Open Access Article
Open Access ArticleLabel-free sensing of virus-like particles below the sub-diffraction limit by wide-field photon state parametric imaging of a gold nanodot array†
Xiao
Jin‡
 a,
Heng
Zhang‡
a,
Bin
Ni
a,
Weiping
Liu
a,
Lianping
Hou
b,
John H.
Marsh
a,
Heng
Zhang‡
a,
Bin
Ni
a,
Weiping
Liu
a,
Lianping
Hou
b,
John H.
Marsh
 b,
Shengwei
Ye
b,
Xiao
Sun
b,
Xiaofeng
Li
c,
Shanhu
Li
d,
Lei
Dong
e,
Jamie Jiangmin
Hou
b,
Shengwei
Ye
b,
Xiao
Sun
b,
Xiaofeng
Li
c,
Shanhu
Li
d,
Lei
Dong
e,
Jamie Jiangmin
Hou
 f,
Ming
Sun
a,
Bin
Xu
a,
Jichuan
Xiong
f,
Ming
Sun
a,
Bin
Xu
a,
Jichuan
Xiong
 *a and
Xuefeng
Liu
*a and
Xuefeng
Liu
 *a
*a
aSchool of Electronic and Optical Engineering, Nanjing University of Science and Technology, Nanjing 210094, P. R. China. E-mail: liuxf_1956@sina.com; jichuan.xiong@njust.edu.cn
bJames Watt School of Engineering, University of Glasgow, Glasgow, G12 8QQ, UK
cState Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Disease, The first affiliated Hospital of Guangzhou Medical University, Guangzhou 510182, P. R. China
dDepartment of Cell Engineering, Beijing Institute of Biotechnology, Beijing, 100850, P. R. China
eSchool of Life Science, Beijing Institute of Technology, Beijing 100081, P. R. China
fThe Royal College of Surgeons of Edinburgh, Nicolson Street, Edinburgh, Scotland EH8 9DW, UK
First published on 23rd October 2021
Abstract
A parallel four-quadrant sensing method utilizing a specially designed gold nanodot array is created for sensing virus-like particles with a sub-diffraction limit size (∼100 nm) in a wide-field image. Direct label-free sensing of viruses using multiple four-quadrant sensing channels in parallel in a wide-field view enables the possibility of high-throughput onsite screening of viruses.
Infections from viruses and other pathogens are a major threat to human health and even to the survival of the human race. The outbreak of SARS-CoV-2 has resulted in millions of deaths worldwide.1 Rapid detection techniques with high throughput and sensitivity which allow onsite operation with minimal requirements for sample preparation and the test environment are essential for the prevention and control of pandemics.2–4
Conventional methods for virus detection include polymerase chain reaction (PCR), serological analysis, and imaging technologies such as computerized tomography (CT) scans.5–9 Various diagnostic sensors have been developed to support standard SARS-CoV-2 diagnostic techniques, which are mainly based on antigenic probes such as whole virus particles and surface antigens (capsids, envelopes, and nucleocapsid proteins) and deliver qualitative outputs (i.e., positive or negative). However, these methods necessitate skilled operators and require hours or even days to perform the analysis. Meanwhile, they are also restricted by the small number of antibodies produced during different phases of infection.10,11
Compared with contemporary virus identification methods, optical imaging is a promising method for fast virus screening,12,13 which includes a visual inspection of cell cultures, fluorescence-based readouts and indirect detection via refractive index changes and other mechanisms.14–16 Challenges faced in direct optical imaging of viruses include the diffraction limit size of particles and the need for sophisticated labeling.17,18
In order to improve the sensitivity of optical imaging, research efforts have been made to develop various labeling and signal amplification methods for virus imaging and sensing. The utilization of nanoparticles, including quantum dots, magnetic particles, polymer nanoparticles, and noble metal nanoparticles, has led to significant enhancement in the detection sensitivity of viruses.19–24 Among various types of nanoparticles, metallic nanoparticles show the distinct feature of localized surface plasmon resonance (LSPR), induced by the interaction of the free-electron cloud of metal nanoparticles with the incident electromagnetic field. Gold nanoparticles have attracted much attention due to their easy modification with biomolecules, allowing tight attachment of the aforementioned antibody or nucleic acid.25–27 These strategies mainly rely on exploiting the spectral response of gold nanoparticles decorated with virus-specific antigens, which needs relatively complicated sample preparation. Moreover, their single-channel operation manner limits their application to high-throughput virus screening.
In our recent studies, gold nanoparticles (AuNPs) were used to amplify the scattering signal of viruses in photon state parametric imaging, which suggests the possibility of wide-field sensing of virus particles.28 It is found that antigen-decorated AuNPs attach to virus particles and enhance the optical scattering signal due to localized plasmons. However, stray AuNPs would randomly aggregate with pathogens, forming different morphologies at random locations in the field of view of the microscope. The random distribution of AuNPs degrades the connection efficiency of viruses to antigens, and also means that the virus-specific signal is obscured by noise scattered from other locations. Furthermore, similar sample preparation is required to that in the gold spectrum-based techniques.
To overcome the limitations of current techniques which will be difficult to use in onsite virus screening, in this paper, a virus-like particle sensing method is proposed that operates in a parallel manner with wide-field optical imaging. This is achieved by utilizing grouped gold nanodots as sensing channels in a specifically designed gold nanodot matrix on a Si substrate. A well-regulated optical distribution is formed in the field of view by the scattering of each dot in the matrix. The gold nanodots also demonstrate high efficiency in capturing the virus, allowing label-free sensing of virus-like particles under the sub-diffraction limit with a simple optical setup. In addition, sample preparation processes are minimized and can be used easily in a drop-and-observe manner, which makes the technique attractive for large-scale onsite virus screening. It should be noted that the process of optical scattering in our method is independent of labels, which provides various label-free applications like water impurity detection or concentration and particle size analysis for a known sample.
The layout of the gold nanodot array is shown in Fig. 1. The fabrication process of the gold nanodot array is given as follows (more details in the ESI and Fig. S2†): The surface of the silicon substrate was first cleaned with acetone and isopropyl alcohol (IPA). After that, a 200 nm thick polymethyl methacrylate (PMMA) photoresist layer was spun on the substrate. Then the gold nanodot array was defined using electron beam lithography (EBL) and such a pattern was subsequently transferred onto PMMA by development. Next, a 10 nm thick titanium layer for enhanced adhesion and a 70 nm thick gold layer were deposited successively by employing the electron beam evaporation method. Followed by lift-off and cleaning procedures, the desired gold nanodot array was finally realized. Here, the diameter of a single gold disk is 110 nm, with a height of 70 nm. To avoid light coupling between adjacent nanodots, the spacing between adjacent disks in the horizontal and vertical directions was 1 μm. The gold nanodot array with a spacing of 0.5 μm was also fabricated and tested as a reference result to confirm the influence of light coupling. Since every group of four neighboring nanodots in the array will be utilized as a sensing channel for virus detection, their detailed geometry plays an important role in determining the image quality and detection sensitivity. The nanodots were made as round disks with dimensions close to the size of the virus-like particles. The circular shape of the disks guarantees an equal probability of virus attachment in any direction around the nanodot. The diameter of the gold disk was chosen to enhance photon scattering at the wavelength of 554 nm, which lies in the spectral range of the camera with a relatively high quantum efficiency.
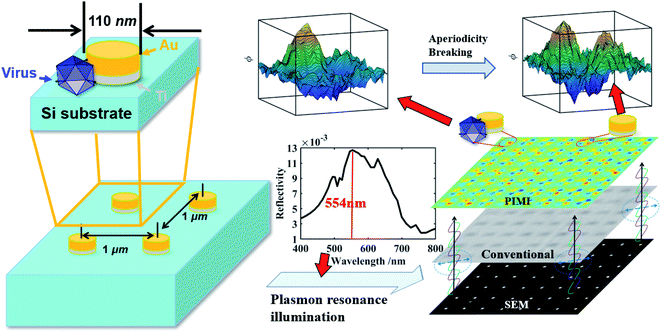 | ||
| Fig. 1 Schematic of virus-like particle detection and dimensions of gold nanodot arrays on a silicon substrate. | ||
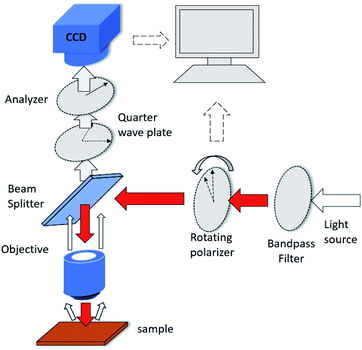
The photon state parametric images of the sample, i.e., sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and ϕ, were obtained using the polarization indirect microscopic imaging (PIMI) technique previously developed by our group (see the ESI†).29,30 The experimental system is based on a conventional reflection microscope (Olympus BX51), where a 554 nm filter is placed in front of the light source. A highly sensitive CCD (PiA2400-17gm, Basler) with 5 million pixels was used in combination with a 100× objective lens, making it possible to obtain images in which the pixel size corresponds to 34.5 nm.
δ and ϕ, were obtained using the polarization indirect microscopic imaging (PIMI) technique previously developed by our group (see the ESI†).29,30 The experimental system is based on a conventional reflection microscope (Olympus BX51), where a 554 nm filter is placed in front of the light source. A highly sensitive CCD (PiA2400-17gm, Basler) with 5 million pixels was used in combination with a 100× objective lens, making it possible to obtain images in which the pixel size corresponds to 34.5 nm.
The schematic of the PIMI system is shown in Fig. 2. In our PIMI measurement, the light reflected by the sample was passed through a quarter-wave plate and a linear analyzer was placed in front of the image plane with their fast axes oriented at 45° and 90° respectively relative to the x-axis.
The incident polarized light was rotated from 0° to 180° in steps of 18°, and the resulting 10 images were collected and fitted to a sine curve at every pixel (more details in the ESI and Fig. S1†), according to the formula:
Ii = 1/2 × I0[1 + sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ δ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(θi − ϕ)]. 2(θi − ϕ)]. | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and ϕ were obtained which respectively represent the phase difference between the two orthogonal polarization components of the scattered light and the polarization ellipse orientation angle of the beam reflected from the sample.
δ and ϕ were obtained which respectively represent the phase difference between the two orthogonal polarization components of the scattered light and the polarization ellipse orientation angle of the beam reflected from the sample.
The polarization states of photons scattered by deep sub-wavelength structures or particles heavily depend on the coupling between the incident photons and the optical anisotropy of the sample at the nanoscale. Calculations of the polarization states of scattered photons in the far-field demonstrate that parametric images can have a resolving power beyond the diffraction limit of conventional microscopy. This advantage was exploited in this work to determine whether virus-like particles were attached to the nanodot array. The lower right part of Fig. 1 displays images from a scanning electron microscope (SEM), conventional microscope and PIMI of the same region on the gold nanodot array. An intact virus particle typically has a dimension of 100 nm, close to the size of a single nanodot and below the optical diffraction limit. As shown in Fig. 1, the point spread function of conventional microscopy smeared out any structural features of the nanodots, making it impossible to recover information about possible virus-like particles that may have attached to the gold nanodots. The PIMI images in the upper right part of Fig. 1, however, show that the scattering photon state pattern of the nanodots exhibits high sensitivity to the attachment of virus particles.
The virus (obtained from Hanbio Technology Co., Ltd), was dissolved in phosphate buffer saline (PBS). After being diluted to a concentration of 1 ng mL−1, a microdroplet of around 1 μL was dropped directly on the gold nanodot array. Virus-like particles in the droplet randomly attach to the gold disk due to Brownian motion and surface adsorption, which breaks the periodic structure of nanodots and results in a local variation of the scattering photon state distribution in the PIMI images. To quantitively evaluate the influence of virus particles attached the gold nanodisk, the finite difference time domain (FDTD) method was used to simulate PIMI images for the experimental configuration. For comparison, every simulation image is down-sampled to the same pixel density as the experimental image.
Every four neighboring nanodots were grouped as a four-quadrant sensing channel and the purpose of such a four-quadrant sensing configuration is to provide a self-reference of the scattering pattern in each group, suppressing the effects of minor variations in morphology between individual nanodots in the quantitative analysis. Furthermore, this four-quadrant sensing channel will provide a mechanism for fast confirmation of the virus particle location in the whole sensing array, by repetitively analyzing each four-nanodot lattice, as shown in later parts.
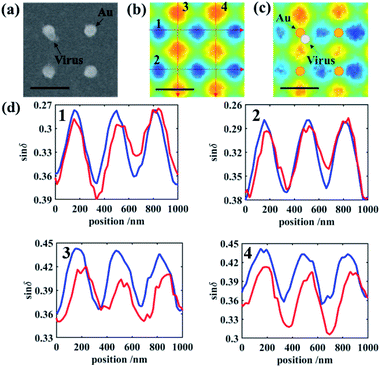
For the gold nanodots with an interval of 1 μm, Fig. 3 shows the comparison between the four bare disks and four gold disks with a virus particle located between them. When the virus particle is in the vicinity of the gold disk, the symmetry of the localized surface plasmon resonance (LSPR) of the gold disk and the symmetry of the scattered photon state distribution are both broken, which is shown clearly in Fig. 3b. This change is barely visible in the conventional microscopy images (Fig. 3a) because the attached virus results in only a small variation in the number of scattered photons (and hence intensity) and cannot be distinguished from the variation caused by background impurities on the substrate or irregularities in the shape of the gold nanodots. On the other hand, polarization state distribution of the scattered photons was drastically changed by the attached virus, not only due to the symmetry breaking of the morphological structure of the gold nanodisk, but also due to the plasmonic emission conditions which are associated with collective electron oscillation in the gold disk. The disturbance of the plasmonic emission further enhances the asymmetry of the photon state distribution induced by the structure of the gold disk–virus combination. As shown in Fig. 3b and c, the simulation and experimental PIMI images sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and ϕ show a significant variation due to the disturbance of the structural periodicity of the nanodots.
δ and ϕ show a significant variation due to the disturbance of the structural periodicity of the nanodots.
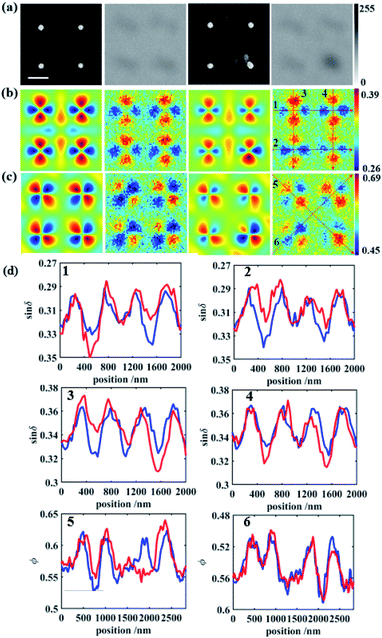
Characteristic curves were plotted along the trajectory shown in Fig. 3b and c, to show the detailed features of the PIMI images. In each figure, regularly appearing peaks in the curve of bare gold disks were disturbed by the attached virus, as indicated by the difference between the red and blue curves in Fig. 3d. In the characteristic curve along certain directions, the difference was significant due to the drastic change of the scattering photon states caused by the virus, e.g., the disappearance of the third peak in the red curve of 5 in Fig. 3d. This change of the characteristic curve can also be utilized to identify which nanodot was attached to the virus and its attaching direction, i.e., from the upper left corner of the attached nanodot.
To quantify the aperiodicity caused by virus, two characteristic parameters were defined to describe the aperiodicity of sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and ϕ at every nanodisk site. As shown in Fig. 4a, the photon state distribution at each site is split into four parts, which correspond to the four peaks of the scattering light distribution. We sum the pixel intensities in the four parts and define as follows:
δ and ϕ at every nanodisk site. As shown in Fig. 4a, the photon state distribution at each site is split into four parts, which correspond to the four peaks of the scattering light distribution. We sum the pixel intensities in the four parts and define as follows:
Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ = A/B + B/A + C/D + D/C δ = A/B + B/A + C/D + D/C | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ represents the aperiodicity of the sin
δ represents the aperiodicity of the sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ image, and A, B, C, and D represent the sum of the pixel intensities in the four parts. Apϕ represents the aperiodicity factor of the ϕ image, and shares the same formula as that in eqn (2) for Apsin
δ image, and A, B, C, and D represent the sum of the pixel intensities in the four parts. Apϕ represents the aperiodicity factor of the ϕ image, and shares the same formula as that in eqn (2) for Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ. For a 6 × 8 array, Apsin
δ. For a 6 × 8 array, Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and Apϕ are shown in Fig. 4c and d, respectively. We can therefore make a preliminary connection between a gold nanodot and virus and values of Apsin
δ and Apϕ are shown in Fig. 4c and d, respectively. We can therefore make a preliminary connection between a gold nanodot and virus and values of Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ, and Apϕ with a higher value being associated with a higher probability of the presence of the virus.
δ, and Apϕ with a higher value being associated with a higher probability of the presence of the virus.
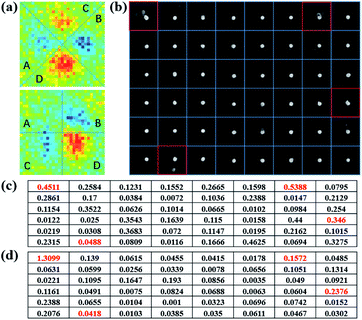 | ||
Fig. 4 (a) Partition of PIMI sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ (top) and ϕ images (bottom). (b) Separation of every site in the SEM image. (c) Apsin δ (top) and ϕ images (bottom). (b) Separation of every site in the SEM image. (c) Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ on the array surface. (d) Apϕ on the array surface. δ on the array surface. (d) Apϕ on the array surface. | ||
However, although there was a virus present in the square of row 6, and column 2, its Apsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ and Apϕ values were relatively low, i.e., 0.0488 and 0.0418, respectively. This is because the virus was too far away from the gold nanodot to modify the scattered photon distribution. The broken LSPR caused by virus-like particles only occurs at a distance smaller than half wavelength, which means that the period of gold nanodots determined the ratio of the detection area. In this paper, a series of distances between the virus and gold nanodots are simulated to determine the detection radius of nanodots, which is written as Rdet. After evaluating the values of the corresponding aperiodicity, Rdet is estimated to be around 150 nm (ESI and Fig. S5†).
δ and Apϕ values were relatively low, i.e., 0.0488 and 0.0418, respectively. This is because the virus was too far away from the gold nanodot to modify the scattered photon distribution. The broken LSPR caused by virus-like particles only occurs at a distance smaller than half wavelength, which means that the period of gold nanodots determined the ratio of the detection area. In this paper, a series of distances between the virus and gold nanodots are simulated to determine the detection radius of nanodots, which is written as Rdet. After evaluating the values of the corresponding aperiodicity, Rdet is estimated to be around 150 nm (ESI and Fig. S5†).
Here, we can also calculate the detection limit of the virus concentration. If N viruses are randomly distributed on the substrate and at least one virus is detected by the gold array, the minimum number of viruses should obey the inequality below:
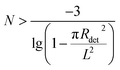 | (3) |
However, the denser gold nanodots will also lead to the overlap and merging of LSPR between two nanodots, which causes more complex conditions. An interval of 1 μm is the acceptable distance to distinguish the scattering field of two neighboring gold nanodots. To clarify the influence of the interval distance of the nanodisk, another gold nanodot array with a 0.5 μm interval is fabricated, and in such an interval, the light coupling between adjacent dots could not be ignored (Fig. S4†). In this interval, the PIMI ϕ image is rambling and cannot be used to accurately distinguish the virus-like particles. The scattering field of δ from neighboring nanodots merged into one and every four nanodots were grouped as a four-quadrant sensor. As shown in Fig. 5, a virus-like particle will always fall within the sensing area of a certain group of four nanodots and affect its scattering distribution. To find a parameter to define and quantify the aperiodicity in the whole field, such as the condition of 1 μm interval, is difficult to achieve. Here four other characteristic curves are utilized to quantify the influence of virus particles in the vicinity of the gold nanodots in the same region in Fig. 5b and c.
The characteristic curves for sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ images before and after deposition of the virus are shown in Fig. 5d. In general, the amplitude of characteristic curves decreases, due to the reduction of polarization characteristics caused by the virus-like particles. Characteristic curves before deposition mainly possess regular peaks with the same amplitude. After deposition, however, curves crossing the center of the disk connected with the virus generate a height difference among peaks. It is clear that in the curve of 1 and 3 in Fig. 5d, the virus raises the local aperiodicity and hardly influences the peaks in curves 2 and 4 in Fig. 5d. The results of the gold nanodots with an interval of 0.5 μm verify the detection feasibility at a higher ratio of the detection area. However, we can hardly conduct a quantitative analysis at every site and obtain similar tables of aperiodicity in Fig. 4 due to interaction within adjacent gold nanodots.
δ images before and after deposition of the virus are shown in Fig. 5d. In general, the amplitude of characteristic curves decreases, due to the reduction of polarization characteristics caused by the virus-like particles. Characteristic curves before deposition mainly possess regular peaks with the same amplitude. After deposition, however, curves crossing the center of the disk connected with the virus generate a height difference among peaks. It is clear that in the curve of 1 and 3 in Fig. 5d, the virus raises the local aperiodicity and hardly influences the peaks in curves 2 and 4 in Fig. 5d. The results of the gold nanodots with an interval of 0.5 μm verify the detection feasibility at a higher ratio of the detection area. However, we can hardly conduct a quantitative analysis at every site and obtain similar tables of aperiodicity in Fig. 4 due to interaction within adjacent gold nanodots.
Conclusions
In conclusion, we have demonstrated a method for label-free sensing of virus-like particles using a wide-field optical imaging system with a specially designed gold nanodot array. The scattering spatial spectra of photon states was found to be highly sensitive to the presence of virus particles attached to or close to gold nanodots. The presence and location of virus-like particles below the diffraction limit can be sensed by quantitative evaluation of the scattering pattern. This technique has the advantages of requiring minimal sample preparation, purification, and labeling, and eliminates the need for stringent and time-consuming image processing. Furthermore, the gold nanodot array provides a platform with the potential of high throughput parallel sensing in wide-field images, employing each nanodot group in the field of view to act as a sensing channel for viruses. Gold nanoarrays on a silicon substrate can be fabricated using the standard CMOS foundry process or nanoimprint technology, which makes it easy to be fabricated on a large scale at a relatively low cost. Hopefully, this can be developed to handle the complex detection process for infectious diseases. We did not give a complete system with a set judging condition or detection rate. This work currently only confirms the single-particle level detection ability of our method. Also, it should be noted that we used samples with known adenovirus particles to prove the capability of this method as a fast onsite screening technique before detailed analysis of the sample using lab-based techniques such as PCR. In future work, antibody modification of the nanodots will be employed to achieve virus-specific capturing and sensing, allowing accurate screening of samples with unknown particles and contaminations with a similar size and morphology as the target virus particles.Author contributions
Xuefeng Liu, Jichuan Xiong and Ming Sun designed and oversaw the project. Shengwei Ye and Xiao Sun fabricated the nanodot arrays with assistance from Lianping Hou and John H. Marsh. Weiping Liu designed the nanodot array with assistance from Bin Ni and Bin Xu. Xiao Jin and Heng Zhang performed the measurement of PIMI images and analysis of the experimental results. Heng Zhang prepared the virus particles with the assistance of Xiaofeng Li, Shanhu Li, and Lei Dong. Xiao Jin and Heng Zhang wrote the manuscript with the help of Jamie Jiangmin Hou. All the authors contributed to the discussion and revision of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Major Scientific Instruments and Equipment Development Project (No. 61827814), Postdoctoral Foundation of Jiangsu Province (No. 2020Z331), Beijing Natural Science Foundation (No. Z190018), the National Natural Science Foundation of China (No. 61627802), Fundamental Research Funds for the Central Universities (No. 30920010011), the Ministry of Education Collaborative Project (B17023), and UK Engineering and Physical Sciences Research Council (EP/R042578/1).References
- S. Lambert-Niclot, A. Cuffel, S. L. Pape, C. Vauloup-Fellous, L. Morand-Joubert, A. Roque-Afonso, J. L. Goff and C. Delaugerre, J. Clin. Microbiol., 2020, 58, e00977 CrossRef CAS PubMed.
- J. Tsou, H. Liu, S. A. Stass and F. Jiang, Biomedicines, 2021, 9, 239 CrossRef CAS PubMed.
- F. U. H. Muhammad, S. S. Bukhari, R. Ejaz, F. U. Zaman, K. Rajan Sreejith, N. Rashid, M. Umer and N. Shahzad, Virus Res., 2021, 32, 198484 Search PubMed.
- B. Reynés, F. Serra and A. Palou, BioTechniques, 2021, 70, 4 CrossRef PubMed.
- T. Suo, X. Liu, J. Feng, M. Guo, W. Hu, D. Guo, H. Ullah, Y. Yang, Q. Zhang, X. Wang, M. Sajid, Z. Huang, L. Deng, T. Chen, F. Liu, K. Xu, Y. Liu, Q. Zhang, Y. Liu, Y. Xiong, G. Chen, K. Lan and Y. Chen, Emerging Microbes Infect., 2019, 9, 1259 CrossRef PubMed.
- H. Shi, X. Han, Y. Cao, O. Alwalid and C. Zheng, Lancet Infect Dis., 2020, 20, 1011 CrossRef CAS PubMed.
- F. Carinci, G. Moreo, L. Limongelli, T. Testori and D. Lauritano, Appl. Sci., 2020, 10, 4506 CrossRef CAS.
- Y. Takeuchi, M. Furuchi, A. Kamimoto, K. Honda, H. Matsumura and R. Kobayashi, J. Oral Sci., 2020, 62, 350 CrossRef CAS PubMed.
- S. Dowlatshahi, E. Shabani and M. J. Abdekhodaie, Arch. Virol., 2021, 166, 1 CrossRef PubMed.
- N. Renard, S. Daniel, N. Cayet, M. Pecquet, F. Raymond, S. Pons, J. Lupo, C. Tourneur, C. Pretis, G. Gerez, S. Pons, J. Lupo, C. Tourneur, C. Pretis, G. Gerez, P. Blasco, M. Combe, I. Canova, M. Lesenechal and F. Berthier, J. Clin. Microbiol., 2021, 59, e02292 CrossRef CAS PubMed.
- T. Yamanaka, M. Nemoto, H. Bannai, K. Tsujimura, T. Kondo, T. Matsumura, T. Q. H. Fu, C. J. Fernandez, S. Gildea and A. Cullinane, J. Vet. Med. Sci., 2017, 79, 1061 CrossRef CAS PubMed.
- S. Enoki, R. Lion, K. Kaihatsu, S. Sakakihara, N. Kato and H. Noji, PLoS One, 2012, 7, e49208 CrossRef CAS PubMed.
- M. V. Maigler, M. Holgado, M. F. Laguna, F. J. Sanza, B. Santamaría, A. Lavin and R. L. Espinosa, IEEE Trans. Instrum. Meas., 2019, 68, 3193 CAS.
- C. Y. Lee, I. Degani, J. Cheong, J. H. Lee, H. J. Choi, J. Cheon and H. Lee, Biosens. Bioelectron., 2021, 178, 113049 CrossRef CAS PubMed.
- A. E. Ekpenyong, M. M. Si, S. Achouri, C. E. Bryant, J. Guck and K. J. Chalut, J. Biophotonics, 2013, 6, 393 CrossRef PubMed.
- P. Y. Liu, L. K. Chin, W. Ser, T. C. Ayi and Y. L. Wang, 2015 Conference on Lasers and Electro-Optics (CLEO), IEEE, 2015 Search PubMed.
- Y. T. Yeh, K. Gulino, Y. Zhang, A. Sabestien, T. W. Chou, B. Zhou, Z. Lin, I. Albert, H. Lu, V. Swaminathan, E. Ghedin and M. Terrones, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895 CrossRef CAS PubMed.
- X. Li, J. Liu, Q. Liu, L. Yu and X. Yin, Chin. J.Biotechnol., 2020, 36, 732 Search PubMed.
- H. Yoo, J. Shin, J. Sim, H. Cho and S. Hong, Biosens. Bioelectron., 2020, 168, 112561 CrossRef CAS PubMed.
- M. Xiao, L. Huang, X. Dong, K. Xie, H. Shen, C. Huang, W. Xiao, M. Jin and Y. Tang, Anal., 2019, 114, 2594 RSC.
- J. Zhong, E. L. Rsch, T. Viereck, M. Schilling and F. Ludwig, ACS Sens., 2021, 6, 976 CrossRef CAS PubMed.
- T. T. S. Lew, K. M. M. Aung, S. Y. Ow, S. N. Amrun and X. Su, ACS Nano, 2021, 15, 12286 CrossRef CAS PubMed.
- Y. Li, P. Ma, Q. Tao, H. J. Krause, S. Yang, G. Ding, H. Dong and X. Xie, Sens. Actuators, B, 2021, 337, 129786 CrossRef CAS PubMed.
- A. Idris, A. Davis, A. Supramaniam, D. Acharya and K. V. Morris, Mol. Ther., 2021, 29, 2219 CrossRef CAS PubMed.
- J. Liu, M. Jalali, S. Mahshid and S. Wachsmann-Hogiu, Analyst, 2020, 145, 364 RSC.
- Y. Xia, Y. Chen, Y. Tang, G. Cheng, X. Yu, H. He, G. Cao, H. Lu, Z. Liu and S. Y. Zheng, ACS Sens., 2019, 4, 3298 CrossRef CAS PubMed.
- H. Zhang, L. Xue, F. Huang, S. Wang, L. Wang, N. Liu and J. Lin, Biosens. Bioelectron., 2019, 127, 142 CrossRef CAS PubMed.
- H. Zhang, X. Jin, H. Zhao, Y. Lin, X. Li, L. Hou, J. Marsh, L. Dong, D. Shi, W. Liu, B. Xu, B. Xu, J. Liu, J. Xiong and X. Liu, IEEE Photonics J, 2021, 13, 3900111 CAS.
- K. Ullah, X. Liu, M. Habib and Z. Shen, Acs Photonics, 2018, 5, 1388 CrossRef CAS.
- K. Ullah, B. Garcia-Camara, M. Habib, N. P. Yadav and X. Liu, J. Quant. Spectrosc. Radiat. Transfer, 2018, 213, 35 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00603g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |