An electricity- and instrument-free infectious disease sensor based on a 3D origami paper-based analytical device†
Abstract
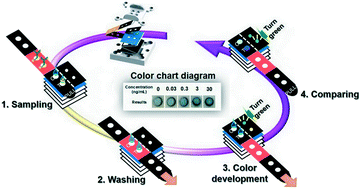
Infectious diseases cause millions of deaths annually in the developing world. Recently, microfluidic paper-based analytical devices (μPADs) have been developed to diagnose such diseases, as these tests are low cost, biocompatible, and simple to fabricate. However, current μPADs are difficult to use in resource-limited areas due to their reliance on external instrumentation to measure and analyze the test results. In this work, we propose an electricity and external instrumentation-free μPAD sensor based on the colorimetric enzyme-linked immunosorbent assay (ELISA) for the diagnosis of infectious disease (3D-tPADs). Designed based on the principle of origami, the proposed μPAD enables the sequential steps of the colorimetric ELISA test to be completed in just ∼10 min. In addition, in order to obtain an accurate ELISA result without using any instrument, we have integrated an electricity-free “timer” within the μPAD that can be controlled by the buffer viscosity and fluid path volume to indicate the appropriate times for washing and color development steps, which can avoid false positive or false negative results caused by an extended or shortened amount of washing and development times. Due to the low background noise and high positive signal intensity of the μPAD, positive and negative detection results can be distinguished by just the naked eye. Furthermore, the ELISA result can be semi-quantified by comparing the results shown on the μPAD with a color chart diagram with a detection limit of HIV type 1(HIV-1) p24 antigen as low as 0.03 ng mL−1. These results demonstrate the proposed sensor can perform infectious disease diagnosis without external instrumentation or electricity, extending the application of the μPAD test for on-site detection and use in resource-limited settings.
- This article is part of the themed collection: Lab on a Chip HOT Articles 2021


 Please wait while we load your content...
Please wait while we load your content...