Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unlocking the potential of marine biodiscovery
Julia D.
Sigwart
 *ab,
Robert
Blasiak
*ab,
Robert
Blasiak
 c,
Marcel
Jaspars
c,
Marcel
Jaspars
 *d,
Jean-Baptiste
Jouffray
*d,
Jean-Baptiste
Jouffray
 c and
Deniz
Tasdemir
c and
Deniz
Tasdemir
 ef
ef
aSenckenberg Research Institute, Frankfurt, Germany. E-mail: julia.sigwart@senckenberg.de
bQueen's University Belfast, Marine Laboratory, Portaferry, Northern Ireland
cStockholm Resilience Centre, Stockholm University, Sweden
dMarine Biodiscovery Centre, Department of Chemistry, University of Aberdeen, Old Aberdeen, Scotland, UK
eGEOMAR Centre for Marine Biotechnology, Research Unit Marine Natural Product Chemistry, GEOMAR Helmholtz Centre for Ocean Research Kiel, Germany
fFaculty of of Mathematics and Natural Sciences, Kiel University, Germany
First published on 8th December 2020
Abstract
The tremendous diversity of life in the ocean has proven to be a rich source of inspiration for drug discovery, with success rates for marine natural products up to 4 times higher than other naturally derived compounds. Yet the marine biodiscovery pipeline is characterized by chronic underfunding, bottlenecks and, ultimately, untapped potential. For instance, a lack of taxonomic capacity means that, on average, 20 years pass between the discovery of new organisms and the formal publication of scientific names, a prerequisite to proceed with detecting and isolating promising bioactive metabolites. The need for “edge” research that can spur novel lines of discovery and lengthy high-risk drug discovery processes, are poorly matched with research grant cycles. Here we propose five concrete pathways to broaden the biodiscovery pipeline and open the social and economic potential of the ocean genome for global benefit: (1) investing in fundamental research, even when the links to industry are not immediately apparent; (2) cultivating equitable collaborations between academia and industry that share both risks and benefits for these foundational research stages; (3) providing new opportunities for early-career researchers and under-represented groups to engage in high-risk research without risking their careers; (4) sharing data with global networks; and (5) protecting genetic diversity at its source through strong conservation efforts. The treasures of the ocean have provided fundamental breakthroughs in human health and still remain under-utilised for human benefit, yet that potential may be lost if we allow the biodiscovery pipeline to become blocked in a search for quick-fix solutions.
1 Introduction
Life in the ocean has existed for some 3.7 billion years, three times as long as life on land, and resulted in far greater evolutionary diversity.1 Of the 34 major animal phyla, only 12 are found on land, while 33 are found in the ocean. With up to 90% of marine species undescribed, marine diversity offers tremendous opportunities for research, and many novel solutions to life's problems. We believe that developing a strategy to address the untapped potential of marine biodiscovery must go well beyond scientific solutions, and instead represents a challenge at the interface of science, policy and practice.Around half of the clinically approved drugs, including antibiotics and anticancer agents, are based on natural products.2 These organic molecules are produced by living organisms, and while over 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 have been identified, less than 10% are from marine organisms.3,4 Nonetheless, marine natural products have up to 4 times higher rates of successful drug discovery than other naturally derived compounds. Around 1 in 5000–10
000 have been identified, less than 10% are from marine organisms.3,4 Nonetheless, marine natural products have up to 4 times higher rates of successful drug discovery than other naturally derived compounds. Around 1 in 5000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tested compounds lead to applications, but this rises to 1 in 2550 for marine natural products.4,5 With some 35
000 tested compounds lead to applications, but this rises to 1 in 2550 for marine natural products.4,5 With some 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 marine natural products already identified, and over 1500 more described each year,4,6 these figures suggest the vast potential health benefits of reducing barriers to ocean biodiscovery. Many chemical products represent multiplied developments from previously-established organismal systems. Our focus in this viewpoint is to illustrate how strategic investment in the early stages of the biodiscovery pipeline could provide exponentially increasing benefits for society.
000 marine natural products already identified, and over 1500 more described each year,4,6 these figures suggest the vast potential health benefits of reducing barriers to ocean biodiscovery. Many chemical products represent multiplied developments from previously-established organismal systems. Our focus in this viewpoint is to illustrate how strategic investment in the early stages of the biodiscovery pipeline could provide exponentially increasing benefits for society.
2 Recognizing the biomedical benefits of marine natural products
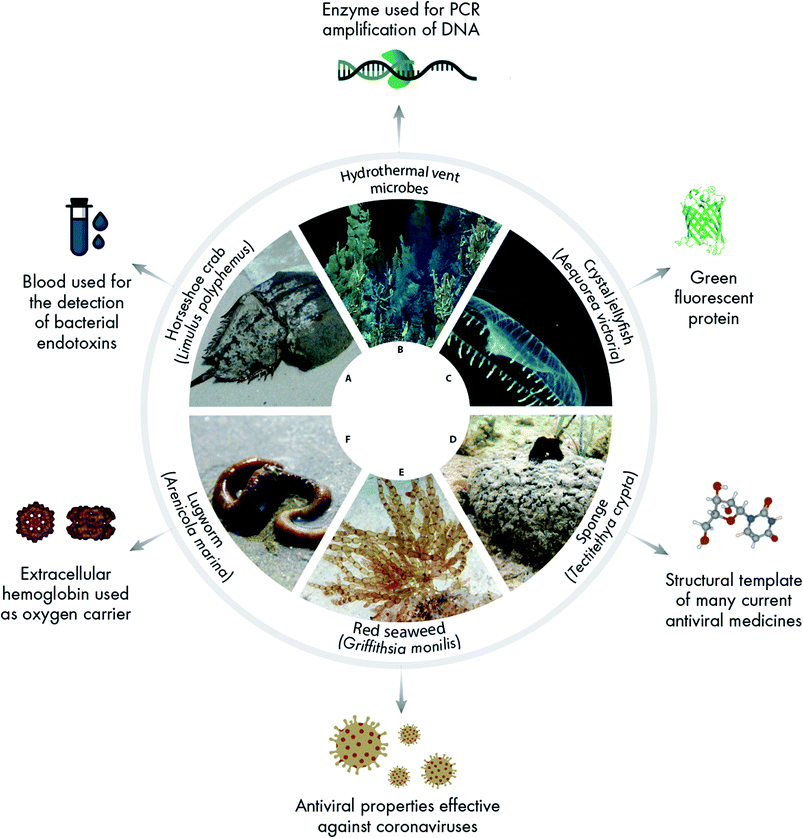
A rich catalogue of natural products from marine invertebrates and algae has contributed fundamentally to modern medicine (Fig. 1), and shows the potential for further diversification. For instance, the blood of the horseshoe crab Limulus polyphemus (one of only four living species of horseshoe crabs worldwide) is used to test all injectables and ensure safe intravenous delivery.7 Enzymes used in PCR amplification of DNA were first isolated from the microbe Thermus aquaticus in freshwater hot springs, but more recently commercially viable alternatives isolated from marine hydrothermal vents have demonstrated higher performance than traditional Taq polymerase.8 Green fluorescent protein (GFP) first isolated from the jellyfish Aequorea victoria is a crucial tool for protein tagging and a wide range of biomedical applications.1 Many other marine species are autofluorescent but unstudied so far. Diversifying both reagent properties and production pathways is fundamental to building an agile research landscape resilient to the logistical challenges in crisis situations, such as a pandemic.Marine sponges provided the first marine natural products,9 and the discovery of nucleoside spongouridine in the 1950s led to the very first antiviral drug Ara-A (Vidarabine®). The antiviral activity of spongouridine was first described in 1964 and later work showed clinical effectiveness against Herpes infections in newborns and immunosuppressed patients.10 Although it has been superseded by newer antiviral drugs, Vidarabine stands out as the ancestor of all antiviral drugs used today. Sponges have been the richest primary source of identified marine natural products,11 yet the basic biology of sponges remains poorly studied,12 including bioactivity in sponges and their microbiomes.13
Another relatively recent development, Halaven® is an approved anti-cancer drug whose structure is based on that of the compound halichondrin B. The structure of this compound, isolated from a Japanese deep-water sponge, Halichondria okadai, was first reported in 1986,14 although its potential anti-cancer activity had been known since the early 1980s. Difficulty in producing sufficient material for preclinical and clinical trials led to the isolation of the compound from a large scale collection of the New Zealand sponge Lissodendoryx sp. with support from the US National Cancer Institute, but this was not sustainable and aquaculture approaches were not economical. Subsequently, total chemical synthesis was achieved in 1992,15 followed by the generation of over 200 simplified analogues in 2005 by the Japanese pharmaceutical company Eisai.16 One of these, E7389/Eribulin, was approved as Halaven® in 2010 for the treatment of metastatic breast cancer. Sales of Halaven® are now in excess of US$500 M per year.
The early stage, upstream bottlenecks are a common theme in blocking the development of therapeutics from natural products, including the organismal identification and extraction. The red seaweed Griffithsia has outstanding antiviral properties, especially effective against coronaviruses, preventing viral entry.17,18 The first studies of novel biochemistry properties in Griffithsia started in the 1950s, and the potent antiviral protein griffithsin has also been shown to be effective against HIV, hepatitis, and encephalitis.19 Although some clinical trials had begun in 2019, and there is a clear need to accelerate effective treatments against novel coronaviruses, there is as yet no commercial development.20
The lugworm Arenicola marina is another marine organism that received particular attention in the wake of the COVID-19 pandemic, when one commercial company offered to test the use of the worm's hemoglobin as a “molecular respirator” to lessen reliance on intensive care equipment. Unlike human hemoglobin, an iron containing oxygen-transport metalloprotein in the red blood cells, that of Arenicola is extracellular, compatible with all blood types, and 250 times smaller than an erythrocyte. It can transport 40 times more oxygen than human hemoglobin and infiltrate deep into places with reduced circulation, making it a promising blood substitute for a range of applications.21,22
These are only few of many examples illustrating the potential of marine organisms for biomedical applications and how productive current research is. Yet why has the development of promising compounds like griffithsin faltered, and why does so much of marine biodiversity remain under-investigated? The research landscape of the past, in which academic institutions were expected to shoulder the risks of discovery-driven research, seems incompatible with a future of ever-increasing pressure to align research with commercial potential. Here we discuss what can be done to explore and unlock the full social and economic potential of the ocean's genetic resources.
3 The marine biodiscovery pipeline
Marine biodiscovery starts with novel biodiversity, based on the assumption that this will provide novel chemistry and hence novel biological activity. There is a long and complex process of biological research that must be established to enable chemical investigation and exploitation. Previously this was limited to larger organisms that would provide kilogram-scale samples, but new technology may require only grams of tissue from a target organism, opening vast new frontiers. Chemical talent can be assessed using classical natural product chemistry, now aided by chemoinformatic tools such as mass spectrometry-based molecular networking,23 or via the use of genome scanning to discover biosynthetic gene clusters that may direct the production of novel metabolites. Further screening gives detailed information on the activity and mechanism of action. These three “filters” (biodiversity, chemistry, and bioactivity) are often applied asynchronously, with the most successful approaches encompassing aspects of all three to prioritise compounds. Once potent and selective activity of a novel compound is identified, it must be isolated from the biomass, after which the initial bioactivity is verified and mechanism of action of the molecule is determined. At this point, a medicinal chemistry approach may optimise and facilitate larger scale production before eventually selecting compounds for preclinical and clinical trials.Marine natural products are ultimately drawn from unique adaptations of a species or evolutionary group, and so our intention is in part to refocus the attention of chemists onto this biological background. Replicability rests on the ability to reliably identify and re-collect or re-cultivate source organisms.24 This requires taxonomic expertise for storage, curation, and definitive identification of voucher samples. Advances in technology can produce increasing volumes of data, but research unavoidably depends on a relatively small number of experts to make the data useful. The current delay between species discovery and the formal publication of a scientific name is on average around 20 years,24,25 although this accelerates when the research is well supported. Moving from the identification of an organism with novel bioactive compounds to the development of a pure compound involves many challenges in detecting, isolating, characterizing, and scaling up the production of bioactive metabolites. Academic studies, particularly those carried out on naturally harvested macroorganisms, generally end up with at most a few milligrams of compound per kg of organism (yields of 1–10 ppm), which needs to be chemically characterized and tested for bioactivity.
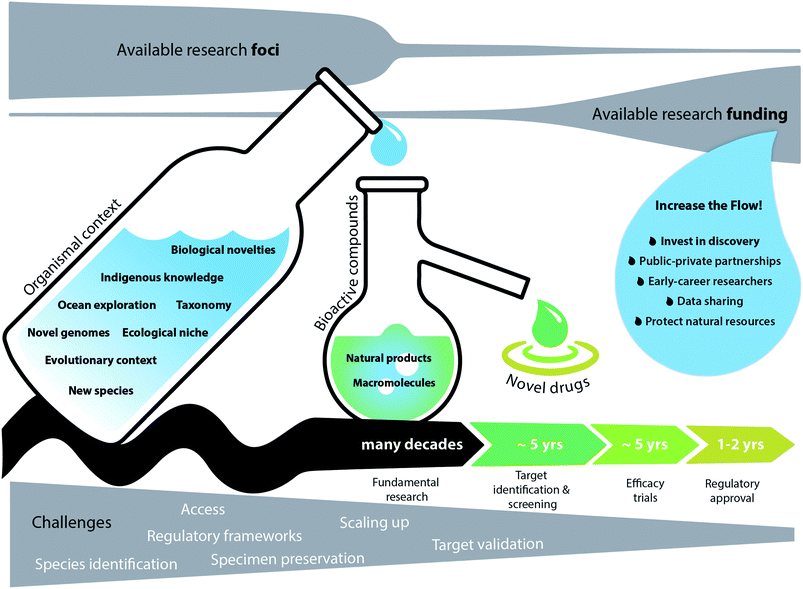
The process of investigating new marine natural solutions to human's problems is challenging, risky, and lengthy, and certainly ill-matched to the 3–4 year life cycle of a typical research grant (Fig. 2). Drug discovery research is “high risk” in these crucial early stages, and therefore a low priority for research funding from either public or industry sources. Novel problems require novel solutions, but these depend on ongoing, long-term support for discovery-driven work on taxonomy and exploration of the ocean.
4 Five ways to broaden the biodiscovery pipeline
4.1 Reward strategic risk taking
The increasing focus of research funding bodies on prioritising “impact” tilts research activity severely toward “low risk” projects, to the detriment of “edge research” with potential for genuinely novel breakthroughs.26 There are a few programmes that support “high risk” research, such as the European Research Council, but the highly competitive modern research landscape necessarily drives research toward established organismal systems that can be expected to produce high impact results, since developing new systems is time consuming and demands a discomforting level of uncertainty. High risk or fundamental science may be seen as a luxury, especially in developing economies, but strategic risk taking can produce substantial rewards. A few funding agencies support high risk projects, but these are currently exceptions to the norm. Important areas remain untapped because many researchers are effectively not permitted to work on them, in a culture that favours immediate and commercially relevant results. The level of research funding for fundamental organismal biology is chronically low, as are employment and retention opportunities, and is eroding the foundation underlying the development of marine natural products.The sheer scale of potential and how much biodiversity remains undiscovered is difficult to effectively communicate.27 There is a whole research pipeline to working with organisms, including finding and identifying specimens, and naming new species, all of which requires highly technical skills that are not accessible to medical researchers. Conversely, experts in biodiversity should not be required to predict which systems may yield optimal drug precursors. Recognising that novel biodiversity is the starting point to future biomedical applications, government agencies urgently need to support fundamental research on marine organisms for its own sake. We recommend that major funding agencies supporting both biology and chemistry research should earmark funding for discovery driven work on non-model marine organisms.
For example, in spite of the known threat from novel coronaviruses such as MERS, SARS, and COVID-19, antivirals effective against coronaviruses (including griffithsin) were not developed, because there was insufficient commercial incentive to do so.28 The primary, perpetual health threat associated with coronaviruses was the common cold, a set of nonlethal symptoms that is well known to be caused by multiple pathogens including coronaviruses and others. Thus, an antiviral would not necessarily produce an effective “cure” to the everyday malady. Nonetheless, the threat of a pandemic caused by novel coronaviruses has been well known to epidemiologists and was played out in spectacular form in 2020, but without a solid research basis that would enable the rapid deployment of effective, targeted antivirals.
4.2 Engage public-private partnerships
Interest and investment by the pharmaceutical industry in natural products has waned in recent years, with major companies stepping back from the lengthy and high-risk process of natural product discovery.2 Academic–industry relationships are complex, with companies often being hesitant to commit funding and likely to rather prioritise their in-house research activities. However, collaboration between academics and industry, particularly in the early stages, would improve success rates. The National Cooperative Drug Discovery Grants (NCDDG) programme of the U.S. National Cancer Institute enabled public-private partnerships to conduct innovative research and led to four marketed anticancer drugs from 1984–2004.29 European Commission funding has provided highly competitive but valuable support in discovery of marine natural products and support for public-private collaboration on other marine issues. Many funding programs have generally sponsored “low hanging fruit” sub-sectors of marine biotechnology such as cosmetics, nutraceuticals and other areas with shorter product development pipelines and less arduous legislation procedures. These industry projects are generally limited to researchers in a few large economies, but should depend on collaboration with global biodiversity experts.Commitment from industry partners to support the early stages of the discovery pipeline would dramatically increase the diversity of products available for preclinical and clinical testing. We recommend that robust industry-science dialogue should embrace the support and involvement of intermediaries with broad experience who can help bridge the differing perspectives and identify common ground. Of vital importance to the success of this is the task of “translators” who can connect academics with interested industry partners. Examples of such translators are the UK Knowledge Transfer Network (https://ktn-uk.org/) and institutions such as the Industrial Biotechnology Innovation Centre (http://www.ibioic.com/).
4.3 Nurture early-career researchers (ECRs)
All science needs the right proportion of specialists and generalists, where the specialists are experts in one area of research or methods and where the generalists understand several areas in lesser detail but can translate concepts from one discipline to another. Whereas large industries often require people who are “deep” specialists, small and medium sized enterprises need people who are multiskilled and can take on research tasks one day and perhaps marketing the next. Including industry partners in the training of PhD students can no doubt be highly beneficial, but this entirely skips the initial taxonomic and biological bottlenecks noted above, and increasing the number or training opportunities for PhD students is not a sufficient solution to the multi-factor challenges of modern research.30 Relatively short-term funding cycles require academic researchers to change research direction every few years and this is a barrier to development of specialists in novel systems. Given that detailed understanding can take decades to acquire, there is a role for long-term support to ensure that this research ripens to yield new knowledge. Although mobility has a potential positive outcome in allowing ECRs to apply their skills in a new research area, often knowledge and momentum are lost when an ECR moves from one field to the next in order to remain funded. Agency funding and support for high-risk, discovery-driven research is especially needed to support ECRs and researchers from under-represented groups who diversify the scientific workforce; many people, especially those with less career security, are often advised to develop skills in well-trodden systems with higher employment potential. That may be good advice for career stability, but a loss of creative talent needed to develop novel solutions.4.4 Share global data
Museum collections and biobanks both aid biodiscovery, by encouraging active participation of global partners, preventing oversampling of marine biodiversity, and ensuring access to specimens with legal certainty. This research pipeline is unimaginably long, and most of it is detached from direct conflict with intellectual property issues. Government support is needed to establish and maintain more collections and data resources in the countries of origin. The Australian Griffith University Nature Bank has specimens from a large proportion of Australian terrestrial and marine biodiversity and makes this available to researchers for the discovery of new products and processes.6 Researchers can access the original material, extracts, and fractions or pure compounds for testing under the terms of a legal agreement. The U.S. National Cancer Institute Natural Products Branch has instigated a global collection programme and makes its extract collections available via the Open Repository programme. Many global museums maintain frozen tissue and DNA extract collections in addition to physical specimen collections. The use of established biodiversity data networks and standards such as the Global Biodiversity Information Facility (GBIF) could not only give researchers access to materials that would otherwise be hard to obtain, but also help prevent unnecessary duplicate collection. Likewise, multilateral sharing of data supports the discovery of new species in less developed economies, where many undescribed species are found but where reference collections are lacking to confirm identification. Collections of microorganisms are also being linked via initiatives such as the EU's initiative for facilitating access to microorganism libraries31 while Global Natural Product Social Molecular Networking (GNPS), an open-access knowledge base shares community-curated fragmentation mass spectrometry (MS/MS) data of thousands of natural products to aid their identification in all stages of discovery.23 Finally, established high-capacity bioactivity screenings platforms such as EU OPENSCREEN are amending their protocols to incorporate natural products, as well as extracts and fractions for the future.4.5 Protect marine resources
Marine natural products can inspire new discoveries and underscore the necessity for conservation. For instance, knowledge of Australia's sponge diversity increased more than threefold after intensive biodiscovery started in the 1970s, providing an improved baseline for conservation efforts.12,32 Clarity about the scope of human benefits can provide compelling arguments for the designation of protected areas as storehouses of genetic diversity, and incentives for governments to safeguard the resilience of marine ecosystems in the context of climate change, pollution and potential future ocean uses such as commercial mining of the international seabed. Industry partners, who benefit from downstream development of natural solutions, should be more vocal advocates for marine conservation and contribute directly to the UN Sustainable Development Goals as part of relevant corporate responsibility.33In almost all cases, biodiscovery aims to discover knowledge that can be applied to a problem (e.g. the treatment of disease) and does not require the resource to be harvested to make the eventual commercial product. Although commercial development can be transferred to a laboratory setting, it is critical to involve diverse scientists and partners from the country of origin of novel materials. Genetic reference databases have grown as a result of expanding exploration paired with taxonomy, enabling non-invasive assessments (e.g. using environmental DNA – eDNA) of distributions of endangered and threatened species and helping to track the spread of invasive species and pathogens. However, these techniques depend entirely on the reliability of reference databases, which are widely known to be incomplete, and contain a large proportion of inaccurate identifications especially for under-studied species.34 Genetic diversity is still largely overlooked in marine conservation planning, and amplifying the synergies between marine biodiscovery and conservation can result in benefits to both efforts.35
5 Conclusions
We argue here that the value of the ocean for biomedical advances remains underdeveloped, and that the time has come to take purposeful and systematic steps to eliminate bottlenecks along the marine biodiscovery pipeline. Although the current funding climate and relative pessimism of the pharmaceutical industry towards natural products may seem obstacles to transformational change, the mobilization of unprecedented resources to address global medical challenges, such as Ebola, or the COVID-19 pandemic, has underscored which timelines can be abbreviated, and which cannot.36 Chartering research vessels, collection of specimens, description of new species, identification of novel bioactive compounds, and registration of these in relevant databases all involve timescales incompatible with a crisis response. Yet this response is reliant on decades of effort spanning exactly such activities. While the current pandemic spurred researchers to screen available compound libraries for properties that were not previously commercially interesting, these libraries from decades of work still only scratch the surface. Our ability to respond rapidly to future crises depends on improving long-term, continuous investment in research capacity.We can imagine the economic and humanitarian toll of the next global crisis, and we also imagine a crisis averted by building a diverse and robust tool kit of natural products that can treat diseases not yet imagined. Several actions can and should be taken immediately, with global participation. Government funding agencies, even in difficult times, must earmark funding for discovery-driven research and taxonomy, and regard it as investment in intellectual infrastructure. The research landscape must nurture ECRs, share data, and embrace open dialogue that helps develop industry-academic partnerships that are genuinely mutually beneficial, including sharing the risks and uncertainty of new frontiers. It is also important for industry partners to join forces and make meaningful commitments to marine conservation and stewardship.
As the United Nations Decade of Ocean Science for Sustainable Development 2021–2030 begins, it is prudent to underscore not only the need for science–industry partnerships and interdisciplinary collaborations, but also the imperative of ensuring that resources are dedicated to ocean exploration and taxonomy, as well as improved bioinformatics tools. Collectively, such investments will provide a deeper understanding of the vast biomedical potential resting in the ocean, and a stronger insurance against crises to come.
6 Conflicts of interest
M. J. is founder of, consultant to, and shareholder in, GyreOx Therapeutics, a company which uses marine-derived enzymes to develop pharmaceutical candidates.7 Acknowledgements
This work was in supported by the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou).8 References
- R. Blasiak, R. Wynberg, K. Grorud-Colvert, S. Thambisetty, N. M. Bandarra, A. V. M. Canário, J. da Silva, C. M. Duarte, M. Jaspars, A. Rogers, K. Sink and C. C. C. Wabnitz, Nat. Sustain., 2020, 3, 588–596 CrossRef.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS.
- M. Sorokina and C. Steinbeck, J. Cheminf., 2020, 12, 20 CAS.
- J. W. Blunt and M. H. G. Munro, MarinLit. A database of the literature on marine natural products for use on a macintosh computer prepared and maintained by the Marine Chemistry Group, Department of Chemistry, University of Canterbury, Canterbury, New Zealand, http://pubs.rsc.org/marinlit/ Search PubMed.
- A. M. S. Mayer, Clinical Pipeline, https://www.midwestern.edu/departments/marinepharmacology/clinical-pipeline.xml, accessed June 2, 2020 Search PubMed.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2020, 37, 175–223 RSC.
- J. Levin, in Endotoxin Detection and Control in Pharma, Limulus, and Mammalian Systems, ed. K. L. Williams, Springer International Publishing, Cham, 2019, pp. 3–16 Search PubMed.
- L. L. Ling, P. Keohavong, C. Dias and W. G. Thilly, Genome Res., 1991, 1, 63–69 CrossRef CAS.
- W. Bergmann and R. J. Feeney, J. Org. Chem., 1951, 16, 981–987 CrossRef CAS.
- H. Field and E. De Clercq, Microbiol. Today, 2004, 31, 58–61 Search PubMed.
- S. Sagar, M. Kaur and K. P. Minneman, Mar. Drugs, 2010, 8, 2619–2638 CrossRef CAS.
- C. Morrow and P. Cárdenas, Front. Zool., 2015, 12, 7 CrossRef.
- M. J. Bertin, S. L. Schwartz, J. Lee, A. Korobeynikov, P. C. Dorrestein, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2015, 78, 493–499 CrossRef CAS.
- Y. Hirata and D. Uemura, Pure Appl. Chem., 1986, 58, 701–710 CAS.
- T. D. Aicher, K. R. Buszek, F. G. Fang, C. J. Forsyth, S. H. Jung, Y. Kishi, M. C. Matelich, P. M. Scola, D. M. Spero and S. K. Yoon, J. Am. Chem. Soc., 1992, 114, 3162–3164 CrossRef CAS.
- M. J. Yu, Y. Kishi and B. A. Littlefield, Discovery of E7389, a fully synthetic macrocyclic ketone analog of Halichondrin B, in Anticancer Agents from Natural Products, ed. G. M. Cragg, D. G. I. Kingston and D. J. Newman, Taylor & Francis, Washington, D.C, 2011 Search PubMed.
- B. R. O'Keefe, B. Giomarelli, D. L. Barnard, S. R. Shenoy, P. K. S. Chan, J. B. McMahon, K. E. Palmer, B. W. Barnett, D. K. Meyerholz, C. L. Wohlford-Lenane and P. B. McCray, J. Virol., 2010, 84, 2511–2521 CrossRef.
- A. Zumla, J. F. W. Chan, E. I. Azhar, D. S. C. Hui and K.-Y. Yuen, Nat. Rev. Drug Discovery, 2016, 15, 327–347 CrossRef CAS.
- S. Lusvarghi and C. Bewley, Viruses, 2016, 8, 296 CrossRef.
- L. C. Griffithsin, Mar. Drugs, 2019, 17(10), 567 CrossRef.
- M. Rousselot, E. Delpy, C. Drieu La Rochelle, V. Lagente, R. Pirow, J.-F. Rees, A. Hagege, D. Le Guen, S. Hourdez and F. Zal, Biotechnol. J., 2006, 1, 333–345 CrossRef CAS.
- Y. Le Meur, L. Badet, M. Essig, A. Thierry, M. Büchler, S. Drouin, C. Deruelle, E. Morelon, F. Pesteil, P. Delpech, J. Boutin, F. Renard and B. Barrou, Am. J. Transplant., 2020, 20, 1729–1738 CrossRef CAS.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. B. P., D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. O. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS.
- J. D. Sigwart, What species mean: a user's guide to the units of biodiversity, CRC Press, Taylor & Francis Group, Boca Raton, 2018 Search PubMed.
- B. Fontaine, A. Perrard and P. Bouchet, Curr. Biol., 2012, 22, R943–R944 CrossRef CAS.
- M. Packalen and J. Bhattacharya, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 12011–12016 CrossRef CAS.
- J. D. Sigwart, K. D. Bennett, S. M. Edie, L. Mander, B. Okamura, K. Padian, Q. Wheeler, J. E. Winston and N. W. Yeung, Integr. Comp. Biol., 2018, 56(18), 1111–1117 Search PubMed.
- A. Zumla, J. F. W. Chan, E. I. Azhar, D. S. C. Hui and K.-Y. Yuen, Nat. Rev. Drug Discovery, 2016, 15, 327–347 CrossRef CAS.
- Y. Hallock and G. Cragg, Pharm. Biol., 2003, 41, 78–91 CrossRef.
- Nature, 2016, 538, 427, https://www.nature.com/news/early-career-researchers-need-fewer-burdens-and-more-support-1.20863 Search PubMed.
- M. Schüngel and E. Stackebrandt, Biotechnol. J., 2015, 10, 17–19 CrossRef.
- R. W. M. Van Soest, N. Boury-Esnault, J. Vacelet, M. Dohrmann, D. Erpenbeck, N. J. De Voogd, N. Santodomingo, B. Vanhoorne, M. Kelly and J. N. A. Hooper, PLoS One, 2012, 7, e35105 CrossRef CAS.
- R. Blasiak, J.-B. Jouffray, C. C. C. Wabnitz, E. Sundström and H. Österblom, Sci. Adv., 2018, 4, eaar5237 CrossRef.
- M. Steinegger and S. L. Salzberg, Genome Biol., 2020, 21, 115 CrossRef CAS.
- L. Laikre, F. W. Allendorf, L. C. Aroner, C. S. Baker, D. P. Gregovich, M. M. Hansen, J. A. Jackson, K. C. Kendall, K. Mckelvey, M. C. Neel, I. Olivieri, N. Ryman, M. K. Schwartz, R. S. Bull, J. B. Stetz, D. A. Tallmon, B. L. Taylor, C. D. Vojta, D. M. Waller and R. S. Waples, Conserv. Biol., 2010, 24, 86–88 CrossRef.
- E. Callaway, H. Ledford and S. Mallapaty, Nature, 2020, 583, 178–179 CrossRef.
| This journal is © The Royal Society of Chemistry 2021 |