Open Access Article
Open Access ArticleUpscaling of pneumatic membrane valves for the integration of 3D cell cultures on chip
Nina
Compera
a,
Scott
Atwell
a,
Johannes
Wirth
a,
Bernhard
Wolfrum
b and
Matthias
Meier
 *ac
*ac
aHelmholtz Pioneer Campus, Helmholtz Zentrum München GmbH, German Research Center for Environmental Health, Munich, Germany. E-mail: matthias.meier@helmholtz-muenchen.de
bNeuroelectronics – Department of Electrical and Computer Engineering, Technical University of Munich, Germany
cTUM School of Medicine, Technical University of Munich, Munich, Germany
First published on 18th June 2021
Abstract
Microfluidic large-scale integration (mLSI) technology enables the automation of two-dimensional (2D) cell culture processes in a highly parallel manner. Despite the wide range of biological applications of mLSI chips, manufacturing limitations of the central functional element, the pneumatic membrane valve (PMV), make the technology inaccessible for integrating tissue cultures and organoids with dimensions larger than tens of microns. In this study, we developed microtechnology processes to upscale PMVs for mLSI chips by combining 3D printing and soft lithography. Therefore, we developed a robust soft lithography protocol for the production of polydimethylsiloxane chips with PMVs from 3D-printed acrylate and wax molds. While scaled-up PMVs manufactured from acrylate-printed molds exhibited channel profiles with staircases, owing to the inherent 3D stereolithography printing process, PMVs manufactured from reflowed wax molds exhibited a semi-half-rounded channel profile. PMVs with different channel profiles showed closing pressures between 130 and 22.5 kPa, respectively. We demonstrated the functionality of the scaled-up PMVs by forming and maintaining 3D cell cultures from mouse fibroblasts (NIH3T3), human induced pluripotent stem cells (hiPSCs), and human adipose-derived adult stem cells (hASCs), with a narrow size distribution between 124 and 136 μm. Further, parallel and serial design of PMVs on an mLSI chip is used to first form and culture 3D cell cultures before fusing them within a defined flow process. Unit cell designs with upscaled PMVs enabled parallel formation, culturing, trapping, retrieval, and fusion of 3D cell cultures. Thus, the presented additive manufacturing strategy for mLSI chips will foster new developments for highly parallel 3D cell culture screening applications.
Introduction
Microfluidic large-scale integration (mLSI) enables the automation of cell culture processes in a highly parallel manner, with precise spatiotemporal control of the cellular microenvironment.1–3 mLSI platforms have been utilized for various cell culture processes, including high-throughput single-cell analysis,4 standardized stem cell differentiation,5 protein signaling analysis,6 conditional transcriptomics,7 and cell-to-cell interaction analysis.8 To date, the majority of mLSI applications utilize traditional two-dimensional (2D) monolayer cell cultures, which provide robust, cost-effective, and repeatable access to cell screening assays. For the integration of 2D cell cultures on mLSI platforms, the surface chemistry and microenvironmental parameters, including oxygen, pH, and nutrients, must be controlled. The channel design elements and corresponding chip production technology for the integration of 2D cell cultures rapidly evolved because the working dimensions of microfluidics (tens of microns) matched with the needs of the 2D cell culture. With the increasing progression of cell culture technology, three-dimensional (3D) cell cultures have gained more attention because of their close similarity to in vivo tissue microenvironments.9–11 Although mLSI chip platforms have recently been used for the analysis of 3D cell cultures on-chip,12 the technology is hampered because of a lack of production methods for scaling up flow channel heights above 100 μm and, thus, to sizes required for processing 3D cell cultures.The central functional element of the mLSI chip technology is the integrated pneumatic membrane valve (PMV),13 which enables the development of complex flow logic. PMVs are built by two orthogonally intersecting channels, namely, one flow channel and one control channel, separated by a thin flexible membrane. Through pressurization of the dead-end control channel, the thin membrane deflects, which restricts the fluid flow in the corresponding fluidic channel. PMVs can be manufactured in push-up or push-down configurations. Push-down PMVs offer the possibility of biochemically functionalizing the substrate surface before mounting the microfluidic device, while push-up PMVs have the advantage of lower closing pressures.
Regardless of the valve configuration, a half-rounded cross-sectional geometry of the flow channel is required for leak-free valving. Multi-layer soft lithography13 with polydimethylsiloxane (PDMS) has evolved as the preferred fabrication strategy for mLSI chips. It comprises photolithography processes for the fabrication of replica molds, which are subsequently used for casting separate chip layers with PDMS. However, photolithographic techniques only enable the production of rectangular channel profiles. To yield half-rounded cross-sectional shapes, photoresists were reflowed at increased temperatures in a post-processing step. Only positive photoresists can be subjected to a reflow process.14 However, this material property is accompanied by several limitations regarding the chemical and thermal stabilities restricting achievable channel heights to a maximum of a few tens of micrometers in traditional processes.15–17 Therefore, the fabrication of replication molds for complex mLSI chip platforms that incorporate various channel heights and cross-sectional geometries is limited by the inherent material properties of available photoresists.16 The channel height limitation of the photolithography production method for mLSI chips has been addressed by manufacturing PDMS casting molds using e.g. backside photolithography,14 deflection of flexible membranes,15 or micro-milling.12 However, design alterations, prolonged production times, and availability of high-precision mills restrict these approaches. Thus, new production methods are required to enable the accessibility of mLSI technology for 3D cell culture, including organoids and whole tissue structures with sizes higher than 50 μm.18,19
Additive production technology is an attractive alternative fabrication method for soft lithography replica molds20–22 because of its high design flexibility, rapid prototyping, and achievable aspect ratios of up to 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.23 Standard commercial 3D printers in the digital light processing (DLP) mode achieve X/Y pixel resolutions of 25–40 μm, which makes them applicable for microfluidic mold fabrication24–26 and for the direct printing of functional components, such as PMVs.22,27,28 In contrast to the direct printing approach, 3D printing of molds provides numerous advantages of 3D printing technology while maintaining the desirable material properties of PDMS, such as biocompatibility and oxygen permeability. Early development has focused on the implementation of single-layered PDMS-based microfluidic devices with 3D-printed molds.21,24 Despite all the technological advances achieved by DLP 3D printers, the mutual dependence of the building size and pixel resolution remains. While 3D printers, based on two-photon polymerization, exhibit sub-micrometer pixel resolution, their building sizes are low. In summary, for intermediate resolutions attributed to microfluidic chip technologies for 3D cell culture studies, optimization of 3D printing processes is required.
1.23 Standard commercial 3D printers in the digital light processing (DLP) mode achieve X/Y pixel resolutions of 25–40 μm, which makes them applicable for microfluidic mold fabrication24–26 and for the direct printing of functional components, such as PMVs.22,27,28 In contrast to the direct printing approach, 3D printing of molds provides numerous advantages of 3D printing technology while maintaining the desirable material properties of PDMS, such as biocompatibility and oxygen permeability. Early development has focused on the implementation of single-layered PDMS-based microfluidic devices with 3D-printed molds.21,24 Despite all the technological advances achieved by DLP 3D printers, the mutual dependence of the building size and pixel resolution remains. While 3D printers, based on two-photon polymerization, exhibit sub-micrometer pixel resolution, their building sizes are low. In summary, for intermediate resolutions attributed to microfluidic chip technologies for 3D cell culture studies, optimization of 3D printing processes is required.
In this study, we developed an additive production process for fabricating mLSI casting molds. This process enables the upscaling of PMVs for channels with heights higher than 50 μm. To do this, we first established robust soft-lithography workflows to produce multi-layer mLSI chips using 3D-printed molds, including surface coatings. Surface staircase effects due to the inherent layer-printing processes with limited Z resolutions of DLP printers diminished the operating range of the PMVs. By systematically investigating grayscale light exposure for acrylate-based printing resins and a reflow process for wax-based molds, the staircase effect was reduced. The closing pressure and leakage rates of the PMVs fabricated from acrylate and wax molds were systematically characterized. In the following, the upscaled PMVs are exploited to design mLSI unit operations for the formation, trapping, retrieving, and fusing of 3D cell cultures.
Experimental section
Master mold fabrication
Flow master molds were 3D-printed using a DLP stereolithography printer (Pico2HD, Asiga, Australia). Molds were printed with the commercially available resins PlasGRAY or SuperWAX (Asiga, Australia), with printing parameters (e.g. light intensity, exposure time) according to the manufacturer's material file. The printing layer thickness and post-processing protocols were adjusted to achieve optimal PDMS molding results with each of the two resins. PlasGRAY molds were printed as negative molds (extruding channels) with a layer thickness of 10 μm. For post-processing, the PlasGRAY parts were cleaned with isopropyl alcohol and cured in a flash-curing device (Otoflash G171, NK-Optik, Germany), according to the manufacturer's recommendations.SuperWAX molds were printed as positive molds (intruding channels) with a layer thickness of 25 μm, which was the minimal possible layer thickness for the resin. The SuperWAX molds were cleaned to remove the residual uncured resin by sonication in prewarmed isopropyl alcohol (30–35 °C) for 20 s, followed by rinsing with distilled water. Once dried, the molds were placed on a hotplate set to 50 °C for 3 min to reflow the channels. Subsequently, a negative mold was manufactured by casting Durosil® (Detax GmbH & Co. KG, Germany) using a SuperWAX mold. Durosil silicone was cured for 24 h at room temperature before being released from the SuperWAX mold.
Master molds for the control layer of the mLSI chips were fabricated according to the standard SU-8 (SU-8 3050; Microresist Technology, Germany) photolithography protocols.13 To prevent PDMS adhesion onto PlasGRAY, Durosil®, or SU-8 molds, their surfaces were permanently coated with CYTOP™ (AGC Chemicals, Japan), which is an amorphous inert fluoropolymer. Molds were either spin-coated (SU-8 molds) or dip-coated (Durosil® and PlasGRAY molds) on the surface with a thin film of CYTOP (<1 μm). The CYTOP-coated SU-8 mold was heated to 160 °C for 1 h, while the CYTOP-coated PlasGRAY and Durosil® molds were incubated on a hotplate for 8 h at 80 °C to evaporate the CYTOP solvent.
Chip fabrication
All mLSI platforms were fabricated using multilayer soft lithography13 using 3D-printed flow and SU-8 silicon control molds. The upper flow layer was manufactured by casting a thick PDMS (Sylgard® 184, Dow Corning, MI, USA) layer (ratio 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of base material to curing agent) onto the 3D-printed mold, while the lower control layer was fabricated by spin-coating a thin PDMS layer (20
1 of base material to curing agent) onto the 3D-printed mold, while the lower control layer was fabricated by spin-coating a thin PDMS layer (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) onto the SU-8 mold. Both layers were partially cured for 20 min at 80 °C. The flow layer was released from the mold and aligned with the control layer. The assembled mLSI device was post-baked for 45 min at 80 °C to enable off-ratio PDMS bonding. Finally, the mLSI device was sealed with a cover glass after oxygen plasma activation and baked for another 60 min at 80 °C.
1 ratio) onto the SU-8 mold. Both layers were partially cured for 20 min at 80 °C. The flow layer was released from the mold and aligned with the control layer. The assembled mLSI device was post-baked for 45 min at 80 °C to enable off-ratio PDMS bonding. Finally, the mLSI device was sealed with a cover glass after oxygen plasma activation and baked for another 60 min at 80 °C.
