Open Access Article
Open Access ArticleMagnetic polymer nanomaterials for sample pretreatment in proteomics†
Bin
Luo
a,
Lingzhu
Yu
ab,
Jia
He
a,
Zhiyu
Li
a,
Fang
Lan
 *ab and
Yao
Wu
*ab and
Yao
Wu
 *ab
*ab
aNational Engineering Research Center for Biomaterials, Sichuan University, Chengdu 610064, P. R. China. E-mail: fanglan@scu.edu.cn; wuyao@scu.edu.cn
bCollege of Biomedical Engineering, Sichuan University, Chengdu 610064, P. R. China
First published on 18th February 2021
Abstract
Protein post-translational modifications (PTMs) play important roles in the regulation of protein functions and cellular processes, which cannot be easily analyzed due to a large number of obstacles, such as the substoichiometric level of PTM proteins, the diversity and complexity of PTM proteins, and the high-intensity interference from ordinary proteins. Therefore, the selective and effective separation of trace amounts of PTM peptides from complex samples is the first crucial step for PTMs. In order to meet the aforesaid requirements, magnetic polymer nanomaterials have emerged as highly effective agents for sample pretreatment in PTM proteomics. In the present review, the recent design of magnetic polymer nanomaterials and their applications in two typical types of protein PTMs (glycosylation and phosphorylation) are summarized. In addition, the development trend of magnetic polymer nanomaterials in PTM proteomics is discussed.
1. Introduction
In spite of great advancements in genetics, the complex living systems are still far from complete understanding, and the main reason behind it is that living systems are far more complex than those predicted by nucleotides. In the complex process of transcription to translation, dozens of proteins can be produced at a single gene site through multiple expression controls, such as RNA splicing and selective translation initiation.1,2 In addition, some new regulatory mechanisms, such as the regulation of small RNAs, are also involved in complex gene expression processes, producing more unpredictable proteins.3 Therefore, as the direct executor of living systems, proteins have attracted great attention in bioscience research. As an important complement to genomics, proteomics is developed to comprehensively explore the biological processes at the level of proteins. Proteomics essentially involves studying the characteristics of proteins on a large scale, including the expression level of proteins, post-translational modifications (PTMs), and protein–protein interactions.4,5In comparison to ordinary proteomics, PTM proteomics provides a better understanding of physiological and pathological processes in organisms.6,7 More than 300 different types of protein PTMs, such as glycosylation, acetylation, phosphorylation, and ubiquitination, have been confirmed till now.8 Protein PTMs are closely related to various cellular processes, gene expression regulation, signal transduction pathway regulation, and protein kinases activation.9–11 Among them, protein glycosylation and phosphorylation are widely studied because of the close relationship between their abnormalities and diseases.12 Vasaikar et al. employed phosphorylated proteomics analysis to prove that RB1 protein hyperphosphorylation was associated with increased proliferation and decreased apoptosis of colon cancer cells, which explained why the tumor suppressor gene RB1 was amplified in colon tumors.13 Recently emerging evidence suggests that phosphoprotein stathmin is highly expressed in different types of human tumor tissues, such as breast cancer, oral carcinoma, liver cancer, and lung cancer.14 For protein glycosylation, various glycoproteins have been confirmed as cancer biomarkers. For example, glycoprotein carbohydrate antigen 15-3 (CA15-3) is the routine biomarker for the screening and monitoring of breast cancer.15 A high level of carbohydrate antigen 12-5 (CA12-5) is required for gastric cancer and ovarian cancer.16 The combined detection of CA12-5 and CA15-3 is essential for the diagnosis of primary ovarian cancer.17 Recently, the researchers illuminated the role of protein glycosylation in the heterogeneity of serous ovarian cancer (HGSC), showing the close relationship between the N-glycan structure and tumor molecular subtypes.18
In recent years, advanced mass spectrometry instruments and technologies have been used to achieve fast, accurate, sensitive, and high-throughput protein detection, providing a solid foundation for proteomics research.19,20 However, mass spectrometry-based PTM proteomics still faces various obstacles and challenges. The first obstacle originates from the complexity of protein samples. The charge state, molecular size, hydrophilicity, and conformation of proteins cause difficulties in mass detection. In addition, proteins often coexist with other biological macromolecules, such as DNA and fatty acid.21 The second obstacle occurs due to the low abundance of PTM proteins and the high abundance of interfering proteins in real biological samples. For example, 22 high-abundance proteins account for 99% of total serum proteins.22 Therefore, it is difficult to efficiently identify target PTM proteins by mass spectrometry. The third obstacle comes from the diversity and complexity of PTM proteins. For example, reversible phosphorylation and multiple phosphorylation sites make the detection difficult.23 Various glycan molecules modified on proteins make glycosylation more complicated than protein phosphorylation. Hence, the elimination of interfering substances and the improvement of the purity and relative abundance of PTM proteins are the main prerequisites for MS-based proteomics. In recent years, nanomaterials combined with the solid-phase extraction technique have emerged as a highly effective means for the separation and enrichment of proteins. Due to their high specific surface area and abundant affinity sites, nanomaterials play an important role in proteomics research.
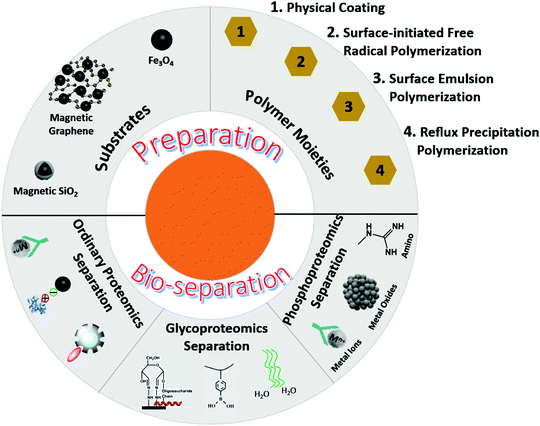
In order to meet the requirements of biological separation, nanomaterials generally include two parts—substrates and functional moieties (Fig. 1). Substrates are responsible for biological separations and act as carriers for functional molecules. Functional moieties are designed to capture targets. Magnetic nanoparticles, especially iron oxides, are widely used for magnetic resonance imaging, sensing, targeted drug delivery, and biological separation.24 Due to their low toxicity, fast magnetic response, stable physical and chemical properties, and flexible surface functionalization, magnetic Fe3O4 nanoparticles are most commonly used in proteomics by introducing different feasible modifications on their surfaces. Polymers are ideal and excellent Fe3O4 modifiers for bio-separation applications. Polymers possess the following inherent characteristics: (1) high surface area, (2) abundant affinity sites, (3) adjustable flexible and rigid structure, (4) flexible modification, and (5) effective interaction space with target molecules. Therefore, magnetic polymer nanomaterials have emerged as highly effective agents for sample pretreatment in proteomics.25 In addition, the magnetic smart polymer nanomaterials provide a series of material platforms by integrating different functional monomers into polymer backbones, and these polymers can serve as stimulus-responsive units or biomolecular recognition units. Through the conformational transformation of polymer chains and the synergistic interactions among functional molecules, guest biomolecules, and polymer backbones, weak biomolecular recognition signals can be captured and recognized.26
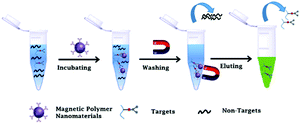
 | ||
| Fig. 1 Schematic illustration of the preparation and bioseparation application of magnetic polymer nanomaterials. | ||
In the current review, the design and application of magnetic polymer nanomaterials for sample pretreatment in proteomics are analyzed. First, different preparation methods of magnetic polymer nanomaterials are summarized, and their capture performance in ordinary protein separation is discussed. Second, the applications of magnetic polymer nanomaterials in glycoproteomics and phosphoproteomics are enumerated based on their enrichment and separation mechanisms. Finally, the prospects of magnetic polymer nanomaterials in proteomics are delineated.
2. Preparation methods of magnetic polymer nanomaterials
Magnetic polymer nanomaterials usually consist of magnetic matrices and polymer moieties, and their preparation strategies mainly include (i) physical coating method, (ii) surface-initiated free radical polymerization, (iii) emulsion polymerization, and (iv) reflux precipitation polymerization.2.1 Physical coating
Magnetic nanoparticles and polymers are prepared separately, and then two moieties are combined through electrostatic interactions, hydrogen bonding, van der Waals force, and cross-linkers (Fig. 2). Yang et al. prepared a new type of magnetic polymer nanomaterial based on an improved emulsion cross-linking method. Fe3O4 nanoparticles and carboxymethyl chitosan (CMCS) were ultrasonically dispersed in water and mixed with paraffin wax. Fe3O4/CMCS composite nanospheres were obtained through the participation of the surfactant and the cross-linking agent (genipin).27 Masoudi et al. synthesized Fe3O4/PEG nanoparticles by adding magnetic iron oxide nanoparticles into a polyethylene glycol (PEG) solution under mechanical stirring for 24 hours.28 According to application requirements, different polymers, such as polyvinyl alcohol (PVA), polyvinyl pyrrolidone (PVP), and polyacrylic acid (PAA), can be modified on magnetic nanoparticle surfaces by physical coating. However, due to the non-covalent bindings of the coated materials, the prepared magnetic polymer nanomaterials are easily affected by complex application environments, such as pH and temperature. | ||
| Fig. 2 Schematic illustration of the preparation of magnetic polymer nanomaterials by a physical coating method. | ||
2.2 Surface-initiated free radical polymerization
Since 1995, atom transfer radical polymerization (ATRP) has become the common method to synthesize polymers with precisely controlled molecular structures. Performing ATRP polymerization on the surface of the substrate is known as surface-initiated ATRP. Recently, the surface-initiated ATRP method has become a popular approach to prepare magnetic polymer nanomaterials. First, the magnetic nanoparticles are modified with an initiator to become macromolecular initiators, and then dispersed in the required monomer solution to initiate the polymerization reaction with suitable catalysts. Based on the principle of monomer polymerization, the magnetic polymer nanomaterials were successfully prepared, which can effectively control the molecular weight of the polymer. Notably, initiators on the surface of magnetic nanoparticles are the most important agents in surface-initiated ATRP. In order to introduce an initiator, surfaces of magnetic nanoparticles are generally coated with CMCS, silicon dioxide (SiO2), and polydopamine (PDA). Wu et al. anchored chloroacetic acid (initiator) on Fe3O4 nanospheres through coordination exchange and then modified polyacrylic acid polymer brushes on the nanospheres using acrylic acid to obtain magnetic polymer nanomaterials.29 Yu et al. reported a new type of Fe3O4@PDA@PMAA nanosphere. First, magnetic nanoparticles prepared by the solvothermal method were coated with PDA, which provided active sites to introduce the initiator. Through surface-initiated free radical polymerization, nanoparticles were then modified with PMAA brushes.30 Luo et al. also exploited a similar material covered with PDA before polymerization. Through the self-polymerization of dopamine, surface-initiated free radical polymerization, and modification after polymerization, Fe3O4/PDA/PAMA-Arg nanospheres were finally obtained.31 However, the severe anaerobic and water-free conditions of the surface-initiated ATRP method have affected its wide range of applications for preparing magnetic polymer nanomaterials.2.3 Emulsion polymerization
Water and oil phases containing monomers and magnetic nanoparticles form emulsion droplets in the presence of surfactants under the shear force. Polymerization reactions proceed in emulsion droplets and generate magnetic polymer nanomaterials. It contains these characteristics: there will be two phases of water and oil in the system to form emulsion droplets; the process requires surfactants and shear force; the polymerization reaction takes place in the emulsion droplets. For example, magnetic nanoparticles prepared by pyrolysis and methyl methacrylate (MMA) were dispersed in n-hexane, and sodium dodecylbenzene sulfonate (SDBS) and PVA were dissolved in the aqueous phase. The mixture was sonicated and emulsified to form one small emulsion droplet. The polymerization reaction was initiated by ammonium persulfate (APS) at 80 °C and continued for five hours. In comparison to the conventional double mini-emulsion polymerization, this method can form uniform magnetic polymer nanospheres with high saturation magnetization.32 Lan et al. used Tween-80 as the surfactant and replaced PVA with triethylene glycol (TEG) to prepare another kind of magnetic polymer nanomaterial.33 Notably, the surfactants or stabilizers used in the emulsion polymerization could be difficult to remove, which will affect the subsequent application of the prepared magnetic polymer nanomaterial to a certain extent.2.4 Reflux precipitation polymerization
Unlike emulsion polymerization, reflux precipitation polymerization (RPP) takes advantage of polymers free of any surfactant or stabilizer. In RPP, under a reflux system, monomers and initiators are dissolved in a solvent to form a homogeneous system for polymerization reactions, and subsequently, the as-generated polymer chains precipitated in the continuous medium. A large amount of monomers is generally used to form polymers in the RPP method. As RPP is an easy, fast, efficient, and stable fabrication process, it is an effective approach to prepare different types of polymer-based composites. RPP is also widely employed to prepare magnetic polymer nanomaterials by combining polymers with magnetic nanoparticles. Jia et al. fabricated a Zn(II)-immobilized polyvinyl imidazole (PVIM) shell-coated magnetic microsphere (dubbed Fe3O4@PVIM@Zn(II)) through RPP and the metal-ion affinity strategy. The core–shell magnetic microsphere had a 20 nm-thick polymer layer and high magnetic responsiveness (32.6 emu g−1).34 Yang et al. first prepared poly(AA-co-DMA) nanohydrogels by RPP and then modified the surface with Fe3O4 and DOX to form magnetic DOX-Fe3O4@NG nanocomposites. The as-prepared nanocomposites with surface-tailorable functionalities were used for target drug delivery systems.35 Therefore, the RPP method can also be adopted to prepare magnetic polymer nanomaterials with different structures. However, the reflux operation and post-treatment process make the RPP method slightly cumbersome.3. Magnetic polymer nanomaterials in ordinary proteomics applications
Magnetic polymer nanomaterials manifest superior performance in protein separation (Fig. 3). In ordinary proteomics, the adsorption mechanisms of proteins mainly include electrostatic interactions, molecularly imprinted technique, metal ion coordination, and other strategies.Electrostatic interactions are the main driving forces for ordinary proteome separation. Calatayud et al. performed the in-situ coating of magnetic nanoparticles (MNPs) with polyethyleneimine (PEI) (PEI-MNPs) and poly(acrylic acid) (PAA-MNPs) by a modified oxidative hydrolysis method.36 The PEI-MNP surface had a positive charge, whereas the PAA-MNP surface had a negative charge. These positively and negatively charged MNPs were immersed in a protein-rich biological medium, and the protein was well adsorbed on nanoparticle surfaces through charge interactions. Yang et al. had proposed a modified inverse emulsion cross-linking approach using genipin as a cross-linking agent to prepare uniform Fe3O4/carboxymethyl chitosan (CMCS) composite nanospheres with high saturation magnetization. The surface potential of the nanospheres began to change with the change of pH. Near the isoelectric point of lysozyme (Lyz), negatively charged Fe3O4/CMCS adsorbed positively charged Lyz through charge interactions.27 Similarly, many other researchers have also used negatively charged materials to adsorb positively charged proteins.37–39 It is worth noting that although the adsorption process of proteins using charge interactions is relatively simple, the specific adsorption effect is not good. Impurities are often adsorbed on material surfaces due to charge interactions.
Molecularly imprinted polymers (MIPs) are also commonly used in protein adsorption. An MIP possesses imprinting sites in the synthetic polymer matrix of complementary functional groups and structures and exhibits many significant advantages.40 Li et al. used BSA, N-isopropylacrylamide (NIPAM), methacrylic acid (MAA; functional monomer), and N,N′-methylenebisacrylamide (MBA; cross-linking agent) to form a complex on the surface of magnetic nanoparticles and then removed BSA to form a molecularly imprinted composite.41 The as-prepared composite had a good adsorption capacity and specific recognition for template proteins. Lan et al. synthesized two molecularly imprinted materials for Lyz adsorption using poly(dihydroxyphenylacetic acid) (PDOPA) and polydopamine (PDA) as the polymer matrix.33 These Lyz-imprinted materials had a good imprinting effect on single protein solutions and also exhibited excellent ability to selectively recognize and capture Lyz from protein mixtures. Molecularly imprinted magnetic polymer nanomaterials with extreme specificity and universality are promising agents for capturing ordinary proteins; however, their preparation steps are complicated.
Moreover, the interactions between metal ions and proteins can also be effectively used in protein adsorption. Shukoor et al. reported a new type of Ni+-modified magnetic polymer nanomaterial for His-tagged protein separation.42 Due to the high-affinity binding interactions between Ni+ ions and His-tagged proteins, the selective adsorption of His-tagged proteins was achieved by Ni+-modified magnetic polymer nanomaterials. Xie et al. used polymer spheres as the core, performed the surface modification of Fe3O4 nanoparticles, grew polymer brushes on the modified surface, and finally performed Ni+ modification. The above materials were effective in adsorbing His-tagged proteins.43 Some common strategies have also been designed to capture proteins.44 The structure-selective protein adsorption strategy has also been employed to capture proteins. For example, Yang et al. used ligand-free Fe3O4/CMCS nanoclusters to selectively capture BHB from a model protein mixture. These ligand-free magnetic nanoclusters displayed an excellent structure-selective protein adsorption ability to capture other proteins structurally similar to BHB, such as LYZ and CTP.45
4. Magnetic polymer nanomaterials in glycoproteomics
As one of the most common and critical protein PTMs, glycosylation is involved in multiple biological activities and cellular processes.46,47 Abnormal glycosylation processes, such as aberrant glycan structures and mismatched glycosylation sites, are closely related to cancer and diabetes.48,49 In order to meet the demands of mass spectrometry analyses, the pre-enrichment of glycoproteins or glycopeptides from complicated systems is imperative. Numerous approaches, such as hydrazide chemistry, boronic acid affinity chromatography, hydrophilic interaction liquid chromatography (HILIC), and molecular imprinting, have been developed for glycoprotein/glycopeptide enrichment. In the current section, magnetic polymer nanomaterials designed for glycoprotein/glycopeptide enrichment are discussed.4.1 Boronic-acid-based magnetic polymer nanomaterials in glycoproteomics
Boronic acid groups and cis-diol-containing glycoproteins/glycopeptides can form stable and reversible covalent bonds through pH adjustment.50,51 Based on this mechanism, boronic acids can be used to fabricate affinity materials for the enrichment of glycoproteins/glycopeptides. Among them, phenylboronic acid (PBA) is most widely used in glycoproteomics. However, conventional PBA-based materials prepared using the small molecule modification strategy face many problems, such as low adsorption capacity, poor selectivity, and low sensitivity.52–54 Boronic acid-based magnetic polymer nanomaterials, possessing abundant affinity sites and high specific surface area for interactions, can significantly increase enrichment efficiency. Therefore, various PBA-functionalized magnetic polymer nanomaterials have been developed in recent years. Generally, two strategies are adopted to prepare PBA-functionalized magnetic polymer nanomaterials: (1) polymerization of PBA-containing monomers on the surface of magnetic nanoparticles and (2) modification of PBA on polymers. Our group prepared a novel Fe3O4/carboxymethylated chitosan/poly(3-acrylaminophenylboronic acid) (dubbed Fe3O4/CMCS/PAAPBA) nanosphere by the surface-initiated ATRP method (Fig. 4A).55 The prepared Fe3O4/CMCS/PAAPBA nanospheres possessed a uniform morphology and excellent magnetic properties. Due to the introduction of PAAPBA, these pH-responsive Fe3O4/CMCS/PAAPBA nanospheres could reversibly capture and release glycoproteins with high selectivity and recyclability in a pure protein, a model protein mixture, and a biological sample. Recently, Fe3O4@SiO2@poly(2-aminoethyl methacrylate hydrochloride)-4-carboxyphenylboronic acid (Fe3O4@SiO2@PAMA-CPBA) nanoparticles have been prepared via surface-initiated ATRP. Owing to the synergistic effects of intermolecular B–N bond-based phenylboronic acid affinity, electrostatic interaction, and polymer brush structure, Fe3O4@SiO2@PAMA-CPBA nanoparticles exhibited selectively capture capacity toward glycoproteins under neutral or slightly acidic conditions. The captured glycoproteins were released in alkaline solution because electrostatic repulsions between glycoproteins and nanoparticles were larger than phenylboronic acid affinity, showing an excellent adsorption capacity and selectivity for glycoproteins.56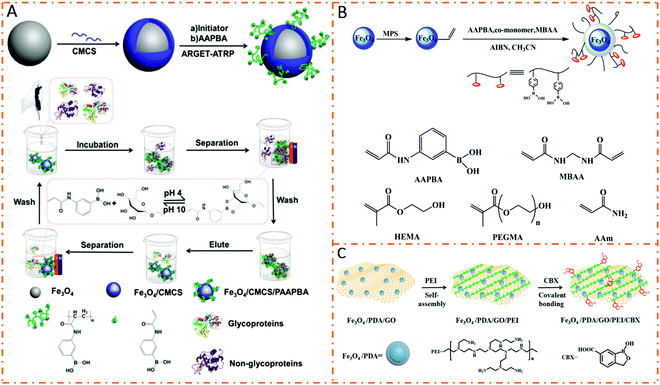 | ||
| Fig. 4 Schematic illustration of the preparation and protein separation procedures of magnetic polymer nanomaterials. (A) Fe3O4/CMCS/PAAPBA. Reproduced from ref. 55 with permission from the Royal Society of Chemistry. (B) Fe3O4@P(AAPBA-co-monomer). Reprinted from ref. 57 with permission from Copyright American Chemical Society. (C) Fe3O4/PDA/GO/PEI/CBX. Reprinted from ref. 61 with permission from Copyright American Chemical Society. | ||
In order to further enhance enrichment capabilities, hydrophilic components are integrated into boronic acid-based magnetic polymer nanomaterials. In the first strategy, boric acid monomers and hydrophilic monomers are copolymerized to enhance the capture performance. Zhang et al. produced hydrophilic Fe3O4@P(AAPBA-comonomer) nanoparticles through copolymerization of two monomers of AAPBA and a hydrophilic functional monomer (Fig. 4B).57 Due to the introduction of hydrophilic molecules, the obtained Fe3O4@P(AAPBA-comonomer) nanoparticles showed better binding capacity for glycoproteins. The other strategy is to introduce hydrophilic substrate materials to form boronic acid-based magnetic polymer nanomaterials. Su et al. used hydrophilic graphene oxide to prepare Fe3O4–GO@PAAPBA as a novel matrix for glycoproteome analyses.58 Large adsorption capacities for glycoproteins OVA and Trf have been achieved.
Recently, boronic acid derivatives have been gradually developed for the enrichment of glycoproteins/glycopeptides. Among them, 3-carboxybenzoboroxole (CBX) having a higher affinity for glycoproteins and a lower pKa value (6.9) is used as a powerful functional molecule to prepare boronate affinity materials. Wu et al. designed and prepared a CBX-modified magnetic dendrimer nanocomposite to enhance glycopeptide enrichment. After enrichment, a total of 1302 glycoproteins with 3185 glycosylation sites were identified in MCF7 cells.59 Wang et al. developed PEI-assisted 3-carboxybenzoboroxole-functionalized magnetic nanoparticles to efficiently capture cis-diol-containing flavonoids under neutral conditions.60 Zhang et al. synthesized CBX-functionalized polyethylenimine (PEI)-modified magnetic graphene oxide nanocomposites (dubbed Fe3O4/PDA/GO/PEI/CBX) for human plasma glycoprotein enrichment under physiological conditions (Fig. 4C).61 Due to the low pKa value (−6.9), the resultant Fe3O4/PDA/GO/PEI/CBX nanocomposites could enrich glycoproteins under physiological conditions even in real human plasma. In addition, the binding capacity of the nanocomposites for HRP glycoprotein was much higher than those of previously reported affinity materials due to the PEI polymer-assisted multivalent binding.
