Open Access Article
Open Access ArticleNanoscopy for endosomal escape quantification
Teodora
Andrian†
 a,
Roger
Riera†
b,
Silvia
Pujals
ac and
Lorenzo
Albertazzi
a,
Roger
Riera†
b,
Silvia
Pujals
ac and
Lorenzo
Albertazzi
 *ab
*ab
aNanoscopy for Nanomedicine, Institute for Bioengineering of Catalonia, Barcelona, Spain. E-mail: l.albertazzi@tue.nl
bDepartment of Biomedical Engineering, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, Netherlands
cDepartment of Electronics and Biomedical Engineering, Faculty of Physics, Universitat de Barcelona, Av. Diagonal 647, 08028, Barcelona, Spain
First published on 27th October 2020
Abstract
The successful cytosolic delivery of nanoparticles is hampered by their endosomal entrapment and degradation. To push forward the smart development of nanoparticles we must reliably detect and quantify their endosomal escape process. However, the current methods employed are not quantitative enough at the nanoscale to achieve this. Nanoscopy is a rapidly evolving field that has developed a diverse set of powerful techniques in the last two decades, opening the door to explore nanomedicine with an unprecedented resolution and specificity. The understanding of key steps in the drug delivery process – such as endosomal escape – would benefit greatly from the implementation of the most recent advances in microscopy. In this review, we provide the latest insights into endosomal escape of nanoparticles obtained by nanoscopy, and we discuss the features that would allow these techniques to make a great impact in the field.
1. Introduction
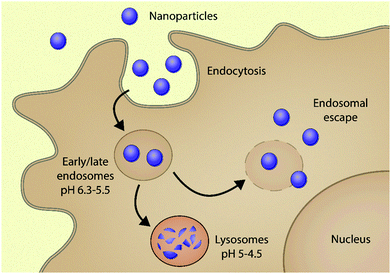
Using nanoparticles (NPs) to deliver drugs to cells (nanomedicine) was foreseen to be a true game-changer of the 21st century in improving the prevention, diagnosis and therapy of various diseases.1–6 The potential of these nanosized carriers in pharmaceutical applications has been envisioned since the 1970's to improve the delivery of therapeutic and imaging agents to specific target sites.7–11 The remarkable interest in NPs is attributed to the plethora of physical and biological advantages they offer in comparison to conventional medicines, such as improved efficacy and safety, enhanced solubility and pharmacokinetic profiles, and increased target selectivity.12–15Although various NP formulations have been marketed,16,17 achieving efficient intracellular delivery still remains a significant challenge.18–22 One of the main culprits is that the majority of NPs – once taken up via endocytosis – are unavoidably distributed in endocytic vesicles. These acidic organelles can degrade the carrier-drug ensemble, reducing its bioavailability in the intracellular environment.23,24 Within these vesicles, pH gradually drops from neutral to acidic because of membrane-incorporated vacuolar-type ATPases. The cargo is first brought into the early endosome (pH ∼ 5.5); then finally the late endosome fuses with the lysosome and the cargo is degraded by hydrolytic enzymes present in the acidic milieu (pH 5–4.5). The recycling endosome may direct some cargo back to the cell surface, whilst the majority remain entrapped in the endo-lysosomal pathway, where they are degraded20,25 (Fig. 1). Endosomal entrapment thus represents one of the main bottlenecks in using NP systems for gene therapy20,26–28 and proteins or small molecular drugs for the treatment of a variety of diseases.20,29–31
Mechanisms through which NPs – and more importantly the therapeutic cargo – can escape these degrading vesicles have become the subject of intense research over the past few decades.19,20,24,32 Inspired by the innate ability of bacterial toxins and viruses to escape endosomal vesicles, various hypothetical endosomal escape mechanisms have been proposed and reviewed in the literature, such as the “proton sponge” effect, membrane fusion, pore formation, membrane disruption, and vesicle budding and collapse.24,27,32–36 Numerous strategies to enhance the escape of NPs have also been suggested, including endosomal buffering agents, membrane fusogenic peptides, lysosomotropic chemical agents,20,23,37–44 morphological-dependent changes45 or external stimuli such as photochemical internalization (PCI).46,47
It is crucial to note that the endosomal escape hypotheses suffer from many inconsistencies. For example, the “proton sponge” hypothesis – based on the buffering capacity of polycations, that are suggested to cause an increase in lysosomal pH – has been heavily disputed in the literature.48,49 As a result, the mechanism of the action of these formulation strategies is generally unknown. This is limiting the development of NPs with efficient endosomal escape, and it is further worsened by the absence of effective methods to detect – and more crucially to quantify – this process. Consequently, it is challenging to determine which strategies are efficient in improving the escape ability of NPs, hindering the development of successful formulations. Additionally, the lack of standardized methods leads to poor comparisons between different endosomal escape studies, leading to contradicting and inconclusive results.48–53
Standard methods used to assess endosomal escape commonly employ fluorescence microscopy, flow cytometry or mass spectrometry. However, fluorescence microscopy cannot be used alone to quantify the total number of particles inside cells, as this requires cumbersome calibration of the fluorescence signal, and lacks the resolution to quantify individual NPs below 250 nm. Flow cytometry measures relative fluorescence intensity rather than individual NPs, and mass spectrometry leads to the loss of spatial information.54 Readers are directed to other available reviews for information on these techniques and how they may compare with nanoscopy methods.24,35,55,56 Studying endosomal escape brings alongside certain challenges; it is a fast process, rare and occurs in the nanoscale. All these techniques have limited spatial resolution, are often poorly quantitative and fail to provide information on endosomal escape at the nanoscale and quantitative level, or with high molecular specificity within the cellular biological environment. Therefore, new, and improved techniques are necessary for the quantification of NP–cell interactions to allow comparison and integration of data and push forward the smart development of NPs.
Here we highlight the most prominent nanoscopy techniques and discuss the features that overcome the limitations of standard methods. We briefly emphasize on how they can be used for quantification of endosomal escape, and we provide a short perspective on how these techniques can help us gain more insight into the process of NP endosomal escape, leading to the development of more effective formulations.
2. Discussion
As previously highlighted, endosomal escape is a process that is fast and rare and occurs at the nanoscale. Here we briefly discuss the pros and cons of various nanoscopy techniques that can be used to quantitatively study this process. For a summary of the techniques discussed see Table 1, and for extra information on how quantification can be achieved using these techniques, see Table 2. We put emphasis on the power of electron microscopy (EM), super-resolution microscopy (SRM) and correlative imaging to answer sought-after questions regarding NP endosomal escape, and ultimately on improving the development of NPs with efficient therapeutic cytosolic delivery.| Technique | Resolution XY | Resolution Z | Live-cell imaging | Multi-colour | Temporal resolution | Quantification | Overall simplicity of technique | References |
|---|---|---|---|---|---|---|---|---|
| a SMLM does not allow live cell imaging in most cases, but there are few examples.87,89 | ||||||||
| Confocal microscopy | ∼200 nm | ∼400 nm | Yes | Yes (3 colours) | ms–s | Worst | Simple | 30,54,56,66 and 144–149 |
| EM | ∼1 nm | NA | No | No | ms | Good | Complex | 18,30,58,60–62,64,66–74,76–78 and 150–153 |
| SMLM | ∼20 nm | ∼80 nm | Noa | Yes (2–124 colours) | min | Good | Complex | 81–85 |
| STED | ∼50 nm | ∼150 nm | Yes | Yes (3 colours) | s | Good | Simple | 98–101 |
| SIM | ∼100 nm | ∼300 nm | Yes | Yes (3 colours) | ms–s | Bad | Simple | 107 and 112–115 |
| CLEM | ∼1 nm (EM) | Dependent on LM technique | No | Dependent on LM technique | Dependent on LM technique | Best | Very complex | 68,72 and 141–143 |
| Technique | Quantification | Quantification process | Throughput | Disadvantages |
|---|---|---|---|---|
| a Quantification is not absolute (not at a single particle level) due to the resolution of the microscope. | ||||
| Confocal | Co-localization | NP endosomal vesicles are tagged with different fluorophores and average fluorescence intensity is calculated (e.g. using ImageJ – Color2/JACoP). Co-localization calculated using Mander's/Pearson's correlation coefficients56,148 | Fairly good throughput – (i.e. a few cells, tens of lysosomes and tens of NP clusters) | -Limited resolution |
| -Cannot resolve individual NPs | ||||
| -Localization precision is affected by resolution | ||||
| Particle tracking | The total number of particles and endosome co-localized NPs is tracked and counted with particle tracking software. As NPs cannot be individually detected, particle events are calculated instead, whereby a single NP event likely corresponds to one vesicle containing NPs75,154 | -Invisible particles (i.e. due to bleaching of fluorophores or de-coupling of fluorescent dyes) | ||
| -Number of particles is underestimated | ||||
| -Choice of fluorophore can influence results (e.g. pH sensitive dyes can have reduced signals in acidic vesicles) | ||||
| -Fluorescence must be quantified in relation to a control to account for fluorophore instability | ||||
| EM | Direct visualization and quantification | The ratio between NPs found in the cytosol and endosomes is calculated148,155,156 | Low throughput – (i.e. one cell, tens of NPs and a few endosomes per field of view) | -Complicated sample preparation |
| Can distinguish between intracellular/extracellular/intramembranous nanoparticles | -Generally, samples are fixed and sectioned (i.e. no living cells) | |||
| Serial sectioning or electron tomography | Imaging in 3D of sequential sample sections. Location, size, and the number of vesicles as well as NPs can be calculated in whole 3D cells74,156 | -Difficulty in distinguishing different intracellular vesicles | ||
| Stereological image analysis | Using the relative particle distribution within cells (RDI). Tests if NPs are localizing randomly or specifically within cellular compartments. The particle density of each compartment is calculated by relating the number of particle events in the specific compartment to the fractional volume of the compartment157 | -Particles must be smaller than the section thickness (∼150 nm) | ||
| Correlating the total number of intracellular particles of a sample with the total cell number of that sample. Using the fractionator principle54,158 | -Quantification from 3D reconstructions is difficult | |||
| The density of intracellular particles is multiplied by the average cell volume to calculate the average number of NPs per cell75 | -Restricted to samples with adequate atomic contrast | |||
| SMLM | Spatial analysis and clustering | Single-molecule localization microscopy techniques produce point cloud data as a result of multiple localizations in time. These data can be analyzed to identify objects, and determine densities or spatial correlations93 | Intermediate – low throughput. The field of view may vary from one to few cells. The imaging time would greatly depend on the specific technique used (seconds to minutes) | -In some cases, there are undesired non-specific interactions or background noise |
| Molecule counting | Single-molecule localization microscopy techniques are based on the identification of individual molecules. Therefore, it is possible to quantify the exact number of molecules on a specific area.159,160 For example, the ligands or proteins on the surface of a nanoparticle161,162 | -High amount of data that can make the quantification process slow | ||
| Stability of NPs and vesicles | The increased resolution and precise molecule counting of super-resolution microscopy allow the determination of the stability of small objects such as nanoparticles and vesicles. It is possible to establish their shape96 and observe the degradation in time99 | |||
| STED/SIM | Size and shape of NPs | The improved resolution of these techniques allows the measurements of the size and shape of smaller objects compared to confocal microscopy109,113,114 | Good throughput – imaging times in the millisecond-second range | -No single-molecule quantification |
| Co-localization | Standard colocalization coefficient calculations are also applied for these techniques, although better resolution yields more precise results109 | |||
| CLEM | Combination of FM and EM techniques | Generally, fluorescence microscopy is carried out prior to EM. Images can be manually aligned using plugins such as eC-CLEM. Quantification can be achieved either via EM or FM, or both.74,163 Detection of ‘invisible particles’ in light microscopy is possible with CLEM, as well as compartment-specific quantification | Low throughput – still limited by EM | -Complex and time-consuming sample preparation |
| -NPs must be detectable using both light and electron microscopes | ||||
| -Alignment mismatch can affect correlation | ||||
2.1. Electron microscopy and cryo-electron microscopy
With a near atomic resolution,57 EM is an irreplaceable tool in studying the physio-chemical properties of NPs and quantifying their voyage through the endo-lysosomal pathway.30,37,58–74 EM can even detect a low number (few hundreds) of single nanoparticles escaping endosomal structures, and since it is a label-free method, it will localise and quantify NPs generally untraceable by standard light microscopy methods. TEM was demonstrated to quantify approximately 150-times more NP/cell compared to NP events/cell using a confocal laser scanning microscope.75 EM allows direct visualisation and quantification of NPs and endosomal compartments and can distinguish between intracellular/extracellular/intramembranous NPs, both in 2D and 3D (Table 2).As a pioneering example, Gilleron et al.37 developed one of the most promising semi-automatic approaches using TEM, quantifying the amount of siRNA-conjugated colloidal gold NPs escaping from various endo-lysosomal compartments (Fig. 2A, top left). The authors developed a gold detection software that automatically detects and quantifies the total number of gold NPs in each image, based on the threshold intensity of gold. Using this, they quantified the ratio of siRNA-gold within the endosomes and in the cytosol to calculate endosomal escape and found that only <2% of siRNA-gold escaped the endosomes in HeLa cells. Furthermore, using distinct mathematical models in combination with a pharmacological blockade of endosomal progression, they observed that release occurs mainly from the early endosome. Additionally, developments in staining methods such as photoconversion of diaminobenzene (DAB) – that allows the conversion of a fluorescent dye into an electron-dense signal – in combination with immunoelectron microscopy demonstrate that EM can be used to examine the interactions of NPs with cellular organelles and to detect if they are intact or degraded after endo-lysosomal breakdown.76,77
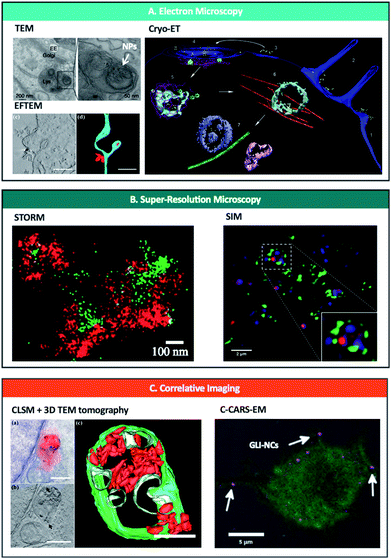 | ||
| Fig. 2 Nanoscopy techniques used to study and/or quantify endo-lysosomal trafficking of nanoparticles. (A) Electron microscopy techniques including TEM (upper left),37 EFTEM (lower left)74 and Cryo-ET (right)69 can be used to track and quantify nanoparticles in intracellular vesicles. Reprinted (adapted) with permission from ref. 37 Copyright © 2013 Nature America, Inc., from ref. 74 © 2019 American Chemical Society and from ref. 69 Copyright © Azubel et al. eLife. (B) Super-resolution microscopy has been used to image nanoparticles bursting out of endosomes with STORM (left)96 and SIM (right).117 Reprinted (adapted) with permission from ref. 96 and 117 Copyright 2018 American Chemical Society. (C) Correlative imaging combines different microscopic techniques such as CLSM and 3D TEM tomography (left)74 or C-CARS and EM (right)70 and offers spatiotemporal localization of labelled NPs and biomolecules with high specificity and sensitivity at a highly subcellular level; quantification possible with CLSM + 3D TEM tomography. Reprinted (adapted) with permission from ref. 74 Copyright © 2019 American Chemical Society and from ref. 70 Copyright © 2018 The Authors. Biotechnology Journal Published by Wiley-VCH Verlag GmbH & Co. KGaA. | ||
One of the limitations of conventional EM is that the image acquired corresponds to a distorted, dehydrated form of the natural specimen, due to the need for drying, staining or plastic embedding the sample. Using cryo-EM, the specimen exists in a near-native frozen-hydrated state, maintaining the structures of interest as they would be in solution.78,79 However, to date, the only paper exploiting cryo-EM to study the trafficking of NPs within the endosomal pathway (albeit indirectly) is by Azubel et al.69 who employed cryo-electron tomography (cryo-ET) to study the endosomal trafficking of fibroblast growth factor 21 tagged to gold NPs (AuNP-FGF21) (Fig. 2A, right). By using 3D tomographic reconstruction, they were able to unequivocally identify gold NPs inside/outside various cellular structures including endosomes. Although the authors did not focus on quantifying the gold NPs, this technique has great potential to quantify the endosomal escape of various inorganic NPs, as well as that of different proteinaceous ligands/protein-based cargo at a single-particle level and with great localisation precision.
Indisputably, EM is an irreplaceable asset in the tracking and quantification of NP endosomal escape. However, it can only be used on fixed or frozen samples and it is inappropriate for studying dynamic changes. Cellular samples must be cut into <200 nm thin sections and exposed to various staining and washing steps that can lead to the loss of NPs.80 Also, at the expense of high resolution, only a small field of view (a few endosomes and a few tens of NPs) can be analysed at one time, making this a low-throughput technique. Lastly, EM has reduced molecular specificity, thus making it difficult to distinguish between different types of vesicles within the endosomal pathway.60
2.2. Super-resolution microscopy or optical nanoscopy
In the history of light microscopy, better lenses were used to improve resolution by focusing more light onto the sample, such as the pinhole in confocal microscopy.81 However, Abbe's diffraction's law82 determines that the ultimate resolution of any light microscope is limited to 200–350 nm due to light diffraction. The advent of SRM83 allows overcoming this limitation combining the advantages of fluorescence microscopy with nanometric resolutions. The specific labelling of proteins, multicolour ability and live-cell imaging at subcellular resolutions transformed this method into a new powerful tool to study endosome escaping.The main advantages of these techniques are that they have an excellent resolution (5–25 nm) to visualize NPs and intracellular vesicles, and they can offer a powerful quantitative tool with single-molecule precision,93i.e. molecular counting. Moreover, they offer multicolour imaging, bringing in the possibility of labelling multiple subcellular structures as well as delivery carriers at the same time. In particular PAINT, by multiple rounds of imaging with different target probes91,94 or by kinetic fingerprinting the binding interaction, has recently achieved 124 colour super-resolution imaging.95 The main disadvantage of SMLM techniques is that they also require long imaging times to reconstruct the final image (few minutes to an hour), making them generally not suitable for live cell imaging.
Recently, STORM has been applied to observe endosomal escape of siRNA polyplexes96 (Fig. 2B). In this study, they imaged polyplexes carrying siRNA in early and late endosomes with 2-colour STORM to directly visualize the rupture of endosomes and the release of polyplexes. They first measured the size of polyplexes in biological environments from 2D STORM images. Then, they observed the shape of individual endosomes and polyplexes inside cells to establish how the endosomal escape process was occurring. Finally, they combined 2-colour STORM images to determine the level of colocalization of polyplexes and endosomes by counting individual polyplexes. In fact, STORM has also been used recently to study the trafficking and stability of NPs in cells.97–99 This and other techniques have shown the capability to image in 3D at the nanometric-scale resolution subcellular structures, such as endosomes and lysosomes, opening the door to a deeper understanding of endosomal escape.100
STED nanoscopy has been applied in internalization and trafficking of NPs.107–109 Specifically, Li Shang and co-workers investigated the internalization of transferrin NPs and measured the size of NP-loaded early endosomes with STED in live cells to conclude that particles were clustered inside the vesicles.109 STED has not been used to date to study endosomal escape of NPs; however, due to the multiple advantages of this technique, we can foresee the potential of STED to contribute to this field.
SIM has had a great impact in studying cell–NP interactions due to its fast imaging speed, live-cell capabilities, and low restrictions on fluorophore selection. It has been applied to investigate NP internalization,111 and trafficking112 – as well as shape113 and degradation114 inside cells – and subcellular dynamic processes at few milliseconds time resolution.115,116 Focusing on endosomal escape, SIM has been recently used to image the rupture of endosomes and the delivery of siRNA into the cytoplasm in breast cancer cells117 (Fig. 2B). Moreover, SIM has revealed that PEI polyplexes are found close to the internal side of the membrane of lysosomes/late endosomes, rather than at a central position in the vesicle.118 These findings prove the potential of SIM to investigate endosome–NP interactions in live cells, where an intermediate resolution is sufficient.
2.3. Frontiers in fluorescence micro/nanoscopy
The field of microscopy is constantly evolving and releasing new tools to tackle the challenges at the micro and nanoscopic scales. Recent developments have proven to be powerful techniques to study NPs in the biological environment offering better resolution and live-cell imaging features. Specifically, AiryScan119 and RESOLFT120 came into play to reduce photobleaching in confocal and STED microscopy respectively, for improved live-cell imaging, dynamic studies and higher throughputs. Moreover, recently developed MINFLUX (minimum photon fluxes) has achieved an outstanding resolution of 1–3 nm with low laser exposure in 3 dimensions.121,1222.4. Dynamic imaging
Endosomal trafficking and escape of NPs not only occur at the nanoscale, but are also a dynamic process. Some microscopy techniques can be combined with other tools to further investigate dynamic processes, such as fluorescence resonance energy transfer (FRET),123–125 fluorescence correlation spectroscopy (FCS)126,127 and single-particle tracking (SPT).128–132 Interestingly, SPT has been extensively used to study the co-localization and quantification of NPs within endocytic vesicles.128,133–139 For instance, Zahid et al.133 used live-cell SPT in combination with multidimensional analysis to characterize the intracellular distributions of quantum dot (QD) properties and to quantify their endosomal escape. The knowledge provided by SPT data analysis – especially when combined with other techniques – can be used to understand the underlying biological mechanisms of what discriminates formulations that achieve endosomal escape from those that cannot.2.5. Correlative imaging
Various papers report the endosomal escape of NPs using several independent microscopic techniques.30,58,68,140,141 However, a correlative approach is more desirable, as it bridges the advantages of two distinct techniques by imaging the same region of interest and overlapping important information from the two methods. Despite a much greater image interpretation confidence – that would not be possible with either of the methods individually142 – there is a very low number of publications in the area. This is probably related to the complex and cumbersome procedures required for sample handling and image aligning.143Correlative light and electron microscopy (CLEM) are perhaps the most explored group of correlated techniques. This combination allows spatiotemporal localization of labelled biomolecules with high specificity and sensitivity (FM), and with (sub)-nanometer resolution and precise subcellular localization of NPs within the cell (EM). In practice, quantification precision can be greatly improved using CLEM, as ‘invisible particles’ (i.e. not labelled with a fluorescent dye/not electron dense enough) can be detected. Also, since it can be difficult to distinguish different endosomal compartments based just on the TEM morphology, correlation with fluorescently labelled compartments in light microscopy can also improve NP localization precision.
To date, the only CLEM approach used to quantitatively study endo-lysosomal tracking of NPs has been developed by Han et al.74 (Fig. 2c, left). Using confocal laser scanning microscopy (CLSM) and 3D TEM tomography, they were able to demonstrate the localization of fNDs within endosomes, lysosomes and autophagosomes. Using the high-resolution TEM tomography results, they precisely quantified single fNDs found in clusters within the endosomal vesicles. However, quantification of single fNDs (not within clusters) was only possible at the single-particle level by using EFTEM (energy filtered TEM) as an additional method (Fig. 2A, bottom left). Furthermore, EFTEM permitted autonomous TEM screening of the whole sample, demonstrating the potential of this technique to precisely identify and quantify intracellular NPs.
Haruta et al.144 used the local surface plasmon resonance (LSPR) of gold NPs as a tag for biological samples in CLEM. To alleviate the problem of the resolution mismatch of several orders of magnitude between the two techniques, EM has also been correlated with SRM. Fluorescent nanodiamonds (FNDs) have been studied at nanometer resolution using STED-TEM145 and integrated light and scanning EM.146 However, in these examples NPs have been used for correlative purposes rather than to quantify or answer specific questions regarding intracellular trafficking. SRM-EM in fact offers a powerful tool to quantify and track endosomal escape and research in this area would benefit greatly the nanomedicine community.
A more distinct approach was achieved by Saarinen et al.70 who used correlative coherent anti-Stokes Raman scattering and TEM (C-CARS-EM) to image glibenclamide-nanocrystals (GLI-NCs) in macrophages (Fig. 2c, right). The combination of a label-free and chemically specific C-CARS technique with the excellent resolution and precision of TEM, allowed precise localization of GLI-NCs within endosomal vesicles. Although not achieved in this work, this technique also has the potential to be quantitative. For example, using 3D information from C-CARS together with precise localization of nanocrystals from EM, one can calculate the ratio between NPs found in the cytosol and in the endosomes (endosomal escape).
3. Conclusions and perspectives
Whilst significant progress has been made on developing a rich formulation databank of NPs for cytosolic delivery,16,17 our understanding of the physiochemical and biological requisites for achieving endosomal escape has been hampered.20,26–31 Our grasp of these mechanisms is hampered by the limitations of the standard techniques used to localise and quantify them. As discussed in this review, nanoscopy techniques – independently, or in correlation – hold the promise of answering some essential questions regarding endosomal escape. Some of these questions include how and which physiochemical properties of NPs influence endosomal escape? Which of the proposed endosomal escape mechanisms stand true and how can we improve the formulation of NPs to exploit them? Can we relate endosomal escape to the time and location at which it occurs intracellularly?Here we have highlighted the relevance of nanoscopy and some of the most recent discoveries in endosomal escape possible only using these methods. With a plethora of advanced microscopic techniques available, it is essential that we weigh the pros and cons of each technique to best suit the scientific question proposed (Tables 1 and 2). For a process such as endosomal escape – that is fast and rare and occurs at the nanoscale – it may seem challenging to answer the various questions projected using individual methods. But as we have seen in this review, we are no longer restricted to a ‘one method at a time’ approach.70,74,142,144–146 The benefits of correlative imaging – especially of SRM-EM – are of tremendous relevance in obtaining quantitative information on NP endosomal trafficking. Furthermore, as the amount of imaging data is increasing, automated quantification is becoming crucial in reducing manual analysis of images (and increasing throughput) and extracting more valuable data found in microscopic images, as well as making these techniques available to a broader research community.
Overall, these new developments in the field of imaging will lead to exciting times ahead for the study of endosomal escape. We prompt the nanomedicine community to adopt the newest techniques available to achieve a better understanding of NP trafficking as well as to facilitate the rational design of NPs with the ability to overcome endosomal barriers.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The project that gave rise to this work received the support of a fellowship from “la Caixa” Foundation (ID 100010434). The fellowship code is LCF/BQ/DI18/11660039. This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 713673.References
- S. Tran, P.-J. DeGiovanni, B. Piel and P. Rai, Cancer nanomedicine: a review of recent success in drug delivery, Clin. Transl. Med., 2017, 6(1), 44 CrossRef.
- Z. Li, S. Tan, S. Li, Q. Shen and K. Wang, Cancer drug delivery in the nano era: an overview and perspectives, Oncol. Rep., 2017, 38(2), 611–624 CrossRef CAS.
- A. Sonawane, Nanoparticle Vaccines Against Infectious Diseases, Front. Immunol., 2018, 9, 16 CrossRef.
- M. Iafisco, A. Alogna, M. Miragoli and D. Catalucci, Cardiovascular nanomedicine: the route ahead, Nanomedicine, 2019, 14(18), 2391–2394 CrossRef CAS.
- V. M. Martín Giménez, D. E. Kassuha and W. Manucha, Nanomedicine applied to cardiovascular diseases: latest developments, Ther. Adv. Cardiovasc. Dis., 2017, 11(4), 133–142 CrossRef.
- K. Smerkova, K. Dolezelikova, L. Bozdechova, Z. Heger, L. Zurek and V. Adam, Nanomaterials with active targeting as advanced antimicrobials, WIRES Nanomed. Nanobi., 2020, e1636 CAS , available from https://onlinelibrary.wiley.com/doi/abs/10.1002/wnan.1636.
- M. Z. El-Readi and M. A. Althubiti, Cancer Nanomedicine: A New Era of Successful Targeted Therapy, J. Nanomater., 2019, 2019, 1–13 CrossRef.
- Y. Wang, P. Yang, X. Zhao, D. Gao, N. Sun and Z. Tian, et al., Multifunctional Cargo-Free Nanomedicine for Cancer Therapy, Int. J. Mol. Sci., 2018, 19(10), 2963 CrossRef.
- S. Katsuki, T. Matoba, J. Koga, K. Nakano and K. Egashira, Anti-inflammatory Nanomedicine for Cardiovascular Disease, Front. Cardiovasc. Med., 2017, 4, 87 CrossRef.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. del P. Rodriguez-Torres and L. S. Acosta-Torres, et al., Nano based drug delivery systems: recent developments and future prospects, J. Nanobiotechnol., 2018, 16(1), 71 CrossRef.
- M. Karimi, H. Mirshekari, M. Aliakbari, P. Sahandi-Zangabad and M. R. Hamblin, Smart mesoporous silica nanoparticles for controlled-release drug delivery, Nanotechnol. Rev., 2016, 5(2), 195–207 CAS , available from https://www.degruyter.com/view/j/ntrev.2016.5.issue-2/ntrev-2015-0057/ntrev-2015-0057.xml.
- J. M. Caster, A. N. Patel, T. Zhang and A. Wang, Investigational nanomedicines in 2016: a review of nanotherapeutics currently undergoing clinical trials: investigational nanomedicines in 2016, WIRES Nanomed. Nanobi., 2017, 9(1), e1416 CrossRef.
- H. Havel, G. Finch, P. Strode, M. Wolfgang, S. Zale and I. Bobe, et al., Nanomedicines: From Bench to Bedside and Beyond, AAPS J., 2016, 18(6), 1373–1378 CrossRef CAS.
- Z. Eskandari, F. Bahadori, B. Celik and H. Onyuksel, Targeted Nanomedicines for Cancer Therapy, From Basics to Clinical Trials, J. Pharm. Pharm. Sci., 2020, 23, 132–157 CAS.
- E. Garbayo, S. Pascual-Gil, C. Rodríguez-Nogales, L. Saludas, A. Estella-Hermoso de Mendoza and M. J. Blanco-Prieto, Nanomedicine and drug delivery systems in cancer and regenerative medicine, WIRES Nanomed. Nanobi., 2020, e1367 Search PubMed , available from https://onlinelibrary.wiley.com/doi/abs/10.1002/wnan.1637.
- C. L. Ventola, Progress in Nanomedicine: Approved and Investigational Nanodrugs, 2017, 42(12), 742–755 Search PubMed.
- F. Farjadian, A. Ghasemi, O. Gohari, A. Roointan, M. Karimi and M. R. Hamblin, Nanopharmaceuticalsand nanomedicines currently on the market: challenges and opportunities, Nanomedicine, 2019, 14(1), 93–126 CrossRef CAS.
- S. Patel, J. Kim, M. Herrera, A. Mukherjee, A. V. Kabanov and G. Sahay, Brief update on endocytosis of nanomedicines, Adv. Drug Delivery Rev., 2019, 144, 90–111 CrossRef CAS.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors, Acc. Chem. Res., 2019, 52(7), 1750–1760 CrossRef CAS.
- L. I. Selby, C. M. Cortez-Jugo, G. K. Such and A. P. R. Johnston, Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles: endosomal escape of polymeric nanoparticles, WIRES Nanomed. Nanobi., 2017, 9(5), e1452 CrossRef.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet and H. F. Dvorak, et al., Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater., 2016, 1(5), 16014 CrossRef CAS.
- D. Rosenblum, N. Joshi, W. Tao, J. M. Karp and D. Peer, Progress and challenges towards targeted delivery of cancer therapeutics, Nat. Commun., 2018, 9(1), 1410 CrossRef.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova and J. J. Senn, et al., A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates, Mol. Ther., 2018, 26(6), 1509–1519 CrossRef CAS.
- S. A. Smith, L. I. Selby, A. P. R. Johnston and G. K. Such, The Endosomal Escape of Nanoparticles: Towards More Efficient Cellular Delivery, Bioconjugate Chem., 2018, 30(2), 263–272 CrossRef.
- M. P. Stewart, A. Lorenz, J. Dahlman and G. Sahay, Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers: challenges in carrier-mediated intracellular delivery, WIRES Nanomed. Nanobi., 2016, 8(3), 465–478 CrossRef.
- M. Durymanov and J. Reineke, Non-viral Delivery of Nucleic Acids: Insight Into Mechanisms of Overcoming Intracellular Barriers, Front. Pharmacol., 2018, 9, 971 CrossRef.
- D. Ma, Enhancing endosomal escape for nanoparticle mediated siRNA delivery, Nanoscale, 2014, 6(12), 6415 RSC.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Delivery materials for siRNA therapeutics, Nat. Mater., 2013, 12(11), 967–977 CrossRef CAS.
- Y. Yang, Z. Wang, Y. Peng, J. Ding and W. Zhou, A Smart pH-Sensitive Delivery System for Enhanced Anticancer Efficacy via Paclitaxel Endosomal Escape, Front. Pharmacol., 2019, 10, 10 CrossRef CAS.
- Y.-W. Lee, D. C. Luther, J. A. Kretzmann, A. Burden, T. Jeon and S. Zhai, et al., Protein Delivery into the Cell Cytosol using Non-Viral Nanocarriers, Theranostics, 2019, 9(11), 3280–3292 CrossRef CAS.
- C. Battistella and H.-A. Klok, Controlling and Monitoring Intracellular Delivery of Anticancer Polymer Nanomedicines, Macromol. Biosci., 2017, 17(10), 1700022 CrossRef.
- T. Bus, A. Traeger and U. S. Schubert, The great escape: how cationic polyplexes overcome the endosomal barrier, J. Mater. Chem. B, 2018, 6(43), 6904–6918 RSC.
- A. Ahmad, J. M. Khan and S. Haque, Strategies in the design of endosomolytic agents for facilitating endosomal escape in nanoparticles, Biochimie, 2019, 160, 61–75 CrossRef CAS.
- D. Pei and M. Buyanova, Overcoming Endosomal Entrapment in Drug Delivery, Bioconjugate Chem., 2019, 30(2), 273–283 CrossRef CAS.
- T. F. Martens, K. Remaut, J. Demeester, S. C. De Smedt and K. Braeckmans, Intracellular delivery of nanomaterials: how to catch endosomal escape in the act, Nano Today, 2014, 9(3), 344–364 CrossRef CAS.
- H. K. Shete, R. H. Prabhu and V. B. Patravale, Endosomal Escape: A Bottleneck in Intracellular Delivery, J. Nanosci. Nanotechnol., 2014, 14(1), 460–474 CrossRef CAS.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico and U. Schubert, et al., Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape, Nat. Biotechnol., 2013, 31(7), 638–646 CrossRef CAS.
- K. K. Hou, H. Pan, P. H. Schlesinger and S. A. Wickline, A role for peptides in overcoming endosomal entrapment in siRNA delivery — A focus on melittin, Biotechnol. Adv., 2015, 33(6), 931–940 CrossRef CAS.
- X. Cheng and R. J. Lee, The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery, Adv. Drug Delivery Rev., 2016, 99, 129–137 CrossRef CAS.
- W. Tai and X. Gao, Functional peptides for siRNA delivery, Adv. Drug Delivery Rev., 2017, 110–111, 157–168 CrossRef CAS.
- H. Wang, Y. Y. C. Tam, S. Chen, J. Zaifman, R. van der Meel and M. A. Ciufolini, et al., The Niemann-Pick C1 Inhibitor NP3.47 Enhances Gene Silencing Potency of Lipid Nanoparticles Containing siRNA, Mol. Ther., 2016, 24(12), 2100–2108 CrossRef CAS.
- S. Ben Djemaa, S. David, K. Hervé-Aubert, A. Falanga, S. Galdiero and E. Allard-Vannier, et al., Formulation and in vitro evaluation of a siRNA delivery nanosystem decorated with gH625 peptide for triple negative breast cancer theranosis, Eur. J. Pharm. Biopharm., 2018, 131, 99–108 CrossRef CAS.
- B. dos Santos Rodrigues, A. Banerjee, T. Kanekiyo and J. Singh, Functionalized liposomal nanoparticles for efficient gene delivery system to neuronal cell transfection, Int. J. Pharm., 2019, 566, 717–730 CrossRef CAS.
- D. Shae, K. W. Becker, P. Christov, D. S. Yun, A. K. R. Lytton-Jean and S. Sevimli, et al., Endosomolytic polymersomes increase the activity of cyclic dinucleotide STING agonists to enhance cancer immunotherapy, Nat. Nanotechnol., 2019, 14(3), 269–278 CrossRef CAS.
- Y. T. Pang, Z. Ge, B. Zhang, P. Xiu, Q. Li and Y. Wang, Pore formation induced by nanoparticles binding to a lipid membrane, Nanoscale, 2020, 12(14), 7902–7913 RSC.
- T. Ohtsuki, S. Miki, S. Kobayashi, T. Haraguchi, E. Nakata and K. Hirakawa, et al., The molecular mechanism of photochemical internalization of cell penetrating peptide-cargo-photosensitizer conjugates, Sci. Rep., 2016, 5(1), 18577 CrossRef.
- L. Martínez-Jothar, N. Beztsinna, C. F. van Nostrum, W. E. Hennink and S. Oliveira, Selective Cytotoxicity to HER2 Positive Breast Cancer Cells by Saporin-Loaded Nanobody-Targeted Polymeric Nanoparticles in Combination with Photochemical Internalization, Mol. Pharmaceutics, 2019, 16(4), 1633–1647 CrossRef.
- R. V. Benjaminsen, M. A. Mattebjerg, J. R. Henriksen, S. M. Moghimi and T. L. Andresen, The Possible “Proton Sponge ” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH, Mol. Ther., 2013, 21(1), 149–157 CrossRef CAS.
- Z. ur Rehman, D. Hoekstra and I. S. Zuhorn, Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis, ACS Nano, 2013, 7(5), 3767–3777 CrossRef.
- V. M. Ngwa, D. S. Axford, A. N. Healey, S. J. Nowak, C. A. Chrestensen and J. L. McMurry, A versatile cell-penetrating peptide-adaptor system for efficient delivery of molecular cargos to subcellular destinations, PLoS One, 2017, 12(5), e0178648 CrossRef.
- J. M. White and G. R. Whittaker, Fusion of Enveloped Viruses in Endosomes: Virus Fusion in Endosomes, Traffic, 2016, 17(6), 593–614 CrossRef CAS.
- C. Wang, Y. Wang, Y. Li, B. Bodemann, T. Zhao and X. Ma, et al., A nanobuffer reporter library for fine-scale imaging and perturbation of endocytic organelles, Nat. Commun., 2015, 6(1), 8524 CrossRef CAS.
- L. M. P. Vermeulen, S. C. De Smedt, K. Remaut and K. Braeckmans, The proton sponge hypothesis: Fable or fact?, Eur. J. Pharm. Biopharm., 2018, 129, 184–190 CrossRef CAS.
- A. Elsaesser, C. A. Barnes, G. McKerr, A. Salvati, I. Lynch and K. A. Dawson, et al., Quantification of nanoparticle uptake by cells using an unbiased sampling method and electron microscopy, Nanomedicine, 2011, 6(7), 1189–1198 CrossRef CAS.
- L. M. P. Vermeulen, T. Brans, S. C. De Smedt, K. Remaut and K. Braeckmans, Methodologies to investigate intracellular barriers for nucleic acid delivery in non-viral gene therapy, Nano Today, 2018, 21, 74–90 CrossRef CAS.
- A. Méndez-Ardoy, I. Lostalé-Seijo and J. Montenegro, Where in the Cell Is our Cargo? Methods Currently Used To Study Intracellular Cytosolic Localisation, ChemBioChem, 2019, 20(4), 488–498 CrossRef.
- R. Erni, M. D. Rossell, C. Kisielowski and U. Dahmen, Atomic-Resolution Imaging with a Sub-50-pm Electron Probe, Phys. Rev. Lett., 2009, 102(9), 096101 CrossRef.
- V. Guglielmi, F. Carton, G. Vattemi, S. Arpicco, B. Stella and G. Berlier, et al., Uptake and intracellular distribution of different types of nanoparticles in primary human myoblasts and myotubes, Int. J. Pharm., 2019, 560, 347–356 CrossRef CAS.
- Z. Chu, K. Miu, P. Lung, S. Zhang, S. Zhao and H.-C. Chang, et al., Rapid endosomal escape of prickly nanodiamonds: implications for gene delivery, Sci. Rep., 2015, 5(1), 11661 CrossRef.
- I. Plaza-GA, V. Manzaneda-González, M. Kisovec, V. Almendro-Vedia, M. Muñoz-Úbeda and G. Anderluh, et al., pH-triggered endosomal escape of pore-forming Listeriolysin O toxin-coated gold nanoparticles, J. Nanobiotechnol., 2019, 17(1), 108 CrossRef.
- M. Costanzo, F. Carton, A. Marengo, G. Berlier, B. Stella and S. Arpicco, et al., Fluorescence and electron microscopy to visualize the intracellular fate of nanoparticles for drug delivery, Eur. J. Histochem., 2016, 60(2), 2640 CAS , available from http://ejh.it/index.php/ejh/article/view/2640.
- N. Hondow, M. R. Brown, T. Starborg, A. G. Monteith, R. Brydson and H. D. Summers, et al., Quantifying the cellular uptake of semiconductor quantum dot nanoparticles by analytical electron microscopy: quantifying the cellular uptake of semiconductor quantum dot nanoparticles, J. Microsc., 2016, 261(2), 167–176 CrossRef CAS.
- I. Cabezón, G. Manich, R. Martín-Venegas, A. Camins, C. Pelegrí and J. Vilaplana, Trafficking of Gold Nanoparticles Coated with the 8D3 Anti-Transferrin Receptor Antibody at the Mouse Blood–Brain Barrier, Mol. Pharmaceutics, 2015, 12(11), 4137–4145 CrossRef.
- C. Iacovita, A. Florea, R. Dudric, E. Pall, A. Moldovan and R. Tetean, et al., Small versus Large Iron Oxide Magnetic Nanoparticles: Hyperthermia and Cell Uptake Properties, Molecules, 2016, 21(10), 1357 CrossRef.
- N. Feng, Y. Liu, M. He, M. Niu, Y. Zhao and Y. Zhu, et al., Delivery of vincristine sulfate-conjugated gold nanoparticles using liposomes: a light-responsive nanocarrier with enhanced antitumor efficiency, Indian J. Nephrol., 2015, 3081 Search PubMed.
- K. V. Kilchrist, B. C. Evans, C. M. Brophy and C. L. Duvall, Mechanism of Enhanced Cellular Uptake and Cytosolic Retention of MK2 Inhibitory Peptide Nano-polyplexes, Cell. Mol. Bioeng., 2016, 9(3), 368–381 CrossRef CAS.
- J. C. Fraire, G. Houthaeve, J. Liu, L. Raes, L. Vermeulen and S. Stremersch, et al., Vapor nanobubble is the more reliable photothermal mechanism for inducing endosomal escape of siRNA without disturbing cell homeostasis, J. Controlled Release, 2020, 319, 262–275 CrossRef CAS.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai and J. P. Griffith, et al., Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun., 2020, 11(1), 983 CrossRef CAS.
- M. Azubel, S. D. Carter, J. Weiszmann, J. Zhang, G. J. Jensen and Y. Li, et al., FGF21 trafficking in intact human cells revealed by cryo-electron tomography with gold nanoparticles, eLife, 2019, 8, e43146 CrossRef.
- J. Saarinen, F. Gütter, M. Lindman, M. Agopov, S. J. Fraser-Miller and R. Scherließ, et al., Cell-Nanoparticle Interactions at (Sub)–Nanometer Resolution Analyzed by Electron Microscopy and Correlative Coherent Anti-Stokes Raman Scattering, Biotechnol. J., 2019, 14(4), 1800413 CrossRef.
- T. Cristofolini, M. Dalmina, J. A. Sierra, A. H. Silva, A. A. Pasa and F. Pittella, et al., Multifunctional hybrid nanoparticles as magnetic delivery systems for siRNA targeting the HER2 gene in breast cancer cells, Mater. Sci. Eng. C, 2020, 109, 110555 CrossRef CAS.
- S. Deshayes, K. Konate, M. Dussot, B. Chavey, A. Vaissière and T. N. N. Van, et al., Deciphering the internalization mechanism of WRAP:siRNA nanoparticles, Biochim. Biophys. Acta, Biomembr., 2020, 1862(6), 183252 CrossRef CAS.
- M. Malatesta, Transmission electron microscopy for nanomedicine: novel applications for long-established techniques, Eur. J. Histochem., 2016, 60(4), 2751 Search PubMed , available from http://www.ejh.it/index.php/ejh/article/view/2751.
- S. Han, M. Raabe, L. Hodgson, J. Mantell, P. Verkade and T. Lasser, et al., High-Contrast Imaging of Nanodiamonds in Cells by Energy Filtered and Correlative Light-Electron Microscopy: Toward a Quantitative Nanoparticle-Cell Analysis, Nano Lett., 2019, 19(3), 2178–2185 CrossRef CAS.
- B. Rothen-Rutishauser, D. A. Kuhn, Z. Ali, M. Gasser, F. Amin and W. J. Parak, et al., Quantification of gold nanoparticle cell uptake under controlled biological conditions and adequate resolution, Nanomedicine, 2014, 9(5), 607–621 CrossRef CAS.
- S. Grecchi and M. Malatesta, Visualizing endocytotic pathways at transmission electron microscopy via diaminobenzidine photo-oxidation by a fluorescent cell-membrane dye, Eur. J. Histochem., 2014, 58(4), 2449 CAS , available from http://ejh.it/index.php/ejh/article/view/2449.
- C. Pellicciari, M. Biggiogera and M. Malatesta, DAB Photo-Oxidation as a Tool for Detecting Low Amounts of Free and Membrane-Bounded Fluorescent Molecules at Transmission Electron Microscopy, 2015, vol. 9 Search PubMed.
- P. L. Stewart, Cryo-electron microscopy and cryo-electron tomography of nanoparticles: cryo-electron microscopy and cryo-electron tomography of nanoparticles, WIRES Nanomed. Nanobi., 2017, 9(2), e1417 CrossRef.
- Y. Cheng, Single-Particle Cryo-EM at Crystallographic Resolution, Cell, 2015, 161(3), 450–457 CrossRef CAS.
- K. Remaut, V. Oorschot, K. Braeckmans, J. Klumperman and S. C. De Smedt, Lysosomal capturing of cytoplasmic injected nanoparticles by autophagy: an additional barrier to non viral gene delivery, J. Controlled Release, 2014, 195, 29–36 CrossRef CAS.
- Basic Confocal Microscopy, ed. W. G. Jerome and R. L. Price, Cham, Springer International Publishing, 2018, available from http://link.springer.com/10.1007/978-3-319-97454-5 Search PubMed.
- E. Abbe, Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung, Archiv für mikroskopische Anatomie, 1873, 9(1), 413–468 CrossRef.
- The Nobel Prize in Chemistry, 2014, NobelPrize.org, available from https://www.nobelprize.org/prizes/chemistry/2014/advanced-information/ Search PubMed.
- S. Bretschneider, C. Eggeling and S. W. Hell, Breaking the Diffraction Barrier in Fluorescence Microscopy by Optical Shelving, Phys. Rev. Lett., 2007, 98(21), 218103 CrossRef.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych and J. S. Bonifacino, et al., Imaging Intracellular Fluorescent Proteins at Nanometer Resolution, Science, 2006, 313(5793), 1642–1645 CrossRef CAS.
- M. J. Rust, M. Bates and X. Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods, 2006, 3(10), 793–796 CrossRef CAS.
- S. T. Hess, T. P. K. Girirajan and M. D. Mason, Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy, Biophys. J., 2006, 91(11), 4258–4272 CrossRef CAS.
- A. Sharonov and R. M. Hochstrasser, Wide-field subdiffraction imaging by accumulated binding of diffusing probes, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(50), 18911–18916 CrossRef CAS.
- M. Heilemann, S. van de Linde, M. Schüttpelz, R. Kasper, B. Seefeldt and A. Mukherjee, et al., Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes, Angew. Chem., Int. Ed., 2008, 47(33), 6172–6176 CrossRef CAS.
- G. Giannone, E. Hosy, F. Levet, A. Constals, K. Schulze and A. I. Sobolevsky, et al., Dynamic Superresolution Imaging of Endogenous Proteins on Living Cells at Ultra-High Density, Biophys. J., 2010, 99(4), 1303–1310 CrossRef CAS.
- R. Jungmann, M. S. Avendaño, J. B. Woehrstein, M. Dai, W. M. Shih and P. Yin, Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT, Nat. Methods, 2014, 11(3), 313–318 CrossRef CAS.
- M. D. Lew, S. F. Lee, J. L. Ptacin, M. K. Lee, R. J. Twieg and L. Shapiro, et al., Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(46), E1102–E1110 CrossRef CAS.
- P. R. Nicovich, D. M. Owen and K. Gaus, Turning single-molecule localization microscopy into a quantitative bioanalytical tool, Nat. Protoc., 2017, 12(3), 453–460 CrossRef CAS.
- S. Agasti, Y. Wang, F. Schueder, A. Sukumar, R. Jungmann and P. Yin, DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging, Chem. Sci., 2017, 8(4), 3080–3091 RSC.
- O. K. Wade, J. B. Woehrstein, P. C. Nickels, S. Strauss, F. Stehr and J. Stein, et al., 124-Color Super-resolution Imaging by Engineering DNA-PAINT Blinking Kinetics, Nano Lett., 2019, 19(4), 2641–2646 CrossRef CAS.
- M. Wojnilowicz, A. Glab, A. Bertucci, F. Caruso and F. Cavalieri, Super-resolution Imaging of Proton Sponge-Triggered Rupture of Endosomes and Cytosolic Release of Small Interfering RNA, ACS Nano, 2019, 13(1), 187–202 CrossRef CAS.
- D. van der Zwaag, N. Vanparijs, S. Wijnands, R. De Rycke, B. G. De Geest and L. Albertazzi, Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking, ACS Appl. Mater. Interfaces, 2016, 8(10), 6391–6399 CrossRef CAS.
- N. Feiner-Gracia, R. A. Olea, R. Fitzner, N. El Boujnouni, A. H. van Asbeck and R. Brock, et al., Super-resolution Imaging of Structure, Molecular Composition, and Stability of Single Oligonucleotide Polyplexes, Nano Lett., 2019, 19(5), 2784–2792 CrossRef CAS.
- R. Riera, N. Feiner-Gracia, C. Fornaguera, A. Cascante, S. Borrós and L. Albertazzi, Tracking the DNA complexation state of pBAE polyplexes in cells with super resolution microscopy, Nanoscale, 2019, 11(38), 17869–17877 RSC.
- T. Kiuchi, M. Higuchi, A. Takamura, M. Maruoka and N. Watanabe, Multitarget super-resolution microscopy with high-density labeling by exchangeable probes, Nat. Methods, 2015, 12(8), 743–746 CrossRef CAS.
- S. W. Hell and J. Wichmann, Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy, Opt. Lett., 1994, 19(11), 780–782 CrossRef CAS.
- T. A. Klar, S. Jakobs, M. Dyba, A. Egner and S. W. Hell, Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(15), 8206–8210 CrossRef CAS.
- R. Foresti, S. Rossi, S. Pinelli, R. Alinovi, C. Sciancalepore and N. Delmonte, et al., In-vivo vascular application via ultra-fast bioprinting for future 5D personalised nanomedicine, Sci. Rep., 2020, 10(1), 3205 CrossRef CAS.
- J.-N. May, S. K. Golombek, M. Baues, A. Dasgupta, N. Drude and A. Rix, et al., Multimodal and multiscale optical imaging of nanomedicine delivery across the blood-brain barrier upon sonopermeation, Theranostics, 2020, 10(4), 1948–1959 CrossRef CAS.
- S. Wäldchen, J. Lehmann, T. Klein, S. van de Linde and M. Sauer, Light-induced cell damage in live-cell super-resolution microscopy, Sci. Rep., 2015, 5(1), 15348 CrossRef.
- N. Kilian, A. Goryaynov, M. D. Lessard, G. Hooker, D. Toomre and J. E. Rothman, et al., Assessing photodamage in live-cell STED microscopy, Nat. Methods, 2018, 15(10), 755–756 CrossRef CAS.
- H. Peuschel, T. Ruckelshausen, C. Cavelius and A. Kraegeloh, Quantification of Internalized Silica Nanoparticles via STED Microscopy, BioMed Res. Int., Hindawi, 2015, 2015, e961208. available from https://www.hindawi.com/journals/bmri/2015/961208/ Search PubMed.
- J. Hanne, H. J. Falk, F. Görlitz, P. Hoyer, J. Engelhardt and S. J. Sahl, et al., STED nanoscopy with fluorescent quantum dots, Nat. Commun., 2015, 6(1), 7127 CrossRef CAS.
- L. Shang, P. Gao, H. Wang, R. Popescu, D. Gerthsen and G. U. Nienhaus, Protein-based fluorescent nanoparticles for super-resolution STED imaging of live cells, Chem. Sci., 2017, 8(3), 2396–2400 RSC.
- M. G. L. Gustafsson, Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy, J. Microsc., 2000, 198(2), 82–87 CrossRef CAS.
- X. Chen, J. Cui, Y. Ping, T. Suma, F. Cavalieri and Q. A. Besford, et al., Probing cell internalisation mechanics with polymer capsules, Nanoscale, 2016, 8(39), 17096–17101 RSC.
- M. H. Teplensky, M. Fantham, P. Li, T. C. Wang, J. P. Mehta and L. J. Young, et al., Temperature Treatment of Highly Porous Zirconium-Containing Metal–Organic Frameworks Extends Drug Delivery Release, J. Am. Chem. Soc., 2017, 139(22), 7522–7532 CrossRef CAS.
- X. Chen, J. Cui, H. Sun, M. Müllner, Y. Yan and K. Fung Noi, et al., Analysing intracellular deformation of polymer capsules using structured illumination microscopy, Nanoscale, 2016, 8(23), 11924–11931 RSC.
- E. Tolstik, L. A. Osminkina, C. Matthäus, M. Burkhardt, K. E. Tsurikov and U. A. Natashina, et al., Studies of silicon nanoparticles uptake and biodegradation in cancer cells by Raman spectroscopy, Nanomedicine, 2016, 12(7), 1931–1940 CrossRef CAS.
- D. Li, L. Shao, B.-C. Chen, X. Zhang, M. Zhang and B. Moses, et al., Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics, Science, 2015, 349(6251), aab3500 CrossRef , available from http://science.sciencemag.org/content/349/6251/aab3500.
- Y. Guo, D. Li, S. Zhang, Y. Yang, J.-J. Liu and X. Wang, et al., Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales, Cell, 2018, 175(5), 1430–1442.e17 CrossRef CAS.
- S. Ben Djemaa, K. Hervé-Aubert, L. Lajoie, A. Falanga, S. Galdiero and S. Nedellec, et al., gH625 Cell-Penetrating Peptide Promotes the Endosomal Escape of Nanovectorized siRNA in a Triple-Negative Breast Cancer Cell Line, Biomacromolecules, 2019, 20(8), 3076–3086 CrossRef CAS.
- T. Bus, C. Englert, M. Reifarth, P. Borchers, M. Hartlieb and A. Vollrath, et al., 3rd generation poly(ethylene imine)s for gene delivery, J. Mater. Chem. B, 2017, 5(6), 1258–1274 RSC.
- J. Huff, The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution, Nat. Methods, 2015, 12(12), i–ii CrossRef CAS.
- M. Hofmann, C. Eggeling, S. Jakobs and S. W. Hell, Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(49), 17565–17569 CrossRef CAS.
- F. Balzarotti, Y. Eilers, K. C. Gwosch, A. H. Gynnå, V. Westphal and F. D. Stefani, et al., Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes, Science, 2017, 355(6325), 606–612 CrossRef CAS.
- K. C. Gwosch, J. K. Pape, F. Balzarotti, P. Hoess, J. Ellenberg and J. Ries, et al., MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells, Nat. Methods, 2020, 17(2), 217–224 CAS.
- Z. Wang, M. Luo, C. Mao, Q. Wei, T. Zhao and Y. Li, et al., A Redox-Activatable Fluorescent Sensor for the High-Throughput Quantification of Cytosolic Delivery of Macromolecules, Angew. Chem., 2017, 129(5), 1339–1343 CrossRef.
- X. Guo, L. Wang, K. Duval, J. Fan, S. Zhou and Z. Chen, Dimeric Drug Polymeric Micelles with Acid-Active Tumor Targeting and FRET-Traceable Drug Release, Adv. Mater., 2018, 30(3), 1705436 CrossRef.
- S. Rayamajhi, J. Marchitto, T. D. T. Nguyen, R. Marasini, C. Celia and S. Aryal, pH-responsive cationic liposome for endosomal escape mediated drug delivery, Colloids Surf., B, 2020, 188, 110804 CrossRef CAS.
- L. Lanzanò, L. Scipioni, M. Di Bona, P. Bianchini, R. Bizzarri and F. Cardarelli, et al., Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS, Nat. Commun., 2017, 8(1), 65 CrossRef.
- E. Sezgin, F. Schneider, S. Galiani, I. Urbančič, D. Waithe and B. C. Lagerholm, et al., Measuring nanoscale diffusion dynamics in cellular membranes with super-resolution STED–FCS, Nat. Protoc., 2019, 14(4), 1054–1083 CAS.
- K. Shin, Y. Song, Y. Goh and K. Lee, Two-Dimensional and Three-Dimensional Single Particle Tracking of Upconverting Nanoparticles in Living Cells, Int. J. Mol. Sci., 2019, 20(6), 1424 CrossRef CAS.
- D. Jin, P. Xi, B. Wang, L. Zhang, J. Enderlein and A. M. van Oijen, Nanoparticles for super-resolution microscopy and single-molecule tracking, Nat. Methods, 2018, 15(6), 415–423 CrossRef CAS.
- M. Gabriel, J. Moya-Díaz, L. I. Gallo, F. D. Marengo and L. C. Estrada, Single particle tracking of internalized metallic nanoparticles reveals heterogeneous directed motion after clathrin dependent endocytosis in mouse chromaffin cells, Methods Appl. Fluoresc., 2017, 6(1), 014003 CrossRef.
- S. Hou, X. Lang and K. Welsher, Robust real-time 3D single-particle tracking using a dynamically moving laser spot, Opt. Lett., 2017, 42(12), 2390 CrossRef CAS.
- E. Zagato, K. Forier, T. Martens, K. Neyts, J. Demeester and S. D. Smedt, et al., Single-particle tracking for studying nanomaterial dynamics: applications and fundamentals in drug delivery, Nanomedicine, 2014, 9(6), 913–927 CrossRef CAS.
- M. U. Zahid, L. Ma, S. J. Lim and A. M. Smith, Single quantum dot tracking reveals the impact of nanoparticle surface on intracellular state, Nat. Commun., 2018, 9(1), 1830 CrossRef.
- M. Liu, Q. Li, L. Liang, J. Li, K. Wang and J. Li, et al., Real-time visualization of clustering and intracellular transport of gold nanoparticles by correlative imaging, Nat. Commun., 2017, 8(1), 15646 CrossRef CAS.
- Q. Li, W. Li, W. Yin, J. Guo, Z.-P. Zhang and D. Zeng, et al., Single-Particle Tracking of Human Immunodeficiency Virus Type 1 Productive Entry into Human Primary Macrophages, ACS Nano, 2017, 11(4), 3890–3903 CrossRef CAS.
- D. Bhatia, S. Arumugam, M. Nasilowski, H. Joshi, C. Wunder and V. Chambon, et al., Quantum dot-loaded monofunctionalized DNA icosahedra for single-particle tracking of endocytic pathways, Nat. Nanotechnol., 2016, 11(12), 1112–1119 CrossRef CAS.
- M. Tiffany and F. C. Szoka, Co-localization of fluorescent labeled lipid nanoparticles with specifically tagged subcellular compartments by single particle tracking at low nanoparticle to cell ratios, J. Drug Targeting, 2016, 24(9), 857–864 CrossRef CAS.
- S. Deville, R. Penjweini, N. Smisdom, K. Notelaers, I. Nelissen and J. Hooyberghs, et al., Intracellular dynamics and fate of polystyrene nanoparticles in A549 Lung epithelial cells monitored by image (cross-) correlation spectroscopy and single particle tracking, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853(10), 2411–2419 CrossRef CAS.
- H. Deschout, T. Martens, D. Vercauteren, K. Remaut, J. Demeester and S. De Smedt, et al., Correlation of Dual Colour Single Particle Trajectories for Improved Detection and Analysis of Interactions in Living Cells, Int. J. Mol. Sci., 2013, 14(8), 16485–16514 CrossRef.
- J. Krawinkel, U. Richter, M. L. Torres-Mapa, M. Westermann, L. Gamrad and C. Rehbock, et al., Optical and electron microscopy study of laser-based intracellular molecule delivery using peptide-conjugated photodispersible gold nanoparticle agglomerates, J. Nanobiotechnol., 2016, 14(1), 2 CrossRef.
- W. Liu, B. Naydenov, S. Chakrabortty, B. Wuensch, K. Hübner and S. Ritz, et al., Fluorescent Nanodiamond–Gold Hybrid Particles for Multimodal Optical and Electron Microscopy Cellular Imaging, Nano Lett., 2016, 16(10), 6236–6244 CrossRef CAS.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek and E. C. Dreaden, et al., Cellular uptake of nanoparticles: journey inside the cell, Chem. Soc. Rev., 2017, 46(14), 4218–4244 RSC.
- K. Anderson, T. Nilsson and J. Fernandez-Rodriguez, Challenges for CLEM from a Light Microscopy Perspective, in Correlative Imaging, ed. P. Verkade and L. Collinson, Wiley, 1st edn, 2019, pp. 23–35, available from https://onlinelibrary.wiley.com/doi/abs/10.1002/9781119086420.ch2 Search PubMed.
- T. Haruta, K. Hasumi, Y. Ikeda, Y. Konyuba, T. Fukuda and H. Nishioka, Local surface plasmon resonance of gold nanoparticles as a correlative light and electron microscopy (CLEM) tag for biological samples, Microscopy, 2019, dfz031 CrossRef.
- N. Prabhakar, M. Peurla, S. Koho, T. Deguchi, T. Näreoja and H.-C. Chang, et al., STED-TEM Correlative Microscopy Leveraging Nanodiamonds as Intracellular Dual-Contrast Markers, Small, 2018, 14(5), 1701807 CrossRef.
- S. R. Hemelaar, P. de Boer, M. Chipaux, W. Zuidema, T. Hamoh and F. P. Martinez, et al., Nanodiamonds as multi-purpose labels for microscopy, Sci. Rep., 2017, 7(1), 720 CrossRef CAS.
- B. Fortuni, T. Inose, M. Ricci, Y. Fujita, I. Van Zundert and A. Masuhara, et al., Polymeric Engineering of Nanoparticles for Highly Efficient Multifunctional Drug Delivery Systems, Sci. Rep., 2019, 9(1), 2666 CrossRef.
- M. Galliani, C. Tremolanti and G. Signore, Nanocarriers for Protein Delivery to the Cytosol: Assessing the Endosomal Escape of Poly(Lactide-co-Glycolide)-Poly(Ethylene Imine) Nanoparticles, Nanomaterials, 2019, 9(4), 652 CrossRef CAS.
- C. A. Combs, Fluorescence Microscopy: A Concise Guide to Current Imaging Methods, Curr. Protoc. Neurosci., 2010, 50(1), Ch. 2, Unit2.1, available from https://onlinelibrary.wiley.com/doi/abs/10.1002/0471142301.ns0201s50 Search PubMed.
- J. A. Kulkarni, M. M. Darjuan, J. E. Mercer, S. Chen, R. van der Meel and J. L. Thewalt, et al., On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA, ACS Nano, 2018, 12(5), 4787–4795 CrossRef CAS.
- M. Yanez Arteta, T. Kjellman, S. Bartesaghi, S. Wallin, X. Wu and A. J. Kvist, et al., Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(15), E3351–E3360 CrossRef.
- C. Sun and R. B. Gennis, Single-particle cryo-EM studies of transmembrane proteins in SMA copolymer nanodiscs, Chem. Phys. Lipids, 2019, 221, 114–119 CrossRef CAS.
- J. Asadi, S. Ferguson, H. Raja, C. Hacker, P. Marius and R. Ward, et al., Enhanced imaging of lipid rich nanoparticles embedded in methylcellulose films for transmission electron microscopy using mixtures of heavy metals, Micron, 2017, 99, 40–48 CrossRef CAS.
- A. D. Lehmann, W. J. Parak, F. Zhang, Z. Ali, C. Röcker and G. U. Nienhaus, et al., Fluorescent-Magnetic Hybrid Nanoparticles Induce a Dose-Dependent Increase in Proinflammatory Response in Lung Cells in vitro Correlated with Intracellular Localization, Small, 2010, 6(6), 753–762 CrossRef CAS.
- N. Hondow, A. Brown, H. D. Summers, M. Rowan Brown, P. Rees and M. D. Holton, et al., Quantifying Nanoparticle–Cell Interactions, Microsc. Microanal., 2014, 20(S3), 1300–1301 CrossRef.
- H. D. Summers, M. R. Brown, M. D. Holton, J. A. Tonkin, N. Hondow and A. P. Brown, et al., Quantification of Nanoparticle Dose and Vesicular Inheritance in Proliferating Cells, ACS Nano, 2013, 7(7), 6129–6137 CrossRef CAS.
- C. Brandenberger, C. Mühlfeld, Z. Ali, A.-G. Lenz, O. Schmid and W. J. Parak, et al., Quantitative Evaluation of Cellular Uptake and Trafficking of Plain and Polyethylene Glycol-Coated Gold Nanoparticles, Small, 2010, 6(15), 1669–1678 CrossRef CAS.
- H. J. G. Gundersen, The smooth fractionator BlackwellScience, Ltd, J. Microsc., 2002, 207, 191–210 CrossRef CAS.
- R. Jungmann, M. S. Avendaño, M. Dai, J. B. Woehrstein, S. S. Agasti and Z. Feiger, et al., Quantitative super-resolution imaging with qPAINT, Nat. Methods, 2016, 13(5), 439–442 CrossRef CAS.
- E. M. Puchner, J. M. Walter, R. Kasper, B. Huang and W. A. Lim, Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(40), 16015–16020 CrossRef CAS.
- P. Delcanale, B. Miret-Ontiveros, M. Arista-Romero, S. Pujals and L. Albertazzi, Nanoscale Mapping Functional Sites on Nanoparticles by Points Accumulation for Imaging in Nanoscale Topography (PAINT), ACS Nano, 2018, 12(8), 7629–7637 CrossRef CAS.
- N. Feiner-Gracia, M. Beck, S. Pujals, S. Tosi, T. Mandal and C. Buske, et al., Super-Resolution Microscopy Unveils Dynamic Heterogeneities in Nanoparticle Protein Corona, Small, 2017, 13(41), 1701631 CrossRef.
- Q. Le Trequesser, G. Devès, G. Saez, L. Daudin, P. Barberet and C. Michelet, et al., Single Cell In Situ Detection and Quantification of Metal Oxide Nanoparticles Using Multimodal Correlative Microscopy, Anal. Chem., 2014, 86(15), 7311–7319 CrossRef CAS.
Footnote |
| † These authors contributed equally to the preparation of this review. |
| This journal is © The Royal Society of Chemistry 2021 |