Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Establishment of a human three-dimensional chip-based chondro-synovial coculture joint model for reciprocal cross talk studies in arthritis research†
Mario
Rothbauer‡
 *ab,
Ruth A.
Byrne‡
bc,
Silvia
Schobesberger‡
*ab,
Ruth A.
Byrne‡
bc,
Silvia
Schobesberger‡
 b,
Isabel
Olmos Calvo
c,
Anita
Fischer
acd,
Eva I.
Reihs
ab,
Sarah
Spitz
b,
Barbara
Bachmann
bef,
Florian
Sevelda
g,
Johannes
Holinka
g,
Wolfgang
Holnthoner
ef,
Heinz
Redl
ef,
Stefan
Toegel
ad,
Reinhard
Windhager
ag,
Hans P.
Kiener
c and
Peter
Ertl
b,
Isabel
Olmos Calvo
c,
Anita
Fischer
acd,
Eva I.
Reihs
ab,
Sarah
Spitz
b,
Barbara
Bachmann
bef,
Florian
Sevelda
g,
Johannes
Holinka
g,
Wolfgang
Holnthoner
ef,
Heinz
Redl
ef,
Stefan
Toegel
ad,
Reinhard
Windhager
ag,
Hans P.
Kiener
c and
Peter
Ertl
 *b
*b
aKarl Chiari Lab for Orthopaedic Biology (KCLOB), Department of Orthopedics and Trauma Surgery, Medical University of Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria. E-mail: mario.rothbauer@meduniwien.ac.at
bFaculty of Technical Chemistry, Vienna University of Technology, Getreidemarkt 9, 1060 Vienna, Austria. E-mail: peter.ertl@tuwien.ac.at
cDivision of Rheumatology, Department of Medicine III, Medical University Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria
dLudwig Boltzmann Institute of Arthritis and Rehabilitation, Vienna, Austria
eAUVA Research Centre, Ludwig Boltzmann Institute for Experimental and Clinical Traumatology, 1200 Vienna, Austria
fAustrian Cluster for Tissue Regeneration, 1200 Vienna, Austria
gDivision of Orthopedics, Department of Orthopedics and Trauma Surgery, Medical University of Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria
First published on 7th September 2021
Abstract
Rheumatoid arthritis is characterised by a progressive, intermittent inflammation at the synovial membrane, which ultimately leads to the destruction of the synovial joint. The synovial membrane as the joint capsule's inner layer is lined with fibroblast-like synoviocytes that are the key player supporting persistent arthritis leading to bone erosion and cartilage destruction. While microfluidic models that model molecular aspects of bone erosion between bone-derived cells and synoviocytes have been established, RA's synovial-chondral axis has not yet been realised using a microfluidic 3D model based on human patient in vitro cultures. Consequently, we established a chip-based three-dimensional tissue coculture model that simulates the reciprocal cross talk between individual synovial and chondral organoids. When co-cultivated with synovial organoids, we could demonstrate that chondral organoids induce a higher degree of cartilage physiology and architecture and show differential cytokine response compared to their respective monocultures highlighting the importance of reciprocal tissue-level cross talk in the modelling of arthritic diseases.
Introduction
In today's ageing society disorders affecting the human musculoskeletal apparatus, also referred to as musculoskeletal diseases (MSD), are of great clinical and scientific interest. In particular, arthritis describes various pathophysiological conditions, including inflammatory arthritis (IA) and osteoarthritis (OA). Types of IA include rheumatoid arthritis (RA) systemic inflammatory autoimmune conditions often found in diarthrodial joints.1 Overall, arthritic diseases frequently lead to swelling, pain and impaired movement of joints, severely affecting the quality of life.2,3Fibroblast-like synoviocytes (FLS) are the primary synovial cell type studied in RA.4 In healthy synovia, FLS lubricate the joint by producing e.g. lubricin or hyaluronan, which ensures homeostasis and function. FLS are also responsible for synovial lining formation and maintaining the inner joint membrane (Fig. 1A, left panel).5 In RA, the synovium is the main joint tissue structure infiltrated by immune cells at the synovial sublining layer and is a critical player in osteoclast-mediated bone erosion and pannus formation as well as enzymatic cartilage degradation.6,7 During the onset and progression of RA, FLS are stimulated by cytokines secreted from activated and migrating immune cells such as macrophages, B cells and T cells (e.g. IL-1β, TNFα, IL-6, IL-17), and in turn also secrete cytokines (e.g. IL-6, IL-15, IL-23), chemokines (e.g. IL-8. CXCL12) and degradative enzymes (various metalloproteases as well as cathepsins).8–15 In this context, IL-6 is a central pro-inflammatory molecule promoting joint inflammation and destruction.16,17
Over the last decade, arthritis research has gradually moved from conventional two-dimensional to more complex three-dimensional cell cultures, which allowed more physiologically relevant interactions mediated by unidirectional as well as reciprocal cell communication and transformation to human arthritic synovial tissue in vitro.18 Starting in 2006, Kiener et al. introduced a 3D synovial micromass organoid model with FLS embedded in Matrigel to form a synovial construct comprising lining and sub-lining layers spontaneously.5 This model has been used to investigate the influence of FLS activity on inflammatory cascades19 and the contribution of the complement system in FLS metabolism.20 Broeren et al. adapted this Matrigel-based primary model by combining FLS and CD14+ monocytes. When stimulated with TNF-α, these micromasses triggered arthritic hallmarks including synovial lining thickening, pro-inflammatory macrophage phenotype (M1) as well as upregulation of pro-inflammatory cytokines.21 Maracle et al. used a 3D drug screening model based on spheroids fabricated from RA-FLS and endothelial cells to recapitulate synovial angiogenesis, which constitutes a detrimental process in RA disease progression.22 Furthermore, Peck et al. successfully engineered a scaffold-free tri-culture drug screening model comprising of FLS (SW982 cell line), lipopolysaccharide (LPS)-activated macrophages and primary isolated chondrocytes from knee-cartilage to investigate cartilage deconstruction and RA reversal in affected porcine joints.23 Our group used a lab-on-a-chip with non-invasive light scattering sensing strategy to rapidly investigate TNFα induced rheumatoid architectural changes in 3D synovial micromass cultures concurring with synovial reorganisation as well as interleukin (e.g. IL-6, IL-8) and MMP (e.g. MMP-1, MMP-3, MMP-13, etc.) molecule secretion.24 More recently, also microphysiological models and organ-on-a-chip devices have been introduced to recapitulate various aspects of arthritic diseases: Ma et al. established a coculture of human FLS (synovial sarcoma SW982 cell line) with murine pre-osteoclasts (RAW264.7) and bone-marrow-derived stem cells (BMSCs) to study FLS migration and invasion-mediated bone erosion in RA.25 Nonetheless, to date most systems rely on non-human or immortalised cell lines to establish these complex 3D cocultures which are less predictive to model arthritic processes in human.
