Open Access Article
Open Access ArticleTumor-targeting inorganic nanomaterials synthesized by living cells
Yuzhu
Yao
 a,
Dongdong
Wang
a,
Dongdong
Wang
 a,
Jun
Hu
a,
Jun
Hu
 *abc and
Xiangliang
Yang
*abc and
Xiangliang
Yang
 *abc
*abc
aNational Engineering Research Center for Nanomedicine, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China
bHubei Key Laboratory of Bioinorganic Chemistry and Materia Medica, School of Chemistry and Chemical Engineering, Huazhong University of Science and Technology, Wuhan 430074, China
cKey Laboratory of Molecular Biophysics of Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China. E-mail: hjun0718@hust.edu.cn; yangxl@hust.edu.cn
First published on 12th April 2021
Abstract
Inorganic nanomaterials (NMs) have shown potential application in tumor-targeting theranostics, owing to their unique physicochemical properties. Some living cells in nature can absorb surrounding ions in the environment and then convert them into nanomaterials after a series of intracellular/extracellular biochemical reactions. Inspired by that, a variety of living cells have been used as biofactories to produce metallic/metallic alloy NMs, metalloid NMs, oxide NMs and chalcogenide NMs, which are usually automatically capped with biomolecules originating from the living cells, benefitting their tumor-targeting applications. In this review, we summarize the biosynthesis of inorganic nanomaterials in different types of living cells including bacteria, fungi, plant cells and animal cells, accompanied by their application in tumor-targeting theranostics. The mechanisms involving inorganic-ion bioreduction and detoxification as well as biomineralization are emphasized. Based on the mechanisms, we describe the size and morphology control of the products via the modulation of precursor ion concentration, pH, temperature, and incubation time, as well as cell metabolism by a genetic engineering strategy. The strengths and weaknesses of these biosynthetic processes are compared in terms of the controllability, scalability and cooperativity during applications. Future research in this area will add to the diversity of available inorganic nanomaterials as well as their quality and biosafety.
Introduction
With the development of nanotechnology, inorganic nanomaterials are widely produced and applied in the biomedical field, as detection, diagnostic, monitoring, and therapeutic tools.1–5 However, inorganic nanomaterials synthesized by conventional methods (such as hydrothermal or solvothermal methods) are far from satisfactory due to their poor biocompatibility, poor targeting effect, and low bioavailability. Inspired by the biosynthesis process in nature, inorganic nanomaterials based on living cell synthesis have attracted researchers' attention.The synthesis of inorganic materials by living cells is widespread in nature. For example, orientationally arranged magnetic nanoparticles (NPs) can be fabricated within magnetotactic bacteria to make them susceptible to the geomagnetic field, which helps them to swim to deep water with relatively low oxygen content.6 Some living plants and microalgae can absorb metal ions in water or soil and then convert them to insoluble nanomaterials to mitigate environmental stress.7,8 Inspired by nature, a variety of inorganic nanomaterials (NMs) including metallic/metallic alloy NMs, metalloid NMs, oxide NMs and chalcogenide NMs could be constructed intracellularly or extracellularly by living cells of bacteria, fungi, animals and plants. For example, Kangpeng Wang et al. reported the biosynthesis of Te nanostructures by growing Bacillus selenitireducens in a lactate-tellurite medium.9 Qing-Ying Luo et al. employed yeast cells to synthesize CdSe quantum dots (QDs) intracellularly.10 Maonan Wang reported the in situ biosynthesis of Au nanoclusters (NCs) using both cultured cancer cells and tumors of an orthotropic liver tumor mouse model.11 Alejandra Arévalo-Gallegos et al. used Botryococcus braunii as a bioreactor for the production of Ag nanoparticles.12
Living cell synthesis can be conducted at room temperature with less energy consumption and negligible hazardous waste generation, and the obtained biosynthetic nanomaterials have the characteristics of good biocompatibility and unique biological properties. The ever-growing interest and significant progress of biosynthetic nanomaterials have witnessed broad prospects in biomedical applications due to their combined advantages of natural substances and nanotechnology.13 For example, engineered bacteria with photothermally controlled TNF-α expression and Au nanoparticles synthesized intracellularly along with near-infrared (NIR) realized efficient tumor-targeting therapy due to their preference for hypoxic and nutrient-rich regions.14In situ biosynthesized Au NCs in tumor cells were proved to suppress cancer development via the inhibition of the P13K-AKT signalling pathway.15
However, compared with chemical synthesis processes, the size and morphology of the synthesized nanomaterials by living cells are usually uncontrollable. As we all know, both the size and morphology have crucial impacts on the properties of nanoparticles. They can be regulated to a certain extent through pH, temperature, precursor type and proportion, and incubation time, such as the pH-controlled morphology of Au nanoparticles synthesized by living plants,16 temperature-adjusted fluorescence intensity of CdSe QDs synthesized by fungus F. oxysporum,17 and incubation time-regulated size and emission wavelength of CdTe QDs synthesized by bacteria.18 But these simple adjustments are far from satisfactory. For the higher controllability of biosynthetic processes, the mechanism of the formation of inorganic nanomaterials within different types of living cells should be clarified first. Bioreduction and detoxification as well as biomineralization are often involved in the mechanism of biosynthesis. Many kinds of intracellular enzymes, reducing molecules such as glutathione and glucose, cell surface expressed polysaccharides and proteins etc. usually participate in these complicated reactions.19–21 By adjusting the expression level of target proteins or peptides by means of genetic engineering, the metabolic pathway, redox state etc. of living cells can be regulated, so as to regulate the composition, morphology, size and yield of synthesized nanomaterials. For instance, the recombinant Escherichia coli (E. coli) coexpressing metallothionein (MT) and phytochelatin synthase (PCS) showed enhanced metal binding capacity and promoted the assembly of diverse metal elements into highly ordered NPs.22 The yield of CdSe QDs was significantly improved by impairing the extracellular electron transfer (EET) ability of the bacteria via the deletion of the CymA-encoding gene.23
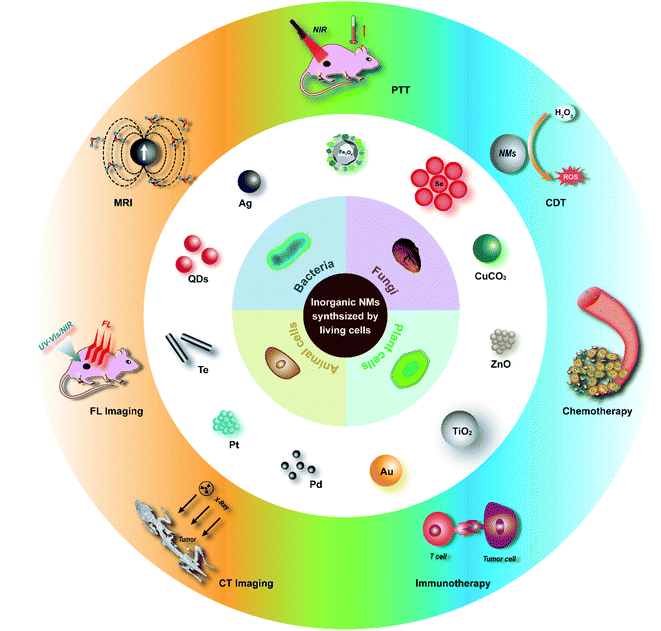
In general, biosynthesis using living cells has emerged as an innovative and attractive green chemical methodology for the preparation of inorganic nanomaterials. In this review, we summarize the biosynthesis of inorganic nanomaterials in different types of living cells including bacteria, fungi, plant cells, and animal cells, while the strengths and weaknesses of the biosynthetic processes are systematically presented, especially for application in tumor-targeting theranostics (Scheme 1). The mechanisms of the biosynthesis are analysed and it is generally accepted that inorganic-ion bioreduction and detoxification as well as biomineralization are usually involved. For controlling the size and morphology of the products, the modulation of precursor ion concentration, pH, temperature, and incubation time, as well as cell metabolism by a genetic engineering strategy is proposed in many cases. However, from the perspective of biomedical application, there are still some issues that need to be addressed which are indicated in the last part of the review. Future research in this area will add to the diversity of available inorganic nanomaterials as well as their quality and biosafety.
Bacterial synthesis
Bacteria, a kind of unicellular prokaryote, have been widely studied in living-cell biosynthesis of inorganic nanomaterials23–25 owing to their characteristics of single cell growth, facile cultivation, rapid proliferation, and easy genetic manipulation.26 Metal ions and some oxyanions can be taken up by bacteria to participate in their metabolic processes to produce metallic/metalloid NMs, oxide NMs and chalcogenide NMs through biomineralization, bioreduction or intracellular detoxification.Metal oxide NMs synthesized through biomineralization
The most famous case of bacterial synthesis is magnetic nanoparticles intracellularly formed by magnetotactic bacteria. In 1975, R. Blackmore for the first time observed and reported the magnetotactic bacteria (MTB), which could swim aligned parallel to the Earth's geomagnetic field.27 The MTB were found to naturally synthesize intracellular chains of magnetic nanoparticles (called magnetosomes), with a structure of one magnetic mineral nanocrystal core and surrounding phospholipids and proteins (Fig. 1A).28–30 The magnetosome biogenesis has been shown to consist of the formation of an empty magnetosome membrane through the invagination of the cytoplasmic membrane, magnetosome protein sorting to the magnetosome membrane, iron transportation into the magnetosome membrane and biomineralization as iron-rich magnetic nanocrystals, and the final assembly and positioning of the magnetosome chain.31 In studies to reveal the mechanism of magnetosome synthesis, researchers found that the protein MamP had unique protein-folding structures called magnetochrome domains which contributed to the attraction for iron.32 MamP can oxidize Fe(II) to Fe(III), and eventually synthesize Fe3O4 containing both Fe(II) and Fe(III). It was verified in vitro that Fe(II) was finally transformed to Fe(III) in the presence of MamP Fe(II). Due to the inherent hypoxia-targeting and magnetic-targeting ability of the MTB and the magnetic and thermal properties of magnetosomes,33 the MTB have been widely used in enhanced magnetic resonance imaging (MRI),34 fluorescence imaging and probing,35–37 as well as tumor-targeting magnetic hyperthermal therapy (MHT)38 and photothermal therapy (PTT).39 Chuanfang Chen et al. reported that the bacterial magnetic nanoparticles (BMPs) isolated from the MTB successfully achieved T2-weighted MRI enhancement and PTT of tumors (Fig. 1A).34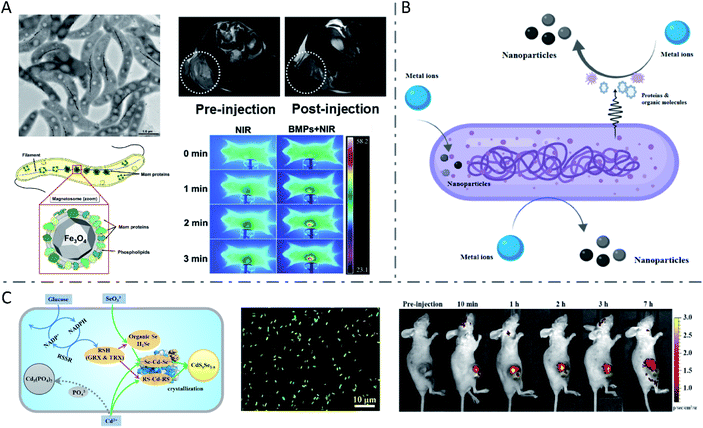 | ||
| Fig. 1 Inorganic nanoparticles synthesized by wild-type bacteria. (A) Biosynthesis of magnetosomes by MTB and application in T2 weighted MRI and PTT of tumors. Reproduced from ref. 28 with permission from American Society for Microbiology, copyright 2019. Reproduced from ref. 34 with permission from Elsevier Ltd., copyright 2016. (B) Biosynthesis mechanism of metal nanoparticles by bacteria. Reproduced from ref. 44 with permission from American Chemical Society, copyright 2020. (C) Metabolism-regulated Se and Cd biotransformation pathways in E. coli, and in vitro and in vivo fluorescence images of the synthesized QDs in glucose-facilitated E. coli cells. Reproduced from ref. 26 with permission from American Chemical Society, copyright 2019. | ||
Metallic NMs synthesized through detoxification
Inspired by the naturally biosynthesized nanomaterials, a variety of bacteria have been explored as biofactories to synthesize a diverse range of nanomaterials. Different from Fe ions, noble metal ions in the environment are usually toxic to bacteria. To mitigate environmental stress, they are commonly reduced to their insoluble form by the bioactive molecules or proteins present inside bacteria (such as nicotinamide dinucleotide (NADH),40 reduced nicotinamide dinucleotide phosphate (NADPH),26,41 glutathione (GSH),41,42 glutathione reductase (GR),41 nitrate reductase (NR),14 fumarate reductase FccA,43etc.), as well as by the cell surface expressed or extracellularly secreted reducing species (Fig. 1B).44 Bioreduction45,46 or intracellular detoxification18,47 is often involved in this process.Various functional groups including thiol, carboxyl, and amino groups present on the surface of bacteria have high affinity for metal ions. Upon reduction with reducing species originating from bacteria or extra added ones, metallic NMs are formed through nucleation and surface growth. For example, E. coli was used as a biofactory to reduce chloroauric acid and modulate the formation of Au NPs on the bacterial surface, in which sugars or enzymes might modulate the biosynthesis.48 Gary Attard et al. demonstrated the synthesis of Pt NPs by E. coli MC4100 with different degrees of metal loading, in which Pt(II) bound on the surface of bacteria was reduced to Pt(0) by sparging hydrogen into the suspension.49 In a recent study, bimetallic Au–Ag nanoparticles were biosynthesized by E. coli, in which Au ions were reduced by the phenolic group of tyrosine residues present in the surface protein of E. coli.50 At basic pH, the phenolic group converted into a negative phenolic ion that can reduce Au ions to Au atoms.51 After the formation of Au NPs, an Au–Ag core–shell nanostructure was obtained with the combination of Ag ions. Compared with single Au or Ag nanoparticles, the biosynthesized Au–Ag bimetallic NPs showed enhanced photothermal therapy performance and antibacterial ability.
Under environmental stress, heavy metal ions can be intracellularly accumulated by bacteria through the transport system and then converted into insoluble zero-valent nanoparticles, such as Au NPs,52 Ag NPs,52,53 and Cu NPs.54 For example, Au, Ag and Au–Ag alloy crystals were synthesized by Lactobacillus strains via incubation with the corresponding precursor ions.52 The bacteria with crystals synthesized remained viable, and the coalescence of the nanoclusters was considered to be protective for the living cells by reducing the surface area of the crystals. In another study, an E. coli strain that carried chromosomally encoded silver resistance determinants was observed to form Ag NPs in the periplasmic space upon exposure to Ag+ ions.53
Metalloid NMs synthesized through bioreduction
Different from the metal detoxification mechanism, for some elements such as Se and Te, their oxyanions could participate in the respiratory chain process as respiratory electron acceptors, and finally could be reduced to a low-valent state. Anaerobic bacterium Bacillus selenitireducens was used to synthesize Te nanostructures at room temperature.9 In another study, intracellular synthesis of Te nanorods (NRs) by living Staphylococcus aureus (S. aureus) cells via intracellular reduction of TeO32− was reported, and the intracellular biochemical pathways were studied.41 The Te(IV) in TeO32− was first reduced to GSTeSG by intracellular GSH, and further reduced to GSTeH by NADPH and GR. The resulting intermediate GSTeH spontaneously decomposed into Te(0), and finally formed Te nanorods. To verify this mechanism, in vitro synthesis of high-quality Te nanorods was successfully achieved using a quasi-biological system containing electrolytes, reduced GSH, reduced NADPH and GR, which were involved in the intracellular reaction. It was further verified that the length of TeNRs can be adjusted both by changing the concentration of NaOH to control the concentration of Te(0) and by changing the concentration of GR to control the generation rate of Te(0). Considering the great photothermal conversion ability of Te55–57 and the tumor-targeting ability and immune response activation of bacteria,58–61 our recent work (unpublished) employed bacteria with Te nanorods synthesized intracellularly as a comprehensive treatment system in tumor-targeting photothermal immunotherapy. Belonging to the same family within the periodic table as Te, Se in Na2SeO3 could also be reduced to low-valence Se by certain bacteria.43,62Shewanella oneidensis (S. oneidensis) MR-1, a model dissimilatory metal-reducing bacterium (DMRB), can respire on multiple metal ions to produce nanoparticles through dissimilatory respiration and energy derivation.43,63,64 Dao-Bo Li et al. reported that fumarate reductase FccA is the terminal reductase of selenite in the periplasm of S. oneidensis, and c-type cytochrome CymA, which is central to respiration, is of vital importance in the reduction process.43Metallic chalcogenides synthesized by bacteria
Combining the two processes mentioned above, a number of metallic chalcogenides can be synthesized by living bacteria, among which the most representative ones are QDs. Quantum dots are a kind of inorganic semiconductor nanocrystal with excellent optical properties, such as tunable emission with size and components in a wide spectral range, high photo stability and fluorescence quantum yield, and a long fluorescence life time, which endow them with great potential in bio-labelling and detection, in vivo imaging and tracking, and tumor-targeting drug delivery and therapy.65–69 Zhang Yi et al. prepared CdSe/CdS QDs with good water solubility and distinct yellow fluorescence using E. coli cells for in vitro Hg2+ detection.70 It was reported that glucose addition to the culture medium of E. coli would change the metabolic route of the Se and Cd from Cd(PO4)2 formation to CdSxSe1−x QDs assembly, yielding fluorescent bio-QDs (Fig. 1C).26 The researchers presented that glucose metabolism improved the NADPH production, thus leading to more reduced thiol groups (RSH), which were critically responsible for Se reduction and Cd binding, thereby significantly raising the CdSxSe1−x QDs synthesis rate and yield. The resulting QDs were successfully applied for imaging tumors. Moreover, S. aureus cells were also used to synthesize CdS0.5Se0.5 QDs with outstanding photostability and high luminance through the “space–time coupling strategy”.47 By adding Na2SeO3 and CdCl2 to S. aureus suspension at different times, the Cd precursor formed by Cd2+ detoxification can react precisely with the low-valence organoselenium compounds resulting from intracellular GR-involved Na2SeO3 reduction, to create CdS0.5Se0.5in vivo, which can never occur in living cells in nature. Due to the specific interaction between the protein A expressed on the S. aureus surface and the Fc fragment domain of antibodies, the constructed cellular beacons (fluorescent cells) can be easily and widely applied as nanobioprobes for pathogen detection in vitro. Another study constructed a system for in vitro detection of human prostate-specific antigens using biosynthesized QDs in a similar way as above.71 Haifeng Bao et al. reported the extracellular biosynthesis of CdTe QDs by E. coli, directly based on secreted proteins.18 It was found that the size of bacteria-synthesized CdTe QDs increased with prolonged incubation time, with a red shift of the absorption edge and emission peak. The obtained QDs with size-tunable optical properties were functionalized with folic acid and employed to image cancer cells in vitro.Engineered bacteria for biosynthesis
Although many species of bacteria can be used to synthesize inorganic nanomaterials, the types of nanomaterials prepared by wild-type (WT) bacteria are still limited, and their size and morphology are often uncontrollable. However, size and morphology have a crucial influence on the properties of nanomaterials. Therefore, genetically engineered bacteria have been used for more extensive and more controlled synthesis of nanomaterials, which is the most significant advantage of bacteria over other living cells. E. coli is one of the widely studied bacterial species for nanomaterial biosynthesis, with finely controllable genomic and metabolic functions.72–75 After being engineered to express MT and/or PCS, recombinant E. coli cells were incubated with various metal ions, including semiconducting (Cd, Se, Zn, Te), alkali-earth (Cs, Sr), magnetic (Fe, Co, Ni, Mn), and noble (Au, Ag) metals and rare-earth fluorides (Pr, Gd), to synthesize the corresponding metal NPs.21 In the synthesis processes, MTs are capable of binding heavy metals (i.e., Cu, Cd, and Zn), while the metal-binding peptide phytochelatin (PC) synthesized by PCS plays a significant role in heavy-metal detoxification processes, as a family of cysteine-rich, thiol-reactive peptides that bind several toxic metals (i.e., Cu, Zn, Cd, Hg, and Pb). Thus, the engineering of E. coli led to the enhanced assembly of diverse metal elements into highly ordered NPs. This was the first time that an engineered bacterium was used for in vivo synthesis of a wide range of functional metal NPs. In the same study, it was demonstrated that the synthesized inorganic nanomaterials after feeding different concentrations of precursor metal ions exhibited various diameters, which further manifested as diverse colors and fluorescence emission wavelengths of QDs. The engineered E. coli cells were also employed to synthesize FeCo nanoparticles, the diameter of which could be tuned from (3.02 ± 1.00) to (5.57 ± 1.04) nm by varying the concentration of Fe2+ (or Fe4+) and Co2+ ions from 0.5 to 2.0 mM. Subsequently, Yoojin Choi et al. scanned the periodic table to select 35 elements and biosynthesized 60 different nanomaterials by employing a recombinant E. coli strain coexpressing MT and PCS (Fig. 2A).40 For most nanomaterials in the study, the biosynthesis begins in the cytoplasm, where MT and PC exist, and then the nanomaterials move to the cell wall to continue the synthesis. Moreover, the metal ions nucleate with PCs as a binding template and nucleation site, and are stabilized by PCs to prevent further aggregation. A Pourbaix diagram that summarized the producibility and crystallinity of a series of synthesized nanomaterials was used to predict the suitable reduction potential (Eh) and pH for nanomaterial biosynthesis. Then, through shifting the initial pH of in vivo reactions from 6.5 to 7.5, various crystalline nanomaterials of previously amorphous or not-synthesized ones were successfully synthesized by living cells of E. coli coexpressing MT and PCS. With respect to QDs, a study on S. oneidensis MR-1 showed that abundant Se and a small amount of CdSe nanoparticles were observed in WT bacterial cells after exposure to Na2SeO3 and CdCl2 (Fig. 2B).23 Then, S. oneidensis mutants with the CymA-encoding gene deleted (ΔcymA) or overexpressed (PYYDT-cymA) were constructed to impair or promote the intrinsic EET ability. The ΔcymA showed high production of CdSe and low production of Se nanoparticles, while the PYYDT-cymA performed oppositely.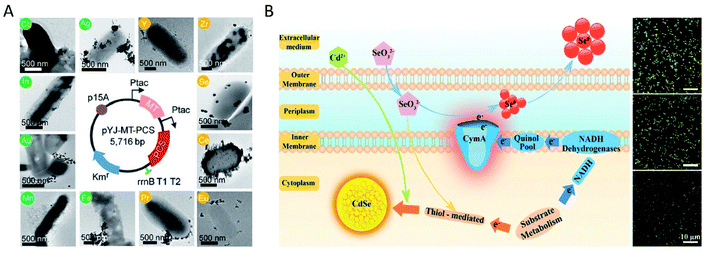 | ||
| Fig. 2 Inorganic nanoparticles synthesized by engineered bacteria. (A) The map of plasmid pYJ-MT-PCS used to construct the recombinant E. coli DH5α strain coexpressing MT and PCS, and TEM images of nanomaterials synthesized in vivo with their corresponding elements labeled in circles. Reproduced from ref. 40 with permission from the National Academy of Sciences, copyright 2018. (B) Schematic diagram of the EET-dependent synthesis of nanoparticles by S. oneidensis MR-1, and fluorescence microscopy images of the biosynthesized nanomaterials in vivo by ΔcymA (above), WT (middle) and PYYDT-cymA (below). Reproduced from ref. 23 with permission from American Chemical Society, copyright 2017. | ||
Applications in tumor-targeting theranostics
Since numerous bacterial strains have an innate ability to target tumors due to their preference for hypoxic and nutrient-rich regions, nanomaterials synthesized in bacterial cells could be delivered to tumors directly with bacteria as carriers.76–79 Moreover, outer membrane proteins expressed on the surface of bacteria could interact with antigen-presenting cells to elicit an immune response to synergize with the nanomaterials to enhance their therapeutic effect.80,81 Additionally, outer membrane proteins can also serve as templates to realize the biomineralization of nanomaterials on the surface of bacteria, retaining the excellent tumor-targeting ability and immunogenicity of bacteria. For example, thermally sensitive programmable bacteria (TPB) expressing therapeutic proteins TNF-α were employed to synthesize Au nanoparticles via electron shuttle enzymatic metal reduction, to obtain TPB@Au.14 The constructed bacteria-based antitumor vehicles along with NIR achieved satisfactory tumor-targeting therapeutic efficiency via oral administration.14 In another study, Pd nanoparticles were biomineralized by S. oneidensis MR-1 on its surface to form the so-called “photothermal bacterium” (PTB).64 The living PTB was further modified with zeolitic imidazole framework-90 (ZIF-90) encapsulating photosensitizer methylene blue (MB), which can selectively release MB in mitochondria. The PTB@ZIF-90/MB showed a significant tumor PTT effect, due to the selective targeting and tumor heat resistance suppression (Fig. 3).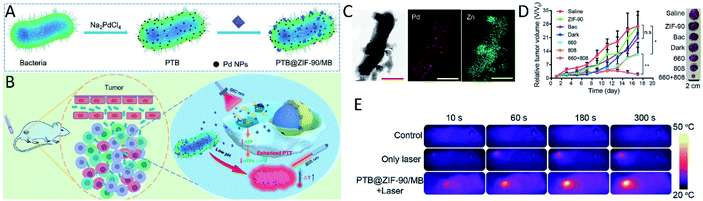 | ||
| Fig. 3 Bacteria with inorganic nanomaterials synthesized on their surface for tumor-targeting therapy. Schematic illustration of the (A) synthetic procedure of PTB and PTB@ZIF-90/MB and (B) application in tumor-targeting PTT. (C) TEM-assisted element mapping of Pd and Zn on PTB@ZIF-90/MB (scale bar: 500 nm). (D) The antitumor efficiency and (E) photothermal property study of PTB@ZIF-90/MB in mice. Reproduced from ref. 64 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2020. | ||
In addition to these mentioned above, some other inorganic nanomaterials such as MnOx nanospindles can be synthesized extracellularly by E. coli to encapsulate the chemotherapeutic agent doxorubicin (DOX) for tumor microenvironment responsive T1-weighted MRI enhancement and chemo-chemodynamic therapy.46 These biosynthetic nanomaterials often show high colloidal stability, owing to the coating of biomolecules. Although WT bacteria have been successfully employed for the synthesis of several types of nanomaterials composed of noble metals, oxides and chalcogenides by using their inherent bioreduction or detoxification processes, the diversity as well as the size and composition of nanomaterials need to be better controlled. Biogenetic engineering seems to be an alternative strategy to address this issue. The production of nanomaterials by bacteria depends on their ability to resist the effects of environmental stress such as toxicity arising from metallic ions.82 High concentration of toxic ions will induce the death of bacteria. Therefore, the efficiency of nanomaterial synthesis by bacteria is always low. Additionally, in terms of biomedical applications, the potential biological toxicity brought by bacteria especially pathogenic bacteria should be considered.83
Fungal synthesis
Fungi, a group of unicellular, multicellular, or syncytial spore-producing eukaryotic organisms, include yeasts, molds, mushrooms, etc.84–86 Similar to bacteria, fungi have the advantages of significant growth rate, simple culture procedures, and facile and economical biomass handling for biosynthesis of nanomaterials.87,88 Furthermore, fungi could tolerate higher metal concentrations, and secrete more extracellular enzymes and proteins than other microorganisms, thus leading to higher productivity of nanomaterials.89–91 However, investigations on fungal synthesis are fewer than those on bacterial synthesis, mainly due to the fact that their structure makes it complicated to characterize the internal nanomaterials for microscopic and mechanistic studies.92Synthesis by yeast cells
Yeasts, a family of various single-celled fungi that reproduce asexually by budding or division, have been broadly used in biosynthesis owing to their rapid proliferation and easy genetic manipulation.93 Due to their strong ability to chelate toxic metals such as Cd, yeasts were extensively utilized for biosynthesis of QDs such as CdS,94 CdSe,10,19,95,96 and CdTe.97 It was first reported in 1989 that CdS crystallites were obtained by incubating the yeasts Candida glabrata and Schizosaccharomyces pombe with cadmium salts.94 When exposed to cadmium ions, the yeast cells produced metal-chelating peptides with a general structure of γ-(Glu-Cys)n-Gly, which in turn bound with cadmium ions and controlled the nucleation and growth of CdS crystallites. Another metal sulphide nanocrystallite, i.e., PbS, synthesized by yeast was reported in 2002.98 Compared with CdS QDs, CdSe or CdTe QDs usually exhibited more excellent optical properties. Through complex intracellular biochemical reactions, Na2SeO3 or Na2TeO3 internalized by yeast cells can be metabolized to low-valence –SeH/–TeH in the cytoplasm. By the detoxification process, Cd2+ can be transformed to more stable [Cd(GS)2]2+ and eventually stored in the vacuoles of yeast cells. –SeH/–TeH and [Cd(GS)2]2+ are actually the precursors for the synthesis of CdSe or CdTe QDs. However, these two reactions are independent of each other in yeast cells. Thus, in order to obtain CdSe or CdTe QDs, it is a key point to make these two unrelated reactions sequentially occur at an appropriate time and space as desired. To address this issue, Ran Cui et al. proposed a “space–time coupling strategy” to synthesize CdSe QDs in living cells of Saccharomyces cerevisiae by feeding the cells with Na2SeO3 and CdCl2 sequentially to collaboratively couple the intracellular metabolism of Na2SeO3 and detoxification of CdCl2 (Fig. 4A).96 Besides, by adjusting the incubation duration of the seleniumized yeast cells with CdCl2 from 10 to 40 hours, CdSe QDs were obtained with tunable emission from green to yellow, then to red, and sizes from 2.69 to 6.34 nm. Se(IV) in SeO32− was first reduced into the selenotrisulfide derivative of glutathione (GSSeSG) in the presence of reduced GSH, and further reduced into low-valence organoselenium compounds by GSH-related enzymes (NADPH and GR), in the cytoplasm and mitochondria of yeast cells. Then, the Cd(SG)2 formed by the reaction of added CdCl2 and GSH at the appropriate moment led to the formation of CdSe QDs in the seleniumized yeast cells. To further verify the significance of GSH, NADPH and GR in QD biosynthesis, the group realized the controlled synthesis of PbSe nanocrystals99 and ultrasmall near-infrared Ag2Se QDs100 using a quasi-biosystem. Na2SeO3, GSH, NADPH and GR were mixed with the precursor of Pb or Ag under inert gas protection, and size-tunable PbSe or Ag2Se QDs were obtained by adjusting the proportion of precursors. Because the fluorescence emission of the prepared Ag2Se QDs was located in the NIR region and they had strong tissue penetration ability, the Ag2Se QDs could be applied in in vivo fluorescence imaging with a high signal-to-noise ratio (Fig. 4B).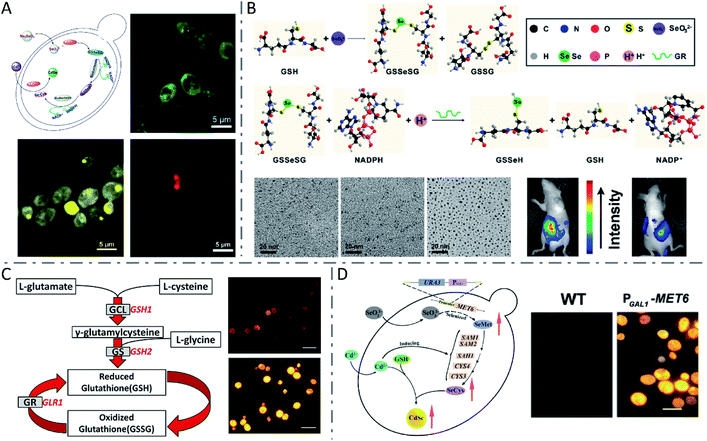 | ||
Fig. 4 QDs synthesized by yeast cells and the quasi-biosystem. (A) Route for unnatural biosynthesis of fluorescent CdSe QDs, and fluorescence microscopy images of seleniumized yeast cells cultured with CdCl2 for 12 h (green), 24 h (yellow) and 40 h (red). Reproduced from ref. 96 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2009. (B) Scheme for the SeO32− reduction process, TEM images of monodisperse Ag2Se QDs with different sizes (synthesized at a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (left), 5 1 (left), 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (middle), and 4 1 (middle), and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (right) molar ratio of the Ag precursor to Se precursor), and fluorescence images of a nude mouse with Ag2Se QDs injected into the abdominal cavity. Reproduced from ref. 100 with permission from American Chemical Society, copyright 2012. (C) The glutathione metabolic pathway in yeast cells and fluorescence microscopy images of WT (above) and PGAL1-GSH1 (below) yeast cells after biosynthesis of CdSe QDs (scale bars: 10 μm). Reproduced from ref. 19 with permission from American Chemical Society, copyright 2013. (D) Mechanism of CdSe QD biosynthesis in yeast cells overexpressing MET6, and fluorescence microscopy images of WT (left) and PGAL1-MET6 cells (scale bar: 5 μm). Reproduced from ref. 95 with permission from Tsinghua University Press and Springer-Verlag GmbH Germany, part of Springer Nature, copyright 2018. 1 (right) molar ratio of the Ag precursor to Se precursor), and fluorescence images of a nude mouse with Ag2Se QDs injected into the abdominal cavity. Reproduced from ref. 100 with permission from American Chemical Society, copyright 2012. (C) The glutathione metabolic pathway in yeast cells and fluorescence microscopy images of WT (above) and PGAL1-GSH1 (below) yeast cells after biosynthesis of CdSe QDs (scale bars: 10 μm). Reproduced from ref. 19 with permission from American Chemical Society, copyright 2013. (D) Mechanism of CdSe QD biosynthesis in yeast cells overexpressing MET6, and fluorescence microscopy images of WT (left) and PGAL1-MET6 cells (scale bar: 5 μm). Reproduced from ref. 95 with permission from Tsinghua University Press and Springer-Verlag GmbH Germany, part of Springer Nature, copyright 2018. | ||
It seems that the GSH metabolic pathway plays an important role in the biosynthesis of QDs in yeast cells. Yong Li and coworkers investigated in detail the vital role of the GSH metabolic pathway in CdSe QD biosynthesis by yeast cells (Fig. 4C), and proved that engineering GSH metabolism could construct a more efficient biofactory for CdSe QD synthesis.19 They found that during the biosynthesis of QDs, the expression of the GSH1 gene was up-regulated and the intracellular content of glutathione was also synergistically regulated. Compared with the WT strain, the glutathione metabolic mutant strains showed a significant decrease in the biosynthetic yield of CdSe QDs and their fluorescence intensity. In turn, inducing the GSH1 gene can significantly promote GSH content in the engineered cells which eventually gave a significantly higher yield of CdSe QDs. Se metabolism is another key factor for biosynthesis of CdSe QDs. Min Shao et al. proved that selenocysteine (SeCys) is the primary Se-precursor in the intracellular biosynthesis of CdSe QDs in yeast cells, and the synthesis yield could be improved by regulating the selenomethionine (SeMet)-to-SeCys pathway (Fig. 4D).95 The production of biosynthesized CdSe QDs was obviously improved by the overexpression of the MET6 gene, which encoded methionine synthase and adjusted Se metabolism. Referring to CdTe QDs, through simply incubating with CdCl2 and Na2TeO3, cells of Saccharomyces cerevisiae were reported to produce CdTe QDs extracellularly with high crystallinity, good water solubility, great stability, and biocompatibility.97 In that work, the formation of QDs was observed to be more rapid at a relatively high temperature (35 °C), and the particle size and emission wavelength could be tuned via simply changing the incubation time.
In addition to QDs, some other nanomaterials were synthesized via biomineralization or direct chemical reaction on the surface of or inside yeast cells. Abundant glycoproteins and peptidoglycans are located on the surface of yeast cells, which can locally accumulate metal ions and serve as reservoirs for the formation of metal–organic framework (MOF) crystals.101 Kang Liang et al. constructed a MOF material (zeolitic imidazolate framework-8 or ZIF-8) mineralized on the surface of living yeast cells under mild conditions (Fig. 5A).102 Later, in another study, the group fabricated a bioactive synthetic porous shell, containing a β-galactosidase (β-gal) layer and an outer MOF coating, to enable yeast cells to survive in nutrient-depleted and inhospitable environments.103 In addition, mesoporous silica nanoparticles (MSNs) were also reversibly encapsulated onto yeast cells, through click reactions between cell-surface polysaccharides containing cis-diols and phenylboronic acid-linked MSNs, to protect the yeast cells in harsh environments (Fig. 5B).104 Xiaoming Ma et al. constructed a living cell-based functional platform with biogenetic intracellular hydroxyapatite nanoscaffolds (nHAP@yeasts) via biomineralization.105 By incubating with Na3PO4 under a basic environment, the yeast cells pretreated with CaCl2 formed HAP nanoparticles intracellularly, maintaining good viability and proliferation ability as well as the naive yeast cells. The constructed nHAP@yeasts with good biocompatibility, hypoxia-targeting ability and high drug loading capacity were further functionalized with a targeting ligand (i.e., folic acid (FA)) and loaded with a therapeutic agent (i.e., DOX). The DOX-nHAP@yeasts-FA showed dual responsive profiles based on the FA dependency of tumors and the pH-sensitivity of HAP nanoparticles, and obviously inhibited tumor growth with little toxicity to normal tissues (Fig. 5C). Moreover, yeast cells have also been utilized to biosynthesize various nanomaterials such as Ag nanoparticles and Se nanoparticles.106
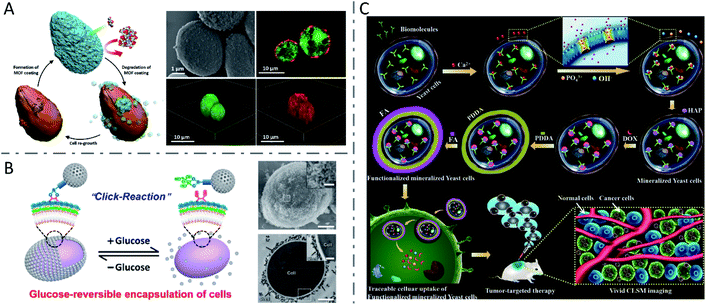 | ||
| Fig. 5 Inorganic nanomaterials synthesized via biomineralization or direct chemical reaction in yeast cells. (A) Biomimetic crystallization of cytoprotective MOF coatings on living cells, and the SEM image and CLSM images of ZIF-8 coated yeast cells. The living yeast cells were labeled with FDA (green) and the ZIF-8 coatings were labeled by infiltration of Alexa Fluor 647 fluorescent dye (red). Reproduced from ref. 102 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2016. (B) Reversible encapsulation of yeast cells with MSNs using a boronic acid vicinal-diol-based click reaction, and the SEM image (scale bars: 1 μm and 200 nm (inset)) and TEM image (scale bars: 1 μm and 300 nm (inset)) of an encapsulated yeast cell. Reproduced from ref. 104 with permission from American Chemical Society, copyright 2019. (C) Formation mechanism of nHAP mineralized yeast cells and functionalized mineralized yeast cells, and their potential application in tumor-targeting delivery. Reproduced from ref. 105 with permission from The Royal Society of Chemistry, copyright 2018. | ||
Synthesis by other fungi
In addition to yeasts, many other fungi have also demonstrated the ability to synthesize inorganic nanomaterials. CdSxSe1−x QDs with a uniform spherical shape of 3.22 ± 0.07 nm diameter were synthesized by Phomopsis sp. XP-8 in a Na2SeO3 and CdCl2 aqueous solution within 6 h (Fig. 6A).107 Several fungi including Coriolus versicolor,90Phanerochaete chrysosporium,108 and Trametes versicolor91 have been employed to synthesize CdS QDs, while Aspergillus flavus89 and Fusarium oxysporum (F. oxysporum)109 have been used to fabricate ZnS QDs, and also F. oxysporum to produce CdSe QDs.17 When incubated with CdCl2 and Na2SeO3 in equimolar concentrations from 0.25 to 5 mM, F. oxysporum produced CdSe QDs in the mycelial cells with a dose-dependent increase in fluorescence intensity, while decreased fluorescence intensity was observed at concentrations higher than 5 mM.17 Smaller increases of reactant concentrations led to an increasing nanoparticle yield, but high amounts of metal ions might suppress the metabolism of F. oxysporum and inhibit biosynthesis. Besides, when cultured in solutions with pH varying from 4.0 to 9.0, the mycelial cells of F. oxysporum synthesized CdSe QDs with the highest fluorescence intensity at pH 7.5. Moreover, CdSe QDs with a higher fluorescence intensity were also obtained when F. oxysporum was incubated at 25 °C, rather than 30 or 37 °C (Fig. 6B). The researchers proposed that higher temperature induced faster precipitation speed of CdSe in the solution, thus forming smaller particles which showed lower fluorescence intensity, or the higher temperature may have influenced the metabolism of F. oxysporum and inhibited biosynthesis. Although biosynthesis of QDs has been demonstrated extensively in both bacteria and fungi, it seems that fungi prefer to synthesize nanomaterials extracellularly, due to their high amounts of bioactive secretion proteins.108–110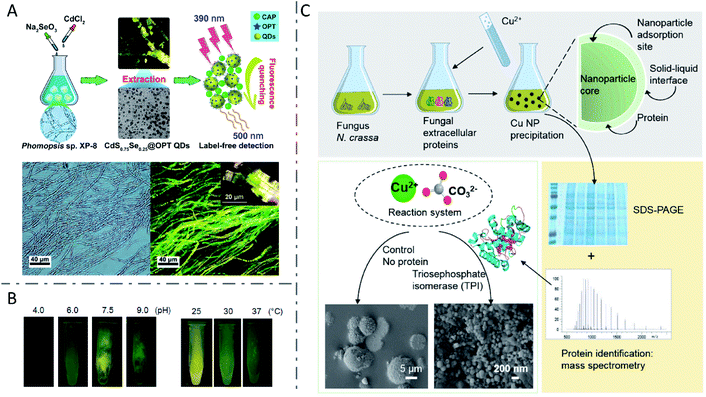 | ||
| Fig. 6 Inorganic nanomaterials synthesized by fungi. (A) Biosynthesis of fluorescent CdSxSe1−x QDs by Phomopsis sp. XP-8 and application in label-free detection. Reproduced from ref. 107 with permission from American Chemical Society, copyright 2020. (B) CdSe QDs with tunable fluorescence intensity by changing pH and temperature. Reproduced from ref. 17 with permission from MDPI, copyright 2016. (C) The function of fungal protein TPI in controlling the morphology and structure of biosynthesized CuCO3 nanoparticles via biomineralization. Reproduced from ref. 111 with permission from Elsevier Inc., copyright 2020. | ||
Recently, Feixue Liu et al. revealed the vital function of the fungal protein triosephosphate isomerase (TPI) in controlling the morphology and structure of biosynthesized CuCO3 nanoparticles via a biomineralization process (Fig. 6C).111 Fungus F. oxysporum was investigated extensively for the biosynthesis of nanomaterials such as crystalline magnetite particles,112 ZrO2 nanoparticles,113 SiO2 nanoparticles,114 TiO2 nanoparticles,114 and Ag nanoparticles.115,116 It was reported that the production of Ag nanoparticles by F. oxysporum was related to NR and a shuttle quinone extracellular process,116 as well as the enzymatic generation of extracellular superoxides,115 suggesting that multiple mechanisms may be involved. Besides, miscellaneous nanomaterials including Au,117 Ag,87,110,118,119 alloy-type Au/Ag nanoparticles,88 TiO2 nanoparticles,120 and Te and Se nanoparticles121 have been manufactured by fungi such as Penicillium brevicompactum (P. brevicompactum),117Cladosporium cladosporioides,110Catharanthus roseus,118Phenerochaete chrysosporium,119Neurospora crassa,88Aspergillus flavus,120Aureobasidium pullulans, Mortierella humilis, Trichoderma harzianum and Phoma glomerata.121
Since most fungi are multicellular organisms with relatively complex structures, yeasts with a simple unicellular structure are more similar to bacteria with the advantages of facile cultivation and easy genetic manipulation. It's also easier to isolate and characterize the synthesized NMs by unicellular organisms. Both bacteria and fungi can biosynthesize a variety of metallic and metalloid nanomaterials as well as their oxides and chalcogenides with a reported similar mechanism. However, in terms of scale up, fungi seem to be better candidates, because they can tolerate higher metal concentrations than bacteria and secrete a large amount of extracellular proteins and enzymes, resulting in higher production of inorganic nanomaterials.122 Furthermore, the untapped biodiversity of fungi endows them with potential to provide different kinds of nanomaterials. In the field of tumor-targeting therapy, the capping agents of the synthesized nanomaterials originating from fungi are less likely to induce an immune response like that induced by bacteria as mentioned above.92
Synthesis by animal cells
The animal cell is another kind of biofactory for inorganic nanomaterial manufacturing. Compared to bacteria and fungi, nanomaterials synthesized by animal cells are scarcely investigated, due to their relatively low proliferation rate, laborious and time-consuming culture procedures, and higher cultivation cost. However, since balancing the efficacy and safety of microbial products remains a big challenge, nanomaterials synthesized by animal cells are much safer due to the same species of origin as treated subjects. Besides, the biosynthesized nanomaterials always showed unique characteristics related to the original cells, which is beneficial for tumor-targeting theranostics.Synthesis by cancer cells
In 2005, Anshup et al. reported the intracellular synthesis of nanoparticles using mammalian cells for the first time.123 They synthesized Au nanoparticles of 20–100 nm diameter using human embryonic kidney cell HEK-293 and human cancer cells (i.e. HeLa, SiHa and SKNSH cells) with their cell membranes remaining intact. Subsequently, several cancer cell lines, such as HeLa,124 HepG2,125 MCF-7,126 B16F10,127 A549,15 HCT-116 (ref. 15) and U87 (ref. 128), were used to study the biosynthesis of various nanomaterials. By incubating with AgNO3 and Na2S for only 20 minutes separately in sequence, followed by an aging process, HepG2 cells successfully synthesized Ag2S QDs intracellularly.125 It was noted that the intracellular synthetic yield grew with the prolongation of aging time, reached the maximum at 16 h, and then levelled off. Results also showed that the formation of QDs was accompanied by the consumption of intracellular GSH. Through a so-called spatiotemporal coupling strategy, Ling-Hong Xiong et al. employed living MCF-7 cancer cells to synthesize fluorescent CdSe QDs intracellularly.126 They proposed a similar mechanism to bacterial and fungal biosynthesis involving selenite reduction metabolism and Cd2+ detoxification. Following a budding process, illuminated cell-derived microvesicles were obtained with QDs inside. Extracellular vesicles derived from mammalian cells have excellent stability, biocompatibility, lower immunogenicity and unique bioinformation from their parent cells, and have been widely employed for tumor-targeting drug delivery and anti-tumor immunotherapy.129–134 In a recent study, AuNPs were intracellularly synthesized in melanoma B16F10 cells, and further shed to the extracellular environment with tumor antigens retained (AuNP@B16F10).127 The AuNP-trapped vesicles were further introduced into dendritic cells (DCs), thus generating DC-derived Au nanoparticles (AuNP@DCB16F10) with improved immunological properties, enhanced biocompatibility and stealth. With NIR laser irradiation, the constructed AuNP@DCB16F10 could realize an efficient combination of AuNP-mediated PTT and tumor antigen-based immunotherapy, with a significant effect on the elimination of primary tumors, as well as the inhibition of tumor recurrence and metastasis (Fig. 7).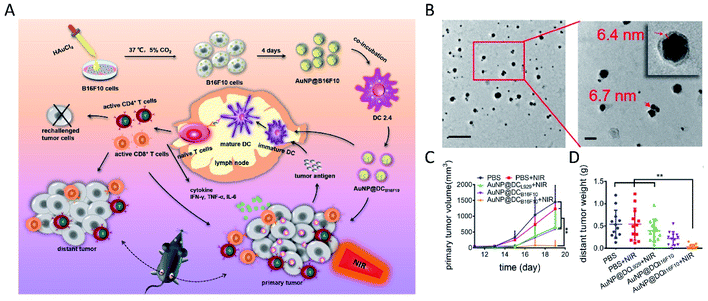 | ||
| Fig. 7 Immunological Au nanoparticles synthesized by animal cells for combinatorial photothermal therapy and immunotherapy against tumors. (A) Schematic preparation of AuNP@DCB16F10 and the mechanism of AuNP@DCB16F10-mediated combinational treatment modality. (B) TEM images of AuNP@DCB16F10 (scale bars: 200 and 50 nm, respectively). (C) Growth curves of the primary tumor. (D) Weight of the distant tumor harvested on 19th day. Reproduced from ref. 127 with permission from American Chemical Society, copyright 2019. | ||
In addition, a variety of nanoclusters synthesized by animal cells have also been reported. Donghua Chen et al. demonstrated that cancerous cells rather than noncancerous cells spontaneously synthesized luminescent Pt NCs upon incubation with H2PtCl6 solution.15 Specific fluorescence of Pt NCs synthesized in xenografted tumors after subcutaneous injection or intravenous injection of H2PtCl6 solutions was also observed. When combined with porphyrin derivatives as photothermal agents, the in situ synthesized Pt NCs realized effective fluorescence imaging and guided photothermal therapy of tumors. Later, with the addition of Eu(NO3)3 solution, a fluorescent Eu complex was also found to be spontaneously biosynthesized in both cancerous cells and tumors.135 After entering the cells through calcium ion channels, Eu(III) ions were reduced to a low-valence state under the relatively high level of oxidative stress and the increased level of GSH in the special microenvironment of tumor cells (Fig. 8A). GR and NADPH might also be involved as reducing equivalents. Moreover, metallic nanomaterials such as Au NCs,11,128,136 Ag NCs124 and Cu NCs137 have also been synthesized by cancer cells and applied for tumor-targeting imaging or/and therapy. In another study, cultured cancerous cells such as HeLa, U87 and HepG2, as well as the tumors of xenografted nude mice could synthesize fluorescent ZnO NCs and superparamagnetic Fe3O4 NCs when incubated or injected with zinc gluconate and FeCl2 solution, while the noncancerous cell L02 or normal tissues could not, which can be explained by the specific redox homeostasis of cancer cells.138 The relatively high concentrations of reactive oxygen species (ROS) such as H2O2 in cancer cells might cause the partial oxidation of Fe2+ and lead to the formation of Zn and Fe oxides, which could induce the specific fluorescence signal, enhanced MRI contrast and X-ray computed tomography (CT) contrast in tumors of xenografted tumor mice (Fig. 8B). It was demonstrated that in situ biosynthesis of iridium and iron oxide nanoclusters in cancer cells or tumor tissue could be realized in a similar way for tumor-targeting multimodal bioimaging.139 In particular, they found exosomes with internalized IrO2 and Fe3O4 NCs from the serum of xenograft mice after injection, which could be used as cancer biomarkers. In addition, the in situ synthesis of inorganic nanomaterials such as fluorescent ZnO NCs140,141 and magnetic Fe3O4 NCs141 was also reported in lesion areas of Alzheimer's mice, for the rapid and early diagnosis of Alzheimer's disease.
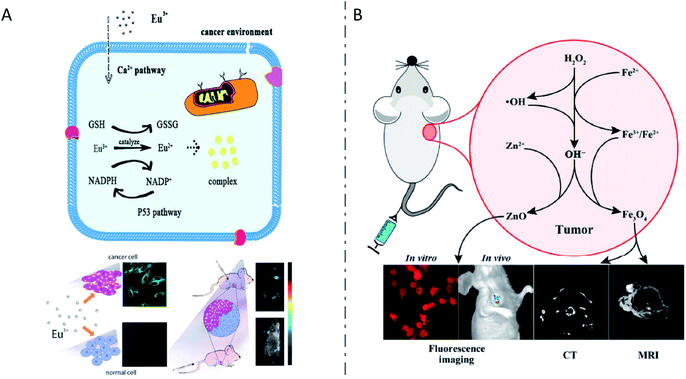 | ||
| Fig. 8 Inorganic nanomaterials synthesized by animal cells. (A) The biological pathways of the biosynthesized fluorescent Eu complex by cancer cells and application in tumor bioimaging. Reproduced from ref. 135 with permission from The Royal Society of Chemistry, copyright 2016. (B) In situ biosynthesis of Zn & Fe oxide nanoclusters and subsequent in vivo multimodal bioimaging of cancer cells. Reproduced from ref. 138 with permission from Tsinghua University Press and Springer-Verlag Berlin Heidelberg, copyright 2017. | ||
Synthesis by normal cells
Besides cancer cells, some normal cells such as human mesenchymal stem cells (MSCs)142 and platelets143 are also reported to be utilized for biosynthesis of inorganic nanomaterials with the assistance of engineering technology or external energy stimulation. After being transfected with the mms6 gene derived from Magnetospirillum magneticum AMB-1, the MSCs were found to successfully synthesize magnetic nanoparticles intracellularly, without any deleterious effects.142 In a study on platelets, Au nanoparticles were synthesized in living platelets exposed to HAuCl4, with the assistance of ultrasound energy and addition of reducing agents (NaBH4 and sodium citrate).143Considering the toxicity of bacterial and fungal products, nanomaterials synthesized by animal cells could be safer and more efficient for in vivo tumor-targeting applications due to the homology and homing effects. However, the large-scale preparation of nanomaterials synthesized by animal cells still faces great challenges, such as the high-cost, time-consuming and laborious cell cultivation and the low synthesis efficiency.144
Synthesis by plant cells
Many living plants can absorb metal ions and then convert them to their insoluble form to mitigate environmental stress, thus resulting in biosynthesized NMs.Synthesis by higher plants
In most cases, the metal ions were reduced to metallic nanomaterials within living plants. Therefore, researchers gave intensive attention to nanoparticle biosynthesis by living plant cells, among which Au nanoparticles16,145 and Ag nanoparticles146 were the majority. In 2002, alfalfa plants were reported to produce crystalline Au nanoparticles when grown on a gold-enriched agar system, which was the first demonstration of Au nanoparticles synthesized by living plants.147 Then, the same group reported the first biosynthesis of Ag nanoparticles by living alfalfa plants in 2003.148 Sharma et al. observed the intracellular formation and growth of spherical Au nanoparticles with diameters of 6–20 nm in Sesbania drummondii (S. drummondii) exposed to KAuCl4 solution (Fig. 9A), which could be explained by the reduction and stabilization by alkaloids or other secondary metabolites.149 In a study on Au nanoparticles synthesized by diverse plant species, it was found that changes in pH of culture media had little influence on the average sizes of the synthesized Au nanoparticles, but increases in the amounts of triangular and hexagonal nanoparticles were observed at low (3.8) and high (7.8) pH, respectively.16 Besides, lower temperature induced larger particle size and higher percent of gold nanorectangle formation with the temperature varying from 15 to 37 °C, which might be related to the effects of temperature on the growth responses and nutrient uptake by plants.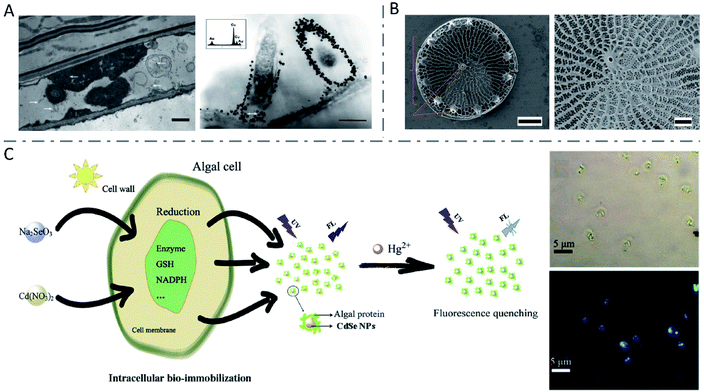 | ||
| Fig. 9 Inorganic nanomaterials synthesized by plant cells. (A) TEM images of S. drummondii root cells loaded with gold nanoparticles (scale bars: 1 μm and 500 nm). The inset shows the EDS spectrum with gold and copper peaks (copper peaks arise from the copper grid that holds the plant tissue). Reproduced from ref. 149 with permission from American Chemical Society, copyright 2007. (B) SEM images of wild-type Thalassiosira pseudonana diatom (scale bars: 1 μm and 300 nm, respectively). Reproduced from ref. 155 with permission from Springer Nature, copyright 2019. (C) The synthesis of CdSe NPs by algal cells and application in Hg2+ detection. Reproduced from ref. 166 with permission from Elsevier B.V., copyright 2019. | ||
The reduction of metal ions by the reductive components of living plants is the commonly accepted molecular mechanism for the formation of nanomaterials. Therefore, the contents of some reducing sugars and antioxidant compounds also played key roles in the construction of synthesized nanomaterials. Isabel R. Beattie and Richard G. Haverkamp demonstrated the synthesis of Au and Ag nanoparticles in all the tissues of Brassica juncea (B. juncea), including leaves, stems, and roots, after growing in AuNO3 or HAuCl4 solution.20 They found that the nanoparticles preferred to grow in or around chloroplasts, the production center and a repository of reducing sugars (glucose and fructose) produced by photosynthesis. The quantity of these sugars in the plant seemed to influence the metal reduction processes and found to be responsible for the amount of metallic nanoparticles formed in total. However, another research study revealed that the contents of reducing sugars and antioxidant compounds, proposed to be involved in the biosynthesis of metallic nanoparticles, were quite different among three species of plants which could synthesize Ag nanoparticles, leading to the concept that there is no substance that is solely responsible for the process.146 It is thought that there are synergistic effects among different substances, such as polysaccharides, proteins, flavonoids and terpenoids. In addition, the formations of Cu nanoparticles150 and Au–Ag–Cu alloy151 in living plants were also studied in a few articles. Given the complexity of plant metabolism, the synthesis of nanomaterials in whole living plants is usually uncontrollable. More reports have focused on biosynthesis using plant extracts.152–154
Synthesis by algae cells
Algae, simple plants with no real leaves, stems or roots, have been investigated for nanoparticle biosynthesis as well.7 Algae are easy to harvest and to culture, they grow fast, and few produce toxins like bacteria and fungi.8 As is well known, diatoms have an innate ability to synthesize nanoscale SiO2 to construct cell walls (Fig. 9B).155–157 Similar to higher plants, various algae such as Chlamydomonas reinhardtii (C. reinhardtii),158,159Cystophora moniliformis,160 and Botryococcus braunii (B. braunii)12 have been used to biosynthesize Ag nanoparticles, while algae such as Klebsormidium flaccidum,161Nostoc ellipsosporum,162Rhizoclonium fontinale,163 and Euglena gracilis164 have been used to synthesize Au nanomaterials. What's different, algal resources including cell-free extracts, broths, isolated proteins and whole living cells can be used in the biosynthesis of metallic-, chalcogenide- and oxide-based nanomaterials.8 Living cells of C. reinhardtii were employed to produce composition-controllable Ag–Au alloy nanoparticles with a uniform spherical shape.165 Composition-tunable bimetallic alloys could be synthesized by simply changing the stoichiometric ratio of metal salts added to the cultures. The researchers described the extracellular alloy nanoparticle creation processes as three stages: cellular uptake of metal ions and reduction-mediated formation of nanoparticles, stabilization of the nanoparticles by the extracellular matrix around, and release of the nanoparticles to the culture medium. Similar to other species, fluorescent CdSe QDs could be efficiently synthesized by algae cells, by incubating algae cells with immobilized Cd ions with different types of selenium sources.166,167 The possible mechanisms included the internalization of Na2SeO3 by an osmotic process, the generation of selenium precursors by the reduction reaction in the thylakoid membranes or cytoplasm within the electron transport system (ETS), the combination of selenium precursors and Cd(SG)2 under the catalysis of relative reductase to form CdSe nanoparticles with protective protein molecules, and the accumulation in the algal cytoplasm or release into culture media.166 Since the obtained QDs showed obvious fluorescence quenching by Hg2+, they were successfully employed for Hg2+ detection (Fig. 9C). In addition, some articles also demonstrated the biofabrication of Cu and CuO nanoparticles,168 superparamagnetic 2-line ferrihydrite nanoparticles169 and Fe-based magnetic nanomaterials170 by algae cells.The synthesis of nanomaterials in whole living plants is usually uncontrollable and inhomogeneous, and thus difficult in practical application. Algae, which are generally unicellular, colonial or constructed of filaments or composed of simple tissues, have the advantages of being easy to harvest and to culture, and produce no toxins, making it possible for scalable, sustainable and eco-friendly bioproduction of safer nanomaterials.8
Viral synthesis
Viruses are nanosized non-cellular organisms, typically consisting of self-assembled capsid proteins (CPs) surrounding an RNA or DNA core of genetic material.171 Since viruses have no cellular structure, the biosynthesis of nanoparticles in them is distinct from that in living cells discussed above, and cannot be veritably called “living cell synthesis”. In general, viruses serve as templates or scaffolds for metal nanoparticle biomineralization, because of their extremely tiny sizes and bioactive protein surfaces.172 Besides, genetic modification173 and chemical modification174 can be used to functionalize the obtained virus-based nanomaterials. M13 filamentous bacteriophages have been widely used to synthesize a variety of inorganic nanoparticles, such as Au nanostructures,175–177 Ag nanoparticles,173 Cu nanostructures178 and Co–Pt alloy nanoparticles.179 Furthermore, tobacco mosaic viruses,180,181 potato virus X182 and cowpea chlorotic mottle viruses183 have also been utilized in templated synthesis of nanomaterials.Conclusions
In this review, we summarized the biosynthesis of inorganic nanomaterials in different types of living cells including bacteria, fungi, plant cells and animal cells, accompanied by their application in tumor-targeting theranostics (Table 1). The mechanisms involving inorganic-ion bioreduction and detoxification as well as biomineralization are emphasized. Based on the mechanisms, we describe the size and morphology control of the products via the modulation of precursor ion concentration, pH, temperature, and incubation time, as well as cell metabolism by a genetic engineering strategy (Table 2). Compared to the conventional chemical synthesis, living cell synthesis has the advantages of easy preparation and environmental friendliness. Meanwhile, the obtained biosynthetic nanomaterials have the characteristics of good biocompatibility and unique biological properties. When it comes to bacteria, characteristics of single cell growth, facile cultivation, rapid proliferation, and particularly easy genetic manipulation make it possible to synthesize designed nanomaterials under mild conditions. The inherent tumor-targeting ability and immune response activation of bacteria also contribute to the targeted diagnosis and treatment of tumors by bacteria-synthesized nanomaterials. Compared with other microorganisms, fungi are more tolerant to metal ions, and secrete more extracellular enzymes and proteins, resulting in higher yields of nanomaterials and easier scale-up production. Considering the toxicity of bacterial and fungal products, nanomaterials synthesized by animal cells could be safer and efficient for in vivo tumor-targeting applications due to the homology and homing effects. Referring to synthesis by plant cells, few produce toxins like bacteria and fungi, leading to scalable, sustainable and eco-friendly bioproduction of nanomaterials. Despite the emerged limitations, such as the controllability of the biosynthetic process and safety of biological components, inorganic nanomaterials synthesized by living cells show a great promise for biomedical applications.| Species | Cells | Inorganic NMs | Applications | Reference |
|---|---|---|---|---|
| Bacteria | Magnetospirillum magneticum AMB-1 | Fe3O4 NPs | T2-weighted MRI and PTT of tumors | 34 |
| Magnetospirillum gryphiswaldense MSR-1 | Fe3O4 NPs | MHT on carcinoma cells | 38 | |
| Escherichia coli MG1655 | Au NPs | Photothermally controlled TNF-α therapy | 14 | |
| Bacillus subtilis | Ag NPs | Antibacterial activity | 184 | |
| Shewanella oneidensis MR-1 | Pd NPs | Augmenting photothermal tumor therapy | 64 | |
| Lactobacillus casei ATCC 393 | Se NPs | Antioxidant activity | 62 | |
| Staphylococcus aureus | Te NRs | — | 41 | |
| Escherichia coli | CdSxSe1−x QDs | Bioimaging of cancer cells and tumor tissue of mice | 26 | |
| Staphylococcus aureus | CdS0.5Se0.5 QDs | Pathogen detection | 24 | |
| Escherichia coli | CdTe QDs | — | 18 | |
| Escherichia coli | MnOx nanospindles | MRI and enhanced cancer-specific chemo-chemodynamic therapy | 46 | |
| Yeasts | Candida glabrata and Schizosaccharomyces pombe | CdS crystallites | — | 94 |
| Saccharomyces cerevisiae BY4742 | CdSe QDs | — | 96 | |
| Saccharomyces cerevisiae | CdTe QDs | — | 97 | |
| Saccharomyces cerevisiae | ZIF-8 | Cytoprotective exoskeletons for living cells | 102 | |
| Saccharomyces cerevisiae | Mesoporous silica NPs | Effective living cell protection under harsh conditions | 104 | |
| — | Hydroxyapatite nanoscaffolds | Cell-based drug carrier system for tumor-targeting therapy | 105 | |
| Pichia pastoris | Ag and Se NPs | — | 106 | |
| Multicellular fungi | Phomopsis sp. XP-8 | CdSxSe1−x QDs | Chloramphenicol detection | 107 |
| Phanerochaete chrysosporium BKMF-1767 | CdS QDs | — | 108 | |
| Fusarium oxysporum | ZnS QDs | — | 109 | |
| Neurospora crassa | CuCO3 NPs | — | 111 | |
| Fusarium oxysporum | ZrO2 NPs | — | 113 | |
| Fusarium oxysporum | SiO2 and TiO2 NPs | — | 114 | |
| Penicillium brevicompactum | Au NPs | Cytotoxic effects against cancer cells | 117 | |
| Catharanthus roseus | Ag NPs | Anticancer properties against cancer cells | 118 | |
| Neurospora crassa | Alloy-type Au/Ag NPs | — | 88 | |
| Aureobasidium pullulans, Mortierella humilis, Trichoderma harzianum and Phoma glomerata | Se and Te NPs | — | 121 | |
| Animal cells | Human hepatoma carcinoma cell HepG2 | Ag2S QDs | NIR fluorescence imaging | 125 |
| Mammalian cancer cell MCF-7 | CdSe QDs | Fluorogenic labeling | 126 | |
| Melanoma cell B16F10 | Au NPs | Combinatorial photothermal therapy and immunotherapy against tumors | 127 | |
| Platelets | Au NPs | Dark-field microscopy (DFM)-based imaging and CT imaging | 143 | |
| Human hepatocarcinoma cell HepG2 and lung cancer cell A549 | Pt NCs | Fluorescence imaging and photothermal therapy of tumors | 15 | |
| Human hepatocarcinoma cell HepG2 | Eu complex | Fluorescence imaging of tumors | 135 | |
| Human breast carcinoma cell MDA-MB-231 | Cu NCs | Intracellular temperature nanoprobes | 137 | |
| HeLa, U87, and HepG2 cancer cells | ZnO and Fe3O4 NCs | Fluorescence imaging, CT imaging and MRI of tumors | 138 | |
| HeLa and HepG2 cancer cells | IrO2 and Fe3O4 NCs | Fluorescence imaging, CT imaging and MRI of tumors | 139 | |
| Human mesenchymal stem cells | Fe3O4 NPs | MRI | 142 | |
| Higher plants | Sesbania drummondii | Au NPs | Catalytic function | 149 |
| Brassica juncea | Au and Ag NPs | — | 20 | |
| Phragmites australis and Iris pseudoacorus | Cu NPs | — | 150 | |
| Brassica juncea | Au–Ag–Cu alloy | — | 151 | |
| Algae | Diatom Thalassiosira pseudonana | SiO2 | — | 157 |
| Klebsormidium flaccidum | Au NPs | — | 161 | |
| Cystophora moniliformis | Ag NPs | — | 160 | |
| Chlamydomonas reinhardtii | Ag–Au alloy NPs | — | 165 | |
| Chlorella pyrenoidosa and Scenedesmus obliquus | CdSe QDs | Fluorescent nanoprobe for imatinib detection | 167 | |
| Chlorella kessleri | Cu and CuO NPs | — | 168 | |
| Chlorella sp. HQ | Fe-based NMs | Harvesting of oleaginous microalgae | 170 |
| Cells | Inorganic NMs | Regulated characteristics | Affecting factors | Reference |
|---|---|---|---|---|
| Escherichia coli DH5α | FeCo metal NPs | Size | Concentration of Fe2+ (or Fe4+) and Co2+ metal ions | 21 |
| Escherichia coli DH5α | CdSe NPs | Size, color and fluorescence emission wavelength | Concentration of Cd2+ and Se+ metal ions | 21 |
| Escherichia coli DH5α | ZnO, BaCO3, Eu, Gd NMs, etc. | Producibility and crystallinity | Reduction potential and pH | 40 |
| Escherichia coli K12 | CdTe QDs | Size and fluorescence emission wavelength | Incubation time | 18 |
| Escherichia coli | CdSxSe1−x QDs and Cd3(PO4)2 | Synthesis rate and yield | Glucose metabolism | 26 |
| Shewanella oneidensis MR-1 | CdSe QDs and Se NPs | Synthetic yield | Engineering to regulate extracellular electron transfer ability | 23 |
| Saccharomyces cerevisiae | CdSe QDs | Size and fluorescence wavelength | Incubation time | 96 |
| Saccharomyces cerevisiae | CdTe QDs | Size and fluorescence wavelength; synthesis rate | Incubation time; temperature | 97 |
| Saccharomyces cerevisiae | CdSe QDs | Synthesis yield | Engineering to regulate Se metabolism | 95 |
| Saccharomyces cerevisiae | CdSe QDs | Synthesis yield | Engineering to regulate glutathione metabolism | 19 |
| Fusarium oxysporum | CdSe QDs | Fluorescence intensity, size, synthesis rate and yield | Concentration of CdCl2 and Na2SeO3, pH, and temperature | 17 |
| Human hepatoma carcinoma cell HepG2 | Ag2S QDs | Synthetic yield and fluorescence intensity | Aging time | 125 |
| Medicago sativa (alfalfa) | Au NPs | Shape and size | pH, temperature and light conditions | 16 |
| Botryococcus braunii | Ag NPs | Crystallinity, shape, size, yield and synthetic rate | Concentration of AgNO3 and pH | 12 |
Perspectives
In spite of what is mentioned above, it is still a far way to go until inorganic nanomaterials synthesized by living cells predominate the field of tumor-targeting applications. Researchers should pay more attention to the following aspects. (1) The mechanisms of inorganic nanomaterial synthesis in living cells are reported to be mainly related to inorganic-ion bioreduction, detoxification and biomineralization. Some kinds of intracellular enzymes, reducing molecules such as glutathione and glucose, cell surface expressed polysaccharides and proteins are proved to participate in these biosynthetic processes and play key roles. However, the biochemical reactions involved are so complicated that the mechanisms are not illustrated deeply in most cases. For the purpose of controllable preparation, the mechanisms should be clearly clarified. (2) Referring to the controllability of biosynthesis, it is a more challenging issue. As we all know, the components, size, morphology, purity etc. have crucial impacts on the properties of nanomaterials. Likewise, their surface coating and stability are also important for their bioapplication. How can we make the living cells give us what we want but not what they can? Modulation of precursor ion concentration, pH, temperature, and incubation time, as well as cell metabolism by genetic engineering can help to some extent, but more strategies are urgently needed. (3) Majority of the biosynthetic products are metallic nanoparticles and their oxides or chalcogenides. However, functional inorganic nanomaterials that can be used in biomedicine and other fields are far more diverse than these. Considering the diversity of living cells, the bioconstruction of many other types of nanomaterials would become possible. (4) Although there are many advantages of biosynthesis, high-quality inorganic nanomaterials are usually prepared by chemical processes. Under mild reaction conditions, the crystal orientation of the available nanomaterials will not be as perfect as those fabricated at high temperature. The presence of defects will degrade the performance of the nanomaterials. (5) Biosynthesis is considered as a cost-effective and eco-friendly approach for inorganic nanomaterial preparation. It is highly possible to scale up the synthesis. But before that, the process parameters should be optimized and the preparation technology needs to be deeply investigated. (6) The capping agents on the surface of nanomaterials originating from living cells during the biosynthesis endow them with high stability and good biocompatibility. Sometimes, they can synergize with the nanomaterials to enhance their therapeutic effect, for example, the immune response activation by lipopolysaccharides on the outer membrane of bacteria. On the other hand, as exogenous biomolecules, their potential toxicity should be considered. (7) When it comes to their commercial application, standards regarding biofabrication, characterization, evaluation and so on of the nanomaterials should be established.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (81771978, 82072066 and 81627901), National Key Research and Development Program of China (2018YFA0208903 and 2020YFA0710700).Notes and references
- Y. Zhang, W. Zhang, K. Zeng, Y. Ao, M. Wang, Z. Yu, F. Qi, W. Yu, H. Mao, L. Tao, C. Zhang, T. T. Y. Tan, X. Yang, K. Pu and S. Gao, Small, 2020, 16, e1906797 CrossRef PubMed.
- Y. Zhang, C. Xu, X. Yang and K. Pu, Adv. Mater., 2020, 32, e2002661 CrossRef.
- X. Zhong, X. Wang, G. Zhan, Y. Tang, Y. Yao, Z. Dong, L. Hou, H. Zhao, S. Zeng, J. Hu, L. Cheng and X. Yang, Nano Lett., 2019, 19, 8234–8244 CrossRef CAS PubMed.
- H. Zhao, J. Xu, W. Huang, G. Zhan, Y. Zhao, H. Chen and X. Yang, ACS Nano, 2019, 13, 6647–6661 CrossRef CAS PubMed.
- Y. Tang, T. Yang, Q. Wang, X. Lv, X. Song, H. Ke, Z. Guo, X. Huang, J. Hu, Z. Li, P. Yang, X. Yang and H. Chen, Biomaterials, 2018, 154, 248–260 CrossRef CAS.
- A. S. Mathuriya, Crit. Rev. Biotechnol., 2016, 36, 788–802 CrossRef CAS PubMed.
- K. Vijayaraghavan and T. Ashokkumar, J. Environ. Chem. Eng., 2017, 5, 4866–4883 CrossRef CAS.
- S. A. Dahoumane, M. Mechouet, K. Wijesekera, C. D. M. Filipe, C. Sicard, D. A. Bazylinski and C. Jeffryes, Green Chem., 2017, 19, 552–587 RSC.
- K. Wang, X. Zhang, I. M. Kislyakov, N. Dong, S. Zhang, G. Wang, J. Fan, X. Zou, J. Du, Y. Leng, Q. Zhao, K. Wu, J. Chen, S. M. Baesman, K. S. Liao, S. Maharjan, H. Zhang, L. Zhang, S. A. Curran, R. S. Oremland, W. J. Blau and J. Wang, Nat. Commun., 2019, 10, 3985 CrossRef PubMed.
- Q.-Y. Luo, Y. Lin, Y. Li, L.-H. Xiong, R. Cui, Z.-X. Xie and D.-W. Pang, Small, 2014, 10, 699–704 CrossRef CAS PubMed.
- M. Wang, Z. Yu, H. Feng, J. Wang, L. Wang, Y. Zhang, L. Yin, Y. Du, H. Jiang, X. Wang and J. Zhou, J. Mater. Chem. B, 2019, 7, 5336–5344 RSC.
- A. Arévalo-Gallegos, J. Saul Garcia-Perez, D. Carrillo-Nieves, R. A. Ramirez-Mendoza, H. M. N. Iqbal and R. Parra-Saldivar, Int. J. Nanomed., 2018, 13, 5591–5604 CrossRef PubMed.
- G. F. Luo, W. H. Chen, X. Zeng and X. Z. Zhang, Chem. Soc. Rev., 2021, 50, 945–985 RSC.
- J.-X. Fan, Z.-H. Li, X.-H. Liu, D.-W. Zheng, Y. Chen and X.-Z. Zhang, Nano Lett., 2018, 18, 2373–2380 CrossRef CAS PubMed.
- D. Chen, C. Zhao, J. Ye, Q. Li, X. Liu, M. Su, H. Jiang, C. Amatore, M. Selke and X. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 18163–18169 CrossRef CAS PubMed.
- D. L. Starnes, A. Jain and S. V. Sahi, Environ. Sci. Technol., 2010, 44, 7110–7115 CrossRef CAS PubMed.
- T. Yamaguchi, Y. Tsuruda, T. Furukawa, L. Negishi, Y. Imura, S. Sakuda, E. Yoshimura and M. Suzuki, Materials, 2016, 9, 855 CrossRef PubMed.
- H. Bao, Z. Lu, X. Cui, Y. Qiao, J. Guo, J. M. Anderson and C. M. Li, Acta Biomater., 2010, 6, 3534–3541 CrossRef CAS PubMed.
- Y. Li, R. Cui, P. Zhang, B.-B. Chen, Z.-Q. Tian, L. Li, B. Hu, D.-W. Pang and Z.-X. Xie, ACS Nano, 2013, 7, 2240–2248 CrossRef CAS PubMed.
- I. R. Beattie and R. G. Haverkamp, Metallomics, 2011, 3, 628–632 CrossRef CAS PubMed.
- T. J. Park, S. Y. Lee, N. S. Heo and T. S. Seo, Angew. Chem., Int. Ed. Engl., 2010, 49, 7019–7024 CrossRef CAS PubMed.
- X. Wang, J. Pu, B. An, Y. Li, Y. Shang, Z. Ning, Y. Liu, F. Ba, J. Zhang and C. Zhong, Adv. Mater., 2018, 30, e1705968 CrossRef PubMed.
- L.-J. Tian, W.-W. Li, T.-T. Zhu, J.-J. Chen, W.-K. Wang, P.-F. An, L. Zhang, J.-C. Dong, Y. Guan, D.-F. Liu, N.-Q. Zhou, G. Liu, Y.-C. Tian and H.-Q. Yu, J. Am. Chem. Soc., 2017, 139, 12149–12152 CrossRef CAS PubMed.
- J.-J. Wang, Y. Lin, Y.-Z. Jiang, Z. Zheng, H.-Y. Xie, C. Lv, Z.-L. Chen, L.-H. Xiong, Z.-L. Zhang, H. Wang and D.-W. Pang, Anal. Chem., 2019, 91, 7280–7287 CrossRef CAS PubMed.
- D. C. Vaigankar, S. K. Dubey, S. Y. Mujawar, A. D'Costa and S. K. Shyama, Ecotoxicol. Environ. Saf., 2018, 165, 516–526 CrossRef CAS PubMed.
- L.-J. Tian, Y. Min, W.-W. Li, J.-J. Chen, N.-Q. Zhou, T.-T. Zhu, D.-B. Li, J.-Y. Ma, P.-F. An, L.-R. Zheng, H. Huang, Y.-Z. Liu and H.-Q. Yu, ACS Nano, 2019, 13, 5841–5851 CrossRef CAS PubMed.
- R. Blakemore, Science, 1975, 190, 377–379 CrossRef CAS PubMed.
- S. Rosenfeldt, C. N. Riese, F. Mickoleit, D. Schüler and A. S. Schenk, Appl. Environ. Microbiol., 2019, 85, e01513–01519 CrossRef CAS PubMed.
- M. A. Carillo, M. Bennet and D. Faivre, J. Phys. Chem. B, 2013, 117, 14642–14648 CrossRef CAS PubMed.
- D. A. Bazylinski and R. B. Frankel, Nat. Rev. Microbiol., 2004, 2, 217–230 CrossRef CAS PubMed.
- R. Uebe and D. Schüler, Nat. Rev. Microbiol., 2016, 14, 621–637 CrossRef CAS PubMed.
- M. I. Siponen, P. Legrand, M. Widdrat, S. R. Jones, W.-J. Zhang, M. C. Y. Chang, D. Faivre, P. Arnoux and D. Pignol, Nature, 2013, 502, 681–684 CrossRef CAS PubMed.
- O. Felfoul, M. Mohammadi, S. Taherkhani, D. de Lanauze, Y. Z. Xu, D. Loghin, S. Essa, S. Jancik, D. Houle, M. Lafleur, L. Gaboury, M. Tabrizian, N. Kaou, M. Atkin, T. Vuong, G. Batist, N. Beauchemin, D. Radzioch and S. Martel, Nat. Nanotechnol., 2016, 11, 941–947 CrossRef CAS PubMed.
- C. Chen, S. Wang, L. Li, P. Wang, C. Chen, Z. Sun and T. Song, Biomaterials, 2016, 104, 352–360 CrossRef CAS PubMed.
- E. Alphandery, D. Abi Haidar, O. Seksek, M. Thoreau, A. Trautmann, N. Bercovici, F. Gazeau, F. Guyot and I. Chebbi, ACS Appl. Mater. Interfaces, 2017, 9, 36561–36572 CrossRef CAS PubMed.
- T. Tang, L. Zhang, R. Gao, Y. Dai, F. Meng and Y. Li, Appl. Microbiol. Biotechnol., 2012, 94, 495–503 CrossRef CAS PubMed.
- C. Lang and D. Schuler, Appl. Environ. Microbiol., 2008, 74, 4944–4953 CrossRef CAS PubMed.
- D. Gandia, L. Gandarias, I. Rodrigo, J. Robles-Garcia, R. Das, E. Garaio, J. A. Garcia, M. H. Phan, H. Srikanth, I. Orue, J. Alonso, A. Muela and M. L. Fdez-Gubieda, Small, 2019, 15, e1902626 CrossRef PubMed.
- A. Plan Sangnier, S. Preveral, A. Curcio, A. K. A. Silva, C. T. Lefèvre, D. Pignol, Y. Lalatonne and C. Wilhelm, J. Controlled Release, 2018, 279, 271–281 CrossRef CAS PubMed.
- Y. Choi, T. J. Park, D. C. Lee and S. Y. Lee, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5944–5949 CrossRef CAS PubMed.
- L.-H. Xiong, R. Cui, Z.-L. Zhang, J.-W. Tu, Y.-B. Shi and D.-W. Pang, Small, 2015, 11, 5416–5422 CrossRef CAS PubMed.
- L.-J. Tian, W.-W. Li, T.-T. Zhu, G.-H. Zhao, X.-W. Liu, J.-C. Dong, P.-F. An, J.-Y. Ma, F. Shen, C. Qian, B. Hu and H.-Q. Yu, J. Mater. Chem. A, 2019, 7, 18480–18487 RSC.
- D. B. Li, Y. Y. Cheng, C. Wu, W. W. Li, N. Li, Z. C. Yang, Z. H. Tong and H. Q. Yu, Sci. Rep., 2014, 4, 3735 CrossRef PubMed.
- S. Iravani and R. S. Varma, ACS Sustainable Chem. Eng., 2020, 8, 5395–5409 CrossRef CAS.
- H. J. Bai, Z. M. Zhang and J. Gong, Biotechnol. Lett., 2006, 28, 1135–1139 CrossRef CAS PubMed.
- Y. W. Bao, X. W. Hua, J. Zeng and F. G. Wu, Research, 2020, 2020, 9301215 CAS.
- L.-H. Xiong, R. Cui, Z.-L. Zhang, X. Yu, Z. Xie, Y.-B. Shi and D.-W. Pang, ACS Nano, 2014, 8, 5116–5124 CrossRef CAS PubMed.
- L. Du, H. Jiang, X. Liu and E. Wang, Electrochem. Commun., 2007, 9, 1165–1170 CrossRef CAS.
- G. Attard, M. Casadesús, L. E. Macaskie and K. Deplanche, Langmuir, 2012, 28, 5267–5274 CrossRef CAS PubMed.
- X. Jiang, X. Fan, W. Xu, R. Zhang and G. Wu, ACS Biomater. Sci. Eng., 2020, 6, 680–689 CrossRef CAS PubMed.
- X. Jiang, C. Zhao, X. Fan and G. Wu, ACS Omega, 2019, 4, 16667–16673 CrossRef CAS PubMed.
- B. Nair and T. Pradeep, Cryst. Growth Des., 2002, 2, 293–298 CrossRef CAS.
- I. W.-S. Lin, C.-N. Lok and C.-M. Che, Chem. Sci., 2014, 5, 3144–3150 RSC.
- R. L. Kimber, E. A. Lewis, F. Parmeggiani, K. Smith, H. Bagshaw, T. Starborg, N. Joshi, A. I. Figueroa, G. van der Laan, G. Cibin, D. Gianolio, S. J. Haigh, R. A. D. Pattrick, N. J. Turner and J. R. Lloyd, Small, 2018, 14, 1703145 CrossRef PubMed.
- S. Y. Chen, C. Y. Xing, D. Z. Huang, C. H. Zhou, B. Ding, Z. H. Guo, Z. C. Peng, D. Wang, X. Zhu, S. Z. Liu, Z. Cai, J. Y. Wu, J. Q. Zhao, Z. Z. Wu, Y. H. Zhang, C. Y. Wei, Q. T. Yan, H. Z. Wang, D. Y. Fan, L. P. Liu, H. Zhang and Y. H. Cao, Sci. Adv., 2020, 6, 11 Search PubMed.
- T. Yang, H. T. Ke, Q. L. Wang, Y. A. Tang, Y. B. Deng, H. Yang, X. L. Yang, P. Yang, D. S. Ling, C. Y. Chen, Y. L. Zhao, H. Wu and H. B. Chen, ACS Nano, 2017, 11, 10012–10024 CrossRef CAS PubMed.
- W. Huang, Y. Huang, Y. You, T. Nie and T. Chen, Adv. Funct. Mater., 2017, 27, 1701388 CrossRef.
- L. L. Shi, J. Y. Sheng, M. L. Wang, H. Luo, J. Zhu, B. X. Zhang, Z. Liu and X. L. Yang, Theranostics, 2019, 9, 4115–4129 CrossRef CAS PubMed.
- J.-X. Fan, M.-Y. Peng, H. Wang, H.-R. Zheng, Z.-L. Liu, C.-X. Li, X.-N. Wang, X.-H. Liu, S.-X. Cheng and X.-Z. Zhang, Adv. Mater., 2019, 31, 1808278 CrossRef PubMed.
- W. Chen, Y. Wang, M. Qin, X. Zhang, Z. Zhang, X. Sun and Z. Gu, ACS Nano, 2018, 12, 5995–6005 CrossRef CAS PubMed.
- L. Shi, J. Sheng, G. Chen, P. Zhu, C. Shi, B. Li, C. Park, J. Wang, B. Zhang, Z. Liu and X. Yang, J. Immunother. Cancer., 2020, 8, e000973 CrossRef PubMed.
- C. Xu, L. Qiao, Y. Guo, L. Ma and Y. Cheng, Carbohydr. Polym., 2018, 195, 576–585 CrossRef CAS PubMed.
- V. K. Nguyen, W. Choi, Y. Ha, Y. Gu, C. Lee, J. Park, G. Jang, C. Shin and S. Cho, J. Ind. Eng. Chem., 2019, 78, 246–256 CrossRef CAS.
- Q.-W. Chen, X.-H. Liu, J.-X. Fan, S.-Y. Peng, J.-W. Wang, X.-N. Wang, C. Zhang, C.-J. Liu and X.-Z. Zhang, Adv. Funct. Mater., 2020, 30, 1909806 CrossRef CAS.
- C.-Y. Wen, J. Hu, Z.-L. Zhang, Z.-Q. Tian, G.-P. Ou, Y.-L. Liao, Y. Li, M. Xie, Z.-Y. Sun and D.-W. Pang, Anal. Chem., 2013, 85, 1223–1230 CrossRef CAS PubMed.
- J. Hu, C.-Y. Wen, Z.-L. Zhang, M. Xie, J. Hu, M. Wu and D.-W. Pang, Anal. Chem., 2013, 85, 11929–11935 CrossRef CAS PubMed.
- X. Wu, J. Hu, B. Zhu, L. Lu, X. Huang and D. Pang, J. Chromatogr., 2011, 1218, 7341–7346 CrossRef CAS PubMed.
- J. Hu, M. Xie, C.-Y. Wen, Z.-L. Zhang, H.-Y. Xie, A.-A. Liu, Y.-Y. Chen, S.-M. Zhou and D.-W. Pang, Biomaterials, 2011, 32, 1177–1184 CrossRef CAS PubMed.
- M. Xie, J. Hu, Y.-M. Long, Z.-L. Zhang, H.-Y. Xie and D.-W. Pang, Biosens. Bioelectron., 2009, 24, 1311–1317 CrossRef CAS PubMed.
- Y. Zhang, J.-Y. Xiao, Y. Zhu, L.-J. Tian, W.-K. Wang, T.-T. Zhu, W.-W. Li and H.-Q. Yu, Anal. Chem., 2020, 92, 3990–3997 CrossRef CAS PubMed.
- W. Wang, Y. Liu, T. Shi, J. Sun, F. Mo and X. Liu, Anal. Chem., 2020, 92, 1598–1604 CrossRef CAS PubMed.
- M. Wu, W. Wu, Y. Duan, X. Li, G. Qi and B. Liu, Chem. Mater., 2019, 31, 7212–7220 CrossRef CAS.
- S.-B. Wang, X.-H. Liu, B. Li, J.-X. Fan, J.-J. Ye, H. Cheng and X.-Z. Zhang, Adv. Funct. Mater., 2019, 29, 1904093 CrossRef.
- D.-W. Zheng, Y. Chen, Z.-H. Li, L. Xu, C.-X. Li, B. Li, J.-X. Fan, S.-X. Cheng and X.-Z. Zhang, Nat. Commun., 2018, 9, 1680 CrossRef PubMed.
- S. Yan, X. Zeng, Y. Wang and B.-F. Liu, Adv. Healthcare Mater., 2020, 9, 2000046 CrossRef CAS PubMed.
- W. Park, S. Cho, D. Kang, J. H. Han, J. H. Park, B. Lee, J. Lee and D. H. Kim, Adv. Healthcare Mater., 2020, 9, e1901812 CrossRef PubMed.
- S. Suh, A. Jo, M. A. Traore, Y. Zhan, S. L. Coutermarsh-Ott, V. M. Ringel-Scaia, I. C. Allen, R. M. Davis and B. Behkam, Adv. Sci., 2019, 6, 1801309 CrossRef PubMed.
- Y. Luo, D. Xu, X. Gao, J. Xiong, B. L. Jiang, Y. Zhang, Y. T. Wang, Y. Tang, C. Chen, H. Qiao, H. N. Li and J. Z. Zou, Biochem. Biophys. Res. Commun., 2019, 514, 1147–1153 CrossRef CAS PubMed.
- R. J. Li, L. Helbig, J. Fu, X. Y. Bian, J. Herrmann, M. Baumann, A. F. Stewart, R. Muller, A. Y. Li, D. Zips and Y. M. Zhang, Res. Microbiol., 2019, 170, 74–79 CrossRef CAS PubMed.
- L. Liu, H. He, Z. Luo, H. Zhou, R. Liang, H. Pan, Y. Ma and L. Cai, Adv. Funct. Mater., 2020, 30, 1910176 CrossRef CAS.
- W. F. Chen, Z. F. Guo, Y. N. Zhu, N. Qiao, Z. R. Zhang and X. Sun, Adv. Funct. Mater., 2020, 30, 1906623 CrossRef CAS.
- S. Iravani and R. S. Varma, Green Chem., 2019, 21, 4583–4603 RSC.
- M. Sedighi, A. Z. Bialvaei, M. R. Hamblin, E. Ohadi, A. Asadi, M. Halajzadeh, V. Lohrasbi, N. Mohammadzadeh, T. Amiriani, M. Krutova, A. Amini and E. Kouhsari, Cancer Med., 2019, 8, 3167–3181 Search PubMed.
- P. Bonfante, F. Venice and L. Lanfranco, Curr. Opin. Microbiol., 2019, 49, 18–25 CrossRef CAS PubMed.
- H. P. Grossart, S. van den Wyngaert, M. Kagami, C. Wurzbacher, M. Cunliffe and K. Rojas-Jimenez, Nat. Rev. Microbiol., 2019, 17, 339–354 CrossRef CAS PubMed.
- M. V. Powers-Fletcher, B. A. Kendall, A. T. Griffin and K. E. Hanson, Microbiol. Spectr., 2016, 4, DMIH2-0002-2015 Search PubMed.
- N. Feroze, B. Arshad, M. Younas, M. I. Afridi, S. Saqib and A. Ayaz, Microsc. Res. Tech., 2020, 83, 72–80 CrossRef CAS PubMed.
- E. Castro-Longoria, A. R. Vilchis-Nestor and M. Avalos-Borja, Colloids Surf., B, 2011, 83, 42–48 CrossRef CAS.
- P. Uddandarao and R. B. Mohan, Mater. Sci. Eng., B, 2016, 207, 26–32 CrossRef CAS.
- R. Sanghi and P. Verma, Chem. Eng. J., 2009, 155, 886–891 CrossRef CAS.
- Z. Qin, Q. Yue, Y. Liang, J. Zhang, L. Zhou, O. B. Hidalgo and X. Liu, J. Biotechnol., 2018, 284, 52–56 CrossRef CAS PubMed.
- M. Kitching, M. Ramani and E. Marsili, Microb. Biotechnol., 2015, 8, 904–917 CrossRef CAS PubMed.
- J. Guo, M. Suástegui, K. K. Sakimoto, V. M. Moody, G. Xiao, D. G. Nocera and N. S. Joshi, Science, 2018, 362, 813–816 CrossRef CAS PubMed.
- C. T. Dameron, R. N. Reese, R. K. Mehra, A. R. Kortan, P. J. Carroll, M. L. Steigerwald, L. E. Brus and D. R. Winge, Nature, 1989, 338, 596–597 CrossRef CAS.
- M. Shao, R. Zhang, C. Wang, B. Hu, D. Pang and Z. Xie, Nano Res., 2018, 11, 2498–2511 CrossRef CAS.
- R. Cui, H.-H. Liu, H.-Y. Xie, Z.-L. Zhang, Y.-R. Yang, D.-W. Pang, Z.-X. Xie, B.-B. Chen, B. Hu and P. Shen, Adv. Funct. Mater., 2009, 19, 2359–2364 CrossRef CAS.
- H. Bao, N. Hao, Y. Yang and D. Zhao, Nano Res., 2010, 3, 481–489 CrossRef CAS.
- M. Kowshik, W. Vogel, J. Urban, S. K. Kulkarni and K. M. Paknikar, Adv. Mater., 2002, 14, 815–818 CrossRef CAS.
- R. Cui, Y.-P. Gu, Z.-L. Zhang, Z.-X. Xie, Z.-Q. Tian and D.-W. Pang, J. Mater. Chem., 2012, 22, 3713–3716 RSC.
- Y. P. Gu, R. Cui, Z. L. Zhang, Z. X. Xie and D. W. Pang, J. Am. Chem. Soc., 2012, 134, 79–82 CrossRef CAS PubMed.
- W. Li, Y. Zhang, Z. Xu, Q. Meng, Z. Fan, S. Ye and G. Zhang, Angew. Chem., 2016, 55, 955–959 CrossRef CAS PubMed.
- K. Liang, J. J. Richardson, J. Cui, F. Caruso, C. J. Doonan and P. Falcaro, Adv. Mater., 2016, 28, 7910–7914 CrossRef CAS PubMed.
- K. Liang, J. J. Richardson, C. J. Doonan, X. Mulet, Y. Ju, J. Cui, F. Caruso and P. Falcaro, Angew. Chem., Int. Ed. Engl., 2017, 56, 8510–8515 CrossRef CAS PubMed.
- W. Geng, N. Jiang, G.-Y. Qing, X. Liu, L. Wang, H. J. Busscher, G. Tian, T. Sun, L.-Y. Wang, Y. Montelongo, C. Janiak, G. Zhang, X.-Y. Yang and B.-L. Su, ACS Nano, 2019, 13, 14459–14467 CrossRef CAS PubMed.
- X. Ma, P. Liu, Y. Tian, G. Zhu, P. Yang, G. Wang and L. Yang, Nanoscale, 2018, 10, 3489–3496 RSC.
- F. Elahian, S. Reiisi, A. Shahidi and S. A. Mirzaei, Nanomedicine, 2017, 13, 853–861 CrossRef CAS PubMed.
- X. Xu, Y. Yang, H. Jin, B. Pang, R. Yang, L. Yan, C. Jiang, D. Shao and J. Shi, ACS Sustainable Chem. Eng., 2020, 8, 6806–6814 CrossRef CAS.
- G. Chen, B. Yi, G. Zeng, Q. Niu, M. Yan, A. Chen, J. Du, J. Huang and Q. Zhang, Colloids Surf., B, 2014, 117, 199–205 CrossRef CAS PubMed.
- S. Mirzadeh, E. Darezereshki, F. Bakhtiari, M. H. Fazaelipoor and M. R. Hosseini, Mater. Sci. Semicond. Process., 2013, 16, 374–378 CrossRef CAS.
- D. S. Balaji, S. Basavaraja, R. Deshpande, D. B. Mahesh, B. K. Prabhakar and A. Venkataraman, Colloids Surf., B, 2009, 68, 88–92 CrossRef CAS PubMed.
- F. Liu, D. S. Shah and G. M. Gadd, Curr. Biol., 2021, 31, 358–368 CrossRef CAS PubMed.
- A. Bharde, D. Rautaray, V. Bansal, A. Ahmad, I. Sarkar, S. M. Yusuf, M. Sanyal and M. Sastry, Small, 2006, 2, 135–141 CrossRef CAS PubMed.
- V. Bansal, D. Rautaray, A. Ahmad and M. Sastry, J. Mater. Chem., 2004, 14, 3303–3305 RSC.
- V. Bansal, D. Rautaray, A. Bharde, K. Ahire, A. Sanyal, A. Ahmad and M. Sastry, J. Mater. Chem., 2005, 15, 2583–2589 RSC.
- Y. G. Yin, X. Y. Yang, L. G. Hu, Z. Q. Tan, L. X. Zhao, Z. Zhang, J. F. Liu and G. B. Jiang, Environ. Sci. Technol. Lett., 2016, 3, 160–165 CrossRef CAS.
- N. Durán, P. D. Marcato, O. L. Alves, G. I. H. D. Souza and E. Esposito, J. Nanobiotechnol., 2005, 3, 8 CrossRef PubMed.
- A. Mishra, S. K. Tripathy, R. Wahab, S.-H. Jeong, I. Hwang, Y.-B. Yang, Y.-S. Kim, H.-S. Shin and S.-I. Yun, Appl. Microbiol. Biotechnol., 2011, 92, 617–630 CrossRef CAS PubMed.
- T. Akther, M. Vabeiryureilai, K. Nachimuthu Senthil, M. Davoodbasha and H. Srinivasan, Environ. Sci. Pollut. Res., 2019, 26, 13649–13657 CrossRef CAS PubMed.
- M. Saravanan, S. Arokiyaraj, T. Lakshmi and A. Pugazhendhi, Microb. Pathog., 2018, 117, 68–72 CrossRef CAS PubMed.
- G. Rajakumar, A. A. Rahuman, S. M. Roopan, V. G. Khanna, G. Elango, C. Kamaraj, A. A. Zahir and K. Velayutham, Spectrochim. Acta, Part A, 2012, 91, 23–29 CrossRef CAS PubMed.
- X. Liang, M. A. M.-J. Perez, K. C. Nwoko, P. Egbers, J. Feldmann, L. Csetenyi and G. M. Gadd, Appl. Microbiol. Biotechnol., 2019, 103, 7241–7259 CrossRef CAS PubMed.
- M. Shah, D. Fawcett, S. Sharma, S. K. Tripathy and G. E. J. Poinern, Materials, 2015, 8, 7278–7308 CrossRef CAS PubMed.
- Anshup, J. S. Venkataraman, C. Subramaniam, R. R. Kumar, S. Priya, T. R. S. Kumar, R. V. Omkumar, A. John and T. Pradeep, Langmuir, 2005, 21, 11562–11567 CrossRef CAS PubMed.
- S. Gao, D. Chen, Q. Li, J. Ye, H. Jiang, C. Amatore and X. Wang, Sci. Rep., 2014, 4, 4384 CrossRef PubMed.
- L. Tan, A. Wan and H. Li, ACS Appl. Mater. Interfaces, 2014, 6, 18–23 CrossRef CAS PubMed.
- L.-H. Xiong, J.-W. Tu, Y.-N. Zhang, L.-L. Yang, R. Cui, Z.-L. Zhang and D.-W. Pang, Sci. China: Chem., 2020, 63, 448–453 CrossRef CAS.
- D. Zhang, T. Wu, X. Qin, Q. Qiao, L. Shang, Q. Song, C. Yang and Z. Zhang, Nano Lett., 2019, 19, 6635–6646 CrossRef CAS PubMed.
- C. Zhao, T. Du, F. U. Rehman, L. Lai, X. Liu, X. Jiang, X. Li, Y. Chen, H. Zhang, Y. Sun, S. Luo, H. Jiang, M. Selke and X. Wang, Small, 2016, 12, 6255–6265 CrossRef CAS PubMed.
- Z. Wei, X. Zhang, T. Yong, N. Bie, G. Zhan, X. Li, Q. Liang, J. Li, J. Yu, G. Huang, Y. Yan, Z. Zhang, B. Zhang, L. Gan, B. Huang and X. Yang, Nat. Commun., 2021, 12, 440 CrossRef CAS PubMed.
- T. Yong, D. Wang, X. Li, Y. Yan, J. Hu, L. Gan and X. Yang, J. Controlled Release, 2020, 322, 555–565 CrossRef CAS PubMed.
- T. Yong, X. Li, Z. Wei, L. Gan and X. Yang, J. Controlled Release, 2020, 328, 562–574 CrossRef CAS PubMed.
- D. Wang, Y. Yao, J. He, X. Zhong, B. Li, S. Rao, H. Yu, S. He, X. Feng, T. Xu, B. Yang, T. Yong, L. Gan, J. Hu and X. Yang, Adv. Sci., 2020, 7, 1901293 CrossRef CAS.
- T. Yong, X. Zhang, N. Bie, H. Zhang, X. Zhang, F. Li, A. Hakeem, J. Hu, L. Gan, H. A. Santos and X. Yang, Nat. Commun., 2019, 10, 3838 CrossRef PubMed.
- Q. Liang, N. Bie, T. Yong, K. Tang, X. Shi, Z. Wei, H. Jia, X. Zhang, H. Zhao, W. Huang, L. Gan, B. Huang and X. Yang, Nat. Biomed. Eng., 2019, 3, 729–740 CrossRef CAS PubMed.
- J. Ye, J. Wang, Q. Li, X. Dong, W. Ge, Y. Chen, X. Jiang, H. Liu, H. Jiang and X. Wang, Biomater. Sci., 2016, 4, 652–660 RSC.
- J. Wang, G. Zhang, Q. Li, H. Jiang, C. Liu, C. Amatore and X. Wang, Sci. Rep., 2013, 3, 1157 CrossRef PubMed.
- J. Ye, X. Dong, H. Jiang and X. Wang, J. Mater. Chem. B, 2017, 5, 691–696 RSC.
- T. Du, C. Zhao, F. ur Rehman, L. Lai, X. Li, Y. Sun, S. Luo, H. Jiang, M. Selke and X. Wang, Nano Res., 2017, 10, 2626–2632 CrossRef CAS.
- S. Shaikh, F. U. Rehman, T. Du, H. Jiang, L. Yin, X. Wang and R. Chai, ACS Appl. Mater. Interfaces, 2018, 10, 26056–26063 CrossRef CAS PubMed.
- L. Lai, C. Zhao, M. Su, X. Li, X. Liu, H. Jiang, C. Amatore and X. Wang, Biomater. Sci., 2016, 4, 1085–1091 RSC.
- L. Lai, X. Jiang, S. Han, C. Zhao, T. Du, F. U. Rehman, Y. Zheng, X. Li, X. Liu, H. Jiang and X. Wang, Langmuir, 2017, 33, 9018–9024 CrossRef CAS PubMed.
- A. Elfick, G. Rischitor, R. Mouras, A. Azfer, L. Lungaro, M. Uhlarz, T. Herrmannsdoerfer, J. Lucocq, W. Gamal, P. Bagnaninchi, S. Semple and D. M. Salter, Sci. Rep., 2017, 7, 39755 CrossRef CAS PubMed.
- J. Jin, T. Liu, M. Li, C. Yuan, Y. Liu, J. Tang, Z. Feng, Y. Zhou, F. Yang and N. Gu, Colloids Surf., B, 2018, 163, 385–393 CrossRef CAS PubMed.
- A. Rahman, J. Lin, F. E. Jaramillo, D. A. Bazylinski, C. Jeffryes and S. A. Dahoumane, Molecules, 2020, 25, 3246 CrossRef CAS PubMed.
- E. Rodriguez, J. G. Parsons, J. R. Peralta-Videa, G. Cruz-Jimenez, J. Romero-Gonzalez, B. E. Sanchez-Salcido, G. B. Saupe, M. Duarte-Gardea and J. L. Gardea-Torresdey, Int. J. Phytorem., 2007, 9, 133–147 CrossRef CAS PubMed.
- L. Marchiol, A. Mattiello, F. Poscic, C. Giordano and R. Musetti, Nanoscale Res. Lett., 2014, 9, 101 CrossRef PubMed.
- J. L. Gardea-Torresdey, J. G. Parsons, E. Gomez, J. Peralta-Videa, H. E. Troiani, P. Santiago and M. J. Yacaman, Nano Lett., 2002, 2, 397–401 CrossRef CAS.
- J. L. Gardea-Torresdey, E. Gomez, J. R. Peralta-Videa, J. G. Parsons, H. Troiani and M. Jose-Yacaman, Langmuir, 2003, 19, 1357–1361 CrossRef CAS.
- N. C. Sharma, S. V. Sahi, S. Nath, J. G. Parsons, J. L. Gardea-Torresdey and T. Pal, Environ. Sci. Technol., 2007, 41, 5137–5142 CrossRef CAS PubMed.
- A. Manceau, K. L. Nagy, M. A. Marcus, M. Lanson, N. Geoffroy, T. Jacquet and T. Kirpichtchikova, Environ. Sci. Technol., 2008, 42, 1766–1772 CrossRef CAS PubMed.
- R. G. Haverkamp, A. T. Marshall and D. van Agterveld, J. Nanopart. Res., 2007, 9, 697–700 CrossRef CAS.
- M. Anandan, G. Poorani, P. Boomi, K. Varunkumar, K. Anand, A. A. Chuturgoon, M. Saravanan and H. G. Prabu, Process Biochem., 2019, 80, 80–88 CrossRef CAS.
- M. N. Borovaya, A. P. Naumenko, N. A. Matvieieva, Y. B. Blume and A. I. Yemets, Nanoscale Res. Lett., 2014, 9, 686 CrossRef PubMed.
- P. Prakash, P. Gnanaprakasam, R. Emmanuel, S. Arokiyaraj and M. Saravanan, Colloids Surf., B, 2013, 108, 255–259 CrossRef CAS PubMed.
- A. A. Trofimov, A. A. Pawlicki, N. Borodinov, S. Mandal, T. J. Mathews, M. Hildebrand, M. A. Ziatdinov, K. A. Hausladen, P. K. Urbanowicz, C. A. Steed, A. V. Ievlev, A. Belianinov, J. K. Michener, R. Vasudevan and O. S. Ovchinnikova, npj Comput. Mater., 2019, 5, 67 CrossRef.
- N. Kröger, R. Deutzmann and M. Sumper, Science, 1999, 286, 1129–1132 CrossRef PubMed.
- N. Javaheri, R. Dries, A. Burson, L. J. Stal, P. M. A. Sloot and J. A. Kaandorp, Sci. Rep., 2015, 5, 11652 CrossRef CAS PubMed.
- A. Rahman, S. Kumar, A. Bafana, S. A. Dahoumane and C. Jeffryes, Molecules, 2019, 24, 98 CrossRef PubMed.
- I. Barwal, P. Ranjan, S. Kateriya and S. C. Yadav, J. Nanobiotechnol., 2011, 9, 56 CrossRef CAS PubMed.
- T. N. V. K. V. Prasad, V. S. R. Kambala and R. Naidu, J. Appl. Phycol., 2013, 25, 177–182 CrossRef CAS.
- S. A. Dahoumane, C. Djediat, C. Yepremian, A. Coute, F. Fievet, T. Coradin and R. Brayner, Biotechnol. Bioeng., 2012, 109, 284–288 CrossRef CAS PubMed.
- D. Parial, H. K. Patra, P. Roychoudhury, A. K. Dasgupta and R. Pal, J. Appl. Phycol., 2012, 24, 55–60 CrossRef CAS.
- D. Parial and R. Pal, J. Appl. Phycol., 2015, 27, 975–984 CrossRef CAS.
- S. Amar Dahoumane, C. Yepremian, C. Djediat, A. Coute, F. Fievet, T. Coradin and R. Brayner, J. Nanopart. Res., 2016, 18, 79 CrossRef.
- S. A. Dahoumane, K. Wijesekera, C. D. M. Filipe and J. D. Brennan, J. Colloid Interface Sci., 2014, 416, 67–72 CrossRef CAS PubMed.
- Z. Zhang, K. Yan, L. Zhang, Q. Wang, R. Guo, Z. Yan and J. Chen, J. Hazard. Mater., 2019, 374, 420–427 CrossRef CAS PubMed.
- Z. Zhang, J. Chen, Q. Yang, K. Lan, Z. Yan and J. Chen, Sens. Actuators, B, 2018, 263, 625–633 CrossRef CAS.
- G. Salas-Herrera, S. Gonzalez-Morales, A. Benavides-Mendoza, A. O. Castaneda-Facio, F. Fernandez-Luqueno and A. Robledo-Olivo, J. Appl. Phycol., 2019, 31, 2437–2447 CrossRef CAS.
- R. Brayner, T. Coradin, P. Beaunier, J.-M. Greneche, C. Djediat, C. Yepremian, A. Coute and F. Fievet, Colloids Surf., B, 2012, 93, 20–23 CrossRef CAS PubMed.
- P.-R. Liu, Z.-Y. Yang, Y. Hong and Y.-L. Hou, Algal Res., 2018, 31, 173–182 CrossRef.
- K. B. Narayanan and S. S. Han, Adv. Colloid Interface Sci., 2017, 248, 1–19 CrossRef CAS PubMed.
- M. Alarcón-Correa, J.-P. Günther, J. Troll, V. M. Kadiri, J. Bill, P. Fischer and D. Rothenstein, ACS Nano, 2019, 13, 5810–5815 CrossRef PubMed.
- X. Dong, P. Pan, D.-W. Zheng, P. Bao, X. Zeng and X.-Z. Zhang, Sci. Adv., 2020, 6, eaba1590 CrossRef CAS PubMed.
- Y. S. Nam, H. Park, A. P. Magyar, D. S. Yun, T. S. Pollom and A. M. Belcher, Nanoscale, 2012, 4, 3405–3409 RSC.
- N.-D. Tam-Triet, Z. Alibay, J. M. Plank, J. E. Cheeney and E. D. Haberer, ACS Appl. Mater. Interfaces, 2020, 12, 126–134 CrossRef PubMed.
- N.-D. Tam-Triet, J. M. Plank, G. Chen, R. E. S. Harrison, D. Morikis, H. Liu and E. D. Haberer, Nanoscale, 2018, 10, 13055–13063 RSC.
- P.-Y. Chen, X. Dang, M. T. Klug, J. Qi, N.-M. Dorval Courchesne, F. J. Burpo, N. Fang, P. T. Hammond and A. M. Belcher, ACS Nano, 2013, 7, 6563–6574 CrossRef CAS PubMed.
- J. C. Zhou, C. M. Soto, M.-S. Chen, M. A. Bruckman, M. H. Moore, E. Barry, B. R. Ratna, P. E. Pehrsson, B. R. Spies and T. S. Confer, J. Nanobiotechnol., 2012, 10, 18 CrossRef CAS PubMed.
- S.-K. Lee, D. S. Yun and A. M. Belcher, Biomacromolecules, 2006, 7, 14–17 CrossRef CAS PubMed.
- S. Li, M. Dharmarwardana, R. P. Welch, C. E. Benjamin, A. M. Shamir, S. O. Nielsen and J. J. Gassensmith, ACS Appl. Mater. Interfaces, 2018, 10, 18161–18169 CrossRef CAS PubMed.
- O. O. Adigun, G. Novikova, E. L. Retzlaff-Roberts, B. Kim, J. T. Miller, L. S. Loesch-Fries and M. T. Harris, J. Colloid Interface Sci., 2016, 483, 165–176 CrossRef CAS PubMed.
- K. L. Lee, A. A. Murray, D. H. T. Le, M. R. Sheen, S. Shukla, U. Commandeur, S. Fiering and N. F. Steinmetz, Nano Lett., 2017, 17, 4019–4028 CrossRef CAS PubMed.
- J. M. Slocik, R. R. Naik, M. O. Stone and D. W. Wright, J. Mater. Chem., 2005, 15, 749–753 RSC.
- K. I. Alsamhary, Saudi J. Biol. Sci., 2020, 27, 2185–2191 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |