MS approach exhibits unparalleled sensitivity and specificity, it involves collecting samples to be analysed before transferring to a laboratory for quantification, rendering this approach laborious, time-consuming, confined to a sophisticated laboratory space and requiring very high capital investment and well-trained operators. Magnetic and plasmonic analysis techniques are some of the other methods implemented in laboratories for rapid and sensitive trace analytes detection.6 However, these techniques are time-consuming, economically unviable, possess low sensitivity and have a noisy background. Therefore, there is a need for a more compact and rapid analyte detection approach at the point-of-sample with minimal steps to meet these challenges.In the last decade, surface-enhanced Raman spectroscopy (SERS)-based ultrasensitive sensing platform has gained much attention as a promising methodology to detect targeted molecules as it offers high specificity, high sensitivity and a quick readout. When an analyte is excited by photon absorption, the Raman-scattered photons shift to lower energy, while the molecule gains its unique vibrational energy. The Raman analytical method involves measuring the Raman-scattered photons to produce a molecular fingerprint in the form of a Raman spectrum. Nevertheless, the Raman effect is intrinsically weak and not sufficient to detect low concentrations of the analyte. The Raman effect can be amplified in the orders of 104–1010 when the analyte is adsorbed onto metallic nanostructures, a phenomenon known as SERS. This can be attributed to the transfer of induced energy from the metal surface to the adsorbed analyte, enhancing the scattering of the analyte.7 Thus, SERS is capable of detecting trace concentrations of specific analytes provided that the molecules are in close vicinity to metallic nanostructures (∼few nm) on SERS substrate platforms. Typically, a droplet of liquid sample is deposited onto a rigid silicon or glass substrate functionalized with a plasmonic nanostructured surface followed by the adsorption of analyte molecules onto the surface as the sample dries.
SERS has been exploited in various measurement methodologies in analytical and biological chemistry since its first reported use as a spectro-chemical approach.8 SERS-based analytical strategies can be categorized as label-mediated or label-free methodologies. The former involves detecting the analyte indirectly whereby the SERS active molecule is functionalized as a label or a reporter molecule onto the capturing moiety and the resultant SERS signal of the reporter is detected as a surrogate measure of the analyte, enhancing the interface specificity of detection. This approach has demonstrated its capabilities in SERS-based immunoassays9 whereby detection of analytes is possible in multiplex environments. Direct label-free detection of analytes, however, mandates the analyte to be SERS-active and its environment free of any interfering molecules to maintain high specificity. Meanwhile, the optical properties of a SERS substrate affect the detection sensitivity.
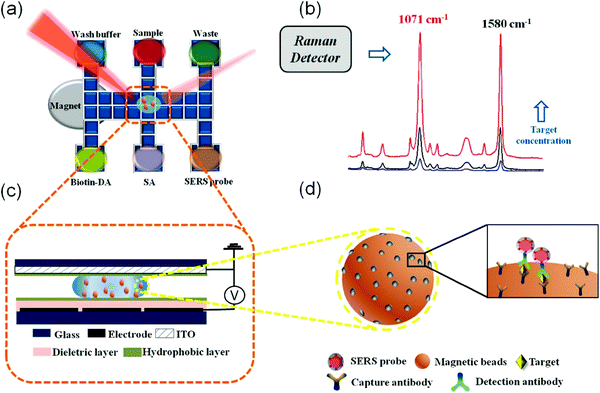
Point-of-care or POC SERS analysis is one emerging application where SERS measurements are acquired on-site as opposed to conventional laboratory-based testing using benchtop SERS instruments. POC, as originally mandated by the World Health Organization (WHO) for HIV testing, has a generic guideline with the criteria of ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable to end-users), especially for low-resource settings.10 Most POC detection methods adopt a lateral fluidic immunoassay (LFIA) platform, such as paper-based glucose strips or pregnancy tests, which do not require formal training for home use. However, they often exhibit low sensitivities. SERS with significant signal enhancement via “hot spots” creation11 shows immense potential to address the sensitivity limitation, with POC SERS arousing more attention. On the other hand, due to the limitation of current Raman instrumentation development and the SERS substrate, POC SERS raised some specific issues. The design of POC SERS is especially centred on the Raman reader (whether it is portable, handheld or movable) and one issue is the integration of a portable Raman reader into a general diagnostic system for on-site and rapid detection. Another issue is the SERS substrate, where various flexible materials have been exploited in SERS substrates for SERS-based POC diagnostics,12 including paper,13 flexible polymers,14 graphene15 and nanowires,16 each contributing different applications to the substrate by virtue of the materials' characteristics. These materials allow the production of SERS substrates to be affordable, disposable and scalable for mass production. To preserve the high specificity of the SERS technique, careful consideration has to be undertaken in the molecular functional interface of these flexible substrates to interact specifically with the analytes in the sample environment. Thus, the key parameters of POC SERS include portability (for the whole sensor system including the Raman detector) and affordability (in favour of mass production and low resource settings) besides sensitivity and specificity. Common forms of the POC-SERS technique adopt LFIA formats but with Raman as the signal output (Scheme 1). A rapid POC SERS sensor with these characteristics is highly desirable for end-users of POC diagnostics.
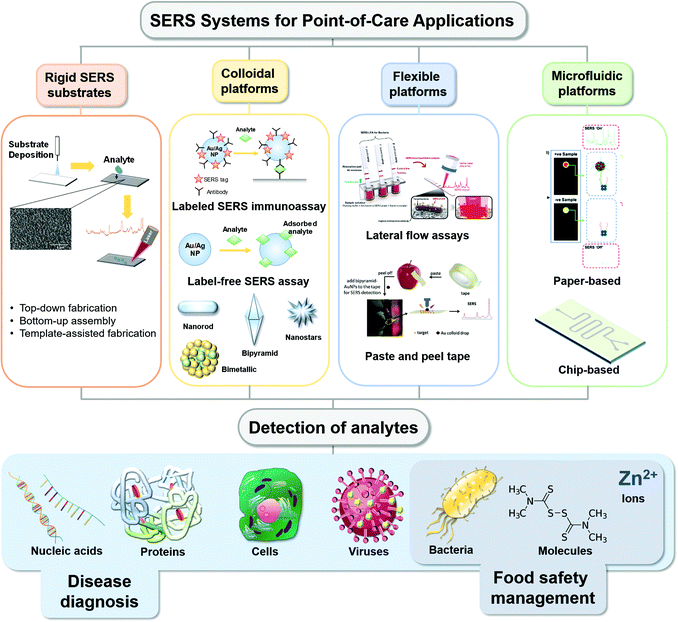 | ||
| Scheme 1 Schematic illustration of reported various types of point-of-care SERS platforms and the different types of analytes detected for various applications. Reproduced with permission from ref. 51. Copyright 2019, Elsevier. Reproduced with permission from ref. 98. Copyright 2020, Royal Society of Chemistry. Reproduced with permission from ref. 139. Copyright 2017; Elsevier. | ||
Several interesting review articles have come out recently, selectively focused on the use of SERS POC type devices for different kinds of biochemical applications such as colloidal nanoparticles with immunoassay techniques17 or flexible substrates for bio-sensing14 and nano-colloids for pathogen diagnostics.18 Seeing the value presented by SERS-based POC analytical strategies, we herein present a review and comprehensive discussion of several approaches in recent practical applications in biomedical diagnostics, detection of infectious agents, food science applications, etc. (Scheme 1). Our review comprises various substrate media that were used from colloidal, rigid planar, flexible planar to microfluidics-based surfaces that are being explored for different applications. In order to show the growing interest in the SERS space for biomedical and agri-food domains over the years, we have plotted a graph showing that the number of publications of SERS in these applications has increased steadily in recent years, evident from the number of hits achieved from Web of Science with the keywords “SERS sensor”, “SERS biosensor” and “SERS for food safety” (Fig. 1). We also highlight the limitations and future directions in challenges faced in the paradigm shift of the SERS-based analytical tool from the laboratory to POC applications.
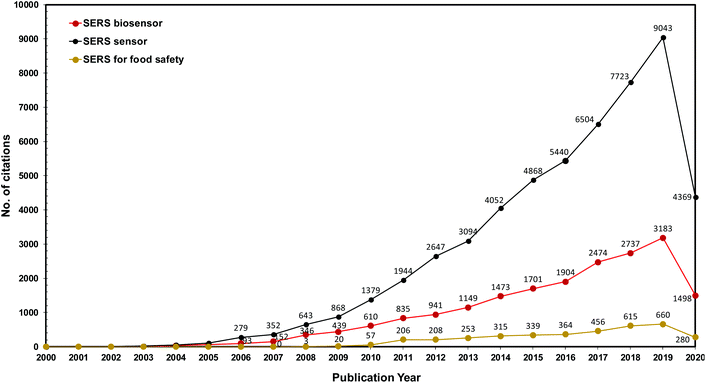 | ||
| Fig. 1 The number of citations found in the Web of Science® database using the keywords “SERS sensor”, “SERS biosensor” and “SERS for food safety” from 2000 to June 2020. | ||
2. SERS-based applications in agri-food analysis
In agricultural and food safety analysis, the gold standard methods for analyte detection are chromatographic-based “wet chemistry” techniques such as gas chromatography (GC) and high-performance liquid chromatography (HPLC) followed by mass spectroscopy (MS). The major limitations with these methods are the requirement for dedicated lab space, costly systems with expensive upkeep and the need for highly trained operators. Additionally, they involve cumbersome analyte extraction and time-consuming sample preparation steps. Our proposed economical POC SERS method offers the possibility for short sample processing time, ease of operation and minimally invasive, on-site detection by virtue of portable hand-held Raman devices. In this section, we will cover an overview of SERS techniques for various agri-food applications that result in reliable quantitative and qualitative analysis.2.1 Detection of nutrients and plant stress
Radu et al.19 adopted a SERS-based method for the simultaneous detection of two variants of vitamin B (riboflavin and cyanocobalamin) in cereal fortified by vitamins by fabricating SERS active substrates in-house using e-beam lithography on a silicon wafer followed by evaporation of the gold layer. Fig. 2 shows the extraction procedures used for the separation of vitamin B from a complex fortified cereal product, which can be further adapted and extended to other food products. One of the key findings was to maintain a fixed pH (i.e. pH 7) for obtaining optimum SERS spectral features under a confocal Raman microscope, as the detection sensitivity was greatly affected in other pH conditions. They were successful in detecting two of the vitamin B variants with a limit of detection (LOD) of 100 nM by maintaining the experimental parameters such as pH and method of digesting the food to analyse for its individual components. The time taken for SERS analysis was within 7 h compared with the lengthy combination of HPLC and microbiological assay which often requires more than 26 h.
 | ||
| Fig. 2 Schematic shows the process of bringing the analytes into the liquid medium for analysis. The extraction protocol involves mincing the sample and dissolving it into suitable solvents to extract the analytes into the liquid medium before the analytes are chemisorbed onto the SERS enhancing platform. Reproduced with permission from ref. 19. Copyright 2016, Elsevier. | ||
In another study,20 a label-free SERS spectroscopy technique was developed for white wine characterization. One hundred and eighty wine samples were selected for testing, consisting of white wine varieties namely Sauvignon Blanc, Ribolla Gialla and Friulano, collected from three different Italian producers in the northeastern Italy region. Sample testing was done using citrate-capped Ag colloidal nanoparticles and a portable Raman device with a 785 nm excitation wavelength as a laser source. Spectral differences between the samples arose from changes in the relative ratios of metabolites (adenine, adenosine, carboxylic acids and glutathione) present, depending on the type and production method of the wines. In addition, the results were obtained with the multivariate predictive model using the relative concentration of metabolites to classify the wine based on its specific variety and its geographical origin with up to 93% high accuracy.
Similarly, other nutrient components such as anti-oxidants and flavonoids have been studied using the SERS platform. Aguilar-Hernández et al.21 used silver colloids as an enhancing medium for SERS detection of phenolic antioxidants, caffeic acid (CA), ferulic acid (FA), p-coumaric acid (4CA) and sinapic acid (SA). Chemometric analysis was performed with the principal component analysis (PCA) method to classify the antioxidant analytes based on nanoparticle size and analyte concentration. It was established that the ideal pH condition of the colloidal silver medium for highest SERS enhancement was between pH 5 and 6.5, because changes in the pH affected the analyte molecular conformation and how it reacts with the silver colloid particles under a confocal Raman microscope. Antioxidants CA and 4CA were reported to have a LOD of 2.5 × 10−9 M. Even though low concentration detections of some of the antioxidants were demonstrated in this study, further investigations are warranted to validate the reproducibility of LODs and to achieve acceptable sensitivity below permissible detection limits.
SERS sensors are not only limited to analysing liquid analytes; with the appropriate design of SERS sensors, they could potentially be employed for the detection of gaseous substances. Park et al.22 developed a novel SERS substrate by depositing a layer of silver-nanosphere (AgNS) on a Tenax-TA polymer film to detect the volatile organic compounds (VOCs) emitting from living or preserved plant materials. In this study, the film-based SERS substrate was able to adsorb and simultaneously detect naturally evaporated VOCs from three validated VOC standards, enabling the discrimination between the three standards while detecting the difference in VOC emission at collection times. The capability of this SERS method was demonstrated through the detection and differentiation of three types of tea blends using a benchtop Raman system, and the differentiation between healthy and caterpillars infested cotton plants based on their VOC emission in which the wavenumber regions in the SERS spectra contributing in the classification process were between 1550 and 1750 cm−1. Cotton plants infected with caterpillars emitted additional VOCs such as (E)-2-hexenal, (E)-2-hexenyl acetate, caryophyllene, humulene and isoamyl acetate, whereas α-pinene was found in both healthy and infected plants.
SERS has proven to be a poor choice of an analytical method for inorganic molecular species due to their intrinsically low Raman cross-sections and a lack of vibrational modes in their atomic systems.23 However, in an interesting study, Brackx et al.24 reported a SERS-based method to detect Zn2+ ions in water as a surrogate measure of water contamination. The simple and frugal SERS method firstly involves randomly aggregating colloidal silver particles upon addition of a complexometric marker, acquiring a SERS spectrum with a portable Raman spectrometer and assessing data employing multivariate methods. Partial least square (PLS) regression has been employed as the multivariate method, allowing large datasets to be acquired simultaneously. This sensor had a detectable range between 160 and 2230 nM in pure water with an accuracy of 96% and fidelity of 4%. While the LOD is higher than that of gold standard methods for the detection of inorganic molecules, its sensitivity is sufficient enough to detect relevant concentrations of Zn in water bodies. Shi et al.25 developed a unique miniaturized silicon SERS analytical sensing method by combining with an internal standard. The resultant sensor was capable of multiplexing, with sensitive and reliable detection of heavy metallic ions present in industrial wastewater demonstrated in real-life systems. The SERS sensor was composed of a silicon wafer with dense packing of Ag NPs along with functionalization with 4-ATP (internal standard molecule) and Pb2+/Hg2+ responsive DNA strands. LODs of 9.9 × 10−11 M for Pb2+ and 8.4 × 10−10 M for Hg2+ were reported respectively, under a confocal Raman microscope. The LOD obtained in the current study was close to two orders of magnitude lower than the maximum residue limit (MRL) defined by US-EPA.
These investigations show the potential for molecularly mediated SERS to characterize food products in a fast turnaround time and with superior sensitivity. In the future, it would be desirable to develop SERS sensors with a short SERS analysis time by avoiding lengthy extraction steps along with the use of stable colloidal nanoparticles that are inert to pH changes for analyte detection. Although only a few SERS-based studies have been reported in this space, more development is needed to establish sensitive detection methods that are rapid and effortless. A SERS sensor for nutrient detection in the food space will likely gain major traction in the coming years with the availability of cheap, stable and high laser power portable Raman systems along with a more reliable and reproducible SERS enhancement medium.
Zhang et al.26 reported a swift SERS-based analysis of residual synthetic cytokinins i.e. 6-benzylaminopurine (6-BAP) using gold colloids and a portable Raman spectrometer for readout. 6-BAP is generally used for stimulating plant growth and growth regulator in stress responses. The major bands in the fingerprint region for 6-BAP were at 738, 1002, 1318 and 1336 cm−1 wavenumber, which represents the vibrational modes of adenine ring. From the acquired SERS spectra, the presence of 6-BAP in sprout extract was demonstrated with the Raman band at 1002 cm−1, increasing in intensity as the 6-BAP concentration increases. A LOD of 0.33 μg mL−1 for 6-BAP detection was reported with the linear correlation in the concentration range 0.1–5.0 μg mL−1.
Anthocyanins are plant pigments that are natural compounds responsible for the various non-green colours such as blue, purple and red. They are mainly derivatives of 2-phenylbenzopyrylium salts, which exist as glycosylated molecules in nature. Anthocyanins are produced in plants in reaction to abiotic stress factors, e.g. drought, high salinity, excess light and cold. Zaffino et al.27 developed a SERS sensor for the detection of anthocyanidins. SERS spectral characterization and pH dependence of anthocyanidins were investigated. The SERS spectra of six derivatives of anthocyanidins: cyanidin, delphinidin, pelargonidin, peonidin, malvidin and petunidin, exhibited distinct differences despite the slight disparities in molecular structures. SERS has presented itself as an attractive option to do a rapid label-free detection of the derivatives of anthocyanidins. A micro-Raman system was used for SERS spectral measurements.
Yang et al.28 developed innovative SERS techniques for real-time monitoring of pesticide penetration or diffusion inside tomato plant tissues. Various concentrations of a pesticide, thiabendazole (TBZ), were introduced into hydroponic systems to be feed to tomato plants. While SERS detection of TBZ has been established before, a unique SERS peak at 737 cm−1 on the plant tissue at day 4 and 6 was additionally detected under a confocal Raman microscope. This peak is thought to correspond to adenine-moiety by virtue of the plant's response to toxic pesticides.
Wang et al.29 reported a SERS technique employing an Ehrlich test followed by mixing with gold nanoparticles (AuNPs) to generate hotspots for aromatic plant hormone detection. Plant hormones mediate the plants’ tolerance to stress and regulate their response. The authors demonstrated that p-(dimethylamino) benzaldehyde (PDAB) undergoes selective reaction with indole-3-butyric acid (IBA) over indole-3-acetic acid (IAA) to form the effective IBA-PDAB cation due to high selectivity and preference for IBA over that of IAA. Fig. 3A represents the schematic illustration for the detection process of IBA in mung bean sprout. Results from the SERS method were consistent with that of the HPLC results. As shown in Fig. 3C, the Raman band at the wavenumber region 1498 cm−1, under a confocal Raman microscope, was of high intensity at both the hypocotyl and root tip (see the red line) compared with very weak Raman signal intensity in the cotyledon. The endogenous IBA concentration showed an increasing trend from cotyledon < leaf < hypocotyl < root tip in which the IBA level was found to be 14.8, 63.7, 154 and 181 ng g−1, respectively. The reported LOD for IBA was 2.0 nM. This label-free SERS method showed high potential for the screening of plant hormones in real-time.
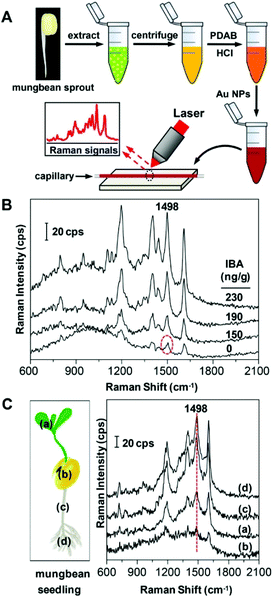 | ||
| Fig. 3 (A) Schematic drawing for the detection of IBA via SERS. (B) SERS spectra of the mung bean sprout extract spiked with IBA. (C) SERS spectra of various parts of mung bean seedling extract: (a) leaf, (b) cotyledon, (c) hypocotyl and (d) root tip. Reproduced with permission from ref. 29. Copyright 2017, ACS Publications. | ||
A field-ready and ultrasensitive SERS assay for the detection of dipicolinic acid (DPA), which was a biomarker for bacterial spores and especially Bacillus anthracis, has been reported.30 Plant and microbiome interactions are thought to ameliorate abiotic and biotic stress. The study employed environmentally safe mercury(II) ions to aid the sensing approach for rationally designed SERS-active AuNPs, which led to the indirect detection of DPA, controllable aggregation of special Au colloids for the detection of Hg2+ ions and DPA with a concentration range (1 nM–8 μM) under a confocal Raman microscope. The authors were able to achieve a LOD for the indirect SERS method and were able to detect DPA as low as a LOD of 0.01 ppb.
Notably, the major limitation of these previously mentioned SERS methods involves destroying the plant samples to detect the analyte of interest efficiently. To address this limitation, there are reported imaging-based methods integrated with SERS allowing longitudinal real-time measurements to monitor the health of the plants at different stages of their growth. For example, Crawford et al. demonstrated the use of the plasmonic nanoprobes based multimodal method for in vivo imaging and biosensing of microRNA biotargets within whole plant leaves.31 In another study, Wang et al. integrated three different and complementary techniques: SERS, X-ray fluorescence (XRF) and plasmonics-enhanced two-photon luminescence (TPL) for in vivo functional imaging of the target nucleic acid with an excellent spatio-temporal resolution of 200 μm and 30 min, respectively.32
2.2 Detection of contaminants and chemicals
Various SERS techniques for pesticide detection make use of principle sampling methods such as swabbing via flexible-SERS substrates, use of cotton swabs and SERS active tape for “press and peel-off”.46–48 As an example, a gecko-inspired nanotentacle substrate was developed for easy sampling and direct detection of pesticides, giving a LOD for thiram, a fungicide, on apple peels of 1.6 ng cm−2.47 Similarly, Chen et al.49 reported a jellylike, flexible silver nanocellulose (Ag-NC) as a highly sensitive SERS substrate for pesticide detection, granting an improved thiram LOD of 0.5 ng cm−2. In this study, fungicides thiabendazole (TBZ) and thiram solutions were combined with the Ag-NC substrates before adsorbing to aluminium plates while apple peels tainted with the two fungicides were prepared with the same substrates for SERS measurements using portable Raman spectrometer. The prominent Raman bands in the SERS spectra for TBZ detection were found to be 783, 884 and 1010 cm−1, whereas the representative Raman bands for thiram were at 564, 938, 1148, 1386 and 1514 cm−1.
As an alternative to conventional thermoplastic thin and flexible SERS substrates, sticky tapes were effective in the analyte collection from any type of sample surface from flat to curved structures by means of a simple “press and peel off” technique. Chen et al.50 developed adhesive tape decorated with gold colloidal particles as a SERS tape sensor for the detection of pesticides in fresh produce. The SERS tape offers flexibility for full contact to touch the surface of produce and considerable adhesion to extract analytes efficiently. The SERS tape has been demonstrated to acquire distinct SERS spectra of the pesticides parathion-methyl, thiram and chlorpyrifos in produce such as green vegetables, cucumber, orange and apple under a confocal Raman microscope. Gong et al.33 similarly demonstrated the capability of the adhesive tape-based SERS platform for the direct detection of pesticide residue from the surface of fruit peels with a portable Raman spectrometer. Different types of adhesive tapes were tested on various surfaces such as aluminium foil, glass and fruit peels for their effectiveness and collection efficiency. The highest collection efficiency of 60.2 ± 7.6% was observed for glass followed by aluminium foil and fruit peel with 54.3 ± 5.0% and 52.3 ± 9.0%, respectively. After the analyte collection, silver nanoparticles were used for SERS enhancement. A LOD of 0.0225 mg kg−1 was reported for triazophos pesticide, which is below the MRL of 0.2 mg kg−1. In another study by Wu et al.,51 colloidal bipyramid gold nanoparticles (BP-AuNPs) were used instead of the conventional spherical or star-shaped gold colloids as an enhancing medium for the target analyte. Fig. 4 shows the schematic of the BP-AuNPs based SERS adhesive tape sensor for trace pesticide detection on the surfaces of products using a portable Raman spectrometer. In this study, SERS detection of trace levels of methyl parathion on the peels of apples, cucumbers and tomatoes were demonstrated, granting LODs of 98.63 ng cm−2 for cucumber, 31.56 ng cm−2 for tomatoes and 36.58 ng cm−2 for apples.51 Flexible SERS sensors could also detect TBZ fungicide down to sub μM 1 μM (0.2 ppm) on orange peel in situ which was much lower than the permissible MRL specified by the EU under a confocal Raman microscope, as demonstrated by Wang et al.52 The detection limit for TBZ was further improved by using gold nanorods (AuNRs) as the SERS enhancing medium in apple samples – down to 0.06 ppm.53 Alsammarraie et al.54 demonstrated the rapid identification of TBZ pesticides liquid samples (lemon, carrot and mango juice) by exploiting gold nanorods assembled onto the silicon surface as the SERS substrate under a confocal Raman microscope. Partial least squares analysis was employed to accurately detect TBZ with R values of ≥0.98 for all the juice samples. The LOD for TBZ in lemon was reported as 149 μg L−1, while carrot and mango juice LODs were 216 and 179 μg L−1, respectively.
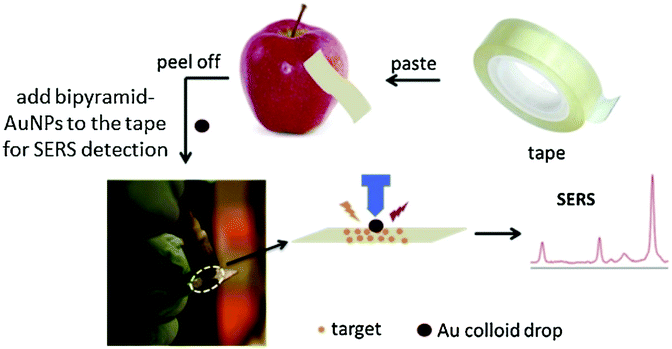 | ||
| Fig. 4 Detection of pesticides using SERS substrates. The schematic shows the use of modified adhesive tape modified with bipyramid-gold nanoparticles (AuNPs) as a SERS sensor for pesticide residue detection on the surface of fruit peels. Reproduced with permission from ref. 51. Copyright 2019, Elsevier. | ||
The presence of graphene in the SERS substrate can assist in the enrichment of pesticide analytes due to its high adsorption capability for organic aromatic compounds by virtue of the π–π interaction as demonstrated by Sun et al.55 They presented a PMMA/Ag NPs/graphene-based flexible SERS 2D substrate to detect trace levels of thiram in apple juice with a LOD of 0.24 ppm under a confocal Raman microscope, which was clearly below the MRL of 7 ppm as prescribed by US-EPA. The use of graphene for analyte enrichment has a great potential for in situ detection of residual pesticide in food products.52
An alternative substrate medium other than flexible adhesive tapes and polymer thin films can include textile fibres with liquid-crystal polymer (LCP) coated with silver nanoparticles (Ag NP).56 The substrate was capable of detecting trace quantities of thiram that were spiked onto real samples with a LOD of 0.024 ppm, which was more than an order lower than the permitted MRL of 5 ppm in fruits. Similarly, the pesticide carbendazim, which is a potential carcinogen, was detected in oolong tea with a LOD of 0.1 ppm, with gold colloids as an enhancing medium.57 Polymer films as SERS substrate can also be advantageous as they cover the produce surface while promoting the self-assembly of AuNPs and immobilizing them on the film. The SERS sensor developed by Wu et al.58 was tested for the suitability on the field with a portable Raman spectrometer and it was able to detect thiram with its representative Raman bands at 551 cm−1 and 1378 cm−1 from the SERS spectra.
Knowledge of the penetration profiles of pesticides in fresh produce is just as important to reduce systemic pesticide residues in food. Yang et al.34 showed the use of a SERS mapping method with AuNPs as permeating probes to monitor the penetration profile of TBZ in spinach leaves under a confocal Raman microscope after removing residues of it on its surface. The same technique was also applied to detect the penetration of multiclass pesticides in additional products such as apples and grapes.59 These studies were done with non-systemic pesticides as controls, revealing that systemic pesticides penetrate deeper into fresh produce compared with non-systemic pesticides.
| Chemical/pesticide | Category of pesticide | Laser wavelength | SERS enhancement medium | Sample type | LODs | Ref. |
|---|---|---|---|---|---|---|
| Pesticide | ||||||
| Thiram | Dithiocarbamate | 785 nm | Ag NPs on Au film-coated nanospheres | Acetone | 0.24 ppm | Guo et al. (2015)39 |
| 632.8 nm | Au@Ag NPs | Orange | 10−7 mol L−1 | Wang et al. (2015)38 | ||
| 0.24 ng cm−2 | ||||||
| 514 nm | AgNRs embedded PDMS | Apple peel | 10−9 g cm−2 | Kumar et al. (2017)60 | ||
| 532 nm | PMMA/Ag NPs/graphene film | Apple juice | 0.24 ppm | Sun et al. (2017)55 | ||
| 785 nm | Ag/NC film | Apple peels, cabbage | 0.5 ng cm−2 | Chen et al. (2019)49 | ||
| 785 nm | AuNPs/PVC film | Apple peel | 10 ng cm−2 | Wu et al. (2019)58 | ||
| Methyl parathion | Organophosphate | 532 nm | AgNC@ PE blended film | Orange | 10 nM | Zhou et al. (2016)37 |
| 633 nm | Au NPs@tape | Vegetables and fruit peel | 26.3 ng cm−2 | Chen et al. (2016)50 | ||
| 785 nm | Bipyramid Au nanoparticles (BP-AuNPs) | Apples, cucumbers, tomatoes | 36.58 ng cm−2, 98.63 ng cm−2, 31.56 ng cm−2 | Wu et al. (2019)51 | ||
| 785 nm | Au nanorod (AuNR) | Lake water, orange peel, apple peel, plant leaves | 1 μM, 0.22 μg cm−2, 0.11 μg cm−2, 0.44 μg cm−2 | Wu et al. (2019)61 | ||
| Thiabendazole | Benzimidazole | 785 nm | Au NP-based ultrafiltration membrane | Orange | 0.125 μg g−1 | Hong et al. (2017)62 |
| 632 nm | Au-coated tattoo copper foil/quartz/glass/Al | Orange peel, sports drink | 0.1 μM, 0.0009 vol% | Wang et al. (2018)52 | ||
| 785 nm | Au nanorods (AuNRs) array | Methanol, apples | 0.037 mg L−1, 0.06 ppm | Fu et al. (2019)53 | ||
| 633 nm | Au@Ag/PMMA/qPCR-PET film chip | Pear juice, orange juice, grape juice | 21 ppb, 43 ppb, 69 ppb | Wang et al. (2019)32 | ||
| Carbaryl (1-naphthyl methylcarbamate) | Carbamate | 785 nm | Standing AuNRs | Orange juice | 509 ppb | Alsammarraie and Lin (2017)63 |
| 785 nm | Standing AuNRs | Grapefruit juice | 617 ppb | |||
| Carbendazim | Benzimidazone | 633 nm | AuNPs | Tieguanyin, oolong tea | 0.1 ppm | Chen et al. (2019)57 |
| Chlorpyrifos | Organophosphate | 632.8 nm | Au nanopopcorn | Pear peel | 0.35 mg kg−1 | Xu et al. (2017)41 |
| Dimethoate | Organophosphate | — | AuNPs | Spinach leaves | 0.1 mg kg−1 | Hou et al. (2017)45 |
| Phosmet | Organophosphate | 785 nm | Functionalized AgNPs | Apples | 0.2 mg kg−1 | Pan et al. (2015)64 |
| Oranges | 0.2 mg kg−1 | |||||
| Triazophos | Organothiophosphate | 785 nm | AgNPs | Apple peel, cherry tomatoes | 25 ng cm−2, 0.0150 mg kg−1 | Gong et al. (2019)33 |
| Contaminant | ||||||
| Melamine and Sudan I | Cyanamide, carcinogenic azo dye | 532 nm | Superhydrophobic-3D Ag nanowire | Water, milk | 0.1 nM, 1 μM | Li et al. (2014)65 |
| Aflatoxin B1 (AFB1) | Mycotoxin | 633 nm | Magnetic beads and silica-encapsulated hollow AuNPs | Tap water | 0.1 ng mL−1 | Ko et al. (2015)66 |
| 633 nm | Au coated glass slide with PET film | TE buffer | 0.4 fg mL−1. | Li et al. (2017)67 | ||
| Microcystin-LR (MC-LR) | Hepatotoxin | 785 nm | Au coated magnetic NPs | Horse blood plasma | 10 fM | Hassanain et al. (2017)68 |
| Bisphenol A (BPA) | Organic synthetic compound, diphenylmethane derivatives | 1064 nm | Au nanostars (AuNSs) | PBS | 0.073 ppb | Lin et al. (2018)69 |
| Okadaic acid (OA), dinophysistoxin-1 (DTX-1)), dinophysistoxin-2 (DTX-2), yessotoxin (YTX) | Marine biotoxins | 532 nm or 785 nm | AgNPs | Shell mussel tissue | 84.6 nM, 0.877 μM, 0.46 μM, 0.259 μM | Pinzaru et al. (2018)70 |
| Tetracycline hydrochloride (TH) | Organic antibiotic | 785 nm | Modified O-g-C3N4 on AgNRs | Water | 7.2 × 10−9 M | Qu et al. (2019)71 |
| Kanamycin (KANA) | Antibiotic | 633 nm | AuNPs | Milk | 0.90 pg mL−1, 0.1 ng mL−1 detected in milk | Jiang et al. (2019)72 |
Mycotoxins, which are of a fungal origin, are toxic secondary metabolites commonly found in contaminated crops during storage processing stages. Grains such as maize, peanut, barley and corn in tropical climates that were stored in humid conditions are prone to toxic fungi growth. Mycotoxins are known carcinogens and cause potential immunotoxicity and neurotoxicity and, therefore, they are detrimental to the wellbeing of the consumers. Aflatoxin B1 (AFB1) is part of the mycotoxin family and is most common in food such as peanuts and maize. AFB1 is produced by a few types of Aspergillus, which are prevalent in hot and humid conditions such as the tropics. Ko et al.66 developed a SERS technique that uses an immuno-analytical method for the rapid and reliable detection of trace levels of AFB1, with the use of magnetic beads for separation and silica-encapsulated hollow gold nanoparticles (SEHGNs) under a confocal Raman microscope. The selectivity performance was evaluated with two toxins, namely fumonisin B (FMB) and ochratoxin A (OTA). These toxins are closely related to AFB1 as they are found along with AFB1 in corn and barley grains. The relative Raman band at 1616 cm−1 was selected as the representative peak for quantitative evaluation. Among AFB1 and its mixtures, the SERS intensity of AFB1 increased with increasing concentrations of AFB1 antigen. In contrast, no such trend was observed for both FMB and OTA mycotoxins. Thus, the SERS-based immuno-analytical detection method was found to be highly selective to AFB1, reporting a LOD of 0.1 ng mL−1 and its sensitivity comparable with the HPLC method. Pan et al.73 developed a simple SERS method for the rapid detection of alternariol (AOH), an Alternaria fungi mycotoxin, in pear fruits using an Ag NPs based substrate. A pyridine modified Ag NPs substrate was used to improve the affinity of AOH and its subsequent detection with a LOD of 1.30 μg L−1 under a confocal Raman microscope.
