Open Access Article
Open Access ArticleAntimicrobial susceptibility testing by measuring bacterial oxygen consumption on an integrated platform†
Yang
Liu
 a,
Thomas
Lehnert
a,
Torsten
Mayr
a,
Thomas
Lehnert
a,
Torsten
Mayr
 b and
Martin A. M.
Gijs
b and
Martin A. M.
Gijs
 *a
*a
aLaboratory of Microsystems, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. E-mail: martin.gijs@epfl.ch
bInstitute of Analytical Chemistry and Food Chemistry, Graz University of Technology, 80 Graz, Austria
First published on 16th July 2021
Abstract
Cellular respiration is a fundamental feature of metabolic activity and oxygen consumption can be considered as a reliable indicator of bacterial aerobic respiration, including for facultative anaerobic bacteria like E. coli. Addressing the emerging global health challenge of antimicrobial resistance, we performed antimicrobial susceptibility testing using the bacterial oxygen consumption rate (OCR) as a phenotypic indicator. We demonstrated that microbial exposure to antibiotics showed systematic OCR variations, which enabled determining minimum inhibitory concentrations for three clinically relevant antibiotics, ampicillin, ciprofloxacin, and gentamicin, within a few hours. Our study was performed by using photoluminescence-based oxygen sensing in a microchamber format, which enabled reducing the sample volume to a few hundred microliters. OCR modeling based on exponential bacterial growth allowed estimating the bacterial doubling time for various culture conditions (different types of media, different culture temperature and antibiotic concentrations). Furthermore, correlating metabolic heat production data, as obtained by nanocalorimetry in the same type of microchamber, and OCR measurements provided further insight on the actual metabolic state and activity of a microbial sample. This approach represents a new path towards more comprehensive microbiological studies performed on integrated miniaturized systems.
Introduction
Emerging antimicrobial resistance (AMR) is evolving into a global public health challenge with the potential to undo decades of medical advancements.1 Better control of unregulated and abusive use of antibiotics has been identified as one of the key measures to expand the lifespan of currently available drugs and to impede AMR spreading.2 In this view, antimicrobial susceptibility testing (AST) enabling decisions on accurate treatment protocols at an early stage of bacterial infection plays an important role. Several innovative and fast AST methods have been proposed, addressing the main drawbacks of clinically established methods.3,4More recently, microfluidic and microtechnology-based approaches opened the way to new AST paradigms. For instance, Baltekin et al. achieved a read-out time of less than 30 min by monitoring directly bacterial growth in a microfluidic array of single-cell traps. The growth rate response to different antibiotics was determined as average over many individual cells.5 Jiang et al. proposed a system for monitoring bacterial growth profiles and rapid phenotypic bacterial screening in milliliter-sized droplets with defined bacteria and antibiotic mixtures. Reliable minimum inhibitory concentrations (MICs) could be obtained within a few hours of incubation.6 Microbial metabolic heat profiles, either recorded in milliliter ampoule-format or with an isothermal nanocalorimeter platform, provide detailed information on the time evolution of the metabolic activity. Such drug-dependent metabolic footprints were used as antibiotic susceptibility indicators.7,8 Kara et al. took advantage of the nanomechanical movement of surface-immobilized bacteria in a microchannel to determine antimicrobial susceptibility.9 Cantilever-based nanomechanical sensors have also been explored as a tool to characterize the metabolism of adsorbed bacteria and to quantitatively screen their response to antibiotics within minutes.10,11
Various traditional methods exist to determine dissolved oxygen (DO) concentrations on a macroscopic scale. Winkler titration is a precise process that is well-suited for calibration, but it cannot monitor DO continuously, as is required for biological studies.12 Also a micro-Winkler titration method has been proposed.13 The emission of short-live radioisotopes allows oxygen sensing with spatial resolution suitable for full-body imaging.14 However, magnetic or electron paramagnetic resonance techniques are expensive, difficult in calibration, and cumbersome, and therefore not suitable to apply on small biological samples.15,16 DO sensing in a microfluidic format requires approaches that are compatible with small-scale integration. Clark-type electrodes allow measuring DO concentrations electrochemically, and have been adapted for miniaturized applications when using platinum thin-film electrodes covered with an oxygen-permeable membrane.17,18 Optical techniques for oxygen sensing, in particular nanomaterial-based sensors for quantifying DO concentrations in a microfluidic system, have competitive advantages over the previously cited methods.19–21 As oxygen is a luminescence quencher, the DO concentration may be correlated to the photoluminescence intensity of the sensing dye. However, photobleaching has to be considered, and recording luminescence decay or the phase shift of excitation/emission pulses are more reliable sensing methods. Compared to a Clark-type sensor, advantages of the optical method include the absence of intrinsic oxygen consumption during the measurements and a higher degree of system integrability, as no direct electrical connections are required for signal read-out. They are particularly important in miniaturized systems with small sample volumes.
Despite the obvious need for new and faster AST techniques, the potential of OCR assays for quantifying metabolic activity has not been investigated to a large extent for the time being. The commercial Seahorse extracellular flux analyzer provides parallelized OCR measurement capabilities in a microplate format with possible injection of compounds. The cellular respirometry assays in this system are based on optical sensors.22 Nevertheless, to the best of our knowledge, AST studies using this system have not been reported yet. Recently, Jewell et al. developed an assay format for monitoring OCR dynamics of Pseudomonas aeruginosa biofilms exposed to antibiotics. A susceptibility study was performed by incorporating an oxygen nanosensor directly into the biofilm.23 Jusková et al. proposed microfluidic chamber arrays, in which oxygen sensing nanoprobes were dispersed in agarose medium and in contact with a small volume of bacterial suspension. Oxygen-dependent nanoprobe luminescence was measured as viability indicator and for performing AST.24 Qiu et al. recently presented a device for AST based on a digital microfluidic platform using a similar principle.25 At least two other earlier studies reported AST of clinical bacterial isolates based on oxygen consumption (OC). Assays were performed in a well plate format with an electrochemical sensor array.26,27 However, electrochemical methods are much more sensitive to culture conditions than photoluminescence-based methods. Most of the previous platforms involve complex fabrication procedures and allow measuring only a single metabolic parameter. A simple integrated analytical system, enabling the assessment of multiple parameters simultaneously and/or under equal experimental conditions would therefore bring about significant advantages for microbial metabolic studies.