Surface measurements
Surface measurements on 3D-printed flow master molds were performed using a confocal laser scanning microscope (VK-X250; Keyence, Japan). Confocal images were then analyzed using MATLAB (R2019a; MathWorks, MA, USA) to obtain the flow channel profiles.PMV design and characterization
The PMVs consisted of two orthogonally intersecting channels separated by a thin PDMS membrane. For this purpose, the PMVs were designed in the push-up configuration. While channels on the upper flow layer had widths and heights of 400 and 200 μm (aspect ratio height to width of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), the channels on the lower control layer had widths and heights of 600 and 100 μm, respectively. The PMV closing pressures decrease with increasing channel cross-section.2 Therefore, the control channels are scaled by a factor of 1.5 compared to the flow channels. The closing pressures of the PMVs were determined by measuring the fluid flow rates under the defined driving pressures. The control channel lines were filled with water and connected through tubing to individual solenoid valves. The solenoid valves were pressurized with defined air pressures (pctrl) between 0 and 200 kPa. The flow inlet ports were connected through Tygon tubing (ND 100-80; Proliquid, Germany) to gas-tight bottles filled with deionized water. Each water bottle was pressurized with air (pfluid) using a flow control unit (Flow EZ™; Fluigent, France). The outlet port was connected to an 80 cm-long tubing (inner diameter: 0.508 mm), which was maintained at atmospheric pressure. The PMV closing pressure was determined by incrementally increasing pctrl, typically by 50 kPa. Concomitantly, the flow rate in the fluid channel was measured using two different methods to screen a large range of flow rates. Flow rates ≥20 μL min−1 were measured using a flow sensor (Flow Unit L, Fluigent). Flow rates <20 μL min−1 were determined by measuring the distance traveled by the meniscus of the liquid in the connected tubing for a specific period. The corresponding volumetric flow rate was then calculated using
2), the channels on the lower control layer had widths and heights of 600 and 100 μm, respectively. The PMV closing pressures decrease with increasing channel cross-section.2 Therefore, the control channels are scaled by a factor of 1.5 compared to the flow channels. The closing pressures of the PMVs were determined by measuring the fluid flow rates under the defined driving pressures. The control channel lines were filled with water and connected through tubing to individual solenoid valves. The solenoid valves were pressurized with defined air pressures (pctrl) between 0 and 200 kPa. The flow inlet ports were connected through Tygon tubing (ND 100-80; Proliquid, Germany) to gas-tight bottles filled with deionized water. Each water bottle was pressurized with air (pfluid) using a flow control unit (Flow EZ™; Fluigent, France). The outlet port was connected to an 80 cm-long tubing (inner diameter: 0.508 mm), which was maintained at atmospheric pressure. The PMV closing pressure was determined by incrementally increasing pctrl, typically by 50 kPa. Concomitantly, the flow rate in the fluid channel was measured using two different methods to screen a large range of flow rates. Flow rates ≥20 μL min−1 were measured using a flow sensor (Flow Unit L, Fluigent). Flow rates <20 μL min−1 were determined by measuring the distance traveled by the meniscus of the liquid in the connected tubing for a specific period. The corresponding volumetric flow rate was then calculated using 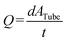 . Here, Q denotes the flow rate (μL min−1), d is the distance traveled by the liquid (mm), ATube is the cross-section of the tubing (0.20268 mm2), and t is the time (typically 30 min) between the start and end of the measurement. The readout accuracy of the flow sensor was ±1.5 μL min−1, while the readout error of the distance measurement was calculated relative to the flow rate (ΔQ = 0.04*Q + 0.002 μL min−1), ranging from 0.002 μL min−1 to 0.8 μL min−1 for flow rates from 0 μL min−1 to 20 μL min−1. The measurements of each valve were performed independently, at least in triplicate (n = 3). Before the start of each measurement, the PMVs were opened and closed at least once to avoid bias due to the remaining pressure in the device.
. Here, Q denotes the flow rate (μL min−1), d is the distance traveled by the liquid (mm), ATube is the cross-section of the tubing (0.20268 mm2), and t is the time (typically 30 min) between the start and end of the measurement. The readout accuracy of the flow sensor was ±1.5 μL min−1, while the readout error of the distance measurement was calculated relative to the flow rate (ΔQ = 0.04*Q + 0.002 μL min−1), ranging from 0.002 μL min−1 to 0.8 μL min−1 for flow rates from 0 μL min−1 to 20 μL min−1. The measurements of each valve were performed independently, at least in triplicate (n = 3). Before the start of each measurement, the PMVs were opened and closed at least once to avoid bias due to the remaining pressure in the device.
Flow analysis of the mLSI cell trapping unit
Flow characterization of the cell trapping unit was performed with the help of a particle tracking experiment. Therefore, 2.55 μm polystyrene beads (PS-F-B237-1; microParticles GmbH, Germany) were flushed through the unit chamber of the mLSI chip. The trapping valve pressure was increased incrementally, typically in steps of 6.9 kPa, and the particle flow was recorded on an AxioObserver (Zeiss, Germany), with a frame rate of 728 fps. Prior to each measurement, the PMVs were opened and closed at least once to avoid bias due to the remaining pressure in the device. A constant fluid forward pressure of 25, 35, or 45 mbar was applied during the measurements. Recorded data were subsequently analyzed in ImageJ (v1.52p), using the TrackMate29 plugin and exported to MATLAB for visualization and flow rate calculations.3D cell culture on-chip
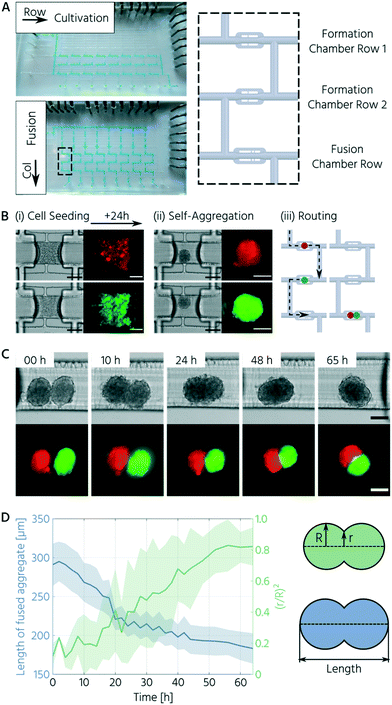
All cell types were maintained in routine 2D cell culture before used for on-chip experiments. To create two differently fluorescently labeled fibroblast cell lines, NIH3T3s were stably transfected with ScaI-digested pCAG Kosak-Venus and pCAG Kosak-Cherry plasmid DNA using Lipofectamine 3000 (Thermo Fisher), according to the manufacturer's instructions. Transfected cells were selected using 1 μg mL−1 puromycin for two weeks before expansion and maintenance in normal growth medium (DMEM + 10% fetal bovine serum (Corning, NY, USA)). Human induced pluripotent stem cells (hiPSCs) and human adipose-derived stem cells (hASCs) were maintained in mTeSR™ Plus (StemCell Technologies, Canada) + 1% penicillin–streptomycin and subcutaneous preadipocyte growth medium (PM-1; ZenBio, NC, USA), respectively. Prior to use in cell experiments, the chip was coated with 10% Pluronic F127 (Sigma Aldrich, MO, USA) for at least 24 h to minimize cell attachment to the PDMS. The cell culture medium was introduced 30 min before seeding to allow for equilibration. To seed cells, 3–5 × 106 cells in approximately 50 μL of the respective culture medium were transferred as a single-cell suspension on-chip. For the fibroblasts, we first seeded mCherry-labeled NIH3T3s in formation chamber row 1, followed by actuation of the trapping valves in row 1 and a medium rinse of all fluidic channels containing the cell material. We then seeded the Venus-labeled NIH3T3s in row 2, actuated these trapping valves, and cleaned all channels of the remaining, non-trapped cells. When hiPSCs or hASCs were used, we seeded them successively in formation chamber rows 1 and 2, before actuating the trapping valves and rinsing all channels with medium. The control pressure for the trapping valves was set to 207 kPa to minimize cell loss during rinsing. Trapped cells were allowed to rest and aggregate for 4 h before the first feeding cycle. During the first feeding, the trapping valve (140–170 kPa) and fluid forward pressure (ranging from 25 to 45 mbar) were adjusted manually based on visual inspection of the cell-trapping site to ensure fluid passage while keeping the spheroid trapped. Thereafter, these parameters were kept constant, and cells were automatically fed every 2–4 h for 10 s, depending on the cell type, using a custom-written MATLAB script. Media bottles were replenished every 24 h. Human iPSC culture media was supplemented with ROCK inhibitor (Y-27632; Apexbio Technology, TX, USA) for the first 24 h, but removed for subsequent maintenance. Other cell types were cultured in standard growth media on-chip.Microscopy and image analysis
Bright-field images were acquired with 2.5× and 10× (Plan-Apochromat) objectives on a Zeiss AxioVert. An epifluorescent microscope (AxioObserver, Zeiss) was used to record the fluorescent images. Image analysis and statistical evaluation were performed using ImageJ and MATLAB. Values are represented as mean ± SD for three independent experiments (n = 3) unless stated otherwise.Results and discussion
Upscaling PMVs by soft lithography using 3D-printed master molds
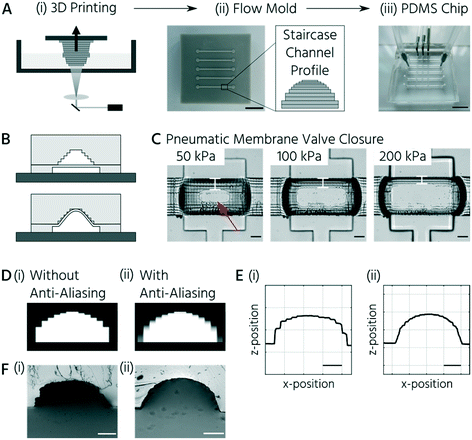
A central requirement for soft lithography using 3D-printed molds is to enable the curing and release of PDMS from mold surfaces. During the direct soft-lithography prototyping of PDMS, using 3D-printed molds, contact inhibition of PDMS curing was observed in this study and previous studies.24,30,31 This problem originates from the non-crosslinked monomers on the surface and in the bulk of the mold.24,32 Postprocessing steps, including bulk material heating and temporary surface coating with anti-adhesive agents, improved the soft lithography of PDMS using the 3D-printed molds. The anti-adhesive coating did not fully overcome the PDMS release problem because molds require frequent re-coating.25,30,33 Thus, we evaluated a simple fluoropolymer surface coating to overcome the contact inhibition of PDMS curing on 3D-printed parts. CYTOP™, an epoxy-based amorphous fluoropolymer, has been developed for coating silicon wafers to enable long-term usage34 and has been successfully applied as a releasing agent for PDMS.35,36 Plasma activation and subsequent dip-coating of the 3D-printed molds in a 2% solution of CYTOP™ (CTL-809M diluted in CT-SOLV180) increased the water contact angle, compared to the non-coated surface from 76° to 88°. The increased surface hydrophobicity of the CYTOP-coated 3D-printed molds enabled complete polymerization and easy removal of the PDMS casts. Notably, the first PDMS cast from the CYTOP-coated mold could not be plasma-bonded. Repetitive usage of the same mold for PDMS casting and demolding up to 30 times led to no noticeable wear-off of the surface coating.The second challenge in manufacturing mLSI chips using 3D-printed molds is the inherent layering nature of DLP printers.37 The flow channels on mLSI chips require a half-rounded channel profile to be closed by PMVs. DLP printers applicable to microfluidics exhibit Z resolutions of approximately 5–50 μm.38,39 At these Z-printing resolutions, the channels on the molds exhibit a step-like rather than a perfectly half-rounded profile (Fig. 1A). Printed features are discretized in slices with defined thicknesses, resulting in the so-called staircase effect on the printed feature surface.31–34 To investigate whether the staircase effect impaired the PMV function (Fig. 1B), we produced a two-layered microfluidic chip with one straight flow channel and three consecutive PMVs (width: 400 μm, length: 600 μm) in the push-up configuration. Bright-field images of the valve compression area at control pressures of 50, 100, and 200 kPa are shown in Fig. 1C. At 50 kPa, the valve membrane closed the flow channel only at the center of the compression area (Fig. 1C, indicated by the red arrow). Meanwhile, at 200 kPa, only thin lines at the sidewalls (Fig. 1C, indicated by white bars) of the channel remained. These lines indicate openings induced by the staircases on the channel surface. A similar observation during the production of sieve valves has been reported, which are PMVs incompletely closing flow channels with a squared channel profile.1
To investigate and mitigate the staircase effect on PMV closure, we used two strategies: (1) reducing the staircase edges by grayscale light exposure (anti-aliasing) during the 3D printing process39–42 and (2) an alternative wax-based printing resin, which can be reflowed in a postprocessing step analogous to positive photoresists in traditional photolithography,2 to obtain perfectly rounded flow channels. Anti-aliasing is a method of encoding individual pixels of a DLP printing slice in grayscale values (Fig. 1D (ii)) instead of the black and white pixels (Fig. 1D (i)). This method leads to a rounding effect on the features during printing. Using anti-aliasing, the flow channel surface was significantly smoothed on the 3D-printed molds (Fig. 1E). Consequently, the staircase effect was minimized (Fig. 1F).
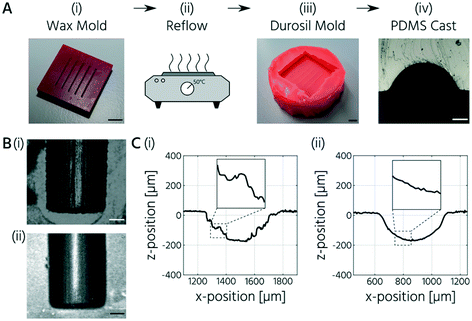
An alternative manufacturing process for upscaled PMVs and removal of the staircase effect is to implement a reflow process analogous to the post-processing of positive photoresists. The acrylate-based printing resin exhibits a glass transition temperature of 84 °C. However, post-backing acrylate 3D-printed parts above their glass transition temperature led only to warpage and deformation of the parts without the reflow of staircases. Therefore, we evaluated the performance of the wax-based printing resin for the fabrication of PDMS replica molds. Wax resins have melting temperatures between 40 and 60 °C (SuperWAX, tmelt = 50 °C) and are, thus, applicable for a reflow process.
To test this approach, we printed a negative mold with a wax resin (SuperWAX, Asiga) and then placed the wax molds (total thickness: 3 mm) for 3 min at 50 °C onto a glass slide to reflow the channels (Fig. 2A (i) and (ii)). Therefore, the printed features faced upward. Indeed, the reflow process resulted in half-rounded channel geometries without an apparent staircase effect (Fig. 2B). However, casting of PDMS using the 3D-printed wax molds was not possible because of the contact curing inhibition reaction between the wax and PDMS. The CYTOP coating of the wax mold did not overcome this problem, presumably owing to the rapid exchange of wax molecules between the bulk and surface areas. Therefore, a double-casting process was introduced (Fig. 2A). First, a positive mold was printed using SuperWAX. The mold was thermally reflowed to remove the staircase effect (Fig. 2C) and then replicated with silicone (Durosil®), which was cured on the wax mold at room temperature for 24 h. The negative silicone mold was then coated with CYTOP and subsequently used to cast the PDMS.
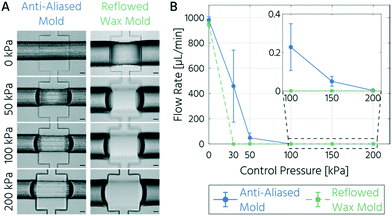
Next, we characterized and compared the closing behavior of upscaled PMVs fabricated using either the anti-aliased acrylate-based or reflowed wax-based molds. Bright-field images of the valve compression area acquired with an increase in the actuation pressure are shown in Fig. 3A. While the PMVs fabricated from the reflowed wax mold visibly closed the flow channel at 50 kPa, the PMVs fabricated from the anti-aliased acrylate mold closed the flow channel at a pressure of 200 kPa. The pressure dependence of the PMV closing state was determined using flow rate measurements (Fig. 3B). The mean flow rates within an unrestricted channel cast from an anti-aliased and reflowed wax mold at a driving pressure of 14 kPa were 982 and 949 μL min−1, respectively. These flow rates evoke flow velocities of about 250 mm s−1 in the fluidic channel, which are at least one order of magnitude higher than those generally required for mLSI cell culture devices.43–45
The closing pressure of the PMVs for a flow channel with a semi-half-round profile, cast from the reflowed wax mold, was 30 kPa at a counteracting flow pressure of 14 kPa. This measured value is slightly higher than the theoretical closing pressure, which can be approximated using a thick spring model.2 Assuming a Young's modulus of 750 kPa46 for the PDMS membrane (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio), the thick spring model predicts a closing pressure of 20.5 kPa. The slight offset can be explained by the boundary conditions of the model, which was developed using the data of mLSI chips with smaller channel dimensions.2 In comparison, the PMVs actuated with a control pressure of 30 kPa on channels with a minimized staircase profile only decreased the flow rate by a factor of 2 (458 μL min−1; Fig. 3B) but did not reach a closed state. Only upon increasing the control pressure to 200 kPa, a closing state with a reduction in the flow rate by a magnitude of 105 compared to the open state was achieved. Thus, at actuation pressures of 200 kPa or higher, the leakage of the PMV fabricated from an anti-aliased mold is negligible, and the valve can be considered fully closed.
1 ratio), the thick spring model predicts a closing pressure of 20.5 kPa. The slight offset can be explained by the boundary conditions of the model, which was developed using the data of mLSI chips with smaller channel dimensions.2 In comparison, the PMVs actuated with a control pressure of 30 kPa on channels with a minimized staircase profile only decreased the flow rate by a factor of 2 (458 μL min−1; Fig. 3B) but did not reach a closed state. Only upon increasing the control pressure to 200 kPa, a closing state with a reduction in the flow rate by a magnitude of 105 compared to the open state was achieved. Thus, at actuation pressures of 200 kPa or higher, the leakage of the PMV fabricated from an anti-aliased mold is negligible, and the valve can be considered fully closed.
PMV-assisted formation of 3D cell culture on mLSI chip platforms
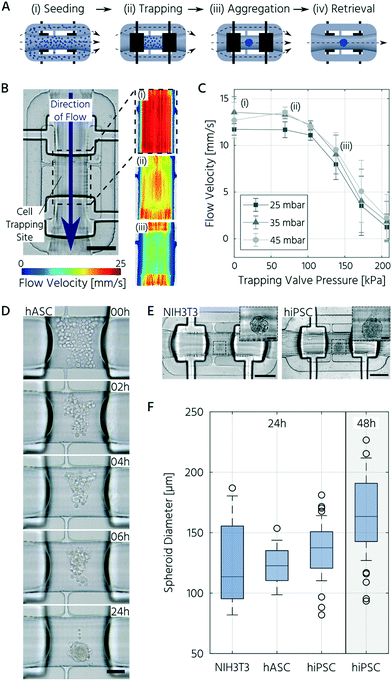
To demonstrate the integration of the upscaled PMVs for 3D cell culturing on mLSI platforms, we first designed and characterized a functional unit cell for the formation, culturing, and trapping of 3D cell cultures. Unit cells can be arrayed for parallel processing on mLSI chips, with standard multiplexing design elements. Pneumatic structures have been exploited for the formation, culturing, and trapping of 3D cell cultures before. The prominent examples for this purpose are U-shaped PMVs,47,48 microwells,49 or bar-shaped gate structures.50 Common in all designs is that they either show a complex operational process or have low application flexibility upon integration into multilayered PDMS platforms. To offer an alternative workflow for handling 3D cell cultures on mLSI, we exploited the sieve-like closing behavior of PMVs on channel profiles, with residual staircase profiles, when actuated with pressures below the closing state. Fig. 4A shows the working principle of the unit cell design. The unit cell comprises three bifurcating flow channels and two PMVs for separating a cell culture volume of 0.1 μL. While the two side channels had widths and heights of 100 μm and 50 μm, respectively, the center channel had a width and height of 300 μm and 200 μm, respectively. The cell culture volume of the unit cell and the cell density of the seeding solution determine the diameter of the resulting 3D cell culture. Here, to avoid quiescent or necrotic zones in the center, the unit cell structure was designed to initiate 3D cell cultures with diameters below 200 μm.52,53For the 3D cell culture formation step, a high-density single-cell suspension was flushed through the unit cell until the center channel was filled. Subsequently, the PMVs were fully closed, and non-trapped cells in the side channels were rinsed out. 3D cell cultures were formed by self-aggregation. For the long-term culturing process, the control pressure of the PMVs was decreased, and the sieve-like behavior of the trapping valves enabled feeding under low shear stress conditions. The open state of the PMVs allowed retrieval of the 3D cell culture.
We measured the flow velocity profile across the cell-trapping site by particle tracking to characterize the unit cell design. Fig. 4B shows the flow profile within the trapping site at changing actuation pressures of the trapping valves and constant fluid pressure. In the PMV open state, a fluid forward pressure of 25 mbar or 45 mbar evokes a mean flow rate at the cell trapping site of 0.6 μL s−1 or 0.7 μL s−1, respectively. At a control pressure of 140 kPa, the flow rate was reduced by approximately 40%. The closed state was reached at a closing pressure of above 200 kPa.
Thereafter, we investigated the homogeneity and reproducibility of the 3D cell culture formation process in the unit cell for three different cell types. All the investigated cell types showed self-aggregation within the first 2–4 h after seeding (Fig. 4D) and formed a compact, spherical morphology with distinct borders within 24 h. Upon reducing the actuation pressure of the trapping PMV (<170 kPa), dead cells were removed, and cell feeding was achieved with the reinstated flow. This process resulted in few to no residual dead cells inside the trapping region (Fig. 4D and E).
With the chosen channel dimensions, we were able to reliably produce 3D cell cultures of NIH3T3 cells, hiPSCs, and hASCs. A boxplot of all spheroid sizes across several technical and biological repetitions is presented in Fig. 4F. The mean diameter of the NIH3T3 spheroids was 125 ± 33 μm (mean ± standard deviation). In comparison, hiPSC and hASC spheroids exhibited mean diameters of 136 ± 22 μm and 124 ± 15 μm, respectively. In addition to the somewhat larger interquartile range for NIH3T3s, the inter-chip variation of the achieved 3D cell culture size, 24 h after seeding, was less than 4% and 15% for the hiPSCs and hASCs, respectively. Further, extending the culture process for the hiPSC led to an increase in the 3D cell culture by approximately 20% within the next 24 h, demonstrating cell growth and viability (Fig. 4F). In summary, we designed a functional unit cell for arraying on an mLSI platform to form 3D cell cultures reliably with homogeneous sizes.
Fusion of 3D cell cultures on an mLSI chip
Upon successfully characterizing the unit cell operation for 3D cell cultures, we built an mLSI platform, in which the unit cell was arrayed in total 24 times. The layout of the platform is shown in Fig. 5A (left side). The primary function of the chip was to form 3D cell cultures in the first step and merge two different 3D cell cultures in the second step. For the implementation of the two operational steps, the unit cell was arrayed in a 3 × 8 (row/column) matrix format. While unit cells in the first two rows are allocated for the formation and cultivation of individually addressable 3D cell cultures, unit cells in the third row were allocated for the fusing operation of 3D cell cultures generated in the upper rows of the same column (Fig. 5A, right side). A multiplexer structure was used to address single-column elements, and individual PMVs were used to address the row elements.To prove the in vitro fusion operation on the chip platform, we used two fluorescent reporter cell lines: mCherry- (red) and Venus-labeled (green) NIH3T3 cells. For cell seeding and formation of the 3D cell cultures, the cells were separately introduced into the first and second row elements (Fig. 5B (i)). Further, 24 hours after seeding, we successively addressed each column to flush the 3D cell cultures sequentially into the unit cell chamber of the third row ((ii)–(iii)). To trap both 3D cell cultures, the entry-trapping PMV was opened while maintaining the PMV at the outlet side in a sieve-like valve state (actuation pressure <170 kPa). Fig. 5C highlights the progression of fusion, based on one representative example. All 3D cell cultures fused within 24 h exhibiting an elongated elliptical morphology and compacted further over the entire time frame of 65 h.
Cell aggregate fusion has been described by the differential adhesion hypothesis wherein the individual surface tensions of the 3D cell cultures in pairs determine cellular rearrangement.51 Based on this, the fusion process has been characterized by an analogy to the coalescence of highly viscous liquid droplets, where coalescence is driven by surface tension and resisted by viscosity.52–54 The progression of fusion can then be quantified by the evolution of the interfacial area (πr2) between the fusing partners, normalized by the initial average cross-sectional area of the aggregates (πR2) over time,54 which is the squared ratio of the radii (r/R)2. Additionally, we characterized morphological changes during fusion by the overall aggregate length.55,56 As the aggregate length decreased, the squared ratio of the radii increased during the 3D cell culture fusion process (Fig. 5D). Both parameters converged to their respective minimal/maximal levels after 60 h. This slow rate is in accordance with the values determined for fusing 3D cell cultures of dermal fibroblasts56 and chondrocytes55 and illustrates high internal cellular cohesive forces. Notably, the two labeled NIH3T3 cell populations did not mix within the given time frame. This finding is consistent with the differential adhesion hypothesis as both populations arise from the same cell type and should, thus, have similar surface tension.
Conclusion
In this study, we developed a 3D printing workflow and design rules for functional upscaled PMVs using 3D-printed soft-lithography molds. Owing to the inherent layering process of 3D printing, the flow channels exhibited a staircase effect on their surface, resulting in the sieve-like behavior of upscaled PMVs. The staircase effect could be significantly minimized by employing an anti-aliasing strategy during mold printing or using a wax-based printing resin. Depending on the selected optimization approach, proper microvalve performance was achieved for a net pressure difference of 130 kPa or 22.5 kPa for the anti-aliased and reflowed wax molds, respectively. The upscaled PMVs could be exploited to design a general unit cell for the formation, culturing, retrieval, and fusion of 3D cell cultures on mLSI chip platforms. Upon integration and arraying of the unit cell on an mLSI platform, we successfully fused 3D cell cultures of two fluorescently labeled NIH3T3 cell lines. With the provided design rules and 3D printing workflow, we enabled 3D cell cultures on mLSI chip platforms. Upscaled PMVs can act on flow channels with heights >150 μm, which ensures unrestricted and non-destructive handling of 3D cell cultures. In the future, we envision mLSI applications that comprise interaction studies of various heterogeneous tissues to assist in in vitro-modeling of more complex developmental processes, such as human embryogenesis,11 organogenesis,57 and brain development.58Author contributions
Conceptualization: NC, BW, MM; investigation: NC, JW; formal analysis: NC, SA; writing: NC, SA, JW, MM; supervision: BW, MM. All authors have approved the final manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Helmholtz Pioneer Campus and the ERC Consolidator Grant (Number 772646).References
- J. Melin and S. R. Quake, Microfluidic large-scale integration: the evolution of design rules for biological automation, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 213–231 Search PubMed.
- P. M. Fordyce, C. A. Diaz-Botia, J. L. DeRisi and R. Gomez-Sjoberg, Systematic characterization of feature dimensions and closing pressures for microfluidic valves produced via photoresist reflow, Lab Chip, 2012, 12(21), 4287–4295 Search PubMed.
- D. Liu, S. Chen and M. W. Naing, A review of manufacturing capabilities of cell spheroid generation technologies and future development, Biotechnol. Bioeng., 2020, 25, bit.27620, DOI:10.1002/bit.27620.
- C. Zhang, et al., Ultra-multiplexed analysis of single-cell dynamics reveals logic rules in differentiation, Sci. Adv., 2019, 5(4), 1–11, DOI:10.1126/sciadv.aav7959_rfseq1.
- M. Tanyeri and S. Tay, Viable cell culture in PDMS-based microfluidic devices, in Methods in Cell Biology, Academic Press, 2018, vol. 148, pp. 3–33 Search PubMed.
- M. Blazek, X. Wu, R. Zengerle and M. Meier, Analysis of fast protein phosphorylation kinetics in single cells on a microfluidic chip, in 18th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2014, 2014, no. 3, vol. 15, pp. 267–269, 10.1039/c4lc00797b.
- H. C. Fan, J. Wang, A. Potanina and S. R. Quake, Whole-genome molecular haplotyping of single cells, Nat. Biotechnol., 2011, 29(1), 51–59, DOI:10.1038/nbt.1739.
- T. Frank and S. Tay, Automated co-culture system for spatiotemporal analysis of cell-to-cell communication, Lab Chip, 2015, 15(10), 2192–2200, 10.1039/c5lc00182j.
- T. Toyoda, et al., Cell aggregation optimizes the differentiation of human ESCs and iPSCs into pancreatic bud-like progenitor cells, Stem Cell Res., 2015, 14(2), 185–197, DOI:10.1016/j.scr.2015.01.007.
- N. Sharon, et al., A Peninsular Structure Coordinates Asynchronous Differentiation with Morphogenesis to Generate Pancreatic Islets, Cell, 2019, 176(4), 790–804.e13, DOI:10.1016/j.cell.2018.12.003.
- S. E. Harrison, B. Sozen, N. Christodoulou, C. Kyprianou and M. Zernicka-Goetz, Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro, Science, 2017, 356(6334), eaal1810, DOI:10.1126/science.aal1810.
- T. S. Santisteban, O. Rabajania, I. Kalinina, S. Robinson and M. Meier, Rapid spheroid clearing on a microfluidic chip, Lab Chip, 2018, 18(1), 153–161 Search PubMed.
- M. A. Unger, H.-P. Chou, T. Thorsen, A. Scherer and S. R. Quake, Monolithic microfabricated valves and pumps by multilayer soft lithography, Science, 2000, 288(5463), 113–116 Search PubMed.
- P. Frank, S. Haefner, G. Paschew and A. Richter, Rounding of negative dry film resist by diffusive backside exposure creating rounded channels for pneumatic membrane valves, Micromachines, 2015, 6(11), 1588–1596, DOI:10.3390/mi6111442.
- D. N. Freitas, A. Mongersun, H. Chau and I. E. Araci, Tunable soft lithography molds enable rapid-prototyping of multi-height channels for microfluidic large-scale integration, J. Micromech. Microeng., 2019, 29(3), aafd9c, DOI:10.1088/1361-6439/aafd9c.
- N. W. Bartlett and R. J. Wood, Comparative analysis of fabrication methods for achieving rounded microchannels in PDMS, J. Micromech. Microeng., 2016, 26(11), 115013, DOI:10.1088/0960-1317/26/11/115013.
- W. Park, S. Han and S. Kwon, Fabrication of membrane-type microvalves in rectangular microfluidic channels via seal photopolymerization, Lab Chip, 2010, 10(20), 2814–2817, 10.1039/c005173j.
- C. Mulas, et al., Microfluidic platform for 3D cell culture with live imaging and clone retrieval, Lab Chip, 2020, 20(14), 2580–2591, 10.1039/D0LC00165A.
- K. Moshksayan, et al., Spheroids-on-a-chip: Recent advances and design considerations in microfluidic platforms for spheroid formation and culture, Sens. Actuators, B, 2018, 263, 151–176, DOI:10.1016/j.snb.2018.01.223.
- J. Shrestha, et al., A Rapidly Prototyped Lung-on-a-chip Model Using 3D-Printed Molds, Organs-on-a-Chip, 2020, 100001, DOI:10.1016/j.ooc.2020.100001.
- K. I. Kamei, et al., 3D printing of soft lithography mold for rapid production of polydimethylsiloxane-based microfluidic devices for cell stimulation with concentration gradients, Biomed. Microdevices, 2015, 17(2), 36, DOI:10.1007/s10544-015-9928-y.
- Y.-S. Lee, N. Bhattacharjee and A. Folch, 3D-printed Quake-style microvalves and micropumps, Lab Chip, 2018, 18(8), 1207–1214, 10.1039/C8LC00001H.
- A. P. Kuo, N. Bhattacharjee, Y.-S. Lee, K. Castro, Y. T. Kim and A. Folch, High-Precision Stereolithography of Biomicrofluidic Devices, Adv. Mater. Technol., 2019, 1800395, DOI:10.1002/admt.201800395.
- S. Razavi Bazaz, et al., Rapid Softlithography Using 3D-Printed Molds, Adv. Mater. Technol., 2019, 1900425, DOI:10.1002/admt.201900425.
- G. Comina, A. Suska and D. Filippini, PDMS lab-on-a-chip fabrication using 3D printed templates, Lab Chip, 2014, 14(2), 424–430, 10.1039/c3lc50956g.
- T. Dahlberg, et al., 3D printed water-soluble scaffolds for rapid production of PDMS micro-fluidic flow chambers, Sci. Rep., 2018, 8(1), 3372, DOI:10.1038/s41598-018-21638-w.
- S. J. Keating, M. I. Gariboldi, W. G. Patrick, S. Sharma, D. S. Kong and N. Oxman, 3D printed multimaterial microfluidic valve, PLoS One, 2016, 11(8), e0160624, DOI:10.1371/journal.pone.0160624.
- H. Gong, A. T. Woolley and G. P. Nordin, High density 3D printed microfluidic valves, pumps, and multiplexers, Lab Chip, 2016, 16(13), 2450–2458, 10.1039/c6lc00565a.
- J. Y. Tinevez, et al., TrackMate: An open and extensible platform for single-particle tracking, Methods, 2017, 115, 80–90, DOI:10.1016/j.ymeth.2016.09.016.
- H. N. Chan, Y. Chen, Y. Shu, Y. Chen, Q. Tian and H. Wu, Direct, one-step molding of 3D-printed structures for convenient fabrication of truly 3D PDMS microfluidic chips, Microfluid. Nanofluid., 2015, 19(1), 9–18, DOI:10.1007/s10404-014-1542-4.
- Y. Hwang, O. H. Paydar and R. N. Candler, 3D printed molds for non-planar PDMS microfluidic channels, Sens. Actuators, A, 2015, 226, 137–142 Search PubMed.
- A. J. Capel, R. P. Rimington, M. P. Lewis and S. D. R. Christie, 3D printing for chemical, pharmaceutical and biological applications, Nat. Rev. Chem., 2018, 1, DOI:10.1038/s41570-018-0058-y.
- C. C. Glick, et al., Rapid assembly of multilayer microfluidic structures via 3D-printed transfer molding and bonding, Microsyst. Nanoeng., 2016, 2(1), 16063, DOI:10.1038/micronano.2016.63.
- D. H. Yoon, K. Kobayashi, D. Tanaka, T. Sekiguchi and S. Shoji, Simple microfluidic formation of highly heterogeneous microfibers using a combination of sheath units, Lab Chip, 2017, 17(8), 1481–1486, 10.1039/c7lc00157f.
- H. Kimura, T. Yamamoto, H. Sakai, Y. Sakai and T. Fujii, An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models, Lab Chip, 2008, 8(5), 741–746, 10.1039/b717091b.
- T. Yang, J. Choo, S. Stavrakis and A. de Mello, Fluoropolymer-Coated PDMS Microfluidic Devices for Application in Organic Synthesis, Chem. – Eur. J., 2018, 24(46), 12078–12083, DOI:10.1002/chem.201802750.
- W. Oropallo and L. A. Piegl, Ten challenges in 3D printing, Eng. Computation, 2016, 32(1), 135–148, DOI:10.1007/s00366-015-0407-0.
- S. M. Hampson, W. Rowe, S. D. R. Christie and M. Platt, 3D printed microfluidic device with integrated optical sensing for particle analysis, Sens. Actuators, B, 2018, 256, 1030–1037, DOI:10.1016/j.snb.2017.10.041.
- S. C. Ligon, R. Liska, J. Stampfl, M. Gurr and R. Mülhaupt, Polymers for 3D Printing and Customized Additive Manufacturing, Chem. Rev., 2017, 117(15), 10212–10290, DOI:10.1021/acs.chemrev.7b00074.
- C. Arnold, D. Monsees, J. Hey and R. Schweyen, Surface quality of 3D-printed models as a function of various printing parameters, Materials, 2019, 12(12), 1–15, DOI:10.3390/ma12121970.
- K. Mostafa, A. J. Qureshi and C. Montemagno, Tolerance Control Using Subvoxel Gray-Scale DLP 3D Printing, in Advanced Manufacturing, 2017, no. 1, vol. 2, pp. 1–7, DOI:10.1115/IMECE2017-72232.
- X. Kuang, et al., Grayscale digital light processing 3D printing for highly functionally graded materials, Sci. Adv., 2019, 5(5), 1–10, DOI:10.1126/sciadv.aav5790.
- Y. Jun, et al., In vivo–mimicking microfluidic perfusion culture of pancreatic islet spheroids, Sci. Adv., 2019, 5(11), eaax4520, DOI:10.1126/sciadv.aax4520.
- M. Beer, et al., A novel microfluidic 3D platform for culturing pancreatic ductal adenocarcinoma cells: Comparison with in vitro cultures and in vivo xenografts, Sci. Rep., 2017, 7(1), 1325, DOI:10.1038/s41598-017-01256-8.
- J. Y. Kim, et al., 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis, J. Biotechnol., 2015, 205, 24–35, DOI:10.1016/j.jbiotec.2015.01.003.
- D. Armani, C. Liu and N. Aluru, Re-configurable fluid circuits by PDMS elastomer micromachining, in Technical Digest. IEEE International MEMS 99 Conference. Twelfth IEEE International Conference on Micro Electro Mechanical Systems (Cat. No.99CH36291), 1999, pp. 222–227, DOI:10.1109/MEMSYS.1999.746817.
- W. Liu, J.-C. Wang and J. Wang, Controllable organization and high throughput production of recoverable 3D tumors using pneumatic microfluidics, Lab Chip, 2015, 15(4), 1195–1204 Search PubMed.
- H.-J. J. Jin, et al., A multicellular spheroid formation and extraction chip using removable cell trapping barriers, Lab Chip, 2011, 11(1), 115–119, 10.1039/C0LC00134A.
- T. Anada, et al., Three-dimensional cell culture device utilizing thin membrane deformation by decompression, Sens. Actuators, B, 2010, 147(1), 376–379, DOI:10.1016/j.snb.2010.01.065.
- H. S. Kim, T. P. Devarenne and A. Han, A high-throughput microfluidic single-cell screening platform capable of selective cell extraction, Lab Chip, 2015, 15(11), 2467–2475, 10.1039/c4lc01316f.
- R. A. Foty and M. S. Steinberg, The differential adhesion hypothesis: A direct evaluation, Dev. Biol., 2005, 278(1), 255–263, DOI:10.1016/j.ydbio.2004.11.012.
- N. V. Kosheleva, et al., Cell spheroid fusion: beyond liquid drops model, Sci. Rep., 2020, 10(1), 1–15, DOI:10.1038/s41598-020-69540-8.
- P. A. Fleming, W. S. Argraves, C. Gentile, A. Neagu, G. Forgacs and C. J. Drake, Fusion of uniluminal vascular spheroids: A model for assembly of blood vessels, Dev. Dyn., 2010, 239(2), 398–406, DOI:10.1002/dvdy.22161.
- E. Flenner, F. Marga, A. Neagu, I. Kosztin and G. Forgacs, Relating Biophysical Properties Across Scales, Curr. Top. Dev. Biol., 2008, 81(07), 461–483, DOI:10.1016/S0070-2153(07)81016-7.
- M. J. Susienka, B. T. Wilks and J. R. Morgan, Quantifying the kinetics and morphological changes of the fusion of spheroid building blocks, Biofabrication, 2016, 8(4), 045003, DOI:10.1088/1758-5090/8/4/045003.
- Z. Hajdu, V. Mironov, A. N. Mehesz, R. A. Norris, R. R. Markwald and R. P. Visconti, Tissue spheroid fusion-based in vitro screening assays for analysis of tissue maturation, J. Tissue Eng. Regener. Med., 2010, 4(8), 659–664, DOI:10.1002/term.291.
- H. Koike, et al., Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary, Nature, 2019, 574, 112–116, DOI:10.1038/s41586-019-1598-0.
- Y. Xiang, et al., Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration, Cell Stem Cell, 2017, 21(3), 383–398.e7, DOI:10.1016/J.STEM.2017.07.007.
| This journal is © The Royal Society of Chemistry 2021 |