Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Proline pre-conditioning of cell monolayers increases post-thaw recovery and viability by distinct mechanisms to other osmolytes†‡
Trisha L.
Bailey
 a,
Juan Ramon
Hernandez-Fernaud
a,
Juan Ramon
Hernandez-Fernaud
 b and
Matthew I.
Gibson
b and
Matthew I.
Gibson
 *ac
*ac
aDepartment of Chemistry, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK. E-mail: m.i.gibson@warwick.ac.uk
bUnidad de Investigación, Hospital Universitario de Canarias, Calle Ofra s/n, La Cuesta, La Laguna, Tenerife, Spain
cWarwick Medical School, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK
First published on 18th May 2021
Abstract
Cell cryopreservation is an essential tool for drug toxicity/function screening and transporting cell-based therapies, and is essential in most areas of biotechnology. There is a challenge, however, associated with the cryopreservation of cells in monolayer format (attached to tissue culture substrates) which gives far lower cell yields (<20% typically) compared to suspension freezing. Here we investigate the mechanisms by which the protective osmolyte L-proline enhances cell-monolayer cryopreservation. Pre-incubating A549 cells with proline, prior to cryopreservation in monolayers, increased post-thaw cell yields two-fold, and the recovered cells grow faster compared to cells cryopreserved using DMSO alone. Further increases in yield were achieved by adding polymeric ice recrystallization inhibitors, which gave limited benefit in the absence of proline. Mechanistic studies demonstrated a biochemical, rather than biophysical (i.e. not affecting ice growth) mode of action. It was observed that incubating cells with proline (before freezing) transiently reduced the growth rate of the cells, which was not seen with other osmolytes (betaine and alanine). Removal of proline led to rapid growth recovery, suggesting that proline pre-conditions the cells for cold stress, but with no impact on downstream cell function. Whole cell proteomics did not reveal a single pathway or protein target but rather cells appeared to be primed for a stress response in multiple directions, which together prepare the cells for freezing. These results support the use of proline alongside standard conditions to improve post-thaw recovery of cell monolayers, which is currently considered impractical. It also demonstrates that a chemical biology approach to discovering small molecule biochemical modulators of cryopreservation may be possible, to be used alongside traditional (solvent) based cryoprotectants.
Introduction
The storage, recovery, and manipulation of viable cells is the key component of all cell biology research, with primary cells being the most rare/valuable, and stem cells are currently emerging for many personalized medical treatments.1,2 Cell cryopreservation can give variable results with the phenotypic impact of frozen/thawed cells still unknown in many cases. Standard protocols use 5–10 wt% of the cryoprotective agent (CPA) dimethyl sulfoxide (DMSO), which is able to enter cells and reduce injury by moderating the increase in solute concentration during freezing.3–5 While suspension freezing in DMSO is highly successful, certain cells such as leukocytes (RAW 264.7) are highly-sensitive to DMSO.6 Additionally, there are some concerns with using DMSO due to its cytotoxicity at high concentrations and/or room temperature.7–11 Freezing with DMSO is the gold standard for suspension cell cryopreservation in nearly every research laboratory but the method does not work well for cell monolayers,12 typically resulting in only around 20–35% recovery, often even lower.13,14 For example, adherent human embryonic stem cells yield extremely low survival rates of <5%.12,13 The ability to cryopreserve monolayered cells would facilitate drug development by providing phenotypically identical cells for assays, reduce the total resource needed for cell culture (e.g. cells are already in format for assay) as well as providing insights into the cryopreservation of more complex biological material such as spheroids or tissues.According to Mazur's two-factor hypothesis, key challenges in cryopreservation are ice formation and osmotic stress due to the addition of cryoprotective agents and the increasing concentration of solutes in the remaining water phase during the freezing process.15,16 Intracellular ice can form when the cooling rate is intermediately high and the cell cannot maintain osmotic equilibrium with the environment, and is almost always lethal.17,18 Controlled-rate freezing ensures movement of water across the plasma membrane in order for osmotic dehydration to reach equilibrium with intracellular and extracellular contents to prevent intracellular ice.19,20 During thawing, ice recrystallization can damage the cells by both mechanical and osmotic stress, and controlling this process by using ice recrystallization inhibitors (IRIs) has been shown to result in enhanced post-thaw recovery.21,22 Antifreeze (glyco)proteins (AF(G)Ps) are potent IRIs and can reduce damage to cell membranes,23–25 however, AF(G)Ps are unsuitable for cryopreservation applications due to their potential toxicity/immunogenicity, their secondary effect of dynamic ice shaping (DIS),21 and their low availability/expense. Considering this, there is significant interest in the discovery of AF(G)P mimics.21,26–28
The most studied IRI active polymer is poly(vinyl alcohol) (PVA), due to its potent IRI, low toxicity, and commercial availability.24,29,30 PVA gives some benefit to the cryopreservation of cells in suspension,31–33 but has not been explored for cell monolayer cryopreservation, although polyproline (a weak IRI) was reported to enhance monolayer recovery when used in conjunction with the protective osmolyte proline34 and has been applied to oocytes also.35 Without addition of proline, there was limited benefit of the IRI-active polymer, but the functional role of this proline incubation is not fully understood.
Cells almost universally respond to the stress of long-term hyperosmolality, such as during freezing, by accumulating compatible organic osmolytes.36 The major osmolytes in water-stressed eukaryotes include metabolic products such as glycerol and sucrose, amino acids (plus derivatives such as taurine and β-alanine), urea, and methylamines (e.g. tri-methylamine-N-oxide).37 The most widely studied ‘compatible’ solutes are the N-substituted amino acids, particularly betaine and proline.38 Betaine's osmoprotection has been widely observed, such as in the dogfish shark Squalus acanthias promoting betaine efflux during hypotonic conditions.39 Betaine reduces leakage from frozen multilamellar liposomes,40 and can enhance the vitrification of mammalian cells in suspension.41 Proline is membrane permeable regardless of pH,42 and transfer can occur by a range of processes.43 Proline has been implicated as a survival factor that protects the cell against apoptosis and maintains the progression of the cell cycle through a signal recognition function of the transporters that may take part in the control of cell cycle progression and programmed cell death.44 Proline has been used with success to cryopreserve mouse oocytes,45 vitrify red blood cells (RBCs) and mammalian cells,46 induce freeze-tolerance in fruit fly larvae,47 and has shown protection for mammalian cell monolayers when used alongside trehalose.14
Considering the above, this study explores the use of proline for the cryopreservation of adherent cell monolayers in comparison to other protective osmolytes. It is shown that incubation of cell monolayers with proline, in advance of cryopreservation, leads to increases in post-thaw recovery. Addition of polymeric IRIs further increased the post-thaw cell yield, but only when used with proline, not without. Evidence is presented that proline prepares the cells for freezing by a temporary slowing of cellular growth which was not seen for other osmolytes and may explain its beneficial role in cell monolayer cryopreservation.
Experimental section
Cell culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 80% hydrolysed), proline, alanine and betaine were obtained from Sigma Aldrich Co Ltd, (Irvine, UK). Solutions for cell experiments were prepared by dissolving the individual compounds in base cell media supplemented with 10% FBS and 1× PSA (solutions used as CPAs did not contain PSA) and sterile filtering prior to use.
000, 80% hydrolysed), proline, alanine and betaine were obtained from Sigma Aldrich Co Ltd, (Irvine, UK). Solutions for cell experiments were prepared by dissolving the individual compounds in base cell media supplemented with 10% FBS and 1× PSA (solutions used as CPAs did not contain PSA) and sterile filtering prior to use.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). The initial cell medium was discarded such that any non-attached cells were not included in the assessment. The percentage of recovered cells was calculated by dividing the number of cells with intact membranes after freezing and thawing by the number of cells present prior to freezing (i.e. after application of pre-treatments), multiplied by 100.
1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). The initial cell medium was discarded such that any non-attached cells were not included in the assessment. The percentage of recovered cells was calculated by dividing the number of cells with intact membranes after freezing and thawing by the number of cells present prior to freezing (i.e. after application of pre-treatments), multiplied by 100.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the sample with 0.4% trypan blue solution. Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the number of cells initially plated.
1 dilution of the sample with 0.4% trypan blue solution. Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the number of cells initially plated.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the number of cells initially plated.
1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the number of cells initially plated.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the average number of cells on day three from the incubation growth assay.
1 dilution of the sample with 0.4% trypan blue solution (Sigma Aldrich). Fold increase of cells was calculated by dividing the number of cells with intact cell membranes by the average number of cells on day three from the incubation growth assay.
Physical and analytical methods
Results and discussion
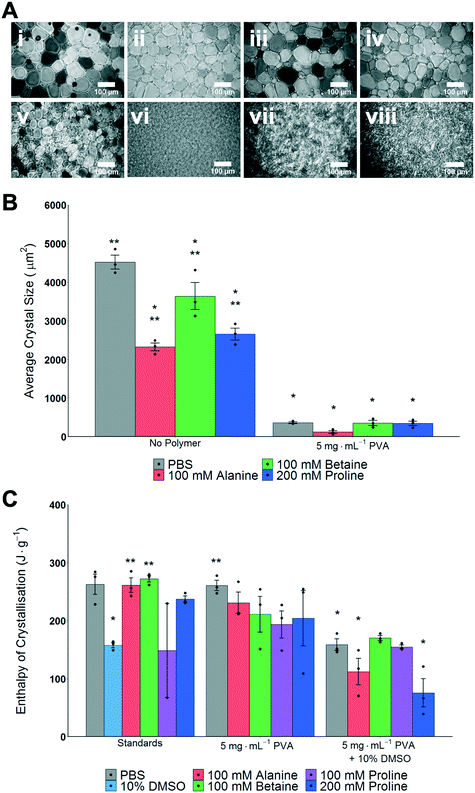
The main aim of this study was to explore the use, and mechanisms, of proline as an additive to enhance the cryopreservation of cell monolayers, in comparison to other osmolytes. A second objective was to explore the synergy of proline with ice recrystallization inhibitors (IRIs) which may also aid cell monolayer cryopreservation. Betaine and alanine were chosen as control osmolytes to be used throughout this study. To evaluate if ice recrystallization inhibition (i.e. preventing ice crystals from growth) occurs in the presence of osmolytes the ‘splat assay’ was used.24,49 This was important, as several studies have shown that the exact ions in solution can impact IRI activity, and any subsequent cryopreservation results can only be interpreted if this effect is known.50,51 The assay involves rapid nucleation of small (<10 μm) ice crystals followed by annealing at −8 °C for 30 minutes. After this time, the wafers were imaged and the size of the ice crystals was assessed; smaller crystals mean more IRI activity, Fig. 1A. For the osmolyte solutions, significantly smaller crystal sizes than PBS alone were observed, yet all also had significantly larger crystal sizes than 5 mg mL−1 PVA in PBS. The data is summarized in Fig. 1B, as average crystal size. Note, mean grain size (MGS), the standard measure52 is not used as the large differences between conditions makes this comparison challenging. It is important to observe that any additive at sufficiently high concentration will slow ice growth, and under these conditions the osmolytes alone would not be considered IRI active.52As high (>100 mM) concentrations of osmolytes will be used in the cryopreservation studies, it was also essential to investigate the impact osmolytes have on total ice formation during cryopreservation conditions. Fig. 1C shows the total enthalpy of crystallization (which is proportional to the total volume of ice formed) for each osmolyte (in PBS), 10% DMSO and with/without PVA as determined by differential scanning calorimetry. When DMSO is used there is an expected decrease in the enthalpy of crystallisation,53 but significant ice is still forming, which confirms the conditions employed here are not leading to vitrification (an alternative method of cryopreservation). Full DSC traces for melting/freezing are shown in Fig. S1 and S2.‡ These results demonstrate that the osmolytes, at the concentrations used, do not significantly impact the physical process of ice formation and growth, which is essential to enable their biochemical roles (below) to be investigated.
Following the above studies on ice formation/growth, A549 (adenocarcinomic human alveolar basal epithelial) cells were employed to test the cytotoxicity of each component, after 24 hours exposure. PVA (polymeric ice recrystallization inhibitor) led to a small reduction in metabolic activity (Fig. S3‡), and there was no significant reduction for any of the osmolytes (proline, betaine, alanine (Fig. S4‡)), and hence cryopreservation studies could be conducted.
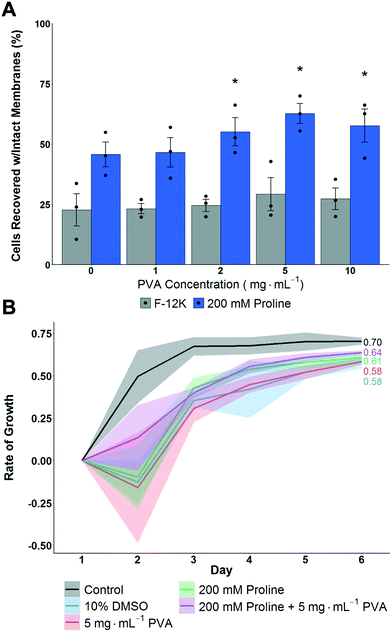
A549 cell monolayers were evaluated for cryopreservation (using 10 vol% DMSO) with and without proline pre-incubation (for 24 hours to allow time for pre-conditioning14,34) and with variable concentrations of PVA as the IRI active agent, Fig. 2A. This was to determine the additive effect of proline when supplemented into standard cryopreservation conditions. It is crucial to note at this point that excess CPA solutions were removed before freezing, thus allowing thawing by addition of pre-warmed media, which also diluted any residual cryoprotectants while ensuring a rapid thawing process. Cells were allowed to recover for 24 hours post-thaw before assessment of recovery. This is important as shorter post-thaw times lead to false positives if cells are evaluated before apoptotic processes have had a chance to complete.13,54 Therefore the number presented is the number of cells after 24 hours, relative to those prior to freezing, and hence includes some time for potential cell growth, but remains a robust measure to avoid over-estimation if shorter times to assay are used.
Cryopreservation of A549 cell monolayers in 10% DMSO alone lead to <25% cell recovery, compared to >60% which is achieved in suspension cryopreservation of A549 cells,55 highlighting the significant challenge of directly cryopreserving cells in this format and the need for disruptive technologies. In contrast to the DMSO only conditions, pre-incubation with proline for 24 hours increased the post-thaw recovery to ∼50%, which is a remarkable increase and in line with previous observations of proline-mediated cryopreservation.14 Additional pre-incubation periods were not evaluated so the potential for further optimisation exists. Addition of PVA, to reduce ice recrystallization, further increased recovery to 60% recovery using 2–5 mg mL−1. Higher concentrations of PVA had no further benefit, as has been previously reported.21,31 Interestingly, without the proline pre-incubation, PVA did not have significantly positive effects on post-thaw recovery, supporting a hypothesis that ice recrystallization is a problem in cell monolayer storage, but that other damage mechanisms are more significant and must be mitigated first.
To gain further insight, the thawed A549 cells were isolated and re-plated at 12.5 × 103 cells per well and their growth rate measured over 6 days. Fig. 2B shows that cells that had been pre-incubated with proline lead to faster growth rates compared to cells without. This shows that not only are more cells recovered, but that they are more viable due to the action of proline. In contrast, PVA increased the number of cells but had no impact on cell growth rate, and hence shows the polymer only has biophysical, not biochemical, function and shows synergistic protective mechanisms are in action. Two other cells lines were also explored, (MC-3T3 and Neuro-2a) where smaller effects were observed which may be due to specific cell differences including size but also their response to hypotonic conditions and contact inhibition (Fig. S5‡). This observation is crucial to highlight how the cell type and format (suspension verses monolayer, freezing rates, etc.) all have major impacts on cryopreservation outcomes and suggests there may not be ‘one size fits all’ solutions.
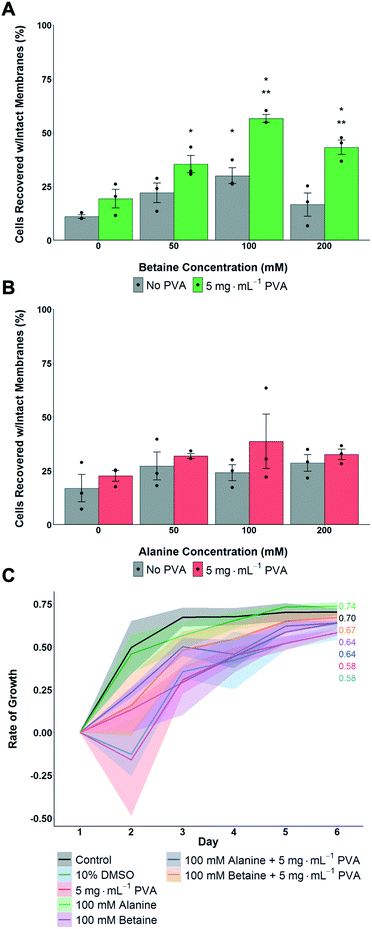
The above data shows that proline had a significant benefit for monolayer cryopreservation, and that recovered cells grew faster, at a rate closer to healthy non-frozen cells. To understand if proline's activity was simply due to it being an osmolyte, or if it had (for example) pre-conditioning effects,56 betaine and alanine were used as control osmolytes. A549 cells were pre-incubated with the osmolytes for 24 hours, then cryopreserved with 5 mg mL−1 PVA + 10% DMSO. Post thaw, the number of cells was determined after 24 hours culture. In the case of betaine, maximum cell recovery was observed with 100 mM, with PVA giving some benefit to cell recovery at all betaine concentrations (Fig. 3A). Alanine showed no significant increase in post-thaw recovery compared to control cells (Fig. 3B). All cells frozen with betaine or alanine, with or without PVA, grew faster post-freeze than cells frozen with only 10% DMSO (Fig. 3C). Betaine's osmoprotection has been previously studied,41 and attributed to the osmoregulated betaine/GABA transporter (BGT1) which couples the transport of betaine to Cl− and Na+ across the cell membrane.57,58 Renomedullary cells grown in hyperosmotic media contained higher concentrations of betaine than cells grown in isotonic media59 and simian-virus-40-transformed Balb/c 3T3 (SV-3T3) cells were resistant to osmotic stress, which typically resulted in inhibition of protein synthesis and decreased proliferation, when treated exogenously with betaine.60 Taken together this data shows that proline and betaine have preferential roles in cryopreservation, as this is not simply an effect of added solutes or osmolarity, and are potent additives for cell monolayer cryopreservation. It is, however, not clear if betaine and proline afford their protection via the same mechanisms.
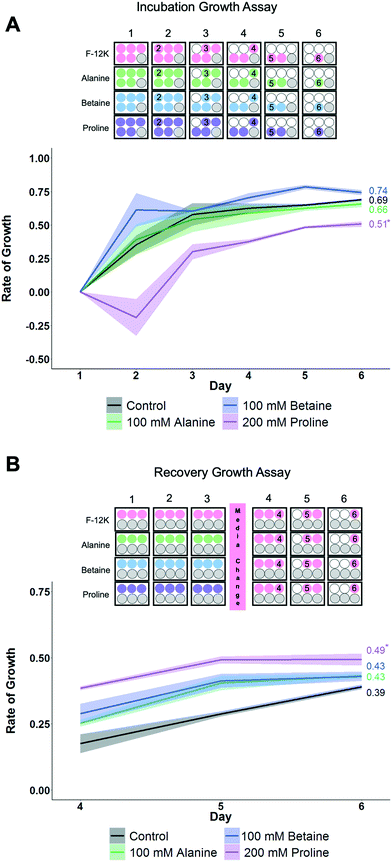
To gain more insight into how each osmolyte functions, growth curves for the cells were monitored as this provides a more rigorous evaluation of health than single point viability measures. Two sets of conditions were evaluated here (Fig. 4) using the concentrations of each osmolyte which gave maximal cell recovery (from Fig. 2 and 3). Firstly, cells were cultured over 6 days in the presence of osmolytes to allow their effects on overall cell growth to be monitored. This experiment is distinct from cytotoxicity assays, in that cells are seeded at low densities so they have space to propagate. Secondly, cells were incubated with osmolytes for 3 days, then exchanged for cell media, and allowed to recover for 3 more days.
When cells were cultured for 6 days in the presence of the osmolytes (at their optimised concentration), proline showed suppression of the growth rate compared to control cells and other osmolytes, which would imply proline has a growth-limiting impact on the cells (Fig. 4A). Next, the cells were incubated with the osmolytes for 3 days, and then the media replaced with standard (no osmolyte) growth media. The proline-treated cells showed a higher fold change in growth rate than other conditions, including control cells, and while the R2 value is lower than control values, it does trend upwards (Fig. 4B). This data shows that proline causes a short-term down-regulation in proliferation, which is rapidly recovered when proline is removed. P493 (B lymphoma) cells incubated in proline have been shown to have growth inhibition, and thus, resulted in the proline biosynthesis (PB) pathway being down-regulated with a downstream effect on the glycolytic pathway.61 This suggests that the lower proliferation rate seen here may not be the proline itself, but rather the effects of the PB pathway not running due to an increase in the available free proline. This down-regulation in growth may be partially responsible for the cryoprotection seen with proline, as metabolic depression and cell stasis are often prerequisites to survival for organisms with natural adaptations to desiccation, freezing temperatures, and anoxia.62 For example, pretreatment with 5-aminoimidazole-4-carboxamide-1-b-D-ribofuranoside (AICAR) can increase cell yields post-thaw, potentially due to metabolic down-regulation (e.g. diapause56).
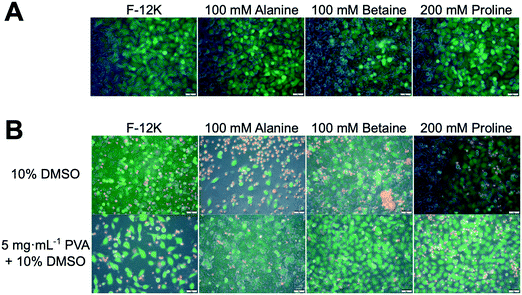
To gain mechanistic insight into the role of proline in cryopreservation, membrane permeabilisation assays were undertaken. Fig. 5A shows results of live/dead staining on A549 cell monolayers after incubation with the osmolytes (green indicates intact membranes, red indicated permeability). In all cases, there was no significant change in membrane permeable (red) cells compared to the controls of media alone (Fig. S6‡). The same assay was undertaken on cryopreserved cells (Fig. 5B) which had been pre-incubated with osmolytes. Minimal differences were seen for cryopreserved cells in any of the osmolyte conditions, with or without PVA, compared to cells frozen with 10% DMSO (Fig. S7‡). This is contrary to the cell recovery data in Fig. 2 and 3, which showed that PVA plus proline or betaine gave higher recovery values than 10% DMSO alone. This suggests that whilst the cryopreserved cells appear to be functionally similar, only cells incubated with proline or betaine and cryopreserved with PVA were able to survive the stress of the subsequent trypsinisation step prior to cell recovery analysis.
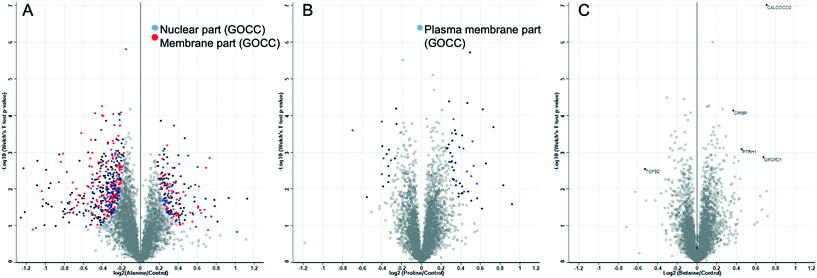
The above data shows that proline has a privileged role in cryopreservation of cell monolayers and acts to slow the growth rate of cells preparing them for cryopreservation without affecting their post-thaw growth rate. This effect is more pronounced than for other osmolytes, and there is no evidence that this is a biophysical process. It is important to again highlight the differential responses seen with other cell lines (Fig. S5‡) to proline pre-incubation. The biochemical mode of action is supported by this differential response and the data presented here does not rule out the use of proline for other cells lines. Further exploration of the large formulation space of concentrations, timings, and cell densities may show a benefit, but highlights that if pre-conditioning (or related strategy) is to be deployed, each line may need individual optimisation. To provide addition insight, whole cell proteomics was undertaken to search for any specific process which might underpin these observations. A549 cells were incubated with the osmolytes for 24 h and then subjected to whole cell proteomic analysis, compared to control cells with the osmolytes. After sample normalisation, 5516 proteins were identified and quantified with a Pearson correlation coefficient >0.99 for all samples (Fig. S8‡). Volcano plots show the distribution of significantly regulated proteins when alanine, proline, and betaine incubated cells were compared against control cells. Gene ontology category Fisher exact test enrichment showed a significant enrichment among the regulated proteins located in the nucleus and membrane when alanine and control conditions were compared. Similar analysis significantly enriched the plasma membrane components when the proline supplemented media was compared to the control (Fig. 6).
Fig. 6 shows that alanine led to more up/down regulation than either proline or betaine, despite resulting in a minimal effect on cryopreservation outcome, compared to the other two osmolytes. Summaries of up/down regulated proteins are shown in the ESI‡ (Fig. S9–S11 and Table S1). For the betaine treated cells, there were 5 significantly different proteins. Upregulation of CALCOCO2 was seen, an effector for clearing bacteria63 with implications in cytoskeleton organization,64 and upregulation was also seen with proline-incubated cells; CIRBP, a cold-inducible protein implicated in the suppression of cell proliferation;65 PTRH1, a t-RNA hydrolase;66 and GPCPD1, a glycerophosphocholine phosphodiesterase.66 Downregulation was seen for TGFB2, a transforming growth factor beta-2 proprotein, a multifunctional protein that regulates numerous processes. With regard to the proline treated cells, there were no clear single up/down regulated proteins which would allow assignment of the biochemical basis of protection, but there were some regulated proteins which could be important and are summarised here. The upregulation seen for the histone RNA hairpin-binding protein (SLBP) may be due to the increase in HIST1H4A and H1F0 as an increase in histones may result in an increase in the protein involved in the processing, translation, and degradation for the mRNAs of histones.67 There was a significant reduction in both SLBP and H1F0 in cells treated with alanine, further suggesting the expression of these proteins is linked. Upregulation in SERPINE1 may provide protection via “wound healing” for cells in a monolayer,68 as this protein would not be expected to be involved in replicative senescence for A549 adenocarcinoma cells as this process protects against oncogenic transformation and requires integrity of the p53 tumour suppressor pathway.69–71 Interestingly, TUT1, a biosensor for the transduction of stress signals within the cell nucleus and also an effector for the fate of target mRNAs,72 was shown to be upregulated in proline treated cells but downregulated in cells treated with alanine.
Pathway enrichment analysis for proline showed that general stress responses are the most frequent changes, suggesting this osmolyte has a broad function on cells which could lead to the observed cryopreservation benefits (Table 1). Proline has been implicated as a survival factor that protects the cell against apoptosis and maintains the progression of the cell cycle.44 This proteomics study identified the cell nucleus and membranes as the principal cell components affected by our treatments and highlighted some stress responses as potential candidates to explain our previous observations in proline pre-treated cells and their enhanced cryopreservation outcomes. This is not intended as a full description of the cells state, but rather to help guide future research in this area to fully understand the biological impact of this pre-conditioning. This may include focusing on some of the candidates described here to identify a target/effect to further explain proline's function and it may be possible to discover target molecules or pathways to help prepare cells for low temperatures, and hence, increase cell yields.
| Pathway name | Entities found | Entities total |
|---|---|---|
| Data analysed online with Reactome open access pathway database. FDR (false discovery rate) = 2.62 × 10−1. | ||
| Apoptosis induced DNA fragmentation | 1 | 13 |
| Cellular responses to stress | 8 | 690 |
| Death receptor signalling | 3 | 157 |
| FGFR2b ligand binding and activation | 1 | 14 |
| TNFR-1-induced proapoptotic signalling | 1 | 14 |
| Chylomicron assembly | 1 | 14 |
| Dissolution of fibrin clot | 1 | 14 |
| Autophagy | 3 | 160 |
Conclusions
Here we have explored the role that proline plays in improving the cryopreservation outcomes of cell monolayers, which currently are very challenging to store frozen and result in low cell recovery rates, compared to suspension cryopreservation. The yield of A549 cell monolayers increased from below 25% to ∼50% by supplementation of proline or betaine, which was further increased to 60% by addition of the polymeric ice recrystallization inhibitor PVA. Without the osmolyte pre-incubation, the PVA had no impact. This demonstrates that IRI is useful to enhance cryopreservation, but other (more significant) sources of cryo-damage must be first mitigated. Post-thaw growth rates of proline and betaine treated cells were also closer to non-frozen controls than other conditions, confirming the beneficial role. It was also observed that other cell lines do not respond in the same manner to the proline treatment, suggesting individual optimisation is required. To understand the impact of the pre-freeze incubation, cell growth rates were examined in the presence of osmolytes. It was shown that cell growth rates were significantly suppressed with proline added, but quickly recovered if the media was exchanged, supporting a hypothesis that proline pre-conditions the cells by slowing growth and preparing the cells for cold stress and agrees with observations that some extremophiles upregulate proline biosynthesis in anticipation of cold stress. Whilst betaine also improved cryopreservation outcomes it did not have the same downregulation on cellular growth rates, providing more evidence that the mechanism of action for proline is unique. Membrane stability assays showed that proline did not impact cell membrane permeability compared to control cells or other osmolytes, and biophysical assays of ice formation/growth also confirm that proline had no biophysical role, and hence its function is purely biochemical. Whole cell proteomics were used to explore protein regulation but did not show conclusively any single pathway which could be attributed to this effect. However, when cells were incubated in proline, the upregulation of key histone proteins was observed (which were downregulated in our alanine samples), along with upregulation of stress and membrane proteins, with lipid and proliferation proteins being downregulated, which may be crucial to proline's role but will require further investigation.Overall this data provides further evidence that pre-conditioning of cell monolayers is a powerful tool to improve post-thaw cell yields, and that proline is an accessible and biocompatible additive that can easily be added into existing protocols. It also suggests that small molecule modulators of cryopreservation could be discovered if the pathways acted upon by proline can be fully elucidated, enabling a chemical biology driven approach towards the discovery of cryoprotective agents/modulators. These findings will help guide processes for improved monolayer storage techniques with subsequent benefits in drug screening, basic research, and improvement for frozen distribution of cells and cell products.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
MG and TB are shareholders in Cryoloyx Ltd.Acknowledgements
This work was supported by the European Research Council (638661 and 789182). We thank the support from a seed grant through the Wellcome–Warwick Quantitative Biomedicine Programme (Institutional Strategic Support Fund: 105627/Z/14/Z). TLB thanks M. Menze for providing the Biocision CoolCell to enable controlled rate freezing. The proteomics research technology platform are acknowledged. Dr Kathryn Murray (UoW) is thanked for critically reading the manuscript.References
- K. E. Healy, T. C. McDevitt, W. L. Murphy and R. M. Nerem, Engineering the Emergence of Stem Cell Therapeutics, Sci. Transl. Med., 2013, 5(207), 207ed17 CrossRef PubMed.
- M. A. Fischbach, J. A. Bluestone and W. A. Lim, Cell-Based Therapeutics: The next Pillar of Medicine, Sci. Transl. Med., 2013, 5(179), 179ps7 CrossRef PubMed.
- X. Stéphenne, M. Najimi and E. M. Sokal, Hepatocyte Cryopreservation: Is It Time to Change the Strategy?, World J. Gastroenterol., 2010, 16(1), 1–14 Search PubMed.
- P. Mazur, Cryobiology: The Freezing of Biological Systems, Science, 1970, 168(3934), 939–949 CrossRef CAS PubMed.
- P. Mazur, J. Farrant, S. P. Leibo and E. H. Chu, Survival of Hamster Tissue Culture Cells after Freezing and Thawing. Interactions between Protective Solutes and Cooling and Warming Rates, Cryobiology, 1969, 6(1), 1–9 CrossRef CAS PubMed.
- M. Timm, L. Saaby, L. Moesby and E. W. Hansen, Considerations Regarding Use of Solvents in in Vitro Cell Based Assays, Cytotechnology, 2013, 65(5), 887–894 CrossRef CAS PubMed.
- J. G. Hengstler, D. Utesch, P. Steinberg, K. L. Platt, B. Diener, M. Ringel, N. Swales, T. Fischer, K. Biefang and M. Gerl, et al., Cryopreserved Primary Hepatocytes as a Constantly Available in Vitro Model for the Evaluation of Human and Animal Drug Metabolism and Enzyme Induction, Drug Metab. Rev., 2000, 81–118 CrossRef CAS PubMed.
- G. M. Fahy, Cryoprotectant Toxicity Neutralization, Cryobiology, 2010, 60(3), S45–S53 CrossRef CAS PubMed.
- G. M. Fahy, The Relevance of Cryoprotectant “Toxicity” to Cryobiology, Cryobiology, 1986, 23(1), 1–13 CrossRef CAS PubMed.
- T. Arakawa, J. F. Carpenter, Y. A. Kita and J. H. Crowe, The Basis for Toxicity of Certain Cryoprotectants: A Hypothesis, Cryobiology, 1990, 27(4), 401–415 CrossRef CAS.
- G. Da Violante, N. Zerrouk, I. Richard, G. Provot, J. C. Chaumeil and P. Arnaud, Evaluation of the Cytotoxicity Effect of Dimethyl Sulfoxide (DMSO) on Caco2/TC7 Colon Tumor Cell Cultures, Biol. Pharm. Bull., 2002, 25(12), 1600–1603 CrossRef CAS PubMed.
- X. Xu, S. Cowley, C. J. Flaim, W. James, L. Seymour and Z. Cui, The Roles of Apoptotic Pathways in the Low Recovery Rate after Cryopreservation of Dissociated Human Embryonic Stem Cells, Biotechnol. Prog., 2010, 26(3), 827–837 CrossRef CAS PubMed.
- C. H. Boon, P. Y. Chao, H. Liu, S. T. Wei, A. J. Rufaihah, Z. Yang, H. B. Boon, Z. Ge, W. O. Hog and H. L. Eng, et al., Loss of Viability during Freeze-Thaw of Intact and Adherent Human Embryonic Stem Cells with Conventional Slow-Cooling Protocols Is Predominantly Due to Apoptosis Rather than Cellular Necrosis, J. Biomed. Sci., 2006, 13(3), 433–445 CrossRef PubMed.
- T. L. Bailey, M. Wang, J. Solocinski, B. P. Nathan, N. Chakraborty and M. A. Menze, Protective Effects of Osmolytes in Cryopreserving Adherent Neuroblastoma (Neuro-2a) Cells, Cryobiology, 2015, 71(3), 472–480 CrossRef CAS PubMed.
- P. Mazur, S. P. Leibo and E. H. Y. A. Chu, Two-Factor Hypothesis of Freezing Injury. Evidence from Chinese Hamster Tissue-Culture Cells, Exp. Cell Res., 1972, 71(2), 345–355 CrossRef CAS PubMed.
- A. Fowler and M. Toner, Cryo-Injury and Biopreservation, Ann. N. Y. Acad. Sci., 2005, 1066, 119–135 CrossRef CAS PubMed.
- P. Mazur, Principles of Cryobiology, ed. B. Fuller, N. Lane and E. E. Benson, CRC Press, Boca Raton, 2004, vol. 17 Search PubMed.
- P. Mazur, Freezing of Living Cells: Mechanisms and Implications, Am. J. Physiol., 1984, 247(3), C125–C142 CrossRef CAS PubMed.
- P. Mazur, R. H. Miller and S. P. Leibo, Survival of Frozen-Thawed Bovine Red Cells as a Function of the Permeation of Glycerol and Sucrose, J. Membr. Biol., 1974, 15(2), 137–158 CrossRef CAS PubMed.
- G. Seth, Freezing Mammalian Cells for Production of Biopharmaceuticals, Methods, 2012, 424–431 CrossRef CAS PubMed.
- H. Chao, P. L. Davies and J. F. Carpenter, Effects of Antifreeze Proteins on Red Blood Cell Survival during Cryopreservation, J. Exp. Biol., 1996, 199, 2071–2076 CrossRef CAS PubMed.
- R. M. F. Tomás, T. L. Bailey, M. Hasan and M. I. Gibson, Extracellular Antifreeze Protein Significantly Enhances the Cryopreservation of Cell Monolayers, Biomacromolecules, 2019, 20(10), 3864–3872 CrossRef PubMed.
- C. A. Knight, J. Hallett and A. L. DeVries, Solute Effects on Ice Recrystallization: An Assessment Technique, Cryobiology, 1988, 25(1), 55–60 CrossRef CAS PubMed.
- C. A. Knight, D. Wen and R. A. Laursen, Nonequilibrium Antifreeze Peptides and the Recrystallization of Ice, Cryobiology, 1995, 32(1), 23–34 CrossRef CAS PubMed.
- C. A. Knight, A. L. DeVries and L. D. Oolman, Fish Antifreeze Protein and the Freezing and Recrystallization of Ice, Nature, 1984, 308(5956), 295–296 CrossRef CAS PubMed.
- J. F. Carpenter and T. N. Hansen, Antifreeze Protein Modulates Cell Survival during Cryopreservation: Mediation through Influence on Ice Crystal Growth, Proc. Natl. Acad. Sci. U. S. A., 1992, 89(19), 8953–8957 CrossRef CAS PubMed.
- A. Eniade, M. Purushotham, R. N. Ben, J. B. Wang and K. A. Horwath, Serendipitous Discovery of Antifreeze Protein-Specific Activity in C-Linked Antifreeze Glycoprotein Analogs, Cell Biochem. Biophys., 2003, 38(2), 115–124 CrossRef CAS PubMed.
- M. I. M. I. Gibson, Slowing the Growth of Ice with Synthetic Macromolecules: Beyond Antifreeze(Glyco) Proteins, Polym. Chem., 2010, 1(8), 1141–1152 RSC.
- T. Inada and S. S. Lu, Inhibition of Recrystallization of Ice Grains by Adsorption of Poly(Vinyl Alcohol) onto Ice Surfaces, Cryst. Growth Des., 2003, 3(5), 747–752 CrossRef CAS.
- C. Budke and T. Koop, Ice Recrystallization Inhibition and Molecular Recognition of Ice Faces by Poly(Vinyl Alcohol), ChemPhysChem, 2006, 7(12), 2601–2606 CrossRef CAS PubMed.
- R. C. R. C. Deller, M. Vatish, D. A. D. A. Mitchell and M. I. M. I. Gibson, Synthetic Polymers Enable Non-Vitreous Cellular Cryopreservation by Reducing Ice Crystal Growth during Thawing, Nat. Commun., 2014, 5, 3244 CrossRef PubMed.
- B. Wowk, E. Leitl, C. M. Rasch, N. Mesbah-Karimi, S. B. Harris and G. M. Fahy, Vitrification Enhancement by Synthetic Ice Blocking Agents, Cryobiology, 2000, 40(3), 228–236 CrossRef CAS PubMed.
- R. C. Deller, J. E. Pessin, M. Vatish, D. A. Mitchell and M. I. Gibson, Enhanced Non-Vitreous Cryopreservation of Immortalized and Primary Cells by Ice-Growth Inhibiting Polymers, Biomater. Sci., 2016, 4(7), 1079–1084 RSC.
- B. Graham, T. L. Bailey, J. R. J. Healey, M. Marcellini, S. Deville and M. I. Gibson, Polyproline as a Minimal Antifreeze Protein Mimic That Enhances the Cryopreservation of Cell Monolayers, Angew. Chem., 2017, 129(50), 16157–16160 CrossRef.
- Q. Qin, L. Zhao, Z. Liu, T. Liu, J. Qu, X. Zhang, R. Li, L. Yan, J. Yan and S. Jin, et al., Bioinspired l -Proline Oligomers for the Cryopreservation of Oocytes via Controlling Ice Growth, ACS Appl. Mater. Interfaces, 2020, 12(16), 18352–18362 CrossRef CAS PubMed.
- M. B. Burg, Molecular Basis of Osmotic Regulation, Am. J. Physiol., 1995, 268(6), F983–F996 CAS.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Living with Water Stress: Evolution of Osmolyte Systems, Science, 1982, 217(4566), 1214–1222 CrossRef CAS PubMed.
- C. F. Higgins, J. Cairney, D. A. Stirling, L. Sutherland and I. R. Booth, Osmotic Regulation of Gene Expression: Ionic Strength as an Intracellular Signal?, Trends Biochem. Sci., 1987, 12, 339–344 CrossRef CAS.
- F. N. Ziyadeh, J. W. Mills and A. Kleinzeller, Hypotonicity and Cell Volume Regulation in Shark Rectal Gland: Role of Organic Osmolytes and F-Actin, Am. J. Physiol., 1992, 262(3), F468–F479 CAS.
- A. W. Lloyd, C. J. Olliff and K. J. A. Rutt, Study of Modified Betaines as Cryoprotective Additives, J. Pharm. Pharmacol., 1994, 46(9), 704–707 CrossRef CAS PubMed.
- J. Yang, N. Cai, H. Zhai, J. Zhang, Y. Zhu and L. Zhang, Natural Zwitterionic Betaine Enables Cells to Survive Ultrarapid Cryopreservation, Sci. Rep., 2016, 6(1), 37458 CrossRef CAS PubMed.
- A. C. Chakrabarti, Permeability of Membranes to Amino Acids and Modified Amino Acids: Mechanisms Involved in Translocation, Amino Acids, 1994, 6(3), 213–229 CrossRef CAS PubMed.
- S. Vilella, G. A. Ahearn, G. Cassano and C. Storelli, Na-Dependent L-Proline Transport by Eel Intestinal Brush-Border Membrane Vesicles, Am. J. Physiol., 1988, 255(4), R648–R653 CAS.
- F. Franěk and K. Srámková, Cell Suicide in Starving Hybridoma Culture: Survival-Signal Effect of Some Amino Acids, Cytotechnology, 1997, 23(1–3), 231–239 CrossRef PubMed.
- L. Zhang, X. Xue, J. Yan, L.-Y. Yan, X.-H. Jin, X.-H. Zhu, Z.-Z. He, J. Liu, R. Li and J. Qiao, Cryobiological Characteristics of L-Proline in Mammalian Oocyte Cryopreservation, Zhonghua Yixue Zazhi, 2016, 129(16), 1963–1968 CAS.
- J. Yang, C. Pan, J. Zhang, X. Sui, Y. Zhu, C. Wen and L. Zhang, Exploring the Potential of Biocompatible Osmoprotectants as Highly Efficient Cryoprotectants, ACS Appl. Mater. Interfaces, 2017, 9(49), 42516–42524 CrossRef CAS PubMed.
- V. Koštál, P. Šimek, H. Zahradníčková, J. Cimlová and T. Štětina, Conversion of the Chill Susceptible Fruit Fly Larva (Drosophila Melanogaster) to a Freeze Tolerant Organism, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(9), 3270–3274 CrossRef PubMed.
- C. A. Schneider, W. S. Rasband and K. Eliceiri, Image to ImageJ: 25 Years of Image Analysis, Nat. Methods, 2012, 9(7), 671–675 CrossRef CAS PubMed.
- T. Congdon, R. Notman and M. I. Gibson, Antifreeze (Glyco)Protein Mimetic Behavior of Poly(Vinyl Alcohol): Detailed Structure Ice Recrystallization Inhibition Activity Study, Biomacromolecules, 2013, 14(5), 1578–1586 CrossRef CAS PubMed.
- R. Surís-Valls and I. K. Voets, The Impact of Salts on the Ice Recrystallization Inhibition Activity of Antifreeze (Glyco)Proteins, Biomolecules, 2019, 9(8), 347 CrossRef PubMed.
- S. Wu, C. Zhu, Z. He, H. Xue, Q. Fan, Y. Song, J. S. Francisco, X. C. Zeng and J. Wang, Ion-Specific Ice Recrystallization Provides a Facile Approach for the Fabrication of Porous Materials, Nat. Commun., 2017, 8(1), 15154 CrossRef PubMed.
- C. I. Biggs, C. Stubbs, B. Graham, A. E. R. Fayter, M. Hasan and M. I. Gibson, Mimicking the Ice Recrystallization Activity of Biological Antifreezes. When Is a New Polymer “Active”?, Macromol. Biosci., 2019, 19(7), 1900082 CrossRef PubMed.
- S. S. N. Murthy, Some Insight into the Physical Basis of the Cryoprotective Action of Dimethyl Sulfoxide and Ethylene Glycol, Cryobiology, 1998, 36(2), 84–96 CrossRef CAS PubMed.
- K. A. Murray and M. I. Gibson, Post-Thaw Culture and Measurement of Total Cell Recovery Is Crucial in the Evaluation of New Macromolecular Cryoprotectants, Biomacromolecules, 2020, 21(7), 2864–2873 CrossRef CAS PubMed.
- T. L. Bailey, C. Stubbs, K. Murray, R. M. F. Tomas, L. Otten and M. I. A. Gibson, Synthetically Scalable Poly(Ampholyte) Which Dramatically Enhances Cellular Cryopreservation, Biomacromolecules, 2019, 20, 3104–3114 CrossRef CAS PubMed.
- M. A. Menze, N. Chakraborty, M. Clavenna, M. Banerjee, X. H. Liu, M. Toner and S. C. Hand, Metabolic Preconditioning of Cells with AICAR-Riboside: Improved Cryopreservation and Cell-Type Specific Impacts on Energetics and Proliferation, Cryobiology, 2010, 61(1), 79–88 CrossRef CAS PubMed.
- A. Yamauchi, S. Uchida, H. M. Kwon, A. S. Preston, R. B. Robey, A. Garcia-Perez, M. B. Burg and J. S. Handler, Cloning of a Na(+)- and Cl(−)-Dependent Betaine Transporter That Is Regulated by Hypertonicity, J. Biol. Chem., 1992, 267(1), 649–652 CrossRef CAS.
- M. B. Burg, J. D. Ferraris and N. I. Dmitrieva, Cellular Response to Hyperosmotic Stresses, Physiol. Rev., 2007, 87(4), 1441–1474 CrossRef CAS PubMed.
- T. Nakanishi, R. S. Balaban and M. B. Burg, Survey of Osmolytes in Renal Cell Lines, Am. J. Physiol., 1988, 255(2), C181–C191 CrossRef CAS PubMed.
- P. G. Petronini, E. M. De Angelis, P. Borghetti, A. F. Borghetti and K. P. Wheeler, Modulation by Betaine of Cellular Responses to Osmotic Stress, Biochem. J., 1992, 282(1), 69–73 CrossRef CAS PubMed.
- W. Liu, C. N. Hancock, J. W. Fischer, M. Harman and J. M. Phang, Proline Biosynthesis Augments Tumor Cell Growth and Aerobic Glycolysis: Involvement of Pyridine Nucleotides, Sci. Rep., 2015, 5(1), 17206 CrossRef CAS PubMed.
- S. Hand, Y. Patil, S. Li, N. Charkraborty, A. Borcar, M. Menze, L. Boswell, D. Moore and M. Toner, Diapause and Anhydrobiosis in Embryos of Artemia Franciscana : Metabolic Depression, LEA Proteins and Water Stress, Cryobiology and Cryotechnology, 2013, 59(1), 41–46 Search PubMed.
- T. L. M. Thurston, M. P. Wandel, N. Von Muhlinen, Á. Foeglein and F. Randow, Galectin 8 Targets Damaged Vesicles for Autophagy to Defend Cells against Bacterial Invasion, Nature, 2012, 482(7385), 414–418 CrossRef CAS PubMed.
- B. Morriswood, G. Ryzhakov, C. Puri, S. D. Arden, R. Roberts, C. Dendrou, J. Kendrick-Jones and F. Buss, T6BP and NDP52 Are Myosin VI Binding Partners with Potential Roles in Cytokine Signalling and Cell Adhesion, J. Cell Sci., 2007, 120, 2574–2585 CrossRef CAS PubMed.
- C. Yang and F. Carrier, The UV-Inducible RNA-Binding Protein A18 (A18 HnRNP) Plays a Protective Role in the Genotoxic Stress Response, J. Biol. Chem., 2001, 276(50), 47277–47284 CrossRef CAS PubMed.
- P. Gaudet, M. S. Livstone, S. E. Lewis and P. D. Thomas, Phylogenetic-Based Propagation of Functional Annotations within the Gene Ontology Consortium, Briefings Bioinf., 2011, 12(5), 449–462 CrossRef PubMed.
- W. F. Marzluff, E. J. Wagner and R. J. Duronio, Metabolism and Regulation of Canonical Histone MRNAs: Life without a Poly(A) Tail, Nat. Rev. Genet., 2008, 9(11), 843–854 CrossRef CAS PubMed.
- K. M. Providence, S. P. Higgins, A. Mullen, A. Battista, R. Samarakoon, C. E. Higgins, C. E. Wilkins-Port and P. J. Higgins, SERPINE1 (PAI-1) Is Deposited into Keratinocyte Migration “Trails” and Required for Optimal Monolayer Wound Repair, Arch. Dermatol. Res., 2008, 300(6), 303–310 CrossRef CAS PubMed.
- A. S. Lundberg, W. C. Hahn, P. Gupta and R. A. Weinberg, Genes Involved in Senescence and Immortalization, Curr. Opin. Cell Biol., 2000, 12(6), 705–709 CrossRef CAS PubMed.
- C. J. Sherr and F. McCormick, The RB and P53 Pathways in Cancer, Cancer Cell, 2002, 2(2), 103–112 CrossRef CAS PubMed.
- J. Massagué, G1 Cell-Cycle Control and Cancer, Nature, 2004, 432(7015), 298–306 CrossRef PubMed.
- W. Li, R. S. Laishram and R. A. Anderson, The Novel Poly(A) Polymerase Star-PAP Is a Signal-Regulated Switch at the 3′-End of MRNAs, Adv. Biol. Regul., 2013, 53(1), 64–76 CrossRef CAS PubMed.
Footnotes |
| † The research data supporting this publication can be found at http://wrap.warwick.ac.uk. |
| ‡ Electronic supplementary information (ESI) available: Experimental procedures and characterisation data are included in the ESI. See DOI: 10.1039/d1md00078k |
| This journal is © The Royal Society of Chemistry 2021 |