In a different study,67 AFB1 was detected using an aptasensor-based DNA as the capturing moiety, which provided high specificity and sensitivity. Several types of aptamer DNA strands were used to capture AFB1. The second type of DNA aptamer was designed as a leaving group that will be released when AFB1 is available for binding. The third type of DNA was hairpin DNA that will be hydrolyzed in the presence of AFB1. Finally, a single-stranded DNA was used to bind the Raman tags. The results were validated by the electrochemical impedance spectroscopy method. The resultant aptasensor exhibited a linear correlation in the concentration range of 1 μg mL−1 to 1 ng mL−1, reporting a very low LOD of 0.4 fg mL−1 under a confocal Raman microscope. Hernández et al.74 developed a label-free SERS method with a gold nanoprisms aptasensor for the detection of OTA, a mycotoxin commonly found in coffee, and alcoholic beverages other than common grains such as corn and maize. They performed an assay optimization with the SERS data of OTA and executed a multivariate analysis using the PLS regression method. This led to the sensitive detection of OTA in a concentration range from 10 to 250 ppb using a portable Raman spectrometer.
Multiplex detection of mycotoxins in maize was reported by Zhang et al.75 that exploited a SERS-based lateral immunosensor approach using a portable Raman spectrometer. The common mycotoxins present include AFB1 and five other biotoxins in the mycotoxin family. The approach involved fixing six capturing moieties dispensed along three test lines of nitrocellulose membrane, enabling the simultaneous detection of multiple toxins in a single test (Fig. 5). The LOD for all the six mycotoxins under investigation was found to be within ng to pg mL−1 range.
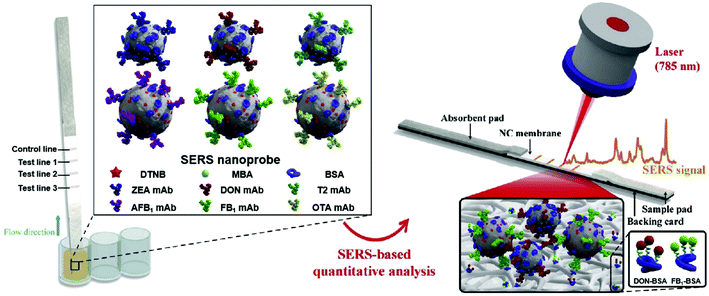 | ||
| Fig. 5 Schematic of multiplex detection of mycotoxins using SERS-based lateral flow immunosensor. Reproduced with permission from ref. 75. Copyright 2020, Elsevier. | ||
Janči et al.76 developed a rapid and simple SERS method for the detection of histamine contamination in fresh fish. For this study, silver colloids were used as the enhancing medium and results were cross-validated with HPLC. The resultant SERS spectral acquisition under a confocal Raman microscope combined with chemometric analysis (PCA plot and PLS regression models) confirmed the reliability of this approach with a detectable concentration range for histamine of up to 200 mg kg−1. Similarly, Lin et al.69 reported a SERS sensing platform developed for bisphenol A (BPA) detection using Au nanostars (AuNS) by means of lateral flow assay. The detectable concentration range for BPA was from 0.05 to 60 ppb, while the LOD for the BPA was approximately 0.073 ppb under a benchtop Raman spectrophotometer.
In a different study done by Pinzaru et al.,70 silver nanoparticles (AgNPs) were used to obtain the characteristic SERS signal for the detection of lipophilic toxins and consolidate a spectral database to better understand the toxins molecular adsorption mechanism. Different types of marine biotoxins were studied in this experiment; three of the toxins are structurally related, namely dinophysistoxin-1 (DTX-1), dinophysistoxin-2 (DTX-2) and okadaic acid (OA), respectively. The other biotoxin, yessotoxin (YTX), was structurally different. They were able to detect and classify structurally similar biotoxins based on the single spectral variation at 1017 cm−1 (corresponding to C-CH3 stretching mode) that was present in DTX-1 and DTX-2 and was absent in OA SERS spectra. A mussel with toxic tissue exhibited strong SERS background compared with that of a healthy mussel. They also reported the detectable concentration range of OA was from 0.81 μM to 84.6 nM. The major advancement in this study was highlighted in the use of a portable Raman system that was capable of reproducing results acquired from laboratory-based benchtop SERS equipment, paving the way for in situ SERS screening for biotoxins in seafood.
Melamine is a nitrogen-rich substance with a white crystalline appearance that exhibits a wide usage in plastic appliances. It received notoriety in a series of food safety incidents as it had been added illegitimately to commonly consumed food items to superficially increase its “protein” content.77 In 2007, melamine was found to be extensively used as an adulterant in wheat gluten and rice protein concentrate and in pet food and feeds, while incidences where milk and infant formula milk were found adulterated with melamine were reported in 2008. Melamine, when ingested, can result in the formation of kidney stones and kidney damage that leads to acute renal failure, and in some cases, even results in infant death. Lin et al.78 paved the way for SERS detection of melamine in several types of food products by comparing its capability to that with HPLC methods by employing the Klarite SERS-active substrate. While both SERS and HPLC can detect trace amounts of melamine, SERS was proven to be 30 times more sensitive than HPLC, giving the LOD at 0.033 μg mL−1 under a confocal Raman microscope. A strategy to negate sample preparation was reported by Betz et al.79 by fabricating silver micro- and nanostructures on both copper tape and coin. This strategy allowed the detection of melamine in infant formula down to 5 ppb using portable Raman spectrometer, while reproducibility was higher for the copper tape substrate than the coin substrate. These substrates may not be superior in terms of sensitivity and specificity to LC/MS but they exhibited a 200-fold enhanced detection than commercial SERS substrates.
Therapeutic drugs commonly used to treat diseases have been exploited illegally for growth-promoting purposes in feeds and animals. Fang et al.80 employed a thin layer chromatography (TLC) separation technique followed by dynamic SERS (DSERS) analysis for the detection of illegal doping of antitussive and antiasthmatic drugs in botanical dietary supplements. The technique was achieved with a portable Raman spectrometer while using 50% glycerol silver colloid as DSERS active substrate. This approach was successful in detecting one sample of botanical dietary supplements tainted with benproperine phosphate and validated with LC methods with its sensitivity superior to the conventional SERS technique.
Use of food colorants are on the rise and they can be characterized as natural or synthetic, the latter of which is categorized into five major types: 1. “azo compounds” based (E102, E110 and E122, etc.); 2. “triarylmethane” based (E131, E133 and E142); 3. “chinophthalon derivative” based (E104); 4. “xanthene/erythrosine” based (E 127); and finally 5. “indigo” based colorants (E 132). The use of synthetic colorants in foods is closely regulated due to food safety incidents in the past. It has been established that food colorants are SERS active and can be distinguished from one another from their unique spectral features.81 Meng et al.82 fabricated a SERS substrate with fabricated Au nanodumbbells as a Raman signal enhancement medium for the ultratrace detection of prohibited colorants in orange juice and cola samples. The food colorants lemon yellow and sunset yellow had a permissible upper limit up to 0.1 g kg−1 in fruit juices and carbonated beverages using a portable Raman spectrometer, according to local regulatory standards, while orange II and chrysoidin have already been prohibited as food colorants. The results showed that the LOD for the four types of adulterants were found to be 10–50 μg mL−1 in orange juice and 1–5 μg mL−1 in aqueous solution, respectively. This SERS method has enabled label-free and rapid analyses without any pre-processing steps. A highly sensitive silver nanorod (Ag NR) array on silicon was fabricated by means of oblique angle deposition (OAD) using E-beam evaporation as the SERS substrate. Allura Red (AR, E129), which is a food colorant most commonly used by the beverage and confectionery industries, was detected using the Ag NR substrate.83 Density functional theory (DFT)-based calculations were used for assigning the characteristic AR SERS peaks. A linear correlation was obtained for AR concentration and its corresponding SERS intensity. Due to the scalable Ag NR fabrication process, rapid and direct detection of AR in various candy samples was demonstrated with a LOD of 0.05 mg L−1 for AR molecules. Flower-shaped silver nanoparticles (flower shaped) (Ag NP) could similarly be employed as a SERS enhancing medium for the direct detection of several food colorants acid red, food blue, sunset yellow and tartrazine.84 A multivariate analysis method like PCA was employed for the detection of different food colorants, at low concentrations ranges. Through this method, LODs of 79.285 μg L−1, 5.3436 μg L−1, 45.238 μg L−1 and 50.244 μg L−1 were obtained for food blue, tartrazine, sunset yellow and acid red colorants, respectively.
Man-made chemical contaminants such as organic antibiotic pollutants, e.g. tetracycline hydrochloride, are also able to easily find their way to animals and humans through the food chain and cause the development of antibacterial resistance in humans. Qu et al.71 fabricated a 3D SERS substrate with the assembly of graphitic carbon nitride on Ag nanorod arrays (Ag NRs) for trace detection of tetracycline hydrochloride, granting a LOD of 7.2 × 10−9 M under a confocal Raman microscope. Kanamycin (KANA), which is an aminoglycoside bactericidal antibiotic, is used to inhibit both Gram-positive and Gram-negative bacteria. However, its overuse without any restrictions can lead to the accumulation of KANA in animal-derived foods such as poultry and dairy products. Jiang et al.72 developed a SERS-based aptasensor for the detection of KANA, which uses a DNA aptamer along with bimetallic gold–silver nanoparticles. This SERS detection platform achieved a LOD of 0.90 pg mL−1 in milk, which was 4 orders lower than the reported MRLs stated by the EU under a confocal Raman microscope. Oxytetracycline (OTC) is a broad-spectrum antibiotic from the tetracycline family. It is effective in controlling Gram-positive and Gram-negative bacteria and other microorganisms by inhibiting the protein synthesis pathway of the bacteria. OTC has been used extensively by apiculture, albeit at the expense of honeybees. Fá et al.85 developed a simple colloidal SERS method that is fast and effective for the detection of trace-level of OTC in honey samples, without any laborious extraction or sample preparation processes. They were able to detect OTC in honey with a LOD of 5 ppb using a portable Raman spectrometer.
Polychlorinated biphenyls (PCBs) are a group of synthetic chemicals with 209 congeners, which are widely used in a range of products such as electrical equipment and plastics. They are classified as organic pollutants exhibiting issues such as toxicity, bioaccumulation and environmental persistence, and pose a significant threat to human health, especially through contamination in food and the environment. SERS stands out as a promising method of detection due to its high detection sensitivity and multiplexing capability, and different strategies towards point-of-care tests have been developed. Templated fabrication of SERS substrates has been reported by depositing metal onto nanostructured support.86–89 One such study of PCB SERS detection was achieved by using DNA aptamer-modified Ag-nanorod arrays, fabricated by anodic aluminium oxide (AAO)-templated deposition.86 By using the PCB-binding aptamer on the SERS substrate under a Raman microscopy, the authors reported the LOD approaching 33 nM in water, and 330 nM in real lake water. The developed arrays exhibited high SERS sensitivity, good reproducibility and high selectivity. Another interesting strategy has been reported to fabricate Ag-nanoplate arrays by PAN-nanopillar templated deposition driven by galvanic cell reaction.87 A detection of 10−6 M PCB-77 in n-hexane has been demonstrated under a confocal Raman microscope in this system. The developed system has advantages such as low cost and green synthesis of the SERS substrate, large scale and tunable SERS intensity. The galvanic cell reaction-driven deposition has also been reported to fabricate gold nanourchin for PCB-77 detection down to 5 × 10−6 M in the n-hexane under a confocal Raman microscope.88 Shanta et al. reported a templated fabrication of nanostructured graphene oxide assemblies with Ag nanoprisms (Fig. 6A).89 By using their non-overlapping SERS molecular fingerprints, a detection of PCB-77 with a LOD of 100 nM, distinct from other aromatic pollutants (i.e. PCB-47 and PCB-52), in a mixture has been demonstrated (Fig. 6B and C) under a confocal Raman microscope. The detection system shows great promise for portable detection, quick processing and good detection limits for environmentally significant pollutants and multiplexing capabilities.
