DOI:
10.1039/D0NA00971G
(Review Article)
Nanoscale Adv., 2021,
3, 1853-1864
Protein-, polymer-, and silica-based luminescent nanomaterial probes for super resolution microscopy: a review
Received
20th November 2020
, Accepted 15th February 2021
First published on 22nd February 2021
Abstract
Super resolution microscopy was developed to overcome the Abbe diffraction limit, which effects conventional optical microscopy, in order to study the smaller components of biological systems. In recent years nanomaterials have been explored as luminescent probes for super resolution microscopy, as many have advantages over traditional fluorescent dye molecules. This review will summarize several different types of nanomaterial probes, covering quantum dots, carbon dots, and dye doped nanoparticles. For the purposes of this review the term “nanoparticle” will be limited to polymer-based, protein-based, and silica-based nanoparticles, including core–shell structured nanoparticles. Luminescent nanomaterials have shown promise as super-resolution probes, and continued research in this area will yield new advances in both materials science and biochemical microscopy at the nanometer scale.
Introduction
Super resolution microscopy methods were developed over the last few decades in order to overcome the optical diffraction limit by Drs Betzig, Hell, and Moerner, who were awarded the 2014 Nobel Prize for their work.1–3 The optical diffraction limit, also called the Abbe diffraction limit,4 limits the size of structures that can be resolved using light microscopy to a few hundred nanometers which is near to or larger than many subcellular structures, such as proteins and vesicles, which are of interest to researchers, as shown in Fig. 1.5 While electron microscopy can also observe length scales down to several nanometers it is incompatible with living systems and thus cannot provide information on biological processes that are of interest to researchers. The diffraction limit, given in the xy plane by eqn (1)5,6 states that the smallest resolvable distance (dx,y) is approximately equal to half of the excitation wavelength (λ) at most numerical apertures (NA); eqn (2) states that the smallest resolvable distance in the z plane (dz) is equal to double the wavelength divided by NA2.4–10| | 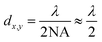 | (1) |
| | 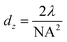 | (2) |
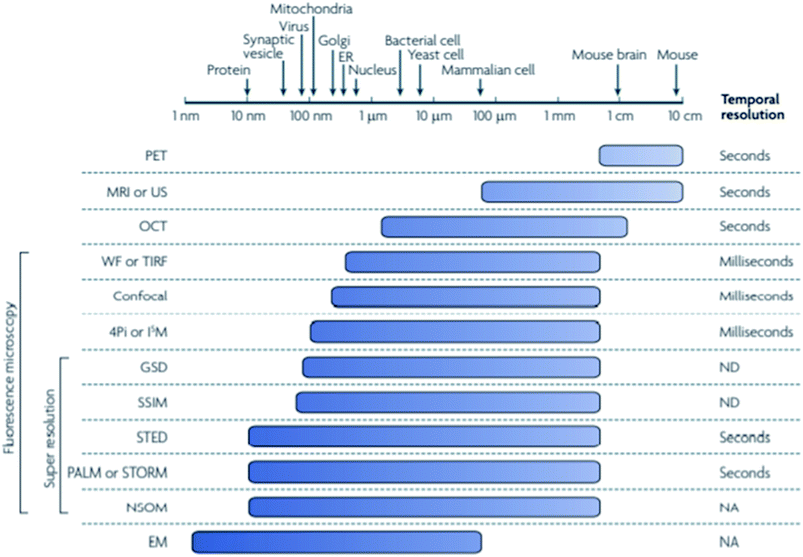 |
| | Fig. 1 The minimum spatial resolution of many techniques currently used to study biological structures is quite large, with even some super resolution techniques unable to resolve structures smaller than the mitochondria. The super resolution techniques stochastic optical reconstruction microscopy (STORM) and stimulated emission depletion microscopy (STED) are shown to be able to resolve structures down to tens of nanometers, making them able to image proteins and other small structures that cannot be resolved using typical fluorescence microscopy. Used with permission.5 | |
Super resolution microscopy is divided into two categories: single molecule localization-based super resolution such as STORM and PALM (photo-activated localization microscopy) – developed separately by Moerner3 and Betzig,2 among others, and patterned excitation super resolution such as STED and (reversible saturable optical fluorescence transitions) RESOLFT – developed by Hell.1 Localization-based super resolution techniques use fluorophores or probes that are capable of switching from dark state to an on state and back, which limits the number of fluorophores available for use. Typically used fluorophores are small molecule fluorophores such as Cy5 and Cy3, and photoswitchable fluorescent proteins such as green fluorescent protein (GFP), although these fluorophores have several disadvantages;8,9,11–15 specific imaging buffers are needed for use with small molecule fluorophores to induce photoswitching, however these buffers are toxic to cells.16,17 Both small molecule fluorophores and photoswitchable fluorescent proteins often have a lower quantum yield and rapid photobleaching. In addition, the number of photons detected positively impacts the localization precision - the more photons collected, the more precisely the fluorophore can be located – making a high quantum yield desirable.7,18–21
Recently the library of fluorescent probes for super resolution microscopy has expanded to include nanomaterials such as carbon dots, quantum dots, and dye doped nanoparticles of varying compositions. Though they are significantly larger than the previously established super resolution fluorophores – single molecule dyes are approximately 0.5 nm and fluorescent proteins, specifically green fluorescent protein (GFP), are approximately 4.2 nm in length and 2.4 nm in diameter and the various nanomaterial probes discussed in this review range in size from approximately 5 nm up to approximately 100 nm – they have been successfully used for super resolution microscopy of both cell surfaces and subcellular structures.16,22–24 These nanomaterial probes show higher quantum yields and better photostability than the previously established probes; GFP for example has a quantum yield ranging from 58–79% depending on the type of GFP being used and the commonly used fluorescent proteins have quantum yields ranging from 7% to 85%, and commonly used single molecule probes have been found to have quantum yields ranging from 23% to 93%, shown in Tables 1 and 2 respectively. Nanoparticles such as quantum dots have quantum yields ranging from 10–85% depending on composition and excitation wavelength and while they have lower quantum yields in the visible light range than fluorescent proteins and single molecule fluorophores they have higher quantum yields in the NIR light range, however carbon dots typically have lower quantum yields around 5% although doping the carbon dots with atoms such as nitrogen(N), sulfur (S), phosphorous (P), and fluorene (F) can increase the quantum yield and recently carbon dots have been developed by Arab et al. that have shown a quantum yield of 85.69%. The quantum yield for dye doped nanoparticles would depend on the dye imbedded in the nanoparticle and the structure of the nanoparticle (mesoporous silica, core–shell, etc.) and no definitive range was found.5,17,23,25–29 In addition, these nanomaterials often exhibit surface chemistry allows them to be modified for the targeting of specific structures, as demonstrated by Dong et al. using Biotin-BSA, Neutravidin, and biotinylated antibodies to modify the surface of Ag@SiO2 nanoparticles to target the surface of cells.17,23,30–32 This review will discuss the recent use of carbon dots and quantum dots, as well as dye doped polymer-, protein-, and silica-based nanoparticles, which include metal-silica core–shell nanoparticles, in super resolution fluorescence microscopy.
Table 1 A table providing information on several fluorescent proteins commonly used for super resolution microscopy. λex is the excitation wavelength, λem is the emission wavelength, εabs is the extinction coefficient, contrast is the fold increase in fluorescence at the emission wavelength after photoactivation, ηfl (%) is the quantum yield, and N is the number of photons detected per single molecule per imaging cycle. NA and ND are not applicable and not determined, respectively. Used with permission5
| Fluorescent protein |
Activating pight |
Quenching pight |
Pre/post colors |
λ
ex (nm) |
λ
em (nm) |
ε
abs (M−1 cm−1) |
η
fl (%) |
Contrast |
N
|
Oligomeric state |
| Irreversible |
Photoactivatable |
Fluorescent |
Proteins |
|
|
|
|
|
|
|
| PA-GFP |
UV-violet |
NA |
Dark/green |
504 |
517 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
79 |
200 |
ND |
Monomer |
| PA-RFP1-1 |
UV-violet |
NA |
Dark/red |
578 |
605 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
8 |
ND |
ND |
Monomer |
| Photoshiftable |
Fluorescent |
Proteins |
|
|
|
|
|
|
|
|
| PS-CFP2 |
UV-violet |
NA |
Cyan/green |
490 |
511 |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
23 |
>2000 |
260 |
Monomer |
| Kaede |
UV-violet |
NA |
Green/orange |
572 |
582 |
60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
33 |
2000 |
−400 |
Tetramer |
| KiKGR |
UV-violet |
NA |
Green/red |
583 |
593 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
65 |
>2000 |
ND |
Tetramer |
| Monometric Eos |
UV-violet |
NA |
Green/orange |
569 |
581 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
62 |
ND |
−490 |
Monomer |
| Dendra-2 |
Blue |
NA |
Green/orange |
553 |
573 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
55 |
4500 |
ND |
Monomer |
| Reversible |
Photoactivatable |
Fluorescent |
Proteins |
|
|
|
|
|
|
|
| FPS95 |
Green |
450 nm |
Dark/red |
590 |
600 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7 |
30 |
ND |
Tetramer |
| Dronpa |
UV-violet |
488 nm |
Dark/green |
503 |
518 |
95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
85 |
ND |
120 |
Monomer |
| Padron |
Blue |
405 nm |
Dark/green |
503 |
522 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
64 |
ND |
ND |
Monomer |
Table 2 A table providing information on several commonly used single molecule probes for super resolution microscopy. Color is the color of light emitted by the fluorescent molecule in the on state, εabs is the extinction coefficient, λex and λem are the excitation and emission coefficients respectively, ηfl (%) is the quantum yield, τfl is the fluorescence lifetime, N is the number of photons detected per molecule per imaging cycle, NA and ND are not applicable and not determined respectively. Used with permission5
|
|
Color |
λ
ex (nm) |
λ
em (nm) |
ε
abs (M−1 cm−1) |
η
fl (%) |
τ
fl (ns) |
N
|
Used for |
|
Photoactivation of these fluorophores is strongly facilitated by the presence of an activator fluorophore, such as Cy2, Cy3 or Alexa Fluor 405, to induce photoswitching.
The extinction coefficient is estimated at physiological pH.
|
|
Regular fluorescent dyes
|
| ATTO532 |
Orange |
532 |
553 |
115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
90 |
3.8 |
ND |
STED |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
Photoswitchers
|
| Rhodamine B |
Red |
530 |
620 |
105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
65 |
ND |
750 |
PALMIRA |
| Alexa Fluor 647a |
Red |
650 |
665 |
240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
33 |
1.0 |
6000 |
STORM |
| Cy5a |
Red |
649 |
664 |
250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
28 |
1.0 |
6000 |
STORM |
| Cy5.5a |
Red |
675 |
694 |
190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
23 |
1.0 |
6000 |
STORM |
| Cy7a |
NearIR |
747 |
767 |
200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
28 |
<0.3 |
∼1000 |
STORM |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
Photocaged fluorophores
|
| Caged Q-rhodamine |
Red |
545 |
575 |
90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
90 |
ND |
ND |
PALM |
| Caged carboxyfluorescein |
Green |
494 |
518 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000b 000b |
93 |
4.6 |
ND |
FPALM |
Super resolution background
Super resolution microscopy can be divided into two categories: super resolution microscopy using spatially patterned excitation, called stimulated emission depletion (STED), reversible saturable optically linear fluorescence transition (RESOLFT), and super resolution microscopy using single molecule/particle localization, called stochastic optical reconstruction microscopy (STORM), photoactivated localization microscopy (PALM), fluorescence photoactivation localization microscopy (FPALM), and point accumulation for imaging in nanoscale topography (PAINT).7,8,28,33–37 Two other techniques, structured illumination microscopy (SIM), and saturated-structured illumination microscopy (SSIM) will also be briefly discussed.37
Patterned excitation-based super resolution microscopy uses two lasers, one with a point of zero intensity in the center giving it a doughnut shape, which is used to switch the peripheral fluorophores into the dark state in order to reduce the effective point spread function (PSF) (Fig. 2).7 The optical resolution of STED and other patterned excitation approaches is represented by the size of the effective PSF created by the doughnut-shaped STED laser; the effective PSF is the area in which the fluorescent probes are allowed to emit fluorescence which is detected by the detector.37,38 STED can achieve spatial resolutions of 50–80 nm with temporal resolution of seconds.36 Data acquisition is relatively fast, on the order of seconds compared to localization-based methods which can take much longer to acquire enough images to construct a single-color composite image, and after acquisition the super resolution image is immediately available, requiring no post processing.6,8 A wider variety of fluorophores can be used for STED microscopy because, due to the relative simplicity of the fluorophore control, fluorophores are not limited to those that can undergo photoswitching, photoconversion, or photoactivation;6 STED has been successfully used with fluorescent labeled antibodies and fluorescent proteins, as well as quantum dots, carbon dots, and nanoparticles, with carbon dots and quantum dots being the most frequently used nanomaterial probes.5,7,16,30,39
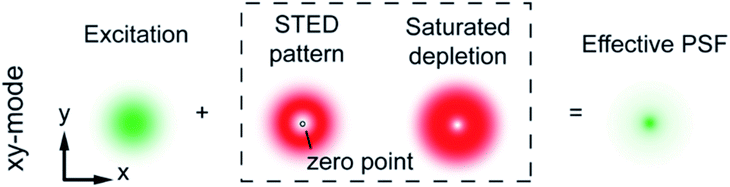 |
| | Fig. 2 A laser is used to excite a set of fluorophores, after which the donut shaped laser is used in the same area to turn the peripheral fluorophores to the dark state, decreasing the size of the effective PSF. Used with permission.7 | |
Structured illumination microscopy (SIM), excites the fluorophores using high intensity periodic line patterns to achieve sub-diffraction limit resolution; the result of this approach is that due to saturation the florescence intensity is not proportional to the excitation source power. SIM uses patterned light to illuminate the sample and measures an interference pattern created by overlaying two grids that either have different mesh sizes or angles – called the Moire pattern.7,16,37,40,41 SIM can achieve a lateral resolution of approximately 125 nm and an axial resolution of approximately 350 nm.36 Saturated structured illumination microscopy (SSIM) uses a strong excitation source in a sinusoidal pattern which allows detection of sub-diffraction limit spatial resolution. The spatial resolution of SSIM is limited by the level of fluorescence saturation rather than by diffraction, as in SIM, and has demonstrated spatial resolution down to 50 nm.7
Localization-based super resolution microscopy is based on the precise localization of individual photoswitchable fluorescent probes in order to build a super resolution image point by point. STORM super resolution uses a system of two lasers, one to activate individual fluorophores and one to image the fluorophores and return them to the dark state; once the fluorophores the area of focus have been imaged the images are processed by a computer program (such as ThunderSTORM) which calculates the center of each fluorescent point and constructs the composite super resolution image.6,13,37,42–44Fig. 3 (ref. 6) illustrates the imaging process of STORM microscopy. The fluorescent probes used for STORM microscopy must be able to photoswitch, or switch between a dark state and an on state, which limits the number and types of fluorescent probes available for use. STORM was first used with the small molecule fluorophores Cy5 and Cy3 in a dye-pair system and has since been used with other small molecule fluorophores as well as fluorescent proteins, carbon dots, quantum dots, and nanoparticles.5,6,13,16,17,23 PAINT methods rely on the diffusion and transient binding of an affinity probe conjugated to a fluorophore; when bound the fluorophore produces a bright spot on the recorded camera frame. The relative brightness of the spot is determined by measuring the brightness of the bright spot compared to the background signal from the unbound fluorophores and can be enhanced by coupling the PAINT set up to a total internal reflection (TIR) set up.45 The resolution of single molecule/particle localization methods is influenced by the labeling density and localization precision of the fluorophores and to date has achieved the highest spatial resolution among super resolution techniques, with spatial resolutions of 20–25 nm being regularly achieved and spatial resolutions as low as 5 nm are possible at the expense of lower temporal resolution (minutes rather than seconds).6,9,36,46
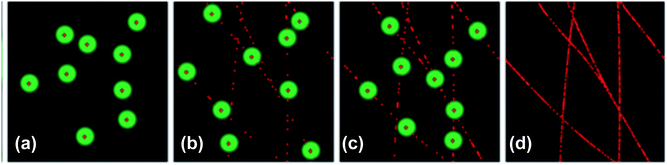 |
| | Fig. 3 All fluorophores are maintained in the dark state and a small selection of fluorophores is excited to the on state by a laser light source before being imaged and returned to the dark state by a second laser. This process is repeated many times and the many images are processed by a computer and a composite super resolution image is constructed. Used with permission.6 | |
A variation on localization-based methods, blink microscopy uses self-blinking probes to perform single-molecule or single-particle localization. The process of image acquisition is similar to STORM and PALM methods, but the self-blinking of the probe requires only a single excitation source and does not require biologically incompatible buffers.23,28,34,47–51
Nanomaterial probes
Carbon dots
Carbon-based nanomaterials, called carbon dots, are luminescent nanomaterials whose optical properties are dependent on their size and the excitation wavelength being used. Carbon dots can be synthesized through top-down or bottom-up methods using various starting materials such as citric acid, glucose, or amino acid. Characterization shows that they can contain significant amounts of oxygen and nitrogen dye to incomplete carbonization of the starting materials; in some instances carbonization of the starting materials may not happen but nanodots are still formed – analysis shows that the primary components of these may be polymers, leading to the name polymer carbon dots (PCD).52 Carbon dots have been shown to be non – toxic, biocompatible, stable in aqueous solution, resistant to photobleaching, allow many possibilities for surface functionalization, are cost effective for large scale production, and also have the potential to fluoresce in multiple colors.16,30,53–55 Originally carbon dots were thought to not exhibit blinking behaviors, however some were later discovered to experience fluorescence fluctuations; this was attributed to energy traps and different surface oxidation states.16 Reversible photoswitching of carbon dots was achieved by alternating the excitation wavelength and it was found that carbon dots exhibit long lived off states when in the presence of an electron acceptor, making carbon dots a suitable fluorescent probe for STORM and STED microscopies.16,30 Compared with typical small molecule super resolution probes carbon dots show increased brightness, greater photostability, and a lower duty cycle, however they are larger than single molecule dyes, with an approximate diameter of 5 nm.16,30
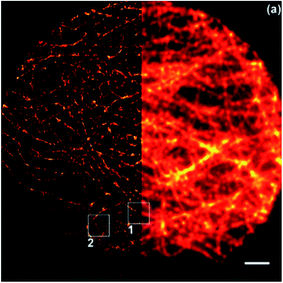
Carbon dots can be effectively used in super resolution imaging of cells and can be made to either bind to the cell membrane or, due to their small size, be taken into a living cell in order to bind to and image specific organelles or structures. Lemenager et al. were able to use carbon dots to obtain STED images the lysosomes of both fixed and living cells, shown in Fig. 4, and were able to show a >6× increase in spatial resolution compared to confocal microscopy.53 Additionally, blinking carbon dots were used by He et al. to perform STORM imaging of subcellular structures such as microtubules (Fig. 5).56 STORM imaging of the cell membrane was also performed in order to study the G protein coupled receptors.56 Khan et al. also had some success in using carbon dots for single molecule imaging of the nucleolus of living cells.57
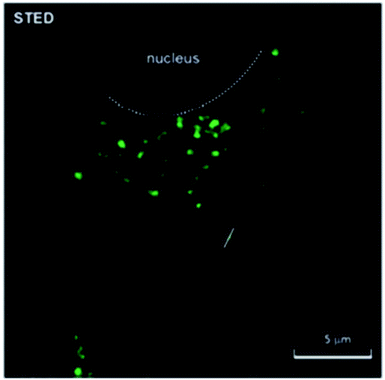 |
| | Fig. 4 STED image of carbon dots inside a MCF7 cell. The cells were incubated in the presence of the fluorescent carbon dots for 48 hours; the carbon dots accumulated in the lysosomes during the incubation period, shown in green. The white dotted line delineates the nuclear region of the cell. Used with permission.53 | |
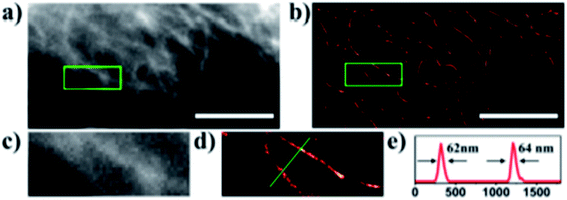 |
| | Fig. 5 (a) Conventional fluorescence image of the carbon dot labeled microtubules. (b) STORM image of the carbon dot labeled microtubules. (c) and (d) close-up of the outlined regions in (a) and (b). (e) Intensity profile of the region shown by the green line in (d). The average width of the microtubules given by the FWHM of the peaks is consistent with previous measurements performed with TEM and STORM using antibody labeling. The scale bars are 10 μm. Used with permission.28,56 | |
Quantum dots
Quantum dots are inorganic metallic nanomaterials composed of semiconducting metals; they are among the earliest reported inorganic fluorescent nanoparticles used for fluorescence labeling and have undergone more development in size, availability, biocompatible surface, and suitability for use with existing microscopy systems.22,30 The quantum dots used in biological studies have a core–shell structure, such as CdSe and ZnS, resulting in narrow emission and wide absorption spectra. Quantum dots are synthesized in organic solvent in coordination with compounds that coat it to make it hydrophobic, thus requiring additional surface modification to become usable for biological studies.58 Their emission color is size dependent, however they are considerably larger than conventional dyes and many are toxic to biological systems.30,59,60 Because the emission wavelength can be changed by adjusting the quantum dot size during synthesis and purification, they are ideal for use in multicolor imaging and super resolution applications. Quantum dots are used with both STED and STORM super resolution microscopies, as well as for other methods such as SOFI (super resolution optical fluctuation imaging) and single particle tracking.30,55,58,61 Besides toxicity, another significant drawback to using quantum dots is that they have a high on–off duty cycle (blinking) that needs to be stabilized before they can be used for super resolution imaging, though this property can be used advantageously for blinking-based super resolution (BBS) microscopy and SOFI.30,58,59
Quantum dots can be used effectively in the super resolution imaging of cells and internal cellular structures, however due to limitations such as blinking behavior and a spectral blue shift (blueing) complicate their use and require some minor modifications to either the quantum dots themselves (to reduce blinking) or to the super resolution technique being used;30,58 an example of the latter is the use of a transmission grating as part of the equipment set up in order to be able to distinguish between neighboring quantum dots that show different degrees of blueing.30 Recent use of quantum dots in super resolution microscopy include the use of quantum dots to image internal cellular structures using both STED and a localization-based super resolution microscopy called 3B by Yang et al.,62 images of which are shown in Fig. 6. Another recent use of quantum dots for multicolor 3D STORM imaging reported by Xu et al.61 a representative microscopy the time the quantum dot spends in the off state must be greater than the time spent in the on state, however quantum dots spend a shorter time in the off state compared to other photoswitchable probes, making them difficult to use for single molecule localization super resolution microscopy; in order to address this problem Xu et al. developed a technique to induce quantum dot blueing that has made quantum dots more suitable for STORM imaging by causing a stochastic shift of the emission to shorter wavelengths and observing the emission through a band pass filter which allows fluorescence from individual quantum dots to be measured (Fig. 7).61
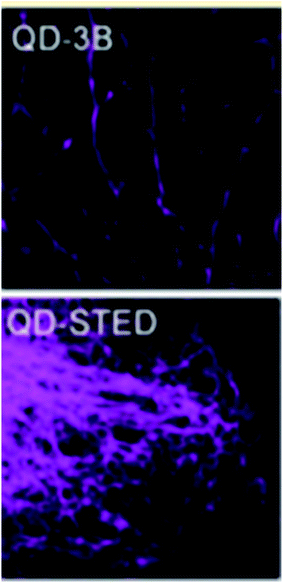 |
| | Fig. 6 Top: A 3B image of subcellular structures of a HeLa cell using quantum dots as fluorescent labels. Bottom: A STED image of a microtubule network of a HeLa cell using quantum dots as fluorescent labels. Used with permission.62 | |
 |
| | Fig. 7 A comparison of STORM (left) and wide field (right) microscopy images of microtubules in a HepG2 cell labeled with quantum dots. The scale bar is 2 μm. Used with permission.61 | |
Other nanoparticles
For the purposes of this review the term “nanoparticle” will refer to any nanomaterial that is not a quantum dot or a carbon dot. The specific nanoparticles discussed here will be dye doped nanoparticles of varying compositions, including silica-based, polymer-based, core–shell, and protein-based. Dye doped nanoparticles have been successfully used in both localization-based super resolution (STORM) and patterned excitation based super resolution (STED). The embedding of dye molecules into the rigid environment of the nanoparticle offers the advantage of reducing photobleaching and increasing the quantum yield; additionally embedding dyes that weakly fluoresce on their own may cause them to exhibit stronger fluorescence, possibly due to suppression of non-radiative deexcitation events.16 The addition of a metal core to the nanoparticle can further alter the behavior of the incorporated dye through metal enhanced fluorescence.17,23,25
Polymer-based nanoparticles, called polymer dots, are nanoparticles composed of mainly π-conjugated polymers and can be dye doped or composed of fluorescent polymers such as CN-PPV or PFBT; a polymer dot can be composed of a single polymer or multiple polymers in specific amounts depending on the needs of the user and have been shown to be bright, photostable, biocompatible, and able to label subcellular structures.10,60 The composition of the polymer dots can influence the optical behavior, producing nanoparticles that show continuous fluorescence, no fluorescence, or photoblinking.16,30 Recently dye doped polystyrene nanoparticles have been successfully used for STORM and STED microscopy, shown in Fig. 8.16,63
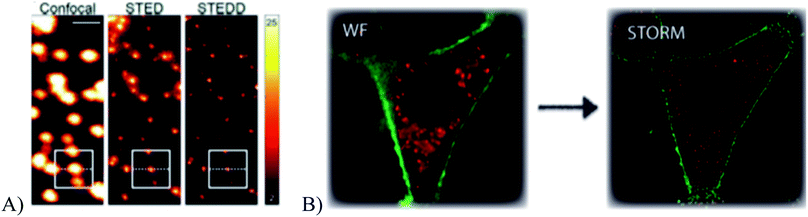 |
| | Fig. 8 (A) A comparison of confocal, STED, and STEDD (a variation of STED; stimulated emission double depletion) microscopy of fluorescent polystyrene polymer dots. Scale bar: 1 μm. (B) A comparison of wide field and STORM images of fluorescent polystyrene polymer dots (red) that have been taken into a HeLa cell. The cell membrane has been labeled separately (green). Used with permission.16,63 | |
Protein-based nanoparticles are composed of proteins such as bovine serum albumin (BSA) or transferrin as well as a variety of other sources including bacteria, viruses, amino acids, and plant or animal cells and are typically used in medicine for drug delivery. Because they are composed of proteins they are non-toxic, highly biocompatible, and can be easily modified for specific targeting; they also have symmetrical structures, uniform size distribution, and high reproducibility.64 These nanoparticles have been used for both localization-based methods (STORM) and patterned excitation methods (STED).16,65 Recently, transferrin-based protein nanoparticles developed by Lin et al.16 have been shown to be able to be taken into cells and were successfully used for STED microscopy of HeLa cells, shown in Fig. 9, and BSA-based protein nanoparticles developed by Zong et al.65 were successfully used for single molecule localization-based super resolution microscopy, shown in Fig. 10.
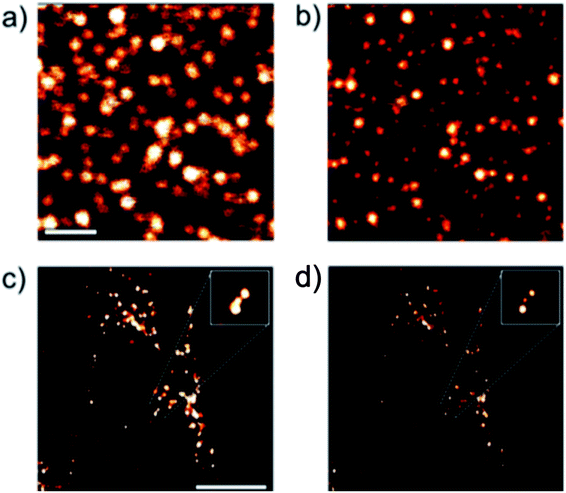 |
| | Fig. 9 (a) and (b) Confocal and STED images of transferrin-based dye doped nanoparticles on a glass surface. Scale bar: 10 μm. (c) and (d) Confocal and STED images of transferrin-based dye doped nanoparticles inside a HeLa cell. Scale bar: 10 μm. Used with permission.16 | |
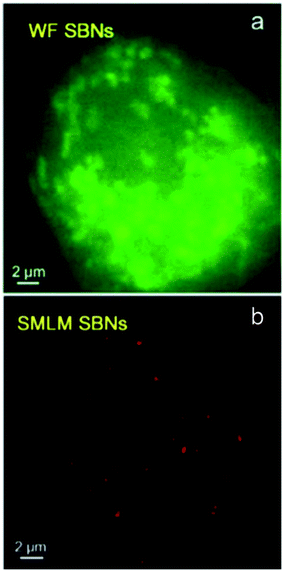 |
| | Fig. 10 Wide field and super resolution images of a SKBR3 cell incubated with dye doped BSA nanoparticles. Used with permission.65 | |
Silica-based nanoparticles, including mesoporous silica nanoparticles and core–shell nanoparticles, have been used for both STORM and STED super resolution microscopy. Silica-based nanoparticles are fairly easy to add dye to and using a core–shell design allows the microscopist to take advantage of metal enhanced fluorescence (MEF) effects, which include increased brightness and photoblinking.17,23,25,39,66,67 Mesoporous silica nanoparticles, which have numerous pores into which various types of molecules can be introduced,67 have been used in STORM microscopy after being taken into cells, shown in Fig. 11.66 Silica nanoparticles with a metal core show increased brightness due to metal enhanced fluorescence (MEF), which may also play a role in causing photoblinking behavior in dyes that do not otherwise exhibit this behavior, as observed by our group23,25 and Lu et al.17 when self-blinking nanoparticles were fabricated. The advantages of these self-blinking nanoparticles are that only a single light source is needed, rather than the two lasers normally needed for single particle localization-based super resolution methods, and that the imaging buffers normally required for these methods are not needed to cause the blinking behavior.17 Lu et al. successfully used these nanoparticles to investigate nanoparticle uptake into live cells using single molecule localization super resolution microscopy, shown in Fig. 12,17 and our group has successfully used our fabricated nanoparticles for single particle localization of individual nanoparticles, shown in Fig. 13.23
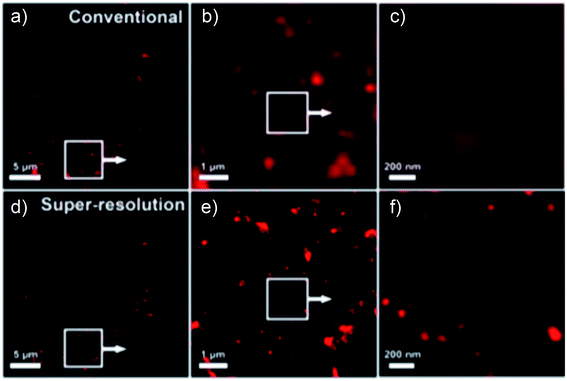 |
| | Fig. 11 (a–c) Progressively zoomed in far field images of dye doped mesoporous silica nanoparticles inside a HeLa cell. (d–f) Progressively zoomed in STORM super resolution images of dye doped mesoporous silica nanoparticles inside a HeLa cell. Used with permission.66 | |
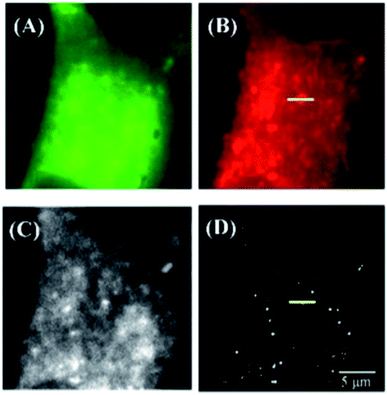 |
| | Fig. 12 (A)–(C) Wide field images of a HeLa cell using (A) FITC, (B) lysotracker deep red, (C) Rose Bengal. (D) Super resolution image of the Au@Ag@SiO2 dye-doped nanoparticles inside a HeLa cell. Used with permission.17 | |
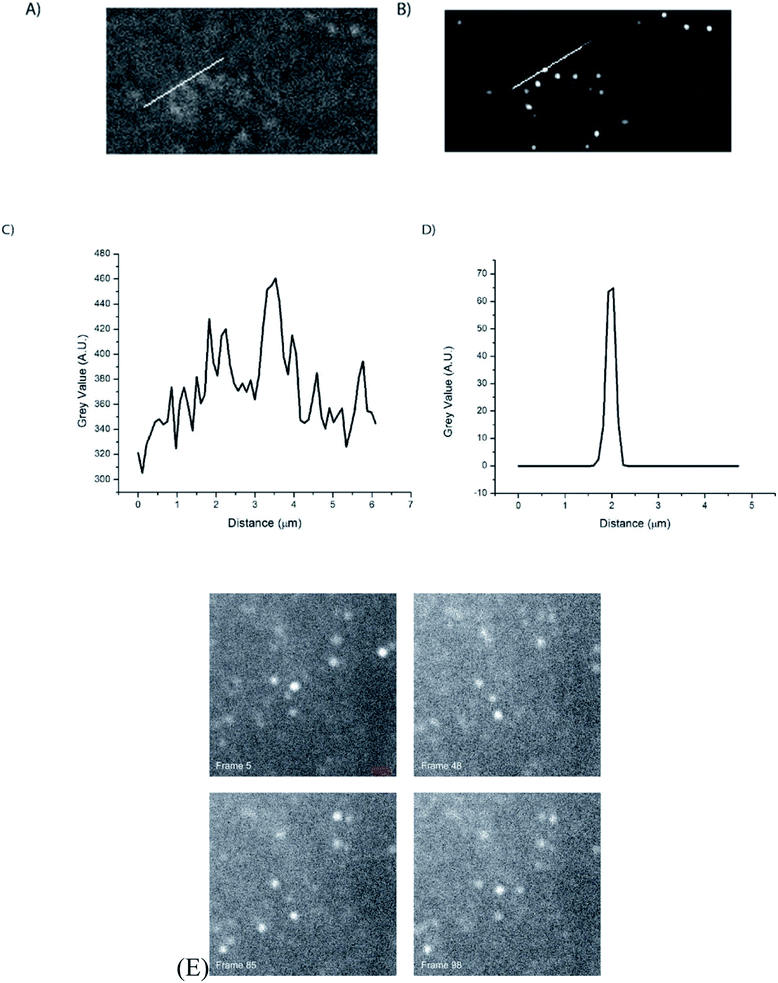 |
| | Fig. 13 (A) Wide field image of dye doped Ag@SiO2 core–shell nanoparticles immobilized on a glass slide. (B) Super resolution image of dye doped core–shell nanoparticles immobilized on a glass slide. (C and D) Graphs of the line traces performed in (A) and (B) demonstrating the increased resolution of super resolution compared to wide field microscopy by measuring the diameter of a single nanoparticle in both images. (E) Four frames from an image sequence of metal-enhanced nanoparticle fluorescence. The “on”–“off” state changes are clearly visible for several nanoparticles. Scale bar = 1.8 μm. Used with permission.23 | |
Conclusions and discussion
Super resolution microscopy is still a relatively new technology that is continuously being modified and improved upon; one such category of improvements is to the fluorescent probes used with super resolution microscopy. The established fluorophores are photoswitchable proteins and small molecule fluorophores, the drawbacks of which are relatively low quantum yield and poor photostability; small molecule fluorophores specifically require the use of buffers which are toxic to living cells and many must be used in combination with another fluorophore in order to obtain the desired behavior.5,16,30 Recently, nanomaterials have been explored for use as fluorescent probes for super resolution microscopy; although they are larger than the established fluorophores they show higher quantum yield and resistance to photobleaching and have no need for toxic buffers or use in combination with other fluorophores. Additionally, the structure of the nanomaterial probes can be modified to produce different optical properties, allowing the user to customize the probe to their specific needs. Nanoparticle probes have been developed that exhibit self-blinking behavior which eliminates the need for the two-laser system normally required for super resolution microscopy, which are used specifically with localization-based super resolution.16,17,23,25,30 These nanomaterials have already been successfully used for super resolution and have potential to become widely used in super resolution due to showing the desired optical properties and the potential to be customizable. Nanoparticles have been used to label the exterior of cells and the smaller nanoparticles have been shown to be able to be taken into the cells and imaged the labeling of subcellular structures such as mitochondria and microtubules is possible and because their surface chemistry allows for easy modification the nanoparticles can be modified in order to bind to specific cell types or organelles. The most significant drawbacks of these nanomaterials are the size and the toxic materials used for quantum dots, both of which are currently being addressed.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
ST and DP would like to acknowledge support from the National Institutes of Health (GM 120669) and National Science Foundation (1849063).
References
- T. A. Klar, S. Jakobs, M. Dyba, A. Egner and S. W. Hell, Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(15), 8206–8210 CrossRef CAS.
- E. Betzig, Proposed method for molecular optical imaging, Opt. Lett., 1995, 20(3), 237–239 CrossRef CAS.
- W. E. Moerner, New directions in single-molecule imaging and analysis, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(31), 12596–12602 CrossRef CAS.
- E. Abbe, Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung, Arch. Mikrosk. Anat., 1873, 9(1), 418–440 CrossRef.
- M. Fernandez-Suarez and A. Ting, Fluorescent probes for super-resolution imaging in living cells, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef CAS.
- J. Tam and D. Merino, Stochastic optical reconstruction microscopy (STORM) in comparison with stimulated emission depletion (STED) and other imaging methods, J. Neurochem., 2015, 135(4), 643–658 CrossRef CAS.
- B. Huang, M. Bates and X. Zhuang, Super-resolution fluorescence microscopy, Annu. Rev. Biochem., 2009, 78, 993–1016 CrossRef CAS.
- B. Huang, Super-resolution optical microscopy: multiple choices, Curr. Opin. Chem. Biol., 2010, 14(1), 10–14 CrossRef CAS.
- A. Oddone, I. V. Vilanova, J. Tam and M. Lakadamyali, Super-Resolution Imaging With Stochastic Single-Molecule Localization: Concepts, Technical Developments, and Biological Applications, Microsc. Res. Tech., 2014, 77(7), 502–509 CrossRef CAS.
- X. Z. Chen, R. Q. Li, Z. H. Liu, K. Sun, Z. Z. Sun, D. N. Chen, G. X. Xu, P. Xi, C. F. Wu and Y. J. Sun, Small Photoblinking Semiconductor Polymer Dots for Fluorescence Nanoscopy, Adv. Mater., 2017, 29(5), 7 Search PubMed.
- P. Bondia, S. Casado and C. Flors, Correlative Super-Resolution Fluorescence Imaging and Atomic Force Microscopy for the Characterization of Biological Samples, Methods Mol. Biol., 2017, 1663, 105–113 CrossRef CAS.
- M. Fernandez-Suarez and A. Y. Ting, Fluorescent probes for super-resolution imaging in living cells, Nat. Rev. Mol. Cell Biol., 2008, 9(12), 929–943 CrossRef CAS.
- G. Patterson, M. Davidson, S. Manley and J. Lippincott-Schwartz, Superresolution imaging using single-molecule localization, Annu. Rev. Phys. Chem., 2010, 61, 345–367 CrossRef CAS.
- J. Vogelsang, C. Steinhauer, C. Forthmann, I. H. Stein, B. Person-Skegro, T. Cordes and P. Tinnefeld, Make them blink: probes for super-resolution microscopy, ChemPhysChem, 2010, 11(12), 2475–2490 CrossRef CAS.
- G. T. Dempsey, M. Bates, W. E. Kowtoniuk, D. R. Liu, R. Y. Tsien and X. W. Zhuang, Photoswitching Mechanism of Cyanine Dyes, Biophys. J., 2010, 98(3), 393A CrossRef.
- Y. H. Lin, K. Nienhaus and G. U. Nienhaus, Nanoparticle Probes for Super-Resolution Fluorescence Microscopy, ChemNanoMat, 2018, 4(3), 253–264 CrossRef CAS.
- J. Lu, S. F. Zong, Z. Y. Wang, C. Chen, Y. Z. Zhang and Y. P. Cui, Yolk-shell type nanoprobe with excellent fluorescence ‘blinking’ behavior for optical super resolution imaging, Nanotechnology, 2017, 28(26), 265701 CrossRef.
- H. Gao, M. Pu, X. Li, X. Ma, Z. Zhao, Y. Guo and X. Luo, Super-resolution imaging with a Bessel lens realized by a geometric metasurface, Optic Express, 2017, 25(12), 13933–13943 CrossRef.
- R. Heintzmann and T. Huser, Super-Resolution Structured Illumination Microscopy, Chem. Rev., 2017, 117(23), 13890–13908 CrossRef CAS.
- Y. S. Hu, H. Cang and B. F. Lillemeier, Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes, Proc. Natl. Acad. Sci. U.S.A., 2016, 113(26), 7201–7206 CrossRef CAS.
- G. T. Dempsey, J. C. Vaughan, K. H. Chen and X. W. Zhuang, Evaluation of Fluorophores for Optimal Performance in Localization-Based Super-Resolution Imaging, Biophys. J., 2012, 102(3), 725A CrossRef.
- D. Y. Jin, P. Xi, B. M. Wang, L. Zhang, J. Enderlein and A. M. van Oijen, Nanoparticles for super-resolution microscopy and single-molecule tracking, Nat. Methods, 2018, 15(6), 415–423 CrossRef CAS.
- S. Thompson and D. Pappas, Core size does not affect blinking behavior of dye-doped Ag@SiO2 core–shell nanoparticles for super-resolution microscopy, RSC Adv., 2020, 10(15), 8735–8743 RSC.
- M. A. Hink, R. A. Griep, J. W. Borst, A. van Hoek, M. H. M. Eppink, A. Schots and A. Visser, Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein, J. Biol. Chem., 2000, 275(23), 17556–17560 CrossRef CAS.
- C. Chakraborty, S. B. Thompson, V. J. Lyons, C. Snoeykink and D. Pappas, Modulation and study of photoblinking behavior in dye doped silver-silica core-shell nanoparticles for localization super-resolution microscopy, Nanotechnology, 2019, 455704 CrossRef CAS.
- R. Y. Tsien, The green fluorescent protein, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Quantum dots versus organic dyes as fluorescent labels, Nat. Methods, 2008, 5(9), 763–775 CrossRef CAS.
- H. He, X. Liu, S. Li, X. Wang, Q. Wang, J. Li, J. Wang, H. Ren, B. Ge, S. Wang, X. Zhang and F. Huang, High-Density Super-Resolution Localization Imaging with Blinking Carbon Dots, Anal. Chem., 2017, 89(21), 11831–11838 CrossRef CAS.
- S. Arab, S. Masoum and E. S. Hosseini, Engineering the Synthesis of Luminescent Carbon Dots With Ultra High-Quantum Yield Using Experimental Design Approach in Order to Develop Sensing Applications, IEEE Sensor. J., 2020, 20(4), 1705–1711 CAS.
- D. Jin, P. Xi, B. Wang, L. Zhang, J. Enderlein and A. M. van Oijen, Nanoparticles for super-resolution microscopy and single-molecule tracking, Nat. Methods, 2018, 415–423 CrossRef CAS.
- M. Dong, Y. Tian and D. Pappas, Facile Functionalization of Ag@SiO2 Core-Shell Metal Enhanced Fluorescence Nanoparticles for Cell Labeling, Anal. Methods, 2014, 6(5), 1598–1602 RSC.
- M. Dong, Y. Tian and D. Pappas, Synthesis of a red fluorescent dye-conjugated Ag@SiO2 nanocomposite for cell immunofluorescence, Appl. Spectrosc., 2015, 69(2), 215–221 CrossRef CAS.
- P. Choudhury and P. K. Das, Carbon Dots-Stimulated Amplification of Aggregation-Induced Emission of Size-Tunable Organic Nanoparticles, Langmuir, 2019, 35(32), 10582–10595 CrossRef CAS.
- B. Zhi, Y. Cui, S. Wang, B. P. Frank, D. N. Williams, R. P. Brown, E. S. Melby, R. J. Hamers, Z. Rosenzweig, D. H. Fairbrother, G. Orr and C. L. Haynes, Malic Acid Carbon Dots: From Super-resolution Live-Cell Imaging to Highly Efficient Separation, ACS Nano, 2018, 5741–5752 CrossRef CAS.
- S. Zong, F. Pan, R. Zhang, C. Chen, Z. Wang and Y. Cui, Super blinking and biocompatible nanoprobes based on dye doped BSA nanoparticles for super resolution imaging, Nanotechnology, 2019, 30(6), 065701 CrossRef CAS.
- S. Pujals, N. Feiner-Gracia, P. Delcanale, I. Voets and L. Albertazzi, Super-resolution microscopy as a powerful tool to study complex synthetic materials, Nat. Rev. Chem., 2019, 3(2), 68–84 CrossRef.
- S. J. Sahl, S. W. Hell and S. Jakobs, Fluorescence nanoscopy in cell biology, Nat. Rev. Mol. Cell Biol., 2017, 18(11), 685–701 CrossRef CAS.
- M. Leutenegger, C. Eggeling and S. W. Hell, Analytical description of STED microscopy performance, Optic Express, 2010, 18(25), 26417–26429 CrossRef CAS.
- N. Prabhakar, T. Nareoja, E. von Haartman, D. Sen Karaman, H. Jiang, S. Koho, T. A. Dolenko, P. E. Hanninen, D. I. Vlasov, V. G. Ralchenko, S. Hosomi, D. I. Vlasov II, C. Sahlgren and J. M. Rosenholm, Core-shell designs of photoluminescent nanodiamonds with porous silica coatings for bioimaging and drug delivery II: application, Nanoscale, 2013, 5(9), 3713–3722 RSC.
- B. O. Leung and K. C. Chou, Review of Super-Resolution Fluorescence Microscopy for Biology, Appl. Spectrosc., 2011, 65(9), 967–980 CrossRef CAS.
- L. Schermelleh, R. Heintzmann and H. Leonhardt, A guide to super-resolution fluorescence microscopy, J. Cell Biol., 2010, 190(2), 165–175 CrossRef CAS.
- M. Ovesny, P. Krizek, J. Borkovec, Z. Svindrych and G. M. Hagen, ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging, Bioinformatics, 2014, 30(16), 2389–2390 CrossRef CAS.
- M. Bates, B. Huang, G. T. Dempsey and X. Zhuang, Multicolor super-resolution imaging with photo-switchable fluorescent probes, Science, 2007, 317(5845), 1749–1753 CrossRef CAS.
- M. J. Rust, M. Bates and X. Zhuang, Stochastic Optical Reconstruction Microscopy (STORM) Provides Sub-Diffraction-Limit Image Resolution, Nat. Methods, 2006, 3(10), 793–795 CrossRef CAS.
-
M. J. Dai, DNA-PAINT Super-Resolution Imaging for Nucleic Acid Nanostructures, in 3d DNA Nanostructure: Methods and Protocols, ed. Y. Ke, P. Wang, 2017, vol. 1500, pp. 185–202 Search PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods, 2006, 3(10), 793–795 CrossRef CAS.
- C. Chakraborty, S. Thompson, V. J. Lyons, C. Snoeyink and D. Pappas, Modulation and study of photoblinking behavior in dye doped silver-silica core-shell nanoparticles for localization super-resolution microscopy, Nanotechnology, 2019, 30(45), 455704 CrossRef CAS.
- X. Chen, Z. Liu, R. Li, C. Shan, Z. Zeng, B. Xue, W. Yuan, C. Mo, P. Xi, C. Wu and Y. Sun, Multicolor Super-resolution Fluorescence Microscopy with Blue and Carmine Small Photoblinking Polymer Dots, ACS Nano, 2017, 11(8), 8084–8091 CrossRef CAS.
- T. Cordes, M. Strackharn, S. W. Stahl, W. Summerer, C. Steinhauer, C. Forthmann, E. M. Puchner, J. Vogelsang, H. E. Gaub and P. Tinnefeld, Resolving Single-Molecule Assembled Patterns with Superresolution Blink-Microscopy, Nano Lett., 2010, 10(2), 645–651 CrossRef CAS.
- P. J. Macdonald, S. Gayda, R. A. Haack, Q. Ruan, R. J. Himmelsbach and S. Y. Tetin, Rhodamine-Derived Fluorescent Dye with Inherent Blinking Behavior for Super-Resolution Imaging, Anal. Chem., 2018, 90(15), 9165–9173 CrossRef CAS.
- S. N. Uno, M. Kamiya, A. Morozumi and Y. Urano, A green-light-emitting, spontaneously blinking fluorophore based on intramolecular spirocyclization for dual-colour super-resolution imaging, Chem. Commun., 2017, 54(1), 102–105 RSC.
- Y. B. Song, S. J. Zhu, J. R. Shao and B. Yang, Polymer Carbon Dots-A Highlight Reviewing Their Unique Structure, Bright Emission and Probable Photoluminescence Mechanism, J. Polym. Sci., Part A: Polym. Chem., 2017, 55(4), 610–615 CrossRef CAS.
- G. Lemenager, E. De Luca, Y. P. Sun and P. P. Pompa, Super-resolution fluorescence imaging of biocompatible carbon dots, Nanoscale, 2014, 6(15), 8617–8623 RSC.
- N. C. Verma, S. Khan and C. K. Nandi, Single-molecule analysis of fluorescent carbon dots towards localization-based super-resolution microscopy, Methods Appl. Fluoresc., 2016, 4(4), 044006 CrossRef.
- Z. H. Liu, J. Liu, X. D. Wang, F. X. Mi, D. Wang and C. F. Wu, Fluorescent Bioconjugates for Super-Resolution Optical Nanoscopy, Bioconjugate Chem., 2020, 31(8), 1857–1872 CrossRef CAS.
- H. He, X. Liu, S. Li, X. J. Wang, Q. Wang, J. Q. Li, J. Y. Wang, H. Ren, B. S. Ge, S. J. Wang, X. D. Zhang and F. Huang, High-Density Super-Resolution Localization Imaging with Blinking Carbon Dots, Anal. Chem., 2017, 89(21), 11831–11838 CrossRef CAS.
- S. Khan, N. C. Verma, Chethana and C. K. Nandi, Carbon Dots for Single-Molecule Imaging of the Nucleolus, ACS Appl. Nano Mater., 2018, 1(2), 483–487 CrossRef CAS.
- T. Ichimura, T. Jin, H. Fujita, H. Higuchi and T. M. Watanabe, Nano-scale measurement of biomolecules by optical microscopy and semiconductor nanoparticles, Front. Physiol., 2014, 5, 273 Search PubMed.
- X. M. Liu, S. Y. Chen, Q. Chen, X. L. Yao, M. Gelleri, S. Ritz, S. Kumar, C. Cremer, K. Landfester, K. Mullen, S. H. Parekh, A. Narita and M. Bonn, Nanographenes: Ultrastable, Switchable, and Bright Probes for Super-Resolution Microscopy, Angew. Chem., Int. Ed., 2020, 496–502 CrossRef CAS.
- C. F. Wu, T. Schneider, M. Zeigler, J. B. Yu, P. G. Schiro, D. R. Burnham, J. D. McNeill and D. T. Chiu, Bioconjugation of Ultrabright Semiconducting Polymer Dots for Specific Cellular Targeting, J. Am. Chem. Soc., 2010, 132(43), 15410–15417 CrossRef CAS.
- J. Q. Xu, K. F. Tehrani and P. Kner, Multicolor 3D Super-resolution Imaging by Quantum Dot Stochastic Optical Reconstruction Microscopy, ACS Nano, 2015, 9(3), 2917–2925 CrossRef CAS.
- X. S. Yang, K. Zhanghao, H. N. Wang, Y. J. Liu, F. Wang, X. Zhang, K. B. Shi, J. T. Gao, D. Y. Jin and P. Xi, Versatile Application of Fluorescent Quantum Dot Labels in Super resolution Fluorescence Microscopy, ACS Photonics, 2016, 3(9), 1611–1618 CrossRef CAS.
- D. van der Zwaag, N. Vanparijs, S. Wijnands, R. De Rycke, B. G. De Geest and L. Albertazzi, Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking, ACS Appl. Mater. Interfaces, 2016, 8(10), 6391–6399 CrossRef CAS.
- E. J. Lee, Recent advances in protein-based nanoparticles, Korean J. Chem. Eng., 2018, 35(9), 1765–1778 CrossRef CAS.
- S. Zong, F. Pan, R. Zhang, C. Chen, Z. Wang and Y. Cui, Super blinking and biocompatible nanoprobes based on dye doped BSA nanoparticles for super resolution imaging, Nanotechnology, 2018, 30, 065701 CrossRef.
- S. S. Chen, J. Wang, B. Xin, Y. B. Yang, Y. R. Ma, Y. Zhou, L. J. Yuan, Z. L. Huang and Q. Yuan, Direct Observation of Nanoparticles within Cells at Subcellular Levels by Super-Resolution Fluorescence Imaging, Anal. Chem., 2019, 91(9), 5747–5752 CrossRef CAS.
- J. Wang, Q. Q. Ma, Y. Q. Wang, Z. H. Li, Z. H. Li and Q. Yuan, New insights into the structure-performance relationships of mesoporous materials in analytical science, Chem. Soc. Rev., 2018, 47(23), 8766–8803 RSC.
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *
*
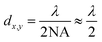
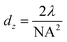

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600
600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000b
000b











