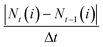Open Access Article
Open Access ArticlePeristaltic on-chip pump for tunable media circulation and whole blood perfusion in PDMS-free organ-on-chip and Organ-Disc systems†
Stefan
Schneider
 a,
Marvin
Bubeck
a,
Marvin
Bubeck
 ab,
Julia
Rogal
ab,
Julia
Rogal
 ac,
Huub J.
Weener
ac,
Huub J.
Weener
 ad,
Cristhian
Rojas
e,
Martin
Weiss
ef,
Michael
Heymann
ad,
Cristhian
Rojas
e,
Martin
Weiss
ef,
Michael
Heymann
 b,
Andries D.
van der Meer
b,
Andries D.
van der Meer
 d and
Peter
Loskill
d and
Peter
Loskill
 *aceg
*aceg
aFraunhofer Institute for Interfacial Engineering and Biotechnology IGB, Stuttgart, Germany
bInstitute of Biomaterials and Biomolecular Systems, University of Stuttgart, Stuttgart, Germany
cDepartment of Biomedical Engineering, Faculty of Medicine, Eberhard Karls University Tübingen, Tübingen, Germany. E-mail: peter.loskill@uni-tuebingen.de
dApplied Stem Cell Technologies, University of Twente, Enschede, The Netherlands
eNMI Natural and Medical Sciences Institute at the University of Tübingen, Reutlingen, Germany
fDepartment of Women's Health, Faculty of Medicine, Eberhard Karls University Tübingen, Tübingen, Germany
g3R-Center for in vitro Models and Alternatives to Animal Testing, Eberhard Karls University Tübingen, Tübingen, Germany
First published on 25th August 2021
Abstract
Organ-on-chip (OoC) systems have become a promising tool for personalized medicine and drug development with advantages over conventional animal models and cell assays. However, the utility of OoCs in industrial settings is still limited, as external pumps and tubing for on-chip fluid transport are dependent on error-prone, manual handling. Here, we present an on-chip pump for OoC and Organ-Disc systems, to perfuse media without external pumps or tubing. Peristaltic pumping is implemented through periodic compression of a flexible pump layer. The disc-shaped, microfluidic module contains four independent systems, each lined with endothelial cells cultured under defined, peristaltic perfusion. Both cell viability and functionality were maintained over several days shown by supernatant analysis and immunostaining. Integrated, on-disc perfusion was further used for cytokine-induced cell activation with physiologic cell responses and for whole blood perfusion assays, both demonstrating the versatility of our system for OoC applications.
Introduction
Microfluidic organ-on-chip (OoC) systems can be used to culture cells under vasculature-like perfusion in a precisely controllable microenvironment to achieve physiological relevant in vitro models of human tissues and organs.1 Several on-chip organ and tissue models have been introduced and provide alternatives to ethically challenging animal testing and non-physiological 2D cell culture, for more efficient drug development and personalized medicine.2–5 Especially OoCs featuring unidirectional, circulatory pumping that recapitulate human vascular flows are highly sought after.External pumps are able to achieve microfluidic, closed-loop perfusion: i) peristaltic pumps allow for a circulatory, unidirectional fluid flow when connected in-line with a shared reservoir for ingoing and outgoing media.6 ii) Pressure pumps that combine multiple external valves and air-pressurized reservoirs can also sustain unidirectional circulation.7,8 However, external pumps are often bulky and require tubing to couple fluids into the chips, which increases the required manual handling, risk of leakage, dead volumes and unspecific surface-binding of small molecules.9
External pumps and tubing can be avoided, for instance, by using gravity-driven pumping, whereby flow is induced by hydrostatic pressure difference of fluid levels in connected reservoirs. Fluid levels and hence the pressure gradient can be maintained over a longer period by tilting of the chip.10 Gravity-driven, unidirectional circulation, however, requires considerable adaptions to the chip and the overall process as e.g. including a bypass between reservoirs,11,12 limiting unidirectional flow to a short-cut channel13 or pumping effluent back to inlet reservoirs.14
Therefore, the integration of peristaltic pumps on-chip is popular when flexibility in chip design and application are important. Here, fluid is displaced by mechanically actuating a film or flexible channel structure. A popular method for actuating on-chip peristaltic pumps is using pneumatics, as e.g. implemented in a perfused multiwell system15 or multi-OoC platform.16,17 Such pneumatic actuation of a flexible film requires multiple lines to supply external pressure to switch periodically between different pressure levels on-chip. Alternatives for pneumatic actuation, directly integrated or in close proximity to the chip, are braille pins,18 piezoelectric discs,19 electromagnets20,21 or permanent magnets.22 Especially magnets are attractive for peristaltic pump actuation, as they allow implementation of relatively simple systems, such as e.g. magnetically dragged steel balls for fluid displacement.23,24
Peristaltic on-chip pumping requires a flexible material, such as the elastomer polydimethylsiloxane (PDMS), to allow for mechanical fluid displacement. PDMS continues to be a popular material for microfluidic devices due to ease of fabrication.25 However, PDMS-related absorption of small hydrophobic molecules,26 high water vapor permeability27 and leaching of uncured PDMS-components28 is popularizing alternative materials, especially for cell-based and OoC applications.29 A promising alternative to PDMS is thermoplastic elastomer (TPE), which combines thermoplastic and elastic characteristics.30 TPE can be structured e.g. by hot embossing31,32 and allows for thermal fusion bonding to other thermoplastic substrates.32–34 TPE subtypes based on styrenic block copolymer formulations without oil additives show reduced absorption of small molecules as well as lower water permeability in comparison to PDMS and provide a biocompatible chip material.32,35
In this study, we present a novel TPE-based on-chip pumping technology for microfluidic cell culture and OoC applications. Advancing the capabilities of the recently introduced organ-on-a-disc technology,36 we integrated a compact peristaltic pump on the “Organ-Disc”. Avoiding suboptimal chip materials such as PDMS and providing compatibility to industry-scale fabrication processes, our disc consists of microstructured, thermoplastic foils and hot embossed, flexible TPE films. For media perfusion through our platform, steel balls compress flexible pump channels in the central TPE pump module and are rolled around the central disc axis by a magnetic actuator underneath the disc. Media is stored in a reservoir on top of the disc, perfused through the disc cell culture channels, and transported back into the same reservoir compartment. Thereby, we introduce an entirely new approach that allows cultivation of microphysiological tissues under a closed-loop media perfusion with tunable, unidirectional flow and continuous access to the supernatant for further analysis. Paving the way for automated workflows and increased throughput, our platform features multiple independent systems per disc that run in parallel and eliminates the need for external pumps and error-prone tubing connections. We demonstrate the utility of this technology through microphysiological perfusion experiments culturing endothelial cells under controllable shear stress. Leveraging the simple access to the circulating media throughout the cultivation, evaporative media loss as well as cell metabolism was monitored over several days. Cytokine-induced activation of endothelial cells resulted in physiologic cell responses and demonstrated the functionality of cells cultured under on-disc perfusion. Finally, we showed the applicability of our platform for perfusion of endothelial lined channels with human whole blood and monitoring of platelet adhesion to the endothelium.
Materials and methods
Disc materials
Discs were fabricated from several layers of approx. 750 μm thick thermoplastic elastomer (TPE; Mediprene OF400M, based on oil-free styrene–ethylene/butylene–styrene elastomer, HEXPOL TPE AB) extruded by an external service provider (Fraunhofer Institute for Process Engineering and Packaging IVV) and 175 μm polymethyl methacrylate foils (PMMA; PLEXIGLAS Film 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524, Röhm). Each disc consists of TPE-based “pump” and “pump support” layers and PMMA-based “port”, “cell channel” and “bottom” layers (Fig. 1a). Reservoirs for media storage were made from polypropylene (PP), fabricated by an external service provider (CNCTeile24) and connected to discs with laser cut (VLS2.30, Universal Laser Systems) biocompatible, double-sided adhesive tape (ARcare 90
524, Röhm). Each disc consists of TPE-based “pump” and “pump support” layers and PMMA-based “port”, “cell channel” and “bottom” layers (Fig. 1a). Reservoirs for media storage were made from polypropylene (PP), fabricated by an external service provider (CNCTeile24) and connected to discs with laser cut (VLS2.30, Universal Laser Systems) biocompatible, double-sided adhesive tape (ARcare 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106, Adhesives Research).
106, Adhesives Research).
Epoxy master fabrication
The epoxy master fabrication started with the generation of SU-8 (SU-8 2075, Kayaku Advanced Materials) microstructures on 150 mm silicon wafers (Siegert Wafer) by standard ultraviolet (UV) light lithography. SU-8 was spin coated with a final speed of 1500 rpm at room temperature and soft baked (7 min at 65 °C then 30 min at 95 °C). Photomasks were designed (CorelCAD 2018, Corel Corporation) and purchased from an external provider (KOPP-desktopmedia). SU-8 layers were exposed to UV light (275 mJ cm−2, ABM Series 60 Exposure Systems, ABM, Inc.) using the photomask, post-exposure baked (5 min at 65 °C then 12 min at 95 °C) developed for 16 min (SU-8 developer, Kayaku Advanced Materials), washed with isopropanol and hard baked (30 min at 160 °C). Subsequently, wafers were treated using 100 μL trichloro(1H,1H,2H,2H-perfluorooctyl)silane (448![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931, Sigma-Aldrich) in a desiccator overnight at reduced pressure.
931, Sigma-Aldrich) in a desiccator overnight at reduced pressure.
Fluoroelastomer-based O-rings (138 × 2 mm FPM 75, Dichtelemente arcus) were placed concentrically on the wafers, both clamped between two 5 mm thick PMMA plates (Oroglas cast acrylic glass, Trinseo) and 50 mL syringes (BD Plastipak, BD) connected to the upper PMMA plate featuring two ports. PDMS (Sylgard 184, Dow Corning) was mixed (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 base
1 base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) curing agent mass ratio), degassed and injected into the cavity above the SU-8 microstructures. For this, PDMS was filled into one syringe without plunger connected to the upper PMMA plate while pulling the plunger of the other syringe and generating a small vacuum for PDMS injection into the cavity. Subsequently, PDMS was cured overnight at 60 °C (Universal Oven UN110, Memmert), peeled off of both PMMA plates and the wafer, cleaned with isopropanol and blow-dried with compressed nitrogen.
curing agent mass ratio), degassed and injected into the cavity above the SU-8 microstructures. For this, PDMS was filled into one syringe without plunger connected to the upper PMMA plate while pulling the plunger of the other syringe and generating a small vacuum for PDMS injection into the cavity. Subsequently, PDMS was cured overnight at 60 °C (Universal Oven UN110, Memmert), peeled off of both PMMA plates and the wafer, cleaned with isopropanol and blow-dried with compressed nitrogen.
An aluminum-based molding tool was designed (Solidworks 2019, Dassault Systèmes) and manufactured by an external service provider (CNCTeile24). The base plate of the tool has an integrated vacuum chuck structure and a hose connector for connecting a vacuum pump (Fig. 1b) allowing for vacuum-assisted fixation of the mold and removal of air bubbles trapped during epoxy injection (Fig. 1c).
For epoxy molding, the replica molded PDMS was placed onto the vacuum chuck with a pump (≥100 mbar max. absolute pressure, LABOPORT N 86 KN.18, KNF) constantly generating a contact vacuum. An aluminum-ring, defining the dimensions of the final epoxy master (8 mm height, 110 mm diameter), was placed onto the PDMS mold and an O-ring (120 × 2 mm FPM 75, Dichtelemente arcus) was placed around the PDMS. A 5 mm PMMA plate (Oroglas cast acrylic glass, Trinseo) with two ports for syringe connection was screwed onto the aluminum ring and sealed with an O-ring (125 × 2 mm FPM 75, Dichtelemente arcus).
A two-component (“part A” and “part B”) epoxy resin (EpoxAcast 670 HT, Smooth-On) modified with an epoxy thinner (Epic Epoxy Thinner, Smooth-on) reducing the viscosity was mixed (part A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) thinner
thinner![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) part B = 100
part B = 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17.6 mass ratio), degassed in a desiccator at reduced pressure for 3 min and placed in an ultrasound bath (JP-031S, RS PRO) for another 10 min.
17.6 mass ratio), degassed in a desiccator at reduced pressure for 3 min and placed in an ultrasound bath (JP-031S, RS PRO) for another 10 min.
Identically to the described PMDS injection, the epoxy mixture was flushed into the tool using syringes connected to the PMMA plate. After partial curing for 24 h at room temperature and constantly applied contact vacuum, the tool was disconnected from the pump and transferred to an oven for 24 h at 60 °C (Universal Oven UN110, Memmert) for complete curing. Afterwards, the cured epoxy was removed from the tool and tempered in an oven (Universal Oven UN30, Memmert) for 2 h at 80 °C followed by 3 h at 150 °C and slowly cooled to room temperature overnight in the closed, switched off, oven.
Vacuum-assisted air bubble removal by gas diffusion through the PDMS mold, was exemplary visualized by filling de-ionized water into the molding cavity with the vacuum pump turned on and imaged with a USB microscope (UM038, Conrad Electronic SE).
TPE hot embossing
TPE layers (11 × 11 cm2) were laminated on a 250 μm thick polytetrafluoroethylene (PTFE) foil (High-tech-flon films and fabrics) as temporary support layer using a handheld pressure roller (Steinel). The epoxy master was placed in the center of the TPE sheet and both were transferred on a 150 mm silicon wafer (Siegert Wafer) into a preheated hot press (LabManual 300, Fontijne Presses) (Fig. 1d).At a plate temperature of 140 °C, the epoxy was pressed into the TPE at a pressure of 0.4 MPa for 10 min, then the plates were water cooled below 40 °C in 4–5 min before opening the press. The TPE was peeled off of the epoxy and PTFE with a few drops of isopropanol. TPE layers were then cut to the respective size, ports were either punched (504![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 647, World Precision Instruments) or drilled (1.2 mm, RoNa Werkzeuge), cleaned with isopropanol and blow-dried with compressed nitrogen.
647, World Precision Instruments) or drilled (1.2 mm, RoNa Werkzeuge), cleaned with isopropanol and blow-dried with compressed nitrogen.
Plotting
PMMA-based channels were fabricated with a vinyl cutter (Graphtec CE6000-40 Plus, Graphtec) by 2D-structuring (Fig. 1e). After plotting, all cutouts were removed and the PMMA layers were cleaned with isopropanol and blow-dried with compressed nitrogen.Thermal fusion bonding
An aluminum-based bonding tool was designed (Solidworks 2019, Dassault Systèmes) and manufactured by an external service provider (MAAS Vorrichtungsbau) (Fig. 1f). Two aluminum plates build the basis of the tool with the bottom plate featuring pins (DIN 427 M5 screws, Reidl) for the alignment of PMMA layers. For a homogenous pressure distribution, 1 mm thick silicone mats (Elastomer plate VMQ 50 Shore A, Angst+Pfister) were placed between aluminum plates and mirror polished stainless steel plates (0.8 mm, TGA GmbH). For bonding, the respective PMMA layers were stacked between the polished steel plates and the tool was transferred into a preheated hot press (LabEcon 150, Fontijne Presses) (Fig. 1g). PMMA layers were bonded in two steps: first 20 min at 103 °C and 0.18 MPa and then 30 min at 103 °C at 1.9 MPa. Afterwards, the tool was removed from the hot press, slowly cooled down to room temperature overnight and then opened for removing the bonded PMMA modules.TPE bonding
Both TPE layers were bonded after each other on the thermal fusion bonded PMMA modules. For each bonding step, the respective TPE layer was oxygen (O2) plasma treated (1 min, <2 mbar, 3.3 sccm O2, 50 W; Zepto, Diener) and laminated on top of PMMA or previously added TPE using a handheld pressure roller (Steinel) (Fig. 1h). Afterwards, a fluoropolymer coated polyester film (3M Scotchpak 1022 Release Liner, 3M) was temporarily laminated on the TPE and a small weight placed (2–5 kg or 6–15 kPa) on top during subsequent bonding at 95 °C for 1 h in an oven (Universal Oven UN30, Memmert).Burst pressure test
For bonding strength assessment, burst pressure tests using compressed nitrogen gas were conducted. Epoxy glue (UHU PLUS ENDFEST, UHU) was used for both attaching luer connectors (BDMFTLL-9, Nordson MEDICAL) to channel inlets and sealing the channel outlets. After at least 24 h for complete curing of the epoxy glue, a nitrogen line was attached to the luer connectors and the disc submerged in water for monitoring gas leakage. A manual pressure controller (DR 022-00-3, Landefeld Druckluft und Hydraulik) was used for increasing the nitrogen gas pressure until either the disc layers delaminated or the maximum output pressure (3.5 bar) of the controller was reached.Microstructure analysis
For microstructure analysis, side cuts from channel structures in various materials (specified in each case) were obtained by cutting with scalpels or scissors. The cross sections were imaged using a stereomicroscope (SteREO Discovery.V8, Carl Zeiss MicroImaging) and analyzed with Fiji (ImageJ version 1.53c).37Pumping setup
For peristaltic pumping, discs were placed on a PMMA rack equipped with a stepper motor (SY42STH38-1684A, Pololu Corporation). A PMMA-based magnet holder with eight magnets (S-10-05-N52N, maximum adhesion 32.4 N, remanence 1.42–1.47 T, Webcraft) was connected to the motor to rotate underneath the mounted disc. The pumping setup was placed in an incubator (Heraeus BBD 6220, Thermo Scientific) for perfused on-disc cell culture. All other electronic components required for controlling the motor remained outside the incubator and were connected with a thin cable to the motor. The stepper motor was controlled by a motor driver (2128, Pololu Corporation) and a microcontroller (ATmega328P Board, Eckstein) with a display (I2C 16 × 2 LCD Display Module, Eckstein) and a rotary encoder (KY-040, reichelt elektronik) user interface to set motor parameters. On top of the disc, eight steel balls (diameter 5 mm, stainless steel 1.4034, HSI-Solutions) were placed above the magnets. A grooved ring of a thrust ball bearing (S51204, CQ GmbH) and another PMMA-based magnet holder with another eight, less strong, magnets (S-05-05-N, maximum adhesion 9.22 N, remanence 1.32–1.37 T, Webcraft) were placed on top of the steel balls. The magnets on top were less strong than the magnets connected to the motor shaft, which provided a suitable compromise between sufficient compression and an acceptable level of friction between steel balls and disc.Flow rate measurements
Flow rate measurements were conducted at room temperature within a period of three days using de-ionized water supplemented with watercolor (Ecoline Liquid Watercolour, Royal Talens) for improved visualization. Luer connectors (BDMFTLL-9, Nordson MEDICAL) were connected to the inlet and outlet using 2 mm thick, laser cut (VLS2.30, Universal Laser Systems) PMMA (Oroglas cast acrylic glass, Trinseo) adapters equipped with adhesive tape (ARcare 90106, Adhesives Research). Syringes with volume scale (1 mL, Omnifix-F, B. Braun Melsungen) were attached to the luer connectors, filled with equal amounts of liquid. Using a 5 s ramp time to the respective, final motor speed, the time needed for changing the volume in both syringes (0.05 mL per measurement) was taken. The volume flow rate was averaged from in total two systems from independent discs, each system was measured three times.Cell culture
Commercially available, cryopreserved human umbilical vein endothelial cells (HUVEC), from pooled donors were acquired from Lonza (C2519A) and cultured in endothelial cell growth medium (EGM-2 BulletKit, CC-3162, Lonza) with 1% (v/v) gentamicin (10 ng mL−1, Gibco). HUVECs at passage three were thawed and seeded in 175 cm2 filter cap cell culture flasks (CELLSTAR, Greiner Bio-One). After 24 h, the cell culture media was exchanged and the HUVECs were expanded for the following 3 days before cell loading into the disc.For generating a cell suspension for subsequent cell loading, adherent cells were washed with PBS (Dulbecco's phosphate buffered saline w/o calcium w/o magnesium, Biowest), detached by a 3 min incubation step at 37 °C using 0.05% (v/v) trypsin (Trypsin-EDTA Solution 10×, SIGMA Life Science) in Versene solution (Versene 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 1×, Gibco). The cell suspension was transferred into a centrifuge tube (50 mL CELLSTAR polypropylene tube, Greiner Bio-One), trypsin inactivated by adding 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) and centrifuged for 5 min at 1000 rpm or 216g (Multifuge 3S-R, Heraeus). Cells were counted using trypan blue (trypan blue 4 g l−1 in aqueous solution, VWR chemicals) using a hemocytometer (C-Chip Neubauer improved DHC-N01, NanoEnTek).
5000 1×, Gibco). The cell suspension was transferred into a centrifuge tube (50 mL CELLSTAR polypropylene tube, Greiner Bio-One), trypsin inactivated by adding 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) and centrifuged for 5 min at 1000 rpm or 216g (Multifuge 3S-R, Heraeus). Cells were counted using trypan blue (trypan blue 4 g l−1 in aqueous solution, VWR chemicals) using a hemocytometer (C-Chip Neubauer improved DHC-N01, NanoEnTek).
Cell loading and cell channel lining
Discs were sprayed with 70% (v/v) ethanol and plasma activated (4 min, <2 mbar, 3.3 sccm O2, 50 W; Zepto, Diener) after drying. Each system of a disc was flushed using 100 μL of 70% (v/v) ethanol and then three times using 100 μL of PBS for the removal of ethanol.For cell adhesion, channels were coated using 100 μL of 0.1 mg mL−1 collagen-I (FibriCol, Catalog #5133, Advanced BioMatrix) in PBS for 1 h at 37 °C. Subsequently, channels were flushed using 100 μL of PBS and afterwards filled with cell culture media.
For cell seeding, pipette tips containing 75 μL of cell suspension with 6 × 106 cells per mL were connected to each cell channel. To prevent cells from entering the pump channel during injection, they were introduced through the outlet of the system in opposite direction of media perfusion. Brightfield microscopy (Leica DMi1, Leica Microsystems) was used to confirm that cells did not enter into the TPE module.
Unless otherwise specified, discs were turned upside down after cell injection for 1 h at 37 °C to promote cell adhesion to the cell channel ceiling. The seeding density for the cell channel with a volume of 15 μL and 84 mm2 large ceiling is approx. 1000 cells per mm2. Afterwards, the discs were turned reservoir side up, and each reservoir compartment was filled with 5 mL cell culture media, sealed with a breathable tape (Z380059, Sigma-Aldrich) and HUVECs were cultured for 72 h at 10 revolutions per hour (rph) motor speed under standard cell culture conditions (37 °C, 95% humidity, 5% CO2, Heraeus BBD 6220, Thermo Scientific) for complete channel lining with cells. Before conducting experiments, the motor was stopped, the tape removed, media exchanged and a new breathable tape applied to seal the reservoir.
Ion, glucose and lactate measurements
Cell loading and initial on-disc culture for cell attachment for solute analysis differed slightly compared to standard cell injection. After collagen-I coating and cell injection (day −1), each reservoir compartment was filled with 5 mL cell culture media, sealed with a breathable tape (Z380059, Sigma-Aldrich) and cells were kept under static conditions for 1 h for cell adherence. Subsequently, the motor was ramped slowly to 100 rph over a period of 4 h. After 24 h (day 0), the cell culture media was exchanged to remove all non-adhering cells and to provide a reference base line for subsequent supernatant monitoring.Subsequently, every 24 h a 110 μL sample from the supernatant of each system was taken and stored at −80 °C until further analysis. The withdrawn sample volume was replaced with an equal amount of cell culture media. Every 72 h (day 3 and day 6), the cell culture media was exchanged completely in all reservoir compartments. For media exchange or sampling, the motor was stopped, the breathable tape replaced and the motor ramped up to 100 rph again in 1 h.
For sample analysis, a bioanalyte analyzer was used (Vi-CELL MetaFLEX, Beckman Coulter): potassium (K+), sodium (Na+), chloride (Cl−) and calcium (Ca2+) concentrations were analyzed in samples from a system without HUVECs (“reference system”) for evaporation monitoring. Glucose and lactate concentrations were analyzed in five systems culturing HUVECs. For day 1 and day 2, one sample was excluded from the analysis due to a sensor response error reported from the bioanalyte analyzer (day 1 and 2: N = 4, all others: N = 5). Fresh cell culture media was identically analyzed for measuring the concentrations of all considered components (N = 4).
CD31/nuclei staining
Each channel was washed three times using 100 μL PBS+ (Dulbecco's phosphate buffered saline with MgCl2 and CaCl2, Sigma-Aldrich). Afterwards, channels were filled with fixation solution (Roti-Histofix 4%, Carl Roth). After 15 min of incubation at room temperature, channels were washed again with PBS.“Permeabilization/diluent buffer” containing 30 mg mL−1 bovine serum albumin (BSA; A9647, Sigma-Aldrich), 0.1% (v/v) Triton X-100 (Sigma-Aldrich) in PBS was used for cell permeabilization, blocking of unspecific binding and as diluent for antibodies and dyes. For cell permeabilization and blocking, channels were filled with this buffer and incubated for 15 min at room temperature. Subsequently, primary CD31 antibody (M0823, mouse anti-human, Agilent Technologies), diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, was injected into cell channels and washed out after 2 h incubation at room temperature using PBS. Staining solution containing secondary antibody (A-11003, goat anti-mouse, Alexa Fluor 546, Invitrogen), diluted 1
50, was injected into cell channels and washed out after 2 h incubation at room temperature using PBS. Staining solution containing secondary antibody (A-11003, goat anti-mouse, Alexa Fluor 546, Invitrogen), diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 1 μg mL−1 of Hoechst (62249, Hoechst 33342, Thermo Scientific) was flushed into cell channels, incubated for 1 h at room temperature and subsequently washed three times using 100 μL of PBS.
100, and 1 μg mL−1 of Hoechst (62249, Hoechst 33342, Thermo Scientific) was flushed into cell channels, incubated for 1 h at room temperature and subsequently washed three times using 100 μL of PBS.
Stained HUVECs were imaged using a confocal laser-scanning-microscope (LSM 710, Carl Zeiss MicroImaging). Tile scan images were stitched with ZEN (ZEN black edition 2.3 SP1, Carl Zeiss Microscopy). Z-stacks were transferred into maximum intensity projections using Fiji (ImageJ version 1.53c)37 and into 3D renders using Fiji in combination with the Volume Viewer plugin (Volume Viewer 2.01.2, https://github.com/fiji/Volume_Viewer).
Live/dead staining
Before live/dead staining, channels were washed three times using 100 μL PBS+. Afterwards, 100 μL of staining solution with 27 μg mL−1 of fluorescein diacetate (FDA; F7378, Sigma-Aldrich) for labeling viable cells and 135 μg mL−1 of propidium iodide (PI; P4170, Sigma-Aldrich) for labeling dead cells in PBS+ was filled into the channels. The staining solution was incubated for 5 min at 37 °C. Chips were washed three times using 100 μL PBS+ and imaged immediately. Images of stained HUVECs were acquired at 37 °C using a fluorescence microscope with heated enclosure (Leica DMi8, Leica Microsystems).Endothelial cell activation and readout of cytokine secretion
After cell loading and 48 h of perfusion at 10 rph motor speed for cell channel lining, media was exchanged to determine baseline cytokine secretion to the supernatant over a 24 h period (“pre-treatment” condition, N = 8). Subsequently, perfusion media was replaced by cell culture media containing either 20 ng mL−1 (“24 h treated” condition, N = 4) or no (“24 h untreated” condition, N = 4) tumor necrosis factor alpha (TNF-α; SRP3177, Sigma-Aldrich) and perfused for another 24 h at 10 rph motor speed.Subsequently, HUVECs were stained for CD106 to visualize endothelial activation. For this, cell channels were first flushed using 100 μL PBS+ with 10 mg mL−1 BSA. Subsequently, channels were filled with conjugated CD106 antibody (130-104-164, CD106 Antibody, anti-human, REAfinity, APC, Miltenyi Biotec), diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in PBS+ with 10 mg mL−1 BSA, and incubated for 30 min at 37 °C. After washing three times using 100 μL PBS+, HUVECs were imaged at 37 °C using a fluorescence microscope with heated enclosure (Leica DMi8, Leica Microsystems).
10 in PBS+ with 10 mg mL−1 BSA, and incubated for 30 min at 37 °C. After washing three times using 100 μL PBS+, HUVECs were imaged at 37 °C using a fluorescence microscope with heated enclosure (Leica DMi8, Leica Microsystems).
To determine cytokine content from supernatants, samples of the supernatant were taken for all conditions at the respective time points, centrifuged for 5 min at 3000 rpm or 1942g (Multifuge 3S-R, Heraeus) for removal of debris and then stored at −80 °C until further analysis. We quantified concentrations of the proinflammatory cytokines interleukin 6 (IL-6), interleukin 8 (IL-8) and angiopoietin-2 (Ang-2) under baseline conditions as well as in response to inflammatory stimulation using a fluorescent bead-based multiplex sandwich immunoassay (LEGENDplex Human Angiogenesis Panel 1, 740697, BioLegend) read by flow cytometry (Guava easyCyte 8HT, Merck) following the manufacturer's manual. In short, the stored supernatants were thawed and, in technical duplicates for each condition, incubated with a cocktail of target-specific capture beads with unique respective sizes and internal fluorescence for 2 h. Subsequently, biotinylated detection antibodies were added for 1 h to allow formation of capture bead-analyte-detection antibody sandwiches. By finally incubating the captured and detected analytes with streptavidin–phycoerythrin, the amount of bound cytokines was quantified via fluorescence signal intensities. To correlate fluorescence signal intensity to cytokine concentration, a standard curve for all analyzed cytokines was generated in the same assay. Flow cytometry data were analyzed using the LEGENDplex cloud-based data analysis software suite (BioLegend). All gates were adjusted manually to find optimal differentiation between capture bead populations, and the same gating strategy applied to all assay runs.
Whole blood perfusion and analysis of platelet adhesion
Collection of whole blood was carried out in accordance with the rules for investigation of human subjects as defined in the Declaration of Helsinki. Patients gave a written agreement according to the permission of the ethical Committee of the Eberhard Karls University Tübingen (Nr. 495/2018-BO02).Blood was collected in tubes with sodium citrate (VACUETTE TUBE 9 mL 9NC Coagulation sodium citrate 3.2%, Greiner Bio-One), supplemented with 1% (v/v) conjugated CD41 antibody (MHCD4104, CD41 Monoclonal Antibody, PE, Invitrogen) for fluorescent platelet labeling and kept at room temperature for 10 min in the dark. Prior to blood perfusion, 1% (v/v) of HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 1 M, Gibco) containing 63.2 mM CaCl2 (Sigma-Aldrich) and 31.6 mM MgCl2 (Invitrogen) was added to the blood sample in order to add ions required for blood coagulation.
Experiments were performed using cell channels lined with HUVECs cultured for 96 h at 10 rph motor speed for media perfusion before blood perfusion experiments. Empty pipette tips (300 μL, Greiner Bio-One) and pipette tips filled with 300 μL blood were connected to the inlet and outlet of the reservoir, respectively. Cell culture media was filled in the reservoir compartments for the subsequent flushing of the blood out of the channels. For linear blood perfusion from the blood-filled pipette tip to the empty pipette tip, the motor was ramped up in 1 min to a final speed of 800 rph maintained for 5 min. Fresh blood was refilled during this initiation step to avoid draining of the blood-filed pipette tip. For flushing blood out from the channels, all pipette tips were removed while the motor was kept running for another 5 min to displace blood with the media previously loaded into the reservoir compartments. During flush-out, 1 mL of cell culture media was exchanged. Subsequently, the motor was stopped, and the channels were additionally flushed manually using PBS+.
After complete blood removal, HUVECs were fixed and stained for CD31 and nuclei as described previously, however, applying overnight incubation at 4 °C for the primary antibody and using a different secondary antibody for fluorescent labeling of CD31 (A32723, goat anti-mouse, Alexa Flour 488 Plus, Invitrogen). After washing with PBS, stained HUVECs and platelets were imaged using a fluorescence microscope (Leica DMi8, Leica Microsystems).
Data presentation and statistical analysis
If not stated otherwise, data is presented as mean ± standard deviation in text and diagrams with sample sizes stated in each case. OriginPro (Version 2021, OriginLab Corporation) in combination with OriginPro's Paired Comparison Plot plugin (version 3.6) was used for testing statistical significance of different cytokine concentrations using Tukey's range test. OriginPro was also used for linear regression of measured flow rates produced by on-disc pumping.Results and discussion
Design, fabrication and pumping mechanism
The peristaltic Organ-Disc features an integrated on-disc pump and is constructed from a total of five polymer layers: TPE pump and pump support layers as well as PMMA port, cell channel and bottom layers (Fig. 1a). Each disc features four individual microfluidic networks symmetrically arranged around its central axis. The TPE pump layer contains 400 μm wide and 100 μm high channels and forms the basis for the integrated peristaltic pump together with the pump support layer (cf. Fig. S1† showing the complete 2D Organ-Disc layout). Three PMMA layers (175 μm thick) enclose the cell channels (1000 μm wide) in a similar configuration as the previously reported centrifugal organ-on-a-disc sytem.36Hot embossing of microstructures into TPE layers uses custom epoxy stamps fabricated by replica molding.38,39 Our custom injection-molding tool allows for the fabrication of temperature stable epoxy stamps via replica molding from PDMS masters.32 Air bubbles trapped in-between microstructures during injection of the epoxy can be removed by vacuum-assisted gas diffusion through the PDMS mold (cf. Video S1† demonstrating the process using water). Side cuts obtained from PDMS-molds of both the initial SU-8 structures and the epoxy stamp as well as the final TPE, confirmed a precise transfer of microstructure dimensions (Fig. 1i). After complete curing and tempering, the epoxy molds can be used for a large number of hot embossing cycles; this is a considerate advantage over hot embossing directly using SU-8 structures, which typically brake off the silicon wafer already after few cycles.
PMMA layers are structured by plotting and thermal fusion bonding. For robust and high-throughput thermal fusion bonding, a custom bonding tool was used to align and bond up to seven stacks of thermoplastic foils (footprint of a 4′′ wafer) simultaneously. A general challenge for thermal fusion bonding of thermoplastics is the optimization of heat, pressure and bonding time. If heat and pressure are too low, an insufficient bonding is achieved, whereas too high temperatures and forces lead to channel distortion.40 For the PMMA used in this study, bonding 10 °C below the material's glass transition (TG = 113 °C) with 1.9 MPa of pressure allows for stable bonds with minimal impact on microchannels: the PMMA bonds withstood a nitrogen pressure of at least 3.5 bar (maximum pressure of the used gas pressure controller) without channel failure (N = 3 discs). After bonding, the channel height was reduced only by 2.7%, from original 175 μm (thickness of the PMMA foil) to 170 ± 15 μm. Similarly, the channel width was only reduced by 3.7% from 1000 μm (CAD design) to 963 ± 28 μm (N = 4 channels).
After thermal fusion bonding of the PMMA layers, the flexible TPE layers were added on top. TPE already displays good but reversible adhesion to PMMA at room temperature. To achieve robust and stable bonding, TPE surfaces were plasma activated41 and bonded to PMMA under heat and pressure (95 °C and 15 kPa for 1 h). Burst pressure tests of fully assembled discs resulted in a failure at the TPE-TPE interface at 3.1 ± 0.3 bar of nitrogen pressure (N = 3 discs; cf. Fig. S2† showing cross sections of a PMMA cell channel and a TPE pump channel after bonding).
To enable on-disc pumping, a peristaltic pumping mechanism was implemented allowing for a closed-loop perfusion through the system (Fig. 2a): medium is pumped from the reservoir into the pumping module, then pushed further into the cell culture channel and subsequently back into the reservoir. Steel balls on top of the pump module are pulled into the pump channels by magnets underneath the disc (Fig. 2b). This results in localized channel collapse and sealing. To perfuse liquid through the channels, the magnets are rotated around the central axis by a stepper motor. Thereby, the steel balls roll over the pump channels and push liquid through the system; whereby, the flow rate is controlled by the rotation speed of the steel balls. An open, PP-based cell culture media reservoir with one compartment for each system is added on top of the disc (Fig. 2c). A grooved ring from a thrust ball bearing and further magnets integrated in a PMMA ring are added on top during operation for better positioning of the steel balls and to sustain higher compression forces.
Closed-loop perfusion for cell culture and cell monitoring
The wall shear stress (WSS) τ generated in the cell channel with width W and height H can be estimated by
Evaporation estimates were based on the average evaporation taking all types of ions into account. Calculated evaporations differed slightly depending on which type of ion was considered. Quantified rates were as high as 6.3% per day for potassium, as low as 2.7% per day for calcium and about 4.2% and 4.3% per day for sodium and chloride respectively. A similar divergence was observed previously when evaporative loss was monitored via salt concentrations during cell culture.44
Condensation on the reservoir sealing tape can also affect cell-culture media concentrations as small volumes are inevitably removed together with the tape during reservoir sampling. Sampling, including removal of the sealing tape, is conducted the same way for all conditions; therefore, the accompanying impact on solute concentrations is comparable. A continued three-day media exchange regiment results in an overall sawtooth profile, around a desired steady state throughout the experimental run.
To derive actual glucose and lactate consumption and production rates, we normalized the measured concentrations against the salt ion derived evaporative water loss quantifications (Fig. 3b and c, central and lower panel). Total amounts of nutrients and metabolites are balanced the same way as for the reference system making the same assumptions of homogenous mixing and representative sampling. Absolute amounts N of solutes i are calculated as
| Nt(i) = Vtct(i) |
results in a production or consumption rate per day. The cumulative production or consumption over time then is
Average glucose consumption was 2.5 ± 2.2 μmol per day, while 7.5 ± 3.5 μmol per day lactate was produced. Each molecule of glucose can at most form two lactate molecules. For HUVECs, a glycolytic index of 1.74 was reported previously, which is the ratio of produced lactate molecules for each consumed glucose molecule.53 Hence, resulting rates are in reasonable agreement, considering the standard deviation. The discrepancy between the calculated consumption and production stems from the measured nutrient and metabolite concentrations. Overall high glucose content in the media results in absolute concentration changes close to the standard deviation, whereas, fresh media is almost lactate-free and, hence, already small changes to lactate content clearly observable. Additionally, the media volume Vt strongly affects the calculated solute amounts as it is also subject to measurement errors and therefore leads to error propagation. However, at the same time, this demonstrates the importance of monitoring evaporation in microfluidic cell culture and organ-on-chip systems.
Confluent endothelial cell lining of disc channels
Endothelial cells (HUVECs) were injected into the disc with sub-confluent seeding densities and cultured for 72 h while gently circulating media (10 rph, corresponding to approx. 30 μL h−1 or 0.02 dyn cm−2). During this period, HUVECs expanded, adhered to all inner channel surfaces and reached confluency.Using the tunable on-disc perfusion, the flow rate was subsequently raised to either 2.6 mL h−1 or 0.32 mL h−1 (shear stress of 1.5 dyn cm−2 or 0.19 dyn cm−2 respectively) for varying shear stress on endothelial cell lined cell channels during another 24 h. Fluorescence microscopy revealed a confluent lining of all channel walls with endothelial layers positive for CD31 (platelet endothelial cell adhesion molecule 1, PECAM1) over the entire length of the channels (Fig. 4a and b). Endothelial layers cultured in the higher shear rate condition (1.5 dyn cm−2) displayed more cell–cell contacts than layers cultured with the 8-fold lower shear stress (Fig. 4b). Nevertheless, under both flow conditions, cells were viable as confirmed by live/dead staining with only minimal amount of PI-positive cells (FDA/PI, Fig. 4c). No distinct cell alignment in flow direction was observed. This is in agreement with previous reports of HUVEC alignment under shear-flows of 7.2 dyn cm−2 and higher.54–56 Flow rates required for shear stress induced HUVEC alignment can in principle be achieved by the peristaltic on-disc pump, either through increased motor speeds or suitably configured pump channel layouts. However, a common problem of peristaltic pumping is the friction associated with high actuation speeds. In our case, pumping at rotation speeds higher than 800 rph resulted in premature TPE degradation and pump failure. Channel designs that increase the actuated volume per duty cycle without increasing mechanical stresses on the disc hence seems to be the more attractive approach to achieve higher flow and shear rates.
Tumor necrosis factor alpha (TNF-α)-induced endothelial cell activation
Physiological function of the endothelium on-disc can be accessed by monitoring the response to stimulation. Thereto, after 72 h of culture with gentle media circulation (approx. 30 μL h−1 or 0.02 dyn cm−2), the HUVEC-lined channels were perfused with 20 ng mL−1 TNF-α for another 24 h without changing the flow rate. TNF-α treated cells show an increased presence of vascular cell adhesion protein 1 (VCAM1, CD106) compared to non-treated cells (Fig. 4d), at comparable channel cell density. This response is in agreement with previous studies showing an increased CD106 expression due to TNF-α treatment.57,58In addition to analysis of CD106 expression, cytokines released into the perfused media in response to endothelial activation were monitored. Analysis of cytokines in the effluent before and after the 24 h TNF-α treatment demonstrated a significant increase in IL-6, IL-8 and Ang-2 levels (Fig. 4e). Compared to untreated systems, the concentration of IL-6 was increased 12-fold, of IL-8 38-fold, and of Ang-2 2.5-fold, respectively. Media from non-treated systems before and after 24 h incubation showed stable levels of all tested cytokines.
Increased IL-6 and IL-8 release from HUVECs upon TNF-α stimulation is well charcterized;59,60 therefore, demonstrating the physiological response to stimulation of an endothelium cultured in our system. Similarly, the upregulation of Ang-2 expression in HUVECs after TNF-α activation has been reported previously.61 Fiedler et al. showed that Ang-2 is stored in Weibel–Palade bodies of endothelial cells and released after stimulation into the media.62 Interestingly, Fiedler et al. did not detect Ang-2 in the supernatant after TNF-α stimulation; however, the authors performed only short-term release experiments. In our case, cytokines were released over 24 h into circulating media containing TNF-α and accumulated over time. Therefore, the disc configuration is suitable to investigate circulating stimuli treatments over longer periods and time-dependent cell response assessments.
On-disc perfusion of whole blood and platelet adhesion
Lining the microchannels with confluent endothelial layers minimizes the exposure of the perfused media to the polymeric chip material and enables artificial, blood-perfused capillaries.63,64 Demonstrating an important step for on-disc blood perfusion, we injected fresh human whole blood into discs featuring HUVEC-lined channels (pre-cultured on-disc for 96 h at approx. 30 μL h−1 or 0.02 dyn cm−2). Immediately prior to injection, whole blood collected in citrate tubes was supplemented with ions required for coagulation processes and CD41 antibody for fluorescent labeling of platelets.In contrast to previous closed-loop media circulation, blood was directly passed from inlet to outlet pipette tips that were attached to the respective ports in the reservoir compartment (Fig. 4f, Video S2†). Subsequent pumping of cell culture media evacuated the perfused blood from the cell culture channel (Video S3†). All four channels remained structurally intact and no blood clots or clogging formed. Antibody staining revealed a strong CD31 signal at cell–cell contacts indicating an intact cell morphology after 96 h of culture and whole blood perfusion (Fig. 4g). Fluorescent CD41 labeling revealed domains of platelets adhering to the endothelium. Platelet adhesion is an important process during thrombosis and stroke.65 The capability of the presented platform to both generate a functional endothelium and to provide an integrated, tunable perfusion paves the way for further microphysiological experiments. As we demonstrated that whole blood perfusion is possible in our systems, future applications could aim at the study of platelet adhesion under blood flows of different shear stress66 and automated platelet aggregation monitoring.67
Conclusion
We engineered an Organ-Disc with integrated peristaltic pumping for parallelized culturing of lumen-like endothelial cell structures. Disc materials and fabrication readily translate to scaled industrial settings. PDMS is completely avoided and TPE is instead used for soft and flexible layers and thermoplastic PMMA as highly transparent, rigid material. The used microstructuring methods, plotting and hot embossing, as well as the bonding processes are precise, parallelizable and result in a highly robust microfluidic system.The integrated, peristaltic pump of our disc achieves accurate perfusion and supersedes expensive, external pumps or problematic tubing connections. Media stored in a reservoir and circulating through the system is easily accessible throughout the whole experiment and allows for user-friendly analysis. Ion, nutrient and metabolite concentrations were assessed over multiple days of on-disc HUVEC culture to monitor cell metabolism as well as evaporation, an important yet often ignored aspect of microfluidic engineering.
The proof-of-concept of our integrated platform technology for Organ-Disc and OoC systems is achieved through generating and culturing lumen-like structures of HUVECs and conducting several on-disc experiments: we demonstrate perfusion under varying shear stress, in vitro recapitulation of endothelial activation and response to inflammatory stimuli, as well as whole blood perfusion. Furthermore, our system allows for versatile adaptions: e.g. transfer to other chip geometries, programmable motor settings for oscillatory flow patterns or co-culture applications. Overall, the presented platform provides a user-friendly and automatable perfusion technology for microphysiological systems and will help to advance the field of OoC research.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The research was supported in part by the Ministry of Science, Research and the Arts of Baden-Württemberg (Az: 33-7542.2-501-1/30/9).References
- S. N. Bhatia and D. E. Ingber, Microfluidic Organs-on-Chips, Nat. Biotechnol., 2014, 32(8), 760–772, DOI:10.1038/nbt.2989.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Advances in Organ-on-a-Chip Engineering, Nat. Rev. Mater., 2018, 3(8), 257–278, DOI:10.1038/s41578-018-0034-7.
- B. Zhang and M. Radisic, Organ-on-a-Chip Devices Advance to Market, Lab Chip, 2017, 17(14), 2395–2420, 10.1039/C6LC01554A.
- A. van den Berg, C. L. Mummery, R. Passier and A. D. van der Meer, Personalised Organs-on-Chips: Functional Testing for Precision Medicine, Lab Chip, 2019, 19(2), 198–205, 10.1039/C8LC00827B.
- N. Franzen, W. H. van Harten, V. P. Retèl, P. Loskill, J. van den Eijnden-van Raaij and M. IJzerman, Impact of Organ-on-a-Chip Technology on Pharmaceutical R&D Costs, Drug Discovery Today, 2019, 24(9), 1720–1724, DOI:10.1016/j.drudis.2019.06.003.
- S. Deinhardt-Emmer, K. Rennert, E. Schicke, Z. Cseresnyés, M. Windolph, S. Nietzsche, R. Heller, F. Siwczak, K. F. Haupt, S. Carlstedt, M. Schacke, M. T. Figge, C. Ehrhardt, B. Löffler and A. S. Mosig, Co-Infection with Staphylococcus Aureus after Primary Influenza Virus Infection Leads to Damage of the Endothelium in a Human Alveolus-on-a-Chip Model, Biofabrication, 2020, 12(2), 025012, DOI:10.1088/1758-5090/ab7073.
- C. G. M. van Dijk, M. M. Brandt, N. Poulis, J. Anten, M. van der Moolen, L. Kramer, E. F. G. A. Homburg, L. Louzao-Martinez, J. Pei, M. M. Krebber, B. W. M. van Balkom, P. de Graaf, D. J. Duncker, M. C. Verhaar, R. Luttge and C. Cheng, A New Microfluidic Model That Allows Monitoring of Complex Vascular Structures and Cell Interactions in a 3D Biological Matrix, Lab Chip, 2020, 20(10), 1827–1844, 10.1039/D0LC00059K.
- T. Qian, D. A. Gil, E. Contreras Guzman, B. D. Gastfriend, K. E. Tweed, S. P. Palecek and M. C. Skala, Adaptable Pulsatile Flow Generated from Stem Cell-Derived Cardiomyocytes Using Quantitative Imaging-Based Signal Transduction, Lab Chip, 2020, 20(20), 3744–3756, 10.1039/D0LC00546K.
- K. Renggli and O. Frey, Chapter 12 - Design and Engineering of Multiorgan Systems, in Organ-on-a-chip, ed. J. Hoeng, D. Bovard and M. C. Peitsch, Academic Press, 2020, pp. 393–427, DOI:10.1016/B978-0-12-817202-5.00012-7.
- C. Lohasz, N. Rousset, K. Renggli, A. Hierlemann and O. Frey, Scalable Microfluidic Platform for Flexible Configuration of and Experiments with Microtissue Multiorgan Models, SLAS Technol., 2019, 24(1), 79–95, DOI:10.1177/2472630318802582.
- M. B. Esch, H. Ueno, D. R. Applegate and M. L. Shuler, Modular, Pumpless Body-on-a-Chip Platform for the Co-Culture of GI Tract Epithelium and 3D Primary Liver Tissue, Lab Chip, 2016, 16(14), 2719–2729, 10.1039/C6LC00461J.
- D. W. Lee, N. Choi and J. H. Sung, A Microfluidic Chip with Gravity-Induced Unidirectional Flow for Perfusion Cell Culture, Biotechnol. Prog., 2019, 35(1), e2701, DOI:10.1002/btpr.2701.
- Y. I. Wang and M. L. Shuler, UniChip Enables Long-Term Recirculating Unidirectional Perfusion with Gravity-Driven Flow for Microphysiological Systems, Lab Chip, 2018, 18(17), 2563–2574, 10.1039/C8LC00394G.
- H. Azizgolshani, J. R. Coppeta, E. M. Vedula, E. E. Marr, B. P. Cain, R. J. Luu, M. P. Lech, S. H. Kann, T. J. Mulhern, V. Tandon, K. Tan, N. J. Haroutunian, P. Keegan, M. Rogers, A. L. Gard, K. B. Baldwin, J. C. de Souza, B. C. Hoefler, S. S. Bale, L. B. Kratchman, A. Zorn, A. Patterson, E. S. Kim, T. A. Petrie, E. L. Wiellette, C. Williams, B. C. Isenberg and J. L. Charest, High-Throughput Organ-on-Chip Platform with Integrated Programmable Fluid Flow and Real-Time Sensing for Complex Tissue Models in Drug Development Workflows, Lab Chip, 2021, 21(8), 1454–1474, 10.1039/D1LC00067E.
- I. Maschmeyer, A. K. Lorenz, K. Schimek, T. Hasenberg, A. P. Ramme, J. Hübner, M. Lindner, C. Drewell, S. Bauer, A. Thomas, N. S. Sambo, F. Sonntag, R. Lauster and U. Marx, A Four-Organ-Chip for Interconnected Long-Term Co-Culture of Human Intestine, Liver, Skin and Kidney Equivalents, Lab Chip, 2015, 15(12), 2688–2699, 10.1039/C5LC00392J.
- K. Domansky, W. Inman, J. Serdy, A. Dash, M. H. M. Lim and L. G. Griffith, Perfused Multiwell Plate for 3D Liver Tissue Engineering, Lab Chip, 2010, 10(1), 51–58, 10.1039/B913221J.
- C. D. Edington, W. L. K. Chen, E. Geishecker, T. Kassis, L. R. Soenksen, B. M. Bhushan, D. Freake, J. Kirschner, C. Maass, N. Tsamandouras, J. Valdez, C. D. Cook, T. Parent, S. Snyder, J. Yu, E. Suter, M. Shockley, J. Velazquez, J. J. Velazquez, L. Stockdale, J. P. Papps, I. Lee, N. Vann, M. Gamboa, M. E. LaBarge, Z. Zhong, X. Wang, L. A. Boyer, D. A. Lauffenburger, R. L. Carrier, C. Communal, S. R. Tannenbaum, C. L. Stokes, D. J. Hughes, G. Rohatgi, D. L. Trumper, M. Cirit and L. G. Griffith, Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies, Sci. Rep., 2018, 8(1), 4530, DOI:10.1038/s41598-018-22749-0.
- W. Gu, X. Zhu, N. Futai, B. S. Cho and S. Takayama, Computerized Microfluidic Cell Culture Using Elastomeric Channels and Braille Displays, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(45), 15861–15866, DOI:10.1073/pnas.0404353101.
- B. Husband, M. Bu, A. G. R. Evans and T. Melvin, Investigation for the Operation of an Integrated Peristaltic Micropump, J. Micromech. Microeng., 2004, 14(9), S64–S69, DOI:10.1088/0960-1317/14/9/011.
- J. R. Coppeta, M. J. Mescher, B. C. Isenberg, A. J. Spencer, E. S. Kim, A. R. Lever, T. J. Mulhern, R. Prantil-Baun, J. C. Comolli and J. T. Borenstein, A Portable and Reconfigurable Multi-Organ Platform for Drug Development with Onboard Microfluidic Flow Control, Lab Chip, 2017, 17(1), 134–144, 10.1039/C6LC01236A.
- V. Tandon, W. S. Kang, T. A. Robbins, A. J. Spencer, E. S. Kim, M. J. McKenna, S. G. Kujawa, J. Fiering, E. E. L. Pararas, M. J. Mescher, W. F. Sewell and J. T. Borenstein, Microfabricated Reciprocating Micropump for Intracochlear Drug Delivery with Integrated Drug/Fluid Storage and Electronically Controlled Dosing, Lab Chip, 2016, 16(5), 829–846, 10.1039/C5LC01396H.
- T. Pan, S. J. McDonald, E. M. Kai and B. Ziaie, A Magnetically Driven PDMS Micropump with Ball Check-Valves, J. Micromech. Microeng., 2005, 15(5), 1021–1026, DOI:10.1088/0960-1317/15/5/018.
- L. Yobas, K.-C. Tang, S.-E. Yong and E. Kye-Zheng Ong, A Disposable Planar Peristaltic Pump for Lab-on-a-Chip, Lab Chip, 2008, 8(5), 660, 10.1039/b720024b.
- M. Du, X. Ye, K. Wu and Z. Zhou, A Peristaltic Micro Pump Driven by a Rotating Motor with Magnetically Attracted Steel Balls, Sensors, 2009, 9(4), 2611–2620, DOI:10.3390/s90402611.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Rapid Prototyping of Microfluidic Systems in Poly(Dimethylsiloxane), Anal. Chem., 1998, 70(23), 4974–4984, DOI:10.1021/ac980656z.
- M. W. Toepke and D. J. Beebe, PDMS Absorption of Small Molecules and Consequences in Microfluidic Applications, Lab Chip, 2006, 6(12), 1484, 10.1039/b612140c.
- Y. S. Heo, L. M. Cabrera, J. W. Song, N. Futai, Y.-C. Tung, G. D. Smith and S. Takayama, Characterization and Resolution of Evaporation-Mediated Osmolality Shifts That Constrain Microfluidic Cell Culture in Poly(Dimethylsiloxane) Devices, Anal. Chem., 2007, 79(3), 1126–1134, DOI:10.1021/ac061990v.
- K. J. Regehr, M. Domenech, J. T. Koepsel, K. C. Carver, S. J. Ellison-Zelski, W. L. Murphy, L. A. Schuler, E. T. Alarid and D. J. Beebe, Biological Implications of Polydimethylsiloxane-Based Microfluidic Cell Culture, Lab Chip, 2009, 9(15), 2132, 10.1039/b903043c.
- S. B. Campbell, Q. Wu, J. Yazbeck, C. Liu, S. Okhovatian and M. Radisic, Beyond Polydimethylsiloxane: Alternative Materials for Fabrication of Organ-on-a-Chip Devices and Microphysiological Systems, ACS Biomater. Sci. Eng., 2021, 7(7), 2880–2899, DOI:10.1021/acsbiomaterials.0c00640.
- E. Roy, J.-C. Galas and T. Veres, Thermoplastic Elastomers for Microfluidics: Towards a High-Throughput Fabrication Method of Multilayered Microfluidic Devices, Lab Chip, 2011, 11(18), 3193, 10.1039/c1lc20251k.
- D. Brassard, L. Clime, K. Li, M. Geissler, C. Miville-Godin, E. Roy and T. Veres, 3D Thermoplastic Elastomer Microfluidic Devices for Biological Probe Immobilization, Lab Chip, 2011, 11(23), 4099, 10.1039/c1lc20714h.
- S. Schneider, E. J. S. Brás, O. Schneider, K. Schlünder and P. Loskill, Facile Patterning of Thermoplastic Elastomers and Robust Bonding to Glass and Thermoplastics for Microfluidic Cell Culture and Organ-on-Chip, Micromachines, 2021, 12(5), 575, DOI:10.3390/mi12050575.
- J. Lachaux, C. Alcaine, B. Gómez-Escoda, C. M. Perrault, D. O. Duplan, P.-Y. J. Wu, I. Ochoa, L. Fernandez, O. Mercier, D. Coudreuse and E. Roy, Thermoplastic Elastomer with Advanced Hydrophilization and Bonding Performances for Rapid (30 s) and Easy Molding of Microfluidic Devices, Lab Chip, 2017, 17(15), 2581–2594, 10.1039/C7LC00488E.
- M. D. Borysiak, K. S. Bielawski, N. J. Sniadecki, C. F. Jenkel, B. D. Vogt and J. D. Posner, Simple Replica Micromolding of Biocompatible Styrenic Elastomers, Lab Chip, 2013, 13(14), 2773, 10.1039/c3lc50426c.
- K. Domansky, J. D. Sliz, N. Wen, C. Hinojosa, G. Thompson, J. P. Fraser, T. Hamkins-Indik, G. A. Hamilton, D. Levner and D. E. Ingber, SEBS Elastomers for Fabrication of Microfluidic Devices with Reduced Drug Absorption by Injection Molding and Extrusion, Microfluid. Nanofluid., 2017, 21(6), 107, DOI:10.1007/s10404-017-1941-4.
- S. Schneider, F. Erdemann, O. Schneider, T. Hutschalik and P. Loskill, Organ-on-a-Disc: A Platform Technology for the Centrifugal Generation and Culture of Microphysiological 3D Cell Constructs Amenable for Automation and Parallelization, APL Bioeng., 2020, 4(4), 046101, DOI:10.1063/5.0019766.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: An Open-Source Platform for Biological-Image Analysis, Nat. Methods, 2012, 9(7), 676–682, DOI:10.1038/nmeth.2019.
- J. S. Jeon, S. Chung, R. D. Kamm and J. L. Charest, Hot Embossing for Fabrication of a Microfluidic 3D Cell Culture Platform, Biomed. Microdevices, 2011, 13(2), 325–333, DOI:10.1007/s10544-010-9496-0.
- E. W. K. Young, E. Berthier, D. J. Guckenberger, E. Sackmann, C. Lamers, I. Meyvantsson, A. Huttenlocher and D. J. Beebe, Rapid Prototyping of Arrayed Microfluidic Systems in Polystyrene for Cell-Based Assays, Anal. Chem., 2011, 83(4), 1408–1417, DOI:10.1021/ac102897h.
- C.-W. Tsao and D. L. DeVoe, Bonding of Thermoplastic Polymer Microfluidics, Microfluid. Nanofluid., 2009, 6(1), 1–16, DOI:10.1007/s10404-008-0361-x.
- S. Yu, S. P. Ng, Z. Wang, C. L. Tham and Y. C. Soh, Thermal Bonding of Thermoplastic Elastomer Film to PMMA for Microfluidic Applications, Surf. Coat. Technol., 2017, 320, 437–440, DOI:10.1016/j.surfcoat.2016.11.102.
- B. J. Chung, A. M. Robertson and D. G. Peters, The Numerical Design of a Parallel Plate Flow Chamber for Investigation of Endothelial Cell Response to Shear Stress, Comput. Struct., 2003, 81(8–11), 535–546, DOI:10.1016/S0045-7949(02)00416-9.
- N. Scott Lynn, C. S. Henry and D. S. Dandy, Evaporation from Microreservoirs, Lab Chip, 2009, 9(12), 1780–1788, 10.1039/B900556K.
- V. Wiegmann, C. B. Martinez and F. Baganz, A Simple Method to Determine Evaporation and Compensate for Liquid Losses in Small-Scale Cell Culture Systems, Biotechnol. Lett., 2018, 40(7), 1029–1036, DOI:10.1007/s10529-018-2556-x.
- M. J. Rogatzki, B. S. Ferguson, M. L. Goodwin and L. B. Gladden, Lactate Is Always the End Product of Glycolysis, Front. Neurosci., 2015, 9, 22, DOI:10.3389/fnins.2015.00022.
- M. G. Vander Heiden, L. C. Cantley and C. B. Thompson, Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation, Science, 2009, 324(5930), 1029–1033, DOI:10.1126/science.1160809.
- K. De Bock, M. Georgiadou, S. Schoors, A. Kuchnio, B. W. Wong, A. R. Cantelmo, A. Quaegebeur, B. Ghesquière, S. Cauwenberghs, G. Eelen, L.-K. Phng, I. Betz, B. Tembuyser, K. Brepoels, J. Welti, I. Geudens, I. Segura, B. Cruys, F. Bifari, I. Decimo, R. Blanco, S. Wyns, J. Vangindertael, S. Rocha, R. T. Collins, S. Munck, D. Daelemans, H. Imamura, R. Devlieger, M. Rider, P. P. Van Veldhoven, F. Schuit, R. Bartrons, J. Hofkens, P. Fraisl, S. Telang, R. J. DeBerardinis, L. Schoonjans, S. Vinckier, J. Chesney, H. Gerhardt, M. Dewerchin and P. Carmeliet, Role of PFKFB3-Driven Glycolysis in Vessel Sprouting, Cell, 2013, 154(3), 651–663, DOI:10.1016/j.cell.2013.06.037.
- J. Börner, S. Buchinger and D. Schomburg, A High-Throughput Method for Microbial Metabolome Analysis Using Gas Chromatography/Mass Spectrometry, Anal. Biochem., 2007, 367(2), 143–151, DOI:10.1016/j.ab.2007.04.036.
- V. Hsiao, E. L. C. de los Santos, W. R. Whitaker, J. E. Dueber and R. M. Murray, Design and Implementation of a Biomolecular Concentration Tracker, ACS Synth. Biol., 2015, 4(2), 150–161, DOI:10.1021/sb500024b.
- F. Pinto, E. L. Thornton and B. Wang, An Expanded Library of Orthogonal Split Inteins Enables Modular Multi-Peptide Assemblies, Nat. Commun., 2020, 11(1), 1529, DOI:10.1038/s41467-020-15272-2.
- F. Seyfarth, S. Schliemann, P. Elsner and U.-C. Hipler, Antifungal Effect of High- and Low-Molecular-Weight Chitosan Hydrochloride, Carboxymethyl Chitosan, Chitosan Oligosaccharide and N-Acetyl-d-Glucosamine against Candida Albicans, Candida Krusei and Candida Glabrata, Int. J. Pharm., 2008, 353(1–2), 139–148, DOI:10.1016/j.ijpharm.2007.11.029.
- B. Müller, P. Sulzer, M. Walch, H. Zirath, T. Buryška, M. Rothbauer, P. Ertl and T. Mayr, Measurement of Respiration and Acidification Rates of Mammalian Cells in Thermoplastic Microfluidic Devices, Sens. Actuators, B, 2021, 334, 129664, DOI:10.1016/j.snb.2021.129664.
- B. Kim, J. Li, C. Jang and Z. Arany, Glutamine Fuels Proliferation but Not Migration of Endothelial Cells, EMBO J., 2017, 36(16), 2321–2333, DOI:10.15252/embj.201796436.
- E. Tkachenko, E. Gutierrez, M. H. Ginsberg and A. Groisman, An Easy to Assemble Microfluidic Perfusion Device with a Magnetic Clamp, Lab Chip, 2009, 9(8), 1085, 10.1039/b812184b.
- E. Tkachenko, E. Gutierrez, S. K. Saikin, P. Fogelstrand, C. Kim, A. Groisman and M. H. Ginsberg, The Nucleus of Endothelial Cell as a Sensor of Blood Flow Direction, Biol. Open, 2013, 2(10), 1007–1012, DOI:10.1242/bio.20134622.
- U. M. Sonmez, Y.-W. Cheng, S. C. Watkins, B. L. Roman and L. A. Davidson, Endothelial Cell Polarization and Orientation to Flow in a Novel Microfluidic Multimodal Shear Stress Generator, Lab Chip, 2020, 20(23), 4373–4390, 10.1039/D0LC00738B.
- L. Osborn, C. Hession, R. Tizard, C. Vassallo, S. Luhowskyj, G. Chi-Rosso and R. Lobb, Direct Expression Cloning of Vascular Cell Adhesion Molecule 1, a Cytokine-Induced Endothelial Protein That Binds to Lymphocytes, Cell, 1989, 59(6), 1203–1211, DOI:10.1016/0092-8674(89)90775-7.
- S. Lechleitner, J. Gille, D. R. Johnson and P. Petzelbauer, Interferon Enhances Tumor Necrosis Factor–Induced Vascular Cell Adhesion Molecule 1 (CD106) Expression in Human Endothelial Cells by an Interferon-Related Factor 1–Dependent Pathway, J. Exp. Med., 1998, 187(12), 2023–2030, DOI:10.1084/jem.187.12.2023.
- S.-I. Yamagishi, Y. Inagaki, K. Nakamura, R. Abe, T. Shimizu, A. Yoshimura and T. Imaizumi, Pigment Epithelium-Derived Factor Inhibits TNF-α-Induced Interleukin-6 Expression in Endothelial Cells by Suppressing NADPH Oxidase-Mediated Reactive Oxygen Species Generation, J. Mol. Cell. Cardiol., 2004, 37(2), 497–506, DOI:10.1016/j.yjmcc.2004.04.007.
- S. Yamagishi, Y. Inagaki, K. Nakamura and T. Imaizumi, Azelnidipine, A Newly Developed Long-Acting Calcium Antagonist, Inhibits Tumor Necrosis Factor-α-Induced Interleukin-8 Expression in Endothelial Cells through Its Anti-Oxidative Properties, J. Cardiovasc. Pharmacol., 2004, 43(5), 724–730 CrossRef CAS PubMed.
- I. Kim, J.-H. Kim, Y. S. Ryu, M. Liu and G. Y. Koh, Tumor Necrosis Factor-α Upregulates Angiopoietin-2 in Human Umbilical Vein Endothelial Cells, Biochem. Biophys. Res. Commun., 2000, 269(2), 361–365, DOI:10.1006/bbrc.2000.2296.
- U. Fiedler, M. Scharpfenecker, S. Koidl, A. Hegen, V. Grunow, J. M. Schmidt, W. Kriz, G. Thurston and H. G. Augustin, The Tie-2 Ligand Angiopoietin-2 Is Stored in and Rapidly Released upon Stimulation from Endothelial Cell Weibel-Palade Bodies, Blood, 2004, 103(11), 4150–4156, DOI:10.1182/blood-2003-10-3685.
- P. F. Costa, H. J. Albers, J. E. A. Linssen, H. H. T. Middelkamp, L. van der Hout, R. Passier, A. van den Berg, J. Malda and A. D. van der Meer, Mimicking Arterial Thrombosis in a 3D-Printed Microfluidic in Vitro Vascular Model Based on Computed Tomography Angiography Data, Lab Chip, 2017, 17(16), 2785–2792, 10.1039/C7LC00202E.
- Y. Zheng, J. Chen, M. Craven, N. W. Choi, S. Totorica, A. Diaz-Santana, P. Kermani, B. Hempstead, C. Fischbach-Teschl, J. A. López and A. D. Stroock, In Vitro Microvessels for the Study of Angiogenesis and Thrombosis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(24), 9342–9347, DOI:10.1073/pnas.1201240109.
- B. Nieswandt, I. Pleines and M. Bender, Platelet Adhesion and Activation Mechanisms in Arterial Thrombosis and Ischaemic Stroke: Platelet Adhesion and Activation Mechanisms, J. Thromb. Haemostasis, 2011, 9, 92–104, DOI:10.1111/j.1538-7836.2011.04361.x.
- D. Meza, S. K. Shanmugavelayudam, A. Mendoza, C. Sanchez, D. A. Rubenstein and W. Yin, Platelets Modulate Endothelial Cell Response to Dynamic Shear Stress through PECAM-1, Thromb. Res., 2017, 150, 44–50, DOI:10.1016/j.thromres.2016.12.003.
- H. J. Albers, R. Passier, A. van den Berg and A. D. van der Meer, Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood, Micromachines, 2019, 10(11), 781, DOI:10.3390/mi10110781.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00494h |
| This journal is © The Royal Society of Chemistry 2021 |