 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring early time points of vimentin assembly in flow by fluorescence fluctuation spectroscopy†
Eleonora
Perego
a and
Sarah
Köster
 *ab
*ab
aInstitute for X-Ray Physics, University of Göttingen, 37077 Göttingen, Germany. E-mail: sarah.koester@phys.uni-goettingen.de
bCluster of Excellence “Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells” (MBExC), University of Göttingen, Germany
First published on 19th January 2021
Abstract
Despite the importance for cellular processes, the dynamics of molecular assembly, especially on fast time scales, is not yet fully understood. To this end, we present a multi-layer microfluidic device and combine it with fluorescence fluctuation spectroscopy. We apply this innovative combination of methods to investigate the early steps in assembly of vimentin intermediate filaments (IFs). These filaments, together with actin filaments and microtubules, constitute the cytoskeleton of cells of mesenchymal origin and greatly influence their mechanical properties. We are able to directly follow the two-step assembly process of vimentin IFs and quantify the time scale of the first lateral step to tens of ms with a lag time of below 3 ms. Although demonstrated for a specific biomolecular system here, our method may potentially be employed for a wide range of fast molecular reactions in biological or, more generally, soft matter systems, as it allows for a precise quantification of the kinetics underlying the aggregation and assembly.
1 Introduction
Molecular assembly is a key mechanism in biological systems,1 and is involved in many cellular processes such as the reconstitution of cytoskeletal filaments and networks during cell division or hemoglobin assembly in red blood cells.2,3 Disordered protein aggregation, by contrast, is related to severe pathological conditions, such as Alzheimer's disease (amyloid-β) or Parkinson's disease (α-synuclein).4–6 The investigation of these aggregation processes requires a spatial and temporal resolution of nanometers and milliseconds, respectively.Fluorescence fluctuation spectroscopy (FFS) is a family of methods capable of detecting single fluorescently labeled objects based on the analysis of fluorescence fluctuations emitted over time within a confined observation volume.7 Fluorescence correlation spectroscopy (FCS) analyzes the time correlations of these fluctuations and provides information about protein mobility and, via the diffusion coefficient, size.8 Alternatively, the frequencies of the fluctuations can be evaluated to investigate the brightness of the fluorescent species, as in photon counting histogram (PCH) or in fluctuation intensity distribution analysis (FIDA).9,10 FFS is often employed to characterize molecular aggregation and protein–protein interactions by brightness analysis,9–13 which allows for distinguishing the signal of a single protein from the signal of proteins complexes.
However, all these techniques lack temporal resolution since long acquisition times are needed for a good signal-to-noise ratio.11,14–16 To overcome this challenge and access the fast dynamics of molecular assembly, laminar flow microfluidics may be employed. The principle has been demonstrated for, e.g., small angle X-ray scattering17–20 and fluorescence microscopy.21–23 The laminar flow ensures that for each position within the flow channel, the reaction state of the aggregating protein is well-defined and does not change in time, thus long exposures lead to ensemble averaging and improve the statistics of the measurement. The time resolution is given merely by the spatial distance of the measuring positions along the channels and unaffected by the exposure time. A variant of the hydrodynamic flow mixer was established by including a “step” in the central inflow channel for combination with X-ray scattering experiments,19,20,24,25 thereby engulfing the protein jet by a buffer layer and preventing clogging of the channels.
Here, we combine FFS with microfluidics to investigate the assembly of vimentin intermediate filament (IF) protein in a time-resolved manner. IFs, together with microtubules and actin filaments constitute the cytoskeleton of eukaryotes.2 They are expressed in a cell-type specific manner, but all share their secondary structure and hierarchical assembly path.26–29 Vimentin is part of this large family of proteins and expressed in cells of mesenchymal origin.30,31 Assembly occurs in at least two distinct steps: lateral assembly of tetramers – the smallest stable subunit in low-salt buffer, such as 2 mM phosphate buffer (PB) – via octamers and 16-mers to so-called unit length filaments (ULFs) of typically 8 tetramers, followed by longitudinal annealing of these ULFs to extended filaments.26,32–34 The process is sketched in Fig. 1.
 | ||
| Fig. 1 Schematic representation of vimentin assembly. Vimentin monomers are labeled with Atto-532 via maleimide chemistry and mixed with unlabeled monomers at labeling ratios between 11% and 41%. Upon dialysis against 2 mM of PB, vimentin monomers assemble laterally in a parallel fashion to form dimers and further laterally in half-staggered configuration to tetramers, which are the starting point for our measurements. Upon the addition of KCl, vimentin tetramers continue to assemble laterally resulting in the formation of ULFs, typically composed of 8 tetramers. Subsequently, ULFs anneal end-to-end and form extended filaments. Micrographs of fluorescent vimentin filaments are shown in Fig. S1a and b.† The assembly steps considered in this work are framed by the red box. | ||
In vitro, assembly is initiated by the addition of salt up to physiological concentrations, such as 100 mM KCl. The time scales of the two assembly steps are distinct: whereas vimentin lateral assembly occurs within less than 1 s,25,35,36 and, as recently reported, on the time scale of 100 ms,37 elongation takes minutes to hours.34,38,39
Our method allows us to determine the increase in label copy number over time, i.e. at different positions of the microfluidic device, and thus follow vimentin assembly, including the two-step process, and precisely determine the time scale of lateral assembly to, on average, 65 ms with a lower bound of 26 ms. More generally, our method allows for studying the influence of reagents, such as multivalent ions or detergents, or of different amounts and types of fluorescent labels on assembly processes.
2 Materials and methods
2.1 Microfluidic device fabrication
We use two different microfluidic devices, which are both fabricated using standard soft lithography methods.40,41 The device for establishing the PCH method at different flow velocities (device A) is a simple straight channel (width 250 μm, height 25 μm, length 2.5 cm). Briefly, SU8-3025 (Kayaku Advanced Materials, Westborough, MA, USA) is spin coated onto a silicon wafer (MicroChemicals GmbH, Ulm, Germany) to a final height of 25 μm, exposed through a photo mask (Selba S. A., Versoix, Switzerland), and developed (master 1).The device for studying protein assembly combines five inlets and one outlet (device B) with a height of 100 μm and a width of 200 μm for the central inlet channel and a height of 200 μm for the remaining channels. The inlet with reduced height is centered with respect to the full channel height by creating steps (height 50 μm) on the coverslip at the bottom as well as in the polydimethylsiloxane (PDMS) replica forming the channels. To achieve this geometry, two different wafers are produced by photolithography, (i) a five-inlet geometry, master 2, and (ii) the step for the protein inlet (200 μm width and 50 μm thickness), master 3. A schematic of the photolithography steps is shown in Fig. S2.† To create the top step on master 2, a 3D structure is created with two layers of SU8-3050 photo resist (150 μm and 50 μm thick). To improve the uniformity of the first layer, the resist is spin coated twice with a height of 75 μm each. At this point, the resist is soft baked for about 2 hours at 95 °C and exposed to UV-light (MJB4 Mask-Aligner, Süss MicroTec SE, Garching, Germany) through a photomask. After a post exposure bake for 20 minutes at 95 °C, the wafer is spin coated with the second layer of SU8-3050 photo resist (thickness 50 μm). The wafer is soft baked again at 95 °C for 30 minutes and exposed to UV light once more, using a second mask, aligned with the first structure. The wafer is baked for 15 minutes at 95 °C and the photo resist is developed. Master 3 is fabricated by spin coating SU8-3050 to a height of 50 μm and exposed to obtain the 200 μm × 50 μm channel. All masters are coated with (heptafluoropropyl)-trimethylsilane (Sigma-Aldrich, Steinheim, Germany) overnight to allow for easier detachment of the cured PDMS.
PDMS (Sylgard 184, Dow Corning, Midland, MI, USA) replicas are fabricated from the master structures (ratio base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cross-linker = 10
cross-linker = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 hour baking time at 65 °C) and holes for connecting the tubing are punched at the inlet positions (biopsy puncher, 0.75 mm diameter, World Precision Instruments, Sarasota, FL, USA). The top part of the step device is integrated in the PDMS replica stemming from master 2, as shown in Fig. S3c.† For the bottom part, the step is created directly on the glass coverslips (number 1, Thermo Fisher Scientific, Wilmington, DE, USA) using master 3. A PDMS replica of master 3 is placed on top of a clean glass coverslip (Fig. 2a) and a drop of liquid adhesive (Norland Optical Adhesive H83, Norland Products Inc., Cranbury, NJ, USA) is used to create a step by micromolding in capillaries (MIMIC).42 The liquid adhesive fills the channel by capillary forces (Fig. 2b). To speed up the process, the coverslip with the liquid adhesive is placed in a desiccator. The liquid adhesive is then cured by UV light (Fig. 2c) at 365 nm for 3 hours. Afterwards, the PDMS channel is removed (Fig. 2d) and the glue is further cured under UV light for 30 minutes. Both the PDMS channel replica and the glass coverslip with the glue step are cleaned with isopropanol, dried with nitrogen and exposed to air plasma (ZEPTO plasma cleaner, Diener electronic GmbH + Co. KG, Ebhausen, Germany) for 12 seconds at setting 40 W. They are aligned under a stereo microscope (Olympus SZ61, Olympus Europa SE & Co. KG, Hamburg, Germany) and pressed together to form a covalent bond (Fig. 2e). The device is left at 95 °C for about 2 hours to improve the strength of the bond. A photograph of the fully assembled device is shown in Fig. S3a.†
1, 1 hour baking time at 65 °C) and holes for connecting the tubing are punched at the inlet positions (biopsy puncher, 0.75 mm diameter, World Precision Instruments, Sarasota, FL, USA). The top part of the step device is integrated in the PDMS replica stemming from master 2, as shown in Fig. S3c.† For the bottom part, the step is created directly on the glass coverslips (number 1, Thermo Fisher Scientific, Wilmington, DE, USA) using master 3. A PDMS replica of master 3 is placed on top of a clean glass coverslip (Fig. 2a) and a drop of liquid adhesive (Norland Optical Adhesive H83, Norland Products Inc., Cranbury, NJ, USA) is used to create a step by micromolding in capillaries (MIMIC).42 The liquid adhesive fills the channel by capillary forces (Fig. 2b). To speed up the process, the coverslip with the liquid adhesive is placed in a desiccator. The liquid adhesive is then cured by UV light (Fig. 2c) at 365 nm for 3 hours. Afterwards, the PDMS channel is removed (Fig. 2d) and the glue is further cured under UV light for 30 minutes. Both the PDMS channel replica and the glass coverslip with the glue step are cleaned with isopropanol, dried with nitrogen and exposed to air plasma (ZEPTO plasma cleaner, Diener electronic GmbH + Co. KG, Ebhausen, Germany) for 12 seconds at setting 40 W. They are aligned under a stereo microscope (Olympus SZ61, Olympus Europa SE & Co. KG, Hamburg, Germany) and pressed together to form a covalent bond (Fig. 2e). The device is left at 95 °C for about 2 hours to improve the strength of the bond. A photograph of the fully assembled device is shown in Fig. S3a.†
2.2 Protein preparation and purification
Human vimentin C328A, carrying three additional amino acids GGC (glycine–glycine–cystein) at the C-terminus (plasmid DNA from Harald Herrmann, Universitätsklinikum Erlangen, Germany), is recombinantly expressed with a protocol adapted from ref. 32, 33 and 43. Vimentin monomers are labeled with Atto532-maleimide (AttoTech GmbH, Siegen, Germany) as described in ref. 44 and 45. Briefly, vimentin is dialyzed against labeling buffer (5 M urea, 50 mM PB, pH 7.5). The dye is dissolved in water-free DMSO to a concentration of 10 mM. It is added in steps of 5 μL to the vimentin solution (concentration 1 g L−1). After two hours of incubation, the free dye is captured by the addition of 1 M cystein and separated from the labeled protein via size exclusion chromatography. Peak fractions are pooled. The labeled protein is dialyzed against the storage buffer (8 M urea in 2 mM PB, pH 7.5) and the protein is stored at −80 °C. Before the experiment, a mixture of labeled and unlabeled vimentin (between 4% to 41% labeled protein) is dialyzed in a step-wise manner (6 M, 4 M, 2 M, 1 M, 0 M urea), against 2 mM PB, pH 7.5 at room temperature, using a 50 kDa cut-off membrane (SpectraPor, Carl Roth GmbH + Co. KG, Karlsruhe, Germany). Afterwards an additional dialysis step against 2 mM PB, pH 7.5 is performed in a coldroom (8 °C) overnight. Finally, the protein is further dialyzed against fresh 2 mM PB, pH 7.5 for 1 hour at room temperature. At the end of the dialysis, vimentin molecules are already assembled into tetramers. The protein concentration and labeling ratio are determined from absorption data at 280 nm (Nanodrop ND-1000, Thermo Fisher Scientific). Bulk assembly is started by mixing the protein solution at a concentration between 0.2 and 0.7 g L−1 at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a salt buffer (2 mM PB with 200 mM KCl, pH 7.5), providing a final salt concentration of 100 mM. In the microfluidic experiments, the starting concentration of vimentin is about 0.003 g L−1 and the assembly is initiated by injecting buffer containing 100 mM KCl from the side channels leading to diffusive mixing. The protein and ion concentrations are chosen to ensure ideal conditions for FFS measurements in flow.7 The flow rates are 12 μL h−1 for the protein solution (central inlet), 10 μL h−1 for the sheath inlets and 190 μL h−1 for the assembly buffer (side inlets). All buffers are degassed prior to the experiments. All measurements are performed at room temperature.
1 with a salt buffer (2 mM PB with 200 mM KCl, pH 7.5), providing a final salt concentration of 100 mM. In the microfluidic experiments, the starting concentration of vimentin is about 0.003 g L−1 and the assembly is initiated by injecting buffer containing 100 mM KCl from the side channels leading to diffusive mixing. The protein and ion concentrations are chosen to ensure ideal conditions for FFS measurements in flow.7 The flow rates are 12 μL h−1 for the protein solution (central inlet), 10 μL h−1 for the sheath inlets and 190 μL h−1 for the assembly buffer (side inlets). All buffers are degassed prior to the experiments. All measurements are performed at room temperature.
2.3 Experimental setup
The setup used is based on an inverted microscope (Olympus IX73, Olympus), which combines confocal and epi-fluorescence modalites. The FFS experiments are performed with the confocal modality using a diode pumped laser with a wavelength 532 nm (Cobolt Samba Cobolt AB, Solna, Sweden) at a power on the sample of 20 μW. The laser is reflected by a dichroic mirror (DualLine zt488/532rpc, AHF Analysentechnik AG, Tübingen, Germany) and focused using a 60× water immersion objective (UPlanApo, NA = 1.2, Olympus). After the emission filter (RazorEdge LP532RU, AHF Analysentechnik AG) and the pinhole (diameter 50 μm, Qioptiq Photonics GmbH & Co. KG, Göttingen, Germany) the fluorescence light is focused on an avalanche photo diode (τ-SPAD, Picoquant GmbH, Berlin, Germany). The τ-SPAD is connected either to a digital correlator card (ALV-7004 USB, ALV-Laser Vertriebsgesellschaft mbH, Langen, Germany) used for autocorrelation measurements, or to an acquisition card (NI-6602, National Instruments, Austin, TX, USA) to access the raw photon arrival times. To access the data from the acquisition card, we use a custom-written program that is based on a python interface.46 The photon arrival times are measured relative to an arbitrary start point with an internal clock of 10 MHz. All the experimental data are then analyzed using self-written Python code (Python Software Foundation, Python Language Reference, version 3.7, available at http://www.python.org). The epi-fluorescence images are acquired with the same setup using a mercury arc lamp (X-Cite 120 PC Q, Excelitas Technologies, Uckfield, United Kingdom) for excitation. Images are acquired using a CCD-camera (Hamamatsu Orca R-2, Hamamatsu Photonics Deutschland GmbH, Herrsching am Ammersee, Germany) controlled by Micro-Manager.47Stationary measurements are performed using 300 μL sample solution placed in an eight-well glass slide (Nunc Lab-Tek Chamber slides, Thermo Fisher Scientific). Flow measurements are performed using the microfluidic device presented in the previous section. Polyethylene tubing (inner diameter 0.38 mm, outer diameter 1.09 mm, Intramedic Clay Adams Brand, Becton Dickinson and Company, Sparks, MD, USA) is connected to 1 mL (for vimentin and PB) or 2.5 mL (for the KCl buffer) glass syringes (Hamilton Gastight, Hamilton Bonaduz AG, Bonaduz, Switzerland) using disposable needles (diameter 0.40 mm, length 20 mm, Sterican, B. Braun AG, Melsungen, Germany). For precise control of the flow, syringe pumps (neMESYS, Cetoni GmbH, Korbußen, Germany) are used to regulate the flow rates of the syringes connected to the inlets. These pumps are particularly suitable for low pressure microfluidic flow. To ensure that our measurements take place exactly at the position of the hydrodynamically focused protein stream, we additionally measure the fluorescence intensity across the channel, perpendicular to the flow direction, and set up our measuring scheme accordingly. To access the different positions in the channel, an automated sample stage (Prior Scientific, Inc., Rockland, MA, USA) is used.
2.4 Data acquisition and analysis
The data for each PCH curve are collected for a total time of 300 s to 800 s, as consecutive 10 s portions, whereas for the FCS curves, we collect data for 30 s. The acquired photon arrival times are binned with a binning time between 2 and 20 μs, stored and used to build the experimental PCH curves. A histogram of the number of counts per bin, k, is created and normalized to a total area of 1. The experimental PCH, p(k) is fitted with the theoretical model described in ref. 9 for n species, with dead-time correction48 and one-photon correction.49 The one-photon-excitation (OPE) correction parameter F for our experiment is fixed to 0.8 and the dead-time is measured to be 86 ns. The autocorrelation functions are fitted using either a single (eqn (S1)†) or a dual component model (eqn (S2)†), depending on the sample measured.7 In the presence of laminar flow the autocorrelation function comprises an additional term that depends on the flow speed50 (eqn (S3)†). A more detailed description of FCS and PCH can be found in the ESI.† Before every experiment, the setup is calibrated measuring rhodamine 6G (Merck KGaA, Darmstadt, Germany) dissolved in water to calculate the beam waist (w0) and the excitation volume. Rarely, high fluorescence intensity peaks, with an amplitude 5 to 10 times higher than the average value, are observed. These spikes are most probably caused by protein aggregates passing the observation volume and are thus removed from the raw data before the analysis. The data are analyzed using a self-written fitting routine employing Python. All correlation curves are fitted with a Levenberg–Marquardt nonlinear least-square procedure. For all flow FFS measurements, the laser is focused in the central z- and y-planes of the microfluidic device to ensure that light is collected from the confocal observation volume in the center of the protein stream.2.5 FEM simulations
To comprehend the flow profile in the microfluidic device, finite element method (FEM) simulations are performed using Comsol Multiphysics 5.5 (COMSOL GmbH, Göttingen, Germany) based on the time-independent Navier–Stokes equation with no-slip boundary conditions. To decrease the computational effort, the simulated channels are shortened and we simulate only a quarter of the device, according to the symmetry conditions. The simulations are performed using an upper limit for the diffusion coefficient for vimentin tetramers51 (Dvim = 24 μm2 s−1) as it decreases along the outlet due to assembly. For the salt, DK+ = 1850 μm2 s−1 is used. The concentrations employed in the simulations reflect the actual concentrations of the experiment. The change in viscosity along the outlet is not included in the simulations, since viscometry measurements show that the viscosity does not significantly increase during the first assembly steps, which we consider here.26,523 Results and discussion
3.1 Device geometry and characterization
In order to access the early time points of protein assembly into supramolecular structures, we employ microfluidics in combination with FFS. Here, we specifically study the first steps of the assembly of vimentin into filaments. Whereas FFS provides a means to measure molecular reactions, microfluidics allows us to initiate the assembly process by diffusive mixing and access different reaction time points.17,18,53 To achieve a controlled starting point of the reaction, a five-inlet geometry, as schematically shown in Fig. 2f, is chosen,54 where the mixing of the two solutions is performed after focusing of the central stream by the side inlets. Thus, hydrodynamic focusing and diffusive mixing are decoupled, ensuring mixing of the ions into the central stream only after the complete focusing of the latter,54 thereby effectively decreasing the mixing time ambiguity. Ions diffuse into the slowly diffusive protein stream in the center of the channel. As the diffusion time scales as t ∝ x2, focusing the vimentin stream from 200 μm (width of the protein inlet) to 6.3 μm in the mixing region, as derived from the FEM simulation, allows for a fast mixing of the solutions in the narrow protein stream shown in Fig. S3b.† The small dimensions of the device and low flow rates employed ensure laminar flow with a Reynolds number on the order of 1 and we can thus investigate reaction kinetics even with inherently “slow” techniques requiring long exposure times, such as FFS. Moreover, since there is no turbulence, the spatial coordinates of each inspected position can be directly converted into time coordinates of the studied reaction once the velocity profile in the channel is known. As consequence, the temporal resolution is not limited by the acquisition time, but only by the beam size and the spacing of the measuring positions in the device. The investigated positions are not equidistantly placed in the device. Close to the cross-section, where we expect rapid changes in the signal, they are spaced by 5 to 50 μm, depending on the device. Further downstream the measuring points are positioned further apart, i.e. 100 to 700 μm, as the brightness is not expected to change much anymore.To characterize the microfluidic device and the flow parameters, FEM simulations are performed using Comsol Multiphysics 5.5 with the same flow rates as used for the experiments, i.e., 190 μL h−1 for the side inlets (PB with KCl), 10 μL h−1 for the sheath inlets (PB) and 12 μL h−1 for the central inlet (vimentin tetramer solution), which corresponds to velocities of 1.5 mm s−1, 0.1 mm s−1 and 0.3 mm s−1, respectively. Vimentin tetramers are hydrodynamically focused by the side inlets as shown in the top part of Fig. 3a, where the concentration of vimentin is displayed in the central x–y plane of the device. The assembly of vimentin is initiated, when KCl, which diffuses into the protein stream from the side inlets as shown in the bottom part of Fig. 3a, reaches a concentration of 10 mM.35,38,39 At our chosen flow rates, the reaction threshold of 10 mM KCl (ref. 35) is reached after 110 μm from the end of the central inlet (yellow–black cross in Fig. 3a), thus this channel position is defined as time point t = 0 and position x = 0, as indicated by the vertical dashed line in Fig. 3d. The salt concentration in the center of the outlet channel increases, as shown in Fig. 3d (blue curve). As a direct consequence of the five inlets merging into one outlet, the velocity is not constant along the entire device, but it increases for the first 250 μm after the channel intersection, as shown in Fig. 3d, red curve. Afterwards, a constant velocity of 5.7 mm s−1 is reached. Therefore, the calculated reaction time, shown in the inset of Fig. 3d, initially is not spaced uniformly with the channel positions.
To prevent clogging of the channels by aggregates adsorbed to the channel walls, a reduction in height of the protein inlet leading to a step is employed.19,24,25 The ability of the step in the central inlet to keep vimentin from touching the channel walls is demonstrated in the x–z central plane of the cross section, as shown in Fig. 3b where the vimentin and KCl concentrations are shown in the top and the bottom part, respectively. The red arrow in the top part of Fig. 3b marks a vimentin concentration of zero at the upper channel wall. Furthermore, all flow lines for vimentin tetramers expand in z-direction (Fig. 3c) after entering the cross section, i.e. behind the step, without reaching the channel wall. We confirm by epi-fluorescence microscopy that no protein aggregates at the channel walls. The flow velocities chosen here ensure that the assembling protein stays distant from the channel walls. As a consequence of the comparatively high flow rates, diffusion and convective flow are overlaid and FCS cannot be employed to measure diffusion. We introduce the flow time  , defined as the ratio between the beam waist w0 and the flow velocity v, and the diffusion time
, defined as the ratio between the beam waist w0 and the flow velocity v, and the diffusion time 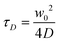 with the diffusion coefficient D to evaluate the two types of molecular motion.7 As a first approximation, τF has to be at least equal to τD to allow for measuring diffusion using FCS. If vimentin tetramers are considered, τD = 937.5 μs, with w0 = 300 nm for our setup and D = 24 μm2 s−1. As a consequence, the maximum possible total flow velocity in the center of the device is ≈ 0.3 mm s−1, which is too slow for the step geometry to come into effect, see Fig. S4.† It was further demonstrated that already at
with the diffusion coefficient D to evaluate the two types of molecular motion.7 As a first approximation, τF has to be at least equal to τD to allow for measuring diffusion using FCS. If vimentin tetramers are considered, τD = 937.5 μs, with w0 = 300 nm for our setup and D = 24 μm2 s−1. As a consequence, the maximum possible total flow velocity in the center of the device is ≈ 0.3 mm s−1, which is too slow for the step geometry to come into effect, see Fig. S4.† It was further demonstrated that already at  , flow dominates over diffusion55 and consequently FCS cannot be employed to measure the dynamic of vimentin assembly. We thus refrain from using FCS to study vimentin assembly in flow.
, flow dominates over diffusion55 and consequently FCS cannot be employed to measure the dynamic of vimentin assembly. We thus refrain from using FCS to study vimentin assembly in flow.
3.2 Fluorescence fluctuation spectroscopy of vimentin tetramers
It is known that in contrast to FCS, PCH is not dependent on the convective flow velocity, as long as undersampling is avoided.55 At our experimental flow rates, τF is approximately 55 μs. To avoid undersampling, a binning time ten times smaller, i.e. 5 μs (ref. 9 and 55) is chosen when measuring vimentin assembly in the microfluidic device. PCH has been employed to measure aggregation14 or assembly11,16 of proteins in vitro but also to measure protein oligomerization in living cells.9–13 Whereas FCS distinguishes different species based on their diffusion properties,7 PCH distinguishes them based on their brightness.9,56 The PCH for a solution of identical molecules is described by two parameters: the average number of fluorescently labeled objects in the observation volume N, and the molecular brightness B, which is the average number of detected photons emitted by a fluorescently labeled object in the center of the observation volume, measured in counts per second (cps).56 To quantify the temporal evolution of vimentin lateral assembly, the label copy number of each vimentin subunit, i.e. the average number of Atto-532 molecules attached to the vimentin assemblies, is followed over time by analyzing the PCHs acquired at different positions along the central channel of the microfluidic device, as sketched in Fig. 2f. As label, we employ Atto-532-maleimide and based on the known brightness of a single dye molecule, the label copy number is calculated by dividing the brightness of the assembling vimentin obtained from the PCHs in the device by the one of a dye molecule.To establish PCH as a valid method for our purpose, we first perform measurements on vimentin tetramers with labeling ratios of 22% and 36% at different protein concentrations without convective flow. The results are shown in Fig. 4a, and the fitting parameters are summarized in Table 1.
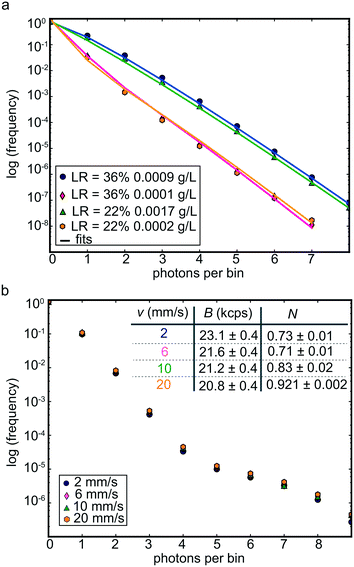 | ||
| Fig. 4 PCHs of vimentin tetramers with and without flow. a) PCHs of vimentin tetramers at different protein concentrations and different labeling ratios measured in bulk. The symbols represent the experimental data and the solid lines the fits to a one-component model. For each PCH curve, the brightness B and the average number N of fluorescent objects in the observation volume is retrieved. The measured brightness values are comparable at the different concentrations, whereas the average numbers of molecules scale roughly as the concentrations, as summarized in Table 1. b) PCHs of vimentin tetramers in microfluidic device A fabricated from master 1 (a single straight channel) at different velocities. The PCHs coincide for all velocities confirming the independence of PCH from the flow velocity. The parameters B and N obtained from the fitting are in agreement (see table, inset top right). Photon counts are acquired for 300 s for each PCH data set. | ||
| LR (%) | c vim (g L−1) | B (kcps) | N PCH |
|---|---|---|---|
| 22 | 0.0017 | 96 ± 3 | 1.36 ± 0.08 |
| 22 | 0.0002 | 100 ± 1 | 0.14 ± 0.02 |
| 36 | 0.0009 | 104 ± 2 | 1.01 ± 0.04 |
| 36 | 0.0001 | 100 ± 2 | 0.14 ± 0.04 |
As expected, the brightness is independent of the protein concentration, confirming the reliability of PCH to quantify the brightness of vimentin tetramers. Between the two labeling ratios, the brightness is similar since the average number of fluorophores per vimentin tetramer is almost identical for both labeling ratios, i.e. 1.4 for 22% and 1.6 for 36%. Note that, intrinsic to our study design, each monomer is either labeled or unlabeled and a tetramer, which represents the smallest stable supermolecular assembly, may contain either 0, 1, 2, 3 or 4 fluorophores. When directly comparing the PCH results to FCS data of the same sample (see Fig. S5 and Table S1†) we obtain agreeing values for the average number of fluorescently labeled objects in the observation volume (NFCS = NPCH).
The independence of PCH from the convective flow55 is confirmed by measuring vimentin tetramers at different flow velocities inside microfluidic device A, which consist of a simple straight channel. At the aspect ratio of device A (height![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) width = 1
width = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), the velocity shows a plug flow profile. Therefore, the velocity is constant along the width of the channel, assuring that all vimentin tetramers in the same z plane move through the observation volume with the same flow velocity. Using device A, no changes in number concentration, e.g. by dilution or assembly, or in brightness are expected, thus the PCHs at different velocities should all be the same, as long as undersampling is avoided. Vimentin tetramers with a concentration of 0.005 g L−1 and a labeling ratio of 3.8%, thus corresponding to a molar dye concentration of 3.5 nM in the solution, are measured at four different velocities (2 mm s−1, 6 mm s−1, 10 mm s−1 and 20 mm s−1). As shown in Fig. 4b the four PCHs all fall on top of each other confirming the independence of PCH from the flow velocity.55 Moreover, the parameters obtained from the PCH fits, N and B at the four velocities are in agreement with each other as shown in the inset table in Fig. 4b. The variability of the fitting parameters is typical for this type of analysis in flow.55,57
10), the velocity shows a plug flow profile. Therefore, the velocity is constant along the width of the channel, assuring that all vimentin tetramers in the same z plane move through the observation volume with the same flow velocity. Using device A, no changes in number concentration, e.g. by dilution or assembly, or in brightness are expected, thus the PCHs at different velocities should all be the same, as long as undersampling is avoided. Vimentin tetramers with a concentration of 0.005 g L−1 and a labeling ratio of 3.8%, thus corresponding to a molar dye concentration of 3.5 nM in the solution, are measured at four different velocities (2 mm s−1, 6 mm s−1, 10 mm s−1 and 20 mm s−1). As shown in Fig. 4b the four PCHs all fall on top of each other confirming the independence of PCH from the flow velocity.55 Moreover, the parameters obtained from the PCH fits, N and B at the four velocities are in agreement with each other as shown in the inset table in Fig. 4b. The variability of the fitting parameters is typical for this type of analysis in flow.55,57
3.3 Combining PCH and microfluidics to quantify vimentin assembly
We use device B, which contains five inlets and a step in the protein inlet channel, to study the assembly kinetics of vimentin. The fluorescence intensity is acquired at typically 30 different positions along the central outlet, as shown schematically in Fig. 2f, for at least 10 minutes each. Knowing the flow velocity profile in the device, each measured position can be converted into the time coordinate of the assembly, see inset of Fig. 3d. From the fluorescence intensity data, PCHs are built for each time point accessed and thus the brightness B and the average number N of fluorescent vimentin assemblies in the observation volume are obtained.The temporal evolution of the label copy number of vimentin, which is obtained by dividing the measured brightness by the brightness of a single molecule of Atto-532, can be compared to the expected label copy number of differently-sized vimentin assemblies. To calculate the expected average number of labels per vimentin assembly (n-mer) we take advantage of the binomial distribution:
 | (1) |
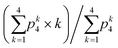 . Note that the average is performed for k ≥ 1 as we cannot detect tetramers without label (k = 0).
. Note that the average is performed for k ≥ 1 as we cannot detect tetramers without label (k = 0).
In a first experiment, to bench mark device B, we do not inject assembly buffer from the side inlets, but just 2 mM PB, pH 7.5, and thus do no initiate assembly, see Fig. 5a. As expected, the brightness as well as the label copy number are constant in time, with an average value of 1.6 ± 0.4 labels per subunit. The brightness of a molecule of Atto-532 is (54 ± 20) kcps, which is measured in a separate experiment performed with the same optical alignment. The average measured label copy number corresponds to the expected value for vimentin tetramers at this particular labeling ratio, 1.9, which is graphically represented by the lowest gray dashed line in Fig. 5a. This confirms that vimentin remains in the tetrameric state when no salt is added and that the in-flow measurements on vimentin tetramers are valid.
Once the assembly buffer is injected from the side inlets, we expect the brightness to increase over time, as during assembly vimentin tetramers associate and form larger subunits. According to the times scales accessible with our microfluidic device, i.e. up to 4.1 s, we focus on the lateral assembly of vimentin tetramers into ULFs. Elongation into extended filaments occurs on longer time scales, i.e. on the order of minutes.37–39,58Fig. 5b shows the brightness and the label copy number plotted against time for a sample of vimentin tetramers with a labeling ratio of 24%. As expected, the brightness increases with time, confirming the assembly of vimentin. At t = 0 s, we observe a label copy number of 2 ± 1, in good agreement with the calculated value for tetramers of 1.5. To quantify the time needed for ULF formation, an exponential growth function with time constant τULF is fitted to the brightness, which is graphically shown as the red dashed line in Fig. 5b. At this labeling ratio, ULFs are formed on a time scale of τULF = (45 ± 10) ms. Note that the error bars of the fitted brightness indicate not only the variability in the assembly dynamics, which might be caused by uncertainties in the flow profile, but mostly the difference in the number of fluorescent labels for each measured vimentin assembly. In principle, a ULF could contain from 1 up to 32 Atto-532 molecules, when each tetramer is labeled with 4 Atto-532 molecules.
When vimentin tetramers with a lower labeling ratio are assembled, the formation of ULFs is faster, as shown in Fig. 5c, where at a labeling ratio of 11%, a time constant of (26 ± 5) ms is obtained by the exponential growth fit. Once the ULFs are formed, the brightness is constant. This is a further demonstration of the two-step process of vimentin assembly: the lateral assembly, which occurs in the first 100 ms is clearly separated from the filament elongation on longer time scales.37,38,44,58 In this example, the brightness at t = 0 s is (41 ± 8) kcps, which is in agreement with the bulk measurement of the same vimentin tetrameric sample (B = (40 ± 5) kcps). The label copy number per vimentin subunit, see cyan diamonds in Fig. 5c, increases with time, starting from a value of 1.2 ± 0.2, which corresponds to the expected value for tetramers (1.2), up to a value of 4.5 ± 0.5, in agreement with the expected value for ULFs (4).
At this comparatively low labeling ratio (11%), the measured label copy number reflects the calculated copy number very accurately. However at higher labeling ratios the measured copy number tends to be lower than the expected values, as shown, for example, in Fig. 5b. In this case, the experimental number of labels per ULF is, on average, 6.3 ± 0.3 while the expected value is 7.5 at a labeling ratio of 24%. The discrepancy at higher labeling ratio might be caused by an effective lower brightness induced by self-quenching of the fluorophores within the ULF. In particular, at a labeling ratio of 24%, on average, 3.5 dye molecules are attached to the tails of vimentin monomers at each side of a single ULF, which has a diameter of 17 nm. Thus, on average, the dye molecules are separated from each other by a distance of 4.8 nm, which is small enough to see quenching effects.59
It has previously been reported that vimentin assembly is influenced by the labeling ratio.44 In fact, as shown in Fig. S1c,† when only labeled vimentin monomers are assembled without the addition of unlabeled protein, filaments are not formed at all. Thus, it is not surprising that vimentin assembles faster at a labeling ratio of 11%, τULF = 26 ms, compared to a labeling ratio of 24%, where the time scale for ULF formation τULF is 45 ms. At higher labeling ratios, the lateral assembly of vimentin is even slower, with τULF = 109 ms measured at a labeling ratio of 40%, as shown in Fig. S6.† In general, a trend of longer lateral assembly time for higher labeling ratios is found within our measured labeling ratios. We assume that at high labeling ratio the lateral assembly is slowed down because the fluorophores interfere with the interaction between two vimentin subunits. On the one hand, this effect might be due to steric hindrance, on the other hand, electrostatic or hydrophobic interactions between the fluorophore and the protein may play a role. Our setup could indeed be used to systematically study the influence of charges and hydrophobic groups carried by varying fluorophores.
If we consider all labeling ratios (varying from 11% to 40%) measured in 9 different experiments, on average we obtain an assembly time scale from tetramers to ULFs of 65 ms, which is in very good agreement with the value of 100 ms measured by static light scattering.37 Mücke et al. measure a peak of the 8mer signal after 7 ms, a peak of the 16mer signal at 66 ms and the signal of ULFs (32 mers) starting from 50 ms. At low labeling ratio, we obtain 8mers after 10 ms, 16 mers after 43 ms and ULFs after 54 ms, thus very much in agreement with the earlier study. The light scattering experiments on unlabeled protein employed a stopped-flow setup to start the assembly. Thus, the mixing strategy differs from our approach. With diffusive mixing, once the assembly is initiated, the positions in the channel can be translated into the time coordinates of the reaction with almost no dead time. However, due to the finite size of the vimentin central stream, uncertainties on the mixing time need to be taken into consideration. Considering the flow rates in our system, the focused vimentin stream has a width of 6 μm, thus the diffusion time of the KCl into the vimentin stream is on the order of 2.5 ms. By contrast, in the stopped-flow measurements, a defined starting point of the assembly process is achieved by turbulent mixing, which, however, introduces a dead time of about 10 ms. Thus, diffusive mixing is comparable to in vitro assembly via dialysis and stopped-flow relates to a “kick start” approach, which might lead to heterogeneities in the assembled vimentin subunits.26,37 In combination with slightly different buffer conditions, small differences in the results can be explained. Overall, both approaches, stopped-flow combined with light scattering and diffusive mixing combined with PCH, are complementary, and the results agree very well.
As the brightness increases, the average number of fluorescent objects in the observation volume, N, decreases, as shown in Fig. 5d. While the increase in brightness, B, can be entirely attributed to the assembly, the decrease of N is caused partly by the assembly and partly by the dilution of the protein stream by the injected buffer from the other inlets. To quantify the temporal evolution of N, we fit the data by an exponential decay. The average number of fluorescent objects in the observation volume decays consistently in different experiments, see orange and green data points in Fig. 5d. The time constants retrieved from the fits are comparable, confirming the reproducibility of our experiments.
All PCHs for vimentin molecules in the step device are analyzed by a two-component fit. The first component corresponds to the data shown in Fig. 5. The second component shows a constant brightness along the channel, as shown in Fig. S7a,† with a value 3 times higher than the expected brightness for ULFs. As is it also present when using the step device on vimentin without assembly buffer injected into the side channels (see, e.g.Fig. 5a), we can exclude that it is due to the assembly reaction. As it is present for flow in channels without the step, we assume it is also not due to the specific channel geometry. Moreover, the second component is not present in PCH measurements in flow of pure dyes, such as rhodamine 6G or Alexa-532 in flow or in vimentin samples measured in bulk at low protein concentrations. We speculate that it could be due to rare large and disordered protein aggregates present even before assembly, as also found by others.37–39 The average number of fluorescent objects in the observation volume for this second component is at least 20 times smaller than the values for the first component, as shown in Fig. S7b.† However, the second component does not compromise our measurements, since the brightness of this second component is very different from the brightness of vimentin assemblies.
4 Conclusion
We present a combination of microfluidics and FFS, adapt it to the investigation of early time points in protein assembly, and demonstrate the utility of our approach on the example of vimentin IF formation. In particular, PCH is exploited to quantify the assembly at molecular resolution, whereas microfluidics allows us to access the temporal information of the process and to initiate the reaction in a highly controlled manner. To realize the method, a multi-layer microfluidic device is designed for fluorescence spectroscopy measurements, with an integrated step in the central channel which provides an intrinsic passivation of the channel walls for the slowly diffusing proteins.With our approach, we are able to resolve the very first time points of assembly, as the lag time needed for mixing is below 3 ms and we clearly confirm that vimentin assembly is a two step process. We determine the time scales for the lateral assembly to be on the order of 65 ms, with a trend for slower assembly when more protein is labeled. Our approach can be generalized to study other biological and soft matter (self-)assembly processes, with a wide range of applications ranging from physiological assembly as in cytoskeletal structures to pathological aggregation processes like α-synuclein aggregation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Susanne Bauch, Harald Herrmann and Norbert Mücke for fruitful discussions and help with the protein preparation, and Don Lamb and Anders Barth for great support and helpful advice concerning the PCH analysis. This work was supported by the German Research Foundation (DFG) in the framework of SFB 1286 “Quantitative Synaptology”, project B02 and under Germany's Excellence Strategy – EXC 2067/1-390729940.Notes and references
- D. Ami, A. Natalello, M. Lotti and S. M. Doglia, Microb. Cell Fact., 2013, 12, 17 CrossRef CAS.
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular Biology of the Cell, Garland Science, New York, 4th edn, 2002 Search PubMed.
- C. E. Walczak and R. Heald, in International Review of Cytology, Elsevier, 2008, pp. 111–158 Search PubMed.
- D. J. Selkoe, Neuron, 1991, 6, 487–498 CrossRef CAS.
- M. G. Spillantini, M. L. Schmidt, V. M.-Y. Lee, J. Q. Trojanowski, R. Jakes and M. Goedert, Nature, 1997, 388, 839–840 CrossRef CAS.
- A. Horwich, J. Clin. Invest., 2002, 110, 1221–1232 CrossRef CAS.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 3rd edn, 2006 Search PubMed.
- E. L. Elson and D. Magde, Biopolymers, 1974, 13, 1–27 CrossRef CAS.
- Y. Chen, J. D. Müller, P. T. C. So and E. Gratton, Biophys. J., 1999, 77, 553–567 CrossRef CAS.
- P. Kask, K. Palo, D. Ullmann and K. Gall, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13756–13761 CrossRef CAS.
- Y. Chen, L.-N. Wei and J. D. Muller, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15492–15497 CrossRef CAS.
- S. Saffarian, Y. Li, E. L. Elson and L. J. Pike, Biophys. J., 2007, 93, 1021–1031 CrossRef CAS.
- M. A. Digman, P. W. Wiseman, C. Choi, A. R. Horwitz and E. Gratton, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2170–2175 CrossRef CAS.
- M. Pitschke, R. Prior, M. Haupt and D. Riesner, Nat. Med., 1998, 4, 832–834 CrossRef CAS.
- C. R. Nayak and A. D. Rutenberg, Biophys. J., 2011, 101, 2284–2293 CrossRef CAS.
- N. Terada, T. Shimozawa, S. Ishiwata and T. Funatsu, Biophys. J., 2007, 92, 2162–2171 CrossRef CAS.
- J. B. Knight, A. Vishwanath, J. P. Brody and R. H. Austin, Phys. Rev. Lett., 1998, 80, 3863–3866 CrossRef CAS.
- L. Pollack, M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W. A. Eaton and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 10115–10117 CrossRef CAS.
- L. Pollack, M. W. Tate, A. C. Finnefrock, C. Kalidas, S. Trotter, N. C. Darnton, L. Lurio, R. H. Austin, C. A. Batt, S. M. Gruner and S. G. J. Mochrie, Phys. Rev. Lett., 2001, 86, 4962–4965 CrossRef CAS.
- R. Russell, I. S. Millett, M. W. Tate, L. W. Kwok, B. Nakatani, S. M. Gruner, S. G. J. Mochrie, V. Pande, S. Doniach, D. Herschlag and L. Pollack, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4266–4271 CrossRef CAS.
- J. P. Brody, P. Yager, R. E. Goldstein and R. H. Austin, Biophys. J., 1996, 71, 3430–3441 CrossRef CAS.
- H. Song and R. F. Ismagilov, J. Am. Chem. Soc., 2003, 125, 14613–14619 CrossRef CAS.
- Z. Wu and N.-T. Nguyen, Microfluid. Nanofluid., 2004, 1, 208–217 CrossRef.
- M. E. Kinahan, E. Filippidi, S. Köster, X. Hu, H. M. Evans, T. Pfohl, D. L. Kaplan and J. Wong, Biomacromolecules, 2011, 12, 1504–1511 CrossRef CAS.
- M. E. Brennich, J.-F. Nolting, C. Dammann, B. Nöding, S. Bauch, H. Herrmann, T. Pfohl and S. Köster, Lab Chip, 2011, 11, 708 RSC.
- H. Herrmann, M. Häner, M. Brettel, S. A. Müller, K. N. Goldie, B. Fedtke, A. Lustig, W. W. Franke and U. Aebi, J. Mol. Biol., 1996, 264, 933–953 CrossRef CAS.
- H. Herrmann and U. Aebi, Curr. Opin. Cell Biol., 2000, 12, 79–90 CrossRef CAS.
- A. A. Chernyatina, S. Nicolet, U. Aebi, H. Herrmann and S. V. Strelkov, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13620–13625 CrossRef CAS.
- A. A. Chernyatina, D. Guzenko and S. V. Strelkov, Curr. Opin. Cell Biol., 2015, 32, 65–72 CrossRef CAS.
- I. Szeverenyi, A. J. Cassidy, C. W. Chung, B. T. Lee, J. E. Common, S. C. Ogg, H. Chen, S. Y. Sim, W. L. Goh, K. W. Ng, J. A. Simpson, L. L. Chee, G. H. Eng, B. Li, D. P. Lunny, D. Chuon, A. Venkatesh, K. H. Khoo, W. I. McLean, Y. P. Lim and E. B. Lane, Hum. Mutat., 2008, 29, 351–360 CrossRef CAS.
- J. E. Eriksson, T. Dechat, B. Grin, B. Helfand, M. Mendez, H.-M. Pallari and R. D. Goldman, J. Clin. Invest., 2009, 119, 1763–1771 CrossRef CAS.
- H. Herrmann, I. Hofmann and W. W. Franke, J. Mol. Biol., 1992, 223, 637–650 CrossRef CAS.
- H. Herrmann, L. Kreplak and U. Aebi, in Intermediate Filament Cytoskeleton, Elsevier, 2004, pp. 3–24 Search PubMed.
- S. Portet, N. Mücke, R. Kirmse, J. Langowski, M. Beil and H. Herrmann, Langmuir, 2009, 25, 8817–8823 CrossRef CAS.
- O. Saldanha, M. E. Brennich, M. Burghammer, H. Herrmann and S. Köster, Biomicrofluidics, 2016, 10, 024108 CrossRef.
- M. Denz, G. Brehm, C. Y. J. Hémonnot, H. Spears, A. Wittmeier, C. Cassini, O. Saldanha, E. Perego, A. Diaz, M. Burghammer and S. Köster, Lab Chip, 2018, 18, 171–178 RSC.
- N. Mücke, L. Kämmerer, S. Winheim, R. Kirmse, J. Krieger, M. Mildenberger, J. Baßler, E. Hurt, W. H. Goldmann, U. Aebi, K. Toth, J. Langowski and H. Herrmann, Biophys. J., 2018, 114, 2408–2418 CrossRef.
- C. G. Lopez, O. Saldanha, K. Huber and S. Köster, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11152–11157 CrossRef CAS.
- C. G. Lopez, O. Saldanha, A. Aufderhorst-Roberts, C. Martinez-Torres, M. Kuijs, G. H. Koenderink, S. Köster and K. Huber, Soft Matter, 2018, 14, 8445–8454 RSC.
- D. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS.
- J. C. McDonald and G. M. Whitesides, Acc. Chem. Res., 2002, 35, 491–499 CrossRef CAS.
- E. Kim, Y. Xia and G. M. Whitesides, J. Am. Chem. Soc., 1996, 35, 5722–5731 CrossRef.
- J. Block, H. Witt, A. Candelli, J. C. Danes, E. J. G. Peterman, G. J. L. Wuite, A. Janshoff and S. Köster, Sci. Adv., 2018, 4, eaat1161 CrossRef.
- S. Winheim, A. R. Hieb, M. Silbermann, E.-M. Surmann, T. Wedig, H. Herrmann, J. Langowski and N. Mücke, PLoS One, 2011, 6(4), e19202 CrossRef CAS.
- B. Nöding and S. Köster, Phys. Rev. Lett., 2012, 108, 088101 CrossRef.
- P. Cladé, PyDAQmx: A Python Interface to the National Instruments DAQmx Driver, http://pythonhosted.org/PyDAQmx/, (accessed: 11.13.2016).
- A. Edelstein, N. Amodaj, K. Hoover, R. Vale and N. Stuurman, Curr. Protoc. Mol. Biol., 2010, 92, 14.20.1–14.20.17 Search PubMed.
- L. N. Hillesheim and J. D. Müller, Biophys. J., 2003, 85, 1948–1958 CrossRef CAS.
- B. Huang, T. D. Perroud and R. N. Zare, ChemPhysChem, 2004, 5, 1523–1531 CrossRef CAS.
- D. Magde, W. W. Webb and E. L. Elson, Biopolymers, 1978, 17, 361–376 CrossRef CAS.
- M. E. Young, P. A. Carroad and R. L. Bell, Biotechnol. Bioeng., 1980, 22, 947–955 CrossRef CAS.
- I. Hofmann, H. Herrmann and W. W. Franke, Eur. J. Cell Biol., 1991, 56, 328–341 CAS.
- M. E. Brennich and S. Köster, Microfluid. Nanofluid., 2013, 16, 39–45 CrossRef.
- H. Y. Park, X. Qiu, E. Rhoades, J. Korlach, L. W. Kwok, W. R. Zipfel, W. W. Webb and L. Pollack, Anal. Chem., 2006, 78, 4465–4473 CrossRef CAS.
- J. Johnson, Y. Chen and J. D. Müller, Biophys. J., 2010, 99, 3084–3092 CrossRef CAS.
- J. D. Müller, Y. Chen and E. Gratton, Biophys. J., 2000, 78, 474–486 CrossRef.
- P. Macdonald, J. Johnson, E. Smith, Y. Chen and J. D. Mueller, in Fluorescence Fluctuation Spectroscopy (FFS), Part A, ed. S. Y. Tetin, Academic Press, 2013, vol. 518, pp. 71–98 Search PubMed.
- R. Kirmse, S. Portet, N. Mücke, U. Aebi, H. Herrmann and J. Langowski, J. Biol. Chem., 2007, 282, 18563–18572 CrossRef CAS.
- W. Chen, L. J. Young, M. Lu, A. Zaccone, F. Ströhl, N. Yu, G. S. K. Schierle and C. F. Kaminski, Nano Lett., 2016, 17, 143–149 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Additional data and detailed methods. See DOI: 10.1039/d0lc00985g |
| This journal is © The Royal Society of Chemistry 2021 |