4.2 Hydrazide-chemistry-based magnetic polymer nanomaterials in glycoproteomics
Hydrazide chemistry is also a specific and highly efficient method for glycoprotein/glycopeptide enrichment. In comparison to boronic acid, hydrazide chemistry can only be applied to N-linked glycoprotein/glycopeptide isolation, resulting in relatively higher specificity.62–64 The enrichment mechanism of hydrazide chemistry proposed by Liu et al. is displayed in Fig. 5A.65 First, cis-diol parts of glycoproteins/glycopeptides are translated into aldehydes by oxidation reactions. Second, aldehydes react with hydrazide groups to form covalent bonds. Third, the captured glycoproteins/glycopeptides are released by PNGase F, which specifically cut N-glycans. Traditional affinity materials prepared by small-molecule modification strategies suffer from the low grafting density of hydrazide units, resulting in poor enrichment performance. In order to increase hydrazide affinity sites, various hydrazide-functionalized magnetic polymer nanomaterials have been developed. Lu et al. prepared a core–shell magnetic nanocomposite, in which both the magnetic core and the polymer shell were modified with abundant hydrazide groups, by reflux precipitation polymerization and a post-synthetic modification strategy (Fig. 5B).66 The as-prepared Fe3O4@PMAH nanocomposites exhibited excellent glycopeptide enrichment performance due to the abundant hydrazide affinity sites. For standard glycopeptide enrichment, the Fe3O4@PMAH nanocomposites improved the signal-to-noise ratio by more than five times that of commercial hydrazide resin.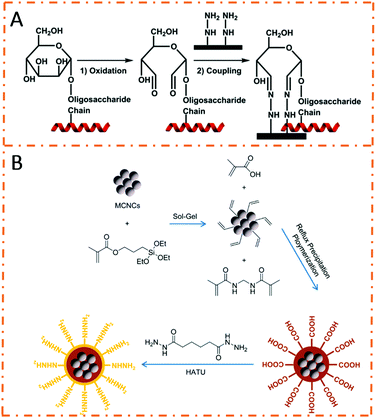 | ||
| Fig. 5 (A) Binding mechanism of hydrazide chemistry for glycopeptide enrichment. Reprinted from ref. 65 with permission from Copyright Wiley-VCH. (B) Schematic illustration of the preparation procedures of Fe3O4@poly(methacrylic hydrazide). Reprinted from ref. 66 with permission from Copyright American Chemical Society. | ||
In addition, after enrichment, 175 unique glycopeptides corresponding to 63 glycoproteins were identified from the serum of a colorectal cancer patient. Qian et al. prepared hydrazide-modified polymer chain-coated magnetic nanoparticles by the surface-initiated ATRP method.67 Thanks to the high efficiency of the ATRP method, the density of hydrazide groups was much higher than that of conventional affinity materials with single-layered hydrazide groups. Therefore, the obtained nanoparticles were endowed with outstanding sensitivity and selectivity toward glycopeptides. A total of 511 unique glycopeptides were detected from mouse liver tissues after enrichment. Generally, hydrazide-based affinity materials possess high specificity for glycopeptide/glycoprotein enrichment. However, hydrazide chemistry is relatively time-consuming and tedious during the pre-enrichment processing of samples as compared to boronic acid chemistry.
4.3 Hydrophilic-interaction-based magnetic polymer nanomaterials in glycoproteomics
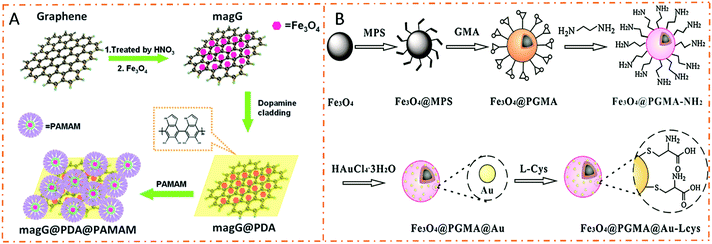
Hydrophilic interaction chromatography (HILIC) is a normal-phase chromatographic mode based on a polar stationary phase and a mobile phase with medium or weak polarity.68 Normally, the polar stationary phase retains the polar solutes due to its higher polarity than the mobile phase. The partitioning mechanism between the stagnant layer of water which results in a hydrated polar stationary phase and the dynamic mobile phase is postulated to be the retention mechanism of HILIC initially. The hydrogen-bonding and electrostatic interactions are discovered to be two main contributors to HILIC retention. Glycopeptides can be easily extracted by HILIC based on different hydrophilic properties of glycopeptides and non-glycopeptides. Therefore, HILIC-based affinity materials are widely employed in glycoproteomics due to their high glycopeptide coverage, excellent reproducibility, and mild enrichment conditions.69–73 Hydrophilic interaction-based magnetic polymer nanomaterials have attracted great attention for glycopeptide enrichment in recent years. Flexible polymeric chains promote the affinity interactions between glycopeptides and polymer molecules, and the adjustable molecular weight and grafting thickness of polymers affect the formation of hydration layers. In addition, various suitable polymer molecules have been developed for HILIC-based affinity materials.74 Among them, carbohydrate and zwitterionic molecules manifest excellent performances in glycopeptide enrichment applications.Poly(ethylene glycol) (PEG) and amino-based polymers are two typical types of hydrophilic polymeric molecules and are widely used in HILIC-based affinity materials. Wang et al. designed and synthesized a kind of dendrimer-modified magnetic graphene@polydopamine@poly(amidoamine) composite (dubbed magG@PDA@PAMAM) for glycopeptide enrichment (Fig. 6A).75 Due to the superhydrophilicity of PAMAM, the resultant magG@PDA@PAMAM composites had outstanding sensitivity (1 fmol mL−1) and high selectivity (HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA molar ratio = 1
BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) for glycopeptide enrichment. Jiang et al. designed and prepared PEG-modified magnetic composites (dubbed GO/Fe3O4/Au/PEG) with high hydrophilic properties and resistance to non-specific adsorption.76 Therefore, these GO/Fe3O4/Au/PEG composites possessed excellent enrichment performance for glycopeptides in complex biological samples. A total of 255 glycopeptides were identified from human plasma after the enrichment process. Recently, Deng et al. prepared PEG-functionalized magnetic nanomaterials through covalent bonding between titanium and sulfur (denoted as Fe3O4@TiO2@PEG).77 Surprisingly, the Fe3O4@TiO2@PEG nanomaterials could enrich 300 intact glycopeptides from only 2 μL of human serum.
100) for glycopeptide enrichment. Jiang et al. designed and prepared PEG-modified magnetic composites (dubbed GO/Fe3O4/Au/PEG) with high hydrophilic properties and resistance to non-specific adsorption.76 Therefore, these GO/Fe3O4/Au/PEG composites possessed excellent enrichment performance for glycopeptides in complex biological samples. A total of 255 glycopeptides were identified from human plasma after the enrichment process. Recently, Deng et al. prepared PEG-functionalized magnetic nanomaterials through covalent bonding between titanium and sulfur (denoted as Fe3O4@TiO2@PEG).77 Surprisingly, the Fe3O4@TiO2@PEG nanomaterials could enrich 300 intact glycopeptides from only 2 μL of human serum.
 | ||
| Fig. 6 Schematic illustration of the preparation procedures of hydrophilic interaction based magnetic polymer nanomaterials in glycoproteomics. (A) magG@PDA@PAMAM. Adapted from ref. 75 with permission from Copyright Royal Society of Chemistry. (B) Fe3O4@PGMA@Au-cys. Adapted from ref. 84 with permission from Copyright Elsevier B.V. | ||
Maltose molecule, as a typical hydrophilic molecule, is widely used to modify magnetic polymer nanomaterials for designing HILIC-based affinity materials. As a consequence, various maltose-functionalized magnetic polymer nanomaterials with excellent performance have been designed for glycopeptide enrichment. Zou et al. prepared maltose-functionalized magnetic nanomaterials (denoted as dM-MNPs) by azide-alkynyl click reactions.78 The dM-MNPs exhibited good performance for glycopeptide enrichment by the HILIC method. Li et al. prepared maltose-functionalized polyethyleneimine (PEI)-coated magnetic nanospheres (dubbed Fe3O4–PEI–maltose) by the “click chemistry” modification method.79 Due to the presence of hydrophilic maltose molecules and branched polyethyleneimine, the Fe3O4–PEI–maltose nanospheres revealed splendid enrichment performances in model proteins. In addition, 1567 unique glycopeptides were identified from 60 μg of a protein sample extracted from mouse liver after enrichment by Fe3O4–PEI–maltose nanospheres. In addition to the above examples, many maltose-modified magnetic polymer nanomaterials are also used for glycopeptide enrichment.80,81
Taking advantage of the superhydrophilicity and ultra-low biofouling capabilities of zwitterionic (ZIC) molecules, thicker hydration layers can be formed around terminal groups of ZIC-based nanomaterials. Therefore, ZIC-HILIC affinity materials exhibit stronger hydrophilic interactions toward glycopeptides. Among them, cysteine and (2-(methacryloyloxy)ethyl)-dimethyl-(3-sulfopropyl)ammonium hydroxide (MSA) are the most widely used zwitterionic molecules for glycopeptide enrichment.82,83 In order to further strengthen the glycopeptide capture effectiveness with multilayer functional molecules, recent research works have revealed the application of ZIC-based magnetic polymer nanomaterials for sample pretreatment in glycoproteomics. Zhao et al. designed and prepared cysteine (Cys)-functionalized magnetic polymer nanomaterials to effectively separate glycopeptides from complex samples. PGMA was prepared on the surface of the Fe3O4 magnetic nanoparticles by one-step reflux–precipitation polymerization. After the modification of Au nanoparticles, zwitterionic cysteine was successfully introduced to obtain Fe3O4@PGMA@Au-cys nanomaterials (Fig. 6B).84 After the treatment with Fe3O4@PGMA@Au-cys nanomaterials, a total of 411 unique glycoproteins were identified from a 75 μg protein sample extracted from mouse liver. Chen et al. reported a kind of PMSA-coated Fe3O4@SiO2 nanosphere for glycopeptide enrichment.85 The as-prepared Fe3O4@SiO2@PMSA nanospheres exhibited high enrichment recovery (73.6%), extremely high sensitivity (0.1 fmol), and binding capacity (100 mg g−1) for glycopeptide enrichment from a tryptic digest of human IgG. In addition, a total of 458 glycoproteins with 905 N-glycosylated sites were identified from mouse liver extracts. Recently, Ji et al. reported another PMSA-based magnetic composite (denoted as Fe3O4@PMSA) for the highly specific enrichment of glycopeptides.86 A summary of HILIC-based magnetic polymer nanomaterials is presented in Table 1. The development and design of various functional molecules with stronger hydrophilic properties will further promote the application of magnetic polymer nanomaterials in glycoproteomics.
| Nanomaterials | Polymer linkers | Hydrophilic moieties | Practical samples | Identified glycoproteins | Identified glycopeptides | Ref. |
|---|---|---|---|---|---|---|
| magG@PDA@PAMAM | PDA | PAMAM | Human serum | — | 11 | 75 |
| GO/Fe3O4/Au/PEG | PEI | PEG | Human serum | 127 | 255 | 76 |
| Fe3O4/TiO2@PEG | — | PEG | Human serum | — | 300 | 77 |
| dM-MNPs | Dendrimers | Maltose | Mouse liver | 418 | 653 | 78 |
| Fe3O4–PEI–maltose | PEI | Maltose | Mouse liver | 684 | 1567 | 79 |
| Fe3O4–PEI-pMaltose | PEI | Maltose | Human renal mesangial cell | 323 | 449 | 80 |
| magG/PDA/Au/L-Cys | PDA | Cys | Human serum | — | 31 | 83 |
| Fe3O4@PGMA@Au-L-cys | PGMA | Cys | Mouse liver | 411 | — | 84 |
| Fe3O4@SiO2@PMSA | — | PMSA | Mouse liver | 458 | — | 85 |
4.4 Molecularly imprinted magnetic polymer manomaterials in glycoproteomics
The molecular imprinting technique, which creates template-shaped cavities with high specificity toward target molecules, is widely used in the fields of separation and purification. Molecularly imprinted polymers (MIPs) are designed and prepared by copolymerizing suitable functional monomers in the presence of templates. MIPs are considered as attractive candidates due to their outstanding advantages, such as high specificity, reversible binding capacity, and applicability in harsh environments. In glycoprotein separation applications, combining molecular imprinting and the boric acid affinity technology is a common strategy to achieve synergistic selectivity toward glycoproteins. Therefore, various molecularly imprinted magnetic polymer nanomaterials with boric acid affinity sites have been reported for glycoprotein separation. Xie et al. prepared magnetic MIP microspheres by the ATRP method using 2-(dimethylamino)ethyl methacrylate and 4-vinylphenylbronic acid as monomers. Meanwhile, the glycoprotein ovalbumin (OVA) was employed as the template (Fig. 7).87 The microspheres showed excellent performance in capturing glycoprotein OVA. Sun et al. synthesized magnetic molecularly imprinted microspheres by employing polydopamine as the polymer matrix and glycoprotein horseradish peroxidase (HRP) and OVA as dual templates.88 The as-prepared microspheres could simultaneously and selectively capture HRP and OVA. Using the same strategy, numerous molecularly imprinted magnetic polymer nanomaterials have been developed to selectively capture glycoproteins.89–91 However, these nanomaterials can only be used to capture a limited amount of glycoproteins. In addition, the molecular imprinting technique is not suitable for glycopeptide enrichment. The lack of universality is a major obstacle to the application of molecular imprinting technology in glycoproteomics.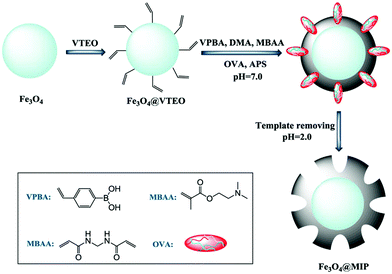 | ||
| Fig. 7 Schematic illustration of the preparation procedures of molecularly imprinted magnetic polymer nanomaterials (Fe3O4@MIP). Reprinted from ref. 87 with permission from Copyright Elsevier B.V. | ||
5. Magnetic polymer nanomaterials in phosphoproteomics
As another ubiquitous PTM, reversible protein phosphorylation plays a vital role in regulating multiple biological processes.92 Abnormal phosphorylation is closely associated with various diseases including cardiovascular diseases, Alzheimer's disease (AD), and cancer.93,94 The effective separation and enrichment of phosphopeptides prior to MS analyses is the prerequisite to analyze phosphoproteomics-related biological and pathological processes. Numerous materials and methods have been developed to selectively enrich phosphopeptides. Affinity methods are mainly divided into immobilized-metal ion affinity chromatography (IMAC), metal oxide affinity chromatography (MOAC), amine-based affinity materials, and some combinations of these methods.95 In order to enhance the enrichment performance of these traditional materials, various IMAC, MOAC, and amine-based magnetic polymer affinity materials have been developed for phosphoproteomics. The present section focuses on the applications of magnetic polymer nanomaterials in phosphoproteomics.5.1 IMAC-based magnetic polymer nanomaterials in phosphoproteomics
IMAC is a widely used enrichment technique in phosphoproteomics. Metal cations interact with phosphate groups through electrostatic interactions and chelation. In order to fabricate IMAC affinity materials, matrix supports carrying chelating ligands are first prepared, and then metal cations are immobilized on chelating ligands. Polymers with abundant functional groups can serve as novel chelating ligands with higher chelating ability, strength, and capacity.PDA is a commendable linker for immobilizing metal ions because it can offer good hydrophilicity, biocompatibility, and abundant active amino and hydroxyl groups.96 Sun et al. developed PDA-coated magnetic graphene/mesoporous silica composites for titanium immobilization (denoted as Ti4+-MGMSs) to selectively enrich phosphopeptides (Fig. 8A).97 The resultant Ti4+-MGMSs presented high enrichment sensitivity (β-casein tryptic digest (0.5 fmol μL−1)) and selectivity (mass ratio of β-casein to BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500)). In addition, Lin et al. reported a kind of magG@PDA-Sn4+ composite by immobilizing Sn4+(IV) ions on polydopamine-coated magnetic graphene.98 The obtained magG@PDA-Sn4+ possessed large surface area, fast magnetic response performance, outstanding hydrophilicity, and strong metal ion bonding ability, leading to high sensitivity (β-casein, 8 fmol) and excellent selectivity (molar ratio of β-casein to BSA (1
500)). In addition, Lin et al. reported a kind of magG@PDA-Sn4+ composite by immobilizing Sn4+(IV) ions on polydopamine-coated magnetic graphene.98 The obtained magG@PDA-Sn4+ possessed large surface area, fast magnetic response performance, outstanding hydrophilicity, and strong metal ion bonding ability, leading to high sensitivity (β-casein, 8 fmol) and excellent selectivity (molar ratio of β-casein to BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000)) toward phosphopeptides. Moreover, magG@PDA-Sn4+ was used to capture endogenous phosphopeptides from human saliva, and a total of 20 phosphopeptides were identified after enrichment. Jiang et al. immobilized different metal ions (Nb5+, Ti4+, Zr4+, Ga3+, Y3+, In3+, Ce4+, and Fe3+) on PDA-coated Fe3O4 (denoted as Fe3O4@PDA-Mn+).99 The microspheres functionalized with different metal ions presented diverse phosphopeptide enrichment efficiencies. Hence, Fe3O4@PDA-Nb5+ and Fe3O4@ PDA-Ti4+ manifested great performances in capturing phosphopeptides with higher selectivity and enhanced sensitivity. Jiang et al. modified the Fe3O4@PDA microspheres with Ti(IV) and Nb(V) ions to prepare Fe3O4@PDA-Ti/Nb composites (Fig. 8B).100 Due to the complementary electrostatic interactions among phosphopeptides, binary Fe3O4@PDA-Ti/Nb microspheres displayed good enrichment efficiency with higher selectivity and sensitivity as compared to single metal ion-loaded microspheres (Fe3O4@PDA-Ti and Fe3O4@PDA-Nb) and their physical mixture.
1000)) toward phosphopeptides. Moreover, magG@PDA-Sn4+ was used to capture endogenous phosphopeptides from human saliva, and a total of 20 phosphopeptides were identified after enrichment. Jiang et al. immobilized different metal ions (Nb5+, Ti4+, Zr4+, Ga3+, Y3+, In3+, Ce4+, and Fe3+) on PDA-coated Fe3O4 (denoted as Fe3O4@PDA-Mn+).99 The microspheres functionalized with different metal ions presented diverse phosphopeptide enrichment efficiencies. Hence, Fe3O4@PDA-Nb5+ and Fe3O4@ PDA-Ti4+ manifested great performances in capturing phosphopeptides with higher selectivity and enhanced sensitivity. Jiang et al. modified the Fe3O4@PDA microspheres with Ti(IV) and Nb(V) ions to prepare Fe3O4@PDA-Ti/Nb composites (Fig. 8B).100 Due to the complementary electrostatic interactions among phosphopeptides, binary Fe3O4@PDA-Ti/Nb microspheres displayed good enrichment efficiency with higher selectivity and sensitivity as compared to single metal ion-loaded microspheres (Fe3O4@PDA-Ti and Fe3O4@PDA-Nb) and their physical mixture.
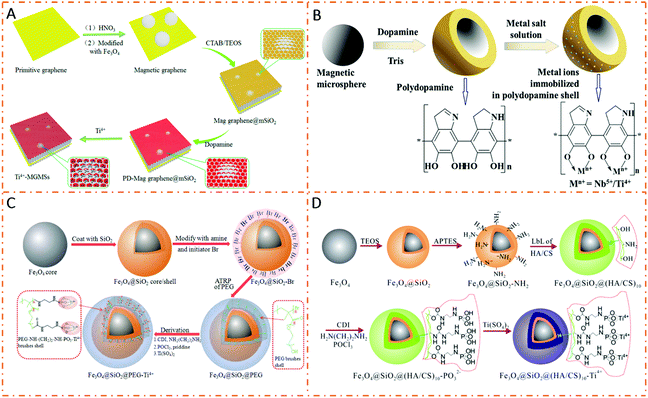 | ||
| Fig. 8 Schematic illustration of the preparation procedures of IMAC-based nanomaterials. (A) Ti4+-MGMSs. Reproduced from ref. 97 with permission from Copyright American Chemical Society. (B) Fe3O4@PDA-Ti/Nb. Reprinted with permission 100. (C) Fe3O4@SiO2@PEG-Ti4+. Adapted from ref. 101 with permission from Copyright Royal Society of Chemistry. (D) Fe3O4@SiO2@(HA/CS)10-Ti4+. Reprinted with permission from ref. 102. | ||
Numerous polymers have also emerged as novel ligands to increase the chelating ability for phosphoproteomics research. Zhao et al. introduced a titanium phosphate moiety on a poly(ethylene glycol)methacrylate (PEG) brush-decorated Fe3O4@SiO2 core–shell nanoparticle (denoted as Fe3O4@SiO2@PEG-Ti4+) (Fig. 8C).101 Due to the enhanced chelating capacity of thick PEG brushes, the Fe3O4@SiO2@PEG-Ti4+ IMAC nanoparticle demonstrated a low limit of detection (β-casein, 0.5 fmol) with an excellent specificity towards a tryptic digest of α-casein and BSA (molar ratio of up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). In the analysis of the real biological samples (Arabidopsis), a superior enrichment performance of Fe3O4@SiO2@PEG-Ti4+ (2447) was achieved compared to those of Fe3O4@SiO2-Ti4+ (1186) and commercial TiO2 microspheres (961). In order to further strengthen the chelation of metal ions, multilayer chelating ligands were developed for metal ion immobilization. For instance, Xiong et al. introduced a titanium phosphate moiety on a cross-linked multilayer polysaccharide (hyaluronate (HA) and chitosan (CS)) to enhance the chelating ability. Meanwhile, the multilayer titanium phosphate modified polysaccharide was coated on the Fe3O4@SiO2 nanoparticle to obtain the Fe3O4@SiO2@(HA/CS)10-Ti4+ nanocomposite (Fig. 8D).102 Due to the hydrophilic properties and high Ti4+ binding capacity of the multilayer chelating ligands, the prepared nanocomposites showed excellent enrichment performance in model proteins. In addition, after treatment with the above nanocomposites, four phosphopeptides from human serum and eleven phosphopeptides from nonfat milk were successfully identified. Ma et al. modified poly(ethylene glycol methacrylate phosphate) (PEGMP) and poly(methylacrylic acid) (PMAA) to a magnetic colloid nanocrystal cluster (MCNC) core through two-step distillation–precipitation polymerization followed by Ti4+ immobilization on the surface of the PEGMP shell (dubbed MCNC@PMAA@PEGMP-Ti4+) for phosphopeptide enrichment.103 Due to the high grafting density of chelating ligands, the MCNC@PMAA@PEGMP-Ti4+ composite microsphere revealed remarkable selectivity (β-casein/BSA molar ratio = 1
2000). In the analysis of the real biological samples (Arabidopsis), a superior enrichment performance of Fe3O4@SiO2@PEG-Ti4+ (2447) was achieved compared to those of Fe3O4@SiO2-Ti4+ (1186) and commercial TiO2 microspheres (961). In order to further strengthen the chelation of metal ions, multilayer chelating ligands were developed for metal ion immobilization. For instance, Xiong et al. introduced a titanium phosphate moiety on a cross-linked multilayer polysaccharide (hyaluronate (HA) and chitosan (CS)) to enhance the chelating ability. Meanwhile, the multilayer titanium phosphate modified polysaccharide was coated on the Fe3O4@SiO2 nanoparticle to obtain the Fe3O4@SiO2@(HA/CS)10-Ti4+ nanocomposite (Fig. 8D).102 Due to the hydrophilic properties and high Ti4+ binding capacity of the multilayer chelating ligands, the prepared nanocomposites showed excellent enrichment performance in model proteins. In addition, after treatment with the above nanocomposites, four phosphopeptides from human serum and eleven phosphopeptides from nonfat milk were successfully identified. Ma et al. modified poly(ethylene glycol methacrylate phosphate) (PEGMP) and poly(methylacrylic acid) (PMAA) to a magnetic colloid nanocrystal cluster (MCNC) core through two-step distillation–precipitation polymerization followed by Ti4+ immobilization on the surface of the PEGMP shell (dubbed MCNC@PMAA@PEGMP-Ti4+) for phosphopeptide enrichment.103 Due to the high grafting density of chelating ligands, the MCNC@PMAA@PEGMP-Ti4+ composite microsphere revealed remarkable selectivity (β-casein/BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), sensitivity (β-casein, 50 fmol), and excellent recovery (87%) toward phosphopeptide enrichment. Poly(vinylphosphonic acid) (PVPA) is also a commonly used polymer for chelating metal ions. For example, Wang et al. introduced poly(vinylphosphonic acid) (PVPA) shells on SiO2-coated Fe3O4 microspheres through a radical polymerization reaction to immobilize Ce(IV) ions (denoted as Fe3O4@SiO2@PVPA-Ce(IV)) for phosphopeptide enrichment.104
500), sensitivity (β-casein, 50 fmol), and excellent recovery (87%) toward phosphopeptide enrichment. Poly(vinylphosphonic acid) (PVPA) is also a commonly used polymer for chelating metal ions. For example, Wang et al. introduced poly(vinylphosphonic acid) (PVPA) shells on SiO2-coated Fe3O4 microspheres through a radical polymerization reaction to immobilize Ce(IV) ions (denoted as Fe3O4@SiO2@PVPA-Ce(IV)) for phosphopeptide enrichment.104
In summary, the inherent performance of magnetic polymer nanomaterials strengthens the chelating ability of metal ions and enhances the enrichment capacity for phosphopeptides based on the IMAC method. The development and design of various chelating ligands with stronger chelating ability will further promote the application of IMAC-based magnetic polymer nanomaterials in phosphoproteomics.
5.2 MOAC-based magnetic polymer nanomaterials in phosphoproteomics
MOAC is another widely used enrichment strategy for phosphopeptides, and it works based on the reversible Lewis acid–base interactions between surface metal cations and phosphate groups. The in situ growth of metal oxides on magnetic polymer surfaces is the main strategy for MOAC-based magnetic polymer nanomaterial design, in which polymers act as growth sites. Liang et al. designed and prepared a functional graphene/Fe3O4/PDA/TiO2 composite for phosphopeptide enrichment.105 The polydopamine layer promoted the growth of TiO2 and decreased the non-specific adsorption to improve selectivity. The material possessed extremely high sensitivity (β-casein, 0.2 amol) and selectivity (β-casein/BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) toward phosphopeptides. Wang et al. prepared FexOy and ZrO2 nanoparticles co-doped layered porous polyimide nanocomposites by a one-step strategy (denoted as PI–FexOy–ZrO2).106 The layered and porous structure of the polymer offered abundant reaction sites for metallic oxide anchoring and provided a large specific surface area. The high doping of ZrO2 facilitated better adsorption performance toward phosphopeptides as compared to commercial titanium dioxide nanoparticles. In addition, multi metal oxides were immobilized on magnetic polymer materials to further improve the ability to capture phosphopeptides. Deng et al. integrated binary metal oxides (ZrO2 and TiO2) into polydopamine-coated magnetic graphene to prepare the magG/PD/(Zr–Ti)O4 nanocomposites (Fig. 9).107 Compared with single metal oxide, the as-prepared magG/PD/(Zr–Ti)O4 composites with binary metal oxides exhibited better enrichment efficiency. This affinity material showed extremely high enrichment performance for phosphopeptides from model proteins and real biological samples. In addition, there are many other MOAC-based magnetic polymer nanomaterials used in phosphoproteomics.108–111 A summary of IMAC/MOAC-based magnetic polymer nanomaterials used in phosphoproteomics is presented in Table 2.
500) toward phosphopeptides. Wang et al. prepared FexOy and ZrO2 nanoparticles co-doped layered porous polyimide nanocomposites by a one-step strategy (denoted as PI–FexOy–ZrO2).106 The layered and porous structure of the polymer offered abundant reaction sites for metallic oxide anchoring and provided a large specific surface area. The high doping of ZrO2 facilitated better adsorption performance toward phosphopeptides as compared to commercial titanium dioxide nanoparticles. In addition, multi metal oxides were immobilized on magnetic polymer materials to further improve the ability to capture phosphopeptides. Deng et al. integrated binary metal oxides (ZrO2 and TiO2) into polydopamine-coated magnetic graphene to prepare the magG/PD/(Zr–Ti)O4 nanocomposites (Fig. 9).107 Compared with single metal oxide, the as-prepared magG/PD/(Zr–Ti)O4 composites with binary metal oxides exhibited better enrichment efficiency. This affinity material showed extremely high enrichment performance for phosphopeptides from model proteins and real biological samples. In addition, there are many other MOAC-based magnetic polymer nanomaterials used in phosphoproteomics.108–111 A summary of IMAC/MOAC-based magnetic polymer nanomaterials used in phosphoproteomics is presented in Table 2.
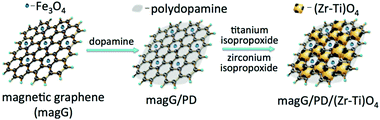 | ||
| Fig. 9 Schematic illustration of the preparation procedures of MOAC-based nanomaterials (magG/PD/(Zr–Ti)O4). Reprinted from ref. 107 with permission from Copyright Wiley-VCH. | ||
| Nanomaterials | Main functional moieties | Linkers | Selectivity | Sensitivity | Practical samples | Identified glycopeptides | Ref. |
|---|---|---|---|---|---|---|---|
| Ti4+-MGMSs | Ti4+ | PDA | — | 0.5 fmol μL−1 | Human saliva | 14 | 97 |
| magG@PDA-Sn4+ | Sn4+ | PDA | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) 1000) |
8 fmol | Human saliva | 20 | 98 |
| Fe3O4@PDA-Mn+ | Nb5+, Ti4+, Zr4+, Ga3+, Y3+, In3+, Ce4+, Fe3+ | PDA | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) 100) |
2 fmol | Nonfat milk | 16 | 99 |
| Fe3O4@PDA-Ti/Nb | Ti4+/Nb5+ | PDA | β-Casein/BSA (mass ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) 1000) |
2 fmol | Nonfat milk | 19 | 100 |
| Fe3O4@SiO2@PEG-Ti4+ | Ti4+ | PEG | α-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) 2000) |
0.5 fmol | Arabidopsis | 2447 | 101 |
| MCNC@PMAA@PEGMP-Ti4+ | Ti4+ | PMAA/PEGMP | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) 500) |
50 fmol | Milk with a fat content of about 3% | 10 | 103 |
| Fe3O4@SiO2@PVPA-Ce(IV) | Ce4+ | PVPA | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) 100) |
500 fmol | Nonfat milk | 9 | 104 |
| PI–FexOy-ZrO2 | ZrO2 | Polyimide | β-Casein/BSA (mass ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) 1000) |
— | Human serum | 4 | 106 |
| MagG/PD/(Zr–Ti)O4 | TiO2/ZrO2 | PDA | β-Casein/BSA (mass ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000) 8000) |
8 fmol | Mouse brain | 622 | 107 |
| MPCS-Ti4+ | Ti4+ | N-Methylene phosphonic chitosan | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) 200) |
— | Human serum | 4 | 108 |
| DFMMOF | Ti4+/Zr–O | PDA | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) 1000) |
5 fmol | Nonfat milk | 15 | 109 |
| MCTiMs | TiO2 | Cellulose | β-Casein/BSA (molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) 1000) |
50 fmol | Human serum | 4 | 110 |
| PEG–Ce/CeO2–Fe3O4 | Ce4+/CeO2 | PEG | α-, β-Casein/BSA (mass ratios = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) 200) |
50 fmol | Nonfat milk | 12 | 111 |
5.3 Amine-based magnetic polymer nanomaterials in phosphoproteomics
Amine-based affinity materials capture phosphopeptides based on the electrostatic attraction and hydrogen bonding interactions between amine groups and phosphate groups. Chen et al. used polyethyleneimine (PEI) to modify magnetic nanoparticles for the preparation of Fe3O4@SiO2@PEI nanoparticles.112 The abundant amine groups on PEI interacted with phosphate groups, leading to high enrichment efficiency toward phosphopeptides. The detection limit for both α-casein and β-casein was 5 fmol, and monophosphopeptides and multiphosphopeptides could be selectively enriched. Recently, our group introduced polyamidoamine dendrimers (PAMAM) to prepare (PAMAM)-grafted poly(methacrylic acid) (PMAA) brush-modified magnetic composite nanospheres (dubbed Fe3O4@PDA@PMAA@PAMAM) (Fig. 10A).30 The extremely abundant amine groups of PAMAM–PMAA brushes and the magnetism of the Fe3O4 core endowed the composite nanospheres with high detection sensitivity (β-casein, 1 fmol mL−1) and excellent selectivity (β-casein/BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500). In addition, the nanospheres exhibited an amazing tunable capturing ability toward global phosphopeptides, monophosphopeptides, or merely multiphosphopeptides based on the tunable bonding strength between PAMAM and phosphate groups.
500). In addition, the nanospheres exhibited an amazing tunable capturing ability toward global phosphopeptides, monophosphopeptides, or merely multiphosphopeptides based on the tunable bonding strength between PAMAM and phosphate groups.
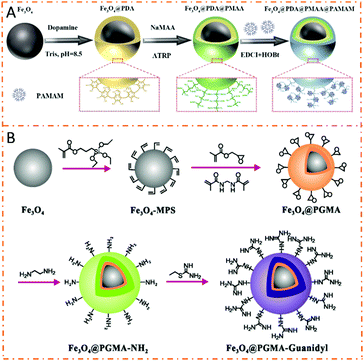 | ||
| Fig. 10 Schematic illustration of the preparation and protein separation procedures of the amine-based magnetic polymer nanomaterials in phosphoproteomics. (A) Fe3O4@PDA@PMAA@PAMAM. Adapted from ref. 30 with permission from Copyright Royal Society of Chemistry. (B) Fe3O4@PGMA-guanidyl. Reprinted with permission 113. | ||
Guanidinium with three amine groups can also strongly interact with phosphate groups by forming steady salt bridge structures. Xiong et al. introduced guanidyl on poly(glycidyl methacrylate) (PGMA)-modified Fe3O4 microspheres (denoted as Fe3O4@PGMA-guanidyl) to selectively enrich global phosphopeptides or only multiphosphopeptides (Fig. 10B).113 Benefitting from the abundant guanidino groups, the composite microspheres achieved high enrichment performance for phosphopeptide enrichment. Notably, the Fe3O4@PGMA-guanidyl possessed a flexible tunable enriching ability for global phosphopeptides or only multiphosphopeptides in both model protein and complex biological samples. Recently, our group fabricated Fe3O4/polydopamine/poly(2-aminoethyl methacrylate hydrochloride)/arginine composite nanospheres (Fe3O4/PDA/PAMA-Arg) that manifested tunable enrichment selectivity toward global phosphopeptides or multiphosphopeptides with extremely high detection sensitivity (β-casein, 0.2 fmol) and selectivity (β-casein/BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) due to the abundant guanidyl and amino groups of PAMA-Arg brushes.31 Employing the materials, 1059 phosphopeptides including 923 monophosphopeptides (87.2%) and 136 multiphosphopeptides (12.8%) were successfully identified from a rat brain lysate digest. It can be seen that the adjustable enrichment performance for phosphopeptides makes the guanidyl-based magnetic polymer nanomaterials possess great application potential.
500) due to the abundant guanidyl and amino groups of PAMA-Arg brushes.31 Employing the materials, 1059 phosphopeptides including 923 monophosphopeptides (87.2%) and 136 multiphosphopeptides (12.8%) were successfully identified from a rat brain lysate digest. It can be seen that the adjustable enrichment performance for phosphopeptides makes the guanidyl-based magnetic polymer nanomaterials possess great application potential.
5.4 Multi-strategy-based magnetic polymer nanomaterials in phosphoproteomics
As different affinity materials exhibit different capturing abilities toward phosphopeptides, the integration of various materials into one system is one of the most interesting topics in phosphoproteomics research. Jiang et al. prepared magnetic hydrazide-functionalized PAMAM dendrimers embedded with TiO2 (denoted as TiHP-MNPs) for phosphopeptide enrichment (Fig. 11).114 The synergistic strength of multiple binding interactions (electrostatic attraction, hydrogen bonding, and Lewis acid–base interaction) contributed to an improved enrichment performance. The specificity toward phosphopeptides was as high as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (mass ratio of β-casein
500 (mass ratio of β-casein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA
BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ovalbumin), and the detection limit of β-casein digests was 0.4 fmol. Recently, our group coated polyvinyl pyrrolidone (PVP) and PEI on Fe3O4 nanoparticles to initiate the epitaxial growth of Zn–MOF and the post-synthetic modification of arginine to prepare the SPIOs@PVP–PEI@MOF@Arg nanospheres (denoted as SPMA nanospheres).115 The integration of multiple affinity sites (Zn–O clusters, NH2, and guanidyl groups) into one material endowed SPMA nanospheres with a high enrichment performance with a low detection limit (β-casein, 10 fmol) and high selectivity (β-casein
ovalbumin), and the detection limit of β-casein digests was 0.4 fmol. Recently, our group coated polyvinyl pyrrolidone (PVP) and PEI on Fe3O4 nanoparticles to initiate the epitaxial growth of Zn–MOF and the post-synthetic modification of arginine to prepare the SPIOs@PVP–PEI@MOF@Arg nanospheres (denoted as SPMA nanospheres).115 The integration of multiple affinity sites (Zn–O clusters, NH2, and guanidyl groups) into one material endowed SPMA nanospheres with a high enrichment performance with a low detection limit (β-casein, 10 fmol) and high selectivity (β-casein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA molar ratio = 1
BSA molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) toward global phosphopeptides. The nanospheres were further used to extract a tryptic digest of rat brain lysate. After enrichment, 1659 unique phosphopeptides (including mono- and multiphosphopeptides) were identified, indicating their good performance toward global phosphopeptides. It can be seen that integrating multiple enrichment methods into a single affinity material will hopefully further improve the enrichment capacity for phosphopeptides.
1000) toward global phosphopeptides. The nanospheres were further used to extract a tryptic digest of rat brain lysate. After enrichment, 1659 unique phosphopeptides (including mono- and multiphosphopeptides) were identified, indicating their good performance toward global phosphopeptides. It can be seen that integrating multiple enrichment methods into a single affinity material will hopefully further improve the enrichment capacity for phosphopeptides.
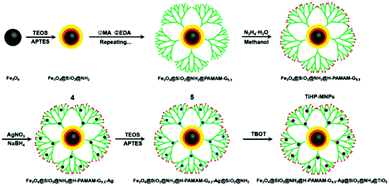 | ||
| Fig. 11 Schematic illustration of the preparation of the magnetic polymer nanomaterials (TiHP-MNPs). Reprinted from ref. 114 with permission from Copyright Elsevier B.V. | ||
5.5 Molecularly imprinted magnetic polymer nanomaterials in phosphoproteomics
Molecularly imprinted polymers (MIPs), as synthetic mimics of antibodies or enzymes, have found important applications in phosphopeptide enrichment. The introduction of MIPs favorably complements the deficiencies of affinity methods and remarkably increases the precise recognition of given phosphorylated amino acid residues (serine, threonine, and tyrosine phosphorylation). Chen et al. developed a dual-template docking-oriented molecular imprinting strategy to prepare molecularly imprinted mesoporous materials (MSNs).116 The resultant MSNs exhibited excellent selectivity, fast binding equilibrium, and large binding capacity toward phosphopeptides. Zhang et al. employed the epitope imprinting strategy to synthesize imprinted mesoporous materials (MIM) with structurally analogous phenylphosphonic acid (acted as the template) and mesoporous materials for the selective recognition of phosphorylated tyrosine.117 The MIM materials had excellent selectivity and a high adsorption capacity toward pTyr peptides from a mixture of angiotensin II peptide and β-casein (pY peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β-casein molar ratio = 1
β-casein molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). However, more smart MIPs and MIPs combined with Fe3O4 are urgently needed to further improve enrichment efficiency.
100). However, more smart MIPs and MIPs combined with Fe3O4 are urgently needed to further improve enrichment efficiency.
5.6 Smart magnetic polymer nanomaterials in phosphoproteomics
Smart polymers can dynamically and reversibly change their structures and functions in response to external stimuli, such as temperature, pH, electric field, and biomolecules. Smart polymers with unique tunability and controllability had a broad prospect in macromolecular separation and purification. Nishio et al. developed dual temperature- and pH-responsive chromatography based on a terpolymer composed of N-isopropylacrylamide, N,N-dimethylaminopropylacrylamide, and butylmethacrylate to analyze phospho-amino acid, phosphopeptide, and oligonucleotides.118 Our group prepared Fe3O4@SiO2@poly(N,N-dimethylamino-2-ethyl methacrylate-co-4-(3-(2-methacryloyloxy)ethyl)-ureido)benzoic acid) composite microspheres through complementary multiple hydrogen-bonding interactions to selectively enrich phosphopeptides (dubbed SPMM) (Fig. 12).119 Driven by specific and tunable hydrogen bonding interactions, smart polymers display tunable adsorption behavior toward phosphorylated and non-modified peptides. The above-mentioned microspheres successfully captured cancer-related phosphopeptides from phosphoprotein Stathmin-1 and identified their relevant phosphorylation sites from the saliva and tissue lysate of oral carcinoma patients, demonstrating their potential toward phosphorylated disease marker detection and discovery. The combination of smart polymers with magnetic Fe3O4 can lead to the development of more intelligent enrichment materials for phosphoproteomics research in the near future.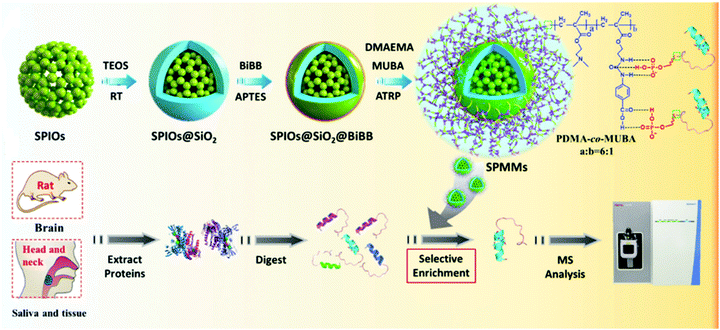 | ||
| Fig. 12 Schematic illustration of the preparation of the magnetic polymer nanomaterials (SPMM) and the general procedure for phosphopeptide extraction, digestion, enrichment, and identification. Adapted from ref. 119 with permission from Copyright Royal Society of Chemistry. | ||
6. Magnetic polymer nanomaterials in multi-PTM proteomics
Generally, a real bio-sample contains multiple PTM proteins. Both protein phosphorylation and glycosylation are closely related to physiological and pathological processes; therefore, it is necessary for proteomics studies to simultaneously obtain information about phosphoproteins and glycoproteins from one sample. Similar to single PTM, the sample pretreatment process for multi-PTMs is indispensable prior to MS analyses. Therefore, the design and preparation of multifunctional nanomaterials to achieve the simultaneous enrichment of phosphopeptides and glycopeptides is an attractive research topic. In recent years, some nanomaterials have been developed to simultaneously capture phosphopeptides and glycopeptides.120–122 Among them, magnetic polymer nanomaterials manifest great advantages. For instance, Hong et al. designed and prepared a kind of microsphere modified with phytic acid (PA) and Ti4+ ions (denoted as Mag-MSMs@PEI–PA–Ti4+) (Fig. 13).123 According to the IMAC and HILIC mechanisms, the as-prepared Mag-MSMs@PEI–PA–Ti4+ microspheres were endowed with excellent properties for the enrichment of phosphopeptides and glycopeptides. For phosphopeptide enrichment, the prepared Mag-MSMs@PEI–PA–Ti4+ microspheres showed outstanding selectivity and high sensitivity in model proteins. After enrichment with the microspheres, a total of 1645 phosphopeptides from 200 μg of HeLa cell extract were successfully detected. For glycopeptide enrichment, the Mag-MSMs@PEI–PA–Ti4+ microspheres also exhibited high selectivity and sensitivity. Meanwhile, 276 N-glycopeptides corresponding to 132 glycoproteins were identified from 2 μL of human serum after the enrichment process. Recently, they also reported another Ti4+-immobilized magnetic graphene nanocomposite (dubbed MagG@PEI@PA–Ti4+) to enrich both phosphopeptides and glycopeptides with outstanding performances.124 Although these affinity materials are endowed with the ability to enrich phosphopeptides and glycopeptides, they cannot achieve the simultaneous enrichment of phosphopeptides and glycopeptides from one buffer solution, leading to several problems including complicated operations, time consumption, and the loss of peptide information. Therefore, the design of magnetic polymer nanomaterials for the simultaneous enrichment of glycopeptides and phosphopeptides may be a key research topic for multi-PTM proteomics in the future.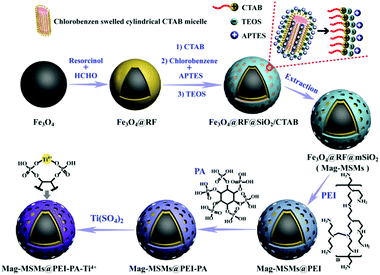 | ||
| Fig. 13 Schematic illustration of the preparation of the IMAC-based magnetic polymer nanomaterials (MagG@PEI@PA-Ti4+). Reprinted from ref. 123 with permission from Copyright Elsevier B.V. | ||
7. Conclusion and perspective
In order to meet the requirements of MS-based proteomics, the purification of target proteins/peptides from complex samples is a crucial prerequisite. Among various affinity materials, magnetic polymer nanomaterials have made great progress in proteomics, especially PTM proteomics. Due to the advantages including mature preparation methods, fast magnetic response, and flexible functionality, magnetic polymer nanomaterials are widely used in the separation and enrichment of proteins/peptides. In the above-discussed review, some most commonly used preparation methods of magnetic polymer nanomaterials are summarized. In ordinary proteomics, magnetic polymer nanomaterials produced by charge interactions and molecular imprinting are mostly used to capture ordinary proteins. In glycoproteomics applications, hydrophilic interaction-driven magnetic polymer nanomaterials manifest great potential in capturing glycoproteins/glycopeptides. It is worth noting that the exploitation of zwitterionic molecules with high hydrophilicity would be a hotspot in material design for glycopeptide enrichment in the future. In phosphoproteomics applications, IMAC/MOAC-based magnetic polymer nanomaterials are mainly used to enrich phosphopeptides. The design of high-performance chelating ligands is the key to ensure the phosphopeptide enrichment ability of IMAC/MOAC-based magnetic polymer nanomaterials. Therefore, magnetic polymer nanomaterials play an important role in sample pretreatment in proteomics.However, there are still numerous challenges involved in designing high-efficiency magnetic polymer nanomaterials for comprehensive analyses of MS-based proteomics. First, the design of novel affinity molecules with high specificity toward target proteins/peptides is highly required. To search for highly specific affinity molecules, large-scale model calculations and molecular docking would be potential techniques. Second, there is currently a lack of multifunctional magnetic polymer nanomaterials to achieve the simultaneous enrichment of multiple PTM peptides. For example it is possible that a bifunctional magnetic polymer nanomaterial could simultaneously enrich glycopeptides and phosphopeptides from one sample. The design and preparation of multifunctional affinity materials will facilitate the comprehensive analysis of proteomics. Third, smart polymers have attracted increasing attention due to their excellent responses to external stimuli and meet the requirements of sample pretreatment in proteomics; thus, smart polymer-based magnetic nanomaterials hold huge potential in proteomics. This review summarizes the applications of magnetic polymer nanomaterials in proteomics and provides references for scientists interested in this field.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the funds of the National Natural Science Foundation of China (No. 31771037), the National Key Research and Development Program of China (No. 2016YFC1100403), and Sichuan International Science and Technology Innovation Cooperation Project (No. 2019YFH0109).References
- M. C. Wahl, C. L. Will and R. Luhrmann, Cell, 2009, 136, 701 CrossRef CAS PubMed.
- P. Van Damme, D. Gawron, W. Van Criekinge and G. Menschaert, Mol. Cell. Proteomics, 2014, 13, 1245 CrossRef CAS PubMed.
- E. Berezikov, G. Van Tetering, M. Verheul, J. van de Belt, E. Wienholds, R. H. Plasterk and E. Cuppen, Genome Res., 2016, 16, 1289 CrossRef PubMed.
- M. Tyers and M. Mann, Nature, 2003, 422, 193 CrossRef CAS PubMed.
- R. E. Banks, M. J. Dunn, D. F. Hochstrasser, J. C. Sanchez, W. Blackstock, D. J. Pappin and P. J. Selby, Lancet, 2000, 356, 1749 CrossRef CAS.
- A. Bah and J. Forman-Kay, J. Biol. Chem., 2016, 291, 6696 CrossRef CAS PubMed.
- V. Csizmok and J. Forman-Kay, Curr. Opin. Struct. Biol., 2018, 48, 58 CrossRef CAS PubMed.
- M. Mann and O. N. Jensen, Nat. Biotechnol., 2003, 21, 255 CrossRef CAS PubMed.
- W. Gruber, T. Scheidt, F. Aberger and C. G. Huber, Cell Commun. Signaling, 2017, 15, 12 CrossRef PubMed.
- E. Amit, R. Obena, Y. T. Wang, R. Zhuravel, A. J. F. Reyes, S. Elbaz, D. Rotem, D. Porath, A. Friedler, Y. J. Chen and S. Yitzchaik, Chem. Sci., 2015, 6, 4756 RSC.
- E. L. Huttlin, M. P. Jedrychowski, J. E. Elias, T. Goswami, R. Rad, S. A. Beausoleil, J. Villen, W. Haas, M. E. Sowa and S. P. Gygi, Cell, 2010, 143, 1174 CrossRef CAS PubMed.
- M. B. Lazarus, Y. S. Nam, J. Y. Jiang, P. Sliz and S. Walker, Nature, 2011, 469, 564 CrossRef CAS PubMed.
- S. Vasaikar, Chen Huang and X. J. Wang, Cell, 2020, 177, 1035 CrossRef PubMed.
- P. Dundr, K. Simon, K. Nemejcova, M. Bartu, I. Ticha, R. Michalkova, R. Jaksa, Z. Veckova and O. Kodet, Melanoma Res., 2019, 29, 157 CrossRef CAS PubMed.
- J. A. Shaw, D. S. Guttery, A. Hills, D. F. Garcia, K. Page, B. M. Rosales, K. S. Goddard, R. K. Hastings, J. L. Luo, O. Ogle, L. Woodley, S. Ali, J. Stebbing and R. C. Coombes, Clin. Cancer Res., 2017, 23, 88 CrossRef CAS PubMed.
- P. Bhatt, I. Vhora, S. Patil, J. Amrutiya, C. Bhattacharya, A. Misra and R. Mashru, J. Controlled Release, 2016, 226, 148 CrossRef CAS PubMed.
- S. Jeonga, M. J. Parka, W. Songa and H. S. Kim, Clin. Biochem., 2020, 78, 43 CrossRef PubMed.
- J. B. Pan, Y. W. Hu and S. S. Sun, Nat. Commun., 2020, 11, 6139 CrossRef CAS PubMed.
- B. Cravatt, G. Simon and J. Yates, Nature, 2007, 450, 991 CrossRef CAS PubMed.
- X. Han, A. Aslanian and J. Yates, Curr. Opin. Chem. Biol., 2008, 12, 483 CrossRef CAS PubMed.
- V. Kolli, K. N. Schumacher and E. D. Dodds, Bioanalysis, 2015, 7, 113 CrossRef CAS PubMed.
- R. Pieper, C. L. Gatlin, A. J. Makusky, P. S. Russo, C. R. Schatz, S. S. Miller, Q. Su, A. M. McGrath, M. A. Estock and P. P. Parmar, Proteomics, 2003, 3, 1345 CrossRef CAS PubMed.
- J. Rush, A. Moritz, K. A. Lee, A. Guo, V. L. Goss, E. J. Spek, H. Zhang, X. M. Zha, R. D. Polakiewicz and M. J. Comb, Nat. Biotechnol., 2005, 23, 94 CrossRef CAS PubMed.
- M. K. Masud, J. Na, M. Younus, M. S. A. Hossain, Y. Bando, M. J. A. Shiddiky and Y. Yamauchi, Chem. Soc. Rev., 2019, 48, 5717 RSC.
- C. Aydoğan, A. Gökaltun, A. Denizli and Z. E. Rassi, J. Sep. Sci., 2019, 42, 962 Search PubMed.
- G. Y. Qing, Q. Lu, X. L. Li, J. Liu, M. L. Ye, X. M. Liang and T. L. Sun, Nat. Commun., 2017, 8, 461 CrossRef PubMed.
- Q. Yang, F. Lan, Z. Y. Liu, S. H. Ma, W. L. Li, Y. Wu and Z. W. Gu, J. Nanosci. Nanotechnol., 2015, 15, 1 CrossRef PubMed.
- A. Masoudi, H. R. Madaah Hosseini, M. A. Shokrgozar, R. Ahmadi and M. A. Oghabian, Int. J. Pharm., 2012, 433, 129 CrossRef CAS PubMed.
- L. Q. Xie, F. Lan, W. L. Li, Z. Y. Liu, S. H. Ma, Q. Yang, Y. Wu and Z. W. Gu, Colloids Surf., B, 2014, 123, 413 CrossRef CAS PubMed.
- L. Z. Yu, B. Luo, Z. Y. Li, J. He, F. Lan and Y. Wu, J. Mater. Chem. B, 2020, 8, 1266 RSC.
- B. Luo, X. Zhou, P. Jiang, Q. Yi, F. Lan and Y. Wu, J. Mater. Chem. B, 2018, 6, 3969 RSC.
- F. Lan, K. X. Liu, W. Jiang, X. B. Zeng, Y. Wu and Z. W. Gu, Nanotechnology, 2011, 22, 225604 CrossRef PubMed.
- F. Lan, S. H. Ma, Q. Yang, L. Q. Xie, Y. Wu and Z. W. Gu, Colloids Surf., B, 2014, 123, 213 CrossRef CAS PubMed.
- Q. Jia, Y. X. Peng, J. M. Pan, X. B. Huang, X. H. Niu and T. Zhang, New J. Chem., 2017, 41, 3308 RSC.
- W. J. Yang, L. j. Liang, X. D. Wang, Y. P. Cao, W. Y. Xu, D. Q. Chang, Y. Gao and L. H. Wang, Biomater. Sci., 2019, 7, 247 RSC.
- M. P. Calatayud, B. Sanza, V. Raffa, C. Riggio, M. R. Ibarra and G. F. Goya, Biomaterials, 2014, 35, 6389 CrossRef CAS PubMed.
- Q. Yang, Y. Wu, F. Lan, S. H. Ma, L. Q. Xie, B. He and Z. W. Gu, Nanotechnology, 2014, 25, 085702 CrossRef CAS PubMed.
- M. M. Zhang, J. Qiao and L. Qi, Anal. Chim. Acta, 2018, 1035, 70 CrossRef CAS PubMed.
- A. Konwar, D. Chowdhury and A. Dan, Mater. Chem. Front., 2019, 3, 716 RSC.
- J. P. Fan, J. X. Yua, X. M. Yang, X. H. Zhang, T. T. Yuan and H. L. Peng, Chem. Eng. J., 2018, 337, 722 CrossRef CAS.
- X. J. Li, B. L. Zhang, W. Li, X. F. Lei, X. L. Fan, L. Tian, H. P. Zhang and Q. Y. Zhang, Biosens. Bioelectron., 2014, 51, 261 CrossRef CAS PubMed.
- M. I. Shukoor, F. Natalio, M. N. Tahir, M. Divekar, N. Metz, H. A. Therese, P. Theato, V. Ksenofontov, H. C. Schröder, W. E. G. Muller and W. Tremel, J. Magn. Magn. Mater., 2008, 320, 2339 CrossRef CAS.
- L. Q. Xie, S. H. Ma, Q. Yang, F. Lan, Y. Wu and Z. W. Gu, RSC Adv., 2014, 4, 1055 RSC.
- Q. Li, B. H. Wang, J. Q. Wang, X. J. Xi, Q. Chu, G. L. Dong and Y. Wei, New J. Chem., 2017, 41, 13673 RSC.
- Q. Yang, Y. Zhu, M. G. Yang, S. H. Ma, Y. Wu, F. Lan and Z. W. Gu, Small, 2016, 12, 2344 CrossRef CAS PubMed.
- N. M. Riley, A. S. Hebert, M. S. Westphall and J. J. Coon, Nat. Chem., 2019, 1311, 5592 Search PubMed.
- M. Y. Chen, X. F. Shi, R. M. Duke, C. I. Ruse, N. Dai, C. H. Taron and J. C. Samuelson, Nat. Commun., 2017, 8, 15487 CrossRef CAS PubMed.
- P. Perego, L. Gatti and G. L. Beretta, Nat. Rev. Cancer, 2010, 10, 523 CrossRef CAS PubMed.
- K. Ohtsubo and J. D. Marth, Cell, 2006, 126, 855 CrossRef CAS PubMed.
- B. Luo, Q. Chen, J. He, Z. Y. Li, L. Z. Yu, F. Lan and Y. Wu, ACS Sustainable Chem. Eng., 2019, 7, 6043 CrossRef CAS.
- M. Y. Wang, X. M. Zhang and C. H. Deng, Proteomics, 2015, 15, 2158 CrossRef CAS PubMed.
- Z. A. Lin, J. N. Zheng, F. Lin, L. Zhang, Z. Cai and G. N. Chen, J. Mater. Chem., 2011, 21, 518 RSC.
- X. Zhang, X. He, L. Chen and Y. Zhang, J. Mater. Chem., 2012, 22, 16520 RSC.
- Y. Zhang, W. Ma, D. Li, M. Yu, J. Guo and C. Wang, Small, 2014, 10, 1379 CrossRef CAS PubMed.
- Q. Yang, Y. Zhu, B. Luo, F. Lan, Y. Wu and Z. W. Gu, J. Mater. Chem. B, 2017, 5, 1236 RSC.
- Z. Y. Li, B. Luo, L. Z. Yu, F. Lan and Y. Wu, J. Mater. Chem. B, 2021, 9, 453 RSC.
- X. H. Zhang, J. W. Wang, X. W. He, L. X. Chen and Y. K. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 24576 CrossRef CAS PubMed.
- J. Su, X. W. He, L. X. Chen and Y. K. Zhang, Talanta, 2018, 180, 54 CrossRef CAS PubMed.
- H. P. Xiao, W. X. Chen, J. M. Smeekens and R. H. Wu, Nat. Commun., 2018, 9, 1692 CrossRef PubMed.
- D. J. Li, Z. Y. Liu, R. M. Song, W. L. Yang, S. M. Zhai and W. H. Wang, RSC Adv., 2019, 9, 38038 RSC.
- Q. Wu, B. Jiang, Y. J. Weng, J. X. Liu, S. W. Li, Y. C. Hu, K. G. Yang, Z. Liang, L. H. Zhang and Y. K. Zhang, Anal. Chem., 2018, 90, 2671 CrossRef CAS PubMed.
- J. h. Huang, H. Wan, Y. t. Yao, J. N. Li, K. Cheng, J. W. Mao, J. Chen, Y. Wang, H. Q. Qin, W. B. Zhang, M. L. Ye and H. F. Zou, Anal. Chem., 2015, 87, 10199 CrossRef CAS PubMed.
- S. S. Sun, P. Shah, S. T. Eshghi, W. M. Yang, N. Trikannad, S. Yang, L. J. Chen, P. Aiyetan, N. Höti, Z. Zhang, D. W. Chan and H. Zhang, Nat. Biotechnol., 2016, 34, 84 CrossRef CAS PubMed.
- H. Zhang, X. J. Li, D. B. Martin and R. Aebersold, Nat. Biotechnol., 2003, 21, 660 CrossRef CAS PubMed.
- G. Y. Qing, Q. Lu, Y. T. Xiong, L. Zhang, H. X. Wang, X. L. Li, X. M. Liang and T. L. Sun, Adv. Mater., 2017, 29, 1604670 CrossRef PubMed.
- L. T. Liu, M. Yu, Y. Zhang, C. C. Wang and H. J. Lu, ACS Appl. Mater. Interfaces, 2014, 6, 7823 CrossRef CAS PubMed.
- Q. C. Cao, C. Ma, H. H. Bai, X. Y. Li, H. Yan, Y. Zhao, W. T. Ying and X. H. Qian, Analyst, 2014, 139, 603 RSC.
- N. P. Dinh, T. Jonsson and K. Irgum, J. Chromatogr. A, 2011, 1218, 5880 CrossRef CAS PubMed.
- B. Luo, J. He, Z. Y. Li, F. Lan and Y. Wu, ACS Appl. Mater. Interfaces, 2019, 11, 47218 CrossRef CAS PubMed.
- Y. F. Ma, L. J. Wang, Y. L. Zhou and X. X. Zhang, Nanoscale, 2019, 11, 5526 RSC.
- J. X. Wang, J. Li, Y. N. Wang, M. X. Gao, X. M. Zhang and P. Y. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 27482 CrossRef CAS PubMed.
- J. X. Wang, J. Li, M. X. Gao and X. M. Zhang, Nanoscale, 2017, 9, 10750 RSC.
- J. N. Li, J. Liu, Z. Y. Liu, Y. Tan, X. Y. Liu and F. J. Wang, Anal. Chem., 2017, 89, 4339 CrossRef CAS PubMed.
- N. R. Sun, H. Wu, H. M. Chen, X. Z. Shen and C. H. Deng, Chem. Commun., 2019, 55, 10359 RSC.
- Y. N. Wang, J. X. Wang, M. X. Gao and X. M. Zhang, J. Mater. Chem. B, 2015, 3, 8711 RSC.
- B. Jiang, Q. Wu, N. Deng, Y. B. Chen, L. H. Zhang, Z. Liang and Y. K. Zhang, Nanoscale, 2016, 8, 4894 RSC.
- J. W. Wang, J. Z. Yao, N. R. Sun and C. H. Deng, J. Chromatogr. A, 2017, 1512, 1 CrossRef CAS PubMed.
- J. N. Li, F. J. Wang, J. Liu, Z. C. Xiong, G. Huang, H. Wan, Z. Y. Liu, K. Cheng and H. F. Zou, Chem. Commun., 2015, 51, 40936 Search PubMed.
- J. N. Li, F. J. Wang, H. Wan, J. Liu, Z. Y. Liu, K. Cheng and H. F. Zou, J. Chromatogr. A, 2015, 1425, 213 CrossRef CAS PubMed.
- C. F. Bi, Y. L. Liang, L. J. Shen, S. S. Tian, K. Zhang, Y. L. Li, X. W. He, L. X. Chen and Y. K. Zhang, ACS Omega, 2018, 3, 1572 CrossRef CAS PubMed.
- H. Wan, J. F. Huang, Z. S. Liu, J. N. Li, W. B. Zhang and H. F. Zou, Chem. Commun., 2019, 51, 9391 RSC.
- Y. L. Wu, H. Z. Lin, Z. X. Xu, Y. Li, Z. B. Chen and C. H. Deng, Anal. Chim. Acta, 2020, 1096, 1 CrossRef CAS PubMed.
- R. Q. Wu, L. T. Li and C. H. Deng, Proteomics, 2016, 16, 1311 CrossRef CAS PubMed.
- Y. M. Zhao, Y. J. Chen, Z. C. Xiong, X. D. Sun, Q. Q. Zhang, Y. Y. Gan, L. Y. Zhang and W. B. Zhang, J. Chromatogr. A, 2017, 1482, 23 CrossRef CAS PubMed.
- Y. J. Chen, Z. C. Xiong, L. Y. Zhang, J. Y. Zhao, Q. Q. Zhang, L. Peng, W. B. Zhang, M. L. Ye and H. F. Zou, Nanoscale, 2015, 7, 3100 RSC.
- Y. S. Ji, R. H. Lv, S. H. Song, J. F. Huang, L. W. Zhang, G. Huang, J. N. Li and J. J. Ou, Anal. Chim. Acta, 2019, 1053, 43 CrossRef CAS PubMed.
- J. F. Xie, G. Q. Zhong, C. Q. Cai, C. Y. Chen and X. M. Chen, Talanta, 2017, 169, 98 CrossRef CAS PubMed.
- X. Y. Sun, R. T. Ma, J. Chen and Y. P. Shi, J. Mater. Chem. B, 2018, 6, 688 RSC.
- Y. Hao, R. X. Gao, D. C. Liu, G. Y. He, Y. H. Tang and Z. J. Guo, Talanta, 2016, 153, 211 CrossRef CAS PubMed.
- D. J. Li, Z. J. Bie, F. F. Wang and E. H. Guo, Analyst, 2018, 143, 4936 RSC.
- M. Zhang, X. H. Zhang, X. W. He, L. X. Chen and Y. K. Zhang, Nanoscale, 2012, 4, 3141 RSC.
- M. Hofweber and D. Dormann, J. Biol. Chem., 2019, 294, 7137 CrossRef CAS PubMed.
- K. Ashman and E. L. Villar, Clin. Transl. Oncol., 2009, 11, 356 CrossRef CAS PubMed.
- J. Zhang, M. J. Guy, H. S. Norman, Y. C. Chen, Q. Xu, X. Dong, H. Guner, S. Wang, T. Kohmoto, K. H. Young, R. L. Moss and Y. Ge, J. Proteome Res., 2011, 10, 4054 CrossRef CAS PubMed.
- Z. G. Wang, N. Lv, W. Z. Bi, J. L. Zhang and J. Z. Ni, ACS Appl. Mater. Interfaces, 2015, 7, 8377 CrossRef CAS PubMed.
- N. R. Sun, H. Wu, X. Z. Shen and C. H. Deng, Adv. Funct. Mater., 2019, 29, 1900253 CrossRef.
- N. R. Sun, C. H. Deng, Y. Li and X. M. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 11799 CrossRef CAS PubMed.
- H. Z. Lin and C. H. Deng, Proteomics, 2016, 16, 2733 CrossRef CAS PubMed.
- J. B. Jiang, X. N. Sun, Y. Lia, C. H. Deng and G. L. Duan, Talanta, 2018, 178, 600 CrossRef CAS PubMed.
- J. B. Jiang, X. N. Sun, X. J. She, J. J. Li, Y. Li, C. H. Deng and G. L. Duan, Microchim. Acta, 2018, 185, 309 CrossRef PubMed.
- L. Zhao, H. Q. Qin, Z. Y. Hu, Y. Zhang, R. A. Wu and H. F. Zou, Chem. Sci., 2012, 3, 2828 RSC.
- Z. C. Xiong, L. Y. Zhang, C. L. Fang, Q. Q. Zhang, Y. S. Ji, Z. Zhang, W. B. Zhang and H. F. Zou, J. Mater. Chem. B, 2014, 2, 4473 RSC.
- W. F. Ma, Y. Zhang, L. L. Li, Y. T. Zhang, M. Yu, J. Guo, H. J. Lu and C. C. Wang, Adv. Funct. Mater., 2013, 23, 107 CrossRef CAS.
- Z. G. Wang, G. Cheng, Y. L. Liu, J. L. Zhang, D. H. Sun and J. Z. Ni, J. Colloid Interface Sci., 2014, 417, 217 CrossRef CAS PubMed.
- Y. L. Liang, X. W. He, L. X. Chen and Y. K. Zhang, RSC Adv., 2014, 4, 18132 RSC.
- X. M. Wang, Z. Y. Guo, Y. Zhang, M. L. Chen and J. H. Wang, Talanta, 2018, 188, 385 CrossRef CAS PubMed.
- M. Y. Wang, X. N. Sun, Y. Li and C. H. Deng, Proteomics, 2016, 16, 915 CrossRef CAS PubMed.
- Z. G. Wang, J. L. Zhang, D. H. Sun and J. Z. Ni, J. Mater. Chem. B, 2014, 2, 6886 RSC.
- J. Q. Zhou, Y. L. Liang, X. W. He, L. X. Chen and Y. K. Zhang, ACS Sustainable Chem. Eng., 2017, 5, 11413 CrossRef CAS.
- J. J. Duan, X. M. He and L. N. Zhang, Chem. Commun., 2015, 51, 338 RSC.
- X. Y. Long, Z. J. Zhang, J. Y. Li, D. Sheng and H. Z. Lian, Chem. Commun., 2017, 53, 4620 RSC.
- C. T. Chen, L. Y. Wang and Y. P. Hou, Anal. Bioanal. Chem., 2011, 399, 2795 CrossRef CAS PubMed.
- Z. C. Xiong, Y. J. Chen, L. Y. Zhang, J. Ren, Q. Q. Zhang, M. L. Ye, W. B. Zhang and H. F. Zou, ACS Appl. Mater. Interfaces, 2014, 6, 22743 CrossRef CAS PubMed.
- D. D. Jiang, X. Q. Li, X. J. Lv and Q. Jia, Talanta, 2018, 185, 461 CrossRef CAS PubMed.
- B. Luo, M. G. Yang, P. P. Jiang, F. Lan and Y. Wu, Nanoscale, 2018, 10, 8391 RSC.
- Y. Chen, D. J. Li, Z. J. Bie, X. P. He and Z. Liu, Anal. Chem., 2016, 88, 1447 CrossRef CAS PubMed.
- G. Y. Zhang, L. Y. Jiang, J. T. Zhou, L. H. Hu and S. H. Feng, Chem. Commun., 2019, 55, 9927 RSC.
- T. Nishio, E. Ayano, Y. Suzuki, H. Kanazawa and T. Okano, J. Chromatogr. A, 2011, 1218, 2079 CrossRef CAS PubMed.
- B. Luo, L. Z. Yu, Z. Y. Li, J. He, C. J. Li, F. Lan and Y. Wu, J. Mater. Chem. B, 2020, 8, 8414 RSC.
- H. J. Zheng, J. X. Jia, Z. Li and Q. Jia, Anal. Chem., 2020, 92, 2680 CrossRef CAS PubMed.
- Q. Lu, C. Chen, Y. T. Xiong, G. D. Li, X. F. Zhang, Y. H. Zhang, D. D. Wang, Z. C. Zhu, X. L. Li, G. Y. Qing, T. L. Sun and X. M. Liang, Anal. Chem., 2020, 92, 6269 CrossRef CAS PubMed.
- N. R. Sun, H. Wu and X. Z. Shen, Microchim. Acta, 2020, 187, 195 CrossRef CAS PubMed.
- Y. Y. Hong, Q. L. Zhan, Y. Zheng, C. L. Pu, H. L. Zhao and M. B. Lan, Talanta, 2019, 197, 77 CrossRef CAS PubMed.
- Y. Y. Hong, H. L. Zhao, C. L. Pu, Q. L. Zhan, Q. Y. Sheng and M. B. Lan, Anal. Chem., 2018, 90, 11008 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma01015d |
| This journal is © The Royal Society of Chemistry 2021 |