Therefore, herein we present a human joint-on-a-chip model based on co-cultivation of two spatially separated hydrogel-based 3D organoid constructs to understand how reciprocal tissue cross talk between synovial and chondral organoids and vice versa stimulates joint (patho)physiology (see Fig. 1B). An essential aspect of our joint-on-a-chip development capable of mimicking the human chondro-synovial niche was relating to improved reproducibility, which is key to all advanced in vitro cell-based assays. Consequently, a range of parameters known to influence organoid formation is investigated, including passaging of isolated synovial cells in 2D, variations of immune cell composition and cytokine secretion. Another focus is the investigation of the effects of chondro-synovial cross talk on tissue-level behaviour. Also, culture conditions including chondrogenic growth factor (TGF-β3) and fetal calf serum (1% or 10% FCS) supplementation were investigated to assess organoid maturation and soluble cross talk changes. Overall, the proposed chip-based organoid coculture model shows promising results due to a high degree of microenvironmental and cellular control. We also demonstrate excellent imaging capabilities of the architectural context and cellular composition with our proposed biochip features, that are impossible for randomly placed conventional 3D micromass and spheroid cultures established within microtiter plates.
Experimental
Clinical specimens
Human synovial tissues from patients were obtained from RA patients undergoing synovectomy (fulfilling the ACR/EULAR classification criteria26) with written informed consent under the terms of the ethics committee of Medical University of Vienna (EK-No. 1065/2011). The medical background of patients was documented and included age (average age: 55, min.: 27, max.: 71), sex (8 female and 2 male), tissue location (carpals: 4, phalanges: 3, elbow joint: 2, tibiofemoral joint: 1), diagnosis (total: 10, rheumatoid arthritis: 9 patients, control: 1 OA patient) and medication (no treatment: 2, Embrel: 2, Arava: 1, cortisone: 3, Salazopyrin: 1, Arava + cortisone: 1). Primary cell populations were isolated and stored in liquid nitrogen, as described earlier.27Cell culture
FLS were thawed at 37 °C and immediately plated in a T75 cell culture flask containing pre-warmed DMEM culture medium supplemented with 10% FCS, 1% MEM NEAA and 1% antibiotics. Cells were split upon reaching 90% confluency. Before passaging, 1 ml of culture medium was collected and frozen for supernatant analysis. For passaging experiments, cells were enzymatically detached, pelleted and resuspended in culture medium. An aliquot of 5 × 105 cells was weekly transferred to a new flask, whereas the residual cells were used for flow cytometry analysis and 3D cultivation. For cross talk experiments, expanded FLS were used between P5 and P7 if not indicated otherwise.Commercial human primary chondrocytes (C-12710; female donor; PromoCell) were maintained in chondrocyte growth medium (411–500; CellApplications Inc.), which was exchanged every other day. Cells were expanded until passage 5 and frozen as aliquots for on-chip cross talk studies.
Synovial micromass culture in well plates
Well plates were coated with 0.03 w/v% ethanolic poly(2-hydroxyethyl methacrylate) (poly-Hema) as described initially for synovial micromass cultures.5 Briefly, after pelleting, the primary FLS cells at a passage number between 1 and 8 were resuspended in ice-cold Matrigel at a concentration of 3000 cells per μl Matrigel. To initiate micromass cultures, a 20 μl drop of the cell-laden Matrigel suspension was pipetted into the centre of a coated well. After 40 min of polymerisation, cell/Matrigel drops were overlaid with 2 ml fully supplemented organoid culture medium containing 1% insulin-transferrin-selenium (ITS) and 0.032 μg ml−1L-ascorbic acid 2-phosphate with a weekly medium exchange.Microfabrication
The two multi-layered designs for either synovial monoculture or chondro-synovial coculture illustrated in ESI† Fig. S1 were fabricated using a CAMM-1 GS-24 vinyl cutter (Roland, Germany). The devices were assembled layer by layer by xurography from two-dimensional CAD templates from 500 μm thick transparent silicone sheets (MVQ Silicone) as described previously.28,29 The basic design principles were adapted from an earlier vascular organoid study,30 and circular medium reservoirs were added to contain a total medium volume of 300 μl. For height variation, the layer numbers of the circular hydrogel compartment were adjusted in the range of 1–4 mm. To prevent cellular outgrowth from within organoids, chip surfaces were coated with 0.5% ethanolic Lipidure® (Amsbio, UK) overnight before UV light sterilisation for 1 hour.Two basic designs with five structured PDMS layers were assembled on object slide base layers to form a joint-on-a-chip (see ESI† Fig. S1A): the monoculture design (left column layers I–V) comprised of a single organoid chamber (Ø 5 mm). In contrast, two opposing organoid chambers allow for soluble tissue-level communication via reciprocal signalling (right column).30,31 To improve medium replacement and sample withdrawal of the optimised design, the medium supply channels interconnect two circular medium reservoirs. To enable reproducible and facile loading of hydrogels, structured layers I and II form a step-like hydrogel stopper feature that prevents hydrogel bleeding into the medium supply channel. Additionally, structure layers III–V form a chamber geometry favouring controlled organoid formation by an anchoring organoid condensation guide (see ESI† Fig. S1B and C for detailed functional and dimensional elaborations on both features).
On-chip organoid culture
For the generation of synovial organoids, a hydrogel suspension containing FLS was loaded via a loading vent into each cell cultivation chamber. To start organoid formation, a total of 45 μl FLS cells in Matrigel were slowly injected at a concentration of 3000 cells per μl through the hydrogel port and was sealed by a small piece of PCR sealing film on ice. After 40 min of polymerisation at 37 °C, 400 μL organoid chip medium was added to start the organoid culture. A previously established cartilage organoid based on TISSEEL fibrin hydrogel (Baxter, Austria) was injected in the chondral compartment as previously described to establish on-chip cocultures.31,32 Briefly, 30 μl of fibrinogen solution at a concentration of 100 mg ml−1 containing 2000 cells μl−1 were mixed with 30 μl 4 U ml−1 thrombin solution supplemented with 40 μM CaCl2 solution and was polymerised for 30 min at 37 °C. For medium composition experiments, TGF-β3 was added to the chip medium at 1 and 10 ng ml−1 in either 1% or 10% FCS-containing organoid culture medium and compared to commercial chondrogenic differentiation medium (411D-250; CELL Applications Inc., USA). For medium replacement, the complete chip reservoir medium of 300 μL was replaced with fresh organoid medium and supernatant was frozen for off-chip supernatant analysis. After cultivation, the top ceiling of the organoid chambers was removed with a scalpel, and the individual organoids were placed in 7% paraformaldehyde for histological preparation or cytometry buffer for flow cytometry analysis as described below.Flow cytometry
Two different staining mixes were prepared on ice, as shown in ESI† Table S1. Aliquots of 1.5 × 105 cells were washed with flow cytometry buffer (PBS with 2% FCS) and pelleted at 4 °C and 1350 rpm for 8 min. After pelleting, cells were resuspended in 50 μl of the staining mix solution, incubated for 20 min, washed and resuspended in 150 μl flow cytometry buffer. Measurements were performed on a BD Canto II flow cytometer using BD FACS Verse™ (note: one measurement cycle lasted either 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells or 2 min). Gating of cell populations was performed as described elsewhere.33,34 Cell populations from organoids were harvested by centrifugal removal after Matrigel liquification for 2 h on ice.
000 cells or 2 min). Gating of cell populations was performed as described elsewhere.33,34 Cell populations from organoids were harvested by centrifugal removal after Matrigel liquification for 2 h on ice.
Confocal microscopy
Imaging was performed using an upright Leica TSC SP5 Microscope (Leica Microsystems) with 20× and 40× water immersion objectives. Imaris software (Bitplane) was used for image processing. Live-cell staining was performed with fluorescent cell tracker dyes (Green CMFDA/Orange CMRA and Deep Red; Thermo Fisher) before pelleting and hydrogel mixing according to the manufacturer's staining protocol and ESI† Table S2.Histology
Tissue and organoid specimens were fixed in 7% paraformaldehyde, dehydrated in ethanol and paraffin-embedded according to standard in-house procedures.35 Paraffin sections of approx. 2.5 μm thickness were stained with hematoxylin and eosin (HE) for morphological evaluations.Morphometric analysis
For morphometric analysis, manual tracing of the organoid boundaries of phase-contrast images of organoids taken at selected time points was used in ImageJ (version 2.0.0.rc.69/1.52.p) in combination with the shape descriptors option of the ‘measure’ as well as the ‘analyse particles’ functions as previously described.36 For synovial network parameter analysis AngioTool (version 0.6a) was used with optimised analysis parameters (vessel diameter: 5; intensity range: 15–100; remove small particles option set to 240).Supernatant protein analysis
For AlphaLISA analysis of IL-6, IL-8, MMP-1 and MMP-3 (AL223C, AL224C, AL284C, AL242C, Perkin Elmer), undiluted supernatants of 2D cultured FLS were analysed according to the manufacturer's protocol using an EnSpire® Multimode plate reader (Perkin Elmer).For cytokine analysis of IL-6, IL-8, MMP-13 and VEGF using human Magnetic Luminex Assay (R&D Systems), supernatants of synovial and chondral organoids in mono and coculture were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in dilution reagent and analysed according to the manufacturer's protocol using a Luminex™ MAGPIX™ Instrument System (Invitrogen). After medium background subtraction, molecule secretion of mono- and cocultures were normalised to 104 cells per mL and day to compensate for differences in total cell numbers between synovial and chondral monocultures as well as cocultures. To reduce overall sample size, occasionally supernatants from n = 3–4 organoids were pooled for analysis of medium compositions (TGF/FCS content) as well as monoculture organoids in the comparative study on mono- and co-culture organoids.
10 in dilution reagent and analysed according to the manufacturer's protocol using a Luminex™ MAGPIX™ Instrument System (Invitrogen). After medium background subtraction, molecule secretion of mono- and cocultures were normalised to 104 cells per mL and day to compensate for differences in total cell numbers between synovial and chondral monocultures as well as cocultures. To reduce overall sample size, occasionally supernatants from n = 3–4 organoids were pooled for analysis of medium compositions (TGF/FCS content) as well as monoculture organoids in the comparative study on mono- and co-culture organoids.
RT-qPCR preparation and analysis
mRNA of snap-frozen patient tissue and organoids using an optimised extraction protocol (Qiagen Easy prep) using Precellys lysing kits (CK25; 3 × 20 s, 5000 rpm, 3 min cooling break) in combination with frozen lysis buffer beads (innuPREP RNA Mini Kit, Analytik Jena) supplemented with 1% beta-mercaptoethanol on a Precellys 24 system. RNA was reverse transcribed into cDNA (Firma und Kit einfügen) and subsequent RT-qPCR was performed on a QuantStudio3 QPCR system (Applied Biosystems, Thermo Fisher) using SensiMix SYBR Green Master Mix as previously described.35 Primer sequences are described in ESI† Table S3.Statistical analysis
Prism software 7 (version 7.0a; GraphPad, USA) was used for statistical analysis of data sets for either t-test analysis (paired and unpaired) or 2-way ANOVA combined with appropriate post-hoc analysis (Tukey, Sidak or Dunnet's test as recommended by the software). Shapiro–Wilk normality test for normal distribution was performed. P values < 0.03 were considered as statistically significant (*p < 0.03, **p < 0.002, ***p < 0002, ****p < 0.0001).Results & discussion
Characterisation of the chip-based three-dimensional synovial organoid model
Initially, individual hydrogel compartments were studied separately to investigate each organoid unit's formation and maturation. In synovial organoid cultures, RA patient-derived primary FLS were mixed into Matrigel before injection and cultured for up to four weeks. It is important to note that during synovial organoid formation, significant hydrogel condensation and remodelling takes place. For instance, Fig. 2A shows the influence of increasing hydrogel volume (note: increasing chamber heights from 1 to 4 mm results in a volume increase from 19.6 to 78.5 μl) on condensation behaviour, where RA-FLS organoid volume gradually declined to between 18–36% after 28 days of organoid cultivation. Interestingly, a chamber height of 1 mm (19.6 μL) was frequently insufficient to generate a single synovial organoid; instead, fragmentation of the organoids and cell dispersion as well as a significantly different condensation behaviour starting at day 6 (p < 0.0001; Tukey's multiple comparisons test) occurred (see also images of ESI† Fig. S2) in comparison to the other hydrogel volumes. In contrast, chamber heights of 1.5, 2.0 and 2.5 mm displayed uniform organoid formation exhibiting an area of 3.8 ± 0.27 mm2 at day 28. Larger volumes (e.g. chamber heights of 3 and 4 mm) produced bigger synovial constructs with 5.7 ± 0.12 mm2 and 7.1 ± 0.09 mm2, respectively. A more detailed morphology analysis revealed that most homogenous synovial organoid formation is achieved using a cultivation chamber with 2 mm height, where almost spherical structures are obtained. Fig. 2B illustrates the time-dependent change of synovial organoid shape from rectangular to spherical throughout 28 days of the cultivation period, which points at increase of organoid sphericity over time as the FLS organoids mature. During early biochip design iterations we have frequently observed that when organoids came in closer contact, they formed direct connections and allowed FLS to migrate towards the chondral unit. As more reproducible spatial control over organoid positioning and maturation was the aim of the current biochip design to enable reciprocal molecular cross talk in the absence of direct cell–cell interactions, it was imperative to guide and spatially control formation of the two organoid types on the biochip using the proposed features. A detailed investigation of overall relative and absolute positioning precision in relation to the hydrogel anchor and the waveguide features was performed as shown in Fig. 2C (for details see also ESI† Table S4). The on-chip positioning features improves the hydrogel positioning of the tissue organoids during the first week of the initial organoid remodelling phase with a lower relative standard deviation of 6.6% compared to 11.9% at day 7 and 32% at day 21 post seeding, which indicates that the integrated structures improve the initial positioning of the hydrogel. During three weeks of cultivation, both positioning parameters aggravated by manual user manipulation (i.e., sample withdrawal and medium exchange). However, positioning accuracy of organoids was not significantly affected by height of the hydrogel compartment (p > 0.1, Tukey's multiple comparisons post hoc test). Next, on-chip synovial condensation dynamics were compared to polyHEMA-coated anti-adhesion well plates (MMC). Results of this comparative study shown in Fig. 3A revealed similar overall architectural features with a significantly higher reproducibility exhibiting a 3.5-fold lower relative standard deviation of 4.4% of organoid biochips (n = 5) compared to 15.5% (n = 3) for MMC cultures generated in anti-adhesion well plates. Moreover, morphometric AngioTool image analysis revealed that synovial network morphology (see Fig. 3B–D) was similar for synovial micromasse and biochip organoids with gradually increasing parameters. The total increase in vessel area 21.45 ± 4.7% and 29.2 ± 0.4%, protrusion length of 119.8 ± 29 μm and 135.6 ± 14 μm, as well as number of junctions of 627 ± 201 and 970 ± 55 junctions per mm2 was observable for biochip in comparison to micromass organoids over the first 7 days of network maturation.Influence of passaging and cultivation on the heterogeneity of patient-specific synovial cell populations
Arthritic synovial tissue comprises, aside from synovial fibroblasts (FLS), a diverse set of immune cells such as macrophages, B cells and T cells. While standard 2D monoculture conditions is known to rapidly deplete immune cells resulting in pure synovial fibroblasts (FLS) using DMEM medium,21 less is known about the effect of this immune cell depletion on the formation of FLS organoids. To evaluate the long-term effect of co-cultivating isolated patient derived FLS and immune cells on synovial tissue heterogeneity, the samples of seven RA donor tissues were characterised using flow cytometry and supernatant analysis. Results shown in ESI† Fig. S3A and B indicate a gradual depletion of leucocytes and selective growth of synovial fibroblast over 5 weeks (5 passages). A high degree of interpatient immune cell variability was evident for CD45+ leucocytes ranging from 22% to 1.1% between individual patient samples. Further analysis showed variations of sub-population of 4.1% to 0.1% CD3+/CD14− T cells, and 3.9% to 0.01% CD19+/CD14− B cells as well as a predominant population of 8.9% to 0.6% CD14+/CD3− macrophages. Similarly, a gradual decrease of pro-inflammatory cytokines (e.g. IL-6 and IL-8) and degradative matrix metalloproteinases (e.g. MMP-1 and MMP-3) in cell culture supernatant was detected over the five-week cultivation period with weekly subcultivations (see ESI† Fig. S3C).Next, the impact of medium composition for on-chip organoid cultivation and 2D/3D cultivation on immune cell survival was investigated in more detail. Initial characterisation of CD45 positive cells of the freshly isolated synovial cell suspensions from donor tissue (n = 4 biological donors) displayed 80.2 ± 3.1% podoplanin positive FLS and 5.7 ± 4.7% CD45 positive leucocytes (i.e., T cells, B cells and macrophages), which was an equal population distribution also found in p1 subcultivations (see ESI† Fig. S3A). Results shown in Fig. 4A revealed a higher degree of leucocyte survival when substituting the DMEM culture medium with leucocyte medium (e.g. RPMI 1640), leading to 4.9, 2.4 and 1.8-fold higher T cell, macrophage and B cell subpopulations from week one to two post-seeding (**p < 0.01; 2-way ANOVA with Sidak's multiple comparisons test). In turn, synovial organoid on-chip cultivation further improved leucocyte survival for both medium types (n = 2; see Fig. 4B) with 11- and a 20-fold increase in leucocyte populations compared to conventional 2D cultivation, which showed a declining population trend. First of all, this means that both the amount and type of immune cells present in rheumatoid arthritis patients vary significantly and need to be accounted for in an in vitro arthritis model. For FLS 2D cultures, immune cell depletion decreases the arthritic sample heterogeneity regarding cell composition and cytokine secretion and favours FLS growth. Furthermore, our chip-based synovial organoid system that features a three-dimensional tissue-like microenvironment protects synovial leucocyte populations from depletion even in unfavourable culture media (e.g. DMEM).
Impact of passaging on the formation of chip-based synovial organoids
To further understand the effects of synovial cell passaging on chip-based 3D synovial cultures, organoid formation and secretion of cytokines were investigated in ensuing experiments. On-chip generated synovial organoids of increasing passages (p1–5) were cultivated over five weeks, and vital organoid parameters including area, solidity, perimeter, roundness and organoid numbers per unit were analysed. Fig. 5A and ESI† Fig. S4 show morphometric analyses at day 28 post-seeding and point at stable organoid formation peaking at 5.5 ± 0.6 mm2 and 5.8 ± 0.06 mm2 of organoid area for later passages in contrast to freshly isolated FLS populations. Early passages (p1) showed a tendency to form smaller cell aggregates ranging from 1.8–4.1 mm2 with higher variability (0.42% RSD). In turn, FLS organoids formed from p5 cells yielded reproducible organoid areas of 5.6 ± 0.67 mm2 (0.01% RSD). The remaining morphometric parameters, including perimeter (0.04% RSD), roundness (0.05% RSD) and solidity (0.01% RSD), further pointed at the highest reproducibility in the presence of higher passage numbers (p3–4). Furthermore, the system's reproducibility to form a single organoid in each organoid compartment scored best for organoids created from cells after 4–5 weeks of passaging. Investigating patient heterogeneity for organoids produced from individual patient-derived synovial tissues (Fig. 5B–F) the overall effect of FLS passaging on organoid formation was still observable. While most parameters including roundness, perimeter and solidity were not affected by patient origin (p > 0.1, 2-way ANOVA with Tukey's posthoc test), organoids from three different RA patient tissue biopsies showed a significant difference in both organoid areas (p < 0.0002) and the number of formed organoids (p < 0.03). These indicators still point out that passaging of FLS populations improves the reproducibility of organoid formation. On-chip cultivation of primary cell in a 3D environment is known for more native tissue-like molecule retention and secretion.37,38 Therefore, next the secretory behaviour for FLS organoids over increasing passages and cultivation time points were monitored as shown for IL-6, IL-8, MMP-13 and VEGF secretion up to passage p5 and week three post-seeding in Fig. 6A–D.FLS organoids overall showed initially higher overall secretory capacity in week 1, which declined significantly over three weeks of cultivation independent of passage number or molecule type (p < 0.002). Considering the passaging effect, RA-FLS organoids showed a considerably higher secretion performance of MMP-13 and VEGF (p < 0.03, 2-way ANOVA with Sidak's posthoc test) with an increase for later passages, which again equalized over three weeks of culture. The passage dependent differences in the early secretory activity of FLS organoids are most probably not caused by immune cell depletion alone as no direct correlation between the number of immune cells, specific secretory molecules (i.e., interleukins or MMPs) were discernible. Furthermore, activated FLS are known for their high secretory capacity.4 This means that other factors such as inflammatory memory or reprogramming20 before isolation are more likely to influence how RA-FLS react and mature during organoid formation in 3D.
Overall, by intentionally reducing cell heterogeneity in the synovial organoid constructs by subcultivation for 5 passages we could increase reproducibility over the cultivation duration by limiting initial cellular variations. Consequently, RA-FLS cells from p5 were used in all subsequent experiments on cross talk studies to reduce sample variability and improve organoid yield and homogeneity.