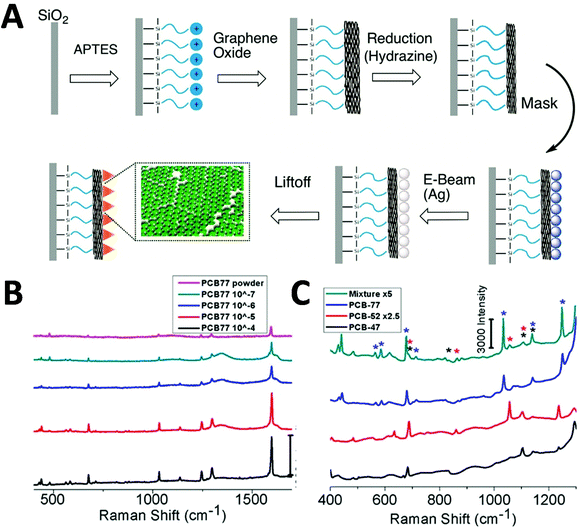 | ||
| Fig. 6 Polychlorinated biphenyl (PCB) detection via SERS. (A) Fabrication of Ag-nanoprisms on top of graphene oxide-covered glass slides. (B and C) Detection of PCB-77 with different concentration, and multiplex detection of PCB-47 (black), PCB-52 (red) and PCB-77 (blue) in a mixture (each 333 μM). Reproduced with permission from ref. 89. Copyright 2017, ACS Publications. | ||
Recently, template-free fabrication of SERS substrates has been reported for PCB detection. By employing Ag nanocube (Ag-NC) self-assembly at the interface of air/water, an Ag-NC monolayer has been obtained.37 After transferring to a transparent polyethylene (PE) film, the Ag-NC@PE film has been used as a flexible cut-and-paste SERS substrate for rapid in situ POC detection of 4-PCB down to 1 μM under a confocal Raman microscope. More recently, a plasmonic vesicle-based 3D platform has also prepared for solution-phase SERS detection of PCB pollutants in soil samples.90 With a small portable Raman spectrometer, the results show that detection of PCB7 from contaminated soil gives an excellent linear SERS response at a wide concentration range (from 10−12 to 10−7 g mL−1). In addition, multiple detections of PCB 7, 77 and 209 have been demonstrated at 10−9 g mL−1. This substrate system was used with a small commercial portable Raman spectrometer and shows potential for in-field food safety analysis and environmental monitoring. Although obvious progress has been made for PCB detection, a practical sensor with high sensitivity, multiplexing and mass-fabrication, while maintaining high reproducibility and repeatability, is still an on-going process towards POCT applications, before a wide adoption in our daily life.
2.3 Detection of foodborne pathogens
As per the WHO's guidelines, foodborne illnesses are diseases instigated by pathogens that infiltrate the body via food consumption. Food becomes the major vehicle of foodborne illnesses when they become in contact with contaminated surfaces, water or handling. Foodborne illnesses are a cause of concern to public health, as it is often under-reported and its source is difficult to establish. In 2015, the WHO reported that foodborne illnesses are caused by 31 pathogens that include bacteria, viruses, chemicals and toxins at global and regional levels.91 The global burden of foodborne illnesses is estimated to affect 600 million people each year, of which 7% die and 30% of these are children under the age of 5 years. Hence, there is a pressing need for simple, reliable and fast screening techniques for the detection and identification of foodborne pathogens in the food chain to ensure food safety. Cho et al.92 developed a SERS membrane filtration-based sensor to detect trace level of Escherichia coli (E. coli) O157:H7 from ground beef by means of bacterial filtration and silver nanoparticle concentration followed by SERS analysis. The target bacteria were isolated from the sample effectively by a pathogen-specific monoclonal antibody conjugated with magnetic nanoparticles and gold nanoparticles with a SERS probe (Fig. 7). Using this immunomagnetic separation technique, E. coli O157:H7 concentration from spiked ground beef as low as ∼10 CFU mL−1 were detected. In this study, possible non-specificity was avoided due to the size of the selective filtration process. While this assay presents a novel way to detect trace level of pathogens, it can be improved if the variability involving this multistep process is addressed along with improved sensitivity and reduced process steps to make it more attractive for POC applications.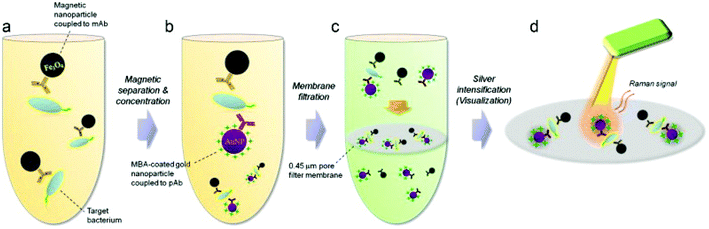 | ||
| Fig. 7 Schematic of the experimental flow for the detection of E. coli O157:H7 in ground beef with membrane filter-based SERS rapid detection method. (a) E. coli is captured using pathogen specific magnetic nanoparticles. (b) Gold nanoparticle coated with Raman probe is conjugated to pathogen-bound magnetic nanoparticles for magnetic separation. (c) Membrane filtration step removes the unbound reactants such as gold and magnetic nanoparticles. (d) Raman probe in the pathogen-nanoparticle sandwich complex undergoes silver intensification process for SERS measurement. Reproduced with permission from ref. 92. Copyright 2015, Elsevier. | ||
In another study, Prakash et al.93 demonstrated SERS label-free detection and discrimination of three types of bacteria: E. coli, Bacillus subtilis and Salmonella Typhimurium. Ag or Au bimetallic nanoparticles (bmNPs), which were positively charged for bacteria interaction, were used as SERS substrates for selective aggregation and the detection of bacteria. Detection and classification of the bacteria were done by the canonical discriminant analysis method under a confocal Raman microscope. The major advantage of this method was the use of positive surface charge on Ag/Au bmNPs for fast aggregation with bacteria and a simplified protocol used for its detection. This SERS method was rapid, sensitive and reproducible due to the use of multivariate analysis techniques and it is well suited in the area of food safety and environmental monitoring.
Enterotoxins are a type of toxins secreted by micro-organisms that are low molecular weight, relatively heat stable and are soluble in water, and can easily find their way into food products. Major types of enterotoxins, if present in food products, can lead to food poisoning, intestinal cramping and diarrheal disease within a few hours of consuming the food. Enterotoxin type Staphylococcal enterotoxin B (SEB) is produced by the Gram-positive Staphylococcus aureus bacteria. These toxins may remain active in food products even if the bacteria are killed by heat or otherwise. Hwang et al.94 developed a novel SERS-based lateral flow immunoassay (LFA) method to address issues arising from conventional LFA strips such as limited quantitative analysis and inferior sensitivity. The SERS probes utilized in these strips were in the form of Raman reporter-labelled hollow gold nanospheres (HGNs) for the capture of the SEB antigen. HGNs clusters were observed in the paper fibre pores only in the presence of the target antigen and this aggregation resulted in the colour change of the strip. When the target antigen was absent, no colour change or SERS signal was observed. This approach of SERS-based LFA trips allowed for highly sensitive quantitative analysis. The detection of SEB with the SERS-based LFA strip was comparable with that of a conventional qualitative LFA strip and ELISA assay kit for high antigen concentration (>10 ng mL−1) under a confocal Raman microscope. Additionally, the SERS-based LFA strip was sensitive and effective even at lower concentration ranges from 0–1 ng mL−1. The LODs for the detection of SEB were 10, 1.0 and 0.001 ng mL−1 for the conventional LFA strip, ELISA assay kit and SERS-based LFA strip, respectively.
In another study, Wang et al.95 demonstrated a label-free detection of foodborne pathogens such as Gram-positive and Gram-negative bacteria by employing plasmon active nanoparticles (nanoplates and nanorod supercrystals) assembled onto a 3D bio-inorganic supercrystal under a confocal Raman microscope. Selectivity of the bacteria was ensured by the use of a vancomycin antibiotic, which exhibits a stronger interaction with Gram-positive bacteria compared with that of Gram-negative bacteria. The resultant SERS spectra with their vibrational band between 600 and 800 cm−1 were considered as a fingerprint region for the identification of bacteria through their unique DNA/RNA composition. High SERS sensitivity was attributed to different amino acids in the following order: adenine > guanine > cytosine > thymine. This study reported a LOD of 103 CFU mL−1 and 102 CFU mL−1 for E. coli (Gram-negative) and S. xylosus (Gram-positive) bacteria, respectively. Similarly, Wu et al.96 reported a sensitive SERS strategy for the detection of a Bacillus thuringiensis special gene fragment with high selectivity. The study employed biotinylated hairpin DNA which was complimentary to target bacterial DNA and was conjugated onto the Au@Ag NRs shell as a signal probe, whereas streptavidin bound magnetic beads will assist in capturing target DNA hybridized Au@Ag NRs by means of the biotin–streptavidin interaction. Strong SERS signals were obtained when the target DNA was absent in the sample, which led to the close packing of Au@Ag NRs with a Raman reporter, and a weak SERS signal resulted in the presence of the target DNA, due to the opening of hairpin DNA. The reported LOD for Bacillus thuringiensis special gene fragment was 0.14 pM. This SERS method was proven to be more efficient and sensitive compared with immunoassay techniques as a result of fewer experimental steps and its non-enzyme based detection method.
In the latest work by Guo et al.,97 a SERS sensor platform (SERS “nose”) was developed by decorating and densely arranging gold nanostars (AuNSs) onto an anodic aluminium oxide membrane with a vacuum filtration process. The resultant AuNSs based membrane SERS substrate acts as a SERS “nose” as it provides strong signal enhancement and high sensitivity to gaseous metabolites from bacteria under a confocal Raman microscope. The application of a SERS “nose” to detect gaseous metabolites of foodborne bacteria (E. coli, Staphylococcus aureus and Pseudomonas aeruginosa) has been reported with high sensitivity. The SERS “nose” sensor was capable of detecting rotten and contaminated food even at an early stage and to monitor its spoilage. This multifunctional SERS sensor can act both as a filter to enrich the pathogens and a sensing platform to detect bacterial strain along with a portable Raman reader, which will be of interest for routine POC food screening to ensure public health.
The new developments in this space have made the SERS sensing platform a more promising food screening tool for the rapid detection of pathogens as SERS provides both high sensitivity and possible quantification, leading to significant potential for the rapid and sensitive detection of target analytes in a faster and simple manner. The previously mentioned biosensors are a step closer to achieving an automated label-free SERS-based biosensor with an AI element for application in portable POC diagnosis in combination with a suitable and highly sensitive portable Raman reader.
Due to continuous development within the photonic space, a sensitive and at the same time portable Raman spectrometer system that provides comparable performance to that of a bulky lab-based system is achievable. The efforts described previously are a testament to SERS as a promising approach for food safety and plant health assessment because it is rapid, sensitive and necessitates minimal sample preparation. There is much room to improve in specific areas such as SERS quantitative analysis which can be addressed by combining chemometrics methods and spectral data analysis. Further research is warranted to establish the optimum settings to acquire reproducible results in complex food matrices. Also, the measurement area for non-destructive in vivo measurement is very small to analyse, i.e. close to 200 μm in area, whereas the detectable scale of the area is always much larger. In addition, there is a need for large volumes of plant tissue extraction to validate a proper diagnosis, as it is very common to note that concentrations of nutrients, toxins and chemicals may not be uniformly distributed along the whole surface of the produce or food grains.
3 SERS-based biomedical applications
In the biomedical domain, conventional detection methods involving bacterial culture, PCR, luminescence and microarray have been used as tools to test clinical analytes, but they suffer from a long processing time, lack of portability, lack of multiplexing and/or low sensitivity. On the other hand, POC SERS offers rapid, on-site, fingerprint multiplexing and high sensitivity. To address the limitations of conventional methods, POC SERS approaches have been developed/adopted for a variety of clinical samples, including nucleic acids, proteins, virus, bacteria, whole cell, ions and small molecules.3.1 Detection of nucleic acids
Nucleic acids as genetic material (e.g. miRNA, RNA, DNA) from organisms constitute an important group of biomarkers in various aspects. Conventional detection methods of nucleic acids include qRT-PCR detection, northern blotting and microarray-based methods. However, they suffer from extensive sample preparation, long assay time, lack of portability, high cost and low sensitivity. To address these limitations, various methods have been developed including electrochemical and optical methods. Among them, SERS is promising as a POC detection strategy, and different SERS detection methods have been developed towards the detection of nucleic acids.98–112A miRNA is a class of small non-coding RNA molecules with an average of 22 nucleotides, which mainly affect post-transcriptional regulation of proteins and can be great candidates as a cancer diagnostic biomarker. The short length of miRNA makes it difficult to be amplified by PCR, and the highly homologous sequence makes specific detection a challenge. To address this limitation, various SERS-based strategies have been developed for clinical miRNA detection.98–102 A dual SERS biosensor assisted by duplex-specific nuclease has been reported.100 Using magnetic concentration with Fe3O4@Ag-SERS tag conjugates together with enzyme-assisted amplification, microRNA-10b in both exosome and residual supernatants of blood plasma have been tested. The results show a LOD of 1 aM with SNP specificity under a portable Raman spectrometer, and significant difference among pancreatic ductal adenocarcinoma, chronic pancreatitis and normal controls. This one-step and one-pot detection without prolonged sample preparation is promising for POC clinical cancer diagnosis. Recently, Graham et al.98 reported a 3D paper-based microfluidics platform for miRNA detection in PBS running buffer by combining both colorimetric and SERS analysis (Fig. 8). Using miR-29a and a negative control (miR-21), the authors achieved the detection of miR-29a with a LOD of 47 pg μL−1 using a handheld Raman spectrometer, and colorimetric differentiation from the control sample (18–360 pg μL−1) with a LOD of >200 pg μL−1. This microfluidic SERS-based detection approach shows its potential as a facile POC device with great portability, fast readout and ease of operation. Further work to improve the sensitivity and specificity is needed along with analysis of a multiplexed real clinical sample.
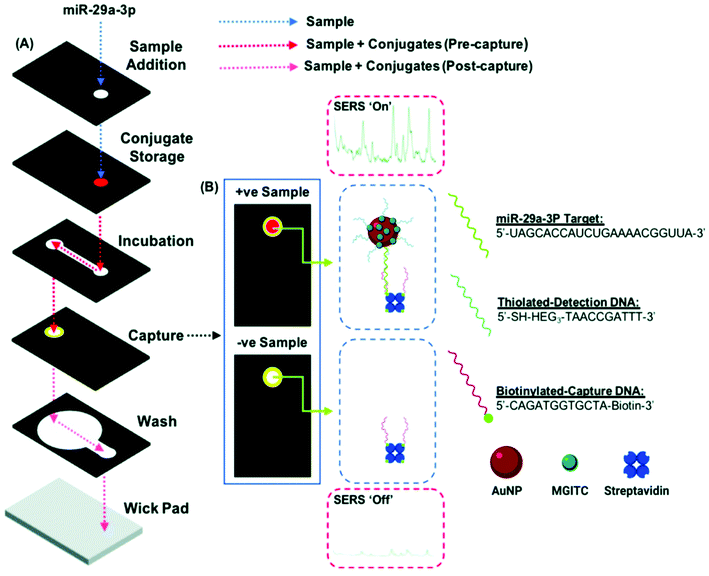 | ||
| Fig. 8 (A) Schematic of duplex detection of microRNA (miR-29a) in a 3D microfluidic paper device, where the image displays how the capture layer is loaded with a handheld Raman reader for SERS detection. The sample pathway is denoted by the dotted lines. (B) Sample readout from combined colorimetric and SERS techniques for samples with miR-29a (+ve) and samples without miR-29a (−ve). Reproduced with permission from ref. 98. Copyright 2020, Royal Society of Chemistry. | ||
On the other hand, longer nucleic acids (relative to miRNA) have also been detected by SERS from both animal and plant pathogen sources, including RNA103,104 and DNA.105–112 For instance, detection of long pathogen RNA has been reported via a SERS-based integrated “lab-in-a-stick” portable device.103 With the integration of SERS nanorattle tags, magnetic separation and enrichment into the portable system, unamplified pathogen RNA in blood lysate has been detected. Results show the detection of RNA with a LOD of 200 fM (2 attomole), and the direct differentiation in infected red blood cells lysate. In addition, the bioassay introduced an automatic washing process for multiple sample handling together with a handheld Raman reader, making it suitable as a POC diagnostic. Innovative SERS-devices have also been reported for the detection of DNA.105 Detection of clinical DNA via a SERS microfluidic chip with molecular beacon probes has been reported. The sensor was fabricated by embedding SERS-active metal NP into a microfluidic chip, which enabled SERS signal turn-off in the presence of a target. The assay detected wild-type KRAS (Kirsten rat sarcoma 2 viral oncogene homolog) DNA with 50 fM LOD and single base mutation differentiation under a confocal Raman microscope system. Parallel differentiation between mutated and wild-type KRAS sequences from clinical tumour cells has also been achieved without amplification in 40 min. With the automated and faster analysis in a microfluidic format, the sensitive and inexpensive chip-based assay shows promise for POC sensor development. Recently, detection of multiplexed DNA has been reported in the lateral flow microarray format by using SERS nanotags.106 The sensor was able to differentiate DNA in a multiplexed sample of 11 respiratory tract infection (RTI) pathogens in a running buffer (Tris and EDTA), with LOD reaching 0.031 pM for influenza A, 0.030 pM for parainfluenza 1, 0.038 pM for parainfluenza 3, 0.038 pM for respiratory syncytial virus, 0.040 pM for coxiella burnetii, 0.039 pM for legionella pneumophila, 0.035 pM for influenza B, 0.032 pM for parainfluenza 2, 0.040 pM for adenovirus, 0.039 pM for chlamydophila pneumoniae and 0.041 pM for mycoplasma pneumonia under a confocal Raman microscope system. The linear dynamic range of nucleic acid detection of the 11 RTI pathogens was 1 pM–50 nM, spanning 5 orders of magnitude.
Different SERS-based nucleic acid detection methods have been demonstrated and show promising results towards POC applications. However, none of them is capable of covering all POC attributes, e.g. being portable, easy sample preparation, clinical sample-compatible, multiplex detection, fast detection, low cost and high sensitivity and selectivity. One possible strategy to address these attributes is the integration of a portable Raman reader and automatic SERS device as an alternative to typical molecular-based amplification assays.
3.2 Detection of proteins
Protein biomarkers in clinical samples have been widely used as surrogate indicators in disease diagnostics and therapeutic treatment monitoring, and POC SERS detection of protein has received increasing attention in this space.113–121 A typical platform is based on POC lateral flow immunoassays (LFAs), where SERS detection has been incorporated with the disposable and portable LFA test strip.113–115 For instance, a rapid SERS-LFA assay has recently been reported to detect hormone human chorionic gonadotropin (hCG).113 Ultrabright SERS nanotag-antibody conjugate has been prepared and used in the sandwich recognition of the hCG from clinical pregnancy serum samples. Results show that hCG is detectable within 2–5 s with approximately 1.6 mIU mL−1 LOD. Importantly, instead of point illumination using raster-scanning Raman microscopy, this SERS-LFA work applied line illumination with a portable Raman reader, and enabled several orders of magnitude faster detection (relative to point illumination) and was 15 times more sensitive than a commercial LFA.Besides the adoption of the SERS-LFA biosensor, integration of digital microfluidics into SERS-based immunoassay has been reported for protein detection.116Fig. 9 illustrates a sensor configuration with a digital microfluidic device processing the SERS probe and magnetic beads with magnetic separation and concentration. The hemagglutinin of avian influenza virus H5N1 in both buffer and human serum has been tested with this platform, and the results show excellent selectivity with a LOD of 74 pg mL−1, a shorter assay time (<1 h) compared with conventional ELISA (6 h) and a lower sample volume required (∼30 μL) relative to the standard ELISA method (650 μL). With a portable Raman spectrometer, the developed sensing system provides a POC SERS sensor for the diagnostic of infectious diseases with low reagent consumption and minimized exposure risk of hazardous samples.
 | ||
| Fig. 9 Schematic of SERS-based digital microfluidic immunoassay (a) with Raman spectra as detection output (b), where a side-view of chip (c) showing a droplet containing immunocomplex magnetic beads functionalized with SERS tags (d). Reproduced with permission from ref. 116. Copyright 2018, ACS Publications. | ||
More recently, a SERS-active photonic crystal fiber (PCF) probe has been employed as a SERS platform for protein biomarker detection.117 In this work, gold nanoparticles were immobilized onto the suspended core of PCF, and cancer biomarker haptoglobin was detected from clinical ovarian cyst fluid under a confocal Raman microscope system. Results show extremely high reproducibility (1.5% relative standard deviation) and repeatability (4.6% relative standard deviation), and the platform can distinguish different ovarian cancer stages from the haptoglobin levels and are in excellent correlation with standard clinical assay results. Due to its unique features, the developed PCF-SERS platform can do both sampling and testing simultaneously with small sample volumes, showing promise for various biomarkers in body fluids for POC detection.
3.3 Detection of viruses
Viruses are responsible for recent pandemic/epidemic disease outbreaks such as COVID-19, and their rapid and reliable detection is key for the prediction of prognosis and monitoring therapeutic treatment. Label-free SERS detection of viruses has been reported on a microfluidic device.122–129 Yeh et al.122 reported a compact microfluidic platform named VIRRION (virus capture with rapid Raman spectroscopy detection and identification), with carbon nanotube arrays of varying filtration permeability for high-throughput virus capture with rapid SERS detection (Fig. 10). More than one type of virus (be it known or unknown) can be captured and detected simultaneously on the chip by SERS under a Raman microscope system and they stay viable on the device for further characterizations. Different avian influenza A virus subtypes and human respiratory viruses from clinical samples were tested on this platform, and the results showed that the viruses were captured and detected within only a few minutes, with a 70-fold enrichment with 102 EID50 per mL (50% egg infective dose per microliter), 90% specificity and the capability of multiplexing and emulating co-infections. In addition, its detection sensitivity is comparable to RT-qPCR results with 70–90% accuracy. This platform holds great promise and can be a boon in outbreak situations, and is capable of rapid tracking and monitoring viral outbreaks in real time.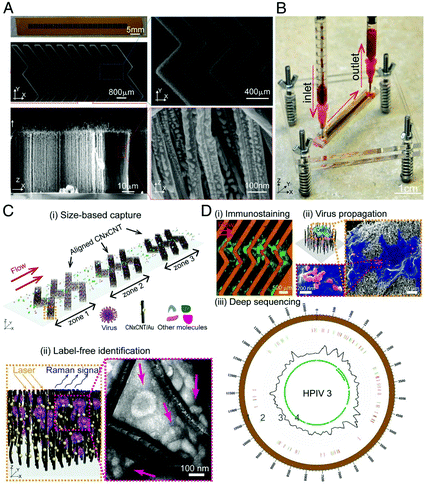 | ||
| Fig. 10 Schematic of the working principle of VIRRION platform, including (A) images of aligned CNTs decorated with gold nanopaticles and (B) assembled VIRRON device, (C) illustration of virus capture and label-free Raman identification, and (D) on-chip analysis and enrichment of virus for NGS. Reprinted with no changes from ref. 122, image licensed under http://creativecommons.org/licenses/by/4.0/. | ||
Labelled SERS detection of a virus could be developed based on typical immunoassays.123–126 One such example is the labelled SERS detection of a virus on the LFA strip based on the antigen–antibody reaction.123 Using a portable Raman reader, wild-type pseudorabies virus (PRV) including clinical samples from various types of pig tissues have been tested on this platform. The process took 15 min, giving 5 ng mL−1 LOD, selective detection of wild-type PRV and quantitatively/semi-quantitatively compatible (detection linear range: 41–650 ng mL−1). The detection results are consistent with the glycoprotein E (gE)-specific PCR detection and could be used to differentiate wild-type PRV and gE-deleted vaccine. Sun et al.124 demonstrated a magnetic immunoassay of virus samples with labelled SERS nanotags. With a portable Raman reader, inactivated virus H3N2 and H1N1 in PBS buffer were tested on the sensor. Through magnetic enrichment and separation of the virus from a complex matrix, results showed a LOD 102 TCID50 per mL for H3N2 (TCID50: tissue culture infection dose at 50% end point), with a good range of linear relationships (102 to 5 × 103 TCID50 per mL). With the efficient, portable and sensitive immunoassay, the SERS-based magnetic detection strategy is promising for POC applications in clinical and diagnostic settings. Recently, a labelled SERS detection combining an LFA strip and magnetic separation and concentration has been reported to detect viruses.127 The magnetic SERS-based LFA strip was decorated with Fe3O4@Ag NPs as magnetic SERS nanotags. Cultured virus samples (HAdV, H1N1 and control samples of influenza B, respiratory syncytial virus and parainfluenza) and human samples without pre-treatment steps, were tested on this SERS LFA platform under a Raman microscope system. Results show a duplexing identification of H1N1 and HAdV with LOD of 50 and 10 pfu mL−1, respectively. The LOD values were 2000 times more sensitive compared with results from the standard colloidal strip detection. The developed virus SERS POCT platform is user-friendly, rapid and stable, which shows potential for high throughput and early screening of virus infections. Although labelled and label-free SERS platforms have been made for POC virus detection, a practical sensor with rapid, sensitive and specific detection with multiple functions (e.g. VIRRON's capture and enrichment of virus, and minimized sample exposure) is highly desirable towards real-world applications.
3.4 Detection of bacteria
The healthcare sector is poised for real time and rapid detection, with an enhanced multiplex capacity of infectious agents for timely therapeutic treatment and controlling the spread of infection. It is critical for any analytical method to have high sensitivity and reliability for success. For a basic understanding of the mechanism behind SERS detection of bacteria, Premasiri et al.130 investigated the vibrational modes of metabolites that contribute to the overall SERS spectra of bacteria. When bacterial cells undergo starvation in pure water treatment, the stress response results in the release of several metabolites, such as adenine, xanthine, guanine, uric acid and adenosine monophosphate, that gives unique SERS spectral features.Dina et al.131 developed a label-free SERS-method for the rapid detection and identification of Gram-positive as well as Gram-negative bacteria. The authors demonstrated the sensitivity of this method to achieve single cell detection. Due to the in situ fabricated silver colloids decorating the bacterial outer membrane that was placed on a polyslide, both 532 nm and 633 nm excitation lasers were compared for a SERS signal with the latter yielding SERS spectra with a better signal to noise ratio under a confocal Raman microscope. The reproducibility of this method was treated with model strains that are common human pathogens.
Kearns et al.132 have developed a SERS sensor that is fast, simple and sensitive, which is capable of multiple detection of bacterial pathogens by employing magnetic separation followed by SERS application. This was achieved by means of a specific antibody immobilized onto lectin functionalized magnetic nanoparticles for the selective enriching of the bacterial strain of interest. Three bacterial pathogens strains were studied, Salmonella Typhimurium, methicillin-resistant Staphylococcus aureus (MRSA) and E. coli, with a LOD of approximately 10 CFU mL−1 successfully under a confocal Raman microscope. The authors also demonstrated that single pathogen detection was feasible with their SERS platform, in addition to its multiplexing capability with specific fingerprint Raman bands corresponding to each bacterium from the combined SERS spectra, which was obtained by mixing three different bacterial strains and by employing a classification method such as principle component analysis (PCA). Similarly, Gao et al.133 developed a SERS method for the direct label-free detection of bacteria that involved the growth of silver nanoparticles selectively around the aptamer conjugated bacterial cell membrane, resulting in silver nanoparticle coated bacteria (bacteria-aptamer@AgNPs). Enhanced SERS signals were generated by bacteria-aptamer@AgNPs which are directly proportional to bacterial concentration, which ranges from 101 to 107 CFU mL−1 with an R2 value of 0.9671 and a LOD of 1.5 CFU mL−1 under a Raman microscope. In another study, Yang et al.134 developed a label-free SERS approach for E. coli detection by incubating the bacteria with silver colloids at balanced incubation conditions such as shaking speed, time and temperature. A visible nanoparticle aggregation process occurred in the presence of E. coli bacteria during the incubation process, resulting in Raman signal enhancement and giving a LOD of 1 × 105 cells per mL under a Raman microscope.
Mühlig et al.135 reported a lab on a chip (LOC) based SERS sensor approach for the detection and identification of tuberculosis bacteria (Mycobacterium tuberculosis, M. tuberculosis) among different strains of bacteria from the Mycobacterium family. Six different species of mycobacteria were identified to be of non-tuberculous mycobacteria (NTM) and M. tuberculosis complex (MTC) bacterial strains. This SERS method exhibited efficient discrimination of bacteria without involving any extraction protocol or sample pre-treatment, with the obtained SERS spectra dominated by the major fatty acid component present in the cell wall called mycolic acids under a confocal Raman microscope. For the classification and identification of the various strains of mycobacteria, a robust dataset was generated with individual strains recorded using a SERS microfluidic device. This database was instrumental in the successful identification of the bacterial strain from the unknown sample, thus showing real-time molecular approaches and the SERS technique can perfectly complement each other with high clinical translatability.
Perumal et al.136 developed a label-free direct SERS detection of mycobacterium by means of their unique cell membrane composition. Mycolic acid (MA) is a lipid that is predominantly present in the mycobacterium cell membrane and is present in three derivative types such as alpha mycolicacid (AMA), methoxy mycolicamcid (MMA) and keto mycolicacid (KMA). Individual and representative SERS spectra for each of the three MA types were identified successfully using silicon nanopillar (SNP) substrates coated with Ag. The relative ratio of different MA forms was used to detect the infectious M. tuberculosis strain under a confocal Raman microscope. With the use of advanced statistical tools to estimate the relative abundance of various MAs, this study reported an easy and cost-effective label-free SERS method for the rapid and direct identification of different MAs from bacteria, without involving any laborious analyte extraction methods.
Classification efforts between bacterial species via SERS-based strategies have been demonstrated. Kotanen et al.137 developed a SERS biosensor for the detection of bacteria in body fluids and its subsequent classification. Lysis filtration was performed on the sample to separate bacteria from serum or other blood-based cellular components. The resultant bacterial samples were treated with silver nanorod substrates for 3 h at 60 °C before acquiring their SERS spectra using a portable spectrometer. The unique SERS spectra of the unknown bacteria were compared with that of known species of bacteria in the database by employing PCA and PLSD type chemometric methods to identify bacterial species level, even in poly-microbial cultures in body fluids. One of the study's limitations is that generation of a robust reference library for various strains of pure culture samples is needed for effective identification of clinically relevant and commonly occurring unknown bacteria strains. It was possible to successfully predict bacterial species with high specificity and sensitivity but differentiation between strains within the same species is still a challenge. In an effort to improve on the classification of bacteria, Lin et al.138 reported a silver nanoparticle (AgNPs)-based SERS biosensor for the detection of microorganisms combined with pattern recognition techniques to identify patterns within SERS spectra (obtained using a portable spectrometer) using machine-learning algorithms. After SERS measurements were acquired, data pre-processing proceeded by standard normal variant (SNV) and exploratory data analysis (EDA), variable selection and reduction. Next, pattern recognition techniques were applied onto the data to differentiate between salmonella, Acinetobacter baumannii and Klebsiella pneumonia. Good predictive classification of up to 100% with SERS spectra was acquired, increasing the specificity of the sensor platform.
The use of LFA strips is well established and they are extensively used as POC diagnostics; they are also easy to use at an affordable cost, but are limited in their reliability and reproducible sensitivity as they provide only qualitative information of the analyte. Development of SERS-based LFA methods is on the rise, as SERS-LFA has been demonstrated to be more sensitive even at low concentration range when compared with standard LFA or ELISA methods. Wang et al.139 developed a SERS-LFA strip for bacterial detection, exploiting a sandwich immunoassay method by using antibody-conjugated gold nanoparticles with Raman probes to bind to the bacteria of interest in the flow. The resultant target-gold nanoparticle complex was captured by pre-immobilized antibodies on the test line of the LFA. Fig. 11 shows the operating principle of the SERS-based LFA sensing platform, showing the accumulation of gold nanoparticles with the Raman reporter Malachite green isothiocyanate (MGITC) exhibiting fingerprint SERS spectra at the test line in the presence of target bacteria and inactive SERS in the absence of the target. This assay can be performed in under 15 min with a small sample volume and with high sensitivity at LODs of 43.4 CFU mL−1, 45.8 CFU mL−1 and 357 CFU mL−1 for Y. pestis, F. tularensis and B. anthracis bacterial strains, respectively. This reported method is several orders of magnitude more sensitive than that achieved with the corresponding LFA-based method. Integrating LFA strips with portable Raman readers is a promising strategy for bedside POC applications, as well as for analytical tools for rapid and low-cost clinical diagnosis.
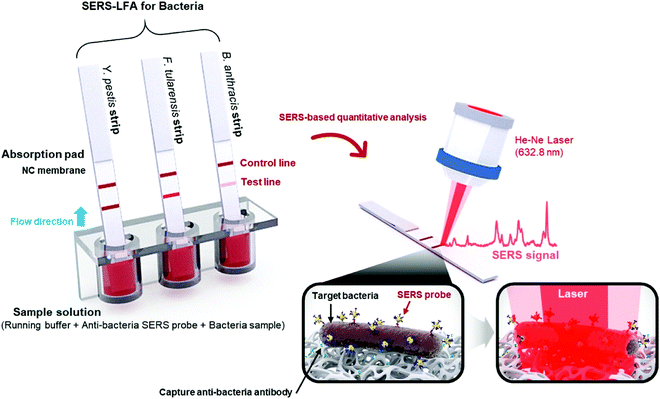 | ||
| Fig. 11 Schematic representation of the experimental flow for detection of Y. pestis, F. tularensis and B. anthracis bacterial strains on the SERS-based LFA sensing platform. The immunocomplexes from bacteria and probe interactions travelled by capillary action towards the test line where SERS measurements were acquired and analysed. Reproduced with permission from ref. 139. Copyright 2017; Elsevier. | ||
3.5 Whole cell
Circulating tumour cells (CTC) in the bloodstream are a possible indicator for the presence of tumours. Hence, detecting CTC early will go a long way towards managing cancer treatment for recovery and survival. CTC-based early cancer diagnosis and the monitoring of cancer treatment involves liquid biopsy sampling, in which blood samples are used instead of invasive tissue biopsy. The major roadblocks for the use of CTC as the preferred method of detection are the technical challenge in isolating CTC from millions of blood cells, its high cost and the poor accuracy of existing methods. Wu et al.140 developed a SERS method with three types of gold nanoparticles, namely, star- (AuNSs), rod- (AuNRs) and spherical- (AuNPs) shaped gold nanoparticles with comparable surface chemistry to detect CTC present in liquid biopsy samples without involving any CTC enrichment procedures under a confocal Raman microscope. The nanoparticles were decorated with 4-mercaptobenzoic acid (4-MBA) as a SERS reporter molecule and the assay stability is warranted by incorporating reductive bovine serum albumin, while folic acid was used to increase high specificity in conjugating with CTC over healthy cells. The spherical nanoparticles displayed the best performance among the gold nanostructures due to their supersensitivity. The performance of this SERS method was satisfactory under a confocal Raman microscope, showing a detection as low as 1–100 cells per mL exhibiting an R2 value of 0.98 of CTC in rabbit blood with cancer cells. Kamińska et al.141 demonstrated the capability of a composite polymer membrane with gold sputtered SERS substrate for dataset generation coupled with the PCA based chemometric classification method for the direct detection and classification of prostate cancer (PC3) and cervical carcinoma (HeLa) cells present in blood samples with high sensitivity and an accuracy of over 95%.Another interesting work was presented using a SERS-based droplet microfluidic sensor that was capable of simultaneous multiplex analyte detection at the single-cell level in a label-free manner.142 The study involved decorating the 400 nm Fe3O4 magnetic nanoparticle surface with 30 nm Ag, resulting in metal-magnetic SERS substrates for the detection of analytes such as pyruvate, adenosine triphosphate and lactate secreted by the CTC. The metal-magnetic substrate granted efficient analyte adsorption, rapid isolation of CTC from multiplex milieus and high SERS sensitivity. The ultrasensitive performance of this SERS method was demonstrated with single cell detection for multiple metabolites. Similarly, Niciński et al.143 developed a photovoltaic (PV)-based SERS microfluidic device for CTC detection from liquid biopsy samples. The detection was achieved by integrating several tools: (i) use of a microfluidic device for the separation of CTC from the rest of the blood components; (ii) a planar SERS substrate made of a integrated PV device to increase plasmonic enhancement; (iii) use of shell-isolated nanoparticles (SHINs) in the successful isolation and detection of CTC with high specificity and sensitivity; and (iv) a chemometric method of analysis for SERS datasets to improve the screening and efficiency of detection. The results were very promising, showing a minimally invasive method of separation and identification of CTC from whole blood.
In a similar study, Girish et al.144 developed a SERS sensor as part of a catheter that was based on TiO2 nanostructures in the shape of a leaf, which was further coated with 30 nm diameter of silver nanoparticles. The authors in this proof-of-concept study analysed clinical samples obtained from 37 subjects with three types of CTC that were related to oral cancer such as oral squamous cell carcinoma (OSCC), verrucous carcinoma and premalignant leukoplakia, along with that of healthy control subjects under a confocal Raman microscope. The cells were detected and classified using the chemometric method of classification with a processing time of under 30 min and an accuracy rate of 97%. Fig. 12 shows the experimental and analytical processes involved in the study. Additionally, the authors were also able to grade the level of malignancy of the tumour under study with their SERS sensing platform. This catheter-based SERS sensor demonstrated rapid CTC detection capabilities, and hence, exhibited great potential as a point-of-care diagnostic tool in the future.
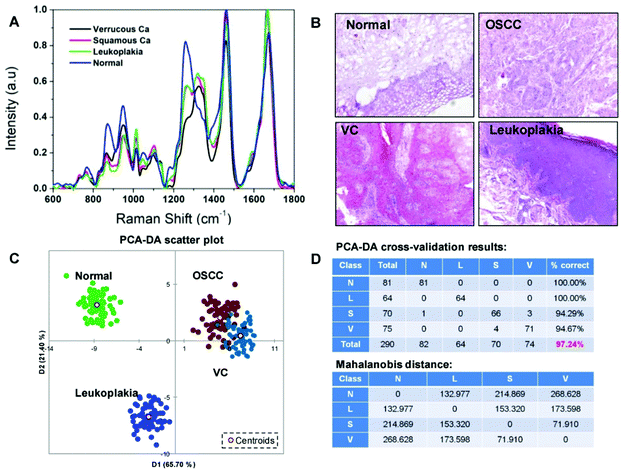 | ||
| Fig. 12 (A) Shows the averaged SERS spectra of the various types of oral cancer malignant CTCs compared with normal cells. (B) Optical images of histology stained tissue from various types of cancer and normal samples. (C) 2D scatter plot showing the efficient classification of various cancers from normal. (D) PCA-DA cross-validation results. Reproduced with permission from ref. 144. Copyright 2019, WILEY-VCH Verlag GmbH. | ||
3.6 Detection of ions and small molecules
Other analytes (e.g. metabolites, drug molecules and ions) from clinical patient samples have additionally been studies towards POC SERS detection,145–153 particularly with combined approaches such as a combination of SERS platform and chemometric analytics,145 a combination of multiple processes146,147 and a combination of multiple optical readings.148Multiplex SERS profiling of urine metabolites from patients by combining a superhydrophobic (SPHB) SERS and chemometric analysis has been reported.145 With the SPHB-mirror SERS platform using silver nanoparticles, the whole screening process took less than 30 min, which includes sample preparation (3 min urine pre-treatment and 25 min drying), SERS screening and analysis (2 min for measurements and chemometric analysis). Target metabolites of interest included 5β-pregnane-3α, 20α-diol-3α-glucuronide and tetrahydrocortisone from clinical urine samples that were tested under a Raman microscope system. With chemometric analysis including PCA and PLS regression, molecular fingerprint information was converted into quantifiable readouts. Results showed a rapid detection (30 min) of metabolites at sub-nM in 10 μL of sample, a 7-order of linear concentration range and metabolite multiplexing at 0.1 nM in biofluids. Importantly, these readouts agreed well with the pregnancy clinical results of 40 patients with symptoms of threatened miscarriage. The reported platform shows rapid non-invasive multiplex SERS detection of urine metabolites, potentially to be applied for clinical POC screening of various diseases. Interestingly, a combined approach of nanopillar-assisted SERS chromatography (NPC-SERS) and a centrifugal microfluidic device has been reported for similar multiplex profiling of analytes in urine samples (Fig. 13).146 Through this two-step process, analytes in complex and multicomponent fluids can be captured and this was tested on a model drug paracetamol in human urine. Results show the successful separation of paracetamol from other human urine components, achieving a quantitative paracetamol detection with 0–500 ppm linear dynamic range in human urine via a Raman microscope system. The novel concept of NPC-SERS in a fully integrated centrifugal microfluidic platform shows general applicability for rapid and quantitative POC detection in complex biofluids. Similarly, a combined centrifugal microfluidic chip with SERS detection has been reported for creatinine in clinical blood samples under a confocal Raman microscope system.147 The microfluidic SERS chip has been used to detect chronic renal failure based on this method and results are consistent with laboratory-based enzymatic methods, showing the potential for quick bedside diagnosis. Recently, a triple combination approach, including colorimetric, UV-visible spectroscopy and SERS via a confocal Raman microscope system, has been reported for the POC diagnosis of anaemia.148 This was achieved by using organic cyanide modified silver nanoparticles (cAgNPs) in a multimodal approach. Analytes tested were Fe3+ and haemoglobin in clinical whole blood samples in both anaemic patients and normal controls, giving a detection LOD of 10 fM and 0.46 μg mL−1 for Fe3+ ions and haemoglobin protein, respectively. This developed combined assay strategy is highly affordable for bulk-samples, fast and POC for on-line practical applications.
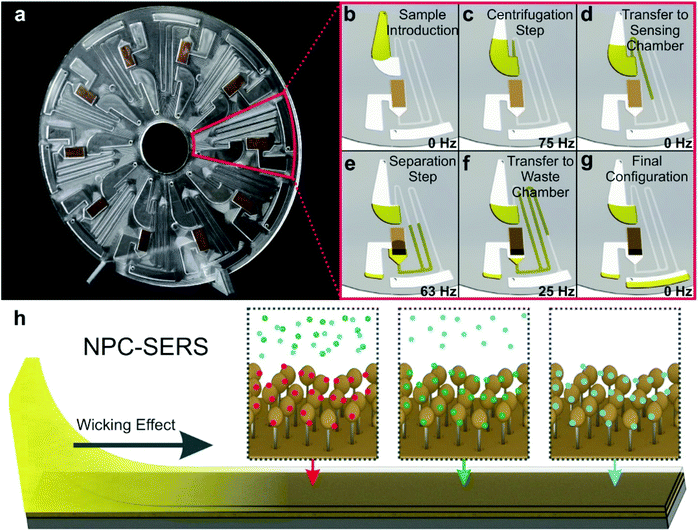 | ||
| Fig. 13 Schematic of nanopillar-assisted SERS chromatography (NPC-SERS) technique for multiplex quantitation of analytes in complex environment fluids. Gold-coated silicon nanopillar (AuNP) SERS substrates and a centrifugal microfluidic platform are strategically combined (a), allowing sample loading into SERS substrate once applying centrifugation (b–g), where the wetting of the stationary phase is by wicking effect (h). Reproduced with permission from ref. 146. Copyright 2018, ACS Publications. | ||
Besides SERS detection of specific biomarkers from complex clinical samples mentioned previously, recent efforts have also been devoted to directly detect whole patient samples to differentiate positive and negative sample for different diseases/cancer types.149–152 One such study involved the label-free on-site SERS detection of breast cancer using human tear fluid from breast cancer patients and control subjects.149 Together with a principal component linear discriminant analysis (PC-LDA) model, the detection employs a portable Raman spectroscopy based on nanosphere SERS substrates for direct sample analysis. Using 2-naphthalenethiol (2-NAT) as a Raman tag, results show the detection of femtomole, and SERS enhancement of giga-fold, and a relative standard deviation of less than 5% for both reliability and reproducibility. Importantly, the developed system employed the leave-one-out cross validation (LOOCV)-assisted PC-LDA identification method and obtained a 92% clinical sensitivity and a 100% specificity to identify asymptomatic breast cancer using tear fluids. The developed label-free SERS screening platform could be applied for the early POC identification of asymptomatic tumours and biomarker discovery. Along with the development of POC SERS detection in biomedical fields, the previously mentioned combination approaches towards multifunction/multimodality for on-site quick screening of clinical samples and for addressing different diagnostic needs are examples of moving SERS testing towards POC applications in the healthcare space.
4 Future perspectives and conclusion
As demonstrated previously, SERS provides a unique advantage in providing tremendously high sensitivity in its embedded rich molecular information due to the vibrational fingerprint spectra that show it to be superior to other conventional optical spectroscopic techniques such as fluorescence and UV/Vis absorption spectroscopy. Furthermore, SERS also provides foolproof multiplex detection of analytes in complex media, which is not possible in many other spectroscopic techniques. We reviewed the recent advancements in the field of SERS as a POC technique for specific biomedical, food and agricultural applications over the last 6 years.In the first part of the review, SERS-based systems and detection methodologies in food analytics, specifically for the detection of nutrients, pesticides, contaminants, toxins, adulterants, pathogens and plant stress markers are analysed in detail. We believe that due to the ongoing pandemic of COVID-19, climate change and other threats to the worldwide food supply chain, it is of great importance to progress towards robust and easy to implement technologies in all aspects of food production, packing and supply. Due to an overwhelming scarcity of nutrient-rich foods and their supply, especially in developing countries, the need to develop technologies and platforms that will propel us towards improved food production and supply is overwhelmingly imperative. In this regard, Singapore's government recently announced a national initiative termed “30 by 30”, implying a 30% self-reliance on all food supplies by the year 2030, a significant deviation from importing over 90% of its food consumption, due to urbanized life and the limitation of land for largescale farming. New solutions to raise productivity include indoor vertical farming and hydroponics, in which POC SERS sensors could play a huge role in monitoring nutrient levels and detecting plant stress at an early stage to improve productivity.
In this context, the “press and peel-off” SERS detection for pesticides is a significant step towards a commercially viable sensor, especially if it can be integrated into a portable, cheap and sensitive Raman spectrometer. As reported by Zhang et al.,75 multiplex detection of mycotoxins in maize using a SERS-based lateral flow immunosensor is a significant step towards realizing a sensor that could be integrated into many other crops with some necessary refinements in the detection protocol and spectral analysis. Another domain where POC SERS sensor could revolutionize food analytics is in the detection of adulterants and chemicals. As reported by Lin et al., SERS detection of melamine in several types of food products gave an excellent sensitivity of about 30 times that of HPLC, giving the LOD at 0.033 μg m−1.78 This platform could be further refined by employing SERS substrates that can be fabricated with simpler procedures, providing greater signal reproducibility. Reported SERS sensors for food analytics will surely gain major traction in the coming years with the availability of cheap, stable and portable Raman systems, along with a more reliable and reproducible SERS substrate platform.
The development of smart nanotechnology-based sensors that have the ability to convert the presence of plant chemicals to digital signals for monitoring devices holds tremendous potential to be integrated along with SERS sensors, introducing a new paradigm in smart plant sensors.154 Furthermore, sensor networks based on flexible wearable nanoelectronic circuits equipped with wireless communication can be engineered to be implanted on plant leaves or fruits, thus playing a significant role in the real-time monitoring of plant nutrients and stress. Such nanoelectronic sensors could play a complementary role in flexible membrane-based SERS sensors. SERS and nanoelectronic or nanophotonics-based integrated sensors could facilitate faster identification of desired crop traits for the high-throughput screening of chemical phenotypes.
In the second part of the review, we analysed the recent advancements in SERS sensors for selected biomedical applications such as the detection of nucleic acids, proteins, pathogens (viruses and bacteria), whole cells (specifically CTC) and small molecules. The most demanding criteria for a biosensor is that it should possess high sensitivity to detect biomarkers of diseases at trace-level concentrations and in some cases even at low sample volumes, which is advantageous for disease prognosis and treatment monitoring purposes. Generally, such a self-contained device/platform that is capable of providing quantitative or semi-quantitative analytical information of a target analyte should possess high specificity, be cheap to produce, have a fast readout and be portable. While commonly used detection methods for nucleic acids such as qRT-PCR, northern blotting or microarray-based methods suffer from long sample preparation time, long assay time, lack of portability and high cost, there exists a pressing unmet need for a faster and sensitive approach for situations such as the detection of positive patients during the on-going COVID-19 pandemic. Though the SERS-based proof of concept studies for the detection of nucleic acids are promising, significant research should be directed towards specifically capturing the target nucleic acids on SERS sensors with high specificity and read out using cheap and portable Raman spectrometers. An easy to implement nano-bio interface should go hand in hand with the sensing to realize this goal.
