Open Access Article
Open Access ArticleMetabolic engineering for plant natural products biosynthesis: new procedures, concrete achievements and remaining limits
Vincent
Courdavault
 *a,
Sarah E.
O'Connor
*a,
Sarah E.
O'Connor
 b,
Michael K.
Jensen
b,
Michael K.
Jensen
 c and
Nicolas
Papon
c and
Nicolas
Papon
 d
d
aUniversity of Tours, EA2106, Faculty of Pharmacy, Tours, France. E-mail: vincent.courdavault@univ-tours.fr
bDepartment of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, 07745 Jena, Germany
cNovo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark
dGroupe d'Etude des Interactions Hôte-Pathogène, GEIHP, EA3142, UNIV Angers, SFR 4208 ICAT, Angers, France
First published on 10th May 2021
Abstract
Microorganisms and plants represent major sources of natural compounds with a plethora of bioactive properties. Among these, plant natural products (PNPs) remain indispensable to human health. With few exceptions, PNP-based pharmaceuticals come from plant specialized metabolisms and display a structure far too complex for a profitable production by total chemical synthesis. Accordingly, their industrial processes of supply are still mostly based on the extraction of final products or precursors directly from plant materials. This implies that particular contexts (e.g. pandemics, climate changes) and natural resource overexploitation are main drivers for the high production cost and recurrent supply shortages. Recently, biotechnological manufacturing alternatives gave rise to a multitude of benchmark studies implementing the production of important PNPs in various heterologous hosts. Here, we spotlight unprecedented advancements in the field of metabolic engineering dedicated to the heterologous production of a prominent series of PNPs that were achieved during the year 2020. We also discuss how the knowledge accumulated in recent years could pave the way for a broader manufacturing palette of natural products from a wide range of natural resources.
1 Introduction
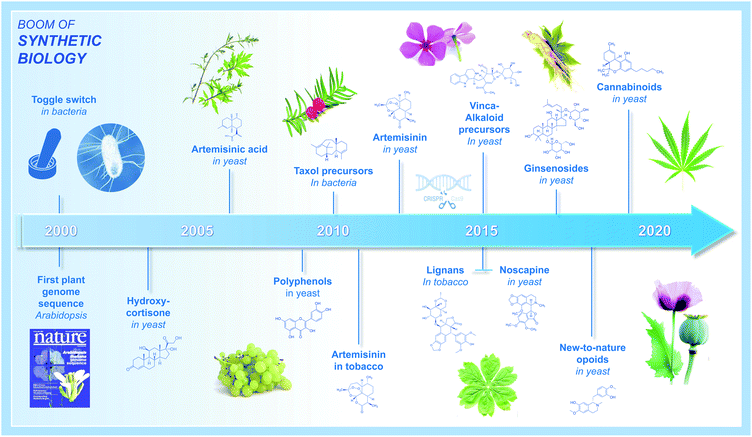
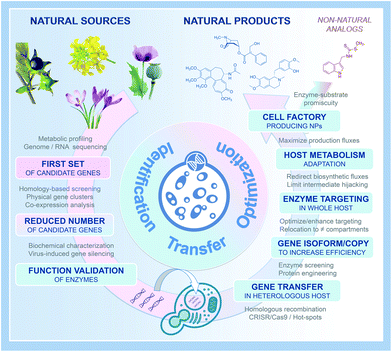
For millennia, bacteria, fungi, and plants have been considered as major sources of natural compounds with a multitude of bioactive properties and associated applications in the fields of nutrition and human health. Among the most valuable plant natural products (PNPs), humans have identified those used to treat serious pathologies such as cancers, infectious illnesses, neuromuscular disorders, or even cardiovascular diseases, and for which selected PNPs have been further processed and manufactured into active pharmaceutical ingredients at industrial scale. With few exceptions, PNP-based pharmaceuticals come from plant specialized metabolism and encompass many different families of complex molecules. Some of the most prominent PNPs, and their associated applications, include wormwood sesquiterpene lactones (antimalarial drugs), tropane alkaloids from Solanaceae (mostly anticholinergics), yew taxane-type terpenoids (anticancer drugs), poppy isoquinoline alkaloids (antitussives, antimicrobials, analgesics, vasodilators), mayapple lignans (anticancer and immunosuppressive drugs), and monoterpene indole alkaloids of Apocynaceae (antihypertensive and anticancer drugs).1While their potency and indispensability to human health is undisputed, a major drawback concerning most PNPs is still the requirement of plant materials used for extraction of final products or precursors (in that case completed by semisynthesis).2 Indeed, most of these molecules display a structure far too complex for a profitable production by total chemical synthesis. Beyond climatic variations (natural disasters) or particular health-care contexts (pandemics) that can drastically impact PNP sourcing from natural resources, the low accumulation of these compounds (or targeted precursors) in planta combined with natural resource overexploitation, is commonly considered to be main drivers for the high production costs of most of these PNPs, while at the same time be the root-cause of recurrent supply shortages.3 With the aim of remediating these limitations, biotechnological PNP manufacturing alternatives began to appear along the emergence of the synthetic biology era in the early 2000's. Initially, this consisted in transferring genes encoding functionally characterized biosynthetic enzymes from plants into genetically tractable hosts (e.g. the bacterium Escherichia coli, the yeast Saccharomyces cerevisiae, or Nicotiana benthamiana leaves) for the heterologous production of corresponding PNPs.4–8 In the past fifteen years, this general strategy has been further refined, benefitting from the tremendous technological and scientific advancements within genome sequencing, genome engineering, plant pathway discovery, and metabolic engineering. This has given rise to a multitude of seminal studies on the heterologous production of important PNPs. A historical summary of the major achievements within the largest PNP families (polyphenols, terpenes, taxanes, lignans, indole alkaloids, opioids, cannabinoids, etc.) is depicted in Fig. 1, starting from the beginning of the 21st century.9–20 While such examples were initially scarce, PNP productions continuously diversified, and notably over the last five years, along with improvements of the molecular toolkit including CRISPR/Cas9 genome editing.21,22 Even more exciting, 2020 stood-out as a milestone providing unprecedented breakthroughs in the field of synthetic biology and metabolic engineering dedicated to the production of plant-derived drugs in heterologous hosts. By highlighting benchmark articles published in this domain during 2020, we summarize here recent key facts concerning innovative tools for (i) accelerating discovery of PNP biosynthetic pathways, (ii) facilitating the transfer of biosynthetic pathways in heterologous hosts, (iii) optimizing PNP biosynthesis up to industrial scale, (iv) and expanding the chemical space of produced PNPs into new-to-nature products. We included concrete examples of authors' discoveries to facilitate understanding and to further illustrate the possible outputs of the described strategies. Finally, we discuss how the knowledge accumulated in recent years could pave the way for rational engineering of cellular metabolism for a broader palette of natural products from a wide range of natural resources.
2 A booming discovery of PNP biosynthetic pathways
As an obvious starting point, transferring biosynthetic pathways in heterologous hosts to create PNP cell factories requires comprehensive knowledge of the enzymatic reactions involved in the biosynthesis of the compound of interest in the native host organisms (i.e. plants). While examples of completely elucidated PNP biosynthetic pathways remained scarce during the 90's and 2000's, the development of new technical approaches recently resulted in a booming discovery of biosynthetic routes of many pharmaceutical compounds from plants. This includes for instance next generation sequencing leading to genomic and massive transcriptomics resources, candidate gene prediction strategies and functional genomic tools.23 Combining or integrating at least a part of these approaches has yielded a benchmark procedure widely used to decipher PNP metabolisms such as syntheses of lignans (podophyllotoxin) and Vinca alkaloids (vindoline, catharanthine).15,24 Besides these latterly published PNP routes, biosynthetic pathways of several other valuable PNPs remain under investigation including notably camptothecin, quinine and taxol. Within this group of remaining PNPs, the recent discovery of the nearly complete pathway of colchicine synthesis by Elizabeth Sattely's research group is a perfect example of a successful combination of multi-omics approaches leading to pathway elucidation without prior knowledge of biosynthetic genes.25Colchicine is a prominent alkaloid mainly found in plants from the Liliaceae family (Colchicum autumnale, Gloriosa superba) and is biogenetically very close to the isoquinoline alkaloids. This age-old drug has been used as a traditional medicine for more than 2000 years and is still of importance in the current human pharmacopeia, as illustrated by its recent approbation as a treatment for acute gout by the U.S Food and Drug Administration. While the current supply of colchicine still relies on the low yield purification from G. superba seeds, pioneering works have initiated pathway discovery by characterizing metabolic intermediates from isotope-labelled feeding studies. The resulting putative biosynthetic scheme thus involves the joining of 4-hydroxydihydrocinnamaldehyde and dopamine through a Pictet–Spengler reaction to form a 1-phenethylisoquinoline scaffold that is further decorated, notably through hydroxylation and methylation. From this hypothesis, the authors initiated the prediction of candidate genes for methylation through a “guilt by association” study. Basically, metabolites accumulated in a tissue-specific manner should be produced by a series of enzymes sharing a similar spatio-temporal expression pattern, which could be identified at the transcript level (Fig. 2A). Since no previously characterized genes could be used to guide candidate gene identification, Nett et al. used existing and newly created RNA-sequencing databases from C. autumnale and G. superba to search for genes displaying a methyltransferase domain and highly expressed in rhizomes that accumulate the highest levels of colchicine. Concretely, the 11 candidate genes thus predicted were functionally assayed through a coupled Agrobacterium-mediated overexpression in leaves of N. benthamiana and a biochemical methyltransferase test with leaf protein crude extracts with 1-phenethylisoquinoline as substrate. This first step led to the identification of both N- and O-methyltransferases from G. superba, named GsNMT and GsOMT1, which was further used to guide the identification of downstream genes of the pathway through measurement of a gene co-expression correlation. Besides identifying two other O-methyltransferases (GsOMT2 and GsOMT3), this allowed the characterization of two cytochrome P450s (GsCYP75A109 and GsCYP75A110) catalyzing complementary hydroxylation and oxidative phenol coupling of biosynthetic intermediates, respectively. Finally, through additional correlation studies and notably hierarchical clustering, authors identified GsCYP71FB1 which ensures the ring expansion of the intermediate O-methylandrocymbine. This ultimately resulted in the formation of the tropolone ring present in colchicine as demonstrated by the transient co-expression of the eight newly identified genes in N. benthamiana leaves yielding N-formyldemecolcine, a direct precursor of colchicine. Undoubtedly, the combination of metabolomics and transcriptomics data exploitation together with functional genomic tools has emerged as a relevant and efficient tool for elucidating PNP pathways over the last years.26Fig. 2A and 3 thus summarizes the first steps of this procedure notably including approaches for candidate gene identification, prioritization, and functional validation.27
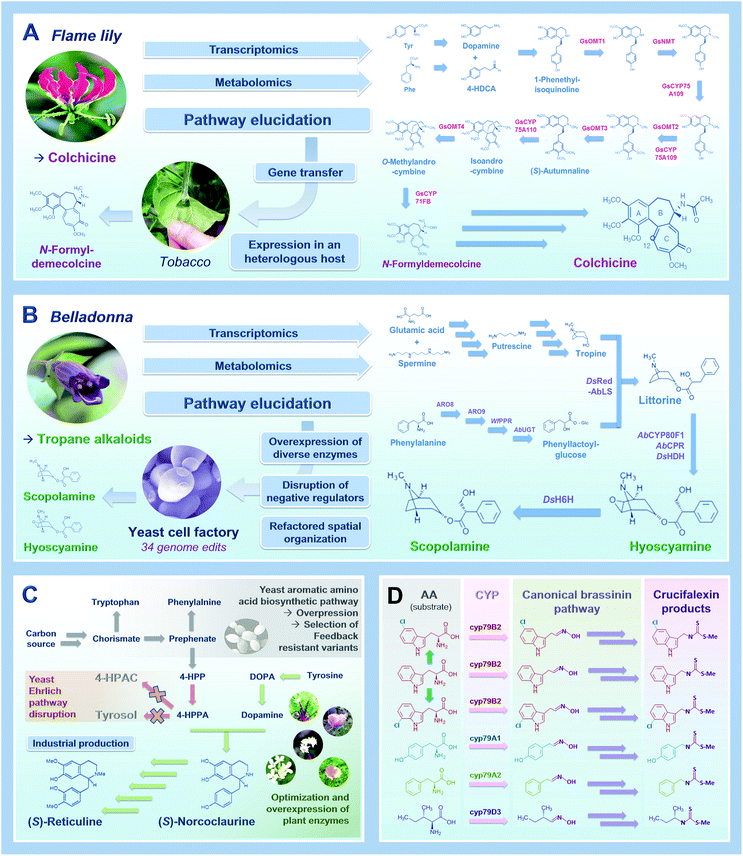 | ||
| Fig. 2 A tale of 4 main stories leading to widening and improvement of metabolic engineering for plant natural products in 2020. (A) The synthesis of a colchicine precursor in tobacco,25 (B) the production of the tropane alkaloid scopolamine and hyoscyamine in yeast,32 (C) the production of the benzylisoquinoline alkaloid intermediate (S)-reticuline at an industrial scale,7 (D) the synthesis of new-to nature PNPs in tobacco.38 | ||
The existence of evolutionary selected biosynthetic modules (P450 and OMT concatenation, for instance) and their conservation in distinct PNP routes are further allies to guide identification of missing biosynthetic steps. In addition, the ongoing accumulation of genomic resources as well as the development of powerful machine learning algorithms has already begun to strengthen our set of pathway discovery tools.28 It is also highly plausible that the increasing efficacy of enzyme prediction will ease identification of novel catalytic reaction as reported by Hafner et al. (2020) in the ATLAS database.29
3 Refactoring PNP biosynthetic pathways in yeast
Once a complete pathway is identified, the transfer of the corresponding biosynthetic genes in heterologous hosts can be initiated to create engineered cell factories heterologously producing PNPs of interest. The presence of membrane-bound proteins or the specific requirement of post-translational protein modifications can orientate the selection of the host organisms towards the use of microbial chassis (Escherichia coli, S. cerevisiae) or plants (N. benthamiana). In this context, S. cerevisiae (bakers' yeast) has progressively gained momentum as an unmissable chassis for fermentation-based production of PNPs. While both E. coli and S. cerevisiae have extensive synthetic biology toolboxes, by which exogenous genes can be readily expressed from auto-replicative plasmids or genomic integrative constructs for fast PNP pathway prototyping,30 yeast is regarded particularly relevant as a chassis for refactoring PNP biosynthetic pathways due to its endomembrane capacity and compartmentalization of organelles (e.g. nucleus, vacuoles). This supports expression of membrane anchored enzymes (e.g. P450s) and intracellular routing of PNP pathway modules, respectively.31 Still, as with any heterologous host, iterative cycles between strain engineering, the pathway identification in plants, and its reconfiguration in yeast is often needed for successful PNP biosynthetic pathway refactoring in yeast. As described in Fig. 2B and 3, improved gene transfer technologies combined with selection of efficient enzyme isoforms and optimized protein subcellular localization are first required. The recent article of Christina Smolke's group, describing the biosynthesis of medicinal tropane alkaloids (TA) in yeast, is one prominent example of this process.32TAs are predominantly synthesized in the Solanaceae plant family (belladonna and mandrakes for instance) with a few exceptions in other clades such as bindweeds and coca plants. Besides the well-known cocaine and atropine, other TAs possess valuable pharmaceutical properties including hyoscyamine and scopolamine used to treat neuromuscular disorders such as Parkinson's disease. However, for many PNPs, the low abundances in native hosts and the complexity of their chemical structures, can raise environmental concerns and prevent total chemical synthesis at competitive costs, respectively, ultimately impacting recurrent PNP supply chain shortages. This has prompted researchers to develop yeast cell factories for an alternative TA supply. By engineering an already existing yeast chassis producing the TA acyl acceptor precursor tropine, the authors implemented three additional biosynthetic modules to synthesize the acyl donor phenylacetic acid glucoside, the acyltransferase reaction, and the final TA scaffold modifications, to generate hyoscyamine and scopolamine.32 Firstly, starting from phenylalanine, the synthesis of phenylacetic acid glucoside was achieved through the concomitant expression of enzymes of both plant and yeast origins. Authors notably over-expressed two endogenous yeast aromatic aminotransferases and compensated the lack of true phenylpyruvate reductase (PPR) activity by expressing a bona fide PPR. While most of the tested PPR from E. coli, Lactobacillus, Atropa belladonna only synthesized trace amounts of the expected product, PPR from the large yeast Wicherhamia fluorescens resulted in an 80-fold increase of its synthesis. The production of phenylacetic acid glucoside also involved the expression of a specific codon-optimized UDP glucosyltransferase from A. belladonna. Interestingly, the accumulation of this intermediate was further improved by the invalidation of yeast endogenous glucosidases hydrolyzing heterologous glucosides. Secondly, reconstituting an active acyltransferase in yeast appeared as the most difficult concern to address for producing the pivotal intermediate littorine. In A. belladonna, the acyltransferase littorine synthase (AbLS) efficiently catalyze this reaction after targeting to the tonoplast. Albeit yeast and plant secretory pathways share roughly similar processing steps, authors established that the lack of AbLS activity in yeast mostly resulted from a targeting default that was fixed by engineering of the N-terminal end of the protein. Increase in the production of the downstream products was also achieved through the concomitant overexpression of intracellular alkaloid transporters ensuring the vacuolar internalization of biosynthetic intermediates. Lastly, the synthesis of hyoscyamine and scopolamine necessitated the identification of the previously unknown hyoscyamine dehydrogenase through (1) an RNA-sequencing procedure using already identified genes as baits for an expression correlation study in A. belladonna, and (2) the search for the most active orthologue in related species.
In sum, through no less than 34 genome edits, Srinivasan and Smolke generated a yeast strain producing two valuable TAs at laboratory scale. Besides further highlighting the importance of transcriptomics for the discovery of enzymes and more active orthologues, this work also demonstrated how control of the spatial organization of refactored biosynthetic pathways in yeast is crucial for efficient PNP production. In this context, a restricted compartmentalization to peroxisomes of norcoclaurine synthase (NCS) that catalyze the first committed steps of benzylisoquinoline alkaloid (BIA) synthesis was recently shown to alleviate NCS toxicity in yeasts. Together with the increase of the peroxisome density triggered by engineered transcription factors, such an approach substantially improved BIA production.33 Similarly, in plants, rerouting the diterpene biosynthesis from the chloroplasts (synthesis through the methylerythritol pathway) to the cytosol (mevalonate pathway) dramatically increased synthesis titers and allowed production of the allopathic momilactone B in N. benthamiana.34
4 Optimizing PNP synthesis in yeast to industrial scale
Up to now, except for a few examples such as artemisinic acid (25 g L−1), the production of PNPs through metabolic engineering in yeast are still mainly represented by proof-of-principle studies conducted at laboratory scale and with titers in the μg to low mg range per liter.16,17,19,20,35 Media optimization, culture conditions (fed-batch, high density cultures) and implementation of bioreactors can increase PNP production. Still, further molecular modifications of yeast host may be required to avoid intermediates and to reroute critical native precursors into PNP biosynthetic pathways without overly impacting cellular fitness as depicted in Fig. 2C.31,36 While we previously discussed the positive effect of expression of more active orthologues and by extension the copy number of genes encoding limiting enzymes, adaptation of the yeast endogenous metabolism is indispensable to reach high production levels. In this regard, Vincent Martin's group developed a yeast platform synthesizing up to 4.6 g L−1 of the central BIA intermediate (S)-reticuline.7Besides identification of more active enzymes and mutagenic optimization of activities, endogenous yeast genes were overexpressed to improve BIA precursor supply. Authors also addressed a recurrent concern in yeast factories, corresponding to the hijacking of some biosynthetic intermediates to other metabolic branches. BIA synthesis relies on the condensation of dopamine with 4-hydroxyphenylacetaldehyde (4-HPAA). However, 4-HPAA can be converted into non-productive acid or alcohol shunt products through the fusel metabolism from the Ehrlich pathway. Since enzymes mediating these redox reactions have evaded from identification so far, a gene deletion screen of more than 20 candidates was performed and ultimately resulted in the identification of the short-chain dehydrogenase Ari1 as one of the main actors of 4-HPAA rerouting. Owing to noticeable redundancy in yeast oxidoreductases, four additional genes encoding NADPH-dependent reductase and dehydrogenases were deleted to inhibit 4-HPAA reduction without affecting yeast fitness. Consequently, the abolishment of this reduction causes a marked increase in 4-HPAA oxidation that further necessitates the invalidation of the aldehyde dehydrogenase Hfd1 naturally oxidizing a structurally related substrate.
Combining all these modifications finally led to a 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold increase in BIA precursor synthesis reaching a threshold titer of 5 g L−1 previously set for commercial-scale production of opioids. By extension, controlling the catabolism of Ehrlich pathway amino-acids could have broad applications, specifically for the synthesis PNPs derived from L-phenylalanine, L-leucine and L-tryptophan building blocks. It is also noteworthy that this work could find implications in food and beverage industries given the key role of fusel products in flavour profiles. Finally, such a high synthesis of BIA related compounds opened the door to a diversification of the produced alkaloids though precursor-directed biosynthesis.
000-fold increase in BIA precursor synthesis reaching a threshold titer of 5 g L−1 previously set for commercial-scale production of opioids. By extension, controlling the catabolism of Ehrlich pathway amino-acids could have broad applications, specifically for the synthesis PNPs derived from L-phenylalanine, L-leucine and L-tryptophan building blocks. It is also noteworthy that this work could find implications in food and beverage industries given the key role of fusel products in flavour profiles. Finally, such a high synthesis of BIA related compounds opened the door to a diversification of the produced alkaloids though precursor-directed biosynthesis.
5 New-to-nature PNPs: a concrete and promising field
If optimized cell factories can provide alternative PNP supplies, they can also offer the possibility to expand the nature of the produced compounds by diversifying/increasing decorations thus resulting in the possible synthesis of non-canonical or new-to-nature NPs (Fig. 2D). For instance, halogenation of NPs, and notably alkaloids, can broaden their properties and be used for further chemical derivatization. While scarce in yeast, such modifications have been already reported in plants through a direct in planta engineering of NP synthesis.37Taking advantage of N. benthamiana transient transformation efficiency, Calgaro-Kozina and co-workers transferred the glucosinolate biosynthetic pathway and expanded upon the diversity of the produced biopesticides.38 Glucosinolate are amino acid-derived compounds with a sulfur bound to glucose and the nitrogen bound to sulfate notably produced in crucifers. They belong to the phytoanticipin class of defense molecules, which correspond to non-toxic phytoalexines activated following herbivory and/or pathogen attack. Glucosinolates are more specifically deglucosylated by a specific β-glucosidase (myrosinase) to yield a reactive isothiocyanate preventing pathogen progression. In this work, authors focused on brassinin, an indole–sulfur phytoalexin found in many cruciferous crop species (e.g. Brassica rapa) and expressed the whole corresponding pathway in N. benthamiana. While brassinin synthesis is normally initiated by the tryptophan-specific CYP79B2, they swapped this enzyme with other phenylalanine-, tyrosine-, isoleucine- or valine-oxidizing cytochrome P450s from distinct plant species. By exploiting the substrate promiscuity of the downstream enzymes, authors extended the set of primary metabolites that can be used by the core pathway to produce brassinin-like and new-to-nature compounds, derived from other amino acids, and named crucifalexins. In addition, they pushed the capacity of the pathway beyond the canonical amino acids to produce halogenated brassinin analogues, known to be more bioactive. A transient expression of the brassinin pathway was performed along with a combination of bacterial halogenases/reductases PyrH/RebF producing 5-chlorotryptophan or RebH/RebF producing 7-chloro and 7-bromotryptophan as already observed for alkaloid diversification in Catharanthus roseus.39 For each specific combination, the formation of 5-chlorobrassinin, 7-chlorobrassinin and even 7-bromobrassin after watering plants with potassium bromide, was monitored thus opening concrete perspectives for the production of halogenated variants in planta.
This enlightening study provides pioneering demonstration of the potential of synthetic biology and metabolic engineering approaches for the biosynthesis of new biopesticides. Beyond this benchmark demonstration, this works shades light on the primordial importance of enzyme substrate promiscuity to produce new-to nature PNPs, as has been brought to light in recent studies carried out in other models (Fig. 2D and 3).7,19,20
6 Concluding remarks
Fifteen years after isolating the first milligrams of artemisinic acid from engineered yeast, who would have imagined that such a spectacular advancement in the development of heterologous platforms producing PNPs could have been reached?10 In recent years, we assisted indeed not only to the expansion of our ability to develop metabolic engineering strategies for a growing range of different PNP classes but also to an acceleration of the implementation processes up to industrial production titers. As illustrated by excellent articles published this year and highlighted is this piece, this tremendous progress first relies on the emergence of omics-based strategies to facilitate the identification of genes from complex PNP biosynthetic metabolic pathways as summarized in Fig. 3. Together with the continuous development of cutting-edge synthetic biology toolboxes and comprehensive genome-scale mechanistic models of metabolism enabling advanced engineering for optimal precursor supplies as well as dynamic regulation of metabolic pathway fluxes, we can now reasonably expect that this booming field will enable true drop-in solutions for marketed PNPs under supply shortages (Fig. 3).40,41Having said this, for microbial production of PNPs to become a relevant supply chain of human therapeutics, efforts (i) to maximise activity of hard-to-express plant enzymes, (ii) relieve effects of toxic PNP intermediates, (iii) limit shunt product formation, (iv) stabilize titers/rates/yields of microbial PNP cell factories over prolonged cultivations, and (v) efficient down-stream processing, are all aspects needing further attention in the near future.
Finally, beyond natural PNPs, we further expect the systematic elucidation of increasingly complex and diversified new-to-nature chemical structures with improved pharmacokinetic properties. By describing concrete results and approaches from each of these articles, we gave an overview of the newly developed benchmark strategies to follow to reach a PNP industrial scale production. Finally, we also anticipate that all these efforts and advancements will benefit to the supply of other difficult-to-access biological compounds, particularly marine natural products (Fig. 3).42,43
7 Conflicts of interest
There are no conflicts to declare.8 Acknowledgements
This research was funded by the EU Horizon 2020 research and innovation program (MIAMi project grant agreement No. 814645), the ARD2020 Biopharmaceutical Program of the Région Centre Val de Loire (BioPROPHARM, CatharSIS, ETOPOCentre projects), La Ligue contre le Cancer and Le Studium (Consortium Fellowship), the Agence Nationale de la Recherche - project MIACYC (ANR-20-CE43-0010).9 References
- D. J. Newman and G. M. Cragg, Natural Products as Sources of New Drugs from 1981 to 2014, J. Nat. Prod., 2016, 79(3), 629–661, DOI:10.1021/acs.jnatprod.5b01055.
- F. Bucar, A. Wube and M. Schmid, Natural product isolation – how to get from biological material to pure compounds, Nat. Prod. Rep., 2013, 30(4), 525–545, 10.1039/c3np20106f.
- N. Gordon, S. M. Stemmer, D. Greenberg and D. A. Goldstein, Trajectories of Injectable Cancer Drug Costs After Launch in the United States, J. Clin. Oncol., 2018, 36(4), 319–325, DOI:10.1200/JCO.2016.72.2124.
- S. Li, Y. Li and C. D. Smolke, Strategies for microbial synthesis of high-value phytochemicals, Nat. Chem., 2018, 10(4), 395–404, DOI:10.1038/s41557-018-0013-z.
- B. J. Kotopka, Y. Li and C. D. Smolke, Synthetic biology strategies toward heterologous phytochemical production, Nat. Prod. Rep., 2018, 35(9), 902–920, 10.1039/c8np00028j.
- J. Nielsen, Cell factory engineering for improved production of natural products, Nat. Prod. Rep., 2019, 36(9), 1233–1236, 10.1039/c9np00005d.
- M. E. Pyne, L. Narcross and V. J. J. Martin, Engineering Plant Secondary Metabolism in Microbial Systems, Plant Physiol., 2019, 179(3), 844–861, DOI:10.1104/pp.18.01291.
- I. Carqueijeiro, C. Langley, D. Grzech, K. Koudounas, N. Papon, S. E. O'Connor and V. Courdavault, Beyond the semi-synthetic artemisinin: metabolic engineering of plant-derived anti-cancer drugs, Curr. Opin. Biotechnol., 2020, 65, 17–24, DOI:10.1016/j.copbio.2019.11.017.
- F. Ménard-Szczebara, C. Chandelier, C. Villeret, A. Masurel, S. Bourot, C. Duport, S. Blanchard, A. Groisillier, E. Testet, P. Costaglioli, G. Cauet, E. Degryse, D. Balbuena, J. Winter, T. Achstetter, R. Spagnoli, D. Pompon and B. Dumas, Total biosynthesis of hydrocortisone from a simple carbon source in yeast, Nat. Biotechnol., 2003, 21(2), 143–149, DOI:10.1038/nbt775.
- D. K. Ro, E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Chang, S. T. Withers, Y. Shiba, R. Sarpong and J. D. Keasling, Production of the antimalarial drug precursor artemisinic acid in engineered yeast, Nature, 2006, 440(7086), 940–943, DOI:10.1038/nature04640.
- E. Trantas, N. Panopoulos and F. Ververidis, Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae, Metab. Eng., 2009, 11(6), 355–366, DOI:10.1016/j.ymben.2009.07.004.
- P. K. Ajikumar, W. H. Xiao, K. E. Tyo, Y. Wang, F. Simeon, E. Leonard, O. Mucha, T. H. Phon, B. Pfeifer and G. Stephanopoulos, Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli, Science, 2010, 330(6000), 70–74, DOI:10.1126/science.1191652.
- M. Farhi, E. Marhevka, J. Ben-Ari, A. Algamas-Dimantov, Z. Liang, V. Zeevi, O. Edelbaum, B. Spitzer-Rimon, H. Abeliovich, B. Schwartz, T. Tzfira and A. Vainstein, Generation of the potent anti-malarial drug artemisinin in tobacco, Nat. Biotechnol., 2011, 29(12), 1072–1074, DOI:10.1038/nbt.2054.
- C. J. Paddon, P. J. Westfall, D. J. Pitera, K. Benjamin, K. Fisher, D. McPhee, M. D. Leavell, A. Tai, A. Main, D. Eng, D. R. Polichuk, K. H. Teoh, D. W. Reed, T. Treynor, J. Lenihan, M. Fleck, S. Bajad, G. Dang, D. Dengrove, D. Diola, G. Dorin, K. W. Ellens, S. Fickes, J. Galazzo, S. P. Gaucher, T. Geistlinger, R. Henry, M. Hepp, T. Horning, T. Iqbal, H. Jiang, L. Kizer, B. Lieu, D. Melis, N. Moss, R. Regentin, S. Secrest, H. Tsuruta, R. Vazquez, L. F. Westblade, L. Xu, M. Yu, Y. Zhang, L. Zhao, J. Lievense, P. S. Covello, J. D. Keasling, K. K. Reiling, N. S. Renninger and J. D. Newman, High-level semi-synthetic production of the potent antimalarial artemisinin, Nature, 2013, 496(7446), 528–532, DOI:10.1038/nature12051.
- W. Lau and E. S. Sattely, Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone, Science, 2015, 349, 1224–1228, DOI:10.1126/science.aac7202.
- S. Brown, M. Clastre, V. Courdavault and S. E. O'Connor, De novo production of the plant-derived alkaloid strictosidine in yeast, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(11), 3205–3210, DOI:10.1073/pnas.1423555112.
- S. Galanie, K. Thodey, I. J. Trenchard, M. Filsinger Interrante and C. D. Smolke, Complete biosynthesis of opioids in yeast, Science, 2015, 349(6252), 1095–1100, DOI:10.1126/science.aac9373.
- Y. Zhuang, G. Y. Yang, X. Chen, Q. Liu, X. Zhang, Z. Deng and Y. Feng, Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme, Metab. Eng., 2017, 42, 25–32, DOI:10.1016/j.ymben.2017.04.009.
- Y. Li, S. Li, K. Thodey, I. Trenchard, A. Cravens and C. D. Smolke, Complete biosynthesis of noscapine and halogenated alkaloids in yeast, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(17), E3922–E3931, DOI:10.1073/pnas.1721469115.
- X. Luo, M. A. Reiter, L. d'Espaux, J. Wong, C. M. Denby, A. Lechner, Y. Zhang, A. T. Grzybowski, S. Harth, W. Lin, H. Lee, C. Yu, J. Shin, K. Deng, V. T. Benites, G. Wang, E. E. K. Baidoo, Y. Chen, I. Dev, C. J. Petzold and J. D. Keasling, Complete biosynthesis of cannabinoids and their unnatural analogues in yeast, Nature, 2019, 567(7746), 123–126, DOI:10.1038/s41586-019-0978-9.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, 337(6096), 816–821, DOI:10.1126/science.1225829.
- E. Deltcheva, K. Chylinski, C. M. Sharma, K. Gonzales, Y. Chao, Z. A. Pirzada, M. R. Eckert, J. Vogel and E. Charpentier, CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III, Nature, 2011, 471(7340), 602–607, DOI:10.1038/nature09886.
- M. H. Medema and A. Osbourn, Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways, Nat. Prod. Rep., 2016, 33(8), 951–962, 10.1039/c6np00035e.
- L. Caputi, J. Franke, S. C. Farrow, K. Chung, R. M. E. Payne, T. D. Nguyen, T. T. Dang, I. Soares Teto Carqueijeiro, K. Koudounas, T. Dugé de Bernonville, B. Ameyaw, D. M. Jones, I. J. C. Vieira, V. Courdavault and S. E. O'Connor, Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle, Science, 2018, 360, 1235–1239, DOI:10.1126/science.aat4100.
- R. S. Nett, W. Lau and E. S. Sattely, Discovery and engineering of colchicine alkaloid biosynthesis, Nature, 2020, 584(7819), 148–153 Search PubMed.
- L. W. Sumner, Z. Lei, B. J. Nikolau and K. Saito, Modern plant metabolomics: advanced natural product gene discoveries, improved technologies, and future prospects, Nat. Prod. Rep., 2015, 32(2), 212–229, 10.1039/c4np00072b.
- T. Dugé de Bernonville, N. Papon, M. Clastre, S. E. O'Connor and V. Courdavault, Identifying Missing Biosynthesis Enzymes of Plant Natural Products, Trends Pharmacol. Sci., 2020, 41(3), 142–146, DOI:10.1016/j.tips.2019.12.006.
- One Thousand Plant Transcriptomes Initiative, One thousand plant transcriptomes and the phylogenomics of green plants, Nature, 2019, 574(7780), 679–685, DOI:10.1038/s41586-019-1693-2.
- J. Hafner, H. MohammadiPeyhani, A. Sveshnikova, A. Scheidegger and V. Hatzimanikatis, Updated ATLAS of Biochemistry with New Metabolites and Improved Enzyme Prediction Power, ACS Synth. Biol., 2020, 9(6), 1479–1482 Search PubMed.
- G. Guirimand, N. Kulagina, N. Papon, T. Hasunuma and V. Courdavault, Innovative Tools and Strategies for Optimizing Yeast Cell Factories, Trends Biotechnol., 2020, S0167-7799(20), 30232–30238, DOI:10.1016/j.tibtech.2020.08.010.
- J. L. Avalos, G. R. Fink and G. Stephanopoulos, Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols, Nat. Biotechnol., 2013, 31(4), 335–341, DOI:10.1038/nbt.2509.
- P. Srinivasan and C. D. Smolke, Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids, Nat. Commun., 2019, 10(1), 3634, DOI:10.1038/s41467-019-11588-w.
- P. S. Grewal, J. A. Samson, J. J. Baker, B. Choi and J. E. Dueber, Peroxisome compartmentalization of a toxic enzyme improves alkaloid production, Nat. Chem. Biol., 2021, 17(1), 96–103, DOI:10.1038/s41589-020-00668-4.
- R. De La Peña and E. S. Sattely, Rerouting plant terpene biosynthesis enables momilactone pathway elucidation, Nat. Chem. Biol., 2021, 17(2), 205–212, DOI:10.1038/s41589-020-00669-3.
- Y. Li and C. D. Smolke, Engineering biosynthesis of the anticancer alkaloid noscapine in yeast, Nat. Commun., 2016, 7, 12137, DOI:10.1038/ncomms12137.
- A. Cravens, J. Payne and C. D. Smolke, Synthetic biology strategies for microbial biosynthesis of plant natural products, Nat. Commun., 2019, 10(1), 2142, DOI:10.1038/s41467-019-09848-w.
- S. A. Bradley, J. Zhang and M. K. Jensen, Deploying Microbial Synthesis for Halogenating and Diversifying Medicinal Alkaloid Scaffolds, Front. Bioeng. Biotechnol., 2020, 8, 594126, DOI:10.3389/fbioe.2020.594126.
- A. Calgaro-Kozina, K. M. Vuu, J. D. Keasling, D. Loqué, E. S. Sattely and P. M. Shih, Engineering Plant Synthetic Pathways for the Biosynthesis of Novel Antifungals, ACS Cent. Sci., 2020, 6(8), 1394–1400 Search PubMed.
- W. Runguphan, X. Qu and S. E. O'Connor, Integrating carbon-halogen bond formation into medicinal plant metabolism, Nature, 2010, 468(7322), 461–464, DOI:10.1038/nature09524.
- F. Monteiro, G. Hubmann, V. Takhaveev, S. R. Vedelaar, J. Norder, J. Hekelaar, J. Saldida, A. Litsios, H. J. Wijma, A. Schmidt and M. Heinemann, Measuring glycolytic flux in single yeast cells with an orthogonal synthetic biosensor, Mol. Syst. Biol., 2019, 15, e9071, DOI:10.15252/msb.20199071.
- J. Zhang, S. D. Petersen, T. Radivojevic, A. Ramirez, A. Pérez-Manríquez, E. Abeliuk, B. J. Sánchez, Z. Costello, Y. Chen, M. J. Fero, H. G. Martin, J. Nielsen, J. D. Keasling and M. K. Jensen, Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism, Nat. Commun., 2020, 11, 4880, DOI:10.1038/s41467-020-17910-1.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Marine natural products, Nat. Prod. Rep., 2020, 37(2), 175–223 Search PubMed.
- F. Zhang, M. Zhao, D. R. Braun, S. S. Ericksen, J. S. Piotrowski, J. Nelson, J. Peng, G. E. Ananiev, S. Chanana, K. Barns, J. Fossen, H. Sanchez, M. G. Chevrette, I. A. Guzei, C. Zhao, L. Guo, W. Tang, C. R. Currie, S. R. Rajski, A. Audhya, D. R. Andes and T. S. Bugni, A marine microbiome antifungal targets urgent-threat drug-resistant fungi, Science, 2020, 370(6519), 974–978, DOI:10.1126/science.abd6919.
| This journal is © The Royal Society of Chemistry 2021 |