Open Access Article
Open Access ArticleThe coenzyme/protein pair and the molecular evolution of life
Andreas
Kirschning

Institut für Organische Chemie und Zentrum für Biomolekulare Wirkstoffchemie (BMWZ), Leibniz Universität Hannover, Schneiderberg 1B, D-30167 Hannover, Germany. E-mail: andreas.kirschning@oci.uni-hannover.de; Fax: +49-511-762-3011; Tel: +49-511-762-4614
First published on 18th November 2020
Abstract
Covering: up to 2020
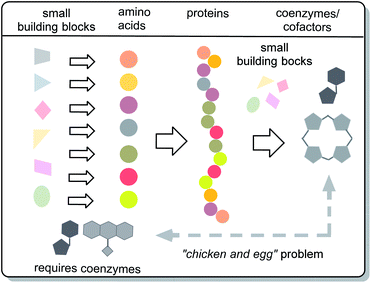
What was first? Coenzymes or proteins? These questions are archetypal examples of causal circularity in living systems. Classically, this “chicken-and-egg” problem was discussed for the macromolecules RNA, DNA and proteins. This report focuses on coenzymes and cofactors and discusses the coenzyme/protein pair as another example of causal circularity in life. Reflections on the origin of life and hypotheses on possible prebiotic worlds led to the current notion that RNA was the first macromolecule, long before functional proteins and hence DNA. So these causal circularities of living systems were solved by a time travel into the past. To tackle the “chicken-and-egg” problem of the protein–coenzyme pair, this report addresses this problem by looking for clues (a) in the first hypothetical biotic life forms such as protoviroids and the last unified common ancestor (LUCA) and (b) in considerations and evidence of the possible prebiotic production of amino acids and coenzymes before life arose. According to these considerations, coenzymes and cofactors can be regarded as very old molecular players in the origin and evolution of life, and at least some of them developed independently of α-amino acids, which here are evolutionarily synonymous with proteins. Discussions on “chicken-and-egg” problems open further doors to the understanding of evolution.
1. Introduction
“If there has been a first man he must have been born without father or mother – which is repugnant to Nature” Aristotle.1The “chicken and egg” problem is the archetypal example of causal circularity found in all living systems. Such scenarios are relevant when we need B for A, but for B first A.2 If we look at the molecular evolution of life, we encounter several such metaphorical paradoxes that deal with this specific problem.3
In molecular evolution, the “chicken-and-egg” problem has been discussed primarily in relation to the major classes of macromolecules: DNA, RNA and proteins (Fig. 1). These are responsible for information storage, replication and transformation into function. It boils down to the question which of these three privileged macromolecules was the first. The RNA world theory4 is not the only one that can explain the origin of life.5 However, no other theory has been subjected to a similar number of experiments to either vary or falsify it. Biosynthetic and probably evolutionary RNA is a precursor to DNA, and if the RNA world theory is correct, the hen's egg dilemma is left to RNA and proteins. It is rather unlikely that these two different types of macromolecules were formed simultaneously.
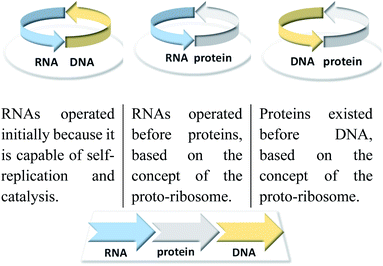 | ||
| Fig. 1 Classical problems of circular causality in biomolecular systems and how this are resolved with reference to the RNA world hypothesis. | ||
The discovery that RNA has catalytic properties6 was used as an argument for a possible solution. Originally, a single macromolecule could have performed both replication and catalysis. However, two further objections7 were raised to the RNA world hypothesis in relation to its catalytic properties. Only long RNA sequences show catalytic properties, and the catalytic repertoire of RNA is small and chemically not diverse.
Nevertheless, it has been postulated that RNA fragments were first formed from simple prebiotic molecules, which led to the early appearance of kind of ribozymes. These might have been involved in the binding and condensation of prebiotically generated amino acids to produce the first peptides and non-coded chains that might be comparable to modern peptidyltransferase centres (PTC). Such ancient complexes have occasionally been referred to as proto-ribosomes.8 Biotically speaking, the highly complicated translation system is necessary for its own formation.
Over time, larger peptides formed a tertiary structure with catalytic properties. Their greater chemical stability might be the reason why the RNA used as catalyst was to a great extent replaced by peptides and hence proteins.
From a chemical point of view, this was the entry into the world of today's enzymes. At that time, DNA formation was possibly facilitated by the occurrence of an iron-dependent ribonucleotide reductase.9 Therefore, the theory of the RNA world assumes that RNA preceded the proteins that preceded DNA (Fig. 1).10
Important chemical players in metabolism and protometabolism – the coenzymes and cofactors – have been largely neglected. These are small organic non-protein molecules that bind specifically to proteins and actively participate in catalytic biotransformations (Fig. 2). This alliance is effective because it is able to promote site-specific oxidations and reductions, group transfer reactions such as acylation, phosphorylation, methylation and formal acylanion transfer reactions. The protein part itself is generally not capable of promoting such reactions, but often are involved in general acid–base catalysis.11 In fact, it can be argued that coenzymes and cofactors are the most chemical species in nature of all molecular architectures, because accept for a few new roles that came into play much later in evolution, their sole purpose is to promote chemical reactions.
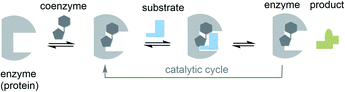 | ||
| Fig. 2 The role of cofactors/coenzymes in enzyme catalysis (the case of reversibly bound coenzymes are shown; prosthetic groups are commonly covalently attached to the protein template). | ||
A closer look at the biosynthetic origin and biological role of coenzymes reveals another biomolecular “chicken and egg” problem, namely the relationship between the pair coenzymes/cofactors and proteins (Fig. 3). In this report it will be shown that an evolutionary view can serve as a basis for solving the dilemma whether coenzymes and cofactors or proteins were the first. It should not be overlooked that there is still a debate about whether the RNA world hypothesis is valid or not.10 Nevertheless, speculations about the origin of life, as covered by the RNA world hypothesis, may point the way that causal cycles in biomolecular systems can be solved if prebiotic milieus are included in the considerations. Biomolecular causal cycles are broken if at least one element of such systems is a molecular remnant from prebiotic time. Based on these fundamental considerations, the “chicken and egg” problem of coenzymes and proteins is discussed here.
2. Coenzymes/cofactors and proteins in the biotic world
2.1 Coenzymes/cofactors – a brief overview
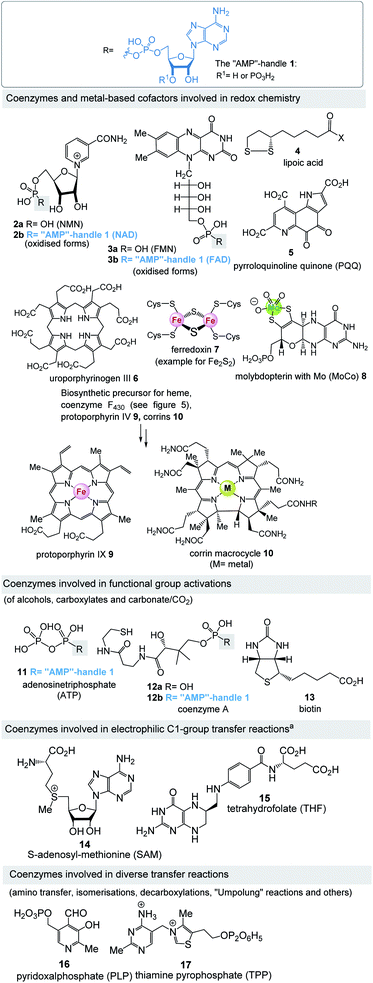
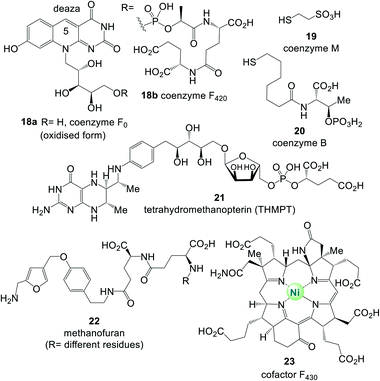
Fig. 4 lists the most important coenzymes and metal-based cofactors 2–17.10 These are categorised according to their chemical properties and their role in metabolism. Most of them are distributed across all phylogenetic kingdoms.Uroporphyrinogen III (6) is the biosynthetic precursor for many macrocyclic ligands such as heme including protoporphyrin IX 9 and cobalamin 10.12 Structurally, several of the coenzymes contain elements of nucleotides, which is manifested in the “AMP handle” 1. This fact was seen as a strong indication that RNA and coenzymes, or simpler analogues derived from them, may have occurred on earth under prebiotic conditions at about the same time.13 Furthermore, methanogens are dependent on several coenzymes that do not occur in other organisms (Fig. 5).14 These prokaryotes belong to the domain of the archaea and are exclusively capable of synthesising methane. As such they are limited to carbon dioxide, formate, methanol, methylamines and acetate as carbon sources. Coenzymes and cofactors are involved in this energy process, such as the 5-deazaflavines, the coenzymes F0 and F420 (18a,b) (structurally related to FMN 3a and FAD 3b), coenzyme M (19), 7-mercaptoheptanoylthreonine phosphate (coenzyme B, 20), tetrahydromethanopterin (THMPT, 21), methanofuran (22) and cofactor F430 (23). Most of them are specialised in their role in methanogenesis.15 Surprisingly none of the coenzymes specific to methanogenesis contains the “AMP handle” 1.
Methanogens are hydrogen-dependent autotrophs that have been suggested as good candidates for the ancestral state of physiology.16
Intriguingly, besides the “AMP-handle” some coenzymes possess an (oligo)-gamma-glutamate handle such as (THF 15, THMPT 21, F42018b, methanofuran 22 and glutathione).
2.2 Casual circularity is a relevant dilemma for the coenzyme/protein pair
At this stage it is useful to ask where the proteinogenic amino acids come from and how they are biosynthesised. The essential information on these questions are summarised in Fig. 6-10. They are grouped in a classical way, and this classification is linked to biosynthetic considerations.19 For example, the members of the pyruvate family of amino acids all use pyruvate as the starting building block for their biosynthesis. In fact, all 20 proteinogenic amino acids use carbohydrate-containing building blocks or carboxylic acids to build their carbon backbones.
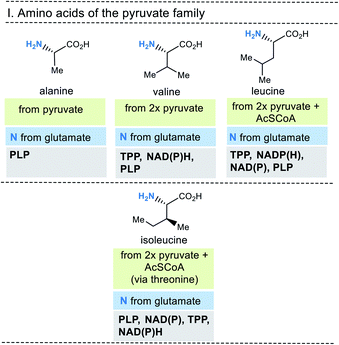 | ||
| Fig. 6 Summary of amino acid biosyntheses of the pyruvate family with reference to starting building blocks and coenzymes. | ||
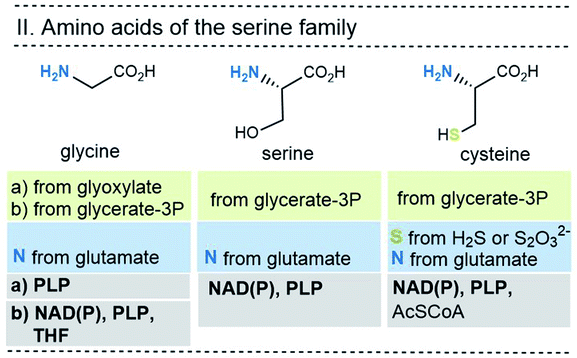 | ||
| Fig. 7 Summary of the amino acid biosyntheses of the serine family with reference to the required starting building blocks and coenzymes. | ||
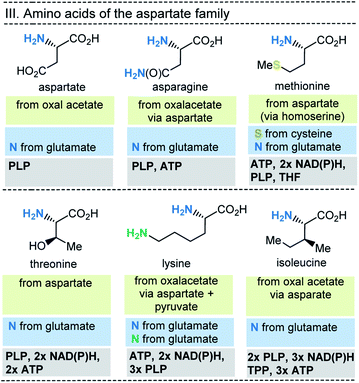 | ||
| Fig. 8 Summary of amino acid biosynthesis of the aspartate family with reference to starting building blocks and coenzymes required. | ||
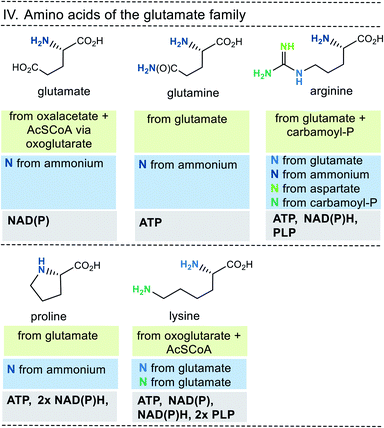 | ||
| Fig. 9 Summary of amino acid biosyntheses of the glutamate family with reference to starting building blocks and coenzymes. | ||
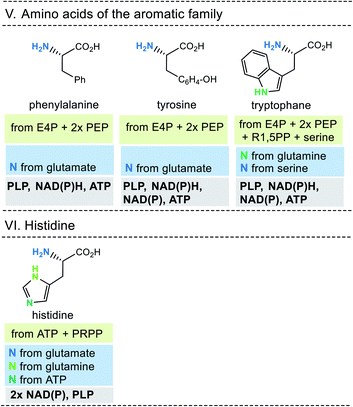 | ||
| Fig. 10 Summary of amino acid biosyntheses of the aromatic family and histidine with reference to starting building blocks and coenzymes required. | ||
In selected cases (e.g. isoleucine, proline, arginine) other amino acids serve as precursors, but these in turn are derived from simpler building blocks. With the exception of glutamate itself, the nitrogen atom of the amino group usually comes from the amino acid glutamate. Finally, the nitrogen atom in glutamate is recruited from ammonium (NH4+). Other nitrogen atoms in amino acids, such as in asparagine, glutamine, arginine, lysine, histidine and tryptophane, are taken from glutamate, glutamine, aspartate or ATP. Finally, the sulfur atoms in cysteine and methionine come from H2S or thiosulfate.
This condensed presentation is far from comprehensive. It does not include the biosyntheses of the building blocks (marked on light green ground) and the question which coenzymes that are required for their biosyntheses. For example, thiamine pyrophosphate (TPP, 17) is a coenzyme that plays an important role in carbohydrate metabolism such as in the pentose phosphate pathway. In addition, the amino acid biosynthetic pathways vary greatly between species. Fig. 6–10 also do not cover the variations found in archaea, bacteria and eukarya, as these details are not essential to convey the message underlying this report. This message is that, based on the listed building blocks, the biosyntheses of all 20 amino acids require different sets of coenzymes. These coenzymes serve as catalytically active units (PLP 16, TPP 17) or as group transfer agents, usually for chemical activation (ATP 11), redox reactions (NAD(P)/NAD(P)H 2) and for the transfer of methyl groups (THF 15). Note, that metal-based cofactors, which are considered evolutionary ancient, are not found in this list.
2.2.2.1 NAD 2b. The past years have seen a revival of NAD 2b. It was found that besides its well-established role in redox biochemistry and energetic metabolism, nicotinamide cofactors can also function as signaling molecules in a variety of cellular processes.20 NAD(P) also serves as substrate in mono- and poly-ADP ribosylation reactions that lead to the covalent modification of proteins.21
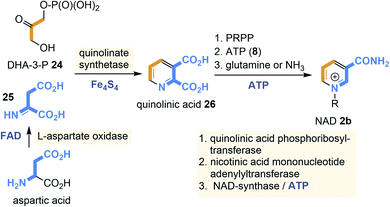
Currently, two different NAD(P) biosyntheses are known with quinolinic acid (26) and niacin being the key intermediates for both pathways (Scheme 1). In bacteria quinolinic acid (26) derives from dihydroxyacetone phosphate (DHA-3-P, 24) and L-aspartate,22 while in plants L-tryptophane is the precursor,23 a route not discussed here. In bacteria, aspartic acid is first oxidised to the corresponding imine 25 by L-aspartase oxidase with FAD 3b as redox coenzyme. In some archaea and thermotogales, this step is catalysed by aspartate dehydrogenase rather than aspartate oxidase.24
Next, condensation with DHA-3-P 24 occurs which is catalysed by the quinolinate synthase. This step is controlled by a [4Fe–4S] cluster that does not act as a redox cofactor but rather as a Lewis acid.25 Decarboxylation provides niacin whose pyridine nitrogen atom is quaternised with 5-phospho-α-D-ribose-1-diphosphate (PRPP) as electrophilic building block to furnish NAD 2b. The amide functionality is introduced in the final step and almost all archaea utilise ammonia as a nitrogen source for this transformation.
2.2.2.2 PLP 16. Pyridoxal phosphate (16), and its “amino sister” pyridoxamine phosphate are coenzymes that promote myriads of biotransformations especially in amino acid metabolism. Typical reactions are transaminations, decarboxylations, racemisations, retro-aldol reactions and Michael additions.26 It also participates as “coworker” in radical mediated reactions with radical SAM, e.g. in the lysine 2,3-aminomutase.27
In nature two principal biosynthetic pathways are found. Here, only the simpler of the two is briefly covered, that starts from glyceraldehyde-3-phosphate (GA3P, 27) and ribose-5-phosphate (28) and e.g. was studied in Bacillus subtilis (Scheme 2). The nitrogen atom is recruited in form of ammonia from glutamine.28 The PLP-synthase that bears an additional glutaminase site for providing ammonia directly condenses 27 and 28 to straightforwardly yield PLP 16. Noteworthy, the whole pathway does not require any additional coenzyme except that ATP is needed for the regeneration of glutamine from glutamate.
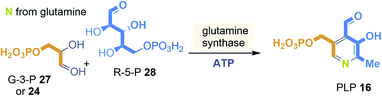 | ||
| Scheme 2 Ribosephosphate-dependent biosynthetic pathway of pyridoxal phosphate (16): building blocks marked in orange, blue and yellow and positions where they end up in PLP 16. | ||
2.2.2.3 Uroporphyrinogen III (6). Uroporphyrinogen is an important biosynthetic precursor for heme, coenzyme F430, cobalamins and protoporphyrin IX (9) and is the last common biosynthetic precursor for all tetrapyrroles.12,29 Porphyrin containing proteins are ubiquitously distributed in all kingdoms of life and among those heme and chlorophylls are the most important ones. Remarkably, cytochrome P450 enzymes, that contain the heme core, are thought to have existed for more than 3.5 billion years.11
The two known biosynthetic pathways to uroporphyrinogen III (6), present in all kingdoms of life including archaea,29,30 utilise 5-amino-levulinic acid (δ-ALA, 29) as linear precursor (Scheme 3). δ-ALA is either biosynthesised from glycine and succinyl-CoA or in a two-step enzymatic process from glutamyl-tRNA. In the first case, ALA synthase catalyses the decarboxylative coupling of glycine to succinyl-CoA catalysed by PLP 16. For the second route NADPH 2b and PLP 16 are coenzyme involved in the biosynthesis of δ-ALA 29. Next, eight molecules of δ-ALA are condensed to yield the macrocycle 6.
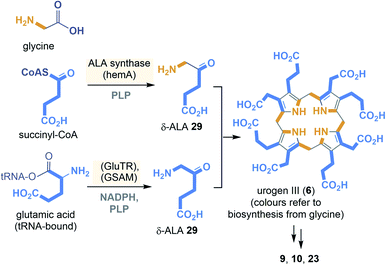 | ||
| Scheme 3 Summary of uroporphyrinogen III (6) biosyntheses: building blocks (orange and blue) (GSAM = glutamyl-tRNA reductase, GluTR = glutamate-L-semialdehyde aminomutase). | ||
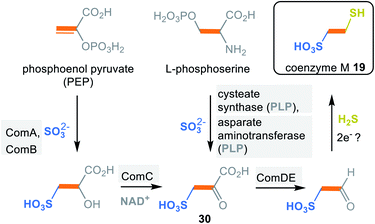
2.2.2.4 Coenzyme M (19). Coenzyme M is found in methanogenic archaea and plays a key role in methane formation.31 The S-methyl derivative is generated from coenzyme M (19) in methyl transfer reactions catalysed by proteins that contain zinc. In 1990 it was reported that coenzyme M is also involved in the bacterial metabolism (e.g. in proteobacterium Xanthobacter autotrophicus) of alkenes and oxiranes, the corresponding oxidation products.32
In methanogens two biosynthetic pathways are known for coenzyme M (19) in which the carbon backbone is derived either from phosphoenolpyruvate (PEP) or L-phosphoserine (Scheme 4).33 The PEP-dependent pathway is initiated by a Michael addition of sulfite. This step is followed by phosphate hydrolysis and oxidation to the α-keto acid 30. Finally, decarboxylation and reductive introduction of H2S furnishes coenzyme M (19). Details about the electron donor involved in this last step have not yet been clarified.
The L-phosphoserine-dependent pathway is based on the concerted elimination of phosphate and the addition of sulfite. The resulting L-cysteate is transaminated to form the joint intermediate sulfopyruvate 30, with α-ketoglutarate serving as co-substrate. From here, the pathway supposedly follows the first one.
Coenzymes, especially PLP, NAD(P), TPP, THF and ATP, are involved in the biosyntheses of all amino acids and consequently for proteins. Proteins are needed for the biosynthesis of coenzymes. At this point, the causal circularity for the coenzyme/protein pair is clearly revealed, and the question arises again: what came first? Proteins or coenzymes?
3. How can this “chicken-and-egg” problem of the protein/coenzyme pair be tackled?
The causal circularity of the three biomacromolecular pairs RNA–DNA, DNA–protein and RNA–protein was solved under the assumption that the RNA world hypothesis is correct by going back in time and making considerations about the origin of life and hypotheses about prebiotic protometabolism (see Fig. 1). It can therefore be assumed that a retrospective approach will help to break the causal circularity of coenzymes and proteins and that time travel either back to the origin of biotic evolution or to prebiotics world can solve this dilemma.3.1 Back in biotic time
According to this idea, viruses did not have to wait for the arrival of bacteria or the archaea; their ancestors could have entered the stage earlier, so that the DNA era was preceded by an epoch of far more primitive, fiercely competing self-replicating RNA chains – in essence the RNA world. Under these circumstances, simple ancestors of the RNA viruses, including retroviruses, could have already appeared in this archaic world. Their independence is manifested in the observation that the vast majority of viral genes are not found in bacteria, plants, animals or any other hosts. Viruses are thus able to create complex genes of their own accord, which are then assembled from other viral pieces. The link is manifested to contemporary protein-free viroids34 often called “living fossils” of primordial RNAs.35,36
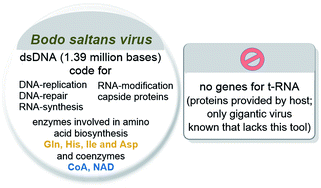
Another line of discussing earliest viral forms of life are nucleocytoplasmic large DNA viruses (NCLDV) and the related giant mimivirus (giant refers to >500 kb). The mimivirus has a capsid diameter of 400 nm, comparable to the size of small intracellular bacteria such as Rickettsia conorii.37 Remarkably, mimivirus contains, among others, genes for sugar, lipid and amino acid metabolism. Details on coded biosynthetic pathways of amino acids and especially of coenzymes have not been published for the mimivirus.37 The major difference in the mimivirus genome compared to small intracellular bacteria is the absence of genes coding for ribosomal proteins. They harbor missing building blocks as incomplete sets not sufficient for independent protein synthesis preventing them from leading an autonomous life.37b,c These giant viruses have been placed at the boundary between living and non-living38a and indicate that the evolutionary transition from virus to cell may have been a continuum (Scheme 6).39,40
Overall, the “virus first” approach does not solve the “chicken-and-egg” problem of the coenzyme/protein pair and does not provide an answer to the question of what came first.
Boyd et al.42 studied the possible origin and evolution of flavin-based electron bifurcating enzymes using a bioinformatics approach.43 Electron bifurcation is called the disproportionation of two electrons at the same redox potential to one electron with a higher and one with a lower redox potential and utilise coenzymes and cofactors such as NAD (2b), FAD (3b), ferredoxins (7), flavodoxin and ubiquinone.
With this mechanism in hand microorganisms generate low-potential electrons for the reduction of ferredoxins and flavodoxins, one central cellular process relevant for anaerobs that inhabit highly reductive environments.44a The phylogenetic analysis of twelve such bifurcating enzymes revealed that these redox systems were not part of LUCA but must have appeared at a later stage of life.
A comprehensive bioinformatic investigation on the (bio)molecular and pysiological basis of LUCA was carried out by Martin et al. They genetically analysed 6.1 million protein-coding genes and 286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 514 protein clusters from sequenced prokaryotic genomes of various phylogenetic trees.44b,c Search for genes was conducted that are involved in the physiology, cells access to carbon, energy and nutrients. 355 Protein clusters were found to be indicative for LUCA's metabolism being likely dominated by iron–sulfur clusters and radical reaction mechanisms.45
514 protein clusters from sequenced prokaryotic genomes of various phylogenetic trees.44b,c Search for genes was conducted that are involved in the physiology, cells access to carbon, energy and nutrients. 355 Protein clusters were found to be indicative for LUCA's metabolism being likely dominated by iron–sulfur clusters and radical reaction mechanisms.45
Cofactor analysis unravelled the presence of biosynthetic pathways for basically all coenzymes and cofactors listed in Fig. 4 and 5 (this list includes those found in the ESI of ref. 44a) (Scheme 7). Important members are pterins such as molybdopterin 8, 5-deazaflavins (coenzyme F420, 18b), S-adenosylmethionine (SAM, 14), coenzymes A (CoA, 12b) and M (19), thiamine pyrophosphate (TPP, 17), ferredoxin (Fe–S proteins, 7), protoporphyrin IX (9) and corrin (10) (see Scheme 3).46 This list of coenzymes and cofactors covers members found in methanogens47 but also bacteria. This comprehensive list means that the biosynthetic machinery of LUCA already utilised all coenzymes and cofactors now found in all kingdoms of life. How can this be rationalised? The authors interpreted the list of cofactors as a strong indication that LUCA must have relied on the Wood–Ljungdahl pathway a noncyclic reductive carbon fixation path from CO2 and other C1 building blocks to (activated) acetic acid.48 A deeper analysis suggests that LUCA could have lived from the gases H2, CO2 and N2.
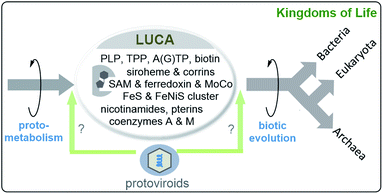 | ||
| Scheme 7 Last unified common ancestor (LUCA) and its coenzymes and cofactors as suggested by bioinformatic analyses disclosed in ref. 44a (including ESI). LUCA is regarded to be the ancestor of bacteria and archaea while eukaryotes arise from archaea (structures of coenzymes and metal cofactors see Fig. 4 and 5).49 the possible role of protoviroids and how there origin may relate to LUCA are included (“virus first” hypothesis).36 | ||
The Wood–Ljungdahl pathway relies on a metalloprotein complex with iron and nickel50 playing a central role as metals. It is composed of a carbon monoxide-dehydrogenase and acetyl-CoA synthase (CODH/ACS). The ferredoxin part promotes the reduction of CO2 to CO.51 In primordial metabolism CO itself could have formed through the gas water shift reaction or by transition metal catalysis. It was also found that LUCA must have contained the reverse gyrase, an enzyme typically associated with hyperthermophiles,48 which supports the assumption that LUCA must have been an autotrophic thermophile.
Returning to the starting point of this article and the “chicken-and-egg” problem, it must be stressed that neither the viral hypothesis nor the current view on the metabolism of LUCA can answer the question of what came first, coenzymes/cofactors or proteins.
Interestingly, Martin and colleagues noted that LUCA could only have had nine nucleotide and five amino acid biosynthetic pathways. The absence of some essential pathways was justified by the possibility that LUCA had not yet developed the genes in question before the phylogenetic separation of bacteria and archaea took place. Instead, the missing products or building blocks could have been provided to LUCA externally from the prebiotic pool.
This statement is important because it shifts the central question of this report further into the past, namely the transitional phase between prebiotic metabolism and the development of LUCA.
Consequently, essential components are provided primarily through cell–cell contacts with the host I. hospitalis (Scheme 8). N. equitans are “incomplete” cells that can exist, provided that the missing nutrients are collected externally from the environment.56 However, it has to be pointed out that parasitic nanoarchaeota has to be regarded as a model for early forms of life but itself is no remnant of life before LUCA. Similar to Martin's analysis they must externally recruit a large number of different metabolites such as amino acids and coenzymes/cofactors.
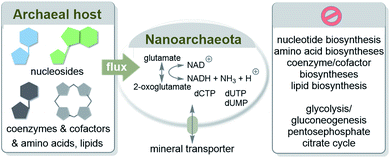 | ||
| Scheme 8 Transfer of biomolecules from the marine hyperthermophilic and chemolithoautotrophic archaea I. hosptalis to N. equitans to supplement its metabolism. Elements of this metabolism are depicted according to ref. 55a. | ||
This recruitment scenario resembles to some degree the retrograde theory of evolution,57 It says that the first living being was a completely heterotrophic entity that reproduced itself at the expense of prebiotically formed organic molecules (I–III) in its environment. The organism will then deplete the environmental reserves of these molecules, e.g. I and exhaust them to a point where growth is limited. In such an environment, any organism that develops an enzyme or catalytic system capable of synthesizing a molecule I from the precursors II and III would have a clear selection advantage and would spread rapidly in the environment.
Horowitz, who suggested this theory, further proposed that next evolution was probably based on the random combination of genes. For example, the simultaneous unavailability of two intermediates (e.g. II and III) would favour a symbiotic association between two mutants, one of which is able to synthesise B and the other able to synthesise C from other precursors in the environment. This would lead to the development of short reaction chains using substances whose synthesis was previously acquired. It is of interest to note that this theory includes the idea of parasitism as a driving force of evolution.
3.2 Back in prebiotic times
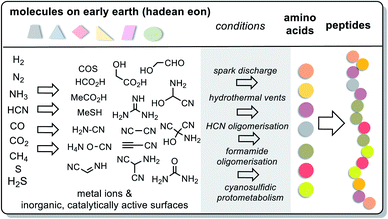
For amino acids, the answer is briefly yes. Amino acids and consequently simple peptides must have formed under different geochemical scenarios as these have been experimentally probed. Conditions include three different electrical charge experiments by Miller and Urey58–60 (spark discharge), the iron–sulfur world of Wächtershäuser and Huber61–63 (hydrothermal vents), oligomerisations of HCN64 and formamide65,66 as well as Sutherland's cyanosulfidic protometabolism (Scheme 9).67 Key reactions are the Strecker reaction and its phosphoro variant68 and the photochemical Kiliani–Fischer reaction.
These experimental designs provided a wide variety of canonical amino acids such as glycine, alanine, valine, leucine, isoleucine, serine, threonine, asparagine, asparagine, glutamate, glutamine, proline methionine, arginine and phenylalanine, as well as various non-canonical amino acids. Di- and tripeptides were also found in experiments mimicking hydrothermal vents.61,62
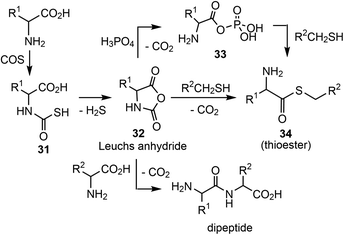
In addition, it has been shown that α-amino acids can be chemically activated by e.g. COS, a product from hydrothermal sources according to Wächtershäuser and Huber.63Via the corresponding thiocarbamates 31 (Scheme 10), carbohydrides 32 (Leuchs anhydride) are formed,69–71 a process that is kinetically accelerated in the presence of metal cations. Leuchs anhydride is also a precursor for highly reactive aminoacyl phosphate anhydrides,33 which are potential precursors for the formation of peptides and thioesters.35
Following this hypothesis, King explained that the earliest biochemicals were not only reproduced by autocatalytic pathways, but that they were actually autocatalytic molecules and that evolution took place through a succession of symbiotic unions.13b
Since arguments for the formation and operation of coenzymes or cofactors did not appear in the experiments listed under “conditions” in Scheme 9, more direct chemical assessments had to be made and fossils collected.72 Since White III first directed a beam of light at coenzymes, various efforts have been made to find and establish conditions for the formation of coenzymes or simplified but functional derivatives under prebiotic conditions.5 Here, too, the focus is on the four representative examples NAD 2b, PLP 16, uroporphyrinogen III (6) and coenzyme M (19), which were already chosen in chapter 2.2.2 with regard to their biotic synthesis.
3.2.2.1 NAD 2b. First attempts to produce the pyridine unit in NAD 2b under ostensibly prebiotic conditions were reported by Orgel et al. The group showed that propiolonitrile 35, propiolaldehyde 36 and ammonia yielded nicotinamide 37 (Scheme 11).73a,b
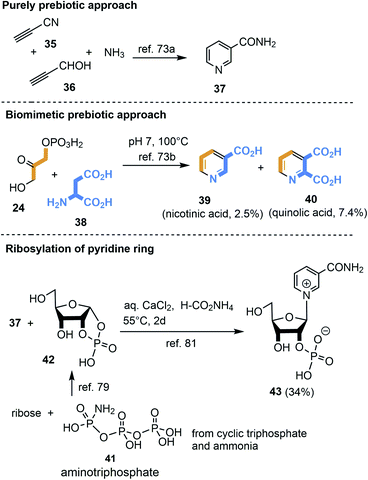 | ||
| Scheme 11 De novo syntheses of nicotinamide 37, nicotinic acid 39, quinolic acid 40 as well as generation of nicotinamide nucleotide 43. | ||
Cleaves and Miller suggested that the prokaryotic biosynthetic pathway20c should in principal be chemically mimicked in a prebiotic environment (Scheme 11).74 Thus, mixing dihydroxyacetone phosphate 24 with aspartic acid 38 (ref. 75) provided nicotinic acid 39 and quinolic acid 40. Is it realistic to assume that phosphorylated small carbohydrate-containing building blocks already existed in such early times? An important finding was that cyclotriphosphate76 reacts with ammonia to aminotriphosphate (41)77 which is a powerful phosphorylating agent e.g. for α-hydroxyaldehydes.78,79 Importantly, water-soluble polyphosphates like cyclotriphosphate are known to be generated in the vicinity of volcanoes.80
Kim and Benner combined the findings of Orgel73a,b and Eschenmoser79 by reacting nicotinamide 37 with ribose-1,2-cyclic phosphate 42 under prebiotically plausible conditions and obtained the nicotinamide nucleotide.81
3.2.2.2 PLP 16. PLP promotes a large and diversified number of biotransformations mainly in amino acid metabolism. Transamination and decarboxylation of α-amino acids are the two key reactions of PLP 16. Due to its central role in the formation of amino acids and especially in the context of this review article, it is worth considering a protometabolic scenario for this coenzyme.
The first example of a simplified pyridoxine derivative formed under putative prebiotic conditions is based on the trimerisation of glycol aldehyde 44 in a heated buffered solution in the presence of ammonia (Scheme 12, top). Glycoladehyde is a small aldose that can undergo aldol reactions forming higher sugars. Against this background, the synthesis to 4-(hydroxymethyl)pyridin-3-ol (45) carries biomimetic elements.82 A prebiotic approach, which is more similar to one of the two principle biosyntheses of PLP, precisely the one that does not require additional coenzymes, is depicted in Scheme 12 (bottom).28 The condensation of ribose or any other pentose with glyceraldehyde-3-phosphate (27) in the presence of ammonia could yield pyridoxine phosphate (46). Finally, the final oxidation to PLP 16 could be carried out by ferric salts such as Fe(CN)6.83
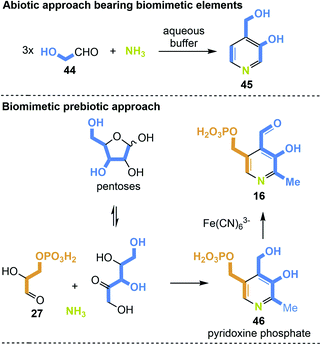 | ||
| Scheme 12 De novo syntheses of 4-(hydroxymethyl)pyridine-3-ol (45) pyridoxine phosphate (46) and PLP 16 under supposedly prebiotic conditions. | ||
3.2.2.3 Uroporphyrinogen III (6). Cofactors of the porphyrin type such as uroporphyrinogen III (6) carry a macrocyclic tetrapyrrole ligand which is suited for the binding of various metals, as is the case with protoporphyrin IX (9) and cofactor F430 (23).15
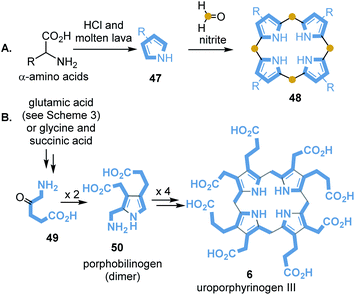
Porphyrin-containing proteins are ubiquitously distributed in the biotic world. It is assumed that some representatives have existed for more than 3.5 billion years, so it can be assumed that they also played a role in the prebiotic world.84 Baker and coworkers reported on first attempts to synthesise porphyrin from pyrrole and formaldehyde under simulated geochemical conditions.85 Interestingly, simple pyrroles 48 can be detected when seawater that contains amino acids is exposed to molten lava. Strasdeit et al. suggested that on primordial volcanic islands the volatile pyrroles and HCl must have condensed in cooler places.
Under these concentration conditions, pyrrole oligomerisation may have taken place.86 It has also been found that 2,4-diethylpyrrole (47) and HCl in the presence of formaldehyde and nitrite provides octaethylporphyrin 48 and other oligomers (Scheme 13A).
Lindsey and collaborators included biosynthetic considerations in their studies on more water-soluble uroporphyrinogen III (6).87 Porphyrinogens are formed by self-condensation from aminoketones 49 and diketones or ketoesters in water under prebiotic conditions (Scheme 13B). Remarkably, these synthetic studies confirmed that dimer 50 is the most important intermediate in this process, similar to uroporphyrinogen III biosynthesis (see Scheme 3).87
It needs to be emphasised that the transfer of basic prebiotic molecules to the starting building blocks, in particular succinic acid and further downstream 5-aminolevulinic acid (49), has not yet been experimentally clarified.
3.2.2.4 Coenzyme M (19). As early as 1993 Miller and coworkers searched for prebiotic conditions that could result in the formation of coenzyme M (19).88 They proposed ethene as a possible C2 building block that can form during prebiotic processes in planetary atmospheres.89 Under photolytic conditions, CS2 or COS serve as a source of 3p-sulfur atoms, which end up in ethylene sulfide (51) after collision with ethene (Scheme 14).90 In the following, cysteamine (52) is generated during the ring opening of ethylene sulfide (51) with ammonia.88 In a similar way, the presence of sulfite leads to the coenzyme M (19).
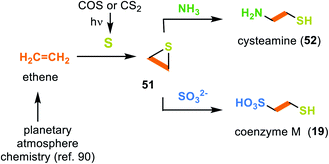 | ||
| Scheme 14 Experimental evidence for the prebiotic formation of cysteamine (52) and coenzyme M (19) from ethene and sulfur. | ||
For a more comprehensive overview of a chemistry that mimics the formation of coenzymes under prebiotic conditions, the reader is referred to ref. 72.
4. Integration of the coenzyme/protein pair in an evolutionary context
It can be concluded from the previous chapters that amino acids and selected coenzymes or simpler analogues must have been present before the first life forms appeared (Scheme 15; early protometabolism). These may have been involved in molecular and biological evolution from the very beginning and have played an active role since then.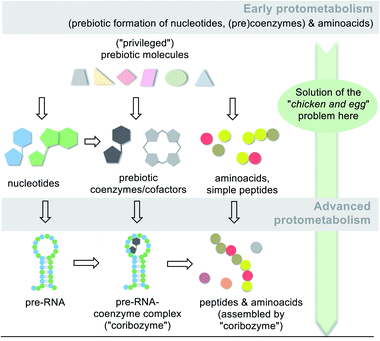 | ||
| Scheme 15 Simplified graphical representation of the protometabolic evolution of life from simple building blocks to key metabolic building blocks (amino acids, nucleotides and coenzymes/cofactors) and finally to biomacromolecules (pre-RNA, peptides,96a RNA/coenzyme–cofactor complexes72). Note, that a network of diverse molecules make up this molecular evolution with RNA playing one key role. | ||
The “chicken-and-egg” problem is solved at this very early stage of chemical evolution, because no circular dependence existed at that time. Both molecular entities could be formed independently of each other under prebiotic conditions. This is because they are the result of the inherent chemical reactivity of molecules that were available under the conditions of the Hadian eon about 4 billion years ago. It has to be stressed that α-amino acids are formed under all the postulated prebiotic scenarios listed in Scheme 9.
Certainly the original evolutionary role of RNA was linked to its catalytic properties. Chemically, however, the known ribozyme-catalysed transformations are rather limited in their diversity. More sophisticated chemical transformations such as redox chemistry, alkylations and C–C bond forming reactions depend on co-catalysts, which, as discussed here, are typically represented by coenzymes and cofactors. To broaden the scope of protometabolism, these co-catalytic small molecules (or simpler analogues) may have bound to RNA that served as a template. Such an association could have taken place via hydrogen bonds and/or electrostatic interactions similar to existing coenzyme/protein complexes (Scheme 15; advanced protometabolism).91 Alternatively, coenzyme-like co-catalysts may be covalently bound to the 5′ terminus of a ribozyme.4a,92
A strong argument for such coenzyme–RNA complexes can already be found in the biotic world. The ability of coenzymes such as TPP 17,93a,b FMN 3a,92c,d SAM 14,93e,f THF 15,93g and adenosylcobalamine (AdoCbl)93h to bind to RNA is found in riboswitches. These short, relatively simple sequences in mRNAs bind metabolites directly and are responsible for regulating gene translation.94 As a result activation or deactivation of gene expression occurs, a role, however, that became relevant later in biotic evolution.
Without explicitly mentioning the coenzymes, Stewart proposed such a scenario by linking the protometabolism to the RNA world.95 The controlled metabolism hypothesis suggests that RNA benefits from the protometabolism by overcoming what he called the cooperation barrier. The RNA would become a manager that manages the metabolism and uses its power to increase its productivity. This corresponds to a trend in the field of molecular evolution of life not to only consider and experimentally verify prebiotic routes towards selected molecules and oligomers but rather to consider cooperative interactions and networks among diverse classes of molecules that include peptides96 and small molecules present in primary metabolisms such as the reversed Krebs cycle.97 What is occasionally framed with the word “system chemistry” has led to a broader focus on cooperative coevolution among the diverse classes of molecules from the earliest times.98
One of several possible geological sites where such an advanced protometabolism could thrive are terrestrial hydrothermal freshwater fields and ponds.99 The conditions in such fields are highly dynamic in that evaporation to dryness occurs either over long periods of time or at high frequency through nearby geysers. Both the concentration of solutions, which preferably contain small molecules and precursors for nucleotides, amino acids and (pre)coenzymes/cofactors,100 and the precipitation on inorganic surfaces and their redilution represent unique changing chemical environments from which more complex peptides, oligonucleotides and coenzyme/cofactor RNA complexes may have formed (Scheme 15). Some evidence has already been collected that wet–dry cycles can drive the polymerization of mononucleotide mixtures and yield polymers 10 to >100 nucleotides in length.99
As soon as nucleotides, amino acids, small peptides and oligonucleotides as well as selected coenzymes and cofactors or their simpler analogues became available, they actively participated in the transition to biotic evolution. The RNA world hypothesis (Scheme 16, advanced protometabolism) assigns an evolutionary leadership position to this polymer, being a part of a collaborative network of diverse molecules including peptides that all coevolved.96 This prominent role has been linked to the ability of RNA to act as a catalyst, as known for ribozymes. From a chemical point of view, however, the variety of RNA-catalysed transformations is rather small, so that, as first suggested by White III,13a it is assumed that co-catalytically active coenzymes (or simpler analogues thereof) must have already been present to form functional co-ribozymes.101 RNA served as a template here to form hydrogen bonds and/or electrostatic interactions with the coenzyme, which are similar to most existing coenzyme/protein complexes.
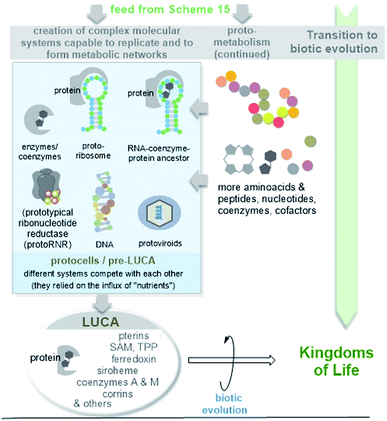 | ||
| Scheme 16 Simplified graphical representation of the evolution of protoviroids, protocells and LUCA (according to ref. 44a) as early life forms. Due to their importance in the evolution of macromolecules, the cofactor-mediated radical transformation of RNA to DNA is highlighted. | ||
Remarkably, RNA can be related to a virus-like state, and the formation of LUCA and modern cells then becomes a subsequent event.102 Viroids, the smallest and simplest replicating RNA molecules known today, have been proposed as subviral descendants of the earliest biomolecules on Earth.103–105 Special features are their small size, the circular structure, high G + C content and lack of protein coding capability compatible with a ribosome-free habitat. Later in evolution, these ancient viroids (protoviroids) may have become parasites, and today they depend on cellular enzymes such as RNA polymerase, RNAaseH and RNA ligase for replication.105 However, protoviroids would always have depended on some kind of ATP-dependent cell metabolism, so that it was speculated that such ancient viroids and protocells likely co-evolved.106
While access to LUCA is possible via a phylogenetic and bioinformatics approach, ideas about the transformation to compartmentalized forms of early life before LUCA appeared on planet Earth are much more speculative. It is likely that LUCA evolved from a confusing variety of different pre-metabolic forms of protocells (pre-LUCA), which are even more difficult to grasp in practice. However, dramatic molecular renewals must have occurred during this transitional phase towards biotic evolution.107 Protoribosomes allowed the controlled synthesis of peptides and from these, enzymes and coenzyme–protein conjugates with extended catalytic potential were formed. The coenzymes and cofactors have changed their macromolecular template (from RNA to protein). And at one point DNA must have appeared on the scene.
Ribonucleotide reduction is a key step in the transformation of the RNA world into a world in which DNA macromolecules became central to information storage. Ribonucleotide reductase (RNR) catalyses the deoxygenation of ribose via a radical process. Lundin et al. drew a picture on the evolution of this process and possible ancestors of the proteins, which they called prototypical ribonucleotide reductase (protoRNR).108 Their analysis revealed that metals or cofactors must have played an important role in this deoxygenation process, initially with a lack of chemoselectivity with respect to the radical abstraction of H from given nucleotides. In anaerobes, the 5′-deoxyadenosyl-5′-radical (dAdo radical) generated by cobalamin-type redox systems or radical SAM/iron–sulfur clusters acts as a redox promoter, and the analysis suggests that these early types of redox systems serve as a raw model for modern RNR.109 In particular, extant B12-dependent class II RNRs have been proposed as the most promising first candidates for modern RNRs that originate from protoRNRs. The discussion about the evolution of this key transformation provides a further argument for the fact that both amino acids/proteins and coenzymes/co-factors must have been present long before the appearance of biotic life in the form of LUCA, which already belonged to the DNA world.
5. Conclusions and outlook
This overview does by no means cover all geological, chemical and biological facets that are important for proposing scenarios on the origin and evolution of life. Indeed, our journey began with the question of what came first: coenzymes or proteins? It turned out that this is a real but overall neglected “chicken-and-egg problem” of the biotic world. Coenzymes or cofactors, which have played the role of small chemical catalysts or promoters of chemical reactions for most of molecular evolution, have often not been included in hypothetical considerations and theories of the origin of life. It is often mentioned that they are part of (pre-)metabolic networks, but their origin has hardly been discussed. But this biorelevant class of molecules is ancient, even in LUCA they are said to have been found,44a but remarkably they have hardly changed their structures and function until today. Coenzymes and co-factors are mainly designed to promote chemical reactions. This was the case in connection with protometabolism and is to a large extent still the case today.The journey took us on a time arrow back to the early days of life with analyses of the existence and role of coenzymes, cofactors and also proteins in viroids, the last common ancestor (LUCA) and nanoarchaeota. While these analyses did not solve the circular dilemma of coenzymes and proteins, they did provide a deeper understanding of coenzymes and cofactors and their role in early life forms, as they are key promoters of metabolism in methanogens, especially the reductive Wood-Ljungdahl pathway. Like proteins, coenzymes and cofactors are old. The “chicken-and-egg” problem can be solved by considering prebiotic formation of amino acids, small peptides and coenzymes.
In the scenario of the RNA world, coenzymes and cofactors were partners of RNA rather than proteins. Only later did it appear that they were brought together in the form of enzyme–coenzyme complexes, and the development of biosynthetic pathways to these led to the circular dilemma that is the starting point of this overview.
This report is intended to provide new ideas and food for thought on the origin of life, which will eventually have to be reinvented, much as A. Eschenmoser would put it.110 Finally, coenzymes and cofactors will hopefully attract more interest as a fourth key player in the molecular evolution of metabolism.
6. Conflicts of interest
There are no conflicts to declare.7. Acknowledgements
I thank Prof. T. Brüser (Institute of Microbiology, Leibniz University Hannover) for fruitful discussions.8. Notes and references
- H. P. Blavatsk, Isis unveiled, 1877, vol. I, p. 428, ISBN 978-0-557-35694-2 Search PubMed.
- Y. Pilpel and O. Rechavi, Biol. Direct, 2015, 10, 34 CrossRef.
- The “chicken-and-egg” problem was also discussed in the context to distinguish between Lamarckian and Darwinian evolution. “Can the phenotype affect the genotype?” or, “can epigenetics translate into genetics”? E. V. Koonin and Y. I. Wolf, Biol. Direct, 2009, 4, 42, DOI:10.1186/1745-6150-4-42.
- (a) W. Gilbert, Nature, 1986, 319, 618 CrossRef; (b) G. F. Joyce, Nature, 2002, 418, 214–221 CrossRef CAS; (c) L. E. Orgel, Crit. Rev. Biochem. Mol. Biol., 2004, 39, 99–123 CrossRef CAS; (d) M. P. Robertson and G. F. Joyce, Cold Spring Harbor Perspect. Biol., 2012, 4, a003608 Search PubMed; (e) D. Penny, Biol. Philos., 2005, 20, 633–671 CrossRef; (f) P. G. Higgs and N. Lehman, Nat. Rev., 2015, 16, 7–17 CrossRef CAS.
- R. Krishnamurthy, Acc. Chem. Res., 2017, 50, 455–459 CrossRef CAS.
- (a) K. Kruger, P.-J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling and T. R. Cech, Cell, 1982, 31, 147–157 CrossRef CAS; (b) X. Chen, N. Li and A. D. Ellington, Chem. Biodiversity, 2007, 4, 633–655 CrossRef CAS.
- S. A. Benner, A. D. Ellington and A. Tauer, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7054–7058 CrossRef CAS.
- (a) C. Davidovych, M. Belousoff, I. Wekselman, T. Shapira, M. Krupkin, E. Zimmerman, A. Bashan and A. Yonath, Isr. J. Chem., 2010, 50, 29–35 CrossRef; (b) S. C. Meyer and P. A. Nelson, BIO-Complexity, 2011, 1–10 Search PubMed; (c) J. C. Bowman, A. S. Petrov, M. Frenkel-Pinter, P. I. Penev and L. D. Williams, Chem. Rev., 2020, 120, 4848–4878 CrossRef CAS.
- Recently, Trapp and coworkers disclosed a prebiotic route to DNA: J. S. Teichert, F. M. Kruse and O. Trapp, Angew. Chem., Int. Ed., 2019, 58, 9944–9947 CrossRef CAS . Biochemically 2-deoxy-D-ribose-5-phosphate aldolase (DERA) is able to generate 2-deoxy-ribose from acetaldehyde and glyceraldehyde-3-phosphate: M. Haridas, E. M. M. Abdelraheem, and U. Hanefeld, Appl. Microbiol. Biotechnol., 2018, 102, 9959–9971.
- It has to be noted that other than the RNA world hypothesis have been proposed as summarised in ref. 5. These included the “protein-first”, the “metabolism-first” or the “lipid world” theories. None of the authors consider coenzymes or cofactors as key players in their origin of life scenarios and cannot help in unravelling the role of these small chemical entities in the evolution of life. “Protein first”: P. Andras and C. Andras, Med. Hypotheses, 2005, 64, 678–688 CrossRef CAS . “Metabolism first”: F. A. Anet, Curr. Opin. Chem. Biol., 2004, 8, 654–659. “Lipid first“: D. Segré, D. Ben-Eli, D. W. Deamer, and D. Lancet, Origins Life Evol. Biospheres, 2001, 31, 119–145.
- (a) M. Richter, Nat. Prod. Rep., 2013, 30, 1324–1345 RSC; (b) J. D. Fischer, G. L. Holliday, S. A. Rahman and J. M. Thornton, J. Mol. Biol., 2010, 403, 803–824 CrossRef CAS; (c) T. P. Begley, Nat. Prod. Rep., 2006, 23, 15–25 RSC; (d) M. E. Webb, A. Marquet, R. R. Mendel, F. Rébeillé and A. G. Smith, Nat. Prod. Rep., 2007, 24, 988–1008 RSC; (e) M. Richter, Nat. Prod. Rep., 2013, 30, 1324–1345 RSC; (f) D. E. Scott, A. Ciulli and C. Abell, Nat. Prod. Rep., 2007, 24, 1009–1026 RSC.
- (a) G. Layer, J. Reichelt, D. Jahn and D. W. Heinz, Protein Sci., 2010, 19, 1137–1161 CrossRef CAS; (b) F. J. Leeper, Nat. Prod. Rep., 1989, 6, 171–203 RSC; (c) D. R. Nelson, T. Kamataki, D. J. Waxman, F. P. Guengerich, R. W. Estabrook, R. Feyereisen, F. J. Gonzalez, M. J. Coon, I. C. Gunsalus, O. Gotoh, K. Okuda and D. W. Nebert, DNA Cell Biol., 1993, 12, 1–51 CrossRef CAS.
- (a) H. B. White III, J. Mol. Evol., 1976, 7, 101–104 CrossRef; (b) G. A. M. King, BioSystems, 1980, 13, 2345 CrossRef.
- A. A. DiMarco, T. A. Bobik and R. S. Wolfe, Annu. Rev. Biochem., 1990, 59, 355–394 CrossRef CAS.
- (a) P. N. Evans, J. A. Boyd, A. O. Leu, B. J. Woodcroft, D. H. Parks, P. Hugenholtz and G. W. Tyson, Nat. Rev. Microbiol., 2019, 17, 219–232 CrossRef CAS; (b) F. Enzmann, F. Mayer, M. Rother and D. Holtmann, AMB Express, 2018, 8, 1 CrossRef CAS.
- W. F. Martin and F. L. Sousa, Cold Spring Harbor Perspect. Biol., 2016, 8, a01812 Search PubMed.
- H. F. Noller, Cold Spring Harbor Perspect. Biol., 2012, 4, a003681 Search PubMed.
- M. Root-Bernstein and R. Root-Bernstein, J. Theor. Biol., 2015, 367, 130–158 CrossRef CAS.
- H. E. Umbarger, Annu. Rev. Biochem., 1978, 47, 533–606 CrossRef CAS.
- (a) A. Volbeda, C. Darnault and J.-C. Fontecilla-Camps, J. Am. Chem. Soc., 2016, 138, 11802–11809 CrossRef CAS; (b) S. O. de Choudens, L. Loiseau, Y. Sanakis, F. Barras and M. Fontecave, FEBS Lett., 2005, 79, 3737–3743 CrossRef; (c) T. Begley, C. Kinsland, R. Mehl, A. Osterman and P. Dorrestein, Vitam. Horm., 2001, 61, 103–119 CrossRef CAS.
- (a) H. J. Cleaves and S. L. Miller, J. Mol. Evol., 2001, 52, 73–77 CrossRef CAS; (b) H. Kim and S. A. Benner, Chem.–Eur. J., 2018, 24, 581–584 CrossRef CAS.
- (a) S. K. Spaans, R. A. Weusthuis, J. van der Oost and S. W. M. Kengem, Front. Microbiol., 2015, 6, 742 Search PubMed; (b) F. Gazzaniga, R. Stebbins, S. Z. Chang, M. A. McPeek and C. Brenner, Microbiol. Mol. Biol. Rev., 2009, 73, 529–541 CrossRef CAS; (c) Evolutionary view on both pathways: W. C. Lima, A. M. Varani and C. F. M. Menck, Mol. Biol. Evol., 2009, 26, 399–406 CrossRef CAS.
- (a) M. Fischer and A. Bacher, Nat. Prod. Rep., 2005, 22, 324–350 RSC; (b) A. Bacher, S. Eberhardt, M. Fischer, K. Kis and G. Richter, Annu. Rev. Nutr., 2000, 20, 153–167 CrossRef CAS; (c) M. Mack and S. Grill, Appl. Microbiol. Biotechnol., 2006, 71, 265–275 CrossRef CAS.
- Z. Yang, A. Savchenko, A. Yakunin, R. Zhang, A. Edwards, C. Arrowsmith and L. Tong, J. Biol. Chem., 2003, 278, 8804–8808 CrossRef CAS.
- (a) M. Zhang, L. Wang and D. Zhong, Arch. Biochem. Biophys., 2017, 632, 158–174 CrossRef CAS; (b) A. Sancar, Biochemistry, 1994, 33, 2–9 CrossRef CAS; (c) C. A. Brautigam, B. S. Smith, Z. Ma, M. Palnitkar, D. R. Tomchick, M. Machius and J. Deisenhofer, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 12142–12147 CrossRef CAS.
- A. C. Eliot and J. F. Kirsch, Annu. Rev. Biochem., 2004, 73, 383–415 CrossRef CAS.
- P. A. Frey, Annu. Rev. Biochem., 2001, 70, 121–148 CrossRef CAS.
- B. Richts, J. Rosenberg and F. M. Commichau, Front. Mol. Biosci., 2019, 6, 32 CrossRef CAS.
- (a) H. A. Dailey, T. A. Dailey, S. Gerdes, D. Jahn, M. Jahn, M. R. O'Brian and M. J. Warren, Microbiol. Mol. Biol. Rev., 2017, 81, e0048-1–6 CrossRef; (b) T. Fan, B. Grimm and G. Layer, Adv. Bot. Res., 2019, 91, 89–131 CAS.
- (a) S. I. Beale, EcoSal Plus, 2007, 2(2) DOI:10.1128/ecosalplus.3.6.3.11; (b) S. Storbeck, S. Rolfes, E. Raux-Deery, M. J. Warren, D. Jahn and G. Layer, Archaea, 2010, 175050 Search PubMed.
- (a) A. A. DiMarco, T. A. Bobik and R. S. Wolfe, Annu. Rev. Biochem., 1990, 59, 355–394 CrossRef CAS; (b) R. K. Thauer, A.-K. Kaster, M. Goenrich, M. Schick, T. i. Hiromoto and S. Shima, Annu. Rev. Biochem., 2010, 79, 507–536 CrossRef CAS; (c) P. N. Evans, J. A. Boyd, A. O. Leu, B. J. Woodcroft, D. H. Parks, P. Hugenholtz and G. W. Tyson, Nat. Rev. Microbiol., 2019, 17, 219–232 CrossRef CAS.
- (a) J. R. Allen, D. D. Clark, J. G. Krum and S. A. Ensign, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 8432–8437 CrossRef CAS; (b) X. Liu and T. E. Mattes, Appl. Environ. Microbiol., 2016, 82, 3269–3279 CrossRef CAS.
- S. E. Partovi, F. Mus, A. E. Gutknech, H. A. Martinez, B. P. Tripet, B. M. Lange, J. L. DuBois and J. W. Peters, J. Biol. Chem., 2018, 293, 5236–5246 CrossRef CAS.
- J. Chela-Flores, J. Theor. Biol., 1994, 166, 163–166 CrossRef CAS.
- T. O. Diener, Biol. Direct, 2016, 11, 15 CrossRef.
- (a) L. P. Villarreal, Viruses and the Evolution of Life, ASM Press, 2005, ISBN 978-1555813093 CrossRef; (b) P. Forterre and D. Prangishvili, Ann. N. Y. Acad. Sci., 2009, 1178, 65–77 CrossRef CAS; (c) K. Moelling and F. Broecker, Front. Microbiol., 2019, 10, 523 CrossRef; (d) F. Broecker and K. Moelling, Ann. N. Y. Acad. Sci., 2019, 1447, 53–68 CrossRef.
- (a) M. Suzan-Monti, B. La Scola and D. Raoult, Virus Res., 2006, 117, 145–155 CrossRef CAS; (b) B. La Scola, S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J.-M. Claverie and D. Raoult, Science, 2003, 299, 2033 CrossRef CAS; (c) B. La Scola, C. Desnues, I. Pagnier, C. Robert, L. Barrassi, G. Fournous, M. Merchat, M. Suzan-Monti, P. Forterre, E. Koonin and D. Raoult, Nature, 2008, 455, 100–104 CrossRef CAS.
- (a) P. Forterre, Intervirology, 2010, 53, 362–378 CrossRef; (b) A. Nasir and G. Caetano-Anollés, BMC Evol. Biol., 2012, 12, 156 CrossRef CAS; (c) A. Nasir and G. Caetano-Anollés, BMC Evol. Biol., 2015, 156, 1471–2148 Search PubMed; (d) J. Durzynska and A. Gozdzicka-Jozefiak, Virol. J., 2015, 12, 169 CrossRef.
- J. M. Claverie and C. Abergel, Adv. Virus Res., 2013, 85, 25–56 CAS.
- E. V. Koonin and N. Yutin, Adv. Virus Res., 2019, 103, 167–202 CAS.
- (a) E. V. Koonin, Nat. Rev. Microbiol., 2003, 1, 127–136 CrossRef CAS; (b) C. A. Ouzounis, V. Kunin, N. Darzentas and L. A. Goldovsky, Res. Microbiol., 2006, 157, 57–68 CrossRef CAS; (c) L. Kannan, H. Li, B. Rubinstein and A. Mushegian, Biol. Direct, 2013, 8, 32 CrossRef; (d) L. A. Hug, B. J. Baker, K. Anantharaman, C. T. Brown, A. J. Probst, C. J. Castelle, C. N. Butterfield, A. W. Hernsdorf, Y. Amano, K. Ise, Y. Suzuki, N. Dudek, D. A. Relman, K. M. Finstad, R. Amundson, B. C. Thomas and J. F. Banfield, Nat. Microbiol., 2016, 16048 CrossRef CAS; (e) J. L. Yuly, C. E. Lubner, P. Zhang, D. N. Beratan and J. W. Peters, Chem. Commun., 2019, 55, 11823–11832 RSC.
- S. Poudel, E. C. Dunham, M. R. Lindsay, M. J. Amenabar, E. M. Fones, D. R. Colman and E. S. Boyd, Front. Microbiol., 2018, 9, 1762 CrossRef.
- (a) G. Herrmann, E. Jayamani, G. Mai and W. Buckel, J. Bacteriol., 2008, 190, 784–791 CrossRef CAS; (b) W. Buckel and R. K. Thauer, Biochim. Biophys. Acta, 2013, 1827, 94–113 CrossRef CAS; (c) J. W. Peters, A. F. Miller, A. K. Jones, P. W. King and M. W. Adams, Curr. Opin. Chem. Biol., 2016, 31, 146–152 CrossRef CAS; (d) W. Buckel and R. K. Thauer, Front. Microbiol., 2018, 9, 401 CrossRef; (e) J. W. Peters, D. N. Beratan, G. J. Schut and M. W. W. Adams, Chem. Commun., 2018, 54, 4091–4099 RSC; (f) V. Müler, N. P. Chowdury and M. Basen, Annu. Rev. Microbiol., 2018, 72, 331–353 CrossRef; (g) J. L. Yuly, C. E. Lubner, P. Zhang, D. N. Beratan and J. W. Peters, Chem. Commun., 2019, 55, 11823–11832 RSC.
- (a) M. C. Weiss, F. L. Sousa, N. Mrnjavac, S. Neukirchen, M. Roettger, S. Nelson-Sathi and W. F. Martin, Nat. Microbiol., 2016, 16116 CrossRef CAS; (b) J. B. Broderick, B. R. Duffus, K. S. Duschene and E. M. Shepard, Chem. Rev., 2014, 114, 4229–4317 CrossRef CAS; (c) A. Benjdia, C. Balty and O. Bertea, Front. Chem., 2017, 5, 87 CrossRef.
- Martin's bioinformatics analysis lack information on which biosynthetic pathways of coenzymes and cofactors were of relevance in LUCA because nature has developed independent pathways for several of them e.g. for NAD, PLP, TPP and heme.
- (a) S. W. Ragsdale, Ann. N. Y. Acad. Sci., 2008, 1125, 129–136 CrossRef; (b) S. W. Ragsdale and E. Pierce, Biochim. Biophys. Acta, 2008, 1784, 1873–1898 CrossRef CAS.
- G. Fuchs, Annu. Rev. Microbiol., 2011, 65, 631–658 CrossRef CAS.
- A. C. Déclais, J. Marsault, F. Confalonieri, C. B. La Tour de and M. Duguet, J. Biol. Chem., 2000, 275, 19498–19504 CrossRef.
- (a) G. E. Fox, E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen and C. R. Woese, Science, 1980, 209, 457–463 CrossRef CAS; (b) S. Jheeta, Life, 2017, 7, 27 CrossRef.
- S. W. Ragsdale, J. Biol. Chem., 2009, 284, 18571–18575 CrossRef CAS.
- T. Shanmugasundaram and H. G. Wood, J. Biol. Chem., 1992, 261, 897–900 CrossRef.
- (a) C. Petitjean, P. Deschamps, P. López-García and D. Moreira, Genome Biol. Evol., 2015, 7, 191–204 CrossRef; (b) H. Huber and L. Kreuter, The Prokaryotes – Other Major Lineages of Bacteria and the Archaea, ed. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, 2014, vol. 23, pp. 311–318 Search PubMed; (c) C. Brochier, S. Gribaldo, Y. Zivanovic, F. Confalonieri and P. Forterre, Genome Biol., 2005, 6, R42 CrossRef; (d) M. J. Hohn, B. P. Hedlund and H. Huber, Syst. Appl. Microbiol., 2002, 25, 551–554 CrossRef CAS.
- H. Huber, S. Burggraf, T. Mayer, I. Wyschkony, R. Rachel and K. O. Stetter, Int. J. Syst. Evol. Microbiol., 2000, 50, 2093–2100 CrossRef.
- H. Huber, M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer and K. O. Stetter, Nature, 2002, 417, 27–28 CrossRef.
- (a) R. J. Giannone, H. Huber, T. Karpinetsm, T. Heimerl, U. Küper, R. Rachel, M. Keller, R. L. Hettich and M. Podar, PLoS One, 2011, 6, e22942 CrossRef CAS; (b) E. Waters, M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Söll, K. O. Stetter, J. M. Short and M. Noordewier, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12984–12988 CrossRef CAS.
- (a) E. St. John, G. E. Flores, J. Meneghin and A.-L. Reysenbach, Environ. Microbiol. Rep., 2019, 11, 262–270 CrossRef CAS; (b) L. Wurch, R. J. Giannone, B. S. Belisle, C. Swift, S. Utturkar, R. L. Hettich, A.-L. Reysenbach and M. Podar, Nat. Commun., 2016, 7, 12115 CrossRef CAS.
- N. H. Horowitz, Proc. Natl. Acad. Sci. U. S. A., 1945, 31, 153 CrossRef CAS.
- (a) S. L. Miller, Science, 1953, 117, 528–529 CrossRef CAS; (b) S. L. Miller and H. C. Urey, Science, 1959, 130, 245–251 CrossRef CAS.
- (a) F. Tian, J. F. Kasting and K. Zahnle, Earth Planet. Sci. Lett., 2011, 308, 417–423 CrossRef CAS; (b) A. Lazcano and J. L. Bada, Origins Life Evol. Biospheres, 2004, 33, 235–242 CrossRef; (c) J. L. Bada, Chem. Soc. Rev., 2013, 42, 2186–2196 RSC.
- (a) J. L. Bada, Chem. Soc. Rev., 2013, 42, 2186–2196 RSC; (b) E. T. Parker, H. J. Cleaves, J. P. Dworkin, D. P. Glavin, M. Callahan, A. Aubrey, A. Lazcano and J. L. Bada, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5526–5531 CrossRef CAS; (c) E. T. Parker, H. J. Cleaves, M. P. Callahan, J. P. Dworkin, D. P. Glavin, A. Lazcano and J. L. Bada, Origins Life Evol. Biospheres, 2011, 41, 201–212 CrossRef CAS; (d) A. P. Johnson, H. J. Cleaves, J. P. Dworkin, D. P. Glavin, A. Lazcano and J. L. Bada, Science, 2008, 322, 404 CrossRef CAS; (e) H. J. Cleaves, J. H. Chalmers, A. Lazcano, S. L. Miller and J. L. Bada, Origins Life Evol. Biospheres, 2008, 38, 105–115 CrossRef CAS.
- E. Blöchl, M. Keller, G. Wächtershäuser and K. O. Stetter, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 8117–8120 CrossRef.
- (a) C. Huber and G. Wächtershäuser, Science, 1997, 276, 245–247 CrossRef CAS; (b) G. Wächtershäuser and M. W. W. Adams, in Thermophiles: The Keys to Molecular Evolution and the Origin of Life, ed. J. Wiegel, 1998, pp. 47–57 Search PubMed.
- (a) C. Huber and G. Wächtershäuser, Science, 2006, 314, 630–632 CrossRef CAS; (b) C. Huber and G. Wächtershäuser, Tetrahedron Lett., 2003, 44, 1695–1697 CrossRef CAS; (c) C. Huber and G. Wächtershäuser, Science, 1998, 281, 670–672 CrossRef CAS; (d) C. Huber, W. Eisenreich, S. Hecht and G. Wächtershäuser, Science, 2003, 301, 938–940 CrossRef CAS; (e) L. Leman, L. Orgel and M. R. Ghadiri, Science, 2004, 306, 283–286 CrossRef CAS.
- (a) J. Oró, Biochem. Biophys. Res. Commun., 1960, 2, 407–412 CrossRef; (b) J. Oró and A. P. Kimball, Arch. Biochem. Biophys., 1961, 94, 217–227 CrossRef; (c) J. Oró and S. S. Kamat, Nature, 1961, 190, 442–443 CrossRef; (d) Origins of Prebiological Systems and of Their Molecular Matrices, ed. J. Oró and S. W. Fox, Academic Press, New York, 1967, p. 137 Search PubMed.
- (a) N. Biver, D. Bockelée-Morvan, V. Debout, J. Crovisier, J. Boissier, D. Lis, N. D. Russo, R. Moreno, P. Colom and G. Paubert, Astron. Astrophys., 2014, 566, L5 CrossRef; (b) M. Saitta and F. Saija, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 13768–13773 CrossRef; (c) R. Saladino, E. Di Mauro and J. M. García-Ruiz, Chem.–Eur. J., 2019, 25, 3181–3189 CrossRef CAS.
- B. M. Bizzarri, L. Botta, M. I. Pérez-Valverde, R. Saladino, E. Di Mauro and J. M. García-Ruiz, Chem.–Eur. J., 2018, 24, 8126–8132 CrossRef and references cited therein.
- (a) J. D. Sutherland, Angew. Chem., Int. Ed., 2016, 55, 104–121 CrossRef CAS; (b) C. Anastasi, F. F. Buchet, M. A. Crowe, A. L. Parkes, M. W. Powner, J. M. Smith and J. D. Sutherland, Chem. Biodiversity, 2007, 4, 721–739 CrossRef CAS.
- K. Ashe, C. Fernández-García, M. K. Corpinot, A. J. Coggins, D.-K. Burčar and M. W. Powner, Commun. Chem., 2019, 2, 23 CrossRef.
- H. Baltscheffsky, Acta Chem. Scand., 1967, 21, 1973–1974 CrossRef CAS.
- (a) L. J. Leman, L. Orgel and M. R. Ghadiri, J. Am. Chem. Soc., 2006, 128, 20–21 CrossRef CAS; (b) J. P. Biron and R. Pascal, J. Am. Chem. Soc., 2004, 126, 9198–9199 CrossRef CAS; (c) J. P. Biron, A. L. Parkes, R. Pascal and J. D. Sutherland, Angew. Chem., Int. Ed., 2005, 44, 6731–6734 CrossRef CAS.
- L. Leman, L. Orgel and M. R. Ghadiri, Science, 2004, 306, 283–286 CrossRef CAS.
- (a) A. Kirschning, Angew. Chem., 2020 DOI:10.1002/ange.201914786; (b) L. Holliday, J. M. Thornton, A. Marquet, A. G. Smith, F. Rébeillé, R. Mendel, H. L. Schubert, A. D. Lawrence and M. J. Warren, Nat. Prod. Rep., 2007, 24, 972–987 RSC.
- (a) M. J. Dowler, W. D. Fuller, L. E. Orgel and R. A. Sanchez, Science, 1970, 169, 1320–1321 CrossRef CAS; (b) N. Friedmann, S. L. Miller and R. A. Sanchez, Science, 1971, 171, 1026–1027 CrossRef CAS; (c) H. J. Cleaves and S. L. Miller, J. Mol. Evol., 2001, 52, 73–77 CrossRef CAS.
- H. J. Cleaves and S. L. Miller, J. Mol. Evol., 2001, 52, 73–77 CrossRef CAS.
- Abiotic formation of aspartic acid: S. Civis, L. Juha, D. Babankova, J. Cvacka, O. Frank, J. Jehlicka, B. Kralikova, J. Krasa, P. Kubat, A. Muck, M. Pfeifer, J. Skala and J. Ullschmied, Chem. Phys. Lett., 2004, 386, 169–173 CrossRef CAS.
- (a) M. Karki, C. Gibard, S. Bhowmik and R. Krishnamurthy, Life, 2017, 7, 32 CrossRef; (b) N. M. Chung, R. Lohrmann, L. E. Orgel and J. Rabinowitz, Tetrahedron, 1971, 27, 1205–1210 CrossRef CAS.
- (a) D. Bezold, T. Derr, J. Singh and H. J. Jessen, Chem.–Eur. J., 2020, 26, 2298–2308 CrossRef CAS; (b) M. A. Pasek, Chem. Rev., 2020, 120, 4690–4706 CrossRef CAS.
- O. T. Quimby and T. J. Flautt, Z. Anorg. Allg. Chem., 1958, 296, 220–228 CrossRef.
- (a) R. Krishnamurthy, S. Guntha and A. Eschenmoser, Angew. Chem., Int. Ed., 2000, 39, 2281–2285 CrossRef CAS; (b) R. Krishnamurthy, G. Arrhenius and A. Eschenmoser, Origins Life Evol. Biospheres, 1999, 29, 333–354 CrossRef CAS.
- Y. Yamagata, H. Watanabe, M. Saitoh and T. Namba, Nature, 1991, 352, 516–519 CrossRef CAS.
- H. Kim and S. A. Benner, Chem.–Eur. J., 2018, 24, 581–584 CrossRef CAS.
- S. M. Austin and T. G. Waddell, Origins Life Evol. Biospheres, 1999, 29, 287–296 CrossRef CAS.
- J. Xu, D. J. Ritson, S. Ranjan, Z. R. Todd, D. D. Sasselov and J. D. Sutherland, Chem. Commun., 2018, 54, 5566–5569 RSC.
- D. R. Nelson, T. Kamataki, D. J. Waxman, F. P. Guengerich, R. W. Estabrook, R. Feyereisen, F. J. Gonzalez, M. J. Coon, I. C. Gunsalus, O. Gotoh, K. Okuda and D. W. Nebert, DNA Cell Biol., 1993, 12, 1–51 CrossRef CAS.
- G. W. Hodgson and B. L. Baker, Nature, 1967, 216, 29–32 CrossRef CAS.
- (a) S. Fox and H. Strasdeit, Astrobiology, 2013, 13, 578–595 CrossRef CAS; (b) H. L. Pleyer, H. Strasdeit and S. Fox, Origins Life Evol. Biospheres, 2018, 48, 347–371 CrossRef CAS.
- (a) J. S. Lindsey, M. Ptaszek and M. Taniguchi, Origins Life Evol. Biospheres, 2009, 39, 495–515 CrossRef CAS; (b) J. S. Lindsey, V. Chandrashaker, M. Taniguchi and M. Ptaszek, New J. Chem., 2011, 35, 65–75 RSC; (c) A. R. M. Soares, M. Taniguchi, V. Chandrashaker and J. S. Lindsey, Chem. Sci., 2012, 3, 1963–1974 RSC; (d) A. R. M. Soares, M. Taniguchi, V. Chandrashaker and J. S. Lindsey, New J. Chem., 2013, 37, 1073–1086 RSC; (e) A. R. M. Soares, D. R. Anderson, V. Chandrashaker and J. S. Lindsey, New J. Chem., 2013, 37, 2716–2732 RSC.
- S. L. Miller and G. Schlesinger, J. Mol. Evol., 1993, 36, 302–307 CAS.
- F. Raulin and C. Frère, Grana, 1991, 30, 510–513 CrossRef.
- W. H. Breckenridge and H. Taube, J. Chem. Phys., 1970, 53, 1750–1767 CrossRef CAS.
- (a) G. J. Connell and E. L. Christian, Origins Life, 1993, 23, 291–297 CrossRef CAS; (b) R. R. Breaker and G. F. Joyce, J. Mol. Evol., 1995, 40, 551–558 CrossRef CAS.
- V. R. Jadhav and M. Yarus, Biochimie, 2002, 84, 877–888 CrossRef CAS.
- (a) W. Winkler, A. Nahvi and R. R. Breaker, Nature, 2002, 419, 952–955 CrossRef CAS; (b) M. T. Croft, M. Moulin, M. E. Webb and A. G. Smith, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20770–20775 CrossRef CAS; (c) A. Serganov, L. Huang and D. J. Patel, Nature, 2009, 458, 233–237 CrossRef CAS; (d) D. B. Pedrolli, A. Matern, J. Wang, M. Ester, K. Siedler, R. Breaker and M. Mack, Nucleic Acids Res., 2012, 40, 8662–8673 CrossRef CAS; (e) F. J. Grundy and T. M. Henkin, Mol. Microbiol., 1989, 30, 737–749 CrossRef; (f) W. C. Winkler, A. Nahvi, N. Sudarsan, J. E. Barrick and R. R. Breaker, Nat. Struct. Biol., 2003, 10, 701–707 CrossRef CAS; (g) T. D. Ames, D. A. Rodionov, Z. Weinberg and R. R. Breaker, Chem. Biol., 2010, 17, 681–685 CrossRef CAS; (h) E. E. Regulski, R. H. Moy, Z. Weinberg, J. E. Barrick, Z. Yao, W. L. Ruzzo and R. R. Breaker, Mol. Microbiol., 2008, 68, 918–932 CrossRef CAS.
- (a) P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, RNA, 2017, 23, 995–1011 CrossRef CAS; (b) A. V. Sherwood and T. M. Henkin, Annu. Rev. Microbiol., 2016, 70, 361–374 CrossRef CAS; (c) A. Serganov and D. J. Patel, Annu. Rev. Biophys., 2012, 41, 343–370 CrossRef CAS; (d) A. Serganov, A. Polonskaia, A. Tuan Phan, R. R. Breaker and D. J. Patel, Nature, 2006, 441, 1167–1171 CrossRef CAS.
- J. E. Stewart, Found. Sci., 2019, 24, 171–195 CrossRef.
- (a) M. Frenkel-Pinter, M. Samanta, G. Ashkenasy and L. J. Leman, Chem. Rev., 2020, 120, 4707–4765 CrossRef CAS; (b) R. D. Firn and C. G. Jones, Nat. Prod. Rep., 2003, 20, 382–391 RSC.
- T. Nunoura, Y. Chikaraishi, R. Izaki, T. Suwa, T. Sato, T. Harada, K. Mori, Y. Kato, M. Miyazaki, S. Shimamura, K. Yanagawa, A. Shuto, N. Ohkouchi, N. Fujita, Y. Takaki, H. Atomi and K. Tak, Science, 2018, 359, 559–563 CrossRef CAS.
- (a) D. Kroiss, G. Ashkenasy, A. B. Braunschweig, T. Tuttle and R. V. Ulijn, Chem, 2019, 5, 1917–1920 CrossRef CAS; (b) S. Islam and M. W. Powner, Chem, 2017, 2, 470–501 CrossRef CAS.
- J. H. Munson-McGee, C. Rooney and M. J. Young, J. Virol., 2020, 94, e01213-1–9 Search PubMed.
- A. A. Sharov, BioSystems, 2016, 144, 8–17 CrossRef CAS.
- E. V. Koonin and V. V. Dolja, Curr. Opin. Virol., 2013, 3, 546–557 CrossRef.
- S. Tsukiji, S. B. Pattnaik and H. Suga, J. Am. Chem. Soc., 2004, 126, 5044–5045 CrossRef CAS.
- N. Kovalskaya and R. W. Hammond, Plant Sci., 2014, 228, 48–60 CrossRef CAS.
- T. O. Diener, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 9370–9374 CrossRef CAS.
- R. Flores, S. Gago-Zachert, P. Serra, R. Sanjuán and S. F. Elena, Annu. Rev. Microbiol., 2014, 68, 395–414 CrossRef CAS.
- The conflict between cells and viruses has been (and still is) the major engine of life evolution.
- The occurence of lipoids (fatty acids or isoprenoids) is not discussed in this overview.
- D. Lundin, G. Berggren, D. T. Logan and B.-M. Sjöberg, Life, 2015, 5, 604–636 CrossRef CAS.
- Iron–sulfur clusters 7 are regarded to have been involved in the formation of biological catalysis on the early Earth, and plausible pathways under prebiotic conditions have recently been identified; (a) A. D. Tsaousis, Front. Microbiol., 2019, 10, 2478 CrossRef; (b) C. Bonfio, L. Valer, S. Scintilla, S. Shah, D. J. Evans, L. Jin, J. W. Szostak, D. D. Sasselov, J. D. Sutherland and S. S. Mansy, Nat. Chem., 2017, 9, 1229–1234 CrossRef CAS; (c) Related studies: M. Dörr, J. Käßbohrer, R. Grunert, G. Kreisel, W. A. Brand, R. A. Werner, H. Geilmann, C. Apfel, C. Robl and W. Weigand, Angew. Chem., Int. Ed., 2003, 42, 1540–1543 CrossRef.
- “The origin of Life cannot be discovered, it has to be reinvented” in A. Eschenmoser, Tetrahedron, 2007, 63, 12821–12844 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |