Fungal benzene carbaldehydes: occurrence, structural diversity, activities and biosynthesis
Huomiao
Ran
and
Shu-Ming
Li
 *
*
Institut für Pharmazeutische Biologie und Biotechnologie, Fachbereich Pharmazie, Philipps-Universität Marburg, Robert-Koch-Straße 4, 35037 Marburg, Germany. E-mail: shuming.li@staff.uni-marburg.de
First published on 11th August 2020
Abstract
Covering: up to April 2020
Fungal benzene carbaldehydes with salicylaldehydes as predominant representatives carry usually hydroxyl groups, prenyl moieties and alkyl side chains. They are found in both basidiomycetes and ascomycetes as key intermediates or end products of various biosynthetic pathways and exhibit diverse biological and pharmacological activities. The skeletons of the benzene carbaldehydes are usually derived from polyketide pathways catalysed by iterative fungal polyketide synthases. The aldehyde groups are formed by direct PKS releasing, reduction of benzoic acids or oxidation of benzyl alcohols.
1. Introduction
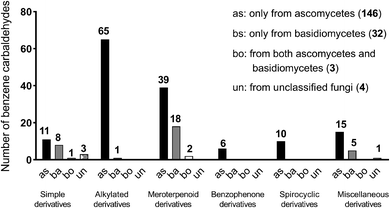
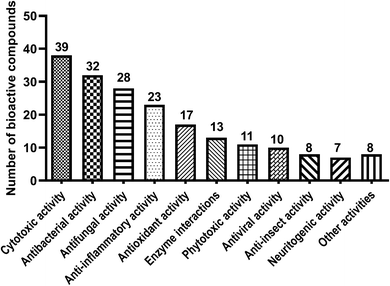
Benzene carbaldehydes, from the simplest benzaldehyde to structural features in relatively complex molecules, are widely distributed in ascomycetes and also found in basidiomycetes (Table 1 and Fig. 1). Their producers include terrestrial, sponge-associated, marine- and mangrove-derived, plant endophytic and pathogenic fungi. The compounds from this family exhibit diverse biological and pharmacological properties. Cytotoxic, antibacterial and antifungal activities have been detected for a large number of benzene carbaldehydes, followed by anti-inflammatory and antioxidant activities (Table 2 and Fig. 2). Since the first report on flavoglaucin and auroglaucin in the fungus Aspergillus glaucus in 1934,1 at least 185 structures including 36 alkylated, 59 prenylated (meroterpenoids) and 30 both alkylated and prenylated derivatives have been described in the literature. 146 of them were isolated from ascomycetes, 32 from basidiomycetes and only three from both ascomycetes and basidiomycetes (Fig. 1). Aspergillus strains with 49 metabolites are clearly the dominant producers of benzene carbaldehydes, followed by Pestalotiopsis, Stachybotrys and Penicillium with 14, 13 and 13 metabolites, respectively (Table 1). Reports on the elucidation of their biosynthetic pathways in fungi have accumulated tremendously in recent years, especially on the backbone assembly by iterative polyketide synthases and the formation of the aldehyde group via different routes. The benzene carbaldehydes act as critical intermediates or end products of various biosynthetic pathways. Furthermore, key pathway-specific enzymes have also been characterized. Up to April 2020, more than 140 publications deal with the producers, isolation and structural elucidation, biological activities and applications as well as biosynthetic origin and pathways of benzene carbaldehydes. However, no systematic review on this natural product family is available in the literature. Therefore, we summarize these data in the present review to fill this gap.| Fungal genera | Simple derivatives | Alkylated derivatives | Meroterpenoid derivatives | Benzophenone derivatives | Spirocyclic derivatives | Miscellaneous derivatives | Total |
|---|---|---|---|---|---|---|---|
| Ascomycetes | |||||||
| Aspergillus | 6 | 29 | 4 | 1 | 7 | 2 | 49 |
| Pestalotiopsis | — | 10 | 1 | — | — | 3 | 14 |
| Stachybotrys | — | — | 13 | — | — | — | 13 |
| Penicillium | 4 | 5 | 1 | 3 | — | 13 | |
| Acremonium | — | — | 8 | — | — | — | 8 |
| Fusarium | — | — | 7 | — | — | — | 7 |
| Torrubiella | — | — | 7 | — | — | — | 7 |
| Colletotrichum | — | — | 6 | — | — | — | 6 |
| Paraphaeosphaeria | — | 6 | — | — | — | — | 6 |
| Pyricularia | — | 5 | — | — | — | 1 | 6 |
| Trichoderma | — | 5 | — | — | — | — | 5 |
| Diaporthe | — | — | — | — | — | 4 | 4 |
| Epicoccum | 1 | — | — | — | — | 2 | 3 |
| Neonectria | — | — | 3 | — | — | — | 3 |
| Chaetomium | 1 | 2 | — | — | — | — | 3 |
| Ascochyta | — | 1 | — | — | 1 | 1 | 3 |
| Daldinia | — | — | — | 2 | — | — | 2 |
| Hymenoscyphus | — | 2 | — | — | — | — | 2 |
| Lasiodiplodia | — | — | — | — | — | 2 | 2 |
| Pestalotia | — | — | — | 2 | — | — | 2 |
| Pyrenula | — | 2 | — | — | — | — | 2 |
| Nalanthamala | — | — | 2 | — | — | — | 2 |
| Zopfiella | — | 2 | — | — | — | — | 2 |
| Amniculicola | — | 1 | — | — | — | — | 1 |
| Cordyceps | — | 1 | — | — | — | — | 1 |
| Diplodia | 1 | — | — | — | — | — | 1 |
| Gelasinospora | — | 1 | — | — | — | — | 1 |
| Phaeomoniella | 1 | — | — | — | — | — | 1 |
| Phoma | 1 | — | — | — | — | — | 1 |
| Sordaria | — | 1 | — | — | — | — | 1 |
| Seiridium | 1 | — | — | — | — | — | 1 |
| Talaromyces | 1 | — | — | — | — | — | 1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Basidiomycestes | |||||||
| Hericium | — | — | 7 | — | — | — | 7 |
| Heterobasidion | — | — | 4 | — | — | — | 4 |
| Albatrellus | — | — | 3 | — | — | — | 3 |
| Bjerkandera | 3 | — | — | — | — | — | 3 |
| Stereum | — | — | 3 | — | — | — | 3 |
| Bondarzewia | — | 1 | 1 | — | — | — | 2 |
| Clitocybe | — | — | — | — | — | 2 | 2 |
| Gloeophyllum | 2 | — | — | — | — | — | 2 |
| Peniophora | 2 | — | — | — | — | — | 2 |
| Sarcodontia | — | — | — | — | — | 2 | 2 |
| Tyromyces | 2 | — | — | — | — | — | 2 |
| Russula | — | — | 2 | — | — | — | 2 |
| Agrocybe | 1 | — | — | — | — | — | 1 |
| Fomitiporia | — | — | — | — | — | 1 | 1 |
| Phlebiopsis | 1 | — | — | — | — | — | 1 |
| Substance class | Biological activities | Compounds |
|---|---|---|
| Simple derivatives | Antiviral activity | — |
| Antifungal activity |
1,1554,1569,1216,2219,2521,2722,2823![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28 28 |
|
| Antibacterial activity |
1,1554,15716,1719![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 25 |
|
| Antioxidant activity |
1,1552,15816![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 19 |
|
| Anti-inflammatory activity |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and 3 8 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 159 159 |
|
| Anti-insect activity |
1,1554,16020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26 26 |
|
| Phytotoxic activity |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
|
| Cytotoxic activity |
4,1619,1310,1416![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 18 |
|
| Enzyme inhibitors, activators and receptors |
2,1623,16316![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 19 |
|
| Anti-nociceptive activity |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
|
| Anti-angiogenic activity |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
|
| Positive modulation of GABAergic neuromodulation |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 158 158 |
|
| Alkylated derivatives | Antiviral activity |
53,4979,4980![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 49 49 |
| Antifungal activity |
24–29,3141,3542,3546,4682![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
|
| Antibacterial activity |
32,3541–44,3546,4656,5657,5660,55,5662,5679,4981,6482,6489![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
|
| Antioxidant activity |
30,3256,53,5457,53,5458,5360,5362,53,5473,5375,5276![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 52 |
|
| Anti-inflammatory activity |
33,3634,3756,36,56,5757,56,5760,5662,5670,3771,3775![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36,56 36,56 |
|
| Anti-insect activity |
47–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 47 |
|
| Phytotoxic activity |
41,4449,16451![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 48 |
|
| Cytotoxic activity |
28,3132,3541–44,3552,3554,5073,6278,6379,4980,4981,6482,6484–86,6989![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
|
| Enzyme inhibitors, activators and receptors |
45,4556,5863![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58 58 |
|
| Immunomodulatory activity |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
|
| Antifouling activity |
36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
|
| Antimalarial activity |
85,6986![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 69 69 |
|
| Meroterpenoid derivatives | Antiviral activity |
119,87134,92135,92137,93138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 93 93 |
| Antifungal activity |
98,7999,79112,79126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79 79 |
|
| Antibacterial activity |
98,7999,79112,91113,91126,91127,78,79,91131,91132,91133,78136,91146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 95 |
|
| Antioxidant activity | — | |
| Anti-inflammatory activity |
126,87127,87132,87136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 87 and 141 87 and 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 87 87 |
|
| Anti-insect activity | — | |
| Phytotoxic activity |
90,7695,7797,3898,7999![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79 79 |
|
| Cytotoxic activity |
97,3899,78127,78129,92131–134,92136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 92 92 |
|
| Enzyme inhibitors, activators and receptors |
98,7899,78114,85127,78134,78133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78 |
|
| Neuritogenic activity |
106–111,83,84116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 86 86 |
|
| Benzophenone | Antibacterial activity |
153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| Enzyme inhibitors, activators and receptors |
154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 101 101 |
|
| Anti-inflammatory activity |
149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 97 and 150 97 and 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 97 97 |
|
| Cytotoxic activity |
153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
|
| Spirocyclic derivatives | Antioxidant activity |
155,104157–160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 105 |
| Cytotoxic activity |
160,106162–164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
|
| Miscellaneous derivatives | Antiviral activity |
169![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 49 and 171 49 and 171![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34 34 |
| Antifungal activity |
165,108166,108179,113180,113182,21183![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
|
| Antibacterial activity |
167,109182,21183![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
|
| Anti-inflammatory activity |
171,34175–178![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112 112 |
|
| Anti-insect activity |
168![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 47 |
|
| Phytotoxic activity |
168![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 and 170 48 and 170![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
|
| Cytotoxic activity |
179,113180,113181![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 114 114 |
2. Occurrence, biological and pharmacological activities
2.1. Simple benzene carbaldehydes
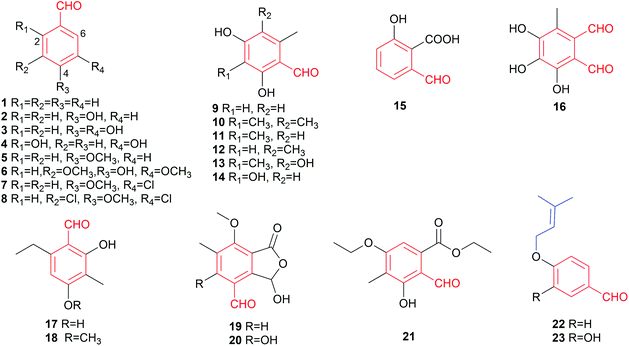
In this review, merely slightly modified like hydroxylated, halogenated, methylated and/or ethylated benzaldehydes are classified as simple benzene carbaldehydes accounting for 23 members (Fig. 3). Despite their simple structures, these compounds also exhibit broad biological and pharmacological activities such as antifungal (eight compounds), antibacterial and cytotoxic activities (Table 2). Hydroxylated and methoxylated simple benzaldehydes are also natural products of plant origin.2 Eleven simple fungal benzene carbaldehydes were isolated from ascomycetes and eight from basidiomycetes (Fig. 1). The main producers are members of the genera Aspergillus, Penicillium and Bjerkandera (Table 1).The simplest member of this family is benzaldehyde 1 without other additional substituents. It is one of the most industrial used chemicals and can be found as a preservative in cosmetics and food as well as in personal care and select car detailing products. Its 4-hydroxylated derivative 2 was identified in plants2 and a wide range of fungi such as the plant pathogens Botryosphaeria obtusa3 and Phaeoacremonium chlamydosporum,4 the endophytic fungi Aspergillus sp. YL-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and Penicillium thiomii6 as well as the brown-rot fungi Tyromyces palustris and Gloeophyllum trabeum.7 In addition to phytotoxicity,32 also possesses anti-angiogenic,8 anti-inflammatory8 and anti-nociceptive8 activities.
5 and Penicillium thiomii6 as well as the brown-rot fungi Tyromyces palustris and Gloeophyllum trabeum.7 In addition to phytotoxicity,32 also possesses anti-angiogenic,8 anti-inflammatory8 and anti-nociceptive8 activities.
Compounds 3–8 are hydroxylated, methoxylated or chlorinated benzaldehyde derivatives. The dihydroxylated benzaldehyde, protocatechuic aldehyde 3, was identified in the aforementioned brown-rot basidiomycetes T. palustris and G. trabeum.7 2,5-Dihydroxylated benzaldehyde 4 and 6-formylsalicylic acid 15 were metabolites of Penicillium patulum.9 Biosynthetic study on the white-rot fungus Bjerkandera adusta led to the identification of compounds 1, 5, 7 and 8.10 Another congener syringaldehyde 6 was obtained from the plant endophytic fungus Phoma sp. YN02-P-3.11
To understand the preventive mechanism of the root rot biocontrol fungus Phlebiopsis gigantea, its chemical constituents were investigated, leading to identification of o-orsellinaldehyde 9 with inhibitory activity against the pathogenic fungi Heterobasidion occidentale and Fusarium oxysporum, and the saprotrophic fungus Penicillium canescens12 as well as cytotoxic activity against the human carcinoma cell line Hep 3B and the lung fibroblast cell line MRC-5.13 The highly decorated and cytotoxic 2,4-dihydroxy-3,5,6-trimethylbenzaldehyde 10 was obtained from the deep sea-derived fungus Aspergillus sydowi.14o-Orsellinaldehyde derivatives 11–14 were identified as biosynthetic precursors in genetically manipulated fungal strains.15,16 The dialdehyde flavipin 16 from Aspergillus,17Chaetomium18,19 and Epicoccum20,21 species was well documented for its antibacterial,17 antifungal,22 antiproliferative18 and antioxidant19 activities as well as inhibitory effect on α-glucosidase, even more potential than the clinically used drug acarbose.19
Benzene carbaldehydes 17 and 18 carrying ethyl groups were isolated from a marine mangrove endophytic fungus.23 Gladiolic acid 19 from Penicillium gladioli24,25 and cyclopaldic acid 20 from Seiridium cupressi26 are hemiacetal lactones and differ from each other just in a hydroxyl group. Chemical investigation on a co-culture broth extract of two marine mangrove pathogenic fungi led to the isolation of the hydroxylated benzaldehyde 21 with both ethyl ether and ester bonds.27 The two antifungal benzene carbaldehydes 22 and 23 with O-prenyl moieties have been isolated from Peniophora polygonia and were demonstrated to strongly inhibit the growth of the aspen decay fungus Phellinus tremulae.28
2.2. Alkylated benzene carbaldehydes
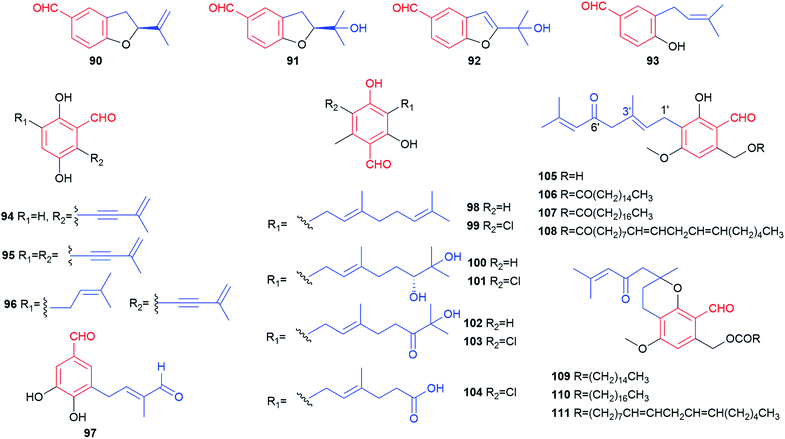
Alkylated derivatives with 66 structures, i.e. more than one-third of the known benzene carbaldehydes, constitute one of the largest classes. In comparison to the simple benzene carbaldehydes, members from this class contain an additional unmodified or modified alkyl chain, which is attached in most cases (94%) to the ortho-position of the formyl group. With the exception for 35 from the basidiomycete Bondarzewia montana, all these fungal products are salicylaldehyde derivatives from ascomycetes (Fig. 1). Their main producers belong to the genera Aspergillus and Pestalotiopsis with 29 and 10 metabolites, respectively (Table 1 and Fig. 1). In addition to their main activities like antibacterial, antifungal and cytotoxic activities, most group members also exhibit anti-inflammatory and antioxidant effects, which were observed only for few members from other classes (Table 2).Biosynthetically, alkylated benzene carbaldehydes are derivatives of aromatic polyketides with different numbers of malonyl-CoA as extension units.29,30 Their alkyl chains differ consequently from each other by numbers of C2 units. Thus, the members of this class can be conveniently subdivided according to the length of the side chains, i.e. C3-, C5-, C7-, C9- and C11-alkylated benzene carbaldehydes.
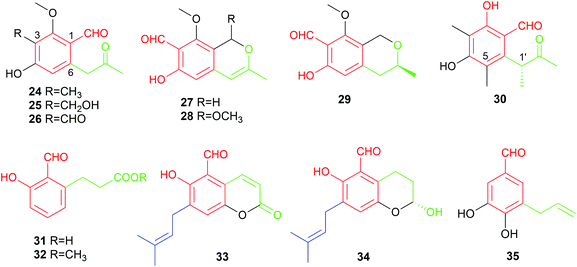
The only basidiomycete-derived metabolite in this group is the dihydroxylated aldehyde 35 from the rare white-rot fungus Bondarzewia montana.38 An alkenyl substitution at the meta-position to the formyl group differs clearly from the ortho-position of other members 24–34 from ascomycetes.
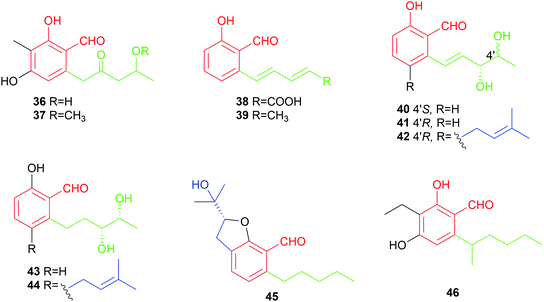
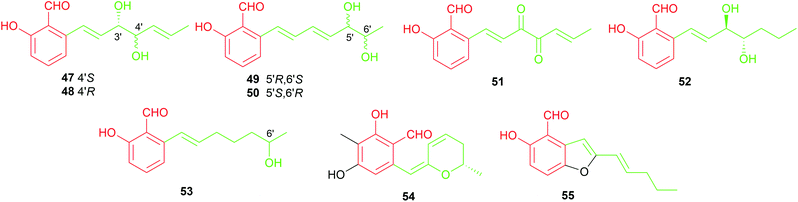
Four salicylaldehydes with a dihydroxyheptyl moiety 47–50 and their oxidised dicarbonyl derivative 51 were obtained from the rice pathogen Magnaporthe grisea.47,48 Two similar metabolites, heterocornol B 52 and pestalol D 53, were isolated from Pestalotiopsis heterocornis and Pestalotiopsis sp. AcBC2, respectively.35,49 Ginsenocin 54 with a substituted 2H-pyran ring resulted from cyclisation on the C7-alkylatd chain was identified as an anti-tumour metabolite in the endophytic fungus Penicillium melinii Yuan-25.50 It shows potent cytotoxicity with IC50 values ranging from 0.49 to 5.03 μg mL−1 to six cell lines including MKN45, LOVO, A549, MDA-MB-435, HepG2 and HL-60. Pyrenulafuran 55, a 2H-benzofuran derivative, was isolated from the cultured lichen mycobionts of Pyrenula sp.51
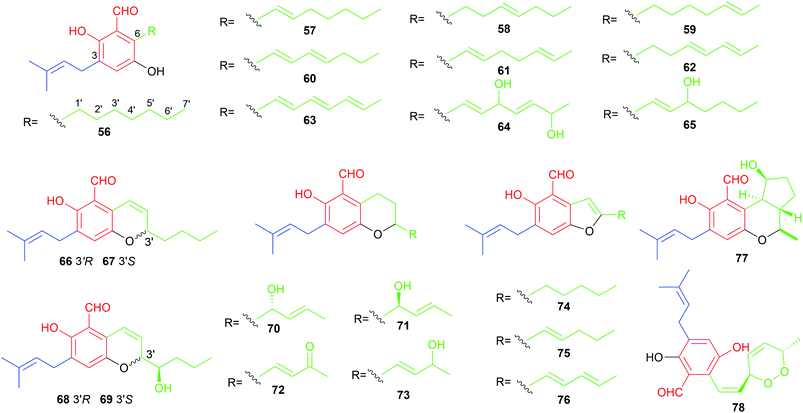
The majority of the C7-alkylated benzene carbaldehydes are 3,6-dihydroxybenzaldehydes with a dimethylallyl moiety at C3 (56–78, Fig. 7). These compounds belong to the groups of flavoglaucins and auroglaucins and were obtained from different Aspergillus/Eurotium species including several mangrove-derived strains. One of the notable features is the presence of a complete saturated (56) or unsaturated (57–63) C7-alkyl chains at C6 of the benzene ring. This set of compounds show broad bioactivities e.g. antioxidant,52–54 antibacterial55,56 and anti-inflammatory activities56,57 as well as binding affinity to human opioid or cannabinoid receptors.58
In the cases of 64 and 65, the alkyl residues are further modified by hydroxylation. Compound 64 with a 3′,6′-dihydroxyhepta-1′,4′-dienyl moiety was identified in the fruit-associated fungus Aspergillus amstelodami.55 The C3′-hydroxylated analogue 65 was isolated from the gorgonian-derived fungus Eurotium sp.59
The alkyl chain has cyclised with the C5-hydroxyl group to a 2H-benzopyran in 66–69, to a dihydrobenzopyran in 70–73, and to a benzofuran ring in 74–76, respectively. A spontaneous intramolecular cyclisation of 65 to an enantiomer pair 66/67 with a 2H-chromene skeleton was observed when it was dissolved in CDCl3.60 Their derivatives 68 and 69 with an additional C4′ hydroxylated group were identified in a gorgonian-derived fungus Eurotium sp. as well.60 Two chromane-5-carbaldehyde isomers, 70 and 71, with opposite configurations of the C4′ hydroxyl group, were characterized from the Antarctic marine-derived fungus Aspergillus sp. SF-5976 and proven to have anti-inflammatory activity.37 Investigation on the chemical constituents of the mangrove endophytic fungus Eurotium rubrum led to the identification of compounds 72–76 and eurotirumin 77 with a cyclopentabenzopyran ring system.61 Among them, chaetopyranin 73 exhibits cytotoxic activity toward several tumour cell lines,62 while compounds 75 and 76 show antioxidant activity.52 The anti-proliferative prenylated benzene carbaldehyde 78 with a rare endo peroxide bond was isolated from the mangrove-derived fungus Aspergillus sp. AV-2.63
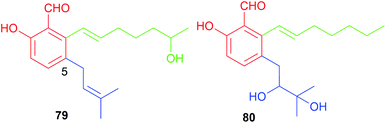
Two rare examples of C5-prenylated and C7-alkylated salicylaldehydes, pestalols B 79 and C 80 (Fig. 8), were obtained from the mangrove endophytic fungus Pestalotiopsis sp. AcBC2 and show stronger anti-influenza virus activity than the non-prenylated precursor 53.49 The dimethylallyl moiety in 79 was further modified by adjunction of two hydroxyl groups in 80.
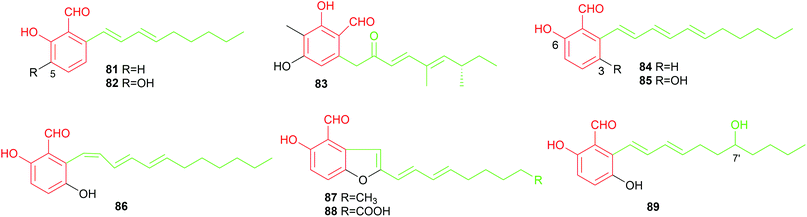
Two reports described the identification of C11-alkylated benzene carbaldehydes. Bioassay-guided constituent investigation of the wood-decay fungus, Hypocrea (syn. Trichoderma) sp. BCC 14122, resulted in the isolation of the C11-alkylated salicylaldehyde 84, gentisaldehyde 85, its isomer 86 with a cis-configured double bond and two benzofuran derivatives 87 and 88.6985 with an additional phenolic hydroxyl group shows stronger cytotoxicity against tumour cell lines KB, BC and NCI-H187 than its non-hydroxylated analogue 84. In 2018, a C7′-hydroxylated congener 89 was obtained from the marine-derived fungus Zopfiella marina. It shows antibacterial activities against Mycobacterium tuberculosis and Bacillus cereus.40
In summary, alkylated benzene carbaldehydes with 66 members contribute not only significantly to the structural diversity, but also to the broad biological activities. They exhibit all the described activities for benzene carbaldehydes with antibacterial, antioxidant, anti-inflammatory and cytotoxic activities as their remarkable features (Table 2).
2.3. Meroterpenoids
Meroterpenoids are hybrid natural products of terpene and other pathways.70 They generally contain a start molecule from the polyketide, alkaloid or shikimate pathway, which is connected with a prenyl moiety of various chain lengths. The attachment of the prenyl moiety to different core structures is usually catalysed by prenyltransferases.71 Several related reviews on meroterpenoids have been published previously.70–74Since the first report on benzaldehyde-containing meroterpenoids by Ellestad et al. in 1969,75 at least 86 metabolites from this class have been isolated from fungal strains. These include structures carrying a dimethylallyl moiety already discussed above, e.g. the simple aldehydes 22 and 23 (2.1), the C3-alkylated benzene carbaldehydes 34 and 35 (2.2.1) and the C7-alkylated derivatives 56–80 (2.2.3).
Therefore, meroterpenoids belong to one of the major benzene carbaldehyde classes and contribute significantly to the structural diversity of these natural products. More than 30% of the mentioned products were isolated from basidiomycetes and 66% from ascomycetes (Fig. 1). The majority of the fungal meroterpenoids have a C5, C10 or C15 terpenoid chain, which is usually connected to meta-position of the formyl group and ortho-position of at least one hydroxyl group or structural feature derived thereof.
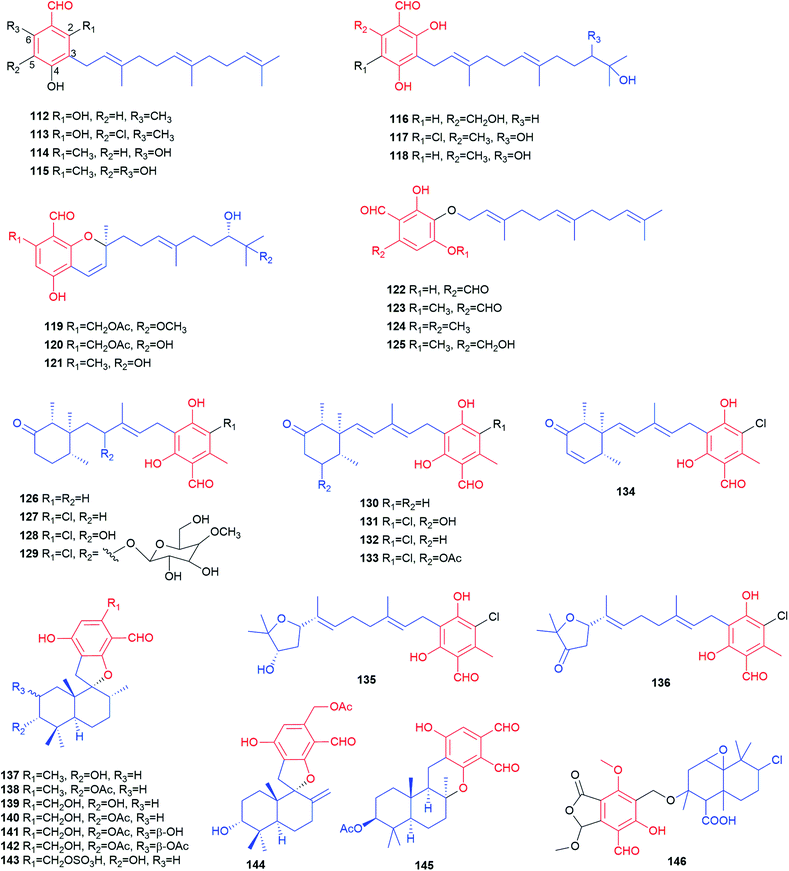
Representatives of the geranylated (C10) meroterpenoids are colletorin B 98 and its chlorinated derivative colletochlorin B 99. They were obtained from several fungi like Nectria galligena,78Fusarium sp.79 and Cephalosporium diospyri.8098 and 99 display moderate herbicidal, antifungal and antibacterial activities against Chlorella fusca, Ustilago violacea and Fusarium oxysporum as well as Bacillus megaterium, respectively.79 They are also regarded as potential drugs for the treatment of Alzheimer's disease due to the inhibitory activities towards β-glucuronidase and acetylcholinesterase (AChE).78 Five structurally-related metabolites with a modified geranyl residue (100–104) were isolated from Colletotrichum nicotianae. Colletochlorins A 101 and C 103 are chlorinated derivatives of colletorins A 100 and C 102, respectively.81 Phytotoxicity tests against Ambrosia artemisifolia and Sonchus arvensis with 100 and 101 as well as their analogues indicated the importance of the stereochemistry at the hydroxylated geranyl chain and the enhancing effect of chlorination.82 Six fatty acid esters hericenones C–H 106–111 bearing a 6′-carbonyl geranyl moiety, were isolated, together with their proposed precursor 105, from the edible mushroom Hericium erinaceum.83,84109–111 can be considered as cyclisation products of 106–108, respectively.
During a screening programme for interacting agents with mammalian CNS receptors, three farnesylated benzene carbaldehydes, LL-Z1272 β 112, ovinal 114 and scutigeral 115, were isolated from an extract of the edible mushroom Albatrellus ovinus by bioassay-guided fractionation of the crude extracts.85 The antibiotic LL-Z1272 α 113, a chlorinated derivative of 112, was isolated from the ascomycete Fusarium sp.75
Three benzene carbaldehydes 116–118 bear a hydroxylated farnesyl moiety. Parvisporin 116 was isolated from the culture broth of Stachybotrys parvispora F4708 and demonstrated to have a weak neuritogenic activity.86 Its analogues chlorocylindrocarpol 117 and cylindrocarpol 118 were later obtained from the sponge-derived fungus Acremonium sp.87 Recently, stachybonoids A–C 119–121, with a benzopyran ring after cyclisation of the farnesyl chain with the salicylic hydroxyl group, were isolated from the crinoid-derived fungus Stachybotrys chartarum 952.88 Compound 119 exhibits an inhibitory activity against the replication of dengue virus.
Asperugin B 122 and A 123, two phthalaldehydes, carrying an intact O-farnesyl moiety were identified as metabolites of a mutated strain of Aspergillus rugulosus.89 Their derivatives 124 and 125 were obtained from a genetically engineered A. nidulans strain as biosynthetic intermediates of aspernidine A 186 (see 3.1.1. for details).90
Structurally, 126–134 are meroterpenoids with a substituted cyclohexone ring by cyclisation within a modified farnesyl chain. To counter antibiotic-resistance bacteria, Mogi et al. screened hundreds of natural products and identified a unique set of active natural products LL-Z1272 β 112, ε 126, δ 127, γ 132, ζ 133, which were isolated originally from Fusarium sp.75,91 Chlorination determines the biological activities. The non-chlorinated derivatives 112 and 126 are active against cytochrome bd, while the chlorinated derivatives 127, 132 and 133 are potent inhibitors of cytochrome bo and trypanosome alternative oxidase.91126 shows very strong antifungal activity against Eurotium repens.79 Compounds 127 and 133 display moderate inhibitory activity towards the enzymes AChE and β-glucuronidase as well as toxicity towards human lung fibroblasts.78
Chemical investigation of the bioactive metabolites in the pathogenic fungus Verticillium hemipterigenum and the sponge-derived fungus Acremonium sp. led to the isolation of deacetylchloronectrin 128,87,92 the glycoside vertihemipterin A 129,92 cylindrol B 130,87 8′-hydroxyascochlorin 131,92 compounds 132 and 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 87,92 as well as ilicicolin E 134.92 Compounds 129 and 131–134 possess remarkable cytotoxicity to several cell lines such as KB, BC-1, NCI-H187 and vero with IC50 values ranging from 0.36 to 19 μg mL−1.92 Two meroterpenoids with a tetrahydrofuran ring at the modified farnesyl chain, ascofuranol 135 and ascofuranone 136, were obtained also from the fungi Verticillium hemipterigenum and Acremonium sp.87,92135 has antiviral potential and cytotoxicity,91 while 136 shows significant anti-inflammatory activity.87
87,92 as well as ilicicolin E 134.92 Compounds 129 and 131–134 possess remarkable cytotoxicity to several cell lines such as KB, BC-1, NCI-H187 and vero with IC50 values ranging from 0.36 to 19 μg mL−1.92 Two meroterpenoids with a tetrahydrofuran ring at the modified farnesyl chain, ascofuranol 135 and ascofuranone 136, were obtained also from the fungi Verticillium hemipterigenum and Acremonium sp.87,92135 has antiviral potential and cytotoxicity,91 while 136 shows significant anti-inflammatory activity.87
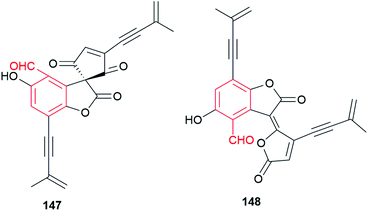
Eight phenylspirodrimane derivatives 137–144 were identified in the fungus Stachybotrys chartarum.88,93 In their structures, a decahydronaphthalene ring system and a fused spiroketal feature are formed within the farnesyl chain. Stachybonoid A 141 exhibits moderate anti-inflammatory activity by inhibiting the production of nitric oxide in lipopolysaccharide-activated RAW264.7 cells with an IC50 value of 27.2 μM.88 Stachybotrysins A 137 and B 138 display antiviral activity.93 Kampanol C 145, a pentacyclic meroterpenoid, was obtained from Stachybotrys kampalensis Hansf.94 Dicarbaldehyde backbone makes it extremely unstable in CD2Cl2, but reasonable stable in acetone. Pestalotiopen A 146 was isolated from the Chinese mangrove-endophytic fungus Pestalotiopsis sp. as an ether of altiloxin A derived from a farnesyl moiety and a highly substituted benzene carbaldehyde (cyclopaldic acid). It shows moderate antibacterial activity against Enterococcus faecalis.95 The rare acetylenic spirodioxolactone ochroleucin A1147 was obtained from the mushrooms Russula ochroleuca and R. viscida after treatment with aqueous KOH. The labile chromogen undergoes easily rearrangement into the isomeric dilactone ochroleucin A2148 (Fig. 12).96
Taking together, meroterpenoid benzene carbaldehydes contain usually C5-, C10-, C15-prenyl moiety or structures derived thereof. Antiviral, antifungal, antibacterial, phytotoxic and cytotoxic activities were determined for many members of this substance class. Furthermore, six compounds act as enzyme inhibitors, activators or receptors (Table 2).
2.4. Benzophenones
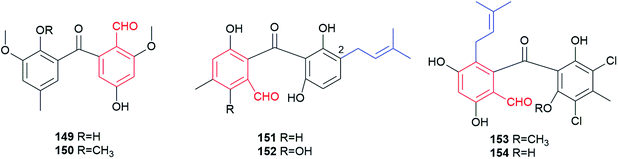
Natural products of this class share a diarylketone skeleton, which can be further modified by hydroxylation, methylation, methoxylation, halogenation or prenylation (Fig. 13). Six such substances were identified in fungi. They are usually oxidative ring opening products of anthrones (see Section 3.2.3 for details).Daldinals A 149 and B 150, benzophenones with two bilateral methoxyl groups, differ from each other by just a hydroxyl or methoxyl group and were isolated from the fungus Daldinia childiae with anti-inflammatory activity.97 The diversity of this group is increased by prenylation on the benzene ring, like the metabolites 151–154. Arugosins I 151 and H 152 with a dimethylallyl moiety at C2 are key intermediates in the shamixanthone biosynthesis and were isolated from the endophytic fungi Penicillum sp. JP-1 and Emericella nidulans var. acristata, respectively.98,99 Co-cultivation of a marine-derived fungus Pestalotia sp. with a unicellular antibiotic-resistant bacterium led to the identification of a chlorinated benzophenone derivative, pestalone 153.100 It shows antibiotic activity against resistant bacteria and moderate cytotoxicity. Its demethylated analogue 154 has been reported for Chrysosporium sp. with inhibitory activity against testosterone-5α-reductase.101
2.5. Spirocyclic benzene carbaldehydes
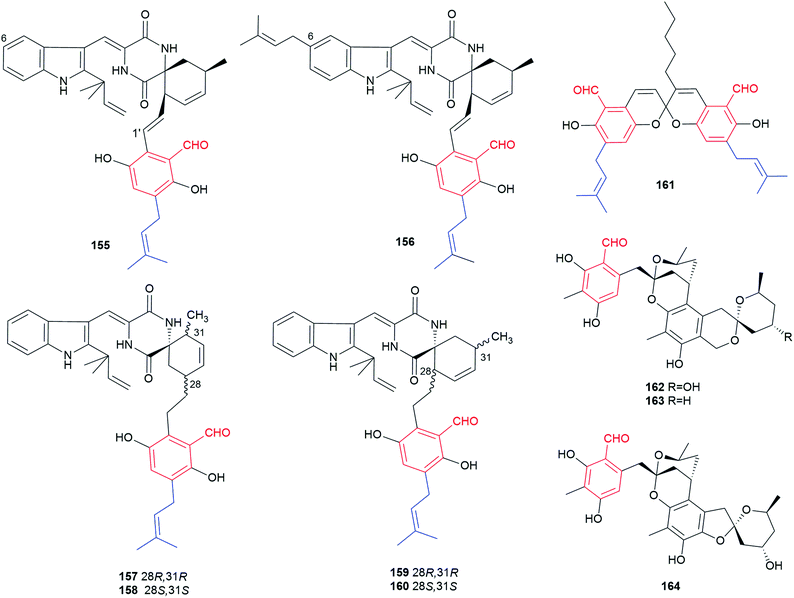
10 spirocyclic benzene carbaldehydes have been found in different Aspergillus, Eurotium and Penicillum species (Fig. 14). The spirocyclic derivatives 155–160 are presumably head-to-tail [4 + 2] Diels–Alder reaction products between the diene feature of a prenylated C7-alkyl benzene carbaldehyde and an enone group of a prenylated diketopiperazine derived from cyclo-Trp-Ala. 155 and 156 have an olefinic bond at C1′ and 156 carries an additional dimethylallyl chain at C6.Cryptoechinuline D 155 and 7-isopentenylcryptoechinuline D 156 were first reported in 1976 from Aspergillus amstelodami and isolated later from the mangrove endophytic fungus Eurotium rubrum.102–104 Eurotinoids A–C 157–159 and dihydrocryptoechinulin D 160 were recently identified as enantiomeric pairs in the marine-derived fungus Eurotium sp. SCSIO F452.105 Compounds 157 and 158 represent two “meta”, while 159 and 160 “ortho” structures, regarding the relative position of the aryl-alkyl substitute to the spiro centre. With the exception for 156, all of the spirocyclic compounds exhibit antioxidant activity.105 Compound 160 also displays cytotoxic activity against two tumour cell lines.106
Four spiroketal benzene carbaldehydes have been until now reported. Cristaldehyde B 161, a spiro dichromene derivative, was isolated from the crinoid-associated fungus Eurotium cristatum.36 Peniciketals A–C 162–164 with two spiroketal features were isolated from the saline soil-derived fungus Penicillium raistrichii and show a selective cytotoxity against HL-60 cell line.107
2.6. Miscellaneous benzene carbaldehydes
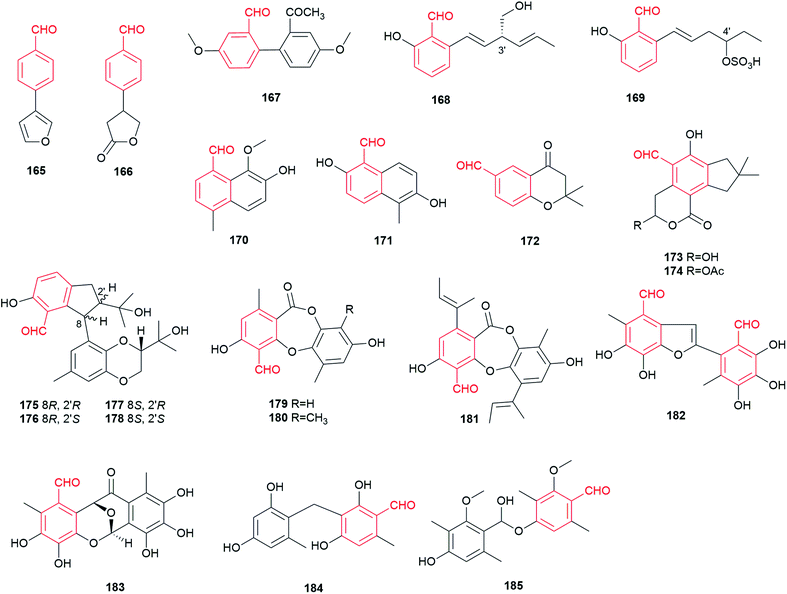
More than 20 fungal benzene carbaldehydes with naphthalene, chromanone or other skeletons cannot be grouped in the classes described above and are listed in this section (Fig. 15). They are usually events of strong rearrangements.Bioactivity-guided fractionation led to isolation of two volatile benzaldehyde derivatives 165 and 166 from an extract of the basidiomycete Sarcodontia crosea (syn. S. setosa).108 They exhibit weak activity against several phytopathogenic fungi including Leptosphaeria maculans and Botrytis cinerea. A biphenyl carbaldehyde 167 with antibacterial activity was obtained from the endophytic fungus Pestalotiopsis zonata.109
Two C6-alkylated salicylaldehydes, pyricuol 168 and pestalol E 169, were obtained from Magnaporthe grisea and Pestalotiopsis sp., respectively.47,49,110 Pyricuol 168 shows a strong nematicidal activity and killed 94.5% of Caenorhabditis elegans at 400 ppm over 24 h. Obviously, the hydroxymethyl group at C3′ in 168 enhances the nematicidal activity, compared to its C7-alkylated analogues 47–50.47 Compound 169 carrying a sulfonic group at C4′ shows inhibitory activity against influenza A and swine flu viruses.
Agropyrenal 170 and vaccinal A 171, two naphthalene carbaldehydes, were isolated from the phytopathogen Ascochyta agropyrina var. nana and the endogenic fungus Pestalotiopsis vaccinii, respectively.34,44 Compound 171 displays anti-enterovirus and anti-inflammatory activities. Three 4/2-chromanone carbaldehydes 172–174 were obtained from the basidiomycetes Fomitiporia punctata and Clitocybe illudens.4,111 Four unusual 2,3-dihydro-1H-indene benzaldehydes bearing a 1,4-benzodioxan moiety, diaporindenes A–D 175–178, were identified in the endophytic fungus Diaporthe sp. SYSU-HQ3.112 They possess significant anti-inflammatory activity against nitric oxide production.
Depsidones 179–181 share a characteristic seven-member ring formed by ester and ether bonds between two benzene rings. They were isolated from the endophyte Botryosphaeria rhodina and the endophytic fungus BCC 8616.113,114 Botryorhodines A 179 and B 180 show cytotoxic and antifungal activities, while compound 181 exhibits only cytotoxic activity.
Secondary metabolite investigation of the endophytic fungus Epicoccum sp. resulted in the isolation of a tetracyclic aromatic benzene carbaldehyde 182 and a 2-phenylbenzofuran carbaldehyde 183, which show potent antibacterial and significant anti-phytopathogenic activities.21 It was proposed that epicoccolides A 182 and B 183 are presumably formed from two molecules of flavipin 16via an unsymmetrical benzoin condensation, which undergoes further modification.21 Two benzene carbaldehydes were identified in the genetically manipulated strains. Compound 184 with a diphenylmethane skeleton, probably derived from o-osellinaldehyde 9, was isolated from a non-reducing polyketide (NR-PKS) heterologous expression host.115 Moreover, deletion of the down-regulator in Aspergillus nidulans led to the discovery of compound 185, a hemiacetal ether from two 3-methylosellinaldehyde 11 molecules.116
3. Formation of benzene carbaldehydes and their involvement in the biosynthesis of fungal metabolites
In the last years, significant progress has been achieved for the understanding of the formation of fungal benzene carbaldehydes. While some of them are formed by direct releasing from non-reducing polyketide synthases (NR-PKSs) with a terminal reductive domain or from highly reducing PKSs (HR-PKSs) by involvement of additional enzymes, other derivatives are formed via modification by tailoring enzymes, e.g. oxidoreductases and NRPS-like enzymes.3.1. Direct releasing from backbone enzymes
Fungal benzene carbaldehydes are often generated by NR-PKSs with acetyl-CoA as the start and malonyl-CoA as the extender unit. The start unit initiates precursor for polyketide synthesis, while the extender units elongate the polyketide backbone to completion.117 The number of extender units determines the length of the polyketide size. A set of grouped catalytic domains control the incorporation of changed or unchanged C2-units into the polyketide backbone. A minimal fungal PKS consists of a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP). Most PKSs also contain accessory domains, such as β-ketoacyl reductase (KR), dehydratase (DH), enoyl reductase (ER), product template (PT), C-methyltransferase (CMeT) and terminal reductase (R).29,118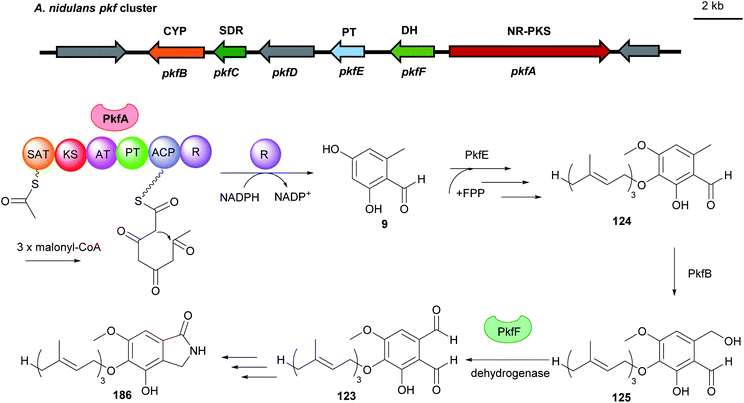 | ||
| Fig. 16 Genetic organisation of the pkf gene cluster in A. nidulans and the simplified postulated biosynthetic pathway of aspernidine A 186 (modified after Yaegashi et al.90). SAT: starter unit, ACP transacylase, CYP: cytochrome P450, SDR: short-chain dehydrogenase, PT: prenyltransferase, DH: dehydrogenase, NR-PKS: non-reducing polyketide synthase. | ||
Similarly, 3-methylosellinaldehyde 11 and redoxcitrinin 30 were detected as the direct releasing products of the NR-PKSs TropA and CitS with R domains (Fig. 17 and 18). In comparison to PkfA, TropA (also known as Tspks1) and CitS (also known as PksCT) contain an additional CMeT domain for methylation during the polyketide chain elongation leading to the formation of the dimethylated benzene carbaldehydes 11 and 30. Heterologous expression of the intronless tropA in the fungal host Aspergillus oryzae led to the identification of the benzaldehyde 11.16 Gene deletion and heterologous expression experiments demonstrated stipitatic acid 187 as the final product of the trop cluster (Fig. 17). Cox and He reported the reconstruction of the biosynthetic gene cluster (BGC) for citrinin 190 from Monascus ruber in A. oryzae.33 The iterative NR-PKS gene citS codes for a redoxcitrinin synthase. Expression of citS alone led to low production of the ketoaldehyde 30 evidently released from the PKS by its terminal reductive R domain. Coexpression of citA coding for a serine hydrolase with citS resulted in a much higher titre of 30. This indicates that cooperation of CitA with the R domain of CitS serves as the release machinery in the native strain (Fig. 18).
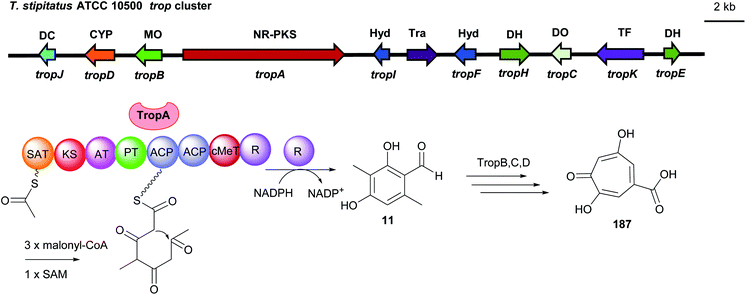 | ||
| Fig. 17 Genetic organisation of the trop gene cluster in T. stipitatus and the simplified postulated biosynthetic pathway of stipitatic acid 187 (modified after Davison et al.16). SAT: starter unit, ACP transacylase, DC: decarboxylase, CYP: cytochrome P450, MO: monooxygenase. Hyd: hydrolase, Tra: transport, DH: dehydrogenase, DO: dioxygenase, TF: transcription factor, NR-PKS: non-reducing polyketide synthase. | ||
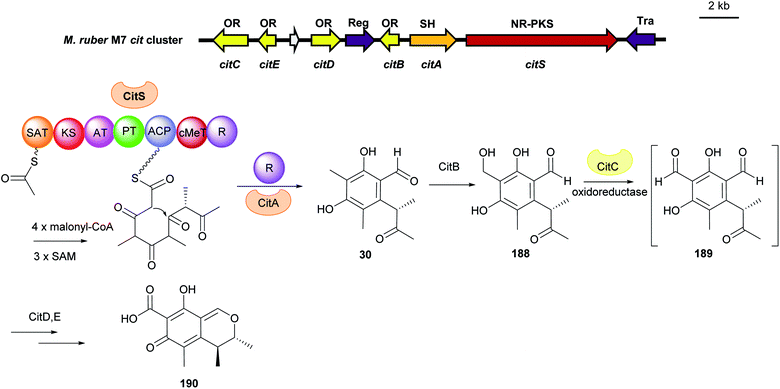 | ||
| Fig. 18 Genetic organisation of the cit gene cluster in M. ruber and the simplified biosynthetic pathway of citrinin 190 (modified after He and Cox33). SAT: starter unit, ACP transacylase, OR: oxidoreductase, Reg: regulator, SH: serine hydrolase, Tra: transporter, NR-PKS non-reducing polyketide synthase. | ||
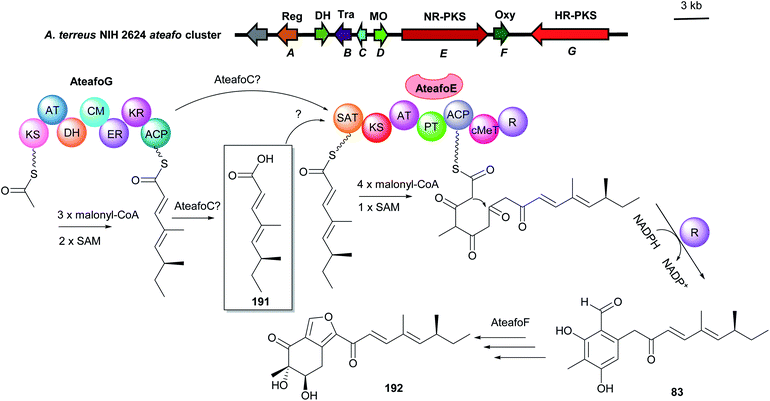 | ||
| Fig. 19 Genetic organisation of the ateafo gene cluster in A. terreus and the simplified postulated biosynthetic pathway of asperfuranone 192 (modified after Chiang et al.120). SAT: starter unit, ACP transacylase, Reg: regulator, DH: dehydrogenasem Tra: transporter, OR: oxidoreductase, MO: monooxygenase, Oxy: oxygenase, NR-PKS: non-reducing polyketide synthase, HR-PKS: highly reducing polyketide synthase. | ||
Yi Tang and coworkers reported a convergent model of dual PKS-containing BGC from A. niger by activation of the silent gene cluster.121 The two PKSs can function independently in parallel to form precursors which can be ultimately connected via accessory enzymes. The NR-PKS AzaA released a C5-alkylated salicylaldehyde 193 by its R domain, which is further reduced to the intermediate 36. The precursor 194 with a pyran ring was then afforded by involvement of the monooxygenase AzaH. In parallel, 2′,4′-dimethylhexanoly CoA 195 as another precursor is synthesized by the HR-PKS AzaB and is proposed to be transferred to the C4-hydroxyl group of 194 to form the key intermediate 196. Further modifications by several tailoring enzymes led to the end product azanigerone C 197 (Fig. 20).
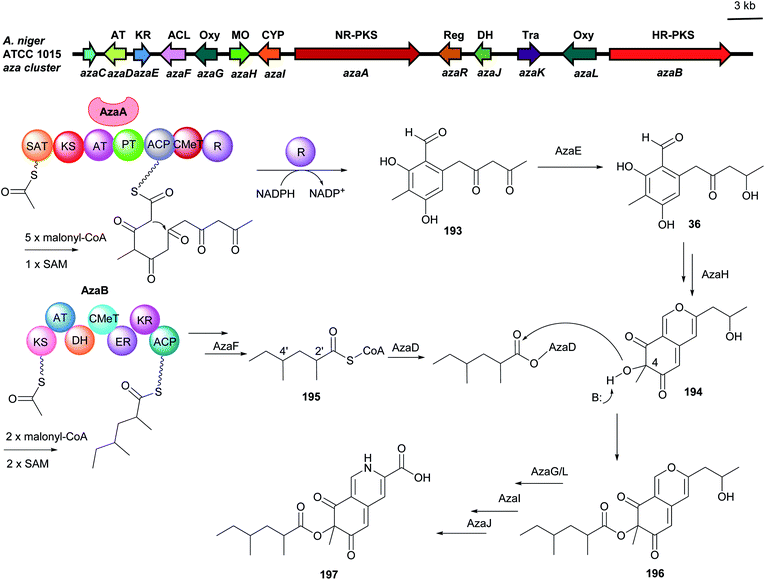 | ||
| Fig. 20 Genetic organisation of the aza gene cluster in A. niger and the simplified postulated biosynthetic pathway of azanigerone 197 (modified after Zabala et al.121). SAT: starter unit, ACP transacylase, AT: acyltransferase, KR: ketoreductase, ACL: acyl:CoA ligase, Oxy: oxygenase, MO: monooxygenase, CYP: cytochrome P450, Reg: regulator, DH: dehydrogenase, Tra: transporter, NR-PKS: non-reducing polyketide synthase, HR-PKS: highly reducing polyketide synthase. | ||
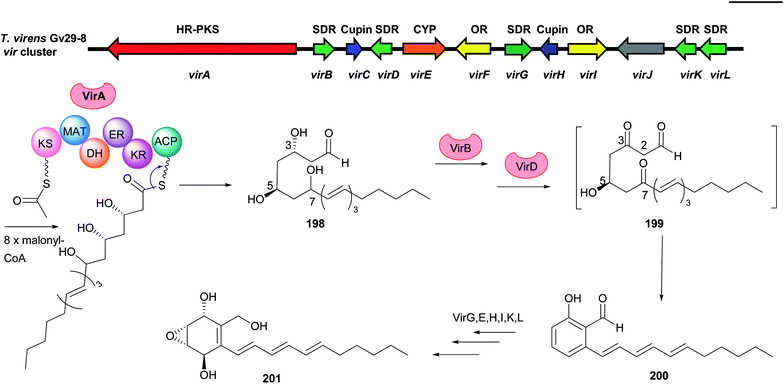 | ||
| Fig. 21 Genetic organisation of the vir gene cluster in T. virens and the simplified postulated biosynthetic pathway of trichoxide 202 (modified after Liu et al.122). SDR: short chain reductase, Cupin: cupin-domain containing protein, CYP: cytochrome P450, OR: oxidoreductase, HR-PKS: highly reducing polyketide synthase. | ||
3.2. Modification by tailoring enzymes
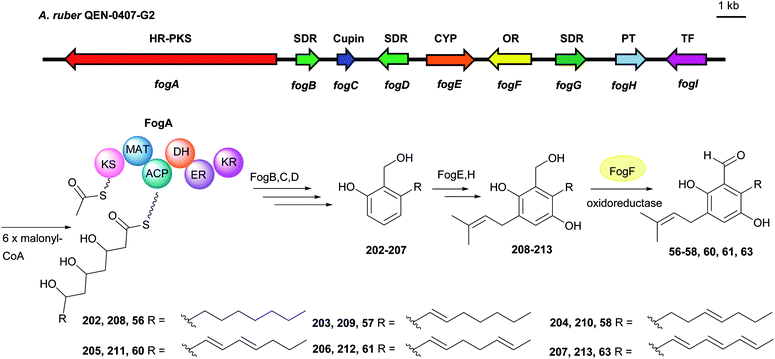 | ||
| Fig. 22 Genetic organisation of the fog gene cluster in A. ruber and the simplified postulated biosynthetic pathway of flavoglaucin 56 and its congeners (modified after Nies et al.123). SDR: short chain reductase, Cupin: cupin-domain containing protein, CYP: cytochrome P450, OR: oxidoreductase, PT: prenyltransferase, TF: transcription factor, HR-PKS: highly reducing polyketide synthase. | ||
In the citrinin biosynthetic pathway, the unstable dialdehyde intermediate 189 was formed via alcohol oxidation catalysed by the nicotinamide-dependent oxidoreductase CitC (also known as Mrl7) (Fig. 18).33 Furthermore, in the biosynthesis of aspernidine A 186, the P450 PkfB introduces a hydroxyl group on the methyl moiety to yield asoernidine E 125, which is proposed to be further oxidised by the choline dehydrogenase PkfF to a reactive dialdehyde 123 (Fig. 16).
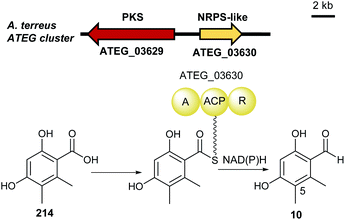 | ||
| Fig. 23 Genetic organisation of the ATEG gene cluster in A. terrus and the proposed biosynthetic pathway of 5-methylosellinaldehyde 10 (modified after Wang et al.15). | ||
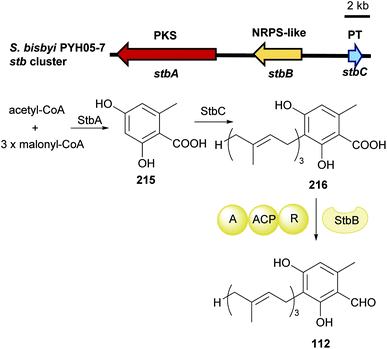 | ||
| Fig. 24 Genetic organisation of the stb gene cluster in Stachybotrys bisbyi and the proposed biosynthetic pathway of LL-Z1272β 112 (modified after Li et al.125). | ||
Investigation on asc cluster in Acremonium egyptiacum shows the aforementioned reduction of 216 to 112 (3.2.2) can also be catalysed by the NRPS-like enzyme AscB, which shares a 59% identity to StbB. This was observed in the biosynthesis of ascochlorin 132 and ascofuranone 136 in Acremonium egyptiacum (Fig. 25).126 Both pathways share the same key precursor ilicicolin A epoxide 217. Cyclisation of 218 catalysed by the terpene cyclase AscF and further dehydrogenation by AscG result in the final product of the ascochlorin pathway. Hydroxylation of 217 by AscH, cyclisation by AscI and oxidation by AscJ complete the ascofuranone pathway. All genes for the ascochlorin biosynthesis are located within the asc-1 cluster, which also contains responsible genes for the common precursors. Additional genes required for the formation of ascofuranone, i.e. ascHIJ were found on the second locus asc-2 (Fig. 25).
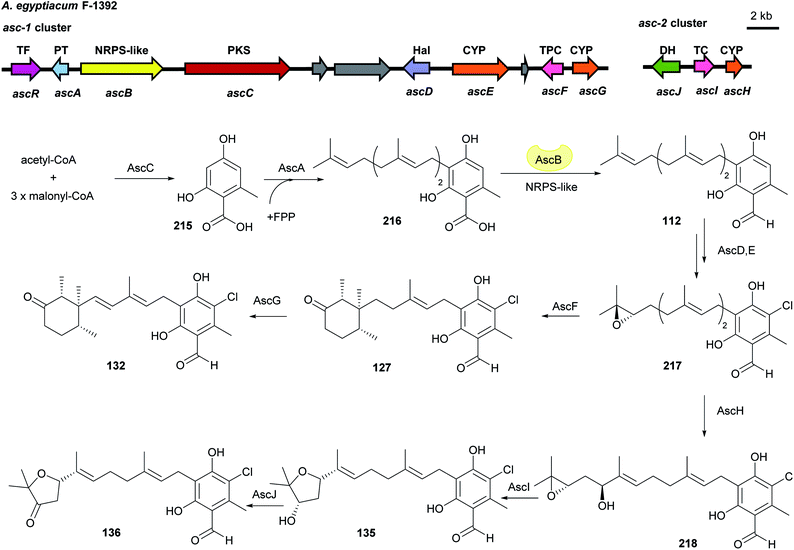 | ||
| Fig. 25 Genetic organisation of the asc gene cluster in A. egyptiacum and the simplified biosynthetic pathways of ascochlorin 132 and ascofuranone 136 (modified after Araki et al.126). TF: transcription factor, PT: prenyltransferase, OR: oxidoreductase, Hal: halogenase, CYP: cytochrome P450, TPC: terpene cyclase, DH: dehydrogenase, PKS: polyketide synthase. | ||
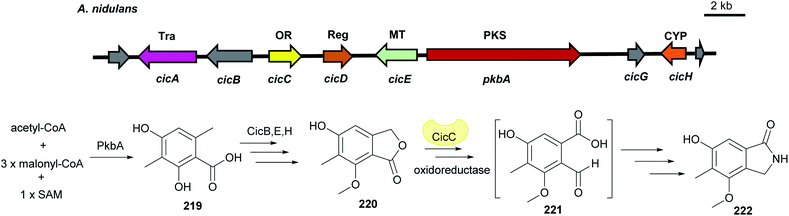 | ||
| Fig. 26 Genetic organisation of the cic gene cluster in A. nidulans and the simplified postulated biosynthetic pathway of cichorine 222 (modified after Sanchez et al.127). Tra: transporter, OR: oxidoreductase, Reg: regulator, MT: methyltransferase, CYP: cytochrome P450, PKS: polyketide synthase. | ||
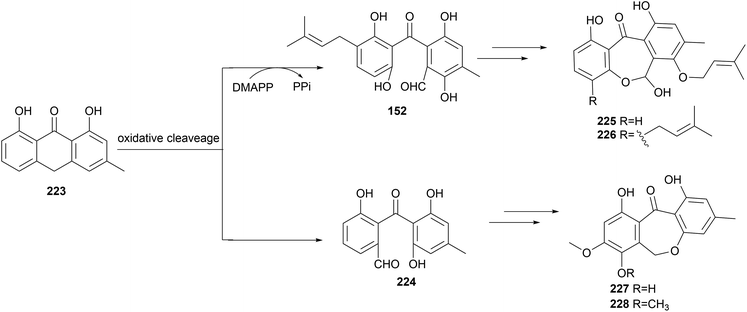
Oxidative cleavage of chrysophanol anthrone 223 was observed in the formation of the benzophenone aldehydes 152 and 224 (Scheme 1). Subsequent intramolecular hemiacetal formation or reduction and ether formation give the dibenzooxepinones 225–228.98,128,129 It is unclear whether enzymes are involved in the transformation.
3.3. Spontaneous reactions
In our previous study, we observed the spontaneous oxidoreduction of the benzoquinone alcohol 229, leading to the formation of the salicylaldehyde 56, the benzyl alcohol 230 and the benzoquinone aldehyde 231.123 A proposed mechanism is given in Scheme 2. Two 229 molecules can act as both oxidant and reductant to form the hydroquinone alcohol 230 and the instable benzoquinone aldehyde intermediate 231, which reacts with a third 229 molecule to form the aldehyde 56. In addition, a set of dimethyl sulfoxide (DMSO) induced oxidations of benzyl alcohol to benzaldehyde were also described in the literature.130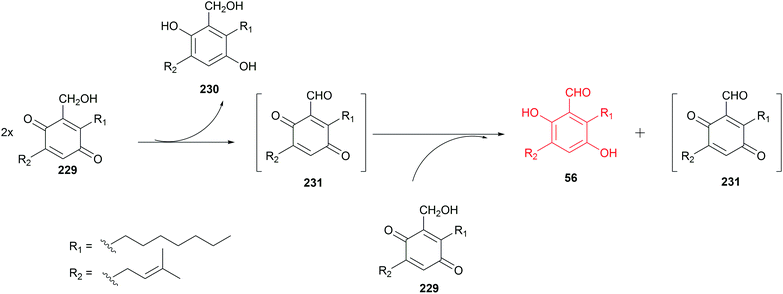 | ||
| Scheme 2 Proposed mechanism of benzoquinone alcohol 229 conversion to salicylaldehyde 56 (modified after Nies et al.123). | ||
4. Conclusions and future perspectives
In this review, we summarized the structural features, distribution, biological activities and applications as well as the origin and biosynthesis of benzene carbaldehydes from fungi. The topic compounds are mainly produced by ascomycetes (79%) and occasionally by basidiomycetes (17%) (Fig. 1). Approx. 51% of the ascomycetes-originated benzene carbaldehydes are from the genera of Aspergillus, Stachybotrys, Penicillium and Pestalotiopsis. For basidiomycetes, the genus of the edible mushroom Hericium contributes to approximate a fifth of benzene carbaldehydes (Table 1). The described biological activities are grouped into eleven categories with cytotoxic, antibacterial and antifungal activities as the top three (Fig. 2).The backbones of the benzene carbaldehydes are usually originated from polyketides assembled by iterative fungal polyketide synthases, although other biosynthetic routes like shikimate or alkaloid pathways also serve as additional possibilities. The key aldehyde functional group can be formed by direct release from the polyketide chain, reduction of carboxylic acids or oxidation of benzyl alcohols. Other procedures such as oxidative ring opening also deliver aldehyde products. The simplest member of these natural products, i.e. benzaldehyde, can be decorated by hydroxylation, alkylation including methylation and ethylation, halogenation, prenylation at the benzene ring. Further modifications include oxidation, reduction and cyclisation. The majority of the compounds mentioned in this review belongs to derivatives of salicylaldehyde from the PKS pathway. Alkylated derivatives with different chain length (C3, C5, C7, C9 or C11) at the ortho-position to the aldehyde group and meroterpenoids containing structural features derived from C5, C10 or C15 prenyl moiety constitute the two large classes of benzene carbaldehydes. Benzene carbaldehydes are accumulated as pathway final products or serve as intermediate for more complex natural products.
As aforementioned, a number of fungal benzene carbaldehydes with interesting biological activities have been discovered in the past decades. However, the studies on structure–activity relationship have been few reported. More information on interactions of benzene carbaldehydes with biological targets will enhance the application potential of these compounds. Furthermore, it became a challenge to get new bioactive natural products under conventional laboratory culture conditions. Therefore, screening microorganisms from less explored or untapped sources, e.g. fungi from extreme environments131–134 like saltern,135 sulfur-rich hydrothermal vents,136 in deep-sea segments,137 hot springs138 and mine area139 becomes more important for bioactive metabolite finding. Symbiotic systems between fungi and bacteria, plants, insect, animals or invertebrate are also less studied promising sources of secondary metabolites.140 Metabolite dereplication, e.g. by library-based LC-MS analysis141,142 and comparative mass spectrometry-based metabolomics143 have been successfully used in the past and will also play an important role in the future to accelerate novel metabolite discovery. Furthermore, the OSMAC (One Strain – Many Compounds) approach144–146 by cultivation under different conditions and co-cultivation with other organisms has also been developed and successfully applied. However, the most putative genes and gene clusters for natural product biosynthesis still remain silent.147 It can be therefore expected that reactivation of such genetic potentials by transcriptional regulator manipulation, promoter engineering and heterologous expression would deliver a large number of structures hidden the silent biosynthetic machinery.148–150 Different new strategies have been published recently for the identification of fungal metabolites and their gene clusters, especially of large clusters. One of such approaches is the fungal artificial chromosomes and metabolic scoring (FAC-MS) strategy, which allows scientists to identify metabolites from complex mixtures after heterologous expression of clusters.151 Metagenomics of uncultivable microbes and reconstruction of biosynthetic pathways provide other possibilities to get new metabolites.152–154 Elucidation of biosynthetic pathways and characterisation of key enzymes would provide another way to create designed molecules by synthetic biology.
5. Conflicts of interest
There are no conflicts to declare.6. Acknowledgements
The researches carried out in the Li's group were funded in part by the Deutsche Forschungsgemeinschaft (DFG) Li844/11-1. H. Ran (201606850085) is a scholarship recipient from the China Scholarship Council.7. References
- B. S. Gould and H. Raistrick, Biochem. J., 1934, 28, 1640–1656 CrossRef CAS
.
- A. Kundu, Planta, 2017, 245, 1069–1078 CrossRef CAS
.
- P. Venkatasubbaiah and W. S. Chilton, J. Nat. Prod., 1990, 53, 1628–1630 CrossRef CAS
.
- R. Tabacchi, A. Fkyerat, C. Poliart and G. M. Dubin, Phytopathol. Mediterr., 2000, 39, 156–161 CAS
.
- R. Gui, L. Xu, Y. Kuang, M. Chung, J. Qin, L. Liu, S. Yang and L. Zhao, J. Plant Interact., 2015, 10, 87–92 CrossRef CAS
.
- M. Shoeb, M. Haque and N. Nahar, J. Nat. Prod. Plant Resour., 2014, 4, 65–70 Search PubMed
.
- F. Kamada, S. Abe, N. Hiratsuka, H. Wariishi and H. Tanaka, Microbiology, 2002, 148, 1939–1946 CrossRef CAS
.
- E. J. Lim, H. J. Kang, H. J. Jung, K. H. Kim, C. J. Lim and E. H. Park, Biomol. Ther., 2008, 16, 231–236 CrossRef CAS
.
- E. W. Bassett and S. W. Tanenbaum, Experientia, 1958, 14, 38–40 CrossRef CAS
.
- H. C. Beck, FEMS Microbiol. Lett., 1997, 149, 233–238 CrossRef CAS
.
- X. Sang, S. Chen, X. An, G. Chen, H. Wang and Y. Pei, J. Asian Nat. Prod. Res., 2017, 19, 436–443 CrossRef CAS
.
- D. Kälvö, A. Menkis and A. Broberg, Molecules, 2018, 23, 1417 CrossRef
.
- J. T. Lin and W. H. Liu, J. Agric. Food Chem., 2006, 54, 7564–7569 CrossRef CAS
.
- L. Tian, S. X. Cai, D. H. Li, Z. J. Lin, T. J. Zhu, Y. C. Fang, P. P. Liu, Q. Q. Gu and W. M. Zhu, Arch. Pharmacal Res., 2007, 30, 1051–1054 CrossRef
.
- M. Wang, M. Beissner and H. Zhao, Chem. Biol., 2014, 21, 257–263 CrossRef CAS
.
- J. Davison, F. A. al, M. Cai, Z. Song, S. Y. Yehia, C. M. Lazarus, A. M. Bailey, T. J. Simpson and R. J. Cox, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7642–7647 CrossRef CAS
.
- A. J. Flewelling, A. L. Bishop, J. A. Johnson and C. A. Gray, Nat. Prod. Commun., 2015, 10, 1661–1662 CrossRef
.
- V. S. Kumar, S. Kumaresan, M. M. Tamizh, M. I. Hairul Islam and K. Thirugnanasambantham, Phytomedicine, 2019, 61, 152830 CrossRef
.
- H. Li, Z. B. Liao, D. Tang, W. B. Han, Q. Zhang and J. M. Gao, J. Antibiot., 2018, 71, 677–681 CrossRef CAS
.
- W. R. Burge, L. J. Buckley, J. D. Sullivan Jr, C. J. McGrattan and M. Ikawa, J. Agric. Food Chem., 1976, 24, 555–559 CrossRef CAS
.
- F. M. Talontsi, B. Dittrich, A. Schüffler, H. Sun and H. Laatsch, Eur. J. Org. Chem., 2013, 2013, 3174–3180 CrossRef CAS
.
- Y. Xiao, H. X. Li, C. Li, J. X. Wang, J. Li, M. H. Wang and Y. H. Ye, FEMS Microbiol. Lett., 2013, 339, 130–136 CrossRef CAS
.
- C. Shao, C. Wang, M. Wei, Z. Jia, Z. She and Y. Lin, Chem. Nat. Compd., 2009, 45, 779–781 CrossRef CAS
.
- J. F. Grove, Biochem. J., 1952, 50, 648–666 CrossRef CAS
.
- J. F. Grove, Biochem. J., 1953, 54, 664–673 CrossRef CAS
.
- A. Cimmino, A. Andolfi, F. Avolio, A. Ali, N. Tabanca, I. A. Khan and A. Evidente, Chem. Biodiversity, 2013, 10, 1239–1251 CrossRef CAS
.
- J. Wang, W. Ding, C. Li, S. Huang, Z. g. She and Y. Lin, Chem. Nat. Compd., 2013, 49, 799–802 CrossRef CAS
.
- L. S. Trifonov, P. Chakravarty, Y. Hiratsuka and W. A. Ayer, Eur. J. For. Pathol., 1992, 22, 441–448 CrossRef
.
- R. J. Cox, Org. Biomol. Chem., 2007, 5, 2010–2026 RSC
.
- J. Staunton and K. J. Weissman, Nat. Prod. Rep., 2001, 18, 380–416 RSC
.
- M. R. de Amorim, F. Hilario, F. M. J. Dos Santos, J. J. Batista, T. M. Bauab, A. R. Araujo, I. Z. Carlos, W. Vilegas and L. C. Dos Santos, Planta Med., 2019, 85, 957–964 CrossRef CAS
.
- D. Zhang, X. Li, J. S. Kang, H. D. Choi, J. H. Jung and B. W. Son, J. Microbiol. Biotechnol., 2007, 17, 865–867 CAS
.
- Y. He and R. J. Cox, Chem. Sci., 2016, 7, 2119–2127 RSC
.
- J. Wang, X. Wei, X. Lu, F. Xu, J. Wan, X. Lin, X. Zhou, S. Liao, B. Yang, Z. Tu and Y. Liu, Tetrahedron, 2014, 70, 9695–9701 CrossRef CAS
.
- H. Lei, X. Lin, L. Han, J. Ma, K. Dong, X. Wang, J. Zhong, Y. Mu, Y. Liu and X. Huang, Phytochemistry, 2017, 142, 51–59 CrossRef CAS
.
- P. Zhang, C. Jia, Y. Deng, S. Chen, B. Chen, S. Yan, J. Li and L. Liu, Phytochemistry, 2019, 158, 120–125 CrossRef CAS
.
- J. Kwon, H. Lee, W. Ko, D. C. Kim, K.-W. Kim, H. C. Kwon, Y. Guo, J. H. Sohn, J. H. Yim and Y. C. Kim, Tetrahedron, 2017, 73, 3905–3912 CrossRef CAS
.
- B. Sontag, N. Arnold, W. Steglich and T. Anke, J. Nat. Prod., 1999, 62, 1425–1426 CrossRef CAS
.
- Z. Huang, X. Nong, Z. Ren, J. Wang, X. Zhang and S. Qi, Bioorg. Med. Chem. Lett., 2017, 27, 787–791 CrossRef CAS
.
- S. Chokpaiboon, P. Unagul, S. Nithithanasilp, S. Komwijit, W. Somyong, T. Ratiarpakul, M. Isaka and T. Bunyapaiboonsri, Nat. Prod. Res., 2018, 32, 149–153 CrossRef CAS
.
- Z. Zhao, Y. Ying, Y. S. Hung and Y. Tang, J. Nat. Prod., 2019, 82, 1029–1033 CrossRef CAS
.
- M. L. Bouillant, J. Favre-Bonvin, N. Salin and J. Bernillon, Phytochemistry, 1988, 27, 1517–1519 CrossRef CAS
.
- H. Fujimoto, T. Fujimaki, E. Okuyama and M. Yamazaki, Chem. Pharm. Bull., 1999, 47, 1426–1432 CrossRef CAS
.
- A. Andolfi, A. Cimmino, M. Vurro, A. Berestetskiy, C. Troise, M. C. Zonno, A. Motta and A. Evidente, Phytochemistry, 2012, 79, 102–108 CrossRef CAS
.
- T. Asai, D. Luo, Y. Obara, T. Taniguchi, K. Monde, K. Yamashita and Y. Oshima, Tetrahedron Lett., 2012, 53, 2239–2243 CrossRef CAS
.
- G. G. Harrigan, B. L. Aremntrout, J. D. Gloer and C. A. Shearer, J. Nat. Prod., 1995, 58, 1467–1469 CrossRef CAS
.
- Y. H. Yang, D. S. Yang, H. M. Lei, C. Y. Li, G. H. Li and P. J. Zhao, Molecules, 2020, 25, 72 CrossRef CAS
.
- K. Tanaka, A. Sasaki, H.-Q. Cao, T. Yamada, M. Igarashi, I. Komine, H. Nakahigashi, N. Minami, S. Kuwahara, M. Nukina and H. Kiyota, Eur. J. Org. Chem., 2011, 2011, 6276–6280 CrossRef CAS
.
- J. Sun, X. Lin, X. Zhou, J. Wan, T. Zhang, B. Yang, X. Yang, Z. Tu and Y. Liu, J. Antibiot., 2014, 67, 451–457 CrossRef CAS
.
- C. Zheng, L. Xu, Y. Y. Li, T. Han, Q. Zhang, Q. Ming, K. Rahman and L. P. Qin, Appl. Microbiol. Biotechnol., 2013, 97, 7617–7625 CrossRef CAS
.
- Y. Takenaka, T. Tanahashi, N. Nagakura, A. Itoh and N. Hamada, Phytochemistry, 2004, 65, 3119–3123 CrossRef CAS
.
- S. W. Sun, C. Z. Ji, Q. Q. Gu, D. H. Li and T. J. Zhu, J. Asian Nat. Prod. Res., 2013, 15, 956–961 CrossRef CAS
.
- Y. Huang, L. Ma, X. Rong, D. Liu, S. Liu and W. Liu, Chin. Tradit. Herb. Drugs, 2012, 43, 837–840 CAS
.
- Y. Miyake, C. Ito, T. Kimura, A. Suzuki, Y. Nishida and M. Itoigawa, Food Sci. Technol. Res., 2014, 20, 139–146 CrossRef CAS
.
- N. Fathallah, M. M. Raafat, M. Y. Issa, M. M. Abdel-Aziz, M. Bishr, M. A. Abdelkawy and O. Salama, Molecules, 2019, 24, 4118 CrossRef CAS
.
- J. Shi, J. Liu, D. Kang, Y. Huang, W. Kong, Y. Xiang, X. Zhu, Y. Duan and Y. Huang, ACS Omega, 2019, 4, 6630–6636 CrossRef CAS
.
- M. D. Wu, M. J. Cheng, S. Y. Hsieh and G. F. Yuan, Chem. Nat. Compd., 2014, 49, 1175–1176 CrossRef CAS
.
- J. Gao, F. León, M. M. Radwan, O. R. Dale, A. S. Husni, S. P. Manly, S. Lupien, X. Wang, R. A. Hill, F. M. Dugan, H. G. Cutler and S. J. Cutler, J. Nat. Prod., 2011, 74, 1636–1639 CrossRef CAS
.
- M. Chen, C.-L. Shao, K.-L. Wang, Y. Xu, Z.-G. She and C.-Y. Wang, Tetrahedron, 2014, 70, 9132–9138 CrossRef CAS
.
- M. Chen, Q. Zhao, J. D. Hao and C. Y. Wang, Nat. Prod. Res., 2017, 31, 268–274 CrossRef CAS
.
- D.-L. Li, X.-M. Li, T.-G. Li, H.-Y. Dang, P. Proksch and B.-G. Wang, Chem. Pharm. Bull., 2008, 56, 1282–1285 CrossRef CAS
.
- S. Wang, X. M. Li, F. Teuscher, D. L. Li, A. Diesel, R. Ebel, P. Proksch and B.-G. Wang, J. Nat. Prod., 2006, 69, 1622–1625 CrossRef CAS
.
- A. M. Elissawy, S. S. Ebada, M. L. Ashour, M. El-Neketi, W. Ebrahim and A. B. Singab, Phytochem. Lett., 2019, 29, 1–5 CrossRef CAS
.
- S. Halecker, F. Surup, H. Solheim and M. Stadler, J. Antibiot., 2018, 71, 339–341 CrossRef CAS
.
- S. Son, S. K. Ko, J. W. Kim, J. K. Lee, M. Jang, I. J. Ryoo, G. J. Hwang, M. C. Kwon, K. S. Shin, Y. Futamura, Y. S. Hong, H. Oh, B. Y. Kim, M. Ueki, S. Takahashi, H. Osada, J. H. Jang and J. S. Ahn, Phytochemistry, 2016, 122, 154–164 CrossRef CAS
.
- K. Matsuzaki, H. Tahara, J. Inokoshi, H. Tanaka, R. Masuma and S. Omura, J. Antibiot., 1998, 51, 1004–1011 CrossRef CAS
.
- J. Arunpanichlert, V. Rukachaisirikul, Y. Sukpondma, S. Phongpaichit, S. Tewtrakul, N. Rungjindamai and J. Sakayaroj, Chem. Pharm. Bull., 2010, 58, 1033–1036 CrossRef CAS
.
- Y. M. Chiang, E. Szewczyk, A. D. Davidson, N. Keller, B. R. Oakley and C. C. Wang, J. Am. Chem. Soc., 2009, 131, 2965–2970 CrossRef CAS
.
- P. Berkaew, N. Soonthornchareonnon, K. Salasawadee, R. Chanthaket and M. Isaka, J. Nat. Prod., 2008, 71, 902–904 CrossRef CAS
.
- R. Geris and T. J. Simpson, Nat. Prod. Rep., 2009, 26, 1063–1094 RSC
.
- Y. Matsuda and I. Abe, Nat. Prod. Rep., 2016, 33, 26–53 RSC
.
- J. W. Blunt, B. R. Copp, M. H. Munro, P. T. Northcote and M. R. Prinsep, Nat. Prod. Rep., 2004, 21, 1–49 RSC
.
- S. N. Sunassee and M. T. Davies-Coleman, Nat. Prod. Rep., 2012, 29, 513–535 RSC
.
- L. A. M. Murray, S. M. K. McKinnie, B. S. Moore and J. H. George, Nat. Prod. Rep., 2020, 37 10.1039/d0np00018c
.
- G. A. Ellestad, R. H. Evans and M. P. Kunstmann, Tetrahedron, 1969, 25, 1323–1334 CrossRef CAS
.
- D. Hansson, A. Menkis, Å. Olson, J. Stenlid, A. Broberg and M. Karlsson, Phytochemistry, 2012, 84, 31–39 CrossRef CAS
.
- G. M. Dubin, A. Fkyerat and R. Tabacchi, Phytochemistry, 2000, 53, 571–574 CrossRef CAS
.
- M. Gutiérrez, C. Theoduloz, J. Rodríguez, M. Lolas and G. Schmeda-Hirschmann, J. Agric. Food Chem., 2005, 53, 7701–7708 CrossRef
.
- H. Hussain, K. H. Drogies, A. Al-Harrasi, Z. Hassan, A. Shah, U. A. Rana, I. R. Green, S. Draeger, B. Schulz and K. Krohn, Asian Pac. J. Trop. Dis., 2015, 5, 186–189 CrossRef CAS
.
- H. Kawagishi, H. Sato, S. Sakamura, K. Kobayashi and U. Tadao, Agric. Biol. Chem., 1984, 48, 1903–1904 CAS
.
- Y. Kosuge, A. Suzuki and S. Tamura, Agric. Biol. Chem., 1974, 38, 1265–1267 CrossRef CAS
.
- G. Marsico, B. A. Pignataro, M. Masi, A. Evidente, F. Casella, M. C. Zonno, J. H. Tak, J. R. Bloomquist, S. Superchi and P. Scafato, Tetrahedron, 2018, 74, 3912–3923 CrossRef CAS
.
- H. Kawagishi, M. Ando, K. Shinba, H. Sakamoto, S. Yoshida, F. Ojima, Y. Ishiguro, N. Ukai and S. Furukawa, Phytochemistry, 1992, 32, 175–178 CrossRef CAS
.
- H. Kawagishi, M. Ando, H. Sakamoto, S. Yoshida, F. Ojima, Y. Ishiguro, N. Ukai and S. Furukawa, Tetrahedron Lett., 1991, 32, 4561–4564 CrossRef CAS
.
- K. Dekermendjian, R. Shan, M. Nielsen, M. Stadler, O. Sterner and M. R. Witt, Eur. J. Med. Chem., 1997, 32, 351–356 CrossRef CAS
.
- Y. Nozawa, K. Yamamoto, M. Ito, N. Sakai, K. Mizoue, F. Mizobe and K. Hanada, J. Antibiot., 1997, 50, 635–640 CrossRef CAS
.
- P. Zhang, B. Bao, H. T. Dang, J. Hong, H. J. Lee, E. S. Yoo, K. S. Bae and J. H. Jung, J. Nat. Prod., 2009, 72, 270–275 CrossRef CAS
.
- P. Zhang, Y. Li, C. Jia, J. Lang, S. I. Niaz, J. Li, J. Yuan, J. Yu, S. Chen and L. Liu, RSC Adv., 2017, 7, 49910–49916 RSC
.
- J. A. Ballantine, V. Ferrito, C. H. Hassall and V. I. P. Jones, J. Chem. Soc. C, 1969, 56–61 RSC
.
- J. Yaegashi, M. B. Praseuth, S. W. Tyan, J. F. Sanchez, R. Entwistle, Y. M. Chiang, B. R. Oakley and C. C. Wang, Org. Lett., 2013, 15, 2862–2865 CrossRef CAS
.
- T. Mogi, H. Ui, K. Shiomi, S. Omura, H. Miyoshi and K. Kita, Biochim. Biophys. Acta, 2009, 1787, 129–133 CrossRef CAS
.
- P. Seephonkai, M. Isaka, P. Kittakoop, U. Boonudomlap and Y. Thebtaranonth, J. Antibiot., 2004, 57, 10–16 CrossRef CAS
.
- J. Zhao, J. Feng, Z. Tan, J. Liu, J. Zhao, R. Chen, K. Xie, D. Zhang, Y. Li, L. Yu, X. Chen and J. Dai, J. Nat. Prod., 2017, 80, 1819–1826 CrossRef CAS
.
- S. B. Singh, D. L. Zink, M. Williams, J. D. Polishook, M. Sanchez, K. C. Silverman and R. B. Lingham, Bioorg. Med. Chem. Lett., 1998, 8, 2071–2076 CrossRef CAS
.
- Y. Hemberger, J. Xu, V. Wray, P. Proksch, J. Wu and G. Bringmann, Chem.–Eur. J., 2013, 19, 15556–15564 CrossRef CAS
.
- B. Sontag, M. Rüth, P. Spiteller, N. Arnold, W. Steglich, M. Reichert and G. Bringmann, Eur. J. Org. Chem., 2006, 2006, 1023–1033 CrossRef
.
- D. N. Quang, L. Harinantenaina, T. Nishizawa, T. Hashimoto, C. Kohchi, G. I. Soma and Y. Asakawa, J. Nat. Med., 2006, 60, 303–307 CrossRef CAS
.
- A. Kralj, S. Kehraus, A. Krick, E. Eguereva, G. Kelter, M. Maurer, A. Wortmann, H. H. Fiebig and G. M. König, J. Nat. Prod., 2006, 69, 995–1000 CrossRef CAS
.
- Z. Lin, T. Zhu, Y. Fang, Q. Gu and W. Zhu, Phytochemistry, 2008, 69, 1273–1278 CrossRef CAS
.
- M. Cueto, P. R. Jensen, C. Kauffman, W. Fenical, E. Lobkovsky and J. Clardy, J. Nat. Prod., 2001, 64, 1444–1446 CrossRef CAS
.
-
Y. Wachi, T. Yamashita, K. Komatsu and S. Yoshida, New Benzophenone Derivative, Its Production and Its Use, JP19930207818 19930823[JKXXAF JP 07061950 A2 19950307], 1995 Search PubMed
.
- G. Gatti, R. Cardillo, C. Fuganti and D. Ghiringhelli, J. Chem. Soc., Chem. Commun., 1976, 435–436 RSC
.
- H.-J. Yan, X.-M. Li, C.-S. Li and B.-G. Wang, Helv. Chim. Acta, 2012, 95, 163–167 CrossRef CAS
.
- D.-L. Li, X.-M. Li, T.-G. Li, H.-Y. Dang and B.-G. Wang, Helv. Chim. Acta, 2008, 91, 1888–1892 CrossRef CAS
.
- W. Zhong, J. Wang, X. Wei, T. Fu, Y. Chen, Q. Zeng, Z. Huang, X. Huang, W. Zhang, S. Zhang, L. Long and F. Wang, Front. Chem., 2019, 7, 350 CrossRef CAS
.
- H. Gao, W. Liu, T. Zhu, X. Mo, A. Mandi, T. Kurtan, J. Li, J. Ai, Q. Gu and D. Li, Org. Biomol. Chem., 2012, 10, 9501–9506 RSC
.
- W. Z. Liu, L. Y. Ma, D. S. Liu, Y. L. Huang, C. H. Wang, S. S. Shi, X. H. Pan, X. D. Song and R. X. Zhu, Org. Lett., 2014, 16, 90–93 CrossRef CAS
.
- T. Kokubun, Z. Rozwadowski and H. Duddeck, J. Nat. Prod., 2007, 70, 1539–1541 CrossRef CAS
.
- X. Yang, S. Zhang, S. Song, Y. Zhang, D. Luo and M. Zhang, Chin. J. Nat. Med., 2011, 9, 101–104 CAS
.
- J. C. Kim, J. Y. Min, H. T. Kim, K. Y. Cho and S. H. Yu, Biosci., Biotechnol., Biochem., 1998, 62, 173–174 CrossRef CAS
.
- M. S. R. Nair, H. Takeshita, T. C. McMorris and M. Anchel, J. Org. Chem., 1969, 34, 240–243 CrossRef CAS
.
- H. Cui, Y. Liu, J. Li, X. Huang, T. Yan, W. Cao, H. Liu, Y. Long and Z. She, J. Org. Chem., 2018, 83, 11804–11813 CrossRef CAS
.
- R. Abdou, K. Scherlach, H. M. Dahse, I. Sattler and C. Hertweck, Phytochemistry, 2010, 71, 110–116 CrossRef CAS
.
- P. Pittayakhajonwut, A. Dramae, S. Madla, N. Lartpornmatulee, N. Boonyuen and M. Tanticharoen, J. Nat. Prod., 2006, 69, 1361–1363 CrossRef CAS
.
- M. Ahuja, Y. M. Chiang, S. L. Chang, M. B. Praseuth, R. Entwistle, J. F. Sanchez, H. C. Lo, H. H. Yeh, B. R. Oakley and C. C. Wang, J. Am. Chem. Soc., 2012, 134, 8212–8221 CrossRef CAS
.
- C. E. Oakley, M. Ahuja, W. W. Sun, R. Entwistle, T. Akashi, J. Yaegashi, C. J. Guo, G. C. Cerqueira, W. J. Russo, C. C. Wang, Y. M. Chiang and B. R. Oakley, Mol. Microbiol., 2017, 103, 347–365 CrossRef CAS
.
- Y. A. Chan, A. M. Podevels, B. M. Kevany and M. G. Thomas, Nat. Prod. Rep., 2009, 26, 90–114 RSC
.
- L. Du and L. Lou, Nat. Prod. Rep., 2010, 27, 255–278 RSC
.
- J. F. Sanchez, A. D. Somoza, N. P. Keller and C. C. Wang, Nat. Prod. Rep., 2012, 29, 351–371 RSC
.
- Y. M. Chiang, C. E. Oakley, M. Ahuja, R. Entwistle, A. Schultz, S. L. Chang, C. T. Sung, C. C. Wang and B. R. Oakley, J. Am. Chem. Soc., 2013, 135, 7720–7731 CrossRef CAS
.
- A. O. Zabala, W. Xu, Y. H. Chooi and Y. Tang, Chem. Biol., 2012, 19, 1049–1059 CrossRef CAS
.
- L. Liu, M. C. Tang and Y. Tang, J. Am. Chem. Soc., 2019, 141, 19538–19541 CrossRef CAS
.
- J. Nies, H. Ran, V. Wohlgemuth, W. B. Yin and S.-M. Li, Org. Lett., 2020, 22, 2256–2260 CrossRef CAS
.
- M. Wang and H. Zhao, ACS Catal., 2014, 4, 1219–1225 CrossRef CAS
.
- C. Li, Y. Matsuda, H. Gao, D. Hu, X. S. Yao and I. Abe, Chembiochem, 2016, 17, 904–907 CrossRef CAS
.
- Y. Araki, T. Awakawa, M. Matsuzaki, R. Cho, Y. Matsuda, S. Hoshino, Y. Shinohara, M. Yamamoto, Y. Kido, D. K. Inaoka, K. Nagamune, K. Ito, I. Abe and K. Kita, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 8269–8274 CrossRef CAS
.
- J. F. Sanchez, R. Entwistle, D. Corcoran, B. R. Oakley and C. C. C. Wang, Med. Chem. Commun., 2012, 3, 997–1002 RSC
.
- D. Pockrandt, L. Ludwig, A. Fan, G. M. König and S.-M. Li, Chembiochem, 2012, 13, 2764–2771 CrossRef CAS
.
- J. Kornsakulkarn, S. Saepua, P. Laksanacharoen, P. Rachtawee and C. Thongpanchang, Tetrahedron Lett., 2016, 57, 305–307 CrossRef CAS
.
- W. W. Epstein and F. W. Sweat, Chem. Rev., 1967, 67, 247–260 CrossRef CAS
.
- R. Chávez, F. Fierro, R. O. Garcia-Rico and I. Vaca, Front. Microbiol., 2015, 6, 903 Search PubMed
.
- Z. E. Wilson and M. A. Brimble, Nat. Prod. Rep., 2020, 37 10.1039/d0np00021c
.
- Z. E. Wilson and M. A. Brimble, Nat. Prod. Rep., 2009, 26, 44–71 RSC
.
- M. Ibrar, M. W. Ullah, S. Manan, U. Farooq, M. Rafiq and F. Hasan, Appl. Microbiol. Biotechnol., 2020, 104, 2777–2801 CrossRef CAS
.
- D. Chung, H. Kim and H. S. Choi, J. Microbiol., 2019, 57, 717–724 CrossRef CAS
.
- W. Jiang, P. Ye, C. T. Chen, K. Wang, P. Liu, S. He, X. Wu, L. Gan, Y. Ye and B. Wu, Mar. Drugs, 2013, 11, 4761–4772 CrossRef CAS
.
- Y. T. Wang, Y. R. Xue and C. H. Liu, Mar. Drugs, 2015, 13, 4594–4616 CrossRef CAS
.
- W. Y. Liao, C. N. Shen, L. H. Lin, Y. L. Yang, H. Y. Han, J. W. Chen, S. C. Kuo, S. H. Wu and C. C. Liaw, J. Nat. Prod., 2012, 75, 630–635 CrossRef CAS
.
- A. A. Stierle and D. B. Stierle, Nat. Prod. Commun., 2014, 9, 1037–1044 CAS
.
- N. Adnani, S. R. Rajski and T. S. Bugni, Nat. Prod. Rep., 2017, 34, 784–814 RSC
.
- H. Mohimani, A. Gurevich, A. Mikheenko, N. Garg, L. F. Nothias, A. Ninomiya, K. Takada, P. C. Dorrestein and P. A. Pevzner, Nat. Chem. Biol., 2017, 13, 30–37 CrossRef CAS
.
- K. F. Nielsen and T. O. Larsen, Front. Microbiol., 2015, 6, 71 Search PubMed
.
- B. C. Covington, J. A. McLean and B. O. Bachmann, Nat. Prod. Rep., 2017, 34, 6–24 RSC
.
- H. B. Bode, B. Bethe, R. Hofs and A. Zeeck, Chembiochem, 2002, 3, 619–627 CrossRef CAS
.
- N. P. Ariantari, G. Daletos, A. Mándi, T. Kurtán, W. E. G. Müller, W. Lin, E. Ancheeva and P. Proksch, RSC Adv., 2019, 9, 25119–25132 RSC
.
- D. M. Selegato, R. T. Freire, A. C. Pilon, C. R. Biasetto, H. C. de Oliveira, L. M. de Abreu, A. R. Araujo, V. da Silva Bolzani and I. Castro-Gamboa, Magn. Reson. Chem., 2019, 57, 458–471 CrossRef CAS
.
- A. A. Brakhage, P. Sprote, Q. Al Abdallah, A. Gehrke, H. Plattner and A. Tuncher, Adv. Biochem. Eng./Biotechnol., 2004, 88, 45–90 CrossRef CAS
.
- M. Zerikly and G. L. Challis, Chembiochem, 2009, 10, 625–633 CrossRef CAS
.
- H. N. Lyu, H. W. Liu, N. P. Keller and W. B. Yin, Nat. Prod. Rep., 2020, 37, 6–16 RSC
.
- A. A. Brakhage, Nat. Rev. Microbiol., 2013, 11, 21–32 CrossRef CAS
.
- K. D. Clevenger, J. W. Bok, R. Ye, G. P. Miley, M. H. Verdan, T. Velk, C. Chen, K. Yang, M. T. Robey, P. Gao, M. Lamprecht, P. M. Thomas, M. N. Islam, J. M. Palmer, C. C. Wu, N. P. Keller and N. L. Kelleher, Nat. Chem. Biol., 2017, 13, 895–901 CrossRef CAS
.
- Y. He, B. Wang, W. Chen, R. J. Cox, J. He and F. Chen, Biotechnol. Adv., 2018, 36, 739–783 CrossRef CAS
.
- M. Katz, B. M. Hover and S. F. Brady, J. Ind. Microbiol. Biotechnol., 2016, 43, 129–141 CrossRef CAS
.
- B. K. Singh and C. A. Macdonald, Drug Discovery Today, 2010, 15, 792–799 CrossRef CAS
.
- I. Ullah, A. L. Khan, L. Ali, A. R. Khan, M. Waqas, J. Hussain, I. J. Lee and J. H. Shin, J. Microbiol., 2015, 53, 127–133 CrossRef CAS
.
- N. C. G. Faria, J. H. Kim, L. A. P. Gonçalves, M. d. Martins, K. L. Chan and B. C. Campbell, Lett. Appl. Microbiol., 2011, 52, 506–513 CrossRef CAS
.
- S. Y. Y. Wong, I. R. Grant, M. Friedman, C. T. Elliott and C. Situ, Appl. Environ. Microbiol., 2008, 74, 5986 CrossRef CAS
.
- J. H. Ha, D. U. Lee, J. T. Lee, J. S. Kim, C. S. Yong, J. A. Kim, J. S. Ha and K. Huh, J. Ethnopharmacol., 2000, 73, 329–333 CrossRef CAS
.
- X. Li, B. Xiang, T. Shen, C. Xiao, R. Dai, F. He and Q. Lin, Int. Immunopharmacol., 2020, 82, 106353 CrossRef CAS
.
- L. G. Malak, M. A. Ibrahim, D. W. Bishay, A. M. Abdel-baky, A. M. Moharram, B. Tekwani, S. J. Cutler and S. A. Ross, J. Nat. Prod., 2014, 77, 1987–1991 CrossRef CAS
.
- F. A. R. Rodrigues, A. C. A. Oliveira, B. C. Cavalcanti, M. P. Costa, C. Pessoa, M. V. N. de Souza and A. C. Pinheiro, Eur. Chem. Bull., 2014, 3, 555–558 CAS
.
- Y. H. Tao, Z. Yuan, X. Q. Tang, H. B. Xu and X. L. Yang, Bioorg. Med. Chem. Lett., 2006, 16, 592–595 CrossRef CAS
.
- B. H. Lee, S. H. Yoon, Y. S. Kim, S. K. Kim, B. J. Moon and Y. S. Bae, Nat. Prod. Res., 2008, 22, 1441–1450 CrossRef CAS
.
- M. Masi, F. Freda, F. Sangermano, V. Calabro, A. Cimmino, M. Cristofaro, S. Meyer and A. Evidente, Molecules, 2019, 24, E1086 CrossRef
.
| This journal is © The Royal Society of Chemistry 2021 |