Detection of biomolecules using a labelled SERS approach, by employing SERS nanotags for readout, is an efficient and sensitive approach that will provide high specificity due to the targeting moieties that are anchored onto the nanoconstructs. SERS nanotags have been extensively used for in vitro and in vivo (primarily in animal models) detection of various biomarkers with high specificity. Labelled SERS detection by employing SERS nanotags revolutionized POC SERS sensing for various proteins. One of the advantages of SERS nanotags is the possibility of using specifically designed and synthesized Raman-reporter labels that could facilitate multiplex detection due to their unique non-overlapping spectral peaks. As demonstrated by Sanjiv Gambhir and co-workers, simultaneous detection of up to ten differently SERS-labelled bio-analytes is possible for passively localized particles,155 and hence this approach could be employed along with a microfluidic platform to develop multiplex detection of biomolecules in complex body fluids such as saliva or blood. Such nanoprobes can find potential applications in the early diagnosis of diseases where simultaneous multiplex detection of biomarkers can be detected at a low concentration. Furthermore, they may also find promising applications in the monitoring of the effectiveness of the therapy.
Another essential ingredient in developing a POC SERS sensor for biomedical applications is a robust microfluidic platform that can be easily integrated into SERS active metallic nanostructures for Raman signal enhancement. Such a microfluidic integrated SERS chip could very well revolutionize the detection of all types of biomolecules. In this regard, the microRNA detection using a 3D microfluidic paper device integrated with a handheld Raman spectrometer is quite promising.99 In a similar way, the innovative SERS device reported for the detection of clinical DNA via a SERS microfluidic chip with molecular beacon has great potential as a POC diagnostic tool.106 The sensor fabricated by incorporating the SERS-active metal NPs into a microfluidic chip, and with the automated and faster analysis, could detect DNA effectively with 50 fM LOD, and with single base mutation differentiation is quite exciting. Similarly, microfluidic-based SERS immunoassay will also be expected to create new paradigms in POC diagnostics in the near future.
The planar or colloidal metallic substrate onto which analyte molecules are directly adsorbed (for label-free detection) or through targeting moieties (such as an antibody, peptides, etc.) forms the foundation of SERS-based sensors. The inherent signal variation from a nano-roughened metal surface owing to irregularities can result in poor reproducibility and repeatability of sensing results. On most occasions, a significant intensity variation of 5–10% from even the best substrates fabricated with sophisticated cleanroom facilities can be a major roadblock in translating this technology for real end-user applications. In this context, a significant advancement is mandatory to realize cheaper but reliable SERS substrate fabrication protocols that will generate confidence to take the technology to the next level. Special interest should be focused on identifying novel materials that can be adopted to develop robust and reproducible flexible SERS substrates. In this context, incorporation and adoption of novel nanophotonic and nanoelectronic materials that could be modified to fit into SERS sensors is a progressive approach. Such an approach could lead to significant advancements in realizing a compact and reliable sensor.
Unprecedented reproducibility and repeatability offered by the PCF-based SERS platform can form one strategy in addressing this limitation. Beffara et al. recently demonstrated that a PCF-SERS sensing platform with suspended core-3 hole PCF could offer unmatched reproducibility and repeatability in measurements with a relative standard deviation of only 1.5% and 4.6%, respectively.117 Such designs are ideal for developing POC sensors for body fluid analysis as they offer superior sensitivity in comparison with planar/colloidal nanoparticle platforms, owing to the increased interaction length between the guided light and analyte, which is inside the microstructed holes of the fibre. With necessary refinement and improved bio-functionalization protocols, an all fibre miniaturized Raman spectrometer could be incorporated for a faster and cost-effective signal readout.
Another roadblock in the advancement of SERS as a POC device is the lack of sensitive yet cheap and portable Raman spectrometers. Thorough research into the development of miniaturized spectrometers and Raman stabilized lasers that can be integrated into an on-chip platform would facilitate a feasible path towards realizing this goal. Moreover, artificial intelligence (AI) can be implemented to analyse the Raman spectra generated from complex samples for disease diagnosis and food analytics. In such complex media, acquired Raman spectra could be spectrally unmixed to estimate the concentration profiles for better quantification. Such an algorithm will offer multidimensional advantages not only to characterize the disease but also to evaluate the adulterants and toxins in food processing as well. Deep learning, a subfield of AI, is one of the highly sought-after data analytic tools and interpretation approaches that has been explored recently. It could automate the approach as it can “learn” the base fingerprint Raman spectra and resolve specific differentiators from representative complex data, outperforming human-operated analysis which could potentially result in more efficient and faster disease diagnosis. Eventually, such data analytics integrated sensors should be translated into a continuous monitoring platform, which could be used in intensive care units, operating theatres or other situations where accurate and real-time monitoring of biological analytes is critical. In a similar way, it could also play a key role in revolutionizing the food supply chain with the aim of providing cleaner and safer food to all.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore under its industry alignment fund pre-positioning programmes, Translational Biophotonics Innovation Platform (TBIP, H19H6a0025) and High Performance Precision Agriculture (HiPPA) system (A19E4a0101). The authors would also like to thank the Biomedical Research Council (BMRC), A*STAR, for the intramural funding support for some aspects of this study. We would also like to thank Ms. Lim Hann Qian for her help in this review.References
- M. Mascini and S. Tombelli, Biomarkers, 2008, 13, 637–657 Search PubMed.
- P. Yáñez-Sedeño, L. Agüí, R. Villalonga and J. Pingarrón, Anal. Chim. Acta, 2014, 823, 1–19 CrossRef.
- C. I. Justino, A. C. Duarte and T. A. Rocha-Santos, Sensors, 2017, 17, 2918 CrossRef.
- Z. Lin and L. He, Curr. Opin. Food Sci., 2019, 28, 82–87 CrossRef.
- W. Niessen and A. Tinke, J. Chromatogr. A, 1995, 703, 37–57 CrossRef CAS.
- J. Mejía-Salazar and O. N. Oliveira Jr., Chem. Rev., 2018, 118, 10617–10625 CrossRef.
- G. McNay, D. Eustace, W. E. Smith, K. Faulds and D. Graham, Appl. Spectrosc., 2011, 65, 825–837 CrossRef CAS.
- T. Vo-Dinh, M. Hiromoto, G. Begun and R. Moody, Anal. Chem., 1984, 56, 1667–1670 CrossRef CAS.
- T. E. Rohr, T. Cotton, N. Fan and P. J. Tarcha, Anal. Biochem., 1989, 182, 388–398 CrossRef CAS.
- G. Wu and M. H. Zaman, Bulletin of the World Health Organization, 2012, 90, 914–920 CrossRef.
- A. Shiohara, Y. Wang and L. M. Liz-Marzán, J. Photochem. Photobiol., C, 2014, 21, 2–25 CrossRef CAS.
- S. Wang, T. Chinnasamy, M. A. Lifson, F. Inci and U. Demirci, Trends Biotechnol., 2016, 34, 909–921 CrossRef CAS.
- E. W. Nery and L. T. Kubota, Anal. Bioanal. Chem., 2013, 405, 7573–7595 CrossRef CAS.
- K. Xu, R. Zhou, K. Takei and M. Hong, Adv. Sci., 2019, 6, 1900925 CrossRef.
- N. Zhang, L. Tong and J. Zhang, Chem. Mater., 2016, 28, 6426–6435 CrossRef CAS.
- G. P. Kumar, J. Nanophotonics, 2012, 6, 064503 CrossRef.
- H. Marks, M. Schechinger, J. Garza, A. Locke and G. Coté, Nanophotonics, 2017, 6, 681–701 CAS.
- J. H. Granger, N. E. Schlotter, A. C. Crawford and M. D. Porter, Chem. Soc. Rev., 2016, 45, 3865–3882 RSC.
- A. I. Radu, M. Kuellmer, B. Giese, U. Huebner, K. Weber, D. Cialla-May and J. Popp, Talanta, 2016, 160, 289–297 CrossRef CAS.
- F. Zanuttin, E. Gurian, I. Ignat, S. Fornasaro, A. Calabretti, G. Bigot and A. Bonifacio, Talanta, 2019, 203, 99–105 CrossRef CAS.
- I. Aguilar-Hernández, N. K. Afseth, T. López-Luke, F. F. Contreras-Torres, J. P. Wold and N. Ornelas-Soto, Vib. Spectrosc., 2017, 89, 113–122 CrossRef.
- J. Park, J. A. Thomasson, C. C. Gale, G. A. Sword, K.-M. Lee, T. J. Herrman and C. P.-C. Suh, ACS Omega, 2020, 5, 2779–2790 CrossRef CAS.
- R. A. Alvarez-Puebla and L. M. Liz-Marzán, Angew. Chem., Int. Ed., 2012, 51, 11214–11223 CrossRef CAS.
- G. Brackx, D. Guinoiseau, L. Duponchel, A. Gélabert, V. Reichel, S. Zrig, J.-M. Di Meglio, M. F. Benedetti, J. Gaillardet and G. Charron, Sci. Rep., 2020, 10, 1–11 CrossRef.
- Y. Shi, N. Chen, Y. Su, H. Wang and Y. He, Nanoscale, 2018, 10, 4010–4018 RSC.
- P. Zhang, L. Wang, X. Wei, T. Lin, H. Wang, X. Liu and D. Zheng, J. Appl. Spectrosc., 2018, 85, 880–884 CrossRef CAS.
- C. Zaffino, B. Russo and S. Bruni, Spectrochim. Acta, Part A, 2015, 149, 41–47 CrossRef CAS.
- T. Yang, J. Doherty, H. Guo, B. Zhao, J. M. Clark, B. Xing, R. Hou and L. He, Anal. Chem., 2019, 91, 2093–2099 CrossRef CAS.
- F. Wang, X. Gu, C. Zheng, F. Dong, L. Zhang, Y. Cai, Z. You, J. You, S. Du and Z. Zhang, Anal. Chem., 2017, 89, 8836–8843 CrossRef CAS.
- X.-R. Bai, Y. Zeng, X.-D. Zhou, X.-H. Wang, A.-G. Shen and J.-M. Hu, Anal. Chem., 2017, 89, 10335–10342 CrossRef CAS.
- B. M. Crawford, P. Strobbia, H.-N. Wang, R. Zentella, M. I. Boyanov, Z.-M. Pei, T.-P. Sun, K. M. Kemner and T. Vo-Dinh, ACS Appl. Mater. Interfaces, 2019, 11, 7743–7754 CrossRef CAS.
- K. Wang, D.-W. Sun, H. Pu, Q. Wei and L. Huang, ACS Appl. Mater. Interfaces, 2019, 11, 29177–29186 CrossRef CAS.
- X. Gong, M. Tang, Z. Gong, Z. Qiu, D. Wang and M. Fan, Food Chem., 2019, 295, 254–258 CrossRef CAS.
- T. Yang, Z. Zhang, B. Zhao, R. Hou, A. Kinchla, J. M. Clark and L. He, Anal. Chem., 2016, 88, 5243–5250 CrossRef CAS.
- R. Hou, Z. Zhang, S. Pang, T. Yang, J. M. Clark and L. He, Environ. Sci. Technol., 2016, 50, 6216–6223 CrossRef CAS.
- R. Hou, S. Pang and L. He, Anal. Methods, 2015, 7, 6325–6330 RSC.
- N. Zhou, G. Meng, Z. Huang, Y. Ke, Q. Zhou and X. Hu, Analyst, 2016, 141, 5864–5869 RSC.
- W. Wang, M. Xu, Q. Guo, Y. Yuan, R. Gu and J. Yao, RSC Adv., 2015, 5, 47640–47646 RSC.
- K. Guo, R. Xiao, X. Zhang, C. Wang, Q. Liu, Z. Rong, L. Ye and S. Chen, Molecules, 2015, 20, 6299–6309 CrossRef CAS.
- T. Tite, N. Ollier, M. C. Sow, F. Vocanson and F. Goutaland, Sens. Actuators, B, 2017, 242, 127–131 CrossRef CAS.
- Q. Xu, X. Guo, L. Xu, Y. Ying, Y. Wu, Y. Wen and H. Yang, Sens. Actuators, B, 2017, 241, 1008–1013 CrossRef CAS.
- J. Zhu, M.-J. Liu, J.-J. Li, X. Li and J.-W. Zhao, Spectrochim. Acta, Part A, 2018, 189, 586–593 CrossRef CAS.
- L. Mandrile, A. Giovannozzi, F. Durbiano, G. Martra and A. Rossi, Food Chem., 2018, 244, 16–24 CrossRef CAS.
- J. Yuan, C. Sun, X. Guo, T. Yang, H. Wang, S. Fu, C. Li and H. Yang, Food Chem., 2017, 221, 797–802 CrossRef CAS.
- R. Hou, M. Tong, W. Gao, L. Wang, T. Yang and L. He, Food Chem., 2017, 237, 305–311 CrossRef CAS.
- V. Suresh and F. L. Yap, RSC Adv., 2015, 5, 61671–61677 RSC.
- P. Wang, L. Wu, Z. Lu, Q. Li, W. Yin, F. Ding and H. Han, Anal. Chem., 2017, 89, 2424–2431 CrossRef CAS.
- X. Wang, X. Zhu, Y. Chen, M. Zheng, Q. Xiang, Z. Tang, G. Zhang and H. Duan, ACS Appl. Mater. Interfaces, 2017, 9, 31102–31110 CrossRef CAS.
- J. Chen, M. Huang, L. Kong and M. Lin, Carbohydr. Polym., 2019, 205, 596–600 CrossRef CAS.
- J. Chen, Y. Huang, P. Kannan, L. Zhang, Z. Lin, J. Zhang, T. Chen and L. Guo, Anal. Chem., 2016, 88, 2149–2155 CrossRef CAS.
- H. Wu, Y. Luo, C. Hou, D. Huo, Y. Zhou, S. Zou, J. Zhao and Y. Lei, Sens. Actuators, B, 2019, 285, 123–128 CrossRef CAS.
- C. M. Wang, P. K. Roy, B. K. Juluri and S. Chattopadhyay, Sens. Actuators, B, 2018, 261, 218–225 CrossRef CAS.
- G. Fu, D.-W. Sun, H. Pu and Q. Wei, Talanta, 2019, 195, 841–849 CrossRef CAS.
- F. K. Alsammarraie, M. Lin, A. Mustapha, H. Lin, X. Chen, Y. Chen, H. Wang and M. Huang, Food Chem., 2018, 259, 219–225 CrossRef CAS.
- H. Sun, H. Liu and Y. Wu, Appl. Surf. Sci., 2017, 416, 704–709 CrossRef CAS.
- S. Fateixa, M. Raposo, H. Nogueira and T. Trindade, Talanta, 2018, 182, 558–566 CrossRef CAS.
- X. Chen, M. Lin, L. Sun, T. Xu, K. Lai, M. Huang and H. Lin, Food Chem., 2019, 293, 271–277 CrossRef CAS.
- P. Wu, L.-B. Zhong, Q. Liu, X. Zhou and Y.-M. Zheng, Nanoscale, 2019, 11, 12829–12836 RSC.
- T. Yang, B. Zhao, R. Hou, Z. Zhang, A. J. Kinchla, J. M. Clark and L. He, J. Food Sci., 2016, 81, T2891–T2901 CrossRef CAS.
- S. Kumar, P. Goel and J. P. Singh, Sens. Actuators, B, 2017, 241, 577–583 CrossRef CAS.
- H. Wu, Y. Luo, C. Hou, D. Huo, W. Wang, J. Zhao and Y. Lei, Talanta, 2019, 200, 84–90 CrossRef CAS.
- J. Hong, A. Kawashima and N. Hamada, Appl. Surf. Sci., 2017, 407, 440–446 CrossRef CAS.
- F. K. Alsammarraie and M. Lin, J. Agric. Food Chem., 2017, 65, 666–674 CrossRef CAS.
- Y. Pan, X. Guo, J. Zhu, X. Wang, H. Zhang, Y. Kang, T. Wu and Y. Du, Microchim. Acta, 2015, 182, 1775–1782 CrossRef CAS.
- X. Li, H. K. Lee, I. Y. Phang, C. K. Lee and X. Y. Ling, Anal. Chem., 2014, 86, 10437–10444 CrossRef CAS.
- J. Ko, C. Lee and J. Choo, J. Hazard. Mater., 2015, 285, 11–17 CrossRef CAS.
- Q. Li, Z. Lu, X. Tan, X. Xiao, P. Wang, L. Wu, K. Shao, W. Yin and H. Han, Biosens. Bioelectron., 2017, 97, 59–64 CrossRef CAS.
- W. A. Hassanain, E. L. Izake, M. S. Schmidt and G. A. Ayoko, Biosens. Bioelectron., 2017, 91, 664–672 CrossRef CAS.
- L.-K. Lin and L. A. Stanciu, Sens. Actuators, B, 2018, 276, 222–229 CrossRef CAS.
- S. C. Pinzaru, C. Müller, I. Ujević, M. M. Venter, V. Chis and B. Glamuzina, Talanta, 2018, 187, 47–58 CrossRef CAS.
- L.-L. Qu, Z.-Q. Geng, W. Wang, K.-C. Yang, W.-P. Wang, C.-Q. Han, G.-H. Yang, R. Vajtai, D.-W. Li and P. M. Ajayan, J. Hazard. Mater., 2019, 379, 120823 CrossRef CAS.
- Y. Jiang, D.-W. Sun, H. Pu and Q. Wei, Talanta, 2019, 197, 151–158 CrossRef CAS.
- T.-t. Pan, D.-W. Sun, H. Pu and Q. Wei, J. Agric. Food Chem., 2018, 66, 2180–2187 CrossRef CAS.
- Y. Hernández, L. K. Lagos and B. C. Galarreta, Sens. Biosensing Res., 2020, 28, 100331 CrossRef.
- W. Zhang, S. Tang, Y. Jin, C. Yang, L. He, J. Wang and Y. Chen, J. Hazard. Mater., 2020, 393, 122348 CrossRef CAS.
- T. Janči, D. Valinger, J. G. Kljusurić, L. Mikac, S. Vidaček and M. Ivanda, Food Chem., 2017, 224, 48–54 CrossRef.
- K. Sharma and M. Paradakar, Food Secur., 2010, 2, 97–107 CrossRef.
- M. Lin, L. He, J. Awika, L. Yang, D. Ledoux, H. a. Li and A. Mustapha, J. Food Sci., 2008, 73, T129–T134 CrossRef CAS.
- J. F. Betz, Y. Cheng and G. W. Rubloff, Analyst, 2012, 137, 826–828 RSC.
- F. Fang, Y. Qi, F. Lu and L. Yang, Talanta, 2016, 146, 351–357 CrossRef CAS.
- J. C. Gukowsky, T. Xie, S. Gao, Y. Qu and L. He, Food Control, 2018, 92, 267–275 CrossRef CAS.
- J. Meng, S. Qin, L. Zhang and L. Yang, Appl. Surf. Sci., 2016, 366, 181–186 CrossRef CAS.
- Y. Yao, W. Wang, K. Tian, W. M. Ingram, J. Cheng, L. Qu, H. Li and C. Han, Spectrochim. Acta, Part A, 2018, 195, 165–171 CrossRef CAS.
- Y.-j. Ai, P. Liang, Y.-x. Wu, Q.-m. Dong, J.-b. Li, Y. Bai, B.-J. Xu, Z. Yu and D. Ni, Food Chem., 2018, 241, 427–433 CrossRef CAS.
- A. G. Fá, F. Pignanelli, I. López-Corral, R. Faccio, A. Juan and M. S. Di Nezio, Trends Anal. Chem., 2019, 121, 115673 CrossRef.
- K. Sun, Q. Huang, G. Meng and Y. Lu, ACS Appl. Mater. Interfaces, 2016, 8, 5723–5728 CrossRef CAS.
- Z. Li, Z. Du, K. Sun, X. He and B. Chen, RSC Adv., 2017, 7, 53157–53163 RSC.
- Z. Li, K. Sun, Z. Du, B. Chen and X. He, Nanomaterials, 2018, 8, 265 CrossRef.
- P. V. Shanta and Q. Cheng, ACS Sens., 2017, 2, 817–827 CrossRef CAS.
- X. Huang, Y. Liu, J. Barr, J. Song, Z. He, Y. Wang, Z. Nie, Y. Xiong and X. Chen, Nanoscale, 2018, 10, 13202–13211 RSC.
- World Health Organization, WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015, World Health Organization, 2015 Search PubMed.
- I.-H. Cho, P. Bhandari, P. Patel and J. Irudayaraj, Biosens. Bioelectron., 2015, 64, 171–176 CrossRef CAS.
- O. Prakash, S. Sil, T. Verma and S. Umapathy, J. Phys. Chem. C, 2019, 124, 861–869 CrossRef.
- J. Hwang, S. Lee and J. Choo, Nanoscale, 2016, 8, 11418–11425 RSC.
- W. Wang, V. Hynninen, L. Qiu, A. Zhang, T. Lemma, N. Zhang, H. Ge, J. J. Toppari, V. P. Hytönen and J. Wang, Sens. Actuators, B, 2017, 239, 515–525 CrossRef CAS.
- L. Wu, X. Xiao, K. Chen, W. Yin, Q. Li, P. Wang, Z. Lu, J. Ma and H. Han, Biosens. Bioelectron., 2017, 92, 321–327 CrossRef CAS.
- J. Guo, Y. Liu, Y. Yang, Y. Li, R. Wang and H. Ju, Anal. Chem., 2020, 92, 5055–5063 CrossRef CAS.
- S. Mabbott, S. C. Fernandes, M. Schechinger, G. L. Cote, K. Faulds, C. R. Mace and D. Graham, Analyst, 2020, 145, 983–991 RSC.
- L. Qi, M. Xiao, X. Wang, C. Wang, L. Wang, S. Song, X. Qu, L. Li, J. Shi and H. Pei, Anal. Chem., 2017, 89, 9850–9856 CrossRef CAS.
- Y. Pang, C. Wang, L. Lu, C. Wang, Z. Sun and R. Xiao, Biosens. Bioelectron., 2019, 130, 204–213 CrossRef CAS.
- Y. Pang, C. Wang, J. Wang, Z. Sun, R. Xiao and S. Wang, Biosens. Bioelectron., 2016, 79, 574–580 CrossRef CAS.
- D. Ma, C. Huang, J. Zheng, J. Tang, J. Li, J. Yang and R. Yang, Biosens. Bioelectron., 2018, 101, 167–173 CrossRef CAS.
- H. T. Ngo, E. Freedman, R. A. Odion, P. Strobbia, A. S. D. S. Indrasekara, P. Vohra, S. M. Taylor and T. Vo-Dinh, Sci. Rep., 2018, 8, 4075 CrossRef.
- S. Han, A. K. Locke, L. A. Oaks, Y.-S. L. Cheng and G. L. Coté, J. Biomed. Opt., 2019, 24, 055001 CAS.
- L. Wu, A. Garrido-Maestu, J. R. Guerreiro, S. Carvalho, S. Abalde-Cela, M. Prado and L. Diéguez, Nanoscale, 2019, 11, 7781–7789 RSC.
- L. H. Di Zhang, B. Liu, Q. Ge, J. Dong and X. Zhao, Theranostics, 2019, 9, 4849 CrossRef.
- D. Lin, Q. Wu, S. Qiu, G. Chen, S. Feng, R. Chen and H. Zeng, Nanomedicine, 2019, 22, 102100 CrossRef CAS.
- Q. Zhou, J. Zheng, Z. Qing, M. Zheng, J. Yang, S. Yang, L. Ying and R. Yang, Anal. Chem., 2016, 88, 4759–4765 CrossRef CAS.
- X. Fu, Z. Cheng, J. Yu, P. Choo, L. Chen and J. Choo, Biosens. Bioelectron., 2016, 78, 530–537 CrossRef CAS.
- X. Li, T. Yang, C. S. Li, L. Jin, H. Lou and Y. Song, Biomed. Opt. Express, 2018, 9, 3167–3176 CrossRef CAS.
- L. M. Freeman, L. Pang and Y. Fainman, Sci. Rep., 2018, 8, 1–7 CrossRef CAS.
- H. Y. Lau, Y. Wang, E. J. Wee, J. R. Botella and M. Trau, Anal. Chem., 2016, 88, 8074–8081 CrossRef CAS.
- V. Tran, B. Walkenfort, M. König, M. Salehi and S. Schlücker, Angew. Chem., Int. Ed., 2019, 58, 442–446 CrossRef CAS.
- M. Sánchez-Purrà, M. Carré-Camps, H. de Puig, I. Bosch, L. Gehrke and K. Hamad-Schifferli, ACS Infect. Dis., 2017, 3, 767–776 CrossRef.
- S.-W. Hu, S. Qiao, J.-B. Pan, B. Kang, J.-J. Xu and H.-Y. Chen, Talanta, 2018, 179, 9–14 CrossRef CAS.
- Y. Wang, Q. Ruan, Z.-C. Lei, S.-C. Lin, Z. Zhu, L. Zhou and C. Yang, Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS.
- F. Beffara, J. Perumal, A. P. Mahyuddin, M. Choolani, S. A. Khan, J. L. Auguste, S. Vedraine, G. Humbert, U. Dinish and M. Olivo, J. Biophotonics, 2020, 13, e201960120 CrossRef.
- Y. Lai, S. Schlücker and Y. Wang, Anal. Bioanal. Chem., 2018, 410, 5993–6000 CrossRef CAS.
- K. Kamil Reza, J. Wang, R. Vaidyanathan, S. Dey, Y. Wang and M. Trau, Small, 2017, 13, 1602902 CrossRef.
- J. Perumal, A. P. Mahyuddin, G. Balasundaram, D. Goh, C. Y. Fu, A. Kazakeviciute, U. Dinish, M. Choolani and M. Olivo, Cancer Manage. Res., 2019, 11, 1115 CrossRef CAS.
- S. Muneer, G. A. Ayoko, N. Islam and E. L. Izake, Talanta, 2020, 208, 120411 CrossRef CAS.
- Y.-T. Yeh, K. Gulino, Y. Zhang, A. Sabestien, T.-W. Chou, B. Zhou, Z. Lin, I. Albert, H. Lu and V. Swaminathan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895–901 CrossRef CAS.
- H. Shen, K. Xie, L. Huang, L. Wang, J. Ye, M. Xiao, L. Ma, A. Jia and Y. Tang, Sens. Actuators, B, 2019, 282, 152–157 CrossRef CAS.
- Y. Sun, L. Xu, F. Zhang, Z. Song, Y. Hu, Y. Ji, J. Shen, B. Li, H. Lu and H. Yang, Biosens. Bioelectron., 2017, 89, 906–912 CrossRef CAS.
- M. Xiao, K. Xie, X. Dong, L. Wang, C. Huang, F. Xu, W. Xiao, M. Jin, B. Huang and Y. Tang, Anal. Chim. Acta, 2019, 1053, 139–147 CrossRef CAS.
- H. J. Park, S. C. Yang and J. Choo, Bull. Korean Chem. Soc., 2016, 37, 2019–2024 CrossRef CAS.
- C. Wang, C. Wang, X. Wang, K. Wang, Y. Zhu, Z. Rong, W. Wang, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 19495–19505 CrossRef CAS.
- W. Wang, R.-l. Dong, D. Gu, J.-a. He, P. Yi, S.-K. Kong, H.-P. Ho, J. F.-C. Loo, W. Wang and Q. Wang, Adv. Med. Sci., 2020, 65, 86–92 CrossRef.
- M. Reyes, M. Piotrowski, S. K. Ang, J. Chan, S. He, J. J. H. Chu and J. C. Y. Kah, Anal. Chem., 2017, 89, 5373–5381 CrossRef CAS.
- W. R. Premasiri, J. C. Lee, A. Sauer-Budge, R. Theberge, C. E. Costello and L. D. Ziegler, Anal. Bioanal. Chem., 2016, 408, 4631–4647 CrossRef CAS.
- N. Dina, H. Zhou, A. Colniţă, N. Leopold, T. Szoke-Nagy, C. Coman and C. Haisch, Analyst, 2017, 142, 1782–1789 RSC.
- H. Kearns, R. Goodacre, L. E. Jamieson, D. Graham and K. Faulds, Anal. Chem., 2017, 89, 12666–12673 CrossRef CAS.
- W. Gao, B. Li, R. Yao, Z. Li, X. Wang, X. Dong, H. Qu, Q. Li, N. Li and H. Chi, Anal. Chem., 2017, 89, 9836–9842 CrossRef CAS.
- D. Yang, H. Zhou, C. Haisch, R. Niessner and Y. Ying, Talanta, 2016, 146, 457–463 CrossRef CAS.
- A. Mühlig, T. Bocklitz, I. Labugger, S. Dees, S. Henk, E. Richter, S. n. Andres, M. Merker, S. Stöckel and K. Weber, Anal. Chem., 2016, 88, 7998–8004 CrossRef.
- J. Perumal, U. Dinish, A. K. Bendt, A. Kazakeviciute, C. Y. Fu, I. L. H. Ong and M. Olivo, Int. J. Nanomed., 2018, 13, 6029 CrossRef CAS.
- C. N. Kotanen, L. Martinez, R. Alvarez and J. W. Simecek, Sens. Biosensing Res., 2016, 8, 20–26 CrossRef.
- C.-C. Lin, C.-Y. Lin, C.-J. Kao and C.-H. Hung, Sens. Actuators, B, 2017, 241, 513–521 CrossRef CAS.
- R. Wang, K. Kim, N. Choi, X. Wang, J. Lee, J. H. Jeon, G.-e. Rhie and J. Choo, Sens. Actuators, B, 2018, 270, 72–79 CrossRef CAS.
- X. Wu, Y. Xia, Y. Huang, J. Li, H. Ruan, T. Chen, L. Luo, Z. Shen and A. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 19928–19938 CrossRef CAS.
- A. Kamińska, T. Szymborski, E. Witkowska, E. Kijeńska-Gawrońska, W. Świeszkowski, K. Niciński, J. Trzcińska-Danielewicz and A. Girstun, Nanomaterials, 2019, 9, 366 CrossRef.
- D. Sun, F. Cao, Y. Tian, A. Li, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 15484–15490 CrossRef CAS.
- K. Niciński, J. Krajczewski, A. Kudelski, E. Witkowska, J. Trzcińska-Danielewicz, A. Girstun and A. Kamińska, Sci. Rep., 2019, 9, 1–14 CrossRef.
- C. M. Girish, S. Iyer, K. Thankappan, G. S. Gowd, S. Nair and M. Koyakutty, Adv. Healthcare Mater., 2019, 8, 1801557 CrossRef.
- Y.-C. Kao, X. Han, Y. H. Lee, H. K. Lee, G. C. Phan-Quang, C. L. Lay, H. Y. F. Sim, V. J. X. Phua, L. S. Ng and C. W. Ku, ACS Nano, 2020, 14, 2542–2552 CrossRef CAS.
- O. Durucan, K. Wu, M. Viehrig, T. Rindzevicius and A. Boisen, ACS Sens., 2018, 3, 2492–2498 CrossRef CAS.
- X. Su, Y. Xu, H. Zhao, S. Li and L. Chen, Talanta, 2019, 194, 903–909 CrossRef CAS.
- P. Muhammad, S. Hanif, J. Yan, F. U. Rehman, J. Wang, M. Khan, R. Chung, A. Lee, M. Zheng and Y. Wang, Nanoscale, 2020, 12, 1948–1957 RSC.
- S. Kim, T. G. Kim, S. H. Lee, W. Kim, A. Bang, S. W. Moon, J. Song, J.-H. Shin, J. S. Yu and S. Choi, ACS Appl. Mater. Interfaces, 2020, 12, 7897–7904 CrossRef CAS.
- A. Stefancu, M. Badarinza, V. Moisoiu, S. D. Iancu, O. Serban, N. Leopold and D. Fodor, Anal. Bioanal. Chem., 2019, 411, 5877–5883 CrossRef CAS.
- V. Moisoiu, A. Stefancu, D. Gulei, R. Boitor, L. Magdo, L. Raduly, S. Pasca, P. Kubelac, N. Mehterov and V. Chiş, Int. J. Nanomed., 2019, 14, 6165 CrossRef CAS.
- W. Kim, S. H. Lee, J. H. Kim, Y. J. Ahn, Y.-H. Kim, J. S. Yu and S. Choi, ACS Nano, 2018, 12, 7100–7108 CrossRef CAS.
- B. Greene, D. Alhatab, C. Pye and C. Brosseau, J. Phys. Chem. C, 2017, 121, 8084–8090 CrossRef.
- J. P. Giraldo, H. Wu, G. M. Newkirk and S. Kruss, Nat. Nanotechnol., 2019, 14, 541–553 CrossRef CAS.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |
