Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rapid quantitative assays for glucose-6-phosphate dehydrogenase (G6PD) and hemoglobin combined on a capillary-driven microfluidic chip†
Marco
Rocca
 ab,
Yuksel
Temiz
ab,
Yuksel
Temiz
 a,
Marie L.
Salva
ab,
Samuel
Castonguay
c,
Thomas
Gervais
a,
Marie L.
Salva
ab,
Samuel
Castonguay
c,
Thomas
Gervais
 cde,
Christof M.
Niemeyer
cde,
Christof M.
Niemeyer
 b and
Emmanuel
Delamarche
*a
b and
Emmanuel
Delamarche
*a
aIBM Research Europe – Zurich, 8803 Rüschlikon, Switzerland. E-mail: emd@zurich.ibm.com
bInstitute for Biological Interfaces (IBG-1) – Karlsruhe Institute of Technology (KIT), 76344 Eggenstein-Leopoldshafen, Germany
cDepartment of Engineering Physics, Polytechnique Montréal, Montréal, Québec, Canada
dInstitut du Cancer de Montréal, Montréal, Québec, Canada
eCentre de Recherche du Centre Hospitalier de l'Université de Montréal (CRCHUM), Montréal, Québec, Canada
First published on 27th July 2021
Abstract
Rapid tests for glucose-6-phosphate dehydrogenase (G6PD) are extremely important for determining G6PD deficiency, a widespread metabolic disorder which triggers hemolytic anemia in response to primaquine and tafenoquine medication, the most effective drugs for the radical cure of malaria caused by Plasmodium parasites. Current point-of-care diagnostic devices for G6PD are either qualitative, do not normalize G6PD activity to the hemoglobin concentration, or are very expensive. In this work we developed a capillary-driven microfluidic chip to perform a quantitative G6PD test and a hemoglobin measurement within 2 minutes and using less than 2 μL of sample. We used a powerful microfluidic module to integrate and resuspend locally the reagents needed for the G6PD assay and controls. We also developed a theoretical model that successfully predicts the enzymatic reactions on-chip, guides on-chip reagent spotting and allows efficient integration of multiple assays in miniaturized formats with only a few nanograms of reagents.
Introduction
This work relates to portable tests for assessing glucose-6-phosphate dehydrogenase (G6PD) deficiency using capillary-driven microfluidic chips. G6PD deficiency is an inherited metabolic disorder of red blood cells (RBCs) affecting more than 400 million people worldwide, mostly in Africa and South Asia.1–4 The spatial geographic distribution of G6PD deficiency consistently overlaps with regions where malaria is endemic,5 and there is strong evidence that G6PD mutations are associated with a lower risk of severe malaria.6,7 Primaquine and the more recent drug tafenoquine are the most preferred drugs for the radical curative treatment of malaria caused by the parasites Plasmodium vivax and Plasmodium ovale. However, as for other 8-aminoquinoline-based drugs, both medications can trigger hemolytic anemia in people with G6PD deficiency.8–10 Hence, testing for G6PD deficiency before deciding which anti-malaria drug to administer can save the life of people affected by malaria.11,12 Being a chromosome X-linked genetic trait, G6PD deficiency causes two discrete ranges of G6PD activities in males (deficient, not deficient), but also an intermediate activity range in females, making it difficult to be identified with a qualitative rapid test.10,13There are two main classes of assays for determining the G6PD status of individuals: genetic assays, which are more suitable for population studies, and phenotypic assays, which are used for making rapid clinical decisions. Most genetic assays employ PCR-based single-nucleotide polymorphism (SNP) analysis.4 However, PCR-SNP analysis only identifies the presence or absence of known genotypes, and therefore it may not identify G6PD deficiency in patients with mutations, which are not included in the reference panel.14 Alternatively, DNA sequencing methods are able to identify all the mutations occurring on the G6PD gene; however, any new mutation identified should be characterized with a phenotypic assay first to determine if it is related with a decrease in the enzymatic activity of G6PD.4 To date, 253 different genetic mutations of the G6PD gene are known.15,16 According to the most recent WHO classification of the variants of the G6PD gene, at least 145 mutations are related to a decrease in the phenotypic activity of G6DP.17,18 Genetic assays are a useful method for population studies because such tests do not require intact enzymatic activity and become cheaper when performed on a larger scale. However, genetic assays are expensive to perform on request, require an equipped laboratory with trained personnel and are not suitable for rapid on-site estimation of the risk associated with G6PD deficiency.
Phenotypic assays are able to determine the activity of G6PD in RBCs and therefore can help physicians in taking fast clinical decisions. Originally, phenotypic assays were only available in newborn-screening facilities and central laboratories. Now, several point-of-care (POC) devices are available on the market and G6PD assays can be performed directly in the field. Phenotypic assays can be classified into three main groups: (i) qualitative assays, (ii) cytochemical assays and (iii) quantitative assays. Qualitative assays only provide a base result of deficient/not deficient and usually diagnose as G6PD-deficient all patients having G6PD activity less than 30% of the normal activity.4 In general, these tests are based on a colorimetric/fluorometric assay, cheap and easy to perform and require only minimal laboratory equipment. The gold standard G6PD assay for a qualitative readout is the fluorescent spot test. It was first established by E. Beutler in the 1960s and relies on the detection of NADPH produced in a dried blood spot mixed with G6P and NADP+. The test readout is obtained using UV light (365 nm).19,20 Brewer et al. developed the methemoglobin reduction test, which relies on the oxidation of hemoglobin to methemoglobin and the subsequent enzymatic reconversion to hemoglobin by TPNH-methemoglobin reductase in the presence of NADPH.21 To date, most of the qualitative assays known in the literature or available on the market are based on a colorimetric assay, including formazan-based methods using either the substrate 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H tetrazolium bromide (MTT) and a hydrogen carrier such as phenazine methosulfate or the substrate 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H tetrazolium monosodium salt (WST-8).22–26 However, qualitative assays are limited in performance since they are not able to distinguish G6PD deficiency in females with an intermediate G6PD activity ranging from ∼30% to 80%.27
Cytochemical assays look at the individual activity of G6PD over a large population of cells and therefore are suited to determine intermediate G6PD deficiency by statistics. In 1968, Fairbanks and Lampe introduced a cytochemical assay based on the reduction of MTT into insoluble tetrazolium salts, which is related to the G6PD activity in RBCs.28 Another cytochemical assay was introduced in 1982 by Van Noorden et al., where the G6PD activity of individual RBCs was analyzed indirectly using the dose-dependent quenching of the autofluorescence of glutaraldehyde by formazan, generated by the activity of G6PD.29,30 Shah et al. proposed a novel cytofluorometric assay to measure G6PD-deficient RBCs through a sequential reaction of cyanide followed by peroxide to generate a fluorescent product (ferryl-Hb) inside RBCs.31 Cytochemical assays provide the most accurate method for determining G6PD deficiency,32 but these assays are expensive, technically challenging and require laboratory equipment, which makes them not suited for implementation on POC diagnostic devices.33,34
Quantitative biochemical assays can measure intermediate levels of G6PD activities and therefore can be used as a universal diagnostic tool for detecting G6PD deficiency also in female subjects. The gold standard for quantitative determination of G6PD activity relies on spectrophotometry, which measures the generation of NADPH by G6PD through an absorbance measurement at 340 nm taken over time. This assay requires 10 μL of blood and takes about 15–20 minutes to be performed.10,35 However, this method requires a kinetic and temperature-controlled spectrophotometer, trained personnel and a well-equipped laboratory. As an alternative, several groups are working on modifying existing personal glucose meters (PGMs) to detect other non-glucose targets. Antibodies, aptamers, or DNA, for example, are coupled to enzymes, such as invertase or glucoamylase, which can convert sugars invisible to the PGM to glucose that is detectable by the PGM. In particular, Zhang et al. investigated the linear response of PGMs to varying concentrations of NADH.36 The same idea can be expanded to measure NADPH as a product from the oxidation of G6P to 6PGL by the activity of G6PD.37 However, using a PGM to measure a non-glucose target suffers from an intrinsic interference from glucose present in blood, which has to be minimized, for example, by introducing additional enzymes. Moreover, since multiple enzymes are employed in this system, the activity of each enzyme must be carefully calibrated. Yu et al. developed an alternative sensor for measuring NADPH:38 in the presence of NADPH, a ligand binds to a receptor and brings a fluorophore close to a NanoLuc luciferase. This leads to increased bioluminescence resonance energy transfer, which is monitored using a digital camera. Such an assay may suffer from interference by endogenous or exogenous substances, such as variations in the concentration of NADPH in blood, inhibitors of luciferase activity, and binding of substances to the sensor (e.g., the antibiotic trimethoprim). A quantitative G6PD assay was also implemented on a digital microfluidic platform, where small droplets can be moved, merged and split arbitrarily using electrowetting phenomena, as explained in detail by Sista et al.39 The fact that this microfluidic droplet assay was released as a commercial product in 2019 by Baebies, Inc. underlines the acute and urgent demand for new quantitative G6PD tests. The droplet platform, however, requires a laborious chip design and fabrication and an electronic peripheral to control the motion of droplets, which might increase the overall cost of the device. Hence, while this device could be a promising solution for newborn screening programs, it might not be affordable to be used as a POC diagnostic device in developing countries.
Phenotypic quantitative biochemical assays are the most promising assay format to be used as a portable diagnostic tool to determine G6PD deficiency. However, a measurement of the activity of G6PD alone, even if very accurate, may be misleading: G6PD is contained in the RBCs, and therefore it is necessary to normalize the activity of G6PD to the number of RBCs to take into account variations in the number of RBCs among different blood samples (e.g., to avoid false positives for anemic people). Standard methods for counting RBCs use a hemocytometer or an automated cell counter40 or else the number of RBCs can also be estimated by measuring the hematocrit and hemoglobin concentrations. A well-suited method for portable diagnostics is the measurement of hemoglobin concentration because it can be performed without bulky equipment and is inexpensive. The most often used methods for this are the hemoglobincyanide assay, the azide-methemoglobin assay, the sodium lauryl sulphate assay and CO-oximetry.41 Other methods are available and were compared by Srivastava et al.42 An interesting alternative is to measure the absorbance of blood at one of its isosbestic points, where the oxidized and non-oxidized state of hemoglobin, which are the most abundant forms of hemoglobin present in blood, have the same extinction coefficient.43 This method does not require additional reactions to convert hemoglobin to a stable colored form, avoids the use of hazardous reagents and can be performed faster than conventional methods (<10 s).
Currently, available POC diagnostic tests for measuring G6PD activity are either qualitative, do not normalize the enzyme activity by the concentration of hemoglobin, or are very expensive.44 Indeed, it is traditionally difficult to combine two classes of assays on a single device.45 Here, we exploit capillary phenomena to control the flow of the sample inside a microfluidic chip to assess G6PD deficiency. A capillary-driven microfluidic chip is a powerful platform, which allows working with just a few microliters of sample and does not require active pumping to move the liquids. We designed multiple flow paths to accommodate the requirements of the hemoglobin measurement and enzymatic assay on a single chip. We used a self-coalescence module (SCM), a simple yet powerful microfluidic element recently developed by our group, for homogenously resuspending reagents in well-defined areas of a microchannel46 to perform a quantitative G6PD test and vicinal positive and negative controls simultaneously on the same chip. We also developed a theoretical model to guide the integration of assay reagents in the SCM by inkjet spotting and to predict their dissolution, diffusion and resulting enzymatic reactions, which generate a fluorescent reporter of G6PD activity.
Results and discussion
Assay workflow and microfluidic chip layout
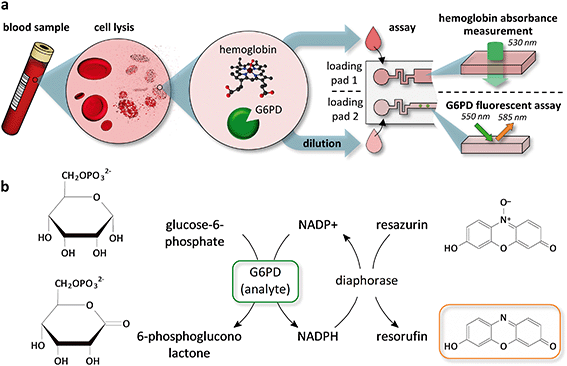
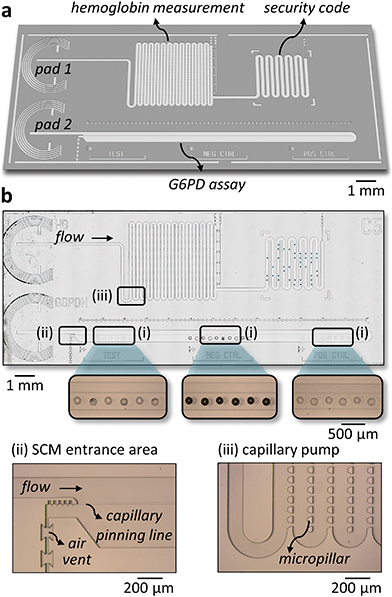
Fig. 1a depicts the workflow we used to combine a G6PD and a hemoglobin assay on a single chip. RBCs from a whole blood sample are lysed chemically using 10 mM Triton X-100, a mild surfactant, to avoid denaturation of proteins in the sample (see ESI† Fig. S1). The concentration of hemoglobin is measured directly from the lysate by an absorbance measurement through a transparent chip at the Hb/HbO2 isosbestic point of 530 nm. It is a common practice to use a diluted sample to perform a G6PD assay to minimize the interference from other molecules present in blood.44 Available POC tests for G6PD deficiency use dilution factors ranging from 8 to 301. In this work, we used a 30-fold dilution, which gives enough dynamic range to distinguish between a G6PD-spiked sample and a control without G6PD (see ESI† Fig. S2). Fig. 1b shows the enzymatic reactions we used in the assay to link the activity of G6PD to the generation of a fluorescent product. G6PD reduces NADP+ to NADPH, and a second enzyme, diaphorase, oxidizes NADPH back to NADP+ while reducing resazurin to the fluorescent product resorufin.The microfluidic chip has two independent flow paths for the hemoglobin concentration measurement and the G6PD assay (Fig. 2a). For the hemoglobin measurement, the lysed blood sample fills a capillary pump with a predefined volume and an absorbance measurement is performed through the chip. The chip is fabricated on a glass substrate, which is transparent in the wavelength needed to perform the hemoglobin measurement. The layout of the micropillars in the pump ensures a steady filling of the capillary pump and avoids trapping air bubbles,47 which may otherwise affect the absorbance measurement (see Fig. 2b and ESI† Movie S1). After the capillary pump, an area having “spots” of inkjet-deposited dyes provides a labile security code area48 (see ESI† Fig. S3), which can be used to protect the tests from counterfeiting and reuse. The reagents for the G6PD assay are integrated into the SCM by spotting two solutions using an alternating pattern (see Fig. 2b). One solution contains the substrates and the cofactors for both enzymes, and the other the section “Experimental”). The reagents are allowed to dry within a few seconds after spotting and the microfluidic chips are sealed.47,49 During an assay, the sample fills the SCM following a specific trajectory that prevents adverse accumulation of reagents and greatly minimizes gradients and dispersion of the reagents due to Taylor–Aris dispersion46 (see ESI† Movie S2). The flow is driven completely by capillarity and controlled by microfluidic structures previously developed by our group.50–53
G6PD assay and measurement of hemoglobin concentration
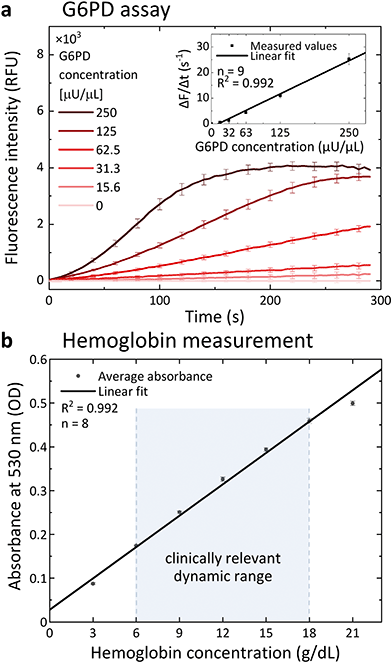
We tested the performance of the G6PD assay using a diluted blood sample spiked with different concentrations of G6PD. Fig. 3a shows the kinetics of the enzymatic assay performed in a microfluidic chip. The assay response was characterized by plotting the slope of the assay kinetics in the linear part of the progress curve against the concentration of G6PD. This is a well-accepted method to measure the activity of enzymes54,55 and allowed us to obtain a quantitative result in less than 1 minute. The assay response is linear over a wide G6PD concentration range (inset in Fig. 3a). Whole blood contains the enzyme 6-phosphogluconate dehydrogenase, which is an additional source of NADPH generation in the pentose phosphate pathway and would increase the generation of the fluorescent product by the enzyme diaphorase. To avoid interference in the assay, 26 mM maleimide was introduced as an inhibitor for the enzyme 6-phosphogluconate dehydrogenase.39,56The concentration of hemoglobin is measured using an absorbance measurement through the transparent chip. The channel height of the chip defines the path length of light through the sample. In order to obtain a signal, which can be easily measured with standard readers (0.1–1 OD), a suitable value for the channel height was estimated using the Beer–Lambert law. Taking two hemoglobin concentrations covering the low range (7 g dL−1, for severe anemia57) and the high range (18 g dL−1, high hemoglobin value58), and the extinction coefficient of hemoglobin at 530 nm (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cm−1 M−1 (ref. 43)), revealed that a channel height between 23 μm and 89 μm would be suitable to measure the concentration of hemoglobin by absorbance. We chose to fabricate the chips with a channel height of 50 μm. The hemoglobin concentration measurement was calibrated using a hemoglobin dilution line ranging from 21 g dL−1 down to 3 g dL−1 (Fig. 3b).
000 cm−1 M−1 (ref. 43)), revealed that a channel height between 23 μm and 89 μm would be suitable to measure the concentration of hemoglobin by absorbance. We chose to fabricate the chips with a channel height of 50 μm. The hemoglobin concentration measurement was calibrated using a hemoglobin dilution line ranging from 21 g dL−1 down to 3 g dL−1 (Fig. 3b).
Modelling and simulations for multiplexing assays in an SCM
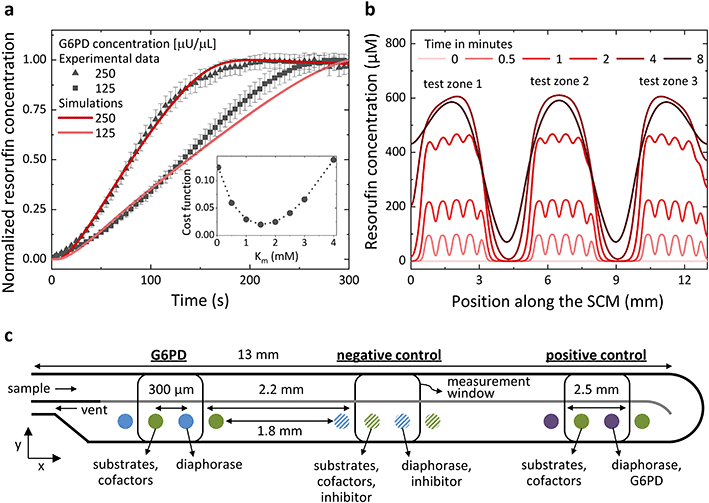
An SCM provides outstanding flexibility for integrating multiple assays on the same chip simply by modifying the spotting pattern of the integrated reagents without changing the chip design, which is laborious and expensive. To perform multiple assays in a single SCM with an economy of footprint and using small volumes of samples, the corresponding test zones need to be spaced close to each other while avoiding interference between the assays. The appropriate distance between the reagents for different assays depends mainly on the assay kinetics, the diffusivity of the reagents involved in the reaction and the assay time. The spotting pattern of the reagents can be optimized by a trial-and-error approach. However, being able to model the assays based on the spotting pattern of the reactants can save time, generalize the method, and make it deterministic. For these reasons, we implemented a theoretical model for predicting the generation of resorufin as a function of G6PD activity and used finite differences (1D in space) to solve numerically a set of 10 reaction–diffusion partial differential equations involving 10 distinct diffusing species and 8 reaction constants (a detailed description of the theoretical model is available in the ESI†). The model accurately predicts the generation of resorufin over time, including the linear regime and saturation of the reaction at longer times (see ESI† Fig. S4 and S5). All parameters were obtained from the literature, except a single one, the Michaelis–Menten constant KM for the conversion of resazurin by diaphorase, which, to the best of our knowledge, is absent from the literature. A single parameter fit nevertheless enabled us to determine it indirectly using our experimental data and to yield an accurate description of the measured production of resorufin (see Fig. 4a). Simulations revealed that with a spacing between the test zones of different assays of 1.8 mm, the fluorescence signal of neighbouring assays overlaps by just 1.23% (>3σ for a Gaussian fit) after 4 minutes (Fig. 4b), which represents 4 times the duration needed for the assay (G6PD assays were achieved in <1 min even for the lower concentration ranges). This spotting pattern allows multiplexing three assays in less than 7 mm2 of chip area. Fig. 4c is a sketch of the spotting pattern used to perform three assays simultaneously in the same SCM, a G6PD test together with a negative and a positive control. The negative control is implemented by co-spotting MgSO4, as an inhibitor for G6PD, together with the reagents needed for the G6PD assay. The positive control is performed by co-spotting G6PD together with diaphorase.Calibration of the G6PD assay to determine G6PD deficiency
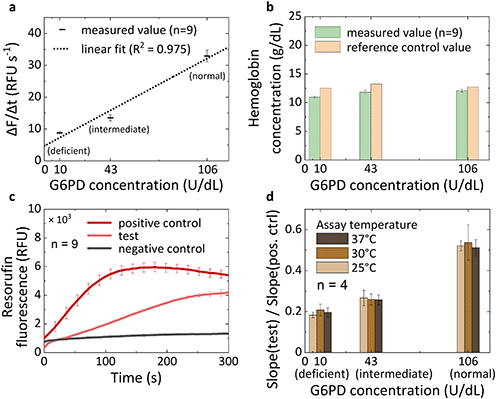
We calibrated the G6PD assay to determine G6PD deficiency using three commercially available reference controls exhibiting normal, intermediate, and deficient G6PD activity, which cover a range of G6PD concentrations from 10 up to 106 U dL−1. The G6PD assay and the measurement of hemoglobin concentration are performed on the same chip and are compared with the values of G6PD activity and hemoglobin concentration provided by the vendor of the reference controls. Fig. 5a shows that we can distinguish the activity of G6PD between the normal, intermediate and deficient samples and that the response of the assay is linear in the clinically relevant dynamic range with an analytical sensitivity of 0.26 (RFU s−1)/(U dL−1). The concentration of hemoglobin could be correctly assessed with an error margin compared to the reference value of 12.6%, 10.6% and 4.9% for the deficient, intermediate and normal samples respectively (Fig. 5b). These variations lie within the error range produced by fabrication variability in the depth of the microfluidic channels at different positions on a wafer and between different wafers (see ESI† Fig. S6). Automatization in the fabrication process would likely yield a more accurate result for the absorbance measurement.A positive and a negative control test are performed simultaneously with the G6PD assay (Fig. 5c), using the spotting pattern shown in Fig. 4c. A high signal in the positive control validates the test and the signal in the negative control outputs the background noise. As expected for any chemical and enzymatic reaction, the activity of G6PD varies with temperature. Here we introduce an alternative method to perform a self-calibration of the assay against temperature variations without the need of additional heating peripherals and temperature sensors by exploiting the signal from the positive control. The variation in the fluorescence signal generation due to temperature between the G6PD assay and the positive control is constant since it is the very same assay simply with different concentrations of G6PD. Therefore, we normalized the signal from the G6PD test to the positive control, which will provide a value that is temperature independent. Fig. 5d shows that by normalizing the signal to the positive control the variations due to temperature in the range between 25 °C and 37 °C are limited to 11.8%.
Comparing our method to the “gold standard” spectrophotometric assay, we note that our test is around 10 times faster (<2 min vs. 20–30 min),59 works with less sample volume (<2 μL vs. 10–500 μL)59,60 and requires fewer steps to be performed by the user. Moreover, our test can measure the concentration of hemoglobin directly on the same chip, while in the gold standard approach, the hemoglobin is measured separately. In the next development iteration of our test, sample preparation steps such as cell lysis and sample dilution could be integrated directly on-chip. One method to perform on-chip lysis of RBCs would be by spotting a surfactant in an additional SCM upstream and releasing a defined lysate volume using passive valves. Alternatively, electrodes could be introduced on-chip to apply a DC or AC electric field and lyse cells by electroporation.61 This method is faster and free of reagents but requires electrodes on-chip and a power source and might generate gas bubbles due to electrolysis. Sample dilution, which is required for our G6PD assay, could be integrated on-chip by adding a blister containing the dilution buffer and a mixing unit. However, since G6PD deficiency tests are mostly performed by trained technicians, we believe that including a kit for off-chip sample dilution is a cheaper and more viable solution and avoids adding complexity and cost to the test. Finally, a miniaturized reader able to perform fluorescence and absorbance measurements would need to be developed to make our test fully portable and affordable at a low price.
Experimental
Chemicals and biochemicals
All reagents were purchased from Sigma-Aldrich unless otherwise indicated. G6PD calibration controls (normal, intermediate, deficient) were from Trinity Biotech (Bray, Ireland). Human serum was purchased from Bioswisstec AB (Schaffhausen, Switzerland). Whole blood was purchased from Interregionale Blutspende SRK (Bern, Switzerland).Microfluidic chip fabrication
Microfluidic chips were fabricated on a 4′′ glass wafer (Planoptik, Elsoff, Germany) using two photolithography steps in a clean room facility. On each wafer, 40 replicates of the chip design were fabricated simultaneously. The mask designs for the lithographic steps were prepared using L-edit from Mentor Graphics (Oregon, USA). The designs were patterned on soda lime masks using a direct laser writer (DWL 2000, Heidelberg Instruments, Heidelberg, Germany). First, the capillary pinning features were patterned using a 10 μm-thick SU-8 layer, then the microchannel walls were patterned using a 50 μm-thick SU-8 layer (SU-8 3010 and SU-8 3050, respectively, MicroChem Corp., Massachusetts, USA). At the end, a thin photoresist (AZ-4562) was deposited everywhere to protect the microstructures from debris during the dicing process and then cleaned in acetone followed by isopropyl alcohol.Reagent integration on-chip
All stock solutions for the integration of reagents were prepared using a 20 mM Tris-HCl buffer (pH 7.8) containing 0.2% BSA. Solutions containing diaphorase and G6PD also contained 3% trehalose. The reagents were integrated in the SCM using a non-contact piezo-dispensing robot (Nanoplotter 2.1 GeSiM, Dresden, Germany) and dried by leaving the solution to evaporate at room temperature. The reagents needed for the G6PD assay were integrated using two distinct solutions. A pattern composed of five spots of solution 1 (0.1 U μL−1 diaphorase) and five spots of solution 2 (200 mM MgCl2, 40 mM G6P, 2 mM NADP+, 26 mM maleimide, and 2 mM resazurin) were spotted in each of the three test zones of the SCM (test, negative control, positive control) with a pitch of 600 μm. Each spot was formed by dispensing eight droplets in the same location (total volume ∼960 pL). The spotting pattern of solution 2 was shifted by 300 μm along the pattern of solution 1, leading to an alternated spotting pattern. To integrate the inhibitor used in the negative control area, solution 3 (2 M MgSO4) was spotted on top of the existing spots of solution 1 and of solution 2 in the negative control test zone, with a pitch of 300 μm (total volume per spot ∼1.2 nL). To integrate the reagent needed for the positive control, five spots of solution 4 (5000 μU μL−1 G6PD) were spotted on top of the spots of solution 1 in the positive control test zone with a pitch of 600 μm (total volume per spot ∼240 pL). The security code was spotted using solution 5 (3.3 mg mL−1 erioglaucine disodium salt containing 10% glycerol). The chips were sealed using a dry film resist (DF-1050, EMS Inc., USA).Assay measurement and software
Images of the microfluidic chips were taken using a Nikon 1 J3 camera connected to a Leica MZ16 stereomicroscope, a slide scanner (VS120, Olympus) or a custom-made microscope equipped with an 8-megapixel CMOS camera and controlled by a Raspberry Pi. The fluorescence and absorbance measurements were performed using a plate reader (Tecan Infinite 200, Männedorf, Switzerland) equipped with a custom-made aluminium chip holder (see ESI† Fig. S7a). A code for evaluating the results and a code for generating a pseudo-random security code were written in Python. The program to run numerical simulations was written in MATLAB.Conclusions
In this work we implemented a quantitative assay for measuring the G6PD activity and the hemoglobin concentration on a single capillary-driven microfluidic chip and we were able to distinguish between deficient, intermediate and normal levels of G6PD activities in less than 1 minute using only 2 μL of sample. This device has several advantages. First, it is a modular and flexible platform, which combines two classes of assays into a single device, namely a hemoglobin concentration measurement and an enzymatic assay. There are various scenarios, where knowing the concentration of hemoglobin together with a diagnostic test can lead to a more accurate diagnosis,4,45,62,63 and this paper showed how this can be useful for G6PD deficiency. Second, we implemented a theoretical model based on the diffusion and reaction of the reagents to predict the generation of the fluorescent reporting molecule for the G6PD assay starting from spotted reagents in an SCM. The main innovation of this diffusion–reaction model is to fully account for the double enzymatic coupling between G6PD and diaphorase, which interact through the exchange of the co-factor NADPH and its oxidized form NADP+. This model can also be used to identify the limiting reagents in the enzymatic reaction and tune accordingly the initial concentrations of the reagents to minimize the amount of reagents required. Such a model is the first of its kind and we believe it will be used as a template for modelling various types of enzymatic reactions and optimizing reagent integration on-chip. Third, this theoretical model provides guidelines for integrating reagents by means of inkjet deposition and allows assays to be multiplexed with a high degree of precision in a very small area of the microfluidic chip. The flexibility in multiplexing assays provided by the SCM outperforms traditional lateral flow assays, where only a single positive control is usually performed together with the test. In this work we demonstrated the integration of a G6PD assay, a positive control and a negative control in less than 7 mm2 of chip area. Such an integration is particularly promising in light of recent developments on integrated, low cost and compact optical and fluorescence readers for mobile diagnostics. For these reasons, we envision that the SCM could be used as a robust and efficient microfluidic platform to perform a variety of solution-based assays, for example, for metabolic disorders, therapy monitoring, or for quality control of drugs in various settings. Fourth, we proposed an alternative method for calibrating an enzymatic assay against temperature variations directly on-chip. Temperature variations can originate, for instance, from ambient conditions and peripheral devices and will inevitably affect the enzymatic activity. The most common way to minimize the effect of temperature on enzymatic assays is by performing the assay at a constant and predefined temperature. This requires a temperature-controlled reader and is usually implemented on clinical analysers in central laboratories. Alternatively, the assay can be calibrated at different temperatures and the enzyme activity retrieved by measuring the ambient temperature. This method, however, requires an additional temperature sensor and electronic circuitry to measure the ambient temperature. Here, we normalized the test signal by the positive control signal and were able to cancel out variations in the output signal due to temperature. Such a feature is a smart alternative to avoid external peripherals and additional electronic circuitry and we were able to implement it thanks to the versatility of the SCM.In summary, this work presents a state-of-the-art experimental approach for performing enzymatic assays in an SCM that is guided by a detailed theoretical model for integrating reagents and predicting the resulting enzymatic reactions. Evidently, the SCM can play an important role in the implementation of solution-based assays, which are performed with very low sample volumes and a limited quantity of reagents. We therefore believe that this work will provide a solid basis for the design and implementation of many types of assays for high-performance point-of-care testing methods.
Author contributions
M. R. and E. D. conceived the research; M. R. performed the experiments; Y. T. gave inputs for experimental work and helped with the optimization of the chip fabrication; T. G. developed the theoretical model; S. C. wrote the software for running simulations; M. L. S. performed the simulations; M. R. and M. L. S. compared experimental data to the simulation results; C. M. N. and E. D. supervised M. L. S. and M. R.; M. R. Y. T. M. L. S. T. G. C. M. N. and E. D. discussed the results; M. R. and E. D. wrote the paper with inputs from all authors.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank A. Kashyap, R. Lovchik, D. Widerker and P. Mathur from IBM for discussions and H. Riel (IBM) and T. Scharnweber (KIT) for their continuous support. We thank DiaLine AG for providing us with G6PD reference control samples. We are grateful to Ina Krebber from Interregionale Blutspende SRK for advice and the procurement of blood samples. This work received funding from the European Union's Horizon 2020 research and innovation program under the Grant agreement No. [764476].Notes and references
- M. Cappellini and G. Fiorelli, Lancet, 2008, 371, 64–74 CrossRef CAS.
- L. Luzzatto and P. Arese, N. Engl. J. Med., 2018, 378, 1066 CrossRef.
- A. Minucci, B. Giardina, C. Zuppi and E. Capoluongo, IUBMB Life, 2009, 61, 27–34 CrossRef CAS.
- WHO, Guide to G6PD deficiency rapid diagnostic testing to support P. vivax radical cure Global Malaria Programme, 2018.
- R. E. Howes, M. Dewi, F. B. Piel, W. M. Monteiro, K. E. Battle, J. P. Messina, A. Sakuntabhai, A. W. Satyagraha, T. N. Williams, J. K. Baird and S. I. Hay, Malar. J., 2013, 12, 418 CrossRef PubMed.
- C. Ruwende, S. C. Khoo, R. W. Snow, S. N. R. Yates, D. Kwiatkowski, S. Gupta, P. Warn, C. E. M. Allsopp, S. C. Gilbert, N. Peschu, C. I. Newbold, B. M. Greenwood, K. Marsh and A. V. S. Hill, Nature, 1995, 376, 246–249 CrossRef CAS PubMed.
- E. F. Roth, C. Raventos-Suarez, A. Rinaldi and R. L. Nagel, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 298–299 CrossRef CAS.
- E. Beutler, Blood, 2008, 111, 16–24 CrossRef CAS PubMed.
- R. Rochford, C. Ohrt, P. C. Baresel, B. Campo, A. Sampath, A. J. Magill, B. L. Tekwani and L. A. Walker, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17486–17491 CrossRef CAS.
- B. Ley, G. Bancone, L. Von Seidlein, K. Thriemer, J. S. Richards, G. J. Domingo and R. N. Price, Malar. J., 2017, 16, 361 CrossRef.
- A. Anderle, G. Bancone, G. Domingo, E. Gerth-Guyette, S. Pal and A. Satyagraha, Int. J. Neonatal Screen., 2018, 4, 34 CrossRef PubMed.
- L. Von Seidlein, S. Auburn, F. Espino, D. Shanks, Q. Cheng, J. McCarthy, K. Baird, C. Moyes, R. Howes, D. Menard, G. Bancone, A. Winasti-Satyahraha, L. S. Vestergaard, J. Green, G. Domingo, S. Yeung and R. Price, Malar. J., 2013, 12, 112 CrossRef CAS PubMed.
- G. J. Domingo, N. Advani, A. W. Satyagraha, C. H. Sibley, E. Rowley, M. Kalnoky, J. Cohen, M. Parker and M. Kelley, Int. Health, 2019, 11, 7–14 CrossRef.
- A. Hirono, H. Fujii, T. Takano, Y. Chiba, Y. Azuno and S. Miwa, Blood, 1997, 89, 4624–4627 CrossRef CAS PubMed.
- T. J. Vulliamy, M. D'Urso, G. Battistuzzi, M. Estrada, N. S. Foulkes, G. Martini, V. Calabro, V. Poggi, R. Giordano, M. Town, L. Luzzatto and M. G. Persico, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 5171–5175 CrossRef CAS PubMed.
- A. Minucci, K. Moradkhani, M. J. Hwang, C. Zuppi, B. Giardina and E. Capoluongo, Blood Cells, Mol., Dis., 2012, 48, 154–165 CrossRef CAS.
- WHO, Updating the WHO G6PD classification of variants and the International Classification of Diseases - 11th Revision, 2019.
- J. Cunningham and A. Bosman, Revision of WHO classification of G6PD variants and International classification of diseases (ICD)-11, 2019.
- E. Beutler and M. C. Baluda, J. Lab. Clin. Med., 1966, 68, 137–141 CAS.
- E. Beutler and M. Mitchell, Blood, 1968, 32, 816–819 CrossRef CAS.
- G. J. Brewer, A. R. Tarlov and A. S. Alving, Bull. W. H. O., 1960, 22, 633–640 CAS.
- V. F. Fairbanks and E. Beutler, Blood, 1962, 20, 591–601 CrossRef CAS.
- H. Fujii, Nippon Ketsueki Gakkai Zasshi, 1984, 47, 185–188 CAS.
- A. Hirono, H. Fujii and S. Miwa, Nippon Nettai Igakkai Zasshi, 1998, 26, 1–4 Search PubMed.
- I. S. Tantular and F. Kawamoto, Trop. Med. Int. Health, 2003, 8, 569–574 CrossRef CAS.
- F. E. Espino, J. A. Bibit, J. B. Sornillo, A. Tan, L. Von Seidlein and B. Ley, PLoS One, 2016, 11, 1–12 CrossRef PubMed.
- K. E. Tinley, A. M. Loughlin, A. Jepson and E. D. Barnett, Am. J. Trop. Med. Hyg., 2010, 82, 210–214 CrossRef CAS PubMed.
- V. F. Fairbanks and L. T. Lampe, Blood, 1968, 31, 589–603 CrossRef CAS.
- C. J. F. Van Noorden, I. M. C. Vogels, J. James and J. Tas, Histochemistry, 1982, 75, 493–506 CAS.
- C. J. F. Van Noorden, F. Dolbeare and J. Aten, J. Histochem. Cytochem., 1989, 37, 1313–1318 CrossRef CAS.
- S. S. Shah, S. A. S. Diakite, K. Traore, M. Diakite, D. P. Kwiatkowski, K. A. Rockett, T. E. Wellems and R. M. Fairhurst, Sci. Rep., 2012, 2, 1–6 CAS.
- D. Nantakomol, R. Paul, A. Palasuwan, N. P. Day, N. J. White and M. Imwong, Malar. J., 2013, 12, 1–8 CrossRef PubMed.
- A. L. Peters and C. J. F. Van Noorden, J. Histochem. Cytochem., 2009, 57, 1003–1011 CrossRef CAS.
- G. Bancone, M. Kalnoky, C. S. Chu, N. Chowwiwat, M. Kahn, B. Malleret, P. Wilaisrisak, L. Rénia, G. J. Domingo and F. Nosten, Sci. Rep., 2017, 7, 1–8 CrossRef.
- N. LaRue, M. Kahn, M. Murray, B. T. Leader, P. Bansil, S. McGray, M. Kalnoky, H. Zhang, H. Huang, H. Jiang and G. J. Domingo, Am. J. Trop. Med. Hyg., 2014, 91, 854–861 CrossRef CAS.
- J. Zhang, Y. Xiang, M. Wang, A. Basu and Y. Lu, Angew. Chem., Int. Ed., 2016, 55, 732–736 CrossRef PubMed.
- Y. Lu, Y. Xiang and J. Zhang, US Pat., 2016/0252515A1, 2016 (PCT US14/64314) Search PubMed.
- Q. Yu, L. Xue, J. Hiblot, R. Griss, S. Fabritz, C. Roux, P. A. Binz, D. Haas, J. G. Okun and K. Johnsson, Science, 2018, 361, 1122–1126 CrossRef CAS.
- R. S. Sista, R. Ng, M. Nuffer, M. Basmajian, J. Coyne, J. Elderbroom, D. Hull, K. Kay, M. Krishnamurthy, C. Roberts, D. Wu, A. D. Kennedy, R. Singh, V. Srinivasan and V. K. Pamula, Diagnostics, 2020, 10, 1–18 CrossRef PubMed.
- M. Buttarello and M. Plebani, Am. J. Clin. Pathol., 2008, 130, 104–116 CrossRef.
- C. Higgins, Hemoglobin and its measurement, available at: https://acutecaretesting.org/~/media/acutecaretesting/files/pdf/hemoglobin-and-its-measurement.pdf.
- T. Srivastava, H. Negandhi, S. Neogi, J. Sharma and R. Saxena, J. Hematol. Transfus., 2014, 2, 1028 Search PubMed.
- S. Prahl, Tabulated Molar Extinction Coefficient for Hemoglobin in Water, available at: https://omlc.org/spectra/hemoglobin/summary.html.
- G. J. Domingo, A. W. Satyagraha, A. Anvikar, K. Baird, G. Bancone, P. Bansil, N. Carter, Q. Cheng, J. Culpepper, C. Eziefula, M. Fukuda, J. Green, J. Hwang, M. Lacerda, S. McGray, D. Menard, F. Nosten, I. Nuchprayoon, N. N. Oo, P. Bualombai, W. Pumpradit, K. Qian, J. Recht, A. Roca, W. Satimai, S. Sovannaroth, L. Vestergaard and L. Von Seidlein, Malar. J., 2013, 12, 1–12 CrossRef.
- T. Guo, R. Patnaik, K. Kuhlmann, A. J. Rai and S. K. Sia, Lab Chip, 2015, 15, 3514–3520 RSC.
- O. Gökçe, S. Castonguay, Y. Temiz, T. Gervais and E. Delamarche, Nature, 2019, 574, 228–232 CrossRef.
- E. Hemmig, Y. Temiz, O. Gökçe, R. D. Lovchik and E. Delamarche, Anal. Chem., 2020, 92, 940–946 CrossRef CAS PubMed.
- O. Gökçe, C. Mercandetti and E. Delamarche, Anal. Chem., 2018, 90, 7383–7390 CrossRef.
- M. L. Salva, M. Rocca, Y. Hu, E. Delamarche and C. M. Niemeyer, Small, 2020, 2005476, 1–8 Search PubMed.
- Y. Arango, Y. Temiz, O. Gökçe and E. Delamarche, Sci. Adv., 2020, 6, 16 Search PubMed.
- M. L. Salva, Y. Temiz, M. Rocca, Y. C. Arango, C. M. Niemeyer and E. Delamarche, Sci. Rep., 2019, 9, 17242 CrossRef PubMed.
- A. Olanrewaju, M. Beaugrand, M. Yafia and D. Juncker, Lab Chip, 2018, 18, 2323–2347 RSC.
- D. Juncker, H. Schmid, U. Drechsler, H. Wolf, M. Wolf, B. Michel, N. De Rooij and E. Delamarche, Anal. Chem., 2002, 74, 6139–6144 CrossRef CAS PubMed.
- J. Boeckx, M. Hertog, A. Geeraerd and B. Nicolai, Plant Methods, 2017, 13, 1–12 CrossRef PubMed.
- H. Bisswanger, Perspect. Sci., 2014, 1, 41–55 CrossRef.
- V. K. Bhutani, M. Kaplan, B. Glader, M. Cotten, J. Kleinert and V. Pamula, Pediatrics, 2015, 136, e1268–e1275 CrossRef.
- Who, Haemoglobin concentrations for the diagnosis of anaemia and assessment of severity, 2011.
- H. H. Billett, in Clinical Methods: The History, Physical, and Laboratory Examinations, ed. H. K. Walker, W. D. Hall and J. W. Hurst, 3rd edn, 1990, vol. 28 Search PubMed.
- PATH, Pointe Scientific Glucose-6 Phosphate Dehydrogenase Quantitative Test: Standard Operating Procedure, available at: https://path.azureedge.net/media/documents/Pointe-Scientific-G6PD-Quantitative-Test_SOP_PATH.pdf.
- S. Pal, P. Bansil, G. Bancone, S. Hrutkay, M. Kahn, G. Gornsawun, P. Penpitchaporn, C. S. Chu, F. Nosten and G. J. Domingo, Am. J. Trop. Med. Hyg., 2019, 100, 213–221 CrossRef CAS.
- L. Nan, Z. Jiang and X. Wei, Lab Chip, 2014, 14, 1060–1073 RSC.
- S. Gupta, U. Jain and N. Chauhan, J. Nanomed. Res., 2017, 5, 1–10 Search PubMed.
- A. Laudisio, S. Bandinelli, A. Gemma, L. Ferrucci and R. A. Incalzi, J. Am. Geriatr. Soc., 2013, 61, 963–968 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00354b |
| This journal is © The Royal Society of Chemistry 2021 |