Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Studying the pharmacogenomic effect of cranberry extract on reducing body weight using collaborative cross mice
Fatima
Amer-Sarsour
,
Rawan
Abu Saleh
 ,
Itzhak
Ofek
and
Fuad A.
Iraqi
,
Itzhak
Ofek
and
Fuad A.
Iraqi
 *
*
Department of Clinical Microbiology and Immunology, Sackler Faculty of Medicine, Tel-Aviv University, Israel. E-mail: fatma.8580@hotmail.com; rona_a.s@hotmail.com; aofek@tauex.tau.ac.il; fuadi@tauex.tau.ac.il
First published on 15th April 2021
Abstract
The non-dialyzable material (NDM) of polyphenol-rich cranberry extract (CRE) powder (NDM-CRE) was studied for its effect of inducing body weight (BW) loss in 13 different mouse lines with well-defined genetically diverse backgrounds, named the collaborative cross (CC). From the age of 8 weeks, the mice were maintained on a high-fat diet (HFD) for 18 weeks, to induce obesity, and BW was measured biweekly. From week 12, CRE was injected intraperitoneally (IP) (50 mg kg−1) 3 times a week per mouse for a 6 week period. Statistical analysis results have shown a significant increase in body weight between week 0 and week 12; the increase in BW of 13 lines of mice on HFD was in the range of 10.41% to 68.65% for males and 9.78% to 64.74% for females. After injecting NDM-CRE extract, our analysis has shown an induced change in BW between week 12 and week 18. In males, NDM-CRE caused a significant decrease in BW of 5 out of the 13 lines in the range of −5.68% to −16.69% and a significant increase of 8.31% in BW of one male line, whereas in seven lines there was no significant decrease (−2.14% to −4.09%). In females, NDM-CRE caused a significant decrease in BW of 5 out of the 13 lines in the range of −3.90% to −11.83%, whereas in eight lines there were no significant changes in BW and it ranged between −1.50% and 4.90%. The broad-sense heritability (H2) and genetic coefficient of variation (CVg) were estimated and found to be between 0.71 and 0.81 for H2, and 0.18 and 0.24 for CVg of females and males, respectively, with respect to the efficacy of NDM-CRE on body weight reduction. Our results have shown that hosts with different genetic backgrounds respond differently to body weight increase, as well as to NDM-CRE treatment for body weight reduction. These results provide a platform for assessing more CC lines and mapping genes underlying the efficacy of the NDM-CRE treatment as a way of understanding pharmacogenomics.
Introduction
Obesity is now considered the greatest threat to human health and as a consequence, accounts for the highest direct and indirect medical cost in most developed and developing countries.1 In addition, it renders individuals susceptible to infectious diseases, as recently reported with regard to Covid-19 infections.2,3 It is probably associated with major changes in the food industry and consumption practices. In fact, over the past few decades, there has been an increase in the consumption of high-calorie food, especially of prepared or partially prepared processed foods. In order to cope with the so-called obesity epidemic, an industry of diets has developed and grown. Most of these diets are aimed to reduce weight by restricting the consumption of high fat and carbohydrate-containing foods. While many diets induce weight loss for a short period of time, the maintenance of weight loss by adhering to a particular diet has been shown to be much more difficult.4,5 Moreover, many individuals often become confused and frustrated with the conflicting information available to them and especially with the outcome. Given the difficulty in sustaining improvements in body weight by adhering to a particular diet, it is not surprising that many studies turn to food supplements, which promise weight loss and are available over the counter.6 Such dietary supplements include herbs, vitamins, amino acids, and certain polyphenol-rich plants such as green tea. The quest for such supplements is reasonable because it negates the necessity to adhere to a particular diet and many of the supplements are believed to be safe. However, none of the over the counter supplements have been proven, with reasonable certainty, to be efficient at reducing weight.6 We reasoned that the failure to ascertain the induction of weight loss by consuming a particular food supplement is that such a function may be limited to individuals with a particular genetic background.The positive health benefits of cranberries and cranberry-derived constituents include improvements in cardiovascular function as measured by decreases in lipid peroxidation, oxidative stress, total and low-density lipoprotein (LDL) cholesterol, and increases in the high-density lipoprotein (HDL) cholesterol level. The non-dialyzable material (NDM) is a fraction of total cranberry extract (CRE), and consists of only the non-dialyzed components of the CRE. All sugar, small components, and dialyzed materials are removed from the CRE during the dialyzation procedure. This fraction has been shown to consist of bioactive components.7
In previously published studies, the assessment of the effect of whole cranberry extract (CRE) on body weight loss was usually performed by using one mouse inbred line (one genetic background), which was not sufficient for providing information about the effect of host genetic background on the efficiency of the treatment.8–10
To test this hypothesis, we undertook the present study using polyphenol-rich cranberry extract powder to induce BW loss in mouse lines of defined diverse genetic backgrounds, namely the collaborative cross (CC) mice. The choice of the usage of non-dialyzable material (NDM) of the cranberry extract (CRE) is driven by numerous studies showing that such extracts are beneficial for health including the treatment of infectious diseases as well as obesity.7–10
The mouse is a powerful animal model for studying human complex traits, phenotypes, and diseases. The majority of the results obtained for mouse models may be translated into humans.11 Nevertheless, unfortunately, some are still not possible to translate. In our recent studies, we have published our successful translational research using the CC mouse population.12,13 The Collaborative Cross (CC) mouse model is a new and genetically diverse reference population.15–17 This unique model is a unique platform for research of complex traits that overcomes the limitation of existing animal models.15,18–20 The CC population is a powerful tool for genetic dissection of different phenotypes. This large, multi-parental, recombinant inbred strain panel exhibits recombinant chromosomal segments of approximately 2–5 Mb size. This model was created by the mating of eight mouse strains, five classical inbred strains: A/J, 57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/H1LtJ, and 3 wild-derived strains: CAST/EiJ, PWK/PhJ, and WSB/EiJ.14,17–19 The eight founder strains capture a much greater level of genetic diversity than existing recombinant inbred line (RIL) panels or other extant mouse genetic resource populations.21 Controlled randomization and minimization of selection during the breeding process recombined the natural genetic variation presented in these inbred strains. The result is a unique collection of RILs exhibiting a large phenotypic and genetic diversity, and bringing the tremendous genetic variation potential of the mouse inbred lines to phenotypic expression.15,22 Complete details of CC lines and their capability of mapping quantitative trait loci (QTL) with host susceptibility to complex traits are presented in various publications.11,18,19,23–32Fig. 1 shows the breeding design of developing a single CC line. Different CC lines with the exclusive genetic background can be generated by changing the crosses between the founders. The CC mice will finally include a set of approximately 75 RILs.
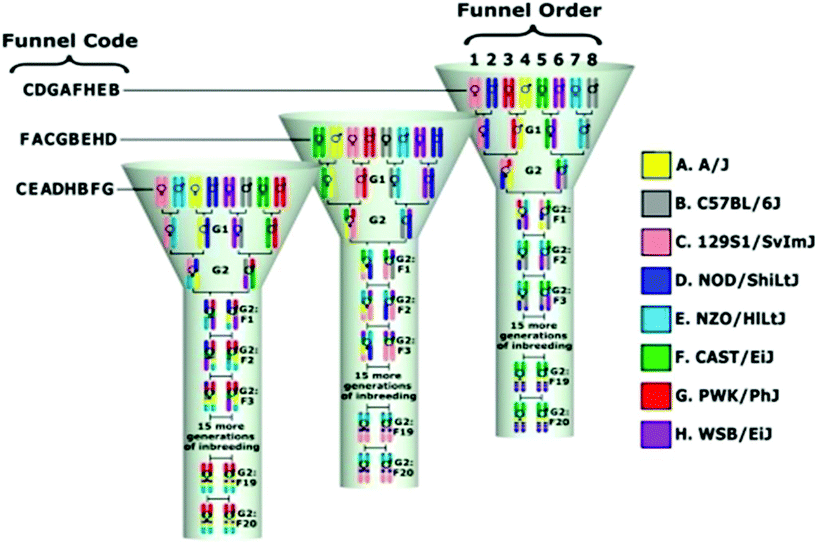 | ||
| Fig. 1 A unique breeding funnel scheme showing the development of the collaborative cross (CC) mouse model. This breeding approach is designed to randomize the genetic makeup of each inbred line. A single breeding funnel results in one CC recombinant inbred line that represents the genomes of the eight CC mice founders. The eight founder strains are arranged in different positions1–8 in each line, i.e. their order is randomized and not repeated across lines and this order determines the funnel code based on a single letter code for each line. In a funnel-breeding scheme, the genetic contributions of all eight-founder strains are incorporated after the G2 generation. A recombinant inbred line is created following 20 generations of inbreeding.18 | ||
Previous studies from our laboratory and collaborators have proven the suitability of the novel and genetically highly diverse CC mouse genetic reference population as a model system for exploring the genetics of complex trait diseases, including diet-induced T2D.28–30 These studies have shown that genetic background plays an important role in the development of T2D and, subsequently in identifying genetic factors, which may underline this variation of T2D development. Furthermore, we showed using HFD-induced T2D and metabolic syndromes (METS) that sex and diet effects were significant among the CC population, where males and females of the different CC lines varied significantly with respect to the development and progression of T2D in response to both types of diets, the CHD and HFD. Additionally, our previous study showed significant diet effects, in which HFD (42% fat) was shown to induce obesity and T2D to greater levels than the standard CHD.26–28 Yet, diet and sex effects varied significantly between the CC lines, i.e. some CC lines were more susceptible than others, and differences based on sex in some CC lines were greater than in others. Altogether, there is significant evidence that sex effects and diet effects among the CC population are complex traits, controlled by multiple genetic factors.25,28–30 Based on these findings, in the current study, we have induced obesity, using a similar dietary challenge for 12 weeks, and thereafter, we have initiated the treatment with NDM of cranberry extract for an additional 6 weeks, to explore treatment efficacy.
Materials and methods
Animals and housing
We employed the CC mouse model which represents a powerful tool for genetic dissection of different phenotypes.12,13,17,18,21–25,29–30 Thirteen different CC mouse lines, with their Tel-Aviv University (TAU) and international designations, and the number of founders (out of the eight founders) contributing to each line are shown in Table 1. The mice were bred and maintained at the Small Animal Facility at Sackler Faculty of Medicine as described in our previous studies.13,14,18,19,23–32| TAU line designation | CC line designation | Number contributors from the eight founders |
|---|---|---|
| IL72 | CC037 | 6 |
| IL111 | TAU unique line | 5 |
| IL521 | CC072 | 6 |
| IL1912 | CC051 | 6 |
| IL5000 | CC010 | 8 |
| IL5001 | CC049 | 8 |
| IL5003 | CC042 | 7 |
| IL5004 | CC039 | 8 |
| IL5008 | CC018 | 8 |
| IL5020 | CC057 | 8 |
| IL5023 | CC061 | 8 |
| IL6009 | CC035 | 8 |
| IL6020 | CC040 | 8 |
This unique model is a platform for research on complex traits that overcomes the limitation of existing animal models.14 The CC population is a powerful tool for genetic dissection of the different phenotypes. This large, multi-parental, recombinant inbred strains panel exhibits recombinant chromosomal segments of approximately 2–5 Mb size. The eight founder strains capture a much greater level of genetic diversity than existing recombinant inbred line (RIL) panels or other extant mouse genetic resource populations.21
The high molecular genomic DNA of the CC lines were genotyped three times, once using 620![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 single nucleotide polymorphism (SNP) markers of mouse diversity array,33 once with mouse universal genotype array (MUGA-7500 markers) and later genotyped using the mouse universal SNP array (MegaMUGA-77800 markers). The updated genotype status of the population has been presented recently.34Fig. 2 shows the genomic reconstruction of two CC lines after genotyping with the mouse diversity array and using HAPPY software,18 exhibiting the diversity of the phenotypic response of the CC lines with complex diseases and their capability for mapping QTL associated with this trait to a small genomic interval less than 0.5 cM, which consists of few genes. Complete genotype data of the CC lines developed at TAU is available at: http://mtweb.cs.ucl.ac.uk/mus/www/preCC/MEGA_MUGA/Mar2015.MEGA+MDA+MUGA/.
000 single nucleotide polymorphism (SNP) markers of mouse diversity array,33 once with mouse universal genotype array (MUGA-7500 markers) and later genotyped using the mouse universal SNP array (MegaMUGA-77800 markers). The updated genotype status of the population has been presented recently.34Fig. 2 shows the genomic reconstruction of two CC lines after genotyping with the mouse diversity array and using HAPPY software,18 exhibiting the diversity of the phenotypic response of the CC lines with complex diseases and their capability for mapping QTL associated with this trait to a small genomic interval less than 0.5 cM, which consists of few genes. Complete genotype data of the CC lines developed at TAU is available at: http://mtweb.cs.ucl.ac.uk/mus/www/preCC/MEGA_MUGA/Mar2015.MEGA+MDA+MUGA/.
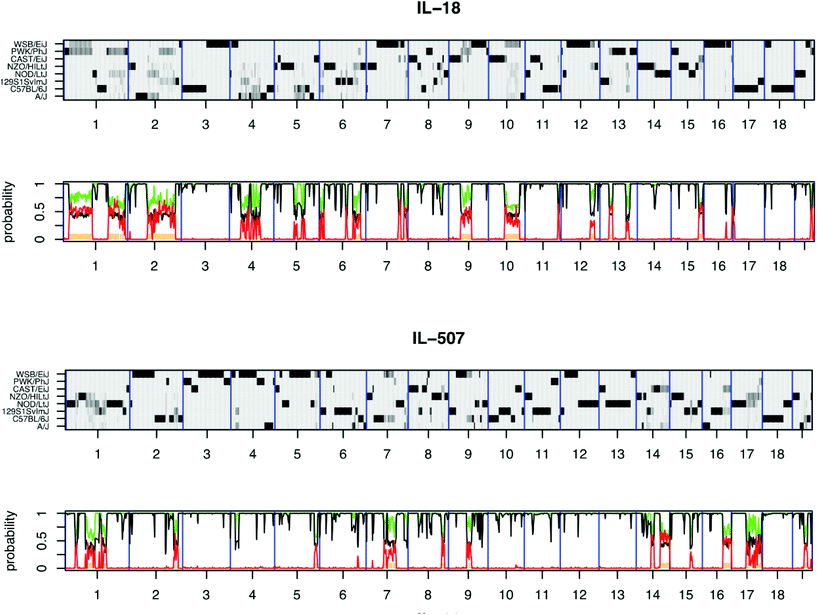 | ||
| Fig. 2 Reconstructions of the genomes of representative CC lines IL-18 and IL-507 from the hidden Markov model (HMM) were implemented using HAPPY. The x-axis shows the 19 autosomes. Each reconstruction is represented by two panels: top panel y-axis shows the 8 CC founders and the probability of descent from a founder at a locus is represented by the shade of grey, with white = 0 and black = 1. Regions where a single haplotype predominates appear as dark horizontal bands; loci with residual heterozygosity or where the founder haplotypes are indistinguishable are paler grey. The lower panel indicates local heterozygosity (red), the posterior probability of the most probable founder (black), and the sum of the most probable pair of founders (green).18 | ||
Mouse experiments
Overall, 156 mice (76 males and 80 females) were analyzed from 13 different CC lines (on average, 6 mice per sex per line) as shown in Tables 2 and 3. We used longitudinal experiment design, where we used a large number of CC lines (13 lines) and a number of mice per sex per line, so we can estimate, accurately, and provide statistical power for calculating the mean value of traits per sex and line. At 3 weeks old, the mice were weaned to separate cages based on sex and CC line, with free access to standard rodent chow diet and water. The experiment started when the mice were 8 weeks old. Throughout the experiment period, which lasts 18 weeks, the mice were maintained on HFD which is considered a Western diet, TD.88137 (Teklad Global, Harlan Inc., Madison, WI, USA), which consists of 42.0% kcal from fat, 15.3% from protein, and 42.7% from carbohydrates. At the end of the experiment, the mice were 26 weeks old. During this period, BW (grams) was measured weekly during the first 12 weeks of the experiment then 3 times per week during the CRE treatment period.| Males | HFD group | BW ± SE | Weeks 0 to 12 | Weeks 12 to 18 | ||||
|---|---|---|---|---|---|---|---|---|
| Line | no of mice | Week 0 | Week 12 | ΔBW(weeks 0–12) | (% change ± SE)s | CRE BW weeks 18 ± SE | ΔCRE BW ± SE (week 12 to week 18) | (% change ± SE)s |
| IL72 | 10 | 23.81 ± 0.26 | 29.91 ± 0.72 | 6.10 ± 0.55 | (25.54 ± 2.14)s | 25.97 ± 0.57 | −3.94 ± 0.51 | (−13.03 ± 1.50)s |
| IL111 | 4 | 21.67 ± 0.18 | 31.21 ± 1.48 | 9.55 ± 1.34 | (43.95 ± 5.81)s | 28.75 ± 0.95 | −2.47 ± 0.59 | (−7.72 ± 1.45)s |
| IL521 | 4 | 19.10 ± 1.24 | 27.26 ± 1.06 | 8.16 ± 1.44 | (44.14 ± 9.14)s | 26.14 ± 0.95 | −1.12 ± 1.24 | (−3.75 ± 4.53)ns |
| IL1912 | 9 | 26.15 ± 0.73 | 40.58 ± 1.64 | 14.43 ± 0.99 | (54.84 ± 2.63)s | 33.70 ± 1.38 | −6.88 ± 1.07 | (−16.69 ± 2.48)s |
| IL5000 | 8 | 26.36 ± 0.48 | 34.02 ± 0.55 | 7.66 ± 0.42 | (29.19 ± 1.84)s | 30.11 ± 0.82 | −3.91 ± 0.47 | (−11.56 ± 1.44)s |
| IL5001 | 7 | 25.09 ± 1.42 | 42.04 ± 1.61 | 16.95 ± 0.50 | (68.65 ± 3.86)s | 40.38 ± 1.32 | −1.65 ± 0.92 | (−3.68 ± 1.95)ns |
| IL5003 | 7 | 22.37 ± 1.11 | 28.54 ± 1.37 | 6.17 ± 1.03 | (28.16 ± 4.83)s | 27.76 ± 0.91 | −0.78 ± 0.76 | (−2.14 ± 2.62)ns |
| IL5004 | 5 | 20.71 ± 0.26 | 22.85 ± 0.38 | 2.14 ± 0.52 | (10.41 ± 2.59)s | 21.54 ± 0.45 | −1.31 ± 0.41 | (−5.68 ± 1.78)s |
| IL5008 | 4 | 20.37 ± 0.90 | 23.39 ± 0.72 | 3.02 ± 0.25 | (15.04 ± 1.90)s | 25.32 ± 0.71 | 1.94 ± 0.13 | (8.31 ± 0.68)s |
| IL5020 | 5 | 31.04 ± 0.53 | 49.21 ± 1.14 | 18.17 ± 1.21 | (58.70 ± 4.28)s | 47.36 ± 2.51 | −1.86 ± 1.80 | (−3.91 ± 3.68)ns |
| IL5023 | 4 | 20.34 ± 0.27 | 32.61 ± 0.81 | 12.28 ± 0.69 | (60.37 ± 3.21)s | 31.29 ± 0.91 | −1.33 ± 0.25 | (−4.09 ± 0.81)ns |
| IL6009 | 4 | 19.22 ± 0.91 | 23.12 ± 0.57 | 3.90 ± 0.60 | (20.74 ± 3.97)s | 23.52 ± 0.60 | 0.40 ± 0.31 | (1.78 ± 1.38)ns |
| IL6020 | 5 | 25.49 ± 2.62 | 38.99 ± 4.14 | 13.50 ± 1.82 | (52.82 ± 5.29)s | 37.47 ± 4.24 | −1.52 ± 0.98 | (−3.91 ± 2.54)ns |
| Total | 76 | 23.76 ± 0.44 | 33.37 ± 0.95 | 9.61 ± 0.62 | (39.56 ± 2.29) | 30.96 ± 0.87 | −2.41 ± 0.34 | (−6.62 ± 0.94) |
| Females | HFD group | BW ± SE | Week 0 to week 12 | Week 12 to week 18 | ||||
|---|---|---|---|---|---|---|---|---|
| Line | n = no of mice | Week 0 | Week 12 | ΔBW (weeks 0–12) | (% change ± SE)s | CRE BW Week 18 ± SE | ΔCRE BW ± SE (week 12 to week 18) | (% change ± SE)s |
| IL72 | 7 | 20.21 ± 0.50 | 24.77 ± 0.83 | 4.56 ± 0.74 | (22.73 ± 3.78)s | 22.02 ± 0.38 | −2.74 ± 0.52 | (−10.75 ± 1.76)s |
| IL111 | 5 | 17.97 ± 0.54 | 23.38 ± 0.57 | 5.41 ± 0.38 | (30.32 ± 2.67)s | 23.79 ± 0.40 | 0.41 ± 0.42 | (1.86 ± 1.83)ns |
| IL521 | 4 | 17.23 ± 0.53 | 28.76 ± 1.95 | 11.53 ± 1.66 | (66.71 ± 9.15)s | 28.99 ± 1.50 | 0.23 ± 0.55 | (1.15 ± 2.02)ns |
| IL1912 | 7 | 18.68 ± 0.65 | 30.65 ± 1.57 | 11.97 ± 1.22 | (64.12 ± 6.12)s | 26.77 ± 0.57 | −3.89 ± 1.08 | (−11.83 ± 2.95)s |
| IL5000 | 4 | 24.03 ± 0.94 | 32.04 ± 1.61 | 8.01 ± 1.31 | (33.49 ± 5.92)s | 28.93 ± 1.70 | −3.11 ± 0.72 | (−9.75 ± 2.12)s |
| IL5001 | 11 | 20.73 ± 0.44 | 34.24 ± 1.71 | 13.50 ± 1.47 | (64.74 ± 6.41)s | 32.70 ± 1.32 | −1.54 ± 0.56 | (−3.90 ± 1.59)s |
| IL5003 | 9 | 17.90 ± 0.60 | 22.19 ± 0.86 | 4.28 ± 0.66 | (24.17 ± 3.67)s | 22.92 ± 0.82 | 0.74 ± 0.42 | (3.55 ± 1.82)ns |
| IL5004 | 4 | 17.86 ± 0.69 | 22.06 ± 1.11 | 4.20 ± 0.93 | (23.67 ± 5.11)s | 21.67 ± 0.73 | −0.39 ± 0.43 | (−1.50 ± 1.83)ns |
| IL5008 | 5 | 16.02 ± 0.81 | 17.85 ± 0.89 | 1.84 ± 0.31 | (11.56 ± 2.13)s | 18.68 ± 0.70 | 0.82 ± 0.30 | (4.90 ± 1.78)ns |
| IL5020 | 5 | 20.36 ± 0.27 | 34.34 ± 2.34 | 13.98 ± 2.14 | (68.31 ± 9.92)s | 31.07 ± 1.70 | −3.27 ± 1.07 | (−8.95 ± 3.21)s |
| IL5023 | 7 | 16.27 ± 1.00 | 22.75 ± 0.81 | 6.49 ± 0.52 | (41.48 ± 5.18)s | 23.64 ± 0.94 | 0.88 ± 0.37 | (3.85 ± 1.60)ns |
| IL6009 | 3 | 19.43 ± 0.60 | 21.34 ± 0.83 | 1.91 ± 0.28 | (9.78 ± 1.24)s | 22.00 ± 1.07 | 0.66 ± 0.24 | (3.01 ± 1.05)ns |
| IL6020 | 9 | 22.96 ± 0.80 | 30.68 ± 1.67 | 7.71 ± 1.44 | (33.81 ± 6.61)s | 30.66 ± 1.55 | −0.02 ± 0.25 | (0.14 ± 0.82)ns |
| Total | 80 | 19.36 ± 0.31 | 27.16 ± 0.71 | 7.81 ± 0.56 | (40.10 ± 2.72) | 26.26 ± 0.58 | −0.90 ± 0.25 | (−2.29 ± 0.83) |
Statistical analysis
Statistical analysis was performed using the software package IBM SPSS Statistics version 24. One-way ANOVA was carried out for testing the significance level of trait variation among CC lines, and two-way ANOVA was performed for testing sex and line, and sex and diet interactions. The analysis revealed a significant (p-value < 0.01) line sex line and sex diet interactions, a p-value of 0.05 or less was considered significant. The Pearson correlation coefficient test was performed to measure the linear correlation between different phenotypes. The t-test was also performed to assess the changes and significance before and after diets as well as before and after NDM-CRE treatment. The post hoc test(s) based on the Bonferroni procedure was/were performed to assess differences between baseline (time 0 of the experiment) and 12 weeks of high-fat diet treatment, and then between 12 and 18 weeks (6 weeks of NDM-CRE treatment).Broad sense heritability and genetic coefficient of variation
The broad-sense heritability and genetic coefficient of variation were estimated as described in detail elsewhere.30 Briefly, heritability (H2) refers to the proportion of variation between individuals in a population that can be influenced by genetic factors. Here, we used the ANOVA output of the phenotypic traits to calculate the heritability (including epistatic, but not dominance effects), using the following equation:| H2 = (Vg/Vg + Ve) |
Results
The increase in body weight between week 0 and week 12 of the 13 lines of mice on HFD was in the range of 10.41% to 68.65% for males and 9.78% to 64.74% for females (Tables 2 & 3). In each of the 13 lines, the increase in the body weight was significant, although the magnitude of the increase was significantly higher for males as compared to females (mean differences in body weight changes of 9.61 and 7.81 in grams, respectively).Effect of NDM-CRE on body weight
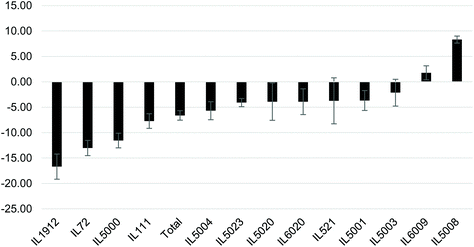
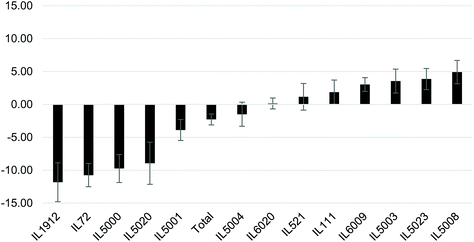
Following the significant increase in BW at week 12, the mice were IP injected 3 times a week for 6 weeks with the NDM cranberry extract (NDM-CRE). Tables 2 & 3 depict the NDM-CRE-induced change in BW between week 12 and week 18. In males (Table 2) CRE caused a significant decrease in BW in 5 out of 13 lines in the range of −5.68% to −16.69% and a significant increase of 8.31% in BW of one male line (IL5008), whereas in seven lines there was no significant decrease (−2.14% to −4.09%).The effect of NDM-CRE in females (Table 3) was different from that in males. It caused a significant decrease in 5 lines and a non-significant decrease or increase in one line and 7 lines, respectively. When the NDM-CRE effect was analyzed for each mouse line, a remarkable difference was noticed between the two sexes. For example, in only 3 lines (IL72, IL1912, and IL5000) NDM-CRE induced a significant decrease of body weight in both females and males of these lines and in one line (IL6009) there was no significant change in both sexes of this line. In contrast, in none of the remaining 9 lines, the magnitude of the change in body weight following NDM-CRE injections was similar in the two sexes of the same line. For example, in one line (IL111) there was a significant decrease in BW of the males and non-significant changes in that of females, in 2 lines IL521 and IL5003 NDM-CRE caused a non-significant increase or decrease of body weight in the males and females of these lines. In two lines (IL5001 and IL5020) NDM-CRE caused a significant decrease in BW of females, but non-significant in males. These remarkable gender-associated differences between males and females of the 13 lines can be appreciated when the pattern of the CRE effects is presented in histograms (Fig. 3 & 4).
Broad sense heritability and genetic coefficient of variation
The H2 and CVg of the CRE effects on BW for females and males of the 13 mice lines are shown in Table 4. For H2, they were estimated to be 0.83 and 0.81 in males before and after the administration of NDM-CRE and 0.69 before and 0.71 after NDM-CRE treatments in females, respectively. In contrast, the CVg values were 0.25 and 0.24 in males before and after NDM-CRE treatments, respectively, and 0.21 before and 0.18 after NDM-CRE treatments in females, respectively. The calculated estimated heritability (H2) was found to be high, which indicates and confirms that these assessed traits are under strong host genetic background influence. Furthermore, the calculated CVg shows that the genetic diversity is high in these tested 13 CC lines and a promising resource for identifying genetic factors underlying these tested traits.| Females | |||
|---|---|---|---|
| Trait | H 2 | Trait mean | CVg |
| BW0 | 0.67 | 19.31 | 0.12 |
| BW12 | 0.69 | 27.16 | 0.21 |
| BW18 | 0.71 | 26.26 | 0.18 |
| Males | |||
|---|---|---|---|
| Trait | H 2 | Trait mean | CVg |
| BW0 | 0.7 | 23.17 | 0.15 |
| BW12 | 0.83 | 33.37 | 0.25 |
| BW18 | 0.81 | 30.96 | 0.24 |
Discussion
Total health care costs attributed to obesity/overweight would double every decade to 860.7–$956 billion U.S. dollars by 2030, which accounts for 16–18 percent of the total US health care cost.35 The development of obesity was shown to be controlled by multiple genetic components (polygenic) and influenced by environmental factors.36–38 Therefore, it is not surprising that most individuals failed to sustain a particular diet aimed at reducing weight. Instead, several investigators studied the effect of food supplements for any diet on weight reduction negating the necessity to adhere to a particular diet.6 In none of these studies, however, was a perfect food supplement found to be reasonably efficient in achieving weight loss.Cranberry derived products can also increase immune function. The American cranberry (Vaccinium macrocarpon) is a particularly rich source of polyphenols, which has been associated in vitro with antibacterial, antiviral, antimutagenic, anticarcinogenic, antitumorigenic, antiangiogenic, anti-inflammatory, and antioxidant properties. Additionally, cranberries are a rich source of phytochemicals, including anthocyanins and other flavonoids that may decrease lipid oxidation and protein glycosylation, and also decrease liver weight and triglyceride accumulation in association with blunted hepatic oxidative stress and inflammation.8,9
Logic states that there is a genetic factor that allows a particular food supplement to function in reducing weight. The present study was undertaken to challenge this assumption by using a genotyped and defined population of CC mouse lines previously shown to allow the determination of the heritability of a test trait employing a relatively low number of mice lines.25,39 Polyphenol-rich cranberry extract was used as a source of food supplement because it holds the premise to influence obesity in a limited study. For example, in one study it was found that the multi-component polyphenol-rich extract obtained from cranberries could have a beneficial effect on the prevention or treatment of obesity via the inhibition of adipogenesis, lipid accumulation, and reactive oxygen species (ROS) production in adipocytes without affecting their viability.9
The results show unequivocally that the reduction in the BW of HFD-induced obesity in 13 CC mice lines following the IP administration of NDM-CRE is a hereditable trait. The high estimated broad-sense heritability (H2) shows the importance of the host genetic background on the performance of these assessed traits, and genetic covariance (CVg) indicates that we have a strong genetic diversity in the assessed population, that will lead to the identification of a variety of factors underlying these traits. The 13 CC strains, which are randomly used in this study are part of a large CC population (75 lines), which is a powerful mouse genetic reference population (GRP) for gene mapping and dissecting complex traits. At this stage, 13 CC lines were assessed for NDM-CRE, just to determine if the host genetic background may define the efficacy of the response to the treatment, which indeed was confirmed in our study. After obtaining these results, we plan to continue the breeding of CC lines and assess them for NDM treatment, so to provide enough information for performing gene mapping. The advantage of the CC lines compared with the eight parental founders is that the new genetic (gene–gene) interactions of the eight genomes, together in the CC lines, may generate new epistatic combinations that may perform better than the parental lines, alone. It is well known that once inbred lines of mice are crossed, the cross progeny often exhibits transgressive variation or presents novel traits that were not present in the parental strains. This indicates that the inbred strains of mice harbor a tremendous amount of natural genetic variation, above what is seen when the lines themselves are compared. This variation has been accessed in part by the construction of recombinant inbred lines (RIL) derived from crosses between mouse pure lines.15,40 It is known that the increase in genetic diversity and variations in the mouse model will increase the possibility of capturing the diversity of phenotypes. The eight founders of the CC mouse population contain 42 million genetic variations (single nucleotide polymorphisms (SNPs)), which is higher than in the human population (20 million SNPs).15,16 Therefore, the CC mouse population has a strong justification to be used for human studies and capture genes which may be translated in humans.
The biological replica (number of mice per line and sex) used in this study was an excellent resource, which allowed performing a strong statistical analysis and calculating heritability and genetic covariance, and met the requirements as suggested earlier.25,39 We anticipate that by increasing the number of assessed CC lines (30 lines and up) with an appropriate number of mice per line,17 and by using the multi-trait and the multi-locus analytical methods developed specifically for this genetically highly diverse reference population,41 we will be able to map a genetic quantitative trait loci (QTL) with unprecedented precision to allow identification of genes associated with the ability of a particular food supplement to function in reducing weight. Because most mouse genes have orthologous genes in humans, we may be able to personalize the use of food supplements to function in reducing BW in humans.
In our study, we injected the NDM into our animals into the peritoneal cavity, which is a common technique in laboratory rodents but is rarely used in larger mammals and humans. Intraperitoneal injection is used for small species for which intravenous access is challenging and it can be used to administer large volumes of fluid safely or as a repository site for surgical implantation of a preloaded osmotic minipump. Although intraperitoneal delivery is considered a parenteral route of administration, the pharmacokinetics of substances administered intraperitoneally are more similar to those seen after oral administration because the primary route of absorption is into the mesenteric vessels, which drain into the portal vein and pass through the liver.42
Therefore, substances administered intraperitoneally may undergo hepatic metabolism before reaching the systemic circulation. In addition, a small amount of intraperitoneal injection may pass directly across the diaphragm through the small lacunae and into the thoracic lymph.43,44
The used dose of NDM (50 mg per kg mouse body weight) in our study was based on previous reports,46,47 which showed that it contains bioactive components and has a significant effect on oral inflammation. Therefore, in this study, we adopted this dose, which indeed, showed an active effect on body weight loss, depending on the host genetic background. Furthermore, in previously published papers, the usual amount of whole cranberry extract was 200 mg per kg mouse body weight, which is four fold what we used in our study.10,45 Our NDM cranberry fraction (which was shown in previous studies to consist of active components against inflammation) is part of the whole cranberry extract, which was used in previous publications.45,46 Therefore, we used ¼ of the usually used amount of cranberry extract, so as to avoid any overdose and unexpected reaction.
The average person can drink one or two glasses per day (e.g. 400–600 ml, which nutritionally also treats urinary tract infection (UTI), for example) of cranberry juice cocktail, which contains 0.2 mg NDM per ml juice meaning 1.07–1.33 mg kg−1 NDM at the most per day by an adult man (weighing 75 kg average). If so, the amount injected is equivalent to the amount of NDM in 2 liters of juice consumed per day by an average man. Therefore, using NDM (4 × 1.07–1.33 = 4.28–5.32 mg NDM per kg per day for humans) can reduce the required drinking volume of whole cranberry juice, which coincides with the United States Food and Drug Association (FDA) recommendations, based on the conversion of the daily dosage found to be effective in mouse models into human equivalent dosage, which should be 0.08 of what is used in the mouse model i.e. 0.08 of 50 mg kg−1 in mouse equal to 4 mg kg−1 in human, may be used.
In this study, IP injection protocol was applied in the administration and introducing the NDM to the assessed mice. However, through both routes of administration, i.e. IP and oral feeding, the drugs enter the systemic circulation mostly through the hepatic portal system. The only difference is in the absorption phase, wherein by oral administration, the substances are absorbed in the gastrointestinal tract and by IP injection, they are diffused across the peritoneal membrane which is a semipermeable membrane, lined with a capillary bed. The blood vessels supplying and draining the abdominal viscera, musculature and mesentery, constitute a blood filled compartment into which drugs can diffuse from the peritoneum. Absorption of intraperitoneally applied drugs occurs rather rapidly but compounds are partially subjected to hepatic first-pass elimination. The effect of biopharmaceutical and biological factors on drug bioavailability is known only to a limited extent. IP administration will result in more bioavailability as it is administered directly into circulation. This leads to an increased absorption rate of the substance after IP administration.48 However, one limitation of this route is the first pass metabolism, as in the case of orally administered drugs, because substances absorbed from the peritoneal cavity end up in the portal vein and pass through the liver.49,50,51
The toxicity effect is a major concern when applying herbal and dietary supplements. Little is known about the toxicity of cranberry extracts but in none of the reports employing various extracts was a toxic effect noted even in some studies where 4 fold (e.g. 200 mg per kg mouse) higher dosage than that used by us was employed.45,46 Although the appropriate dose to be used in humans is out of the scope of the present study, a dose-response to determine the lowest dose of NDM will eventually be determined on those lines bearing the genetic loci responsible for lowering BW. Such a study should form the basis for dosing in humans bearing the orthologous genetic loci. Interestingly, this effect can vary with different host genetic backgrounds (inter individual toxicity responses), as reported by Church et al.52 In Church and colleagues′ study, they showed that within the diversity outcross (DO) population, an equal exposure to green tea extract containing the polyphenol epigallocatechin gallate (EGCG) (50 mg kg−1; daily for three days) was found to be tolerated to hepatotoxicity in the majority of mice; however, a small fraction of the animals (16%; 43/272) exhibited severe hepatotoxicity (10–86.8% liver necrosis), which was assumed to be analogous to the clinical cases. In our study, the toxicity effect of NDM was not assessed, because a study by Hochman et al.,53 reported that at 4 mg per day (160 mg per kg mouse body weight of an average mouse weight of 25 g), toxicity was not observed. However, they stated that using four fold of this amount i.e. 16 g of NDM (640 mg per kg mouse body weight of an average mouse weight of 25 g) was toxic. Although we employed body weight measurements only in the present study, we realize that metabolic syndrome actually includes a number of other metabolic parameters such as fat distribution, liver weight, determination of serum components such as adiponectin, leptin and ghrelin, fasting blood glucose, glucose tolerance (insulin resistance) as well as changes in the microbiome. Indeed, studies are designed to include some of these parameters in the CC mice to better understand the effect of cranberry extracts on metabolic syndrome.
Finally, the results of our study have shown that the amount of NDM used by us shows a significant effect, which is believed to be due to the high purity and concentration of the active component in our NDM extract.
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.Acknowledgements
This report was supported by the core fund from Tel Aviv University.References
- H. Huang, Y. Zhijun, C. Yahong and L. Fangyan, A social contagious model of the obesity epidemic, Sci. Rep., 2016, 6, 37961–37970 CrossRef CAS PubMed.
- N. Finer, P. Sarah, S. P. Garnett and J. M. Bruun, COVID-19 and obesity, Clin. Obes., 2020, 10, e12365 CrossRef PubMed.
- M. Rottoli, P. Bernante, A. Belvedere, F. Balsamo, S. Garelli, M. Giannella, A. Cascavilla, S. Tedeschi, S. Ianniruberto, E. Rosselli Del Turco, T. Tonetti, V. M. Ranieri, G. Poggioli, L. Manzoli, U. Pagotto, P. Viale and M. Bartoletti, How important is obesity as a risk factor for respiratory failure, intensive care admission and death in hospitalised COVID-19 patients? Results from a single Italian centre, Eur. Soc. Endocrinol., 2019, 503–541 Search PubMed.
- J. C. M. Barte, N. C. W. ter Bogt, R. P. Bogers, P. J. Teixeira, B. Blissmer and T. A. Mori, et al., Maintenance of weight loss after lifestyle interventions for overweight and obesity, a systematic review, Obes. Rev., 2011, 11, 899–906 CrossRef PubMed.
- N. S. Mitchell, V. A. Catenacci, H. R. Wyatt and J. O. Hill, Obesity: overview of an epidemic, Psychiatr. Clin. North Am., 2011, 34, 717–732 CrossRef PubMed.
- R. Saper, D. M. Eisenberg and R. S. Phillips, Common Dietary Supplements for Weight Loss, Am. Fam. Physician, 2004, 70, 1731–1738 Search PubMed.
- H. Shmuely, I. Ofek, E. I. Weiss, Z. Rones and Y. Houri-Haddad, Cranberry components for the therapy of infectious disease, Curr. Opin. Biotechnol, 2011, 23, 148–152 CrossRef PubMed.
- K. Kowalska and O. Olejnik, Beneficial effects of cranberry in the prevention of obesity and related complications: Metabolic syndrome and diabetes – A review, J. Funct. Foods, 2016, 20, 171–181 CrossRef CAS.
- K. Kowalska, A. Olejnik, D. Szwajgier and M. Olkowicz, Inhibitory Activity of Chokeberry, Bilberry, Raspberry and Cranberry Polyphenol-Rich Extract towards Adipogenesis and Oxidative Stress in Differentiated 3 T3-L1 Adipose Cells, PLoS One, 2017, 12(11), e0188583 CrossRef PubMed.
- F. F. Anhê, D. Roy, G. Pilon, S. Dudonné, S. Matamoros, T. V. Varin, C. Garofalo, Q. Moine, Y. Desjardins, E. Levy and A. Marette, A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice, Gut, 2015, 64(6), 872–883 CrossRef PubMed.
- K. Schughart, C. Libert, SYSGENET consortium and M. J. Kas, Controlling complexity: the clinical relevance of mouse complex genetics, Eur. J. Hum. Genet., 2013, 21(11), 1191–1196, DOI:10.1038/ejhg.2013.79.
- A. Shusterman, M. Munz, G. Richter, S. Jepsen, W. S. Lieb, B. Krone, P. Hoffman, M. Laudes, J. Wellmann, K. Berger, T. Kocher, S. Offenbacher, K. Divaris, A. Franke, S. Schreiber, H. Dommisch, E. Weiss, A. S. Schaefer, Y. Houri-Haddad and F. A. Iraqi, The PF4/PPBP/CXCL5, Gene Cluster Is Associated with Periodontitis, J. Dent. Res., 2017, 96(8), 945–952 CrossRef CAS PubMed.
- A. Nashef, M. Matthias, E. Weiss, B. G. Loos, S. Jepsen, N. van der Velde, A. G. Uitterlinden, J. Wellmann, K. Berger, P. Hoffmann, M. Laudes, W. Lieb, A. Franke, H. Dommisch, A. Schäfer, Y. Houri-Haddad and F. A. Iraqi, Translation of a mouse model of periodontitis to humans using RNA-sequencing and genetic association studies gives novel insights into the disease aetiology, Sci. Rep., 2020, 10(1), 4892, DOI:10.1038/s41598-020-61819-0.
- N. I. Lorè, B. Sipione, G. He, L. J. Strug, H. J. Atamni, A. Dorman, R. Mott, F. A. Iraqi and A. Bragonzi, Collaborative Cross mice yield genetic modifiers for Pseudomonas aeruginosa, infection in human lung disease, mBio, 2020, 11(2), e00097-20 CrossRef PubMed.
- G. A. Churchill, D. C. Airey and H. Allayee, et al., The Collaborative Cross, a community resource for the genetic analysis of complex traits, Nat. Genet., 2004, 36(11), 1133–1137 CrossRef CAS PubMed.
- T. M. Keane, L. Goodstadt, P. Danecek, M. A. White, K. Wong and B. Yalcin, et al., Mouse genomic variation and its effect on phenotypes and gene regulation, Nature, 2011, 77(7364), 289–294 CrossRef PubMed.
- D. L. Aylor, W. Valdar, W. Foulds-Mathes, R. J. Buus, R. A. Verdugo and R. S. Baric, Genetic analysis of complex traits in the emerging Collaborative Cross, Genome Res., 2011, 21, 1213–1222 CrossRef CAS PubMed.
- C. Durrant, H. Tayem, B. Yalcin, J. Cleak, L. Goodstadt, F. Pardo-Manuel de Villena, R. Mott and F. A. Iraqi, Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection, Genome Res., 2011, 21(8), 1239–1248 CrossRef CAS PubMed.
- G. A. Churchill, Recombinant inbred strain panels: A tool for systems genetics, Physiol. Genomics, 2007, 31, 174–175 CrossRef CAS PubMed.
- H. J. Abu Toamih-Atamni, A. Nashef and F. A. Iraqi, The Collaborative Cross mouse model for dissecting genetic susceptibility to infectious diseases, Mamm. Genome, 2018, 29, 471–487 CrossRef CAS PubMed.
- A. Roberts, F. Pardo-Manuel de Villena, W. Wang, L. McMillan and D. W. Threadgill, The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: Implications for QTL discovery and systems genetics, Mamm. Genome, 2007, 18, 473–481 CrossRef CAS PubMed.
- A. H. Paterson, Molecular dissection of quantitative traits: progress and prospects, Genome Res., 1995, 5, 321–333 CrossRef CAS PubMed.
- F. Iraqi, G. Churchill and R. Mott, The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort, Mamm. Genome, 2008, 19, 379–381 CrossRef PubMed.
- K. Vered, C. Durrant, R. Mott and F. A. Iraqi, Susceptibility to Klebsiella pneumonaie Infection in Collaborative Cross Mice is a Complex Trait Controlled by at Least Three Loci Acting at Different Time Points, BMC Genomics, 2014, 15, 865, DOI:10.1186/1471-2164-15-865.
- F. Iraqi, H. Athamni, A. Dorman, Y. Salymah, I. Tomlinson, A. Nashif and M. Stoller, Heritability and coefficient of genetic variation analysis of phenotypic trait s provide a strong basis for high resolution QTL mapping in. the Collaborative Cross mouse genetic reference population, Mamm. Genome, 2014, 25, 109–119 CrossRef PubMed.
- R. Levy, R. F. Mott, F. A. Iraqi and Y. Gabet, Collaborative cross mice in a genetic association study reveal new candidate genes for bone microarchitecture, BMC Genomics, 2015, 16(1), 1013 CrossRef PubMed.
- A. Dorman, D. Baerr, I. Tomlinson, R. Mott and F. A. Iraqi, Genetic Analysis of Intestinal polyp development in Collaborative Cross mice carrying the ApcMin/+mutation, BMC Genet., 2016, 17, 46 CrossRef PubMed.
- H. J. Atamni, Y. Zinar, R. Mott, L. Wolf and F. A. Iraqi, Glucose tolerance female-specific QTL mapped in collaborative cross mice, Mamm. Genome, 2016c, 1–11 Search PubMed.
- H. J. Atamni, M. Botzman, R. Mott, I. Gat-Viks and F. A. Iraqi, Mapping liver fat female-dependent quantitative trait loci in collaborative cross mice, Mamm. Genome, 2016a, 1–9 Search PubMed.
- H. J. Atamni, R. Mott, M. Soller and F. A. Iraqi, High-fat-diet induced development of increased fasting glucose levels and impaired response to intraperitoneal glucose challenge in the collaborative cross mouse genetic reference population, BMC Genet., 2016a, 17, 10 Search PubMed.
- R. T. Molenhuis, H. Bruining, M. J. V. Brandt, P. E. van Soldt, H. J. Abu-Toamih Atamni, J. Peter H. Burbach, F. A. Iraqi, R. F. Mott and M. J. H. Kas, Modeling the quantitative nature of neurodevelopmental disorders using Collaborative Cross mice, Mol. Autism, 2018, 9, 63, DOI:10.1186/s13229-018-0252-2.
- A. Frishberg, N. Peshes-Yaloz, O. Cohn, D. Rosentul, Y. Steuerman, L. Valadarsky, G. Gal Yankovitz, M. Mandelboim, F. A. Iraqi, I. Amit, L. Mayo, E. Bacharach and I. Gat-Viks, Cell composition analysis of bulk genomics using single cell data, Nat. Methods, 2019, 16, 327–332 CrossRef CAS PubMed.
- H. Yang, Y. Ding, L. N. Hutchins, J. Szatkiewicz, T. A. Bell and B. Paigen, A customized and versatile high-density genotyping array for the mouse, Nat. Methods, 2009, 6, 663–666 CrossRef CAS PubMed.
- Collaborative Cross Consortium, The genome architecture of the Collaborative Cross mouse genetic reference population, Genetics, 2011, 190, 389–401 CrossRef PubMed.
- Y. Wang, M. A. Beydoun and L. Liang, Will all Americans become overweight or obese? Estimating the progression and cost of the U.S. obesity epidemic, Obesity, 2008, 16, 2323–2330 CrossRef PubMed.
- G. Comuzzie and D. B. Allison, The search for human obesity genes, Science, 1998, 280, 1374–1377 CrossRef PubMed.
- H. K. Tiwari and D. B. Allison, Do allelic variants of SLC6A14 predispose to obesity?, J. Clin. Invest., 2003, 112, 1633–1636 CrossRef CAS.
- N. Yi, A. Diament, S. Chiu, K. Kim, D. B. Allison and J. S. Fisler, et al., Characterization of epistasis influencing complex spontaneous obesity in the BSB model, Genetics, 2004, 167, 399–409 CrossRef CAS PubMed.
- M. Soller, H. J. Abu-Toamih Atamni, I. Binenbaum, A. Chatziioannou and F. A. Iraqi, Designing a QTL mapping study for implementation in the realized Collaborative Cross genetic reference population, Curr. Protoc. Mouse Biol., 2019, 9, e66, DOI:10.1002/cpmo.66.
- K. W. Broman, The genome of recombinant inbred lines, Genetics, 2005, 169(2), 133–146 CrossRef PubMed.
- W. K. Broman, M. D. Gatti, P. Simecek, A. N. Furlotte, P. Prins, S. Sen, B. Yandell and A. G. Churchill, R/QTL2: Software for mapping quantitative trait loci with high-dimensional data and multiparental populations, Genetics, 2019, 21, 495–502 CrossRef PubMed.
- G. Lukas, S. D. Brindle and P. Greengard, The route of absorption of intraperitoneally administered compounds, J. Pharmacol. Exp. Ther., 1971, 178, 562–566 CAS.
- M. F. Abu-Hijleh, O. A. Habbal and S. T. Moqattash, The role of the diaphragm in lymphatic absorption from the peritoneal cavity, J. Anat., 1995, 186, 453–467 Search PubMed.
- V. P. Turner, T. Brabb, C. Pekow and A. M. Mary Ann Vasbinder, Administration of Substances to Laboratory Animals: Routes of Administration and Factors to Consider, J. Am. Assoc. Lab. Anim. Sci., 2011, 50(5), 600–613 Search PubMed.
- D. Zafriri, I. Ofek, R. A., M. Pocino and N. Sharon, Inhibitory activity of cranberry juice on adherence of type 1 and type P fimbriated Escherichia coli to eukaryotic cells, Antimicrob. Agents Chemother., 1989, 33, 92–98 CrossRef CAS PubMed.
- C. C. Neto, K. A. Penndorf, M. Feldman, S. Meron-Sudai, Z. Zakay-Rones, D. Steinberg, M. Fridman, Y. Kashman, I. Ginsburg, I. Ofek and E. I. Weiss, Characterization of non-dialyzable constituents from cranberry juice that inhibits adhesion, co-aggregation and biofilm formation by oral bacteria, Food Funct., 2017, 8, 1955–1965 RSC.
- F. F. Anhê, T. R. Nachbar, V. T. Varin, V. Vilela, S. Dudonne, G. Polon, M. Fourinier, M. A. Lecours, Y. Desjardins, D. Roy, E. Levy and A. Marette, A polyphenol-rich cranberry extract reverses insulin resistance and hepatic steatosis independently of body weight loss, Mol. Metab., 2017, 6, 1563–1573 CrossRef PubMed.
- J. B. Blumberg, T. A. Camesano, A. Cassidy, P. Kris-Etherton, A. Howell, C. Manach, L. M. Ostertag, H. Sies, A. Skulas-Ray and J. A. Vita, Cranberries and Their Bioactive Constituents in Human Health, Adv. Nutr., 2013, 4(6), 618–632 CrossRef CAS PubMed.
- J. B. Blumberg, T. A. Camesano, A. Cassidy, P. Kris-Etherton, A. Howell, C. Manach, L. M. Ostertag, H. Sies, A. Skulas-Ray and J. A. Vita, Cranberries and Their Bioactive Constituents in Human Health, Adv. Nutr., 2013, 4(6), 618–632 CrossRef CAS PubMed.
- P. Canal, Y. Plusquellec, E. Chatelut, R. Bugat, J. De Biasi and G. Houin, A pharmacokinetic model for intraperitoneal administration of drugs: application to teniposide in humans, J. Pharm. Sci., 1989, 78(5), 389–392 CrossRef CAS PubMed.
- G. Lukas, S. D. Brindle and P. Greengard, The route of absorption of intraperitoneally administered compounds, J. Pharmacol. Exp. Ther., 1971, 178(3), 562–564 CAS.
- R. J. Church, D. M. Gatti, T. J. Urban, N. Long, X. Yang, Q. Shi, J. S. Eaddy, M. Mosedale, S. Ballard, G. A. Churchill, V. Navarro, P. B. Watkins, D. W. Threadgill and A. H. Harrill, Sensitivity to hepatotoxicity due to epigallocatechin gallate is affected by genetic background in diversity outbred mice, Food Chem. Toxicol., 2015, 76, 19–26 CrossRef CAS PubMed.
- N. Hochman, Y. Houri-Haddad, J. Koblinski, L. Wahl, M. Roniger, A. Bar-Sinai, E. I. Weiss and J. Hochman, Cranberry juice constituents impair lymphoma growth and augment the generation of antilymphoma antibodies in syngeneic mice, Nutr. Cancer, 2008, 60(4), 511–517 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2021 |