Building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection†‡
Aaron
Bivins
a,
Megan
Lott
b,
Marlee
Shaffer
a,
Zhenyu
Wu
 a,
Devin
North
a,
Erin K.
Lipp
a,
Devin
North
a,
Erin K.
Lipp
 b and
Kyle
Bibby
b and
Kyle
Bibby
 *a
*a
aDepartment of Civil & Environmental Engineering & Earth Sciences, University of Notre Dame, 156 Fitzpatrick Hall, Notre Dame, IN 46556, USA. E-mail: kbibby@nd.edu
bDepartment of Environmental Health Science, University of Georgia, USA
First published on 23rd September 2021
Abstract
Wastewater surveillance for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA has demonstrated useful correlation with both coronavirus disease 2019 (COVID-19) cases and clinical testing positivity at the community level. Wastewater surveillance on college campuses has also demonstrated promising predictive capacity for the presence and absence of COVID-19 cases. However, to date, such monitoring has most frequently relied upon composite samplers and reverse transcription quantitative PCR (RT-qPCR) techniques, which limits the accessibility and scalability of wastewater surveillance, particularly in low-resource settings. In this study, we trialed the use of tampons as passive swabs for sample collection and reverse transcription loop-mediated isothermal amplification (RT-LAMP), which does not require sophisticated thermal cycling equipment, to detect SARS-CoV-2 RNA in wastewater. Results for the workflow were available within three hours of sample collection. The RT-LAMP assay is approximately 20 times less analytically sensitive than RT-droplet digital PCR. Nonetheless, during a building-level wastewater surveillance campaign concurrent with independent weekly clinical testing of all students, the method demonstrated a three-day positive predictive value (PPV) of 75% (excluding convalescent cases) and same-day negative predictive value (NPV) of 80% for incident COVID-19 cases. These predictive values are comparable to that reported by wastewater monitoring using RT-qPCR. These observations suggest that even with lower analytical sensitivity the tampon swab and RT-LAMP workflow offers a cost-effective and rapid approach that could be leveraged for scalable building-level wastewater surveillance for COVID-19 potentially even in low-resource settings.
Water impactWastewater has been recognized as a potential information stream regarding human disease occurrence and dynamics, especially in response to the COVID-19 pandemic. Critically, most wastewater surveillance approaches have relied upon centralized sampling and intensive molecular analyses, limiting their potential for decentralized, widespread application. Here, we demonstrate a passive sampling approach coupled with isothermal LAMP SARS-CoV-2 RNA detection that has the potential to enable more widespread application of wastewater surveillance, both for COVID-19 and future infectious disease targets. |
Introduction
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), is often accompanied by shedding of the virus and its genetic material in respiratory fluids, feces,1 saliva,2 and urine.3 Since these body fluids are frequently discharged to wastewater collection networks in domestic sewage, wastewater-based epidemiology (WBE; also called wastewater surveillance) has become a useful tool for assessing community trends of COVID-19.4 SARS-CoV-2 RNA has been detected in untreated wastewater samples throughout the world.5–10 Longitudinal measurements of SARS-CoV-2 RNA in wastewater influent and primary solids at wastewater treatment plants (WWTPs) have been found to correlate with COVID-19 clinical testing metrics in various communities.11–14 In many contexts, increases in SARS-CoV-2 RNA in wastewater or wastewater solids have preceded increases in COVID-19 cases and hospitalizations by days to weeks.15–17 Thus, wastewater monitoring offers a complementary method of assessing COVID-19 trends in communities that is agnostic to care seeking behavior, less resource intensive than clinical testing, and in some contexts leads trends observed by clinical testing.While promising, monitoring SARS-CoV-2 RNA in influent at WWTPs can lack the spatial resolution required to target clinical testing or other public health interventions at fine geographic scales. Building-level surveillance, on the other hand, could inform clinical testing at specific locations on the basis of wastewater data from individual facilities, such as schools18 and skilled nursing facilities.19 Spurbeck et al. used 24-hour wastewater composite samples and RT-qPCR to detect one infection among 60 skilled nursing facility residents.19 Wastewater surveillance for SARS-CoV-2 RNA, including build-level surveillance, is also being used to manage COVID-19 on university campuses throughout the United States.20 At the University of Arizona, wastewater surveillance with serial grab samples identified one symptomatic and two asymptomatic infections in a dorm and provided early warning of infections in a total of 13 dorms over a semester.21 An innovative high-throughput wastewater monitoring platform allowed for the detection of a single case of COVID-19 among 415 residents of a dorm at University of California San Diego.22 And another building-level monitoring effort leveraged composite wastewater samples and RT-qPCR performed three times weekly to identify asymptomatic COVID-19 cases on multiple occasions down to one asymptomatic infection among 150 to 200 dorm residents.23
At universities, student behavior,24 congregate living,25 asymptomatic transmission,26 emerging variants of concern, and breakthrough infections among vaccinated communities may combine to fuel outbreaks. Complicating transmission control are asymptomatic infections, which have been observed to account for 43%27 to 50% of infections28 among adults. Since viral loads have been found to be similar among asymptomatic, pre-symptomatic, and symptomatic patients27,29 and asymptomatic and mild COVID-19 cases have been observed to shed SARS-CoV-2 RNA in stool,30 wastewater surveillance offers a compelling opportunity to screen for COVID-19 cases among building-level populations and identify cases via follow-up clinical testing.31
While wastewater surveillance is compelling, most of the reported efforts have depended on composite samplers to achieve representative samples over a defined time period (usually 24 hours). These samplers can be expensive and difficult to place in building service lines. Other studies have used grab samples, but such samples are “snapshots” and may not afford a reliably representative sample. A few SARS-CoV-2 wastewater surveillance efforts to date, however, have used the Moore Swab, a gauze bundle left suspended in sewers to sorb wastewater and enteric pathogens. This type of “passive” sampling was first used to detect Salmonella paratyphi in 194832 and has also been used to detect Vibrio cholerae33 and enteric viruses34 in wastewater. While passive sampling methods make rigorous quantification of analytes in wastewater difficult due to uncertainties concerning the volume of wastewater sampled and the efficiency of sorption, they can be used to produce useful qualitative data. More recently as reported in a preprint, Moore swabs in combination with RT-qPCR were used to sample wastewater at a university and were able to detect one to two COVID-19 cases in a building.35 The same study found that when used alongside grab samples, the Moore swab allowed a greater sensitivity for SARS-CoV-2 RNA in wastewater from a hospital treating COVID-19 patients.35 Another evaluation of passive samplers (gauze, electronegative filter, and cotton buds) alongside traditional sampling techniques (flow-weighted and time-average composite, and grab samples) found that passive samplers were at least as sensitive over 24-hour deployments and a positive correlation between SARS-CoV-2 RNA concentrations in wastewater and those from passive samplers.36 Several wastewater surveillance teams participating in the COVID-19 wastewater-based epidemiology collaborative (https://www.covid19wbec.org/) have reported experiments with tampons as a form of off-the-shelf passive sampler, but published studies of their performance are lacking.
Passive samplers, such as the Moore swab or tampons, could make wastewater surveillance possible without the use of expensive composite samplers. However, detection and quantification of SARS-CoV-2 RNA in wastewater samples has also required the use of RT-qPCR techniques, which depend on specialized PCR equipment such as thermal cyclers. Reverse transcription loop-mediated isothermal amplification (RT-LAMP)37 offers the potential to detect SARS-CoV-2 RNA in wastewater samples without the use of such equipment. RT-LAMP has been validated for rapid testing of clinical samples including serum, urine, saliva, oropharyngeal swabs, and nasopharyngeal swabs for SARS-CoV-2 RNA.38,39 A colorimetric RT-LAMP kit developed by New England Biolabs using multiplexed primers targeting the N and E regions of the SARS-CoV-2 genome had accuracy (true positive and negative rate) greater than 90% compared to RT-qPCR and a 95% limit of detection of 59 copies per reaction when used to test heat treated saliva samples.40 Multiplexing primers and the addition of guanidine chloride was found to increase the sensitivity five- to tenfold for colorimetric LAMP with the N2 and E1 primers yielding the best performance among seven primer sets.41 A preprint has even reported the use of RT-qLAMP with primers targeting the ORF1a, E, and N genes to test wastewater samples for SARS-CoV-2 RNA without extraction in wastewater volumes up to 9.5 μL.42
During the current study, we trialed the application of colorimetric RT-LAMP to detect SARS-CoV-2 RNA in wastewater from tampon swabs deployed in manholes and primary influent from WWTPs. We assessed the sensitivity, specificity, and limit of detection of RT-LAMP for wastewater samples compared to reverse transcription droplet digital PCR (RT-ddPCR). We then used tampon swabs and RT-LAMP for rapid surveillance of building-level wastewater at the University of Notre Dame (ND) over six weeks in conjunction with ongoing public health