Evaluation of the chondral organoid model in monoculture biochips
Recently our group has studied the impact of biochip culture31 as well as matrix stiffness/composition32 on chondrocyte phenotype. Previous results already indicated that primary chondrocytes embedded in 50 mg ml−1 fibrin hydrogel show good chondrocyte viability, morphology and cartilage-specific functions (i.e., collagen II, cggrecan, etc.) independently of culture technique (i.e., biochip organoid vs. micromass). Before co-cultivation and cross talk experiments on chondro-synovial organoids, chondral organoids comprising of human articular chondrocytes suspended in 50 mg mL−1 fibrin were re-evaluated to demonstrate that the current biochip is in agreement with previous findings. Confocal imaging confirmed the typical round morphology of articular chondrocytes within the Fibrin construct after seeding (see Fig. 7A and ESI† Video S1) with 83.46 ± 16.5% chondrocyte viability at day 21 post-seeding (see Fig. 7B and C). Further histological comparison (see Fig. 7D) revealed that the chip-based chondral organoids can recapitulate the morphological and cell density characteristics of human articular cartilage of superficial tissue zones, which are primarily affected by mechanical wear as well as protease degradation. Furthermore, the chondral organoid derived from a single healthy donor sample indicated comparable ratios of cartilage health indicators Col II/Col I and AGC/Col I ratio similar to values found in native human cartilage (n = 3 donors; see Fig. 7E). Overall these results indicate good agreement with previous studies in terms of native tissue-like properties of fibrin-embedded chondrocytes in vitro.Establishment of a joint-on-chip organoid coculture model for chondro-synovial cross talk studies
Another critical aspect in establishing a functional in vitro arthritis model of the joint deals with proper tissue to tissue interactions between the synovium as inflammatory tissue and the cartilage as target tissue during the onset and progression of the disease. High serum content is a crucial requirement for FLS since it provides and maintains the proper synovial cell function and architecture. However, redifferentiation of expanded chondrocytes is conducted frequently using differentiation media that contain no or low serum concentrations, but TGF-β3 and other chondrogenic supplements to promote a chondrogenic niche.39 Consequently, the impact of TGF-β3 in addition to serum content (e.g., 1 ng ml−1vs. 10 ng ml−1, 1 vs. 10% FCS) was investigated against an unknown commercial chondrogenic medium formulation to identify suitable coculture media formulations for chondro-synovial cross talk studies. Results in Fig. 8A show that the chondrogenic medium significantly improves stable chondral organoid formation retaining over 90% of the initial organoid size up to day 8 with a gradual decline to 60.9 ± 8.8% at day 21 in comparison to the control organoids (p < 0.0001). Even though the presence of 1% FCS combined with 10 ng ml−1 TGF-β3 exhibited comparable values up to day 8, keeping 85 ± 1.28% of the initial chondral organoid area, variations in serum content and TGF-β3 showed no significant difference to untreated control organoids at day 21 (p > 0.1, 2-way ANOVA with Dunnett's post hoc test) and were unable to outperform the commercial formulations.In contrast to chondral organoids, where chondrogenic differentiation medium showed a positive effect on the condensation of chondral organoids in coculture, the synovial organoid formation was severely affected by TGF-β3 in combination with high serum content as well as chondrogenic medium formulation. For instance, 1 and 10 ng ml−1 TGF-β3 with 10% serum significantly reduced initial organoid compaction and cell–cell interaction at day 4 post-seeding generating deformed and often scattered organoids with 42% (p < 0.0002, 2-way ANOVA with Dunnett's post hoc test) and 55% lower initial organoid area (p < 0.0001), respectively. Moreover, commercial chondrocyte medium also significantly impeded proper organoid formation by 32% (p < 0.0001), as RA-FLS formed not even spherical aggregates but a network of tubular structures. This significant effect of serum and growth factor supplementation on organoid formation was observable for the first week of coculture. However, it disappeared over the next two weeks and only synovial organoids in the commercial chondrogenic medium were still significantly impeded (p < 0.0001). Interestingly, the combination of 1% FCS and 10 ng ml−1 TGF-β3 led to a significant increase in synovial organoid condensation (p < 0.0002) similar to their neighbouring chondral counterparts. To find a tentative explanation, secretion analysis (see Fig. 8B) was performed on coculture supernatants throughout three weeks and showed no significant differences in secretion levels of IL-6, MMP-13 or VEGF except for the fact that chondrogenic differentiation medium showed reduced MMP-13 secretion levels and elevated IL-6 and VEGF levels, which were probably caused by the presence of pro-inflammatory cytokine supplements (i.e., IL-6) or corticosteroids in the differentiation kit itself, which are known to boost chondrogenic differentiation further.40 Also, TGF-β3 is frequently used as a trigger in synovial fibrosis models,21 and the effect of enhanced organoid instability with an increase in serum are in line with previous studies on synovial fibroblast models, showing that high serum conditions further exacerbate the aggravating effect of TGF-β3.41 As our results indicated that there is a strong negative synovial performance when using media formulations and additives that are known to improve chondrogenic performance, further experiments concerning chondro-synovial cross talk were conducted in the absence of additives to investigate the baseline communication of primary synovial organoids generated from purified patient FLS with chondral organoids under very basic non-compromising conditions.
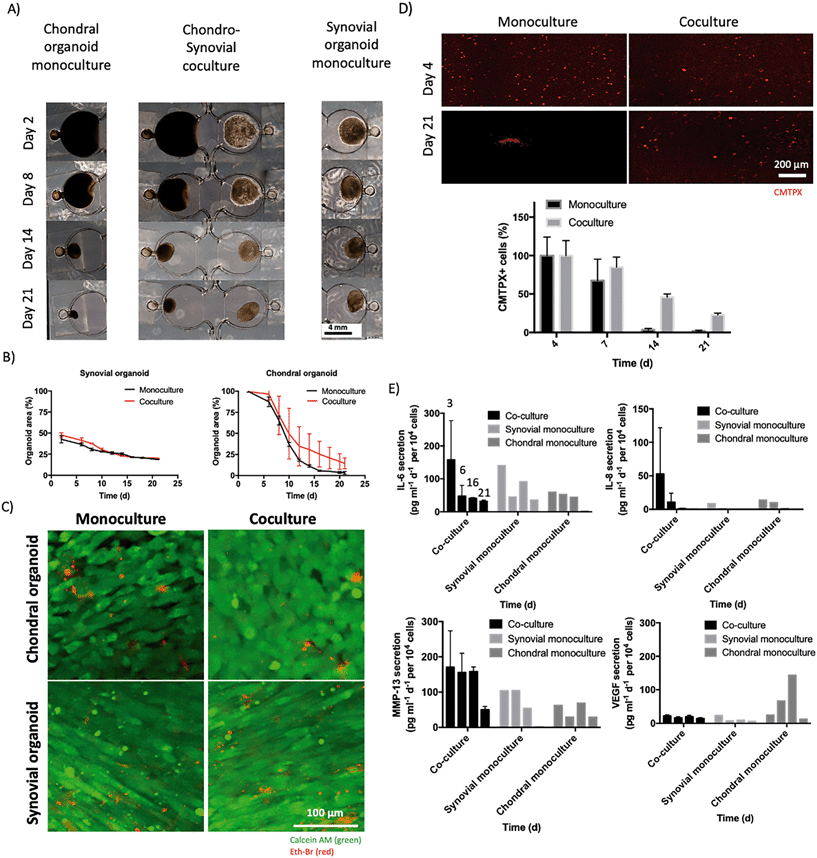
As a final experimental setup, our cocultures of synovial organoids made from patient-derived RA-FLS cultures were interfaced in coculture biochips with chondral organoids from primary human chondrocytes to establish and investigate the molecular effects of chondro-synovial cross talk (see Fig. 9). Chondral organoid monocultures condensed from 18.56 ± 0.45 mm2 to 0.65 ± 0.33 mm2 (2.0 ± 4.3% of the initial organoid area), whereas synovial monocultures retained approx. 44% of the initial organoid area at day 2 from an initial organoid area of 8.72 ± 0.88 mm2 to 3.85 ± 0.03 mm2 throughout 21 days of monoculture (see Fig. 9A and B). Relative to the respective monoculture counterparts, synovial organoids in co-culture showed similar condensation dynamics with an approx. 4-fold decrease of the initial organoid area (p > 0.1, 2-way ANOVA with Sidak's post hoc test) from 9.7 ± 0.6 mm2 at day 2 to 4.14 ± 0.03 mm2 at day 28 post-seeding. Interestingly, the chondral compartment was more affected by tissue–tissue cross talk and showed a tendency to gain approx. 22% organoid area when cocultured with an organoid area of 2.9 ± 1.3 mm2 for cocultured in comparison to 0.65 ± 0.33 mm2 for monoculture chondral organoids at day 21. To investigate whether this effect was caused by alterations in cell viability, a live/dead dye exclusion assay as shown in Fig. 9C was conducted and showed comparable high viability of above 91% for chondral and 84% for synovial organoids independent of the cultivation scenario after three weeks of on-chip cultivation (see ESI† Fig. S5A). While epifluorescence analysis of calcein-AM stained monoculture chondral organoids revealed an overall homogenous population of chondrocytes (see Fig. 7C as reference), higher resolution confocal microscopy at a higher resolution for superficial areas near the organoid surface revealed higher round chondrocyte numbers for cocultured chondral organoids compared to monoculture organoids, that also showed slightly elongated cell subpopulations. To better understand the correlation of this cellular difference with organoid condensation, mono and cocultured organoids were stained with CMTPX cell tracker before hydrogel loading at day 0 and organoids were investigated over 3 weeks. This specific cell tracker dye creates cell-impermeant and red-fluorescent reaction products by intracellular esterase hydrolysis. It provides strong and stable staining for chondrocytes over weeks and months. In contrast, highly proliferative cells such as dermal or synovial fibroblasts in our experience lose their fluorescence signal within a few days, especially when using standard epifluorescence microscopy analysis. Consequently, the loss of fluorescent signal of chondrocytes was investigated to quantify the change in chondrocyte morphology over time directly. Representative epifluorescence micrographs of chondral organoids in mono- and coculture for 21 days (see Fig. 9D) revealed that the initial number of chondrocytes with round morphology at day 4 gradually declined for chondral monocultures, while round chondrocyte subpopulations were significantly more retained in the coculture with synovial organoids (p < 0.005, paired t-test) by 23 ± 2.3%. As shown in ESI† Fig. S5B, dedifferentiating chondrocytes in 2D cultures show approx. 300% increase in resazurin to resorufin conversion within 7 days of culture caused by increased esterase activity and cell proliferation. Chondral organoids showed a 2-fold lower conversion rate over the first seven days of culture, pointing at low proliferation and is in line with previous work comparing chondrocyte culture conditions.31 To investigate the influence of chondro-synovial cross talk on molecule secretion, Luminex analysis was used to compare monoculture with coculture organoids. As shown in Fig. 9E, the chondro-synovial organoid coculture system showed no significant changes in cytokine secretion levels of IL-6, IL-8 and MMP-13 (p > 0.1, 2-way ANOVA with Tukey's post hoc test) at day three with time-dependent a comparable decline similar to previous secretion curves of FLS organoids. However, chondral organoids in coculture, which were characterised before by a more chondrogenic environment with a higher degree of organoid area and chondrocyte roundness as well as lower metabolic actividisplayed significantly reduced VEGF secretion levels compared to chondral monocultures (p < 0.03, 2-way ANOVA with Sidak's post hoc test). This is in line with previous studies on VEGF secretion increase of chondrocytes cultivated under non-physiological conditions.35,36,42
Conclusions
In the current study, we have, for the first time to our knowledge, established a chip-based chondro-synovial dual organoid model based on self-organising patient-derived cells that allow investigating reciprocal tissue–tissue cross talk in orthopaedic/arthritic research. Due to the intrinsic microfluidic design features, the current microfluidic joint-on-a-chip design enables the reliable formation of organoids with high reproducibility and positioning precision, which is crucial for any organ-on-a-chip analysis system used in biomedical and biopharmaceutical research. We also demonstrated that expanded synovial fibroblast cultures generate organoids of higher precision and reproducibility compared to freshly isolated cell populations. This passaging effect that reduces the epigenetic functional characteristics of synovial subpopulations and results in synovial cell assimilation in vitro has been reported for the loss of lining versus sublining characteristics of FLS.43–45 We furthermore investigated chondrogenic and fibroblast medium conditions to rule out the effects of medium type on chondro-synovial cross talk as cocultures require a typical medium formulation. For instance, primary chondrocytes perform best in culture medium supplemented with TGF-β3 at low or no serum conditions as it enhances chondrocyte redifferentiation46 and stimulates extracellular matrix production.47 At the same time, proper cultivation environment for FLS and FLS micromasses encompasses high serum conditions with vitamin C supplementation. However, these chondrogenic factors severely influence synovial fitness (i.e., proliferation) in low concentrations of FBS and high TGF-β3 concentrations and fail to recapitulate healthy synovial tissue function.41 In our current study, we followed a basal medium formulation with 10% FCS for the present proof-of-concept study; however, a more optimised medium formulation, most probably using xeno-free formulations, needs to be investigated to reduce the effects of molecules from the medium formulation for chondro-synovial cross talk. Finally, we could demonstrate that co-cultivation of chondral and synovial organoids improved the phenotype of chondrocytes within chondral organoids in comparison to chondral monocultures in the absence of synovial fibroblasts. We plan to investigate this molecular aspect and more disease-relevant mechanisms (i.e. induction of synovitis and pathway inhibition) in the future and believe that our current dual organoid-based joint-on-a-chip approach highlights the importance of tissue-level cross talk in the context of musculoskeletal diseases. For the current proof-of-principle study, we envision for the future of the chondro-synovial arthritis model to elaborate on the exact molecular cross talk mechanism between synovial and chondral organoids with a higher, statistically more significant patient sample size in combination with genomic and proteomic analysis.Author contributions
AF, EIR, IOC, and SSch: experiments and data analysis; editing and reviewing – original draft. FS, JH and RW: surgical preparation of primary patient tissue; reviewing – original draft. SS, BB, WH and HR: assistance with fibrin-based organoids, editing and reviewing – original draft. HPK: reviewing and editing – original draft. RAB: experiments; data analysis; reviewing and editing – original draft. MR: supervision; experiments; data analysis; methodology; supervision, writing – original draft, review and editing. ST: writing – original draft, review and editing. PE: supervision; writing, review and editing – original draft; funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by the Austrian Federal Ministry of Education, Science and Research (BMBWF; grant agreement number 1612889) and the Vienna Science and Technology Fund (WWTF, project number LS13-092). Figure schemes were created with BioRender.com under paid subscription. The authors acknowledge the TU Wien University Library for financial support through its Open Access Funding Program.Notes and references
-
L. Gettman, M. Max, C. Eisenhower and R. L. Cooke, Musculoskeletal Conditions, 2019 Search PubMed
.
- P. C. Taylor, A. Moore, R. Vasilescu, J. Alvir and M. Tarallo, Rheumatol. Int., 2016, 36, 685–695 CrossRef CAS PubMed
.
- G. R. Burmester and J. E. Pope, Lancet, 2017, 389, 2338–2348 CrossRef
.
- E. H. Noss and M. B. Brenner, Immunol. Rev., 2008, 223, 252–270 CrossRef CAS PubMed
.
- H. P. Kiener, D. M. Lee, S. K. Agarwal and M. B. Brenner, Am. J. Pathol., 2006, 168, 1486–1499 CrossRef CAS PubMed
.
- E. A. Littlejohn and S. U. Monrad, Prim. Care, 2018, 45, 237–255 CrossRef PubMed
.
- M. Asif Amin, D. A. Fox and J. H. Ruth, Semin. Immunopathol., 2017, 39, 385–393 CrossRef CAS PubMed
.
- R. W. Kinne, B. Stuhlmüller and G. R. Burmester, Arthritis Res. Ther., 2007, 9, 224 CrossRef PubMed
.
- I. A. Udalova, A. Mantovani and M. Feldmann, Nat. Rev. Rheumatol., 2016, 12, 472–485 CrossRef CAS PubMed
.
- H.-Y. Yap, S. Tee, M. Wong, S.-K. Chow, S.-C. Peh and S.-Y. Teow, Cell, 2018, 7, 161 CrossRef CAS PubMed
.
- A. P. Cope, H. Schulze-Koops and M. Aringer, Clin. Exp. Rheumatol., 2007, 25, S4–S11 CAS
.
- S. Bugatti, B. Vitolo, R. Caporali, C. Montecucco and A. Manzo, BioMed Res. Int., 2014, 2014, 681678 Search PubMed
.
- A. Kimura and T. Kishimoto, Eur. J. Immunol., 2010, 40, 1830–1835 CrossRef CAS PubMed
.
- A. Alunno, M. Manetti, S. Caterbi, L. Ibba-Manneschi, O. Bistoni, E. Bartoloni, V. Valentini, R. Terenzi and R. Gerli, Mediators Inflammation, 2015, 2015, 751793 Search PubMed
.
- E. H. Noss and M. B. Brenner, Immunol. Rev., 2008, 223, 252–270 CrossRef CAS PubMed
.
- Y. Kaneko and T. Takeuchi, Int. Immunol., 2017, 29, 511–517 CrossRef CAS PubMed
.
- M. Narazaki, T. Tanaka and T. Kishimoto, Expert Rev. Clin. Immunol., 2017, 13, 535–551 CrossRef CAS PubMed
.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed
.
- M. Bonelli, K. Dalwigk, A. Platzer, I. Olmos Calvo, S. Hayer, B. Niederreiter, J. Holinka, F. Sevelda, T. Pap, G. Steiner, G. Superti-Furga, J. S. Smolen, H. P. Kiener and T. Karonitsch, Exp. Mol. Med., 2019, 51, 1–11 CrossRef CAS PubMed
.
- J. Friščić, M. Böttcher, C. Reinwald, H. Bruns, B. Wirth, S.-J. Popp, K. I. Walker, J. A. Ackermann, X. Chen, J. Turner, H. Zhu, L. Seyler, M. Euler, P. Kirchner, R. Krüger, A. B. Ekici, T. Major, O. Aust, D. Weidner, A. Fischer, F. T. Andes, Z. Stanojevic, V. Trajkovic, M. Herrmann, A. Korb-Pap, I. Wank, A. Hess, J. Winter, V. Wixler, J. Distler, G. Steiner, H. P. Kiener, B. Frey, L. Kling, K. Raza, S. Frey, A. Kleyer, T. Bäuerle, T. R. Hughes, A. Grüneboom, U. Steffen, G. Krönke, A. P. Croft, A. Filer, J. Köhl, K. Klein, C. D. Buckley, G. Schett, D. Mougiakakos and M. H. Hoffmann, Immunity, 2021, 54, 1002–1021.e10 CrossRef PubMed
.
- M. G. A. Broeren, C. E. J. Waterborg, R. Wiegertjes, R. M. Thurlings, M. I. Koenders, P. L. E. M. van Lent, P. M. van der Kraan and F. A. J. van de Loo, ALTEX, 2019, 36, 18–28 CrossRef PubMed
.
- C. X. Maracle, P. Kucharzewska, B. Helder, C. van der Horst, P. C. de Sampaio, A. R. Noort, K. van Zoest, A. W. Griffioen, H. Olsson and S. W. Tas, Rheumatology, 2017, 56, 294–302 CrossRef CAS PubMed
.
- Y. Peck, L. T. Leom, P. F. P. Low and D. A. Wang, J. Tissue Eng. Regener. Med., 2018, 12, e237–e249 CrossRef CAS PubMed
.
- M. Rothbauer, G. Höll, C. Eilenberger, S. R. A. Kratz, B. Farooq, P. Schuller, I. Olmos Calvo, R. A. Byrne, B. Meyer, B. Niederreiter, S. Küpcü, F. Sevelda, J. Holinka, O. Hayden, S. F. Tedde, H. P. Kiener and P. Ertl, Lab Chip, 2020, 20, 1461–1471 RSC
.
- H. P. Ma, X. Deng, D. Y. Chen, D. Zhu, J. L. Tong, T. Zhao, J. H. Ma and Y. Q. Liu, R. Soc. Open Sci., 2018, 5(9), 180528 CrossRef PubMed
.
- D. Aletaha, T. Neogi, A. J. Silman, J. Funovits, D. T. Felson, C. O. Bingham, N. S. Birnbaum, G. R. Burmester, V. P. Bykerk, M. D. Cohen, B. Combe, K. H. Costenbader, M. Dougados, P. Emery, G. Ferraccioli, J. M. W. Hazes, K. Hobbs, T. W. J. Huizinga, A. Kavanaugh, J. Kay, T. K. Kvien, T. Laing, P. Mease, H. A. Ménard, L. W. Moreland, R. L. Naden, T. Pincus, J. S. Smolen, E. Stanislawska-Biernat, D. Symmons, P. P. Tak, K. S. Upchurch, J. Vencovský, F. Wolfe and G. Hawker, Arthritis Rheum., 2010, 62, 2569–2581 CrossRef PubMed
.
- H. P. Kiener, G. F. M. Watts, Y. Cui, J. Wright, T. S. Thornhill, M. Sköld, S. M. Behar, B. Niederreiter, J. Lu, M. Cernadas, A. J. Coyle, G. P. Sims, J. Smolen, M. L. Warman, M. B. Brenner and D. M. Lee, Arthritis Rheum., 2010, 62, 742–752 CrossRef CAS PubMed
.
- S. R. A. Kratz, C. Eilenberger, P. Schuller, B. Bachmann, S. Spitz, P. Ertl and M. Rothbauer, Sci. Rep., 2019, 9, 9287 CrossRef CAS PubMed
.
- G. Shabestani Monfared, P. Ertl and M. Rothbauer, Sci. Rep., 2020, 10, 16192 CrossRef CAS PubMed
.
- M. Purtscher, M. Rothbauer, W. Holnthoner, H. Redl and P. Ertl, in IFMBE Proceedings, 2015, vol. 45, pp. 313–317 Search PubMed.
- J. Rosser, B. Bachmann, C. Jordan, I. Ribitsch, E. Haltmayer, S. Gueltekin, S. Junttila, B. Galik, A. Gyenesei, B. Haddadi, M. Harasek, M. Egerbacher, P. Ertl and F. Jenner, Mater. Today Bio, 2019, 4, 100023 CrossRef CAS PubMed
.
- B. Bachmann, S. Spitz, B. Schädl, A. H. Teuschl, H. Redl, S. Nürnberger and P. Ertl, Front. Bioeng. Biotechnol., 2020, 8, 373 CrossRef PubMed
.
- F. Zhang, K. Wei, K. Slowikowski, C. Y. Fonseka, D. A. Rao, S. Kelly, S. M. Goodman, D. Tabechian, L. B. Hughes, K. Salomon-Escoto, G. F. M. Watts, A. H. Jonsson, J. Rangel-Moreno, N. Meednu, C. Rozo, W. Apruzzese, T. M. Eisenhaure, D. J. Lieb, D. L. Boyle, A. M. Mandelin, J. Albrecht, S. L. Bridges, C. D. Buckley, J. H. Buckner, J. Dolan, J. M. Guthridge, M. Gutierrez-Arcelus, L. B. Ivashkiv, E. A. James, J. A. James, J. Keegan, Y. C. Lee, M. J. McGeachy, M. A. McNamara, J. R. Mears, F. Mizoguchi, J. P. Nguyen, A. Noma, D. E. Orange, M. Rohani-Pichavant, C. Ritchlin, W. H. Robinson, A. Seshadri, D. Sutherby, J. Seifert, J. D. Turner, P. J. Utz, B. F. Boyce, E. DiCarlo, E. M. Gravallese, P. K. Gregersen, L. Moreland, G. S. Firestein, N. Hacohen, C. Nusbaum, J. A. Lederer, H. Perlman, C. Pitzalis, A. Filer, V. M. Holers, V. P. Bykerk, L. T. Donlin, J. H. Anolik, M. B. Brenner and S. Raychaudhuri, Nat. Immunol., 2019, 20, 928–942 CrossRef CAS PubMed
.
- L. T. Donlin, D. A. Rao, K. Wei, K. Slowikowski, M. J. McGeachy, J. D. Turner, N. Meednu, F. Mizoguchi, M. Gutierrez-Arcelus, D. J. Lieb, J. Keegan, K. Muskat, J. Hillman, C. Rozo, E. Ricker, T. M. Eisenhaure, S. Li, E. P. Browne, A. Chicoine, D. Sutherby, A. Noma, C. Nusbaum, S. Kelly, A. B. Pernis, L. B. Ivashkiv, S. M. Goodman, W. H. Robinson, P. J. Utz, J. A. Lederer, E. M. Gravallese, B. F. Boyce, N. Hacohen, C. Pitzalis, P. K. Gregersen, G. S. Firestein, S. Raychaudhuri, L. W. Moreland, V. M. Holers, V. P. Bykerk, A. Filer, D. L. Boyle, M. B. Brenner, J. H. Anolik, J. Buckner, D. Todd, M. Weisman, A. Ben Artzi, L. Forbess, J. Bathon, J. Carrino, O. Nwawka, E. Matteson, R. Darnell, D. Orange, R. Satija, D. Horowitz, H. Perlman, A. Mandelin, L. Bridges, L. B. Hughes, A. Ceponis, P. Lowry, P. Emery, A. Zayat, A. Aslaam, K. Salomon-Escoto, D. Fox, R. Ike, A. Cordle, A. Wise, J. Ashton, J. Rangel-Moreno, C. Ritchlin, D. Tabechian, R. Thiele, D. Parks, J. Akinson, C. Putterman, E. Der, E. Massarotti, M. Weisman, D. Hildeman, R. Furie, B. Diamond, M. Petri, D. Kamen, M. Cunningham, J. Buyon, I. Lee, H. Salameh, M. McMahon, K. Kalunian, M. Dall'Era, D. Wofsy, M. Kretzler, C. Berthier, W. McCune, R. Kado, W. Pendergraft, D. Waguespack, Y. Liu, G. Watts, A. Arazi, R. Gupta, H. Maecker, P. Dunn, R. Mao, M. Pichavant, Q. Chen, J. Peyman, E. Goldmuntz, J. Buschman, J. Chi, S. Y. Mao, S. Serrate-Sztein, Y. Wang, T. Tuschl, Y. Lee, C. Fonseka, F. Zhang, I. Korsunskiy, J. A. James and J. Guthridge, Nat. Immunol., 2019, 20(7), 928–942 CrossRef PubMed
.
- M. Elshamly, K. Kinslechner, J. G. Grohs, D. Weinmann, S. M. Walzer, R. Windhager, H. J. Gabius and S. Toegel, J. Orthop. Res., 2019, 37, 2204–2216 CrossRef CAS PubMed
.
- C. Eilenberger, M. Rothbauer and E. K. Ehmoser,
et al.
, Sci. Rep., 2019, 9, 4863 CrossRef PubMed
.
- B. Bachmann, S. Spitz, M. Rothbauer, C. Jordan, M. Purtscher, H. Zirath, P. Schuller, C. Eilenberger, S. F. Ali, S. Mühleder, E. Priglinger, M. Harasek, H. Redl, W. Holnthoner and P. Ertl, Biomicrofluidics, 2018, 12, 042216 CrossRef PubMed
.
- M. Rothbauer, G. Hoell, C. Christoph, S. A. R. Kratz, B. Farooq, P. Schuller, I. Olmos Calvo, R. A. Byrne, B. Meyer, B. Niederreiter, S. Kuepcue, F. Sevelda, J. Holinka, O. Hayden, S. F. Tedde, H. P. Kiener and P. Ertl, Lab Chip, 2020, 20, 1461–1471 RSC
.
- H. Lee, L. Gu, D. J. Mooney, M. E. Levenston and O. Chaudhuri, Nat. Mater., 2017, 16, 1243–1251 CrossRef CAS PubMed
.
- M. Kondo, K. Yamaoka, K. Sakata, K. Sonomoto, L. Lin, K. Nakano and Y. Tanaka, Arthritis Rheumatol., 2015, 67, 1250–1260 CrossRef CAS PubMed
.
- D. Vivien, P. Galéra, E. Lebrun, G. Loyau and J.-P. Pujol, J. Cell. Physiol., 1990, 143, 534–545 CrossRef CAS PubMed
.
- M. C. Honorati, L. Cattini and A. Facchini, Osteoarthr. Cartil., 2004, 12, 683–691 CrossRef PubMed
.
- M. C. Honorati, L. Cattini and A. Facchini, Connect. Tissue Res., 2007, 48, 239–245 CrossRef CAS PubMed
.
- A. P. Croft, J. Campos, K. Jansen, J. D. Turner, J. Marshall, M. Attar, L. Savary, C. Wehmeyer, A. J. Naylor, S. Kemble, J. Begum, K. Dürholz, H. Perlman, F. Barone, H. M. McGettrick, D. T. Fearon, K. Wei, S. Raychaudhuri, I. Korsunsky, M. B. Brenner, M. Coles, S. N. Sansom, A. Filer and C. D. Buckley, Nature, 2019, 570, 246–251 CrossRef CAS PubMed
.
- K. Wei, I. Korsunsky, J. L. Marshall, A. Gao, G. F. M. Watts, T. Major, A. P. Croft, J. Watts, P. E. Blazar, J. K. Lange, T. S. Thornhill, A. Filer, K. Raza, L. T. Donlin, Accelerating Medicines Partnership Rheumatoid Arthritis & Systemic Lupus Erythematosus (AMP RA/SLE) Consortium, C. W. Siebel, C. D. Buckley, S. Raychaudhuri and M. B. Brenner, Nature, 2020, 582, 259–264 CrossRef CAS PubMed
.
- F. L. J. Cals, C. A. Hellingman, W. Koevoet, R. J. Baatenburg de Jong and G. J. V. M. van Osch, J. Tissue Eng. Regener. Med., 2012, 6, 68–76 CrossRef CAS PubMed
.
- P. M. van der Kraan, P. Buma, T. van Kuppevelt and W. B. van Den Berg, Osteoarthr. Cartil., 2002, 10, 631–637 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00130b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |