Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and evaluation of bis(imino)anthracene derivatives as G-quadruplex ligands†
Tomris
Coban
,
Cameron
Robertson
,
Sianne
Schwikkard
,
Richard
Singer
and
Adam
LeGresley
 *
*
LSP&C, SEC Faculty, Kingston University, Kingston-upon-Thames, KT1 2EE, UK. E-mail: a.legresley@kingston.ac.uk
First published on 26th March 2021
Abstract
The synthesis of a small number of bis(imino)anthracene derivatives is reported. They were evaluated via NMR for binding efficacy to the G-quadruplex-forming oligonucleotide sequence (TTGGGTT) and show activity against the HeLa cancer cell line. These novel ligands are compared to previously synthesised G-quadruplex ligands that target telomeres and oncogenes.
Introduction
G-quadruplexes (G4) became biologically relevant in the 1980s mostly based on the seminal work undertaken by Dr. Elizabeth Blackburn which showed telomeric DNA forms non-Watson–Crick guanine–guanine base-paired intramolecular DNA structures.1 This discovery led to increased research interest and initiated interest in the field of G-quadruplex research.2 G-Quadruplex DNA structures heralded a new approach in anticancer drug design and development through the targeting of secondary DNA structures.3 It has been demonstrated that G-quadruplexes are present in genes that have been shown to be overexpressed in several different cancers, the stabilisation of which can effectively act as a switch for these genes.4–8 The first telomere interacting molecular structure was found to be 2,6-diamidoanthraquinone derivatives synthesised by Sun et al.9 These observations have led to an exponential growth in the development of synthetic small molecules for G-quadruplex binding and represent a rational approach to lead molecule design. Despite a large and diverse libraries of potential lead compounds having been reported, the search for G-tetrad selective compounds that can indirectly inhibit telomerase extension of DNA in cancer cells continues.The principle motifs of a selective G-quadruplex ligand have been reported by a number of research groups.10–14 These include the requirement for planar aromatic systems and distal ionisable amine groups to interact with the phosphate backbone of the quadruplex DNA. Our intention was to couple a range of ionisable amine groups a–h to anthracene-9,10-dicarbaldehdye 1 and probe their selectivity for quadruplex over duplex DNA and to test them for cytotoxicity in HeLa cells (Fig. 1).
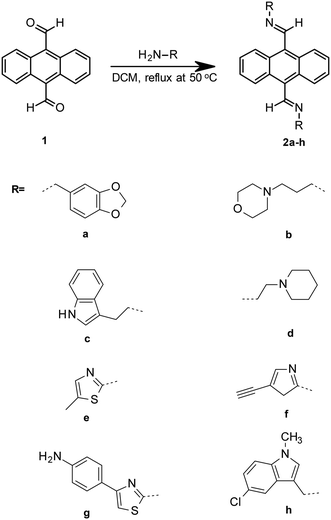 | ||
| Fig. 1 Simplified scheme for imine coupling to acrylate as done for novel imine coupled anthracene derivatives. | ||
The reversible nature of the imine reaction facilitates the future establishment of a dynamic combinatorial library for this class of ligand, which may enable the in situ amplification of a specific (bis)imino compound from our anthracene derivative depending on the thermodynamics associated with exposure to quadruplex over duplex DNA. However, in this manuscript we report the formation and testing of a small static combinatorial library based on 1.
Results
Reaction of a series of amines to 1 was achieved using literature methods.15 The amines were selected owing to their potential for selectivity for the G-quadruplex e.g. Tryptamine is a known monoamino alkaloid which was selected based upon its amino group position in the ring system (see ESI† S1 and S2).From Table 1, we show 3 compounds 2f, 2g and 2h were not successfully synthesised due to solubility issues.
| Compound | Yield (%) | Time (h) |
|---|---|---|
| 2a | 78 | 24 h |
| 2b | 60 | 24 h |
| 2c | 80 | 12 h |
| 2d | 65 | 24 h |
| 2e | 75 | 24 h |
| 2f | — | 48 h |
| 2g | — | 48 h |
| 2h | — | 48 h |
Before biochemical and in vitro analysis, the imines were tested for aqueous stability in Dulbecco's modified Eagle's medium (DMEM). Using an internal standard (TSP), after qNMR analysis at time point 0 and 24 h later no breakdown was observed with no significant concentration changes detected in triplicate experiments.16
Compounds 2a–2e were evaluated for G-quadruplex interaction using NMR titration studies with d(TTGGGTT)4 with intercalation comparisons made against doxorubicin and nemorubicin (see ESI† S1). Previous literature with d(TTGGGTT)4 has demonstrated that G-quadruplex ligands intercalate specifically at the point between G5–T6.17 Large broadening of G5 imino proton peaks, accompanied with an up-field shift, shows this. We also see the peaks for T6 (methyl, aromatic and ribose) shifting to a lower field in accord with the up-field shift of the G5 peaks. This change is explained by the formation of a complex between the ligand and the tetrad formed at G5, which pushes the T6 unit away from the guanine π-system. This intercalation framework agreed with ESI/MS carried out which illustrated a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex.17 We see in Fig. 2, much like previous work, that line broadening and up-field shift occurs at peaks representative of proton G6NH (10.79 ppm) significantly, alongside the significant peak broadening and down field shifting of T6H6/8 and T6CH3 peaks with the addition of proposed ligands.18
1 complex.17 We see in Fig. 2, much like previous work, that line broadening and up-field shift occurs at peaks representative of proton G6NH (10.79 ppm) significantly, alongside the significant peak broadening and down field shifting of T6H6/8 and T6CH3 peaks with the addition of proposed ligands.18
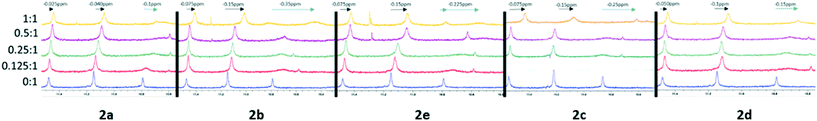 | ||
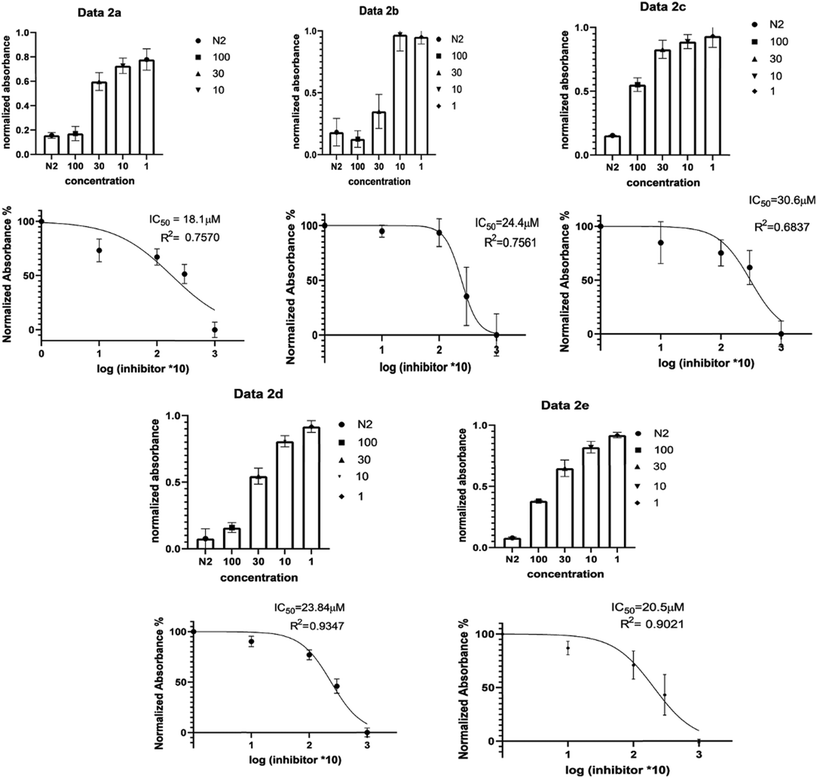
| Fig. 2 1D 1HNMR spectra, referenced to TSP, showing titration of drug compounds and d(TTAGGGT)4. A slow exchange interaction, is illustrated with black arrows and fast exchanging interactions by blue arrows preliminary experiments were done to screen for activity and synthesised compounds that showed activity were run in triplicate to get an average, SD and SEM values, from these results, it was shown that all 5 ligands showed cancer activity with IC50 values ranging from (∼18–31 μM) as illustrated in Fig. 3. | ||
These significant changes demonstrate distinctly different bound and unbound forms through large broadening effects at a ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. There is also subtle line broadening and up-field shifting of G4NH (11.48 ppm) and G5NH (11.15 ppm) for each ligand that shows chemical exchange between two similar environments of bound and unbound ligand. Maximum line broadening occurs at a ratio of 1
1. There is also subtle line broadening and up-field shifting of G4NH (11.48 ppm) and G5NH (11.15 ppm) for each ligand that shows chemical exchange between two similar environments of bound and unbound ligand. Maximum line broadening occurs at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 drug to G-quadruplex and chemical shift difference changing up to 1
1 drug to G-quadruplex and chemical shift difference changing up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with the bound and unbound peaks being present. This matches the effect seen by nemorubicin intercalation with d(TTGGGTT)4 in previous literature suggesting a 1
1 with the bound and unbound peaks being present. This matches the effect seen by nemorubicin intercalation with d(TTGGGTT)4 in previous literature suggesting a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. This investigation showed that the five imine coupled ligands showed G-quadruplex intercalation with 1
1 stoichiometry. This investigation showed that the five imine coupled ligands showed G-quadruplex intercalation with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry matching that of the current cancer drugs doxorubicin and nemorubicin (Table 2).
1 stoichiometry matching that of the current cancer drugs doxorubicin and nemorubicin (Table 2).
| Compound | Difference from 1.58 ppm (ppm) T6CH3 | Difference from 7.23 ppm (ppm) T6H6/8 | Difference from 11.55 ppm (ppm) G3NH | Difference from 11.22 ppm (ppm) G4NH | Difference from 10.86 ppm (ppm) G5NH |
|---|---|---|---|---|---|
| 2a | 0.09 | 0.17 | −0.04 | −0.08 | −0.2 |
| 2b | 0.093 | 0.22 | −0.066 | −0.135 | −0.34 |
| 2e | 0.146 | 0.19 | −0.056 | −0.11 | −0.24 |
| 2c | 0.157 | 0.126 | −0.06 | −0.115 | −0.25 |
| 2d | 0.094 | 0.16 | −0.03 | −0.07 | −0.13 |
Compounds 2a–2e were subsequently tested against a HeLa cancer cell line (HeLa, American Type Culture Collection (ATCC) (ATCC® CCL-2™)) and they were all prepared in 100, 30, 10 and 1 μM concentrations so that IC50 values could be calculated to establish whether drugs were cytotoxic to HeLa cells (see ESI† S1).
Discussion
The ligands have different functional groups present in their side chains; 1,3-benzodioxole, propylmorpholine, 2,5-dimetylthiazole, 3-methyl indole and piperidine respectively, which we suspected stabilise the hydrogen bond between the tertiary amine and oxygens of phosphate. However, all these groups have been shown in previous studies to demonstrate DNA and G-quadruplex intercalation alongside anti-cancer activity. This suggests that each of these groups may have more of an effect on IC50, than solely the effects on hydrogen bond stabilisation.We see that the two groups can be grouped by their absence of oxygen or sulphur, piperidine and 3-methyl indole show quite divergent differences in ppm between bound and unbound forms of the G4 model. These two compounds also have 2 of the higher IC50 values of the 5 intercalating compounds, 24 μM (2d) and 30.5 μM (2c).
Previous studies have demonstrated non-selective binding to DNA and G4 structures where the mode of binding to DNA, like our results, showed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, with Gibbs free energy calculations showing a spontaneous intercalation which suggests a stabilising, energetically favourable reaction.19 It was also shown that using piperidine as a N-propylamino attached to carbazoles gave the highest stabilizing ligands in a study.20 Interestingly, however they did identify that there was an inverse correlation between G-tetrad stabilization and G4 DNA selectivity.21 This is very interesting for our results, as it could begin to explain why we see that the relationship between the change in ppm does not correlate completely with IC50 values, even showing an inverse relationship to what was expected from nemorubicin results.17 It could show that G4 ligands intercalate and stabilise very effectively but IC50 will be increased as G4 ligand is less selective for G4 structures. This could explain why we see piperidine having similar chemical difference effects and stabilization effects to other compounds but has a higher IC50. 3-Methyl indole is less investigated for this purpose because of its role primarily as an odourant. There are however genetic studies that show protective mechanisms against damage by 3-methyl indole derivatives that can induce apoptosis at low concentrations. This was shown in a study by Nichols WK they looked at 3-methyl indole mediated cytotoxicity in human epithelial lung cells, with CYP2F1 over expressed.22 This is relevant to this study as it shows that 3-methyl indole has a structure activity relationship which relates to cytotoxicity of genetically impaired cells, which could possibly explain anti-cancer activity.22 The anti-cancer activity of indoles has also been shown with indole groups in isoxazolo[5′,4′:5,6]pyrido[2,3-b]indoles, which showed significant anticancer activity comparable to standard drugs however the mechanism of action and structure activity relationship is yet to be investigated.23
1 stoichiometry, with Gibbs free energy calculations showing a spontaneous intercalation which suggests a stabilising, energetically favourable reaction.19 It was also shown that using piperidine as a N-propylamino attached to carbazoles gave the highest stabilizing ligands in a study.20 Interestingly, however they did identify that there was an inverse correlation between G-tetrad stabilization and G4 DNA selectivity.21 This is very interesting for our results, as it could begin to explain why we see that the relationship between the change in ppm does not correlate completely with IC50 values, even showing an inverse relationship to what was expected from nemorubicin results.17 It could show that G4 ligands intercalate and stabilise very effectively but IC50 will be increased as G4 ligand is less selective for G4 structures. This could explain why we see piperidine having similar chemical difference effects and stabilization effects to other compounds but has a higher IC50. 3-Methyl indole is less investigated for this purpose because of its role primarily as an odourant. There are however genetic studies that show protective mechanisms against damage by 3-methyl indole derivatives that can induce apoptosis at low concentrations. This was shown in a study by Nichols WK they looked at 3-methyl indole mediated cytotoxicity in human epithelial lung cells, with CYP2F1 over expressed.22 This is relevant to this study as it shows that 3-methyl indole has a structure activity relationship which relates to cytotoxicity of genetically impaired cells, which could possibly explain anti-cancer activity.22 The anti-cancer activity of indoles has also been shown with indole groups in isoxazolo[5′,4′:5,6]pyrido[2,3-b]indoles, which showed significant anticancer activity comparable to standard drugs however the mechanism of action and structure activity relationship is yet to be investigated.23
The three groups that contained oxygen, sulphur and nitrogen, and nitrogen and oxygen, 1,3-benzodioxole, 2,5-dimethylthiazole and N-propylmorpholine respectively, showed a close linear correlation between their ability to stabilise G4 structure and IC50. They also, as a group had the best IC50 values with 1,3-benzodioxole and 2,5-dimethyl thiazole as the best candidates for further testing. Further synthesis and reinforcement of DNA binding potential of 1,3-benzodioxole has been illustrated by a study which showed DNA binding was moderate to mild and suggested that cytotoxic effects were through a different mode of action, which supports the selectivity argument for 1,3-benzodioxole.24 They demonstrated through molecular docking that a benzodioxole derivative had potent telomerase inhibition alongside potent anticancer activity in human gastric and human melanoma cell lines.25 This shows that this group may add additional benefit through telomerase inhibition in a different method of action to the one investigated here with G4 stabilisation. The 2,5-dimethylthiazole has been studied extensively in the form of the fluorescent marker thiazole orange for its intercalation potential. It was shown in early research that thiazole orange binds both as a monomer and dimer to DNA. Monomers of thiazole orange stack between DNA bases. It is also shown as the main way of binding with duplex DNA. Interestingly they also reported binding to poly(dG) which showed binding as a monomer and dimer as well. This supports our results showing intercalation and stabilisation of G4 as seen in our results.26 Selective stabilization of G4 structures by thiazole has further been supported more recently with studies that showed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry between dye and G-tetrads in quadruplex DNA. It also illustrated that there was a tight complex formed between the two. Interestingly they showed that in the presence of K+ ions, this binding seemed to disappear.27 To attempt to rationalise the chemical shift difference and IC50 values observed, computational modelling of the 5 ligands with parallel (PDB entry139d) and antiparallel (PDB entry 6IMS) quadruplex structures was undertaken using Autodock Vina both in conjunction with UCSF Chimera 1.15v and CB-Dock – these values are show in Table 3.28–30 The docking box was chosen within Chimera to encompass the entire quadruplex model, thus allowing Vina to search the whole of the target space. The docking box was larger than the upper size recommended for use with Autodock Vina. Therefore blind docking was also carried out using the CB-Dock web server, which uses cavity detection based on analysis of the solvent accessible surface to identify potential docking sites in the target, followed by the use Autodock Vina to generate and score the ligand poses at the detected dicking sites. Because the same scoring function is used by both methods the docking scores are directly comparable. The results are very close to those from Autodock Vina and Chimera which forgoes the cavity detection step. Details of the parameters and programs used can be found in ESI† S1. Energy minima scores were obtained and are shown against IC50 and Δδ values shown in Table 3. Interestingly, ligand 2a gave the best apparent fit to both quadruplex structures as indicated in Fig. 4. It should be noted that there is a correlation between IC50 and all of the docking data, with the exception of 2c, which appears to be an outlier. There appears to be preferential binding to the end of the quadruplex as opposed to intercalation. There is further evidence of groove binding.
1 stoichiometry between dye and G-tetrads in quadruplex DNA. It also illustrated that there was a tight complex formed between the two. Interestingly they showed that in the presence of K+ ions, this binding seemed to disappear.27 To attempt to rationalise the chemical shift difference and IC50 values observed, computational modelling of the 5 ligands with parallel (PDB entry139d) and antiparallel (PDB entry 6IMS) quadruplex structures was undertaken using Autodock Vina both in conjunction with UCSF Chimera 1.15v and CB-Dock – these values are show in Table 3.28–30 The docking box was chosen within Chimera to encompass the entire quadruplex model, thus allowing Vina to search the whole of the target space. The docking box was larger than the upper size recommended for use with Autodock Vina. Therefore blind docking was also carried out using the CB-Dock web server, which uses cavity detection based on analysis of the solvent accessible surface to identify potential docking sites in the target, followed by the use Autodock Vina to generate and score the ligand poses at the detected dicking sites. Because the same scoring function is used by both methods the docking scores are directly comparable. The results are very close to those from Autodock Vina and Chimera which forgoes the cavity detection step. Details of the parameters and programs used can be found in ESI† S1. Energy minima scores were obtained and are shown against IC50 and Δδ values shown in Table 3. Interestingly, ligand 2a gave the best apparent fit to both quadruplex structures as indicated in Fig. 4. It should be noted that there is a correlation between IC50 and all of the docking data, with the exception of 2c, which appears to be an outlier. There appears to be preferential binding to the end of the quadruplex as opposed to intercalation. There is further evidence of groove binding.
| Compound | Parallel docking score | Antiparallel docking score | ||||
|---|---|---|---|---|---|---|
| Chimera | CB-Dock | Chimera | CB-Dock | Δδ (ppm) | IC50 (μM) | |
| 2c | −6.8 | −7.2 | −7.3 | −7.8 | 0.25 | 30.6 |
| 2b | −6.4 | −6.3 | −6.2 | −6.5 | 0.34 | 24.4 |
| 2d | −6.6 | −6.9 | −6.4 | −6.6 | 0.13 | 23.84 |
| 2e | −6.7 | −6.8 | −6.4 | −7.0 | 0.24 | 20.5 |
| 2a | −8.1 | −8.1 | −7.7 | −7.8 | 0.2 | 18 |
| Doxorubicin17 | 0.31 | 0.374 | ||||
| Nemorubicin17 | 0.31 | 0.08 | ||||
| RHPS4 (ref. 32) | — | 0.2 | ||||
| BRACO19 (ref. 33) | — | 5.25 | ||||
| Quarfloxin34 | — | 4.44 | ||||
| BMH-21 (ref. 34) | — | 0.46 | ||||
| CX-5461 (ref. 34) | — | 6.89 | ||||
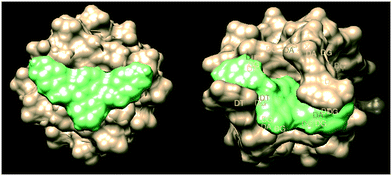 | ||
| Fig. 4 Surface energy model of ligand 2a intercalating parallel (PDB entry139d) and antiparallel (PDB entry 6IMS) G-quadruplex strands (UCSF Chimera 1.15). | ||
Compound 2b, which has a propylmorpholine group has been implemented in the design of compounds for telomeric and oncogenic G-quadruplex affinity with varying degrees of success. It was shown through molecular docking analysis that compounds made with propylmorpholine interact with the groove and or loop of the G-quadruplex, which supports our stacking hypothesis through interaction with these external components. This study also showed inhibition in the growth of A549 cancer cell lines with low cytotoxicity to normal cells. This study concluded that addition of propylmorpholine to naphthalimide supports tumor cell cytotoxicity selectivity.21,31
Looking at Table 3, we see the IC50's of doxorubicin and nemorubicin are much lower than the IC50 values of our synthesised compounds, however we do see that their Δδ is comparable to our two lowest IC50 compounds. This suggests a similar interaction mechanism where a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, with planar stacking on the G6 tetrad.17 This however does show that G-quadruplex binding may start through this stacking and be an initiating step for cytotoxicity, but we see from these results that side chains give comparable chemical shift differences and intercalation stoichiometry but produce very different IC50 values. This however could be explained by selectivity measures. Comparisons of other G-quadruplex ligands and their IC50 values to synthesised ligands shows promise for these compounds as scaffolds for drug discovery as we see the synthesised compounds IC50 values are within the same order as previously synthesised G-quadruplex ligands.
1 stoichiometry, with planar stacking on the G6 tetrad.17 This however does show that G-quadruplex binding may start through this stacking and be an initiating step for cytotoxicity, but we see from these results that side chains give comparable chemical shift differences and intercalation stoichiometry but produce very different IC50 values. This however could be explained by selectivity measures. Comparisons of other G-quadruplex ligands and their IC50 values to synthesised ligands shows promise for these compounds as scaffolds for drug discovery as we see the synthesised compounds IC50 values are within the same order as previously synthesised G-quadruplex ligands.
Conclusions
The promotion of quadruplex DNA formation as observed in solution by NMR, and initial biological testing, suggests the class of G-quadruplex ligand based on imine coupled ligands possess an anticancer activity. Whilst, compared to other synthesised G-quadruplex ligands, we see a poorer IC50 value for these preliminary imine coupled anthracene derivatives, the propensity for intercalation to G-quadruplex and simple synthesis allows for extensive expansion of imine coupled anthracene derivatives as scaffolds for further modification and drug discovery. Whilst this communication expands incrementally on the planar anthracene arrays, already evaluated for G-quadruplex stabilisation, there is a requirement for further investigation to identify the specific G-quadruplexes targeted. E.g. telomere or oncogene promoter regions. However, as discussed in a recent review much of the efforts over the past decade have been focused on achieving selectivity for a specific G-quadruplex.35 Despite this, there is evidence that the most clinically efficacious G-quadruplex ligands are those that target multiple types of G-quadruplexes, such as in the case of triarylpyridine 20A.36Whilst there are a number of G-quadruplex ligands currently in clinical trials, there are, as yet no ligands approved for clinical use. The formation of more G-quadruplex structures during the S phase of the cell cycle in neoplasia continues to endorse the need for a greater range of potential ligands, both to probe selectivity and as lead compounds for therapeutic agents as more is understood about how these structures are modulated in human cells at different points within the cycle, having a diverse range of potential ligands may help to make personalised cancer chemotherapy a reality.
General preparation example – 2c
Tryptamine (390 mg, 0.00245 mol) and 9,10-dibromocarbaldehyde (250 mg) were stirred in dry DCM (40 ml) over molecular sieves. The reaction mixture was refluxed for 12 h under N2 environment. The resultant mixture was monitored via TLC which showed the consumption of both starting materials. After TLC analysis, the crude mixture was left to cool down to room temperature. The resulting yellow solid 2c was filtered and it was dried under vacuum before being recrystallised from methanol to give (0.44 g, 80%) yield. 1H NMR (400 MHz; d6-DMSO) δ = 3.68 (4H, t, J = 6.65 Hz, CH-8) 4.26 (4H, t, J = 8.66 Hz, CH2-7), 7.01 (2H, t, J = 8.76 Hz, CH-6), 7.10 (2H, t, J = 8.78 Hz, CH-5) 7.23 (2H, s, CH-4), 7.39 (2H, dddd, J = 8.0, 1.2, 0.5, 0.5 Hz, H-10), 7.424 (4H, dd, J = 6.63, 2.21 Hz, CH-3), 7.695 (2H, d, J = 7.74 Hz, CH-9), 8.123 (4H, dd, J = 6.97,2.68, CH-2)13C-NMR (600 MHz; d6-DMSO) δ = 22.5, 46.9, 111.6, 112, 118.7, 120.2, 121.7, 122.2, 126.7, 127.5, 127.6, 130.7, 130.8, 136.4, 159.9 IR (ATR): v = 2843.46, 2922.6, 1652.99, 1621.80, 741.77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 755.12 HR MS (ES) m/z = found 519.2541, requires 520.6642 [M + 2H]+.
755.12 HR MS (ES) m/z = found 519.2541, requires 520.6642 [M + 2H]+.
Conflicts of interest
There is no conflict of interest to declare.References
- C. W. Greider and E. H. Blackburn, Identification of a specific telomere terminal transferase activity in Tetrahymena extracts, Cell, 1985, 43, 405–413 CrossRef CAS PubMed.
- E. H. Blackburn, Telomeres and telomerase: their mechanisms of action and the effects of altering their functions, FEBS Lett., 2005, 579, 859–862 CrossRef CAS PubMed.
- J. Zhou, A. Bourdoncle, F. Rosu, V. Gabelica and J. Mergny, Tri-G-Quadruplex: Controlled Assembly of a G-Quadruplex Structure from Three G-Rich Strands, Angew. Chem., Int. Ed., 2012, 51, 11002–11005 CrossRef CAS PubMed.
- E. Y. N. Lam, D. Beraldi, D. Tannahill and S. Balasubramanian, G-quadruplex structures are stable and detectable in human genomic DNA, Nat. Commun., 2013, 4, 1–8 Search PubMed.
- S. A. Ohnmacht, C. Marchetti, M. Gunaratnam, R. J. Besser, S. M. Haider, G. Di Vita, H. L. Lowe, M. Mellinas-Gomez, S. Diocou, M. Robson and J. Šponer, A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer, Sci. Rep., 2015, 5, 11385 CrossRef PubMed.
- C. Leonetti, M. Scarsella, G. Riggio, A. Rizzo, E. Salvati, M. D'Incalci, L. Staszewsky, R. Frapolli, M. F. Stevens, A. Stoppacciaro and M. Mottolese, G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors, Clin. Cancer Res., 2008, 14, 7284–7291 CrossRef CAS PubMed.
- J. L. Mergny and C. Hélène, G-quadruplex DNA: A target for drug design, Nat. Med., 1998, 4, 1366–1367 CrossRef CAS PubMed.
- A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef CAS PubMed.
- D. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, Inhibition of human telomerase by a G-quadruplex-interactive compound, J. Med. Chem., 1997, 40, 2113–2116 CrossRef CAS PubMed.
- K. Shin-ya, K. Wierzba, K. I. Matsuo, T. Ohtani, Y. Yamada, K. Furihata, Y. Hayakawa and H. Seto, Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus, J. Am. Chem. Soc., 2001, 123, 1262–1263 CrossRef CAS PubMed.
- M. Y. Kim, H. Vankayalapati, K. Shin-Ya, K. Wierzba and L. H. Hurley, Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex, J. Am. Chem. Soc., 2002, 124, 2098–2099 CrossRef CAS PubMed.
- A. M. Burger, F. Dai, C. M. Schultes, A. P. Reszka, M. J. Moore, J. A. Double and S. Neidle, The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function, Cancer Res., 2005, 65, 1489–1496 CrossRef CAS PubMed.
- A. Le Gresley, A. Abdullah, D. Chawla, P. Desai, U. Ghosh, U. Gollapalli, M. Kiran, S. Lafon and A. Sinclair, Diacrylamides as selective G-quadruplex ligands in in vitro and in vivo assays, MedChemComm, 2011, 2, 466–470 RSC.
- D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten and J. K. Lim, Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis, Cancer Res., 2009, 69, 7653–7661 CrossRef CAS PubMed.
- A. Le Gresley and N. Kuhnert, The design and synthesis of acrylato and imino derivatives of calix [4] arene for applications in static and dynamic combinatorial libraries, J. Chem. Res., 2010, 34, 61–67 CrossRef CAS.
- F. Malz and H. Jancke, Validation of quantitative NMR, J. Pharm. Biomed. Anal., 2005, 38, 813–823 CrossRef CAS PubMed.
- L. Scaglioni, R. Mondelli, R. Artali, F. R. Sirtori and S. Mazzini, Nemorubicin and doxorubicin bind the G-quadruplex sequences of the human telomeres and of the c-MYC promoter element Pu22, Biochim. Biophys. Acta, Gen. Subj., 2016, 1860, 1129–1138 CrossRef CAS PubMed.
- R. K. Moyzis, J. M. Buckingham, L. S. Cram, M. Dani, L. L. Deaven, M. D. Jones, J. Meyne, R. L. Ratliff and J. R. Wu, A highly conserved repetitive DNA sequence, (TTAGGG) n, present at the telomeres of human chromosomes, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 6622–6626 CrossRef CAS PubMed.
- S. Das, C. J. da Silva, M. D. Silva, M. D. Dantas, Â. de Fátima, A. L. Ruiz, C. M. da Silva, J. E. de Carvalho, J. C. Santos, I. M. Figueiredo and E. F. da Silva-Júnior, Highly functionalized piperidines: Free radical scavenging, anticancer activity, DNA interaction and correlation with biological activity, Arabian J. Chem., 2018, 9, 51–61 CAS.
- A. Głuszyńska and B. Juskowiak, Practical Microwave Synthesis of Carbazole Aldehydes for the Development of DNA-Binding Ligands, Molecules, 2019, 24, 965 CrossRef PubMed.
- A. Ou, A. Guédin, B. W. Skelton, S. Amrane, C. W. Evans, M. Norret, K. S. Iyer, J. L. Mergny and N. M. Smith, Multicarbazole scaffolds for selective G-quadruplex binding, Chem. Commun., 2018, 54, 9647–9650 RSC.
- W. K. Nichols, R. Mehta, K. Skordos, K. Macé, A. M. Pfeifer, B. A. Carr, T. Minko, S. W. Burchiel and G. S. Yost, 3-methylindole-induced toxicity to human bronchial epithelial cell lines, Toxicol. Sci., 2003, 71, 229–236 CrossRef CAS PubMed.
- E. Rajanarendar, K. G. Reddy, S. Ramakrishna, M. N. Reddy, B. Shireesha, G. Durgaiah and Y. N. Reddy, Synthesis and in vitro and in vivo anticancer activity of novel 3-methyl-5H-isoxazolo [5′, 4′: 5, 6] pyrido [2, 3-b] indoles, Bioorg. Med. Chem. Lett., 2012, 22, 6677–6680 CrossRef CAS PubMed.
- S. D. Gupta, G. B. Rao, M. K. Bommaka, N. M. Raghavendra and S. Aleti, Eco-sustainable synthesis and biological evaluation of 2-phenyl 1, 3-benzodioxole derivatives as anticancer, DNA binding and antibacterial agents, Arabian J. Chem., 2016, 9, S1875–S1883 CrossRef.
- M. Baginski and K. Serbakowska, In silico design of telomerase inhibitors, Drug Discovery Today, 2020, 25, 1213–1222 CrossRef CAS PubMed.
- J. Nygren, N. Svanvik and M. Kubista, The interactions between the fluorescent dye thiazole orange and DNA, Biopolymers, 1998, 46, 39–51 CrossRef CAS PubMed.
- I. Lubitz, D. Zikich and A. Kotlyar, Specific high-affinity binding of thiazole orange to triplex and G-quadruplex DNA, Biochemistry, 2010, 49, 3567–3574 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, J. Comput. Chem., 2010, 31, 455–461 CAS.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, UCSF Chimera--a visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- Y. Liu, M. Grimm, W. T. Dai, M. C. Hou, Z. X. Xiao and Y. Cao, CB-Dock: a web server for cavity detection-guided protein–ligand blind docking, Acta Pharmacol. Sin., 2020, 41, 138–144 CrossRef CAS PubMed.
- Z. Ou, Z. Li, Y. Gao, W. Xing, H. Jia, H. Zhang and N. Yi, Novel triazole and morpholine substituted bisnaphthalimide: Synthesis, photophysical and G-quadruplex binding properties, J. Mol. Struct., 2019, 1185, 27–37 CrossRef CAS.
- J. Cookson, F. Dai, V. Smith, R. A. Heald, C. A. Laughton, M. F. Stevens and A. M. Burger, Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor RHPS4 in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents, Mol. Pharmacol., 2005, 68, 1551–1558 CrossRef CAS PubMed.
- A. Guimaraes and D. Oliveira, Effect of long term treatment with BRACO 19 on HeLa proliferation and senescence, Cancer Chemistry, 2017, Abstract 3476 Search PubMed.
- L. E. Kerry, E. E. Pegg, D. P. Cameron, J. Budzak, G. Poortinga, K. M. Hannan, R. D. Hannan and G. Rudenko, Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis, PLoS Neglected Trop. Dis., 2017, 11, e0005432 CrossRef PubMed.
- J. Carvalho, J. L. Mergny, F. Gilmar, F. Salgado, J. A. Queiroz and C. Cruz, G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies, Trends Mol. Med., 2020, 26, 848–861 CrossRef CAS PubMed.
- J. Beauvarlet, P. Bensadoun, E. Darbo, G. Labrunie, B. Rousseau, E. Richard, I. Draskovic, A. Londono-Vallejo, J. W. Dupuy, R. Nath Das, A. Guédin, G. Robert, F. Orange, S. Croce, V. Valesco, P. Soubeyran, K. M. Ryan, J. L. Mergny and M. Djavaheri-Mergny, Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells, Nucleic Acids Res., 2019, 47, 2739–2756 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0md00428f |
| This journal is © The Royal Society of Chemistry 2021 |