In this paper, we present simple and robust microbial OCR measurements in a microfluidic chamber. We analyzed the specific OCR footprints of bacterial growth under different culture conditions, in particular in different culture media and at different temperatures. An OCR model estimating the bacterial doubling time is proposed. Taking advantage of OCR as a microbial viability indicator, we performed a proof-of-concept AST study based on purified bacterial samples, involving three clinically relevant antibiotics, i.e. ampicillin, ciprofloxacin, and gentamicin. Our platform takes advantage of photoluminescence-based oxygen sensing units that are incorporated in a microfluidic system. The sample reservoirs are fabricated by micro-milling in gas non-permeable polycarbonate, as is required for OC measurements. The oxygen sensor units are reusable thus providing an economical solution suitable for array integration with disposable sample units. Compared to existing technologies, our system design paradigm can be applied to a range of different assays. Here we demonstrate simultaneous heat and oxygen measurements in a miniaturized reservoir. By this means, and based on a previous nanocalorimetric study,8 we were able to evaluate the correlation between microbial metabolic heat and OCR. This new approach paves the way toward integrated analytical systems enabling more holistic studies and diagnostics in microbiology.
Materials and methods
Materials and chemicals
E. coli ATCC 25922 strain (E. coli WDCM 00013 Vitroids™), ciprofloxacin (17850, CAS Number 85721-33-1), ampicillin sodium salt (A0166, CAS Number 69-52-3), gentamicin (G1264, CAS Number 1405-41-0), lysogeny broth (L3522), and sodium sulfite (239321, CAS Number 7757-83-7) were purchased from Sigma-Aldrich (Buchs, Switzerland). Mueller Hinton agar (CM0337), Mueller Hinton broth (CM0405), and brain heart infusion broth (CM1135) were purchased from Thermo Scientific (Reinach, Switzerland). RAPICIDE™ OPA/28 high-level disinfectant was purchased from Cantel Medical Corp (NJ, USA).Sample preparation
Assays in this work were carried out using the E. coli ATCC 25922 strain, which is a typical AST control strain. We retrieved bacteria from −20 °C and performed overnight bacterial culture in liquid Mueller Hinton media (MH) with shaking. Subsequently, 10 μL of this suspension was cultured on MH agar plates and diluted by streaking. Single colonies were picked to prepare a bacterial suspension with an optical density value at 600 nm (OD600) of 0.2 (around 0.5 McFarland standard), i.e. a concentration of ∼1 × 108 CFU mL−1. By dilution in pure culture medium we obtained the primary samples for the OCR and heat flow assays with an initial bacterial concentration of 2.8 ± 0.5 × 105 CFU mL−1 for all experiments (confirmed by 3 serial quality control experiments). This concentration is in the recommended range for standard AST.28 A purity, viability and concentration check prior to each measurement series was carried out by inoculation on a non-selective MH agar plate and colony counting after overnight culture. We determined that 1.0 OD600 corresponds to 3.7–5.5 × 108 CFU mL−1 in our case, which is coherent with a recent systematic OD and bacterial number study.29 All bacterial suspensions were stored in a small flacon and used within 15 min. We prepared a sub-culture on a new MH agar plate by picking single colonies from the previous one and cultured overnight to guarantee a fresh sample for the experiments during the next day. This protocol is performed for no more than one week to avoid strain mutation. After one week, a new bacterial culture cycle restarts from the −20 °C stock.Antimicrobial solutions used in this study were prepared according to the recommended protocols. Ampicillin sodium salt and gentamicin are soluble in deionized (DI) water. 1 mol L−1 hydrogen chloride solution was used for ciprofloxacin. Ciprofloxacin, ampicillin, and gentamicin stock solutions were prepared with a concentration of 25 mg mL−1, 50 mg mL−1 and 36 mg mL−1, respectively. Stock solutions were stored at 4 °C and used within one week to avoid a risk of declining activity. Prior to the OCR measurements, stock solutions were thermalized at room temperature and gradually diluted in DI water to prepare a series of concentrations according to the potency of each antibiotic (10 times higher than the final target concentration). Test solution aliquots were prepared by further 10-fold dilution in MH. Target concentrations were chosen according to the MIC range suggested by the European Committee on Antimicrobial Susceptibility Testing (EUCAST).30
Statistical analysis
All experiments shown in this work were carried out by performing each time three nominally identical assays. Statistical analysis and modeling of results were performed using OriginLab 2020 and Python (package: SciPy and pandas).Experimental
OCR measurement set-up
The oxygen sensitive coating was provided by the Institute of Analytical Chemistry and Food Chemistry (Assoc. Prof. T. Mayr, Graz University of Technology, Austria). Phosphorescent sensors spots were prepared by applying a sensor formulation with a microdispenser MDS3200+ from VERMES Microdispensing GmbH, equipped with a 70 μm nozzle and a tungsten tappet with a tip diameter of 0.7 mm onto polymeric substrate as described by Müller et al. onto a standard glass slide.31 Sensor spots have a diameter of ∼1 mm and a thickness of ∼2 μm. The sensor formulation consists of 1.65 mg of platinum(II) meso-tetra(4-fluorophenyl) tetrabenzoporphyrin (PtTPTBPF), 76.7 mg polystyrene (PS, 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Fisher Scientific) and silicone rubber ELASTOIL® E4 (Wacker) dissolved in 500 mg in toluol. The dye was excited at 620 nm and emits near-infrared light at ∼700 nm. The sensing unit worked in the frequency domain, i.e. by correlating the excitation/emission light phase shift with the oxygen concentration.32 Calibration data for the oxygen sensor spot were similar to reported in Ehgarnter et al.33
000 g mol−1, Fisher Scientific) and silicone rubber ELASTOIL® E4 (Wacker) dissolved in 500 mg in toluol. The dye was excited at 620 nm and emits near-infrared light at ∼700 nm. The sensing unit worked in the frequency domain, i.e. by correlating the excitation/emission light phase shift with the oxygen concentration.32 Calibration data for the oxygen sensor spot were similar to reported in Ehgarnter et al.33
A schematic of the central part of the OCR measurement system is shown in Fig. 1a. A cylinder-shaped microincubator was fixed on a glass slide directly over the thin-film optical oxygen sensor spot. The microincubator was fabricated by micro-milling in polycarbonate, which has a low gas permeability.34,35 Samples were injected into the microincubator by air pressure control in the external reservoir. In order to avoid oxygen exchange with the environment during the OCR measurements, the microincubator was isolated by switch valves located in the inlet and outlet pathways, respectively. Optical access to the sensor was provided from the bottom. An optical sensing unit was positioned beneath the sensor glass slide, for light excitation and recording of the near-infrared emission signals with a photodetector (PICO2-OEM, PyroScience GmbH, Germany) (Fig. 1b). Bacterial sample populations sediment on the bottom of the microincubator close to the sensing dye. We therefore expect that variations in the bacterial metabolic activity and corresponding changes of DO concentrations can be measured almost simultaneously. Fig. 1c shows a photo of a microincubator for the OCR measurements. The microincubator and part of the connecting fluidic tubing were implemented in a PID-controlled thermostat for maintaining the culture temperature constant. In terms of system integration, as well as fluidic and thermal control, the OCR platform is based on the same design principles as the nanocalorimetry platform previously developed by our group.8 Actually, we considerably advanced the latter study in the present work by correlating two distinct metabolic parameters, namely OCR and microbial heat production. For the nanocalorimetry measurements the optical sensor/glass substrate was replaced by a thermopile sensor chip (XEN-9924, Xensor Integration, Netherlands, shown in Fig. 1d). The same microincubator format was used for both types of assay, enabling parallel real-time OCR and heat measurements under quasi-identical assay conditions.
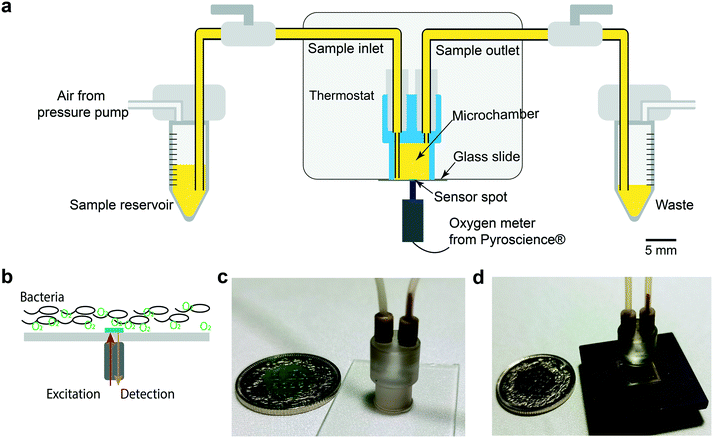 | ||
| Fig. 1 Microfluidic-assisted OCR measurement set-up. (a) Schematic cross-sectional view of the oxygen sensing unit with fluidic connections. A microincubator is attached to the glass slide featuring the sensing spot. The optical device for sensor excitation and capturing the oxygen-dependent luminescence signal is positioned underneath. The sample is injected into the microincubator from an external reservoir by pressure control. The microincubator may be isolated from the environment by valves in the fluidic tubing. Thermal system integration is similar to that used in a nanocalorimetric platform developed by our group.8 (b) The sensor spot location (green, diameter ∼1 mm) and the position of the optical device allows measuring the oxygen concentration of the bacterial population sedimented in the microincubator. (c) Photograph of the microincubator (outer diameter ∼6 mm at the bottom) positioned on the sensor glass slide with fluidic connectors. (d) Photograph of a microincubator positioned on the thin-film thermopile-based heat flow sensor for measuring the metabolic heat production during bacterial growth. | ||
Measurement protocol
The sensor was first calibrated by a 2-point calibration procedure, i.e. by filling the microincubator with two types of liquid. DI water was used to calibrate the DO saturation point at a certain temperature. 0.1 mol L−1 sodium sulfite (Na2SO3) solution was used to calibrate the zero point (fully depleted DO) as it reduces all the oxygen in the solution to O2−. After each OCR measurement, the microincubator and tubing were first cleaned with ethanol (3 times) to remove most of the bacterial suspension residues. Then the chamber was soaked with RAPICIDE™ OPA/28 for 20 min at 37 °C for sterilization. After that the system was eluted with ethanol (3 times) and sterilized DI water (5 times) to remove detergent residues. Sterilization was verified by injecting pure MH broth into the system and overnight incubation. If the oxygen concentration in the solution is maintained at the initial level after this protocol, no bacteria should persist in the microincubator. In this way, the microfluidic chip and the sensing unit can be reused for a new experiment. As a remark, we found that applying 15% H2O2 for sterilizing the system strongly perturbed subsequent measurements, possibly due to decomposition of H2O2 residues into O2 and H2O.Results
Measurement of microbial oxygen consumption and modeling
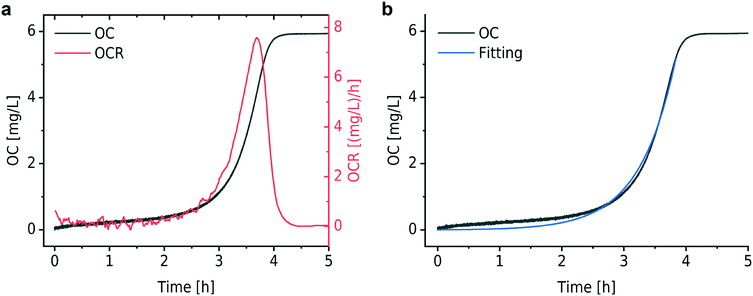
Fig. 2a shows a typical experimental OC and derived OCR curve that were recorded after bacterial inoculum loading into the micro-incubator on the platform (E. coli, ∼3 × 105 CFU mL−1 in MH). The black curve indicates the total amount of oxygen consumed (OC) per volume over time in the microincubator. Similar to bacterial growth curves, the OC curve starts with a slowly rising section before an exponentially increasing phase. OC increased continuously until the DO in the sealed chamber was depleted (at ∼4 h in Fig. 2a). At this point, bacterial metabolism is expected to change from the aerobic to an anaerobic state. OCR for a bacterial culture corresponds to the time-derivative of the recorded OC curve and is shown in Fig. 2a as the red curve. The OCR curve also features a strong exponential increase up to a maximum value, followed by a transition region with decreasing OCR until oxygen depletion.In a nutrient-rich medium, growth of bacteria like E. coli obeys an exponential model with a doubling time tdb, as expressed by the following equation36
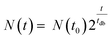 | (1) |
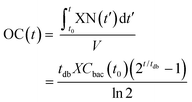 | (2) |
For the modeling of OC(t), e.g. the black curve shown in Fig. 2b (same as in Fig. 2a), we consider two fitting parameters determined by the iterative regression, namely tdb and XCbac(t0), which are highly related to the bacterial metabolic activity. tdb determines the characteristic time constant of the exponential phase in the OC curve. The blue curve in Fig. 2b shows the result of the fitting with our model. A customized Python program was applied for automated fitting in a defined range. As a first step, the program detects the maximum oxygen consumption (OCmax) value, corresponding to the plateau region in the OC curve (Fig. 2a and b, black curve). Then we define a fitting region, where the OC is in the range of 0 to (OCmax − 0.5 mg L−1). This restriction allows OC fitting from the beginning of an experiment at t = 0 h until about 30 min before full oxygen depletion, i.e. we do not consider the transition region, where the increasing lack of oxygen in the microincubator starts influencing bacterial growth. The fitted curve corresponds well to the experimental curve in the region of interest.
Microbial OCR for different culture conditions
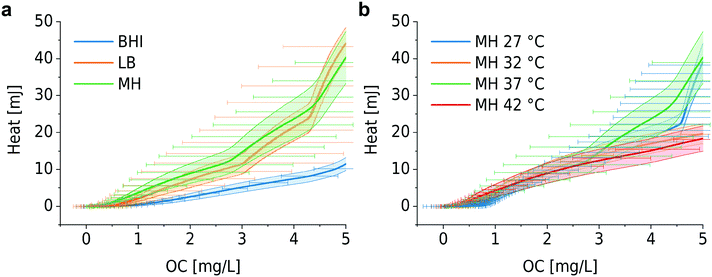
We found that bacterial OCR depends on the nutrient level of the culture medium and the temperature, as shown in Fig. 3a and b. Theses curves have been derived from the corresponding OC curves Fig. S1a and S1b,† respectively. Table S1† provides an overview of the nutrient content of different culture media, indicating that BHI has a higher nutrient level than MH and LB. During incubation in BHI, OC increased faster and the peak in the OCR signal appeared earlier than for the other two media, for which the maximum OCR occurred at the same time. However, this tendency is subjected to certain variations and may be more or less pronounced (as can be seen in the measurement series in Fig. S1a and S2a†). Except for an evident displacement and a variation of the peak amplitude, the underlying shape of the OCR curves in Fig. 3a is similar, indicated by the overlap of the OCR curves for MH and LB in the growing part, for instance. A more systematic variation of the OCR curves was observed for different culture temperatures. The highest OCR peak value was found at 42 °C, and gradually decreasing values were observed as the temperature was lowered (Fig. 3b). The slopes of the OCR curves and peak values also decreased for lower temperatures, accompanied by a shift on the time scale. This evolution reflects the impact of culture temperature on the enzymatic efficiency of cellular metabolic reactions. The OC of the microbial population is clearly slowed down at the more unfavorable growth conditions (27 °C, Fig. 3b). Additional OC measurement series and corresponding OCR curves are shown in Fig. S1 and S2,† respectively. The oxygen sensing method is based on measuring the phase shift between the excitation light and the emission light. The intensity of the emission light, which also influences the accuracy of the phase shift measurement, is inversely proportional to the DO concentration. Consequently, larger signal variations were observed in the initial phase of the OCR curves prior to oxygen depletion (Fig. 3a), or above MIC (purple curves, Fig. 4) for which the DO concentration is maintained at the initial level.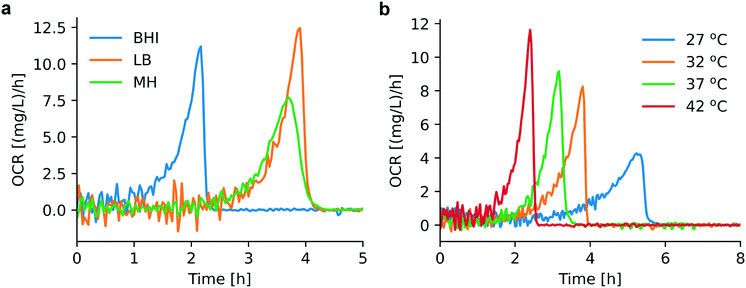 | ||
| Fig. 3 Assessment of OCR under different cultural conditions. (a) OCR for bacteria in different culture media (derived from Fig. S1a,† series 1): BHI (blue), MH (green), LB (orange). Earlier depletion occurs for bacterial culture in BHI compared to MH and LB. (b) OCR for bacterial culture in MH at different temperatures (derived from Fig. S1b,† series 1): 27 °C (blue), 32 °C (orange), 37 °C (green), 42 °C (red). Appropriate culture temperatures (37 °C and 42 °C) promote bacterial growth and consequently OCR increases. | ||
Results of parametric modeling for the bacterial growth in different culture media and at different temperatures are shown in Table 1. We observe that our model allows determining the doubling time tdb and see that, for bacteria cultured at unfavorable conditions (MH at 27 °C), tdb is strongly prolonged (∼71 min) with respect to more suitable conditions (∼24 min, e.g. for 37 °C in MH or LB). The highest XCbac(t0) value was found for BHI at 37 °C, which implies highest OCR per inoculum size. However, in all cases the error range for XCbac(t0) is considerable. We suppose that this is mainly due to the bacterial sample preparation step, as the initial concentration Cbac(t0) can only be controlled within a certain range and the variability for the sample preparation time prior to each measurement may also influence the initial conditions. Our current OCR platform only supports one test of the bacteria concentration at the beginning of the experiment. An improved system supporting successive evaluations of the bacteria concentration may help to determine XCbac(t0) more accurately.
| Condition | Doubling time tdb [min] | XC bac(t0) [mg L−1 min−1] |
|---|---|---|
| MH at 37 °C | 23 ± 1 | 5.7 × 10−4 ± 3.1 × 10−4 |
| BHI at 37 °C | 31 ± 5 | 4.6 × 10−3 ± 1.3 × 10−3 |
| LB at 37 °C | 25 ± 3 | 1.1 × 10−3 ± 8.2 × 10−4 |
| MH at 27 °C | 71 ± 9 | 1.9 × 10−3 ± 1.1 × 10−3 |
| MH at 32 °C | 35 ± 6 | 1.2 × 10−3 ± 6.5 × 10−4 |
| MH at 42 °C | 21 ± 3 | 1.9 × 10−3 ± 9.4 × 10−4 |
OCR for antimicrobial susceptibility testing
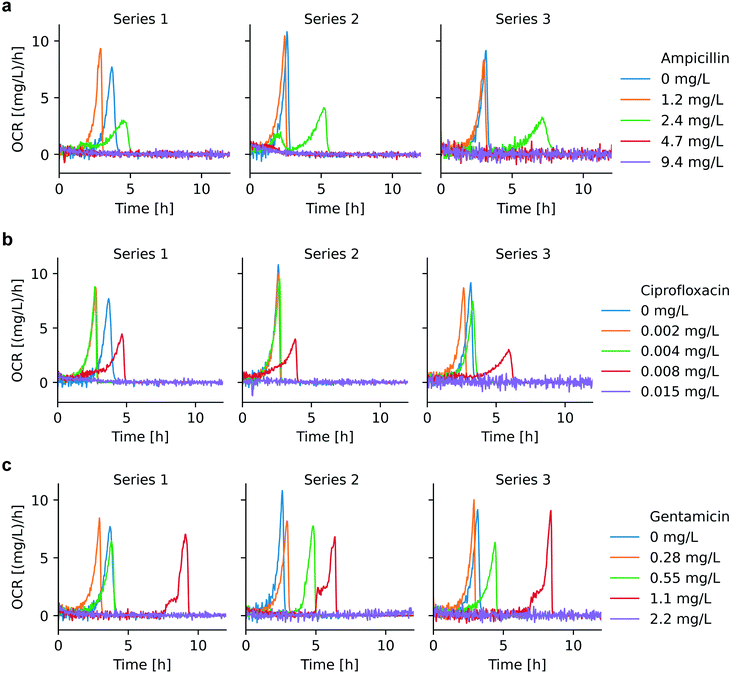
OCR can be applied as a viability indicator for non-anaerobic bacteria to evaluate metabolic activity and thus to perform AST. We have recorded bacterial OC curves for different pharmacological conditions, specifically for culture with different antibiotics in a relevant concentration range. OC curves for ampicillin, ciprofloxacin, and gentamicin are shown in Fig. S3.†Fig. 4 shows the corresponding OCR curves derived for these assays. Bacterial culture with different antibiotics demonstrated specific OCR patterns from which the minimum inhibitory concentration (MIC) could be determined. MIC value ranges, as defined by EUCAST, are indicated in Table S2.† For antibiotic concentrations below MIC, OCR continuously increased due to bacterial growth, followed by a relatively sharp drop until full oxygen depletion occurred. Maximum OCR rates are increasingly delayed for higher antibiotic concentrations. A strong growth inhibition effect, reflected also by longer lag phases, could be observed for ampicillin at 2.4 mg L−1 and ciprofloxacin at 0.008 mg L−1, for instance (green curves in Fig. 4a, and red curves in Fig. 4b, respectively). All 3 antibiotics tested are bactericidal, thus the OCR in the microchamber incubator remained at zero-level above MIC, as was observed for ampicillin at 4.7 mg L−1 and 9.5 mg L−1 (red and purple curves in Fig. 4a, respectively) and for ciprofloxacin at 0.015 mg L−1 (purple curve in Fig. 4b). For ampicillin and ciprofloxacin, OC curves above MIC (Fig. S3,† purple curves) show a slight increase at the beginning of the measurements before reaching a constant level, corresponding to lack of oxygen consumed due to ceasing metabolic activity (the corresponding minor OCR signal increase is hardly visible in the graphs of Fig. 4a and b). We assume that this effect is due to the fact that, in the initial phase of the assays, all bacteria are not yet killed, even by the high antibiotic dose. For example, the action of ciprofloxacin is known to be the blocking of DNA division,37 thus making it impossible for the treated bacteria to replicate, a process that will have some delay with respect to the application of the antibiotic.In the specific case of gentamicin, OCR at subinhibitory concentrations evolves in principle as for ciprofloxacin and ampicillin with pronounced delays close to the expected MIC (e.g. red curve in Fig. 4c, 1.1 mg L−1). OCR remained zero at a concentration of 2.2 mg L−1, i.e. at the upper limit of the suggested MIC value range (1.1 mg L−1 < MIC ≤ 2.2 mg L−1) up to an incubation duration of 12 h in the microincubator. However, more extended OC measurements as shown in Fig. S3c† (purple curves) indicated that OC raised after a more prolonged incubation period (t > ∼10 h to 13 h). This fact suggests that a small population of bacteria was able to survive and starts consuming oxygen after a long lag phase due to a strong inhibition effect at this high gentamicin concentration. The antimicrobial action of gentamicin is based on an oxygen-dependent process.38 It is therefore possible that diffusion of oxygen to bacteria clusters attached on the surface of the oxygen sensing spot is limited thus inhibiting temporarily the action of gentamicin. Additional features of the OCR curves close to MIC, in particular a two-level increase in the red curve in Fig. 4c, suggests further specificities in the mode of action of gentamicin. This crinkle-shaped transition possibly is related to the prominent inhibition effect of gentamicin in the initial region, and the subsequent decreased potency due to the lack of oxygen.
Table 2 shows the OC modeling results based on eqn (2) in the presence of different antibiotics. A clear prolonged doubling time tdb is observed for increasing antimicrobial concentrations. However, whether bacteria exposed to antimicrobial stress are still proliferating in the normal way, i.e. by dividing and cell doubling, needs further investigation. From a microscopic view, antibiotics such as ampicillin may generate a significant filamentation effect, i.e. an elongation of the cell body without division, thus bacteria may accumulate biomass without increasing their number. Similarly, exposure to ciprofloxacin being a genotoxic antibiotic inducing the so-called SOS response, which refers to a DNA repair network, changes the E. coli rod shape into multi-chromosome containing filaments.39 In these cases, the bacterial size distribution is larger than for normal growth conditions, where divided bacteria usually have identical biomass.
| Condition | Doubling time tdb [min] |
|---|---|
| Ampicillin 1.2 mg L−1 | 29 ± 5 |
| Ampicillin 2.4 mg L−1 | 110 ± 30 |
| Ciprofloxacin 0.002 mg L−1 | 27 ± 3 |
| Ciprofloxacin 0.004 mg L−1 | 29 ± 2 |
| Ciprofloxacin 0.008 mg L−1 | 90 ± 10 |
| Gentamicin 0.28 mg L−1 | 32 ± 4 |
| Gentamicin 0.55 mg L−1 | 44 ± 7 |
| Gentamicin 1.1 mg L−1 | 43 ± 4 |
| Gentamicin 2.2 mg L−1 | 180 ± 60 |
Heat vs. oxygen consumption
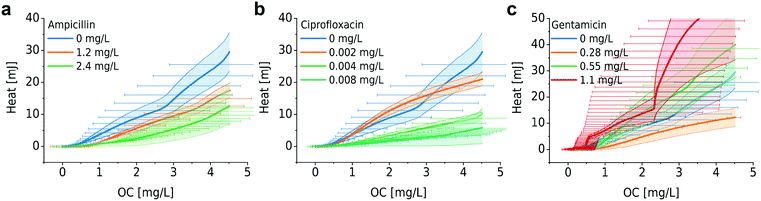
As already explained in the context of Fig. 1, we have previously developed a nanocalorimeter platform.8 Both platforms, the one for OC and for metabolic heat assays, respectively, are based on the same fluidic and thermal integration and control principles. By this means, we were able to study microbial heat production and OC of quasi-identical bacterial samples (within in the range of inoculum variability) under comparable conditions, and we could correlate both metabolic parameters for different assay conditions. Fig. 5 and 6 show the plots of heat vs. OC for the range of experimental conditions described before. With respect to the culture media, MH and LB generated comparable curves (Fig. 5a, orange and green curves), whereas in the richer BHI medium heat-per-OC values are considerably lower (blue curve). Corresponding metabolic heat curves are shown in Fig. S4a.† Variations of the slopes in plots for bacteria growing at different temperatures in MH (Fig. 5b) were less prominent. The total heat generated per amount of consumed oxygen remained at the same level in this region, despite the fact that systematic lag phase variations were observed for OC (Fig. S1b†) and heat production (Fig. S4b†).Furthermore, we investigated the relation between metabolic heat production and OC in the presence of antimicrobials (Fig. 6). Corresponding heat curves are shown in Fig. S5.† In our previous work we have shown that bacteria may produce more heat for the synthesis of the same amount of biomass in the presence of antibiotics (i.e. the same OD600). We called this phenomenon “energy spilling” that revealed that antibiotics do alter the bacterial metabolic activity.8 Considering the heat vs. OC curves, no clear systematic effect was observed. For ciprofloxacin and ampicillin (Fig. 6a and b, respectively), an increasing antimicrobial concentration may lead to a lower heat production per consumed unit amount of oxygen (lower slope of the curves). However, for gentamicin no systematic effect was observed (Fig. 6c). Correlating these two types of measurements results in a big error range and further improvement of the set-ups is required for a more detailed analysis.
Discussion
Oxygen is associated to aerobic metabolic activity. As the main carbohydrate, glucose is converted to pyruvate by the process of glycolysis in the cytoplasm. In eukaryotes, mitochondria couple pyruvate oxidation to the production of a high amount of adenosine triphosphate (ATP) by the electron transport chain.40 Under aerobic respiration, i.e. in the presence of oxygen that serves as the electron acceptor, pyruvate is metabolized and ATP is formed with energy loss in terms of heat. Under anaerobic conditions (without oxygen), pyruvate undergoes a fermentation process in which energy can be produced. In bacteria, depending on the species, different toxic compounds can be generated as well, such as lactic acid, ethanol, or mixed acids. An important aspect of the study of cellular OC is that it helps in understanding mitochondrial activity. Mitochondrial dysfunction in eukaryote cells is highly related to metabolic disorders, e.g. obesity or diabetes mellitus, but also to cardiovascular diseases and neurodegenerative disorders, such as Alzheimer's disease, Parkinson's disease, Huntington's disease and amyotrophic lateral sclerosis.41,42 There is evidence that mitochondria in eukaryotes have bacterial origin,43 thus OC measurements on bacteria may also be considered as a relevant assay for evaluating mitochondrial dysfunction.OCR can be explored as a potential parameter for AST on aerobic bacteria within a time scale of only a few hours and the presented method is also useful to determine antimicrobial susceptibility of facultative anaerobic bacteria. Nevertheless, oxygen depletion in the microchamber must be considered carefully when evaluating the results. In principle, according to the OCR curves shown in Fig. 4a and b, MIC values for ampicillin and ciprofloxacin could be determined on a time scale in the range of 4–7 hours. MIC values obtained with the OCR method are coherent with the suggested EUCAST values for E. coli ATCC 25922 in the case of ampicillin and ciprofloxacin (see Table S2†). However, specific properties of the antibiotic may have an impact on the accuracy of the AST assays. This was observed in the case of gentamicin, where the MIC interval determined with the oxygen-sensing platform was above EUCAST reference values. The oxygen-dependent antimicrobial action of gentamicin may be at the origin of this observation.38 Progressive oxygen depletion in the microincubator during the assay possibly reduced the antimicrobial efficacy of gentamicin, resulting in somewhat enhanced MIC values with respect to conventional protocols. In our system, for gentamicin, longer OCR recording was required (at least 12 h) to determine safely a non-growth condition. In addition, different bacteria types or strains may produce specific OCR patterns. For instance, Klebsiella pneumonia, Acinetobacter baumannii or Pseudomonas aeruginosa have doubling times in the range of 40 to 140 min, i.e. longer than the E. coli strain used in the present study. We therefore expect that longer OCR recording times may be necessary for reliable AST assay outputs in such cases. The recommended AST time window for standard methods in current clinical settings (e.g. broth microdilution) is in the range of 16–24 hours.28 As OCR measurements are not turbidity-dependent, it is possible to perform AST directly on clinical samples, for instance a positive blood sample. In this respect, the OCR technique indeed does enable faster susceptibility testing due to significantly simpler sample preparation protocols. However, we must also consider that a clinical sample may contain a consortium of multiple bacteria. False-negative results, indicating that the antibiotic is ineffective, may be obtained due to observing oxygen consumption from a non-pathogenic species that is resistant/tolerant to the active compound used. OCR is only suitable for non-anaerobic pathogens. For anaerobic bacteria, requiring oxygen-depleted culture media, integrating alternative methods, such as pH sensing, glucose sensing or metabolic heat measurements, are more suitable.
Combining OC measurements with metabolic heat recordings of bacteria provides a more holistic phenotypic characterization of the microbial metabolic activity. Heat production per OC has been reported in some previous works using a high-volume and bulky format.44 For example, the work of Birou et al. still required a volume of ∼1 L. High correlation of direct heat calorimetry and indirect calorimetry based on OC has been demonstrated.45 We observed that oxygen is consumed faster in BHI than in MH and LB (Fig. S1a†). Intuitively, bacterial growth is therefore expected to be faster in BHI. However, as was derived by modeling, the bacterial doubling time tdb in BHI (∼31 min) is slightly longer than for the two other cases (∼24 min). The nutrient composition of MH and LB is quite similar and suitable for the growth of most non-fastidious bacteria, whereas BHI has a higher amount of proteins and other relevant ionic compounds. Based on our results, we conclude that the nutrient composition may play an important role in the heat production-per-OC relationship. We suggest that OC pathways depend on the nutrient components in the culture media, an assumption that was supported by the heat vs. OC curves (Fig. 5a), showing a clear difference of the BHI curve with respect to the MH and LB curves. However, comprehensive chemical screening technologies would be required to unravel such pathways.46 Culture temperature had limited impact on the heat/OC ratio. Although enzymatic efficiency declines at unfavorably low culture temperatures, the gross heat production per OC was at the same level for low and high culture temperatures. In addition to the study of cultures in conventional culture media, investigating bacterial metabolism in media with well-controlled and accurately known nutrient composition would be of great interest.
It has been proved that the presence of antimicrobials alters the metabolic pathways of bacteria.47,48 Yang et al. demonstrated that antibiotic-induced adenine limitation increases ATP demand, which elevates carbon metabolism activity and OC.46 As shown in Fig. 4, OCR curves measured without antibiotics are shifted to the right compared to OCR curves at low antimicrobial concentrations (Fig. 4, series 1 and 3, and occasionally series 2). This effect might be due to the enhanced bacterial respiration in the presence of antimicrobials despite growth inhibition. Although this effect also exists at high concentrations, growth inhibition will be the dominant factor in the latter case. In our case, we also suggest that bacteria that experience antimicrobial stress reveal higher heat/OC ratios, in particular for growth in the presence of ampicillin and ciprofloxacin in the lower concentration range. Systematic errors, especially related to the sample preparation step, led to high standard deviations that especially become prominent when correlating the two measurements. More refined studies of the heat/OC ratio under different conditions should take advantage of a further developed system enabling higher resolution and throughput. As our microchamber-based platforms require only very small sample volumes, implementation of improved multiplexed assay formats can be envisioned.
Unlike metabolic heat measurements, which require a minimum bacterial quantity to reach a certain limit of detection, OCR values in a well-sealed chamber depend only on the bacterial concentration. The latter method thus facilitates system miniaturization. The required sample volume, including the dead volume in the tubing, was around 500 μL in our case and possibly can be further reduced if necessary. Optical oxygen sensing based on luminescence quenching take advantage of a wide range of biocompatible nanomaterials. Integration of small photosensitive spots in microsystems is well suited for assay parallelization.19 OC measurements pose a particular condition on the material used for the microchamber or microfluidic chip: it should enable uncoupling the microchamber oxygen content from environmental oxygen sources. Polydimethylsiloxane (PDMS) is a widely used material for soft lithography in microfluidic chip fabrication due to fast prototyping capabilities, low cost, and biocompatibility.49 However, PDMS is a gas-permeable material that may serve as an oxygen reservoir that replenishes on-chip dissolved oxygen levels.50,51 Therefore, PDMS is not suitable to be used for continuous OCR measurements. On the other hand, most thermoplastic materials, such as poly(methyl methacrylate), cyclic olefin copolymer and polycarbonate can be considered as gas-non-permeable. As we used polycarbonate for the microbial microchamber, which has a permeability that is 200 times lower than PDMS, the bacterial culture was therefore well protected from oxygen diffusion from the surrounding environment during the OCR measurements. In our E. coli study, oxygen is not vital for the growth of these facultative anaerobic bacteria. For cell cultures, strictly aerobic bacterial cultures or culture of other small model organisms, however, the material choice and assay design requirements can be more difficult in order to find a compromise between adequate on-chip growth conditions and accurate OC assessment conditions. PDMS is still the most widely used material for cell culture thanks to its high gas permeability and biocompatibility. Other materials may provide interesting alternatives. Ochs et al. have investigated polymethylpentene, a thermoplastic material with relatively high oxygen permeability, as an alternative option for cell culture.52 Krenger et al. applied OSTE+ for an OC study on C. elegans.53 However, as OSTE+ itself may consume dissolved oxygen,54 the OC baseline had to be considered carefully.
Conclusions
OC is an interesting bacterial phenotype that provides valuable information for a more holistic evaluation of the microbial metabolic state. We implemented bacterial OC assays in a dedicated microchamber-based isothermal platform. An OC model based on the exponential growth behavior of bacteria in rich culture conditions was established. We concluded that under unfavorable conditions, in particular at low incubation temperatures, the microbial doubling time prolongs significantly. In a proof-of-concept study, we demonstrated that OCR measurements can be used for fast AST of facultative anaerobic bacteria, which represent most of clinically relevant bacteria in blood stream infections, for instance. The presence of antimicrobials strongly prolonged the apparent doubling time and lag phases. MIC values obtained with the OCR platform compared well to standard EUCAST values. Correlating microbial metabolic heat production and OC provided an interesting new approach for evaluating metabolic activities from a more holistic perspective and possibly for elucidating potential metabolic pathways. We found that heat-per-OC depends on the available nutrient content in the culture medium but was maintained at the same level for different culture temperatures. We also found that bacteria have a higher heat-per-OC ratio for growth in the presence of low antimicrobial concentrations.Author contributions
Y. L. designed the platform, performed experiments and data processing. Y. L., T. L., T. M. and M. G. wrote and/or revised the manuscript. T. L. and M. G. supervised this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors also gratefully acknowledge EPFL-ATMX and EPFL-AFA for support in microfabrication and 3D printing. Funding: This work was carried out in the frame of the EU project “New Diagnostics for Infectious Diseases” (https://cordis.europa.eu/project/id/675412). This project received funding from the European Union's Horizon 2020 program under the Marie Skłodowska-Curie grant agreement No 675412.Notes and references
- K. Bush, P. Courvalin, G. Dantas, J. Davies, B. Eisenstein, P. Huovinen, G. A. Jacoby, R. Kishony, B. N. Kreiswirth, E. Kutter, S. A. Lerner, S. Levy, K. Lewis, O. Lomovskaya, J. H. Miller, S. Mobashery, L. J. V. Piddock, S. Projan, C. M. Thomas, A. Tomasz, P. M. Tulkens, T. R. Walsh, J. D. Watson, J. Witkowski, W. Witte, G. Wright, P. Yeh and H. I. Zgurskaya, Nat. Rev. Microbiol., 2011, 9, 894–896 CrossRef CAS PubMed.
- A. van Belkum, G. Lüdke, J. G. Lisby, G. Kahlmeter, A. Mohess, K. Becker, J. P. Hays, N. Woodford, K. Mitsakakis, J. Moran-Gilad, J. Vila, H. Peter, J. H. Rex and Wm. M. Dunne, Nat. Rev. Microbiol., 2019, 17, 51–62 CrossRef CAS PubMed.
- M. Benkova, O. Soukup and J. Marek, J. Appl. Microbiol., 2020, 129, 806–822 CrossRef CAS PubMed.
- J. Dietvorst, L. Vilaplana, N. Uria, M.-P. Marco and X. Muñoz-Berbel, TrAC, Trends Anal. Chem., 2020, 127, 115891 CrossRef CAS.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 201708558 Search PubMed.
- L. Jiang, L. Boitard, P. Broyer, A.-C. Chareire, P. Bourne-Branchu, P. Mahé, M. Tournoud, C. Franceschi, G. Zambardi, J. Baudry and J. Bibette, Eur. J. Clin. Microbiol. Infect. Dis., 2016, 35, 415–422 CrossRef CAS PubMed.
- U. von Ah, D. Wirz and A. Daniels, BMC Microbiol., 2009, 9, 106 CrossRef PubMed.
- Y. Liu, T. Lehnert and M. A. M. Gijs, Lab Chip, 2020, 20, 3144–3157 RSC.
- V. Kara, C. Duan, K. Gupta, S. Kurosawa, D. J. Stearns-Kurosawa and K. L. Ekinci, Lab Chip, 2018, 18, 743–753 RSC.
- G. Longo, L. Alonso-Sarduy, L. M. Rio, A. Bizzini, A. Trampuz, J. Notz, G. Dietler and S. Kasas, Nat. Nanotechnol., 2013, 8, 522–526 CrossRef CAS PubMed.
- H. Etayash, M. F. Khan, K. Kaur and T. Thundat, Nat. Commun., 2016, 7, 12947 CrossRef CAS PubMed.
- L. W. Winkler, Ber. Dtsch. Chem. Ges., 1888, 21, 2843–2854 CrossRef.
- I. Helm, L. Jalukse, M. Vilbaste and I. Leito, Anal. Chim. Acta, 2009, 648, 167–173 CrossRef CAS PubMed.
- Y. Watanabe, B. B. Hyllbrant and B. Långström, Biochem. Biophys. Res. Commun., 1997, 231, 131–134 CrossRef CAS PubMed.
- H. Zhou, N. Arias-Ramos, P. López-Larrubia, R. P. Mason, S. Cerdán and J. Pacheco-Torres, in Preclinical MRI: Methods and Protocols, ed. M. L. García Martín and P. López Larrubia, Springer, New York, NY, 2018, pp. 297–313 Search PubMed.
- S. S. Velan, R. G. S. Spencer, J. L. Zweier and P. Kuppusamy, Magn. Reson. Med., 2000, 43, 804–809 CrossRef CAS PubMed.
- C.-C. Wu, T. Yasukawa, H. Shiku and T. Matsue, Sens. Actuators, B, 2005, 110, 342–349 CrossRef CAS.
- S. Iguchi, K. Mitsubayashi, T. Uehara and M. Ogawa, Sens. Actuators, B, 2005, 108, 733–737 CrossRef CAS.
- X. Wang and O. S. Wolfbeis, Chem. Soc. Rev., 2014, 43, 3666–3761 RSC.
- B. Ungerböck and T. Mayr, in Quenched-phosphorescence Detection of Molecular Oxygen, 2018, pp. 278–297 Search PubMed.
- P. Gruber, M. P. C. Marques, N. Szita and T. Mayr, Lab Chip, 2017, 17, 2693–2712 RSC.
- D. Nicholas, E. A. Proctor, F. M. Raval, B. C. Ip, C. Habib, E. Ritou, T. N. Grammatopoulos, D. Steenkamp, H. Dooms, C. M. Apovian, D. A. Lauffenburger and B. S. Nikolajczyk, PLoS One, 2017, 12, e0170975 CrossRef PubMed.
- M. P. Jewell, S. C. Saccomano, A. A. David, J. K. Harris, E. T. Zemanick and K. J. Cash, Analyst, 2020, 145, 3996–4003 RSC.
- P. Jusková, S. Schmitt, A. Kling, D. G. Rackus, M. Held, A. Egli and P. S. Dittrich, ACS Sens., 2021, 6, 2202–2210 CrossRef PubMed.
- W. Qiu and S. Nagl, ACS Sens., 2021, 6, 1147–1156 CrossRef CAS PubMed.
- T. Kitahara, N. Koyama, J. Matsuda, Y. Hirakata, S. Kamihira, S. Kohno, M. Nakashima and H. Sasaki, Biol. Pharm. Bull., 2003, 26, 1229–1234 CrossRef CAS PubMed.
- J. Karasinski, L. White, Y. Zhang, E. Wang, S. Andreescu, O. A. Sadik, B. K. Lavine and M. Vora, Biosens. Bioelectron., 2007, 22, 2643–2649 CrossRef CAS PubMed.
- J. H. Jorgensen and M. J. Ferraro, Clin. Infect. Dis., 2009, 49, 1749–1755 CrossRef CAS PubMed.
- K. Stevenson, A. F. McVey, I. B. N. Clark, P. S. Swain and T. Pilizota, Sci. Rep., 2016, 6, 38828 CrossRef CAS PubMed.
- EUCAST, The European Committee on Antimicrobial Susceptibility Testing. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST, Version 9.0, 2019, http://www.eucast.org, 2019 Search PubMed.
- B. Müller, P. Sulzer, M. Walch, H. Zirath, T. Buryška, M. Rothbauer, P. Ertl and T. Mayr, Sens. Actuators, B, 2021, 334, 129664 CrossRef.
- T. Ladner, D. Flitsch, T. Schlepütz and J. Büchs, Microb. Cell Fact., 2015, 14, 161 CrossRef PubMed.
- J. Ehgartner, M. Strobl, J. M. Bolivar, D. Rabl, M. Rothbauer, P. Ertl, S. M. Borisov and T. Mayr, Anal. Chem., 2016, 88, 9796–9804 CrossRef CAS PubMed.
- V. Siracusa, Int. J. Polym. Sci., 2012, 2012, 1–11 CrossRef.
- S. I. Moon, L. Monson and C. W. Extrand, ACS Appl. Mater. Interfaces, 2009, 1, 1539–1543 CrossRef CAS PubMed.
- J. Monod, Annu. Rev. Microbiol., 1949, 3, 371–394 CrossRef CAS.
- Y. Pommier, E. Leo, H. Zhang and C. Marchand, Chem. Biol., 2010, 17, 421–433 CrossRef CAS PubMed.
- P. D. Walker and S. V. Shah, Am. J. Physiol., 1987, 253, C495–C499 CrossRef CAS PubMed.
- J. Bos, Q. Zhang, S. Vyawahare, E. Rogers, S. M. Rosenberg and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 178–183 CrossRef CAS PubMed.
- K. Carroll, J. Butel and S. Morse, Jawetz Melnick & Adelbergs Medical Microbiology 27 E, McGraw-Hill Education / Medical, New York, 27th edn, 2015 Search PubMed.
- P. Fernyhough, S. K. Roy Chowdhury and R. E. Schmidt, Expert Rev. Endocrinol. Metab., 2010, 5, 39–49 CrossRef CAS PubMed.
- I. L. Ferreira, R. Resende, E. Ferreiro, A. C. Rego and C. F. Pereira, Curr. Drug Targets, 2010, 11, 1193–1206 CrossRef CAS PubMed.
- M. J. Pallen, Trends Microbiol., 2011, 19, 58–64 CrossRef CAS PubMed.
- C. L. Cooney, D. I. C. Wang and R. I. Mateles, Biotechnol. Bioeng., 1969, 11, 269–281 CrossRef CAS PubMed.
- B. Birou, I. W. Marison and U. V. Stockar, Biotechnol. Bioeng., 1987, 30, 650–660 CrossRef CAS PubMed.
- J. H. Yang, S. N. Wright, M. Hamblin, D. McCloskey, M. A. Alcantar, L. Schrübbers, A. J. Lopatkin, S. Satish, A. Nili, B. O. Palsson, G. C. Walker and J. J. Collins, Cell, 2019, 177, 1649–1661.e9 CrossRef CAS PubMed.
- M. A. Lobritz, P. Belenky, C. B. M. Porter, A. Gutierrez, J. H. Yang, E. G. Schwarz, D. J. Dwyer, A. S. Khalil and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8173–8180 CrossRef CAS PubMed.
- J. M. Stokes, A. J. Lopatkin, M. A. Lobritz and J. J. Collins, Cell Metab., 2019, 30, 251–259 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- T. C. Merkel, V. I. Bondar, K. Nagai, B. D. Freeman and I. Pinnau, J. Polym. Sci., Part B: Polym. Phys., 2000, 38, 415–434 CrossRef CAS.
- G. Firpo, E. Angeli, L. Repetto and U. Valbusa, J. Membr. Sci., 2015, 481, 1–8 CrossRef CAS.
- C. J. Ochs, J. Kasuya, A. Pavesi and R. D. Kamm, Lab Chip, 2014, 14, 459–462 RSC.
- R. Krenger, M. Cornaglia, T. Lehnert and M. A. M. Gijs, Lab Chip, 2020, 20, 126–135 RSC.
- D. Sticker, M. Rothbauer, J. Ehgartner, C. Steininger, O. Liske, R. Liska, W. Neuhaus, T. Mayr, T. Haraldsson, J. P. Kutter and P. Ertl, ACS Appl. Mater. Interfaces, 2019, 11, 9730–9739 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00296a |
| This journal is © The Royal Society of Chemistry 2021 |