Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mycoprotein ingredient structure reduces lipolysis and binds bile salts during simulated gastrointestinal digestion
Raffaele
Colosimo
 a,
Ana-Isabel
Mulet-Cabero
a,
Ana-Isabel
Mulet-Cabero
 a,
Frederick J.
Warren
a,
Cathrina H.
Edwards
a,
Frederick J.
Warren
a,
Cathrina H.
Edwards
 a,
Tim J. A.
Finnigan
b and
Pete J.
Wilde
a,
Tim J. A.
Finnigan
b and
Pete J.
Wilde
 *a
*a
aQuadram Institute Bioscience, Norwich Research Park, Norwich, Norfolk NR4 7UQ, UK. E-mail: pete.wilde@quadram.ac.uk
bMarlow Foods Ltd, Station Road, Stokesley, North Yorkshire TS9 7AB, UK
First published on 26th November 2020
Abstract
Mycoprotein is the fungal biomass obtained by the fermentation of Fusarium venenatum, whose intake has been shown to lower blood lipid levels. This in vitro study aimed to understand the mechanisms whereby mycoprotein can influence lipid digestion by reducing lipolysis and binding to bile salts. Mycoprotein at 30 mg mL−1 concentration significantly reduced lipolysis after 60 min of simulated intestinal digestion with oil-in-water emulsion (P < 0.001) or 10 min of incubation with tributyrin (P < 0.01). Furthermore, mycoprotein effectively bound bile salts during simulated small intestinal digestion, but only after being exposed to the acidic environment of the preceding gastric phase. However, the extent of bile salts sequestered by mycoprotein was decreased by pepsin and lipase–colipase activity. Besides, extracted mycoprotein proteins showed bile salt binding activity, and proteins with a molecular weight of ∼37 kDa showed resistance to trypsin hydrolysis. Thus, eleven extracted mycoprotein proteins (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 kDa) were identified by liquid chromatography-tandem mass spectrometry. In addition, the viscosity of mycoprotein digesta appeared to have no impact on bile salt binding since no statistically significant differences were detected between samples exposed or not to the previous gastric step. This study has identified mechanisms by which mycoprotein can reduce blood lipid levels.
37 kDa) were identified by liquid chromatography-tandem mass spectrometry. In addition, the viscosity of mycoprotein digesta appeared to have no impact on bile salt binding since no statistically significant differences were detected between samples exposed or not to the previous gastric step. This study has identified mechanisms by which mycoprotein can reduce blood lipid levels.
1. Introduction
Mycoprotein (MYC) is the common and major ingredient of all Quorn™ meat-free food products. This food ingredient comprises the whole fungal biomass obtained by the continuous fermentation and processing of the filamentous fungus Fusarium venenatum A3/5 (ATCC PTA-2684). The production of MYC includes RNA content reduction, from 10% to <![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% on a dry weight basis (DW), by heating treatment (>
2% on a dry weight basis (DW), by heating treatment (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68 °C) which triggers the action of endogenous RNAse enzymes. This step is followed by heating at 90 °C, centrifugation, and freezing.1 MYC is rich in fibre (24% DW, of which approximately 2/3 are β-glucans and 1/3 is chitin) and protein (44% DW), while it is low in digestible carbohydrate (12% DW mainly glycogen) and fat (11.6% DW).2
68 °C) which triggers the action of endogenous RNAse enzymes. This step is followed by heating at 90 °C, centrifugation, and freezing.1 MYC is rich in fibre (24% DW, of which approximately 2/3 are β-glucans and 1/3 is chitin) and protein (44% DW), while it is low in digestible carbohydrate (12% DW mainly glycogen) and fat (11.6% DW).2
Some types of meats have been associated with the risk of cardiovascular diseases.3 In contrast, plant-based products, as well as edible mushrooms, being rich in fibre and low in saturated fat,4–6 could help in the prevention of coronary heart disease and stroke by reducing blood lipid levels.7,8 There is now accumulating evidence that MYC-based products can elicit similar beneficial effects on blood lipids. For instance, a study by Turnbull et al.9 reported improvements in blood lipid profiles (e.g., lower total cholesterol and low-density lipoprotein cholesterol (LDL) and higher high-density lipoprotein cholesterol (HDL) levels) in participants with slightly raised cholesterol levels following a MYC enriched-diet (three weeks) when compared to a meat-based diet (type not disclosed). A follow-up study on participants with slightly elevated cholesterol levels showed that eight-week supplementation of MYC-enriched cookies reduced total cholesterol and LDL cholesterol levels when compared with nutritionally balanced cookies without MYC added.10 Another study in healthy participants showed that total cholesterol and LDL cholesterol levels were reduced after taking a diet rich in Quorn™ products for six weeks. In contrast, no change was seen in the control group that followed their habitual diet.11 Overall, these studies suggest that MYC-based diets may have similar effects to plant-based diets on blood lipid profiles and may, therefore, have a role in reducing cardiovascular disease risk.
It is also essential to understand the underlying mechanisms by which these dietary components influence lipid markers. From a mechanistic point of view, the blood lipid-lowering effect could be mediated by different biological and physicochemical mechanisms.12 The inhibition of lipase activity and the consequent delay in lipid digestion is considered an important mechanism that can positively influence post-prandial hyperlipidaemia.13 Similarly, the binding of bile salts (BS) by fibre14–16 promotes the faecal excretion of BS and, consequently, the alteration of the bile acid pool that forces the liver to produce more BS from endogenous cholesterol.17 In the same manner, protein from different sources (e.g., milk, soybean seeds, corn, cumin, and lupin) has been reported to bind BS.18–22 The hydrophobicity of protein subunits or amino acids has been linked to BS sequestering activity.18,20,23 Furthermore, BS binding could also be influenced by increased viscosity promoted by dietary fibre24 depending on its composition16 or solubility.25
Considering the various mechanisms proposed above, the aim of this in vitro study was to investigate the physicochemical processes which occur during the digestion of MYC in the gastrointestinal (GI) tract, which may underlie the blood lipid-lowering effect observed after MYC consumption in vivo. Thus, we hypothesised that MYC and its components (e.g., fibre and protein) might reduce lipid digestion by reducing lipase activity, sequestering BS from solution and increasing viscosity during simulated digestion in the human GI tract. To test these hypotheses, an oil in water (O/W) emulsion or a tributyrin (TBT) control was subjected to in vitro digestion in the presence of MYC to understand how MYC affects lipolysis rates during simulated small intestinal digestion. In addition, BS were incubated with different MYC concentrations or extracted MYC proteins (EMPs) to determine BS sequestration levels from isolated supernatants collected after simulated digestion. As BS binding could also be influenced by the increased viscosity promoted by dietary fibre, we also measured the viscosity of MYC digesta to assess its contribution towards lipid digestion and BS binding. This study provides new insight into the digestive fate of MYC, which is relevant to understanding the role of MYC-enriched diets in improving blood lipid profiles and thereby potentially lowering cardiovascular disease risk.
2. Materials & methods
2.1 Materials
2.2 Methods
2.2.1.1 O/W emulsion preparation. An O/W emulsion was prepared by pre-emulsifying 0.4 g of sunflower oil in 9.6 g of whey protein isolate (WPI) solution (1% w/v in water) and stirred for 120 min at room temperature using an Ultra-Turrax T-25 homogeniser (IKA, Staufen, Germany) for 1 min at 13
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm. A WPI-stabilised emulsion was used as it is known to be readily digested during in vitro GI digestion. The pre-emulsion was then sonicated (Branson Digital Sonifier®, Marshall Scientific, New Hampshire, USA) for 2 min at 30% amplitude with 10 s sonication and 10 s of rest cycles. The particle size of the O/W emulsion was measured by static light scattering with a Beckman Coulter LS 13-320 instrument (Beckman Coulter, California, USA), ensuring a mean particle diameter (D4,3) within the range of 1.5–2.5 μm.
500 rpm. A WPI-stabilised emulsion was used as it is known to be readily digested during in vitro GI digestion. The pre-emulsion was then sonicated (Branson Digital Sonifier®, Marshall Scientific, New Hampshire, USA) for 2 min at 30% amplitude with 10 s sonication and 10 s of rest cycles. The particle size of the O/W emulsion was measured by static light scattering with a Beckman Coulter LS 13-320 instrument (Beckman Coulter, California, USA), ensuring a mean particle diameter (D4,3) within the range of 1.5–2.5 μm.
2.2.1.2 Lipolysis during simulated small intestinal digestion. The lipolysis of O/W emulsion in the presence of MYC was measured using a pH-stat device (KEM AT-700, Kyoto electronics, Tokyo, Japan). Briefly, 2.5 mL of MYC at different concentrations (10, 20, or 30 mg mL−1 in the final digesta) was placed in a jacketed reaction vessel warmed to 37 °C. The final composition in the reaction system was 300 mg lipid from the O/W emulsion, 10 mM bile salts, and 100 U mL−1 pancreatin (based on lipase activity) in a final volume of 20 mL. The volumes of simulated intestinal fluid (SIF) and CaCl2·2H2O were added to obtain the concentration of the electrolytes, according to Minekus et al.26 However, in the composition of the SIF, the solution of NaHCO3 was replaced by NaCl in order to avoid CO2 formation, which would have introduced undesirable pH changes as described by Mat et al.27 The pH was first adjusted at 7.0 and then maintained at this value by constant titration with 0.1 M NaOH for 60 min of simulated digestion and continuous stirring (500 rpm). Lipolysis was expressed as volume (mL) of NaOH 0.1 M necessary to maintain pH 7.0 against the decrease of pH promoted by the release of free fatty acids during lipolysis.
To test the effect of MYC on intrinsic lipase activity, TBT was used as a substrate due to its high efficiency to be hydrolysed in the absence of BS. Therefore, 0.5 mL of TBT was digested as described in the pancreatic lipase activity assay by the INFOGEST method26 in the presence or absence of MYC at 30 mg mL−1 in the final volume. The pH was first adjusted at 8.0 and then maintained at this value by constant titration with 0.1 M NaOH for 10 min with constant stirring (500 rpm). Lipolysis was expressed as volume (mL) of NaOH 0.1 M necessary to maintain pH 8.0 against the decrease of pH promoted by the release of free fatty acids during lipolysis.
2.2.2.1 Simulated static GI or only intestinal digestion. GI digestion (120 min of gastric phase followed by 120 min of small intestinal step) or only small intestinal digestion (referred to as I) for 120 min was performed on MYC (10, 20, and 30 mg mL−1) or EMPs (8.8 mg mL−1) according to the INFOGEST standardised method for in vitro food digestion.26
After simulated digestion, MYC digesta (20 mL) was centrifuged (Heraeus Megafuge 16 centrifuge, Hanau, Germany) at 2500 rpm for 5 min at room temperature. In contrast, only 1 mL of EMP digesta was centrifuged (Eppendorf® centrifuge 5424-R, Eppendorf, Hamburg, Germany) under the same conditions as the MYC. The resulting supernatant was collected and used for BS quantification.
2.2.2.2 BS quantification. Bile salt concentration was determined by a colourimetric enzymatic kinetic assay using a total bile acids kit (Dialab, Wiener Neudorf, Austria). Briefly, the supernatants from the in vitro digestion samples (section 2.2.2.1) were diluted 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 with NaCl 0.9%. Then, 5 μL of the sample, blank (NaCl 0.9%), or standard (sodium deoxycholate) were transferred into a 96-well microplate and incubated for 5 min at 37 °C with 240 μL of reagent 1 (buffer, Thio-NAD (>
500 with NaCl 0.9%. Then, 5 μL of the sample, blank (NaCl 0.9%), or standard (sodium deoxycholate) were transferred into a 96-well microplate and incubated for 5 min at 37 °C with 240 μL of reagent 1 (buffer, Thio-NAD (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 mM)). Subsequently, 80 μL of reagent 2 (buffer, 3-α-hydroxysteroid dehydrogenase (>
0.1 mM)). Subsequently, 80 μL of reagent 2 (buffer, 3-α-hydroxysteroid dehydrogenase (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 kU L−1), and NADH (>
2 kU L−1), and NADH (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 mM)) were added to start the reaction. The absorbance was read at 405 nm every 30 s for 19.5 min (40 cycles) with a UV-vis plate reader (Molecular Devices, LLC VersaMax, California, United States). The values were reported as the bile salt concentration (mM) using a calibration curve of a BS standard and subtracting the absorbance from the blank.
0.1 mM)) were added to start the reaction. The absorbance was read at 405 nm every 30 s for 19.5 min (40 cycles) with a UV-vis plate reader (Molecular Devices, LLC VersaMax, California, United States). The values were reported as the bile salt concentration (mM) using a calibration curve of a BS standard and subtracting the absorbance from the blank.
2.2.3.1 SDS-PAGE. The proteins from digested GI samples from MYC and EMPs were separated by SDS-PAGE. Briefly, a sample aliquot of 65 μL was mixed with 25 μL NuPAGE LDS sample buffer and 10 μL reducing agent and heated for 10 min at 70 °C. Then, aliquots of 15 μL of the reduced samples were loaded onto pre-cast 10% bis-tris gels along with 10 μL pre-stained marker. Electrophoresis of the gels was performed for 35 min at 200 V and 350 mA in MES buffer. The gels were incubated overnight with a protein stain (InstantBlue™), washed three times with ultra-pure water to remove excess dye, and finally scanned with a Bio-Rad ChemiDoc MP imaging system (Bio-Rad Laboratories Ltd, Hertfordshire, UK). Images were acquired using analysis software Image Lab version 6.0.1.
2.2.3.2 Preparation of gel slices for LC-MS/MS. In a separate experiment, EMPs underwent GI digestion with pepsin (120 min) and trypsin (120 min) or only intestinal digestion with trypsin (120 min). Then, SDS-PAGE was performed on those samples as previously described. Protein bands that were resistant to trypsin digestion were observed, and gel slices in these bands were cut and prepared, as described below, for LC-MS/MS analysis, which was done at the proteomic facility at the John Innes Centre (Norwich, UK). For gel slice preparation, washing steps were performed for 20 min each (unless stated otherwise) with strong vortex mixing using 1 mL of solvent: gel slices were de-stained with 30% ethanol for 30 min at 65 °C until they were clear, and then washed with NH4HCO3 (50 mM) prepared in 50% acetonitrile (ACN) (NH4HCO3/50%ACN) and incubated with dithiothreitol (DTT) (10 mM) at 55 °C for 30 min. DTT was removed, and an iodoacetamide (IAA) solution (30 mM) was added for another 30 min incubation by vortex mixing at room temperature in the dark. The IAA solution was removed, and the gel slices were washed with NH4HCO3/50%ACN by vortex mixing. Another wash with NH4HCO3 was performed to remove the buffer, and gel slices were transferred individually into a Petri dish for cutting. The volume of each gel slice was measured/estimated before using a scalpel to cut the gel slices into smaller pieces (approximately 1 × 1 mm). The pieces were transferred to a clean tube and washed twice with NH4HCO3/50%ACN and 100% ACN. Finally, the solvent was completely removed by spinning the samples in an Eppendorf concentrator plus a vacuum dry centrifuge (Eppendorf Ltd, Hamburg, Germany) for 30 min. The precipitated and dried gel slice residues were collected and sent for LC-MS/MS analysis.
2.2.3.3 LC-MS/MS analysis. The dried gel slice residuals were digested with chymotrypsin (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 enzyme
20 enzyme![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein) for 8 h at room temperature and pH 7.5. LC-MS/MS analysis was performed with an Orbitrap Eclipse Tribrid Mass Spectrometer (Thermo Fisher Scientific) and a nanoflow high-performance liquid chromatography (HPLC) system (Dionex Ultimate3000, Thermo Fisher Scientific). Other settings were as follows: flow 0.2 μL min−1, solvent A: 0.1% trifluoroacetic acid, solvent B: 80% acetonitrile in 0.1% trifluoroacetic acid; total running time of 118 min (multi-step gradient, including wash and re-equilibration), an ion source voltage of 2300 V (positive mode), operated in the DDA mode, MS1 resolution of 120
protein) for 8 h at room temperature and pH 7.5. LC-MS/MS analysis was performed with an Orbitrap Eclipse Tribrid Mass Spectrometer (Thermo Fisher Scientific) and a nanoflow high-performance liquid chromatography (HPLC) system (Dionex Ultimate3000, Thermo Fisher Scientific). Other settings were as follows: flow 0.2 μL min−1, solvent A: 0.1% trifluoroacetic acid, solvent B: 80% acetonitrile in 0.1% trifluoroacetic acid; total running time of 118 min (multi-step gradient, including wash and re-equilibration), an ion source voltage of 2300 V (positive mode), operated in the DDA mode, MS1 resolution of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and range 300–1800 m/z, RF = 30%, maximum injection time of 50 ms, dynamic exclusion for 15 seconds. MS2 with CID and HCD fragmentation (ion trap, turbo mode), isolation window of 1.6 Da, and collision energy of 20%, maximum injection time of 35 ms. Protein identification was carried out using Scaffold™ software V4.10.0 (Proteome Software Inc, Oregon, USA) and UniProt database.
000 and range 300–1800 m/z, RF = 30%, maximum injection time of 50 ms, dynamic exclusion for 15 seconds. MS2 with CID and HCD fragmentation (ion trap, turbo mode), isolation window of 1.6 Da, and collision energy of 20%, maximum injection time of 35 ms. Protein identification was carried out using Scaffold™ software V4.10.0 (Proteome Software Inc, Oregon, USA) and UniProt database.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 (deg
21 (deg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min
min![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sec). The rheological protocol was set using software Rheology Advantage Instrument Control AR V5.8.2 as follows: continuous ramp from a shear rate of 10 to 1000 1 s−1 on a log scale, for 2 min at 37 °C, 10 points per decade; sample density was estimated as 1.000 g cm−3.
sec). The rheological protocol was set using software Rheology Advantage Instrument Control AR V5.8.2 as follows: continuous ramp from a shear rate of 10 to 1000 1 s−1 on a log scale, for 2 min at 37 °C, 10 points per decade; sample density was estimated as 1.000 g cm−3.
3. Results and discussion
3.1 Lipolysis reduction of O/W in the presence of MYC
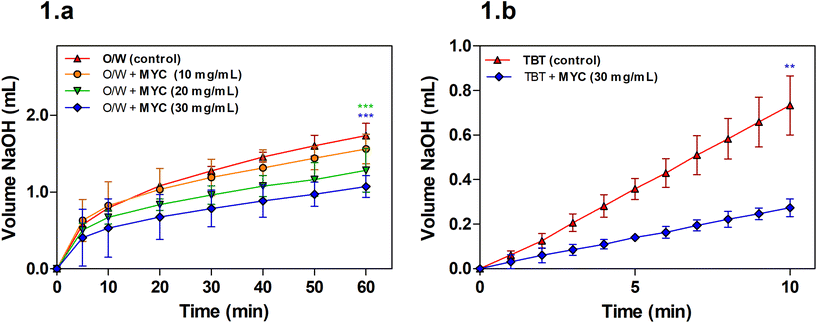
Lipolysis during simulated small intestinal digestion was measured for the O/W emulsion alone (control) or the O/W emulsion in the presence of increasing MYC concentration (10, 20, and 30 mg mL−1) in order to determine the impact of MYC on lipolysis mediated by pancreatic lipase.A concentration of 10 mg mL−1 of MYC was not sufficient to promote a statistically significant decrease in the volume of NaOH required when compared to the control (O/W) at the endpoint of the digestion experiment (60 min). The lipolysis of MYC at 20 and 30 mg mL−1 was statistically lower (P < 0.001) after 60 min of digestion when compared to the control. Visual checks using light microscopy confirmed that MYC did not induce any coalescence or flocculation in the emulsion, which may have interfered with lipid digestion (results not shown). Fig. 1b shows that the lipolysis of TBT in the presence of MYC 30 mg mL−1 was reduced (P < 0.01). This implied that the impact of MYC on lipolysis was mediated by the direct interference with the activity of lipase, as the TBT assay is performed in the absence of BS, sunflower oil emulsion. The reduction in lipolysis mediated by MYC would reduce the rate of lipid digestion and absorption in the small intestine, and could, therefore, be linked with the blood lipid-lowering effect of MYC observed in vivo.9–11 This may be beneficial for the prevention of cardiovascular disease.28 Dietary fibres have been reported to reduce lipase activity,29,30 but the mechanisms that cause the reduction in lipolysis rates are not clear. A possible explanation is the entrapment of the enzyme lipase into the MYC matrix, similar to what we have previously reported during in vitro digestion in the presence of α-amylase.31 The latter remained entrapped within MYC cell walls, and consequently, less starch hydrolysis was observed by removing the enzyme from the solution. Likewise, the entrapment of lipase within the cell wall matrix may result in lower lipolysis. Moreover, the structure of filamentous fungal cells and cell walls, composed of β-glucans/chitin, can have a strong impact on reducing lipolysis. For instance, dietary fibre such as barley β-glucan has been seen to interact differently with lipid droplets depending on the emulsifier type and molecular weight.32
3.2 BS binding by MYC
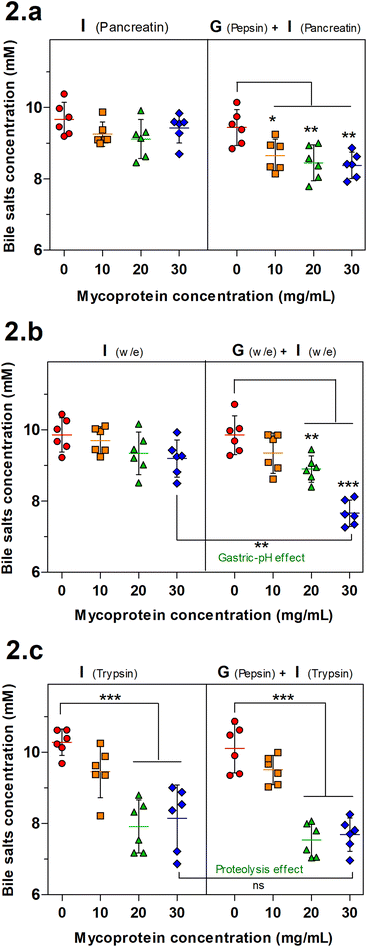
BS binding is another mechanism associated with reduced lipolysis. The BS binding by MYC was determined by both gastric and intestinal (GI) or only intestinal simulation of human digestion that was performed with enzymes (Fig. 2a), without enzymes (Fig. 2b) or with individual enzymes (only trypsin for the intestinal step). BS binding by plant fibre,14,33 and edible mushrooms6 has been reported in the literature. Therefore, a similar effect was expected to be observed with MYC as the mechanism behind the blood lipid-lowering effect reported by in vivo studies.9–11Fig. 2a shows that just the intestinal step was not sufficient to promote significant binding of BS whereas adding the previous gastric incubation facilitated the binding (gastric-pH effect) with a statistically significant decrease of the BS concentration in all the MYC concentrations (P < 0.05 in MYC 10 mg mL−1; P < 0.01 in MYC 20 and 30 mg mL−1). In order to further investigate the limiting factors of the gastric-pH effect, the same previous experimental conditions were adapted in simulated digestion without enzymes (Fig. 2b). Similarly, the gastric plus intestinal digestion of 30 mg mL−1 MYC without enzymes promoted a significant BS binding effect (P < 0.01) when compared to its counterpart incubated without a gastric step. This confirmed the above-mentioned gastric pH effect promoted by the change of pH from the gastric phase (pH 3.0) to the intestinal phase (pH 7.0). The change of pH from acid to neutral may have enhanced the interactions between MYC cell walls and BS. It has been suggested that a chemical alteration of fibres that alters their functionality may occur when exposed to an acidic environment (pH 3.0) followed by washing to neutrality (pH 7.0).34,35 Nevertheless, the gastric pH effect enhanced the BS sequestering effect that is considered crucial in lowering blood lipid levels since the binding, and consequent faecal excretion of BS enhances the hepatic production of BS from endogenous cholesterol, and the expression of the LDL-C receptor to increase lipoprotein uptake from the blood circulation.17,36
Fig. 2c shows the BS binding to MYC when only trypsin is used in the intestinal step. The results of GI digestion were consistent with the gastric pH effect. The highest concentrations of MYC (20 and 30 mg mL−1) were able to promote a statistically significant BS binding effect (P < 0.001). Nevertheless, the intestinal digestion alone showed a significant decrease in BS concentration. Hence, excluding the necessity for the gastric step, this finding suggested that the hydrolytic activity of trypsin towards protein associated with MYC cells could somehow have altered the existing protein/peptides to enhance their BS binding activity (proteolysis effect). In line with this, previous studies have reported the presence of proteins with BS binding properties,18–22,37 and this will be discussed in the next section.
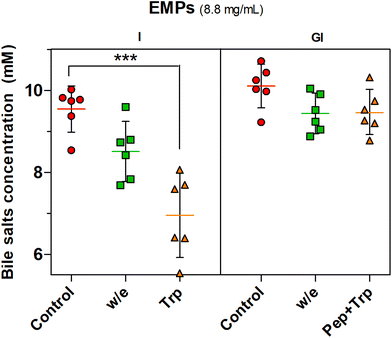
The proteolysis effect of trypsin on BS binding was confirmed by the statistically significant effect (P < 0.001) observed with trypsin digestion (Trp) when only intestinal digestion was performed while the incubation without enzyme (w/e) had no effect. The gastrointestinal incubation without enzyme (w/e) did not promote any BS binding effect with EMPs compared to MYC (Fig. 2). This supported once more the previously described gastric pH effect observation, which suggested that BS binding was promoted by the previous gastric acid incubation that altered the cell wall fibre matrix in the whole MYC. In contrast, gastric pH had no impact on BS binding by EMPs due to the absence of cell walls, thus working as a negative control. Conversely, the previous incubation of EMPs with pepsin followed by trypsin digestion (Pep + Trp) diminished the BS binding effect. It can be suggested that pepsin hydrolysis could reduce the binding between BS and protein by breaking down the primary protein structure38 and, therefore, potentially affecting its binding functionality.
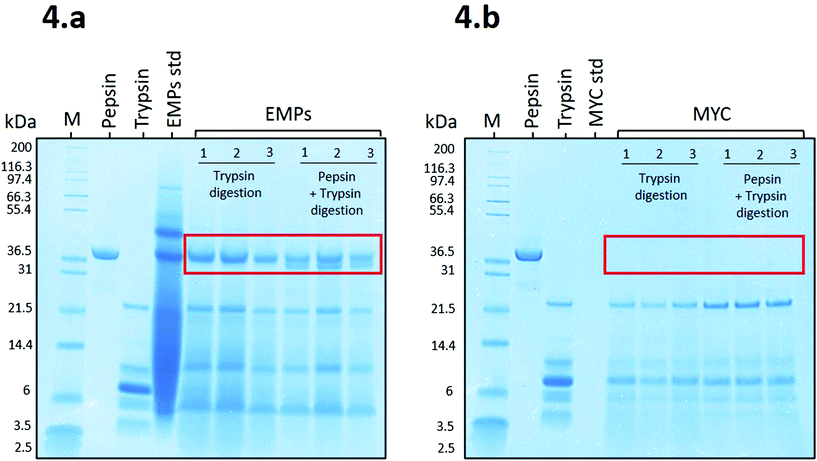
Fig. 4a shows that the proteins belonging to EMPs (EMPs std) appeared as a smeared band, apart from the identifiable protein bands from ∼37 kDa to ∼97.4 kDa, due to a high initial concentration (8.8 mg mL−1). Despite the high concentration, EMPs were effectively digested during the simulated digestion apart from a resistant protein band observed at ∼37 kDa during digestion with trypsin or pepsin plus trypsin. The latter showed a new protein band that appeared to belong to pepsin. Similar to our results, Makino et al.19 reported a protein band protein at ∼37 kDa from soybean that, when in contact with BS anions, was able to modify its secondary structure and bind to BS.
Fig. 4b shows the SDS-PAGE profile related to MYC digestions. In this case, there is no evidence of the presence of resistant proteins, and the only bands recognisable appear to belong to trypsin (from ∼21.5 to 3.5 kDa). This might be due to the presence of fungal cell walls that could limit the release of the intracellular proteins into the extracellular solution and, and thus are not detected on the SDS-PAGE gel. This is supported by a recent in vitro study on MYC, which suggested that the fungal cell wall is permeable to proteases that can diffuse into the cell and hydrolyse the intracellular proteins, which are subsequently released as small peptides or amino acids.39 Similarly, the diffusion of the enzyme α-amylase through the fungal cell wall was also reported.31 We excluded the contribution of residual extracellular proteases from the fungal biomass of MYC in protein hydrolysis. Indeed, the heating treatment for reducing the RNA content (>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68 °C followed by an increase to 90 °C), and subsequent centrifugation, would have been sufficient for denaturing and/or removing most of the extracellular proteases. Furthermore, pepsin was not detected in Fig. 4b. This could be explained by the diffusion of pepsin through the cell wall and the resulting entrapment of the enzyme that was depleted from the solution. Alternatively, trypsin may have digested pepsin when incubated with MYC (the low bioaccessible protein that is encapsulated into cell walls; see MYC standard of Fig. 4b) whereas pepsin remained undigested in the presence of a highly bioaccessible substrate in the EMP digestion (no cell walls; see EMPs std of Fig. 4a).
68 °C followed by an increase to 90 °C), and subsequent centrifugation, would have been sufficient for denaturing and/or removing most of the extracellular proteases. Furthermore, pepsin was not detected in Fig. 4b. This could be explained by the diffusion of pepsin through the cell wall and the resulting entrapment of the enzyme that was depleted from the solution. Alternatively, trypsin may have digested pepsin when incubated with MYC (the low bioaccessible protein that is encapsulated into cell walls; see MYC standard of Fig. 4b) whereas pepsin remained undigested in the presence of a highly bioaccessible substrate in the EMP digestion (no cell walls; see EMPs std of Fig. 4a).
The EMP gel slices in the region of ∼37 kDa digested with trypsin (A in Table 1) and pepsin plus trypsin (B in Table 1) were further investigated by LC-MS/MS. This analysis was performed to address the question of which proteins/peptides were resistant to trypsin digestion and responsible for the BS binding. Proteins and peptides which are resistant to digestion are essential for various physiological outcomes, including nutrient signalling and allergenicity. The identification of these peptides could be crucial for future investigation of their specific functionality, including BS binding activity, and can open the way to other studies, including clinical trials for the development of nutritional supplements with blood-lipid lowering activity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 kDa
37 kDa
| No. | Accession number | Protein | Molecular weight (kDa) | Average ± SD total spectra protein from A (%) | Average ± SD total spectra protein from B (%) |
|---|---|---|---|---|---|
| A refers to EMP trypsin digestion, B refers to EMP pepsin + trypsin digestion. Unpaired t-test with Welch's correction (two-tailed P < 0.05); no statistically significant differences found in the same protein between A and B. Values represent the average ± SD of 3 replicates unless stated (†n = 2). | |||||
| 1 | CEI63049 | Uncharacterised protein | 38 | 0.034 ± 0.018 | 0.039 ± 0.006† |
| 2 | CEI70330 | Uncharacterised protein | 39 | 0.022 ± 0.012 | 0.026 ± 0.010 |
| 3 | CEI38487 | AB hydrolase-1 domain-containing protein | 40 | 0.039 ± 0.019 | 0.012 ± 0.003 |
| 4 | CEI67957 | Uncharacterised protein | 42 | 0.004 ± 0.001† | 0.004 ± 0.001† |
| 5 | CEI66110 | Uncharacterised protein | 45 | 0.056 ± 0.037 | 0.010 ± 0.013 |
| 6 | CEI65138 | Uncharacterised protein | 47 | 0.005 ± 0.005 | 0.009 ± 0.008 |
| 7 | CEI66021 | Peptidase_M14 domain containing protein | 49 | 0.092 ± 0.042 | 0.081 ± 0.014 |
| 8 | CEI61026 | Elongation factor 1-alpha | 51 | 0.006 ± 0.003 | 0.008 ± 0.002 |
| 9 | CEI63178 | Uncharacterised protein | 93 | 0.022 ± 0.013 | 0.022 ± 0.011 |
| 10 | CEI64213 | Uncharacterised protein | 117 | 0.007 ± 0.006 | 0.009 ± 0.005 |
| 11 | CEI64538 | C2H2-type domain-containing protein | 125 | 0.017 ± 0.008† | 0.018 ± 0.002 |
Table 1 shows a list of the 11 proteins that were identified from the SDS-PAGE gel slices (Fig. 4a) with molecular weight >![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 kDa that could have been involved in the binding of bile salts. The raw LC-MS/MS data reported a list of 104 proteins, among which 11 potential proteins were selected after screening and removal of proteins with molecular weight <
37 kDa that could have been involved in the binding of bile salts. The raw LC-MS/MS data reported a list of 104 proteins, among which 11 potential proteins were selected after screening and removal of proteins with molecular weight <![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 kDa, protein contaminants, trypsin-derived peptides, protein structure decoy, and proteins which failed to be identified in at least two replicates. Proteins with a molecular weight of 38, 39, and 40 kDa match the molecular weight of ∼37 kDa observed in the SDS-PAGE gel (see Fig. 4a). However, proteins with a molecular weight >
37 kDa, protein contaminants, trypsin-derived peptides, protein structure decoy, and proteins which failed to be identified in at least two replicates. Proteins with a molecular weight of 38, 39, and 40 kDa match the molecular weight of ∼37 kDa observed in the SDS-PAGE gel (see Fig. 4a). However, proteins with a molecular weight >![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 kDa were also listed since high-molecular-weight proteins may have been hydrolysed into smaller protein fractions at ∼37 kDa during the extraction protocol of the EMPs from MYC by the manufacturer (information not available). Similarly, the trypsin hydrolysis of EMPs with high molecular weights (e.g., 97.4 kDa) could have produced protein fractions in the region of ∼37 kDa (see EMPs std in Fig. 4a). AB hydrolase-1 domain-containing protein (number 3 in Table 1), peptidase_M14 domain-containing protein (number 7 in Table 1), elongation factor 1-alpha (number 8 in Table 1) and C2H2-type domain-containing protein (number 11 in Table 1) were identified whereas other proteins remain uncharacterised. However, none of the listed proteins has been reported in the literature for their BS binding capacity. Hence, further research is required to test the BS binding activity of the listed proteins (Table 1).
40 kDa were also listed since high-molecular-weight proteins may have been hydrolysed into smaller protein fractions at ∼37 kDa during the extraction protocol of the EMPs from MYC by the manufacturer (information not available). Similarly, the trypsin hydrolysis of EMPs with high molecular weights (e.g., 97.4 kDa) could have produced protein fractions in the region of ∼37 kDa (see EMPs std in Fig. 4a). AB hydrolase-1 domain-containing protein (number 3 in Table 1), peptidase_M14 domain-containing protein (number 7 in Table 1), elongation factor 1-alpha (number 8 in Table 1) and C2H2-type domain-containing protein (number 11 in Table 1) were identified whereas other proteins remain uncharacterised. However, none of the listed proteins has been reported in the literature for their BS binding capacity. Hence, further research is required to test the BS binding activity of the listed proteins (Table 1).
The proteolysis effect (Fig. 2c) was observed only when the intestinal phase digestion with the individual enzyme trypsin was performed while the intestinal digestion using pancreatin (mix of proteases, lipase–colipase, and amylase) was not able to promote a similar effect (Fig. 2a). Therefore, the presence of lipase–colipase that digested lipids (lipolysis effect) could have been a limiting factor that diminished the proteolysis effect. Potentially, the lipid component of MYC (12% DW, 3.6 mg mL−1) was digested by lipase–colipase with the help of the surfactant action of BS, which could potentially enter the fungal cell wall. Then, BS solubilised the products of lipolysis to form mixed BS micelles that were detected in the supernatant, regardless of the proteolysis effect promoted by trypsin. This hypothesis was tested and supported by the performance of just the intestinal digestion of MYC (30 mg mL−1) in the presence of trypsin plus lipase–colipase (Trp/Lip–Colip). Fig. 5 illustrates a reversal of the proteolysis effect when lipolysis took place (e.g., the presence of lipase and colipase), similar to what was observed using pancreatin (Fig. 2a).
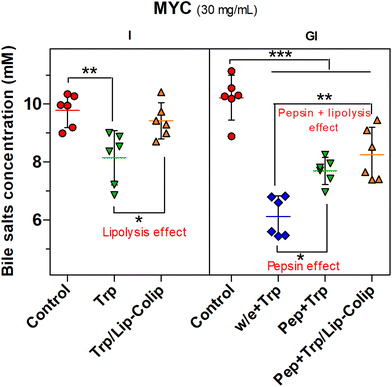
Furthermore, Fig. 5 shows the negative effect of pepsin on BS binding (pepsin effect). Pepsin plus trypsin (Pep + Trp) digestion in MYC (30 mg mL−1) significantly reduced (P < 0.05) the BS binding compared to the gastric step without enzyme (w/e) that was followed by trypsin digestion (w/e + Trp). Besides, the BS binding effect was further decreased (P < 0.01) by the combination of the pepsin and lipolysis effects (Pep + Trp /Lip–Colip), both confirming the negative impact on BS binding.
Therefore, in summary, pepsin and lipase digestion can hinder BS binding that is mainly promoted by the MYC structure per cell wall component as confirmed by the effect of pH change in the absence of digestive enzymes (Fig. 2b), and also by the protein component, as evidenced by the role of the digestive proteases on EMPs (Fig. 3 and 5).
3.3 Effect of digestion conditions on viscosity and particle size of MYC
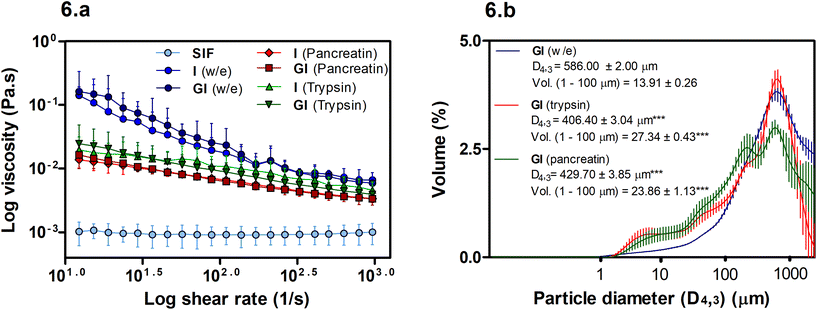
The viscosity of soluble fibres during digestion has been linked with reductions in lipid digestion and absorption. However, in this case, Fig. 6a shows that viscosity may not play an essential role in the BS binding promoted by MYC as the w/e samples have the highest viscosity, yet, the intestinal alone w/e sample had no significant BS binding activity (Fig. 2b). Conversely, the particle size of MYC under different in vitro digestion conditions can influence viscosity (Fig. 6b).Overall, the viscosity of MYC digesta did not differ between GI and only intestinal digestions (Fig. 6a). Hence, the gastric pH effect on BS binding (Fig. 2b) mediated by acid pH was not influenced by viscosity and, therefore, appeared to be unlinked to the BS binding effect of MYC. However, higher viscosity of MYC digested with or without enzymes was observed when compared with SIF (P < 0.001). Thus, an increased viscosity in the small intestine due to MYC particles could potentially entrap BS in a viscous matrix. This might increase the faecal BS excretion and may lead to lower serum cholesterol levels due to the de novo synthesis of BS from cholesterol in the liver similar to that observed with oat β-glucans.40,41
Fig. 6a shows that the use of enzymes (pancreatin or trypsin) reduced the viscosity (P < 0.01) compared to digestion without enzymes. Moreover, the digestion without enzyme had a higher viscosity at low shear that decreased with the increase of the log shear rate. This can be explained by the presence of elongated particles (MYC filamentous cells) that have a high shear-thinning effect and higher viscosity at low shear rates compared to spherical particles.42 Moreover, Fig. 6b shows an overall decrease in the particle size in intestinal digestion with trypsin (D4,3: 406.40 ± 0.34 μm) or pancreatin (D4,3: 429.70 ± 3.85 μm), which were statistically significant (P < 0.001) compared to the incubation without enzymes (w/e) (D4,3: 586.00 ± 2.00 μm). The reduction of particle size is usually associated with an increase in viscosity.42 However, this assumes that the total dispersed phase volume is the same, and the properties of all the particles are identical. The reduced viscosity in the presence of enzymes in Fig. 6a, therefore, suggests that either the dispersed phase volume is reduced, or the properties of the particles are different following digestion. We have previously shown that digestion removes protein and carbohydrates from fungal cells.31,39 Thus, it could be assumed that the properties of the cells, such as deformability and size, may have changed, thus affecting the contribution of the hyphal cells to the viscosity. The increased polydispersity of particles, promoted by enzyme digestion, may also contribute to reducing viscosity.42 For instance, Fig. 6b shows a significant increase (P < 0.001) in the volume (Vol.) of small particles in the region of 1–100 μm in both trypsin and pancreatin digestions compared to the digestion without enzyme.
4. Conclusions
This study aimed to identify the mechanisms by which the complex cellular structure of the mycoprotein ingredients influence lipid digestion. We provide new evidence that MYC reduces lipolysis and can bind BS. The reduction of lipolysis appeared to be mediated by direct interference of the MYC matrix with lipase. At the same time, the binding of BS was promoted both by the impact of pH changes on the structure of MYC cell wall and also by protein activated by trypsin hydrolysis. However, protein-mediated binding was negated following full enzymatic digestion. Moreover, viscosity during simulated GI digestion appeared to be unlinked with the BS binding capacity of MYC. These findings can help in understanding the complex interaction of macronutrients, such as fibre and protein, with digestive components (e.g., enzymes and BS) that subsequently may influence physiological pathways (e.g., de novo cholesterol synthesis) and lipid bioaccessibility. This contributes to a new understanding of the mechanisms by which diets enriched with MYC can improve blood lipid profiles.Author contributions
Raffaele Colosimo: Methodology, investigation, formal analysis, data curation, writing – original draft. Ana-Isabel Mulet-Cabero: Conceptualization, writing – review and editing. Frederick Warren: Conceptualization, supervision, writing – review and editing. Cathrina Edwards: Conceptualization, supervision, writing – review and editing. Tim Finnigan: Conceptualization, resources, supervision. Peter Wilde: Conceptualization, supervision, project administration, writing – review and editing.Abbreviations
| MYC | Mycoprotein |
| DW | Dry weight |
| LDL | Low-density lipoprotein |
| HDL | High-density lipoprotein |
| BS | Bile salts |
| GI | Gastrointestinal |
| O/W | Oil in water emulsion |
| TBT | Tributyrin |
| EMPs | Extracted mycoprotein proteins |
| SDS-PAGE | Sodium dodecyl phosphate-polyacrylamide gel electrophoresis |
| LC-MS/MS | Liquid chromatography with tandem mass spectrometry |
| WPI | Whey protein isolate |
| SIF | Simulated intestinal fluid |
| ACN | Acetonitrile |
| DTT | DL-Dithiothreitol |
| IAA | Iodoacetamide. |
Conflicts of interest
Tim Finnigan is an employee of Marlow Foods Ltd. The other authors have no conflict of interest to declare.Acknowledgements
The authors gratefully acknowledge Dr Louise Salt and Dr Davood Zaeim for proofreading; Dr Carlo De-Oliveira Martins and Dr Gerhard Saalbach at the John Innes Centre for the LC-MS/MS analysis; Marlow Foods Ltd for funding and supporting this study, and the support from the Biotechnology and Biological Sciences Research Council (BBSRC) through the Institute Strategic Programme Food Innovation and Health (BB/R012512/1).References
- M. G. Wiebe, Quorn™ Myco-protein—Overview of a successful fungal product, Mycologist, 2004, 18, 17–20 CrossRef.
- Mycoprotein.org, Nutritional profile of mycoprotein, URL: https://www.mycoprotein.org/health-nutrition/nutritional-composition, Accessed 18/11/2019.
- I. Abete, D. Romaguera, A. R. Vieira, A. L. de Munain and T. Norat, Association between total, processed, red and white meat consumption and all-cause, CVD and IHD mortality: a meta-analysis of cohort studies, Br. J. Nutr., 2014, 112, 762–775 CrossRef CAS.
- A. Denny and J. Buttriss, Plant foods and health: focus on plant bioactives, EuroFIR Synth. Rep., 2007, 4, 1–64 Search PubMed.
- I. Schneider, G. Kressel, A. Meyer, U. Krings, R. G. Berger and A. Hahn, Lipid lowering effects of oyster mushroom (Pleurotus ostreatus) in humans, J. Funct. Foods, 2011, 3, 17–24 CrossRef CAS.
- A. Gil-Ramírez, D. Morales and C. Soler-Rivas, Molecular actions of hypocholesterolaemic compounds from edible mushrooms, Food Funct., 2018, 9, 53–69 RSC.
- J. Lunn and J. L. Buttriss, Carbohydrates and dietary fibre, Nutr. Bull., 2007, 32, 21–64 CrossRef.
- O. Pabois, A. Antoine-Michard, X. Zhao, J. Omar, F. Ahmed, F. Alexis, R. D. Harvey, I. Grillo, Y. Gerelli, M. M. L. Grundy, B. Bajka, P. J. Wilde and C. A. Dreiss, Interactions of bile salts with a dietary fibre, methylcellulose, and impact on lipolysis, Carbohydr. Polym., 2020, 231, 115741 CrossRef CAS.
- W. H. Turnbull, A. R. Leeds and G. D. Edwards, Effect of mycoprotein on blood lipids, Am. J. Clin. Nutr., 1990, 52, 646–650 CrossRef CAS.
- W. H. Turnbull, A. R. Leeds and D. G. Edwards, Mycoprotein reduces blood lipids in free-living subjects, Am. J. Clin. Nutr., 1992, 55, 415–419 CrossRef CAS.
- C. H. Ruxton and B. McMillan, The impact of mycoprotein on blood cholesterol levels: a pilot study, Br. Food J., 2010, 112, 1092–1101 CrossRef.
- P. Gunness and M. J. Gidley, Mechanisms underlying the cholesterol-lowering properties of soluble dietary fibre polysaccharides, Food Funct., 2010, 1, 149–155 RSC.
- S. Adisakwattana, J. Intrawangso, A. Hemrid, B. Chanathong and K. Mäkynen, Extracts of edible plants inhibit pancreatic lipase, cholesterol esterase and cholesterol micellization, and bind bile acids, Food Technol. Biotechnol., 2012, 50, 11 CAS.
- V. Goel, S. K. Cheema, L. B. Agellon, B. Ooraikul, M. I. McBurney and T. K. Basu, In vitro binding of bile salt to rhubarb stalk powder, Nutr. Res., 1998, 18, 893–903 CrossRef CAS.
- A. Lia, G. Hallmans, A. S. Sandberg, B. Sundberg, P. Aman and H. Andersson, Oat beta-glucan increases bile acid excretion and a fiber-rich barley fraction increases cholesterol excretion in ileostomy subjects, Am. J. Clin. Nutr., 1995, 62, 1245–1251 CrossRef CAS.
- J. A. Story and D. Kritchevsky, Comparison of the binding of various bile acids and bile salts in vitro by several types of fiber, J. Nutr., 1976, 106, 1292–1294 CrossRef CAS.
- B. Staels, A review of bile acid sequestrants: potential mechanism (s) for glucose-lowering effects in type 2 diabetes mellitus, Postgrad. Med., 2009, 121, 25–30 CrossRef.
- J. Guerin, A. Kriznik, N. Ramalanjaona, Y. Le Roux and J.-M. Girardet, Interaction between dietary bioactive peptides of short length and bile salts in submicellar or micellar state, Food Chem., 2016, 209, 114–122 CrossRef CAS.
- S. Makino, H. Nakashima, K. Minami, R. Moriyama and S. Takao, Bile acid-binding protein from soybean seed: isolation, partial characterization and insulin-stimulating activity, Agric. Biol. Chem., 1988, 52, 803–809 CrossRef CAS.
- J. U. Kongo-Dia-Moukala, H. Zhang and P. C. Irakoze, In vitro binding capacity of bile acids by defatted corn protein hydrolysate, Int. J. Mol. Sci., 2011, 12, 1066–1080 CrossRef CAS.
- H.-L. Siow, S.-B. Choi and C.-Y. Gan, Structure–activity studies of protease activating, lipase inhibiting, bile acid binding and cholesterol-lowering effects of pre-screened cumin seed bioactive peptides, J. Funct. Foods, 2016, 27, 600–611 CrossRef CAS.
- Y. Yoshie-Stark and A. Wäsche, In vitro binding of bile acids by lupin protein isolates and their hydrolysates, Food Chem., 2004, 88, 179–184 CrossRef CAS.
- K. Iwami, K. Sakakibara and F. Ibuki, Involvement of post-digestion'hydrophobia'peptides in plasma cholesterol-lowering effect of dietary plant proteins, Agric. Biol. Chem., 1986, 50, 1217–1222 CAS.
- C. Zacherl, P. Eisner and K.-H. Engel, In vitro model to correlate viscosity and bile acid-binding capacity of digested water-soluble and insoluble dietary fibres, Food Chem., 2011, 126, 423–428 CrossRef CAS.
- W. Wang, M. Onnagawa, Y. Yoshie and T. Suzuki, Binding of bile salts to soluble and insoluble dietary fibers of seaweeds, Fish. Sci., 2001, 67, 1169–1173 CrossRef CAS.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carriere, R. Boutrou, M. Corredig and D. Dupont, A standardised static in vitro digestion method suitable for food–an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- D. J. Mat, S. Le Feunteun, C. Michon and I. Souchon, In vitro digestion of foods using pH-stat and the INFOGEST protocol: Impact of matrix structure on digestion kinetics of macronutrients, proteins and lipids, Food Res. Int., 2016, 88, 226–233 CrossRef CAS.
- P. McBride, Triglycerides and risk for coronary artery disease, Curr. Atheroscler. Rep., 2008, 10, 386–390 CrossRef CAS.
- V. Balasubramaniam, S. Mustar, N. M. Khalid, A. A. Rashed, M. F. M. Noh, M. D. Wilcox, P. I. Chater, I. A. Brownlee and J. P. Pearson, Inhibitory activities of three Malaysian edible seaweeds on lipase and α-amylase, J. Appl. Phycol., 2013, 25, 1405–1412 CrossRef CAS.
- D. Houghton, M. D. Wilcox, P. I. Chater, I. A. Brownlee, C. J. Seal and J. P. Pearson, Biological activity of alginate and its effect on pancreatic lipase inhibition as a potential treatment for obesity, Food Hydrocolloids, 2015, 49, 18–24 CrossRef CAS.
- R. Colosimo, F. J. Warren, C. H. Edwards, T. J. A. Finnigan and P. J. Wilde, The interaction of α-amylase with mycoprotein: Diffusion through the fungal cell wall, enzyme entrapment, and potential physiological implications, Food Hydrocolloids, 2020, 108, 106018 CrossRef CAS.
- H. Zhai, P. Gunness and M. J. Gidley, Barley β-glucan effects on emulsification and in vitro lipolysis of canola oil are modulated by molecular size, mixing method, and emulsifier type, Food Hydrocolloids, 2020, 103, 105643 CrossRef CAS.
- S. Sayar, J.-L. Jannink and P. J. White, In Vitro Bile Acid Binding of Flours from Oat Lines Varying in Percentage and Molecular Weight Distribution of β-Glucan, J. Agric. Food Chem., 2005, 53, 8797–8803 CrossRef CAS.
- M. Eastwood, R. Anderson, W. Mitchell, J. Robertson and S. Pocock, A method to measure the adsorption of bile salts to vegetable fiber of differing water holding capacity, J. Nutr., 1976, 106, 1429–1432 CrossRef CAS.
- G. Isaksson, I. Lundquist and I. Ihse, Effect of dietary fiber on pancreatic enzyme activity in vitro: the importance of viscosity, pH, ionic strength, adsorption, and time of incubation, Gastroenterology, 1982, 82, 918–924 CrossRef CAS.
- J. Shepherd, C. J. Packard, S. Bicker, T. V. Lawrie and H. G. Morgan, Cholestyramine promotes receptor-mediated low-density-lipoprotein catabolism, N. Engl. J. Med., 1980, 302, 1219–1222 CrossRef CAS.
- T. Takeshita, M. Okochi, R. Kato, C. Kaga, Y. Tomita, S. Nagaoka and H. Honda, Screening of peptides with a high affinity to bile acids using peptide arrays and a computational analysis, J. Biosci. Bioeng., 2011, 112, 92–97 CrossRef.
- J. Adler-Nissen, Enzymic hydrolysis of food proteins, Elsevier Applied Science Publishers, UK, 1986 Search PubMed.
- R. Colosimo, F. J. Warren, T. J. A. Finnigan and P. J. Wilde, Protein bioaccessibility from mycoprotein hyphal structure: in vitro investigation of underlying mechanisms, Food Chem., 2020, 127252 CrossRef CAS.
- EFSA Panel on Dietetic Products, N., Allergies, Scientific Opinion on the substantiation of a health claim related to oat beta glucan and lowering blood cholesterol and reduced risk of (coronary) heart disease pursuant to Article 14 of Regulation (EC) No 1924/2006, EFSA J., 2010, 8, 1885 Search PubMed.
- R. A. Othman, M. H. Moghadasian and P. J. Jones, Cholesterol-lowering effects of oat β-glucan, Nutr. Rev., 2011, 69, 299–309 CrossRef.
- M. Panalytical, Changing the Properties of Particles to Control Their Rheology, URL: https://www.azom.com/article.aspx?ArticleID=12304, Accessed 06/02/2020.
| This journal is © The Royal Society of Chemistry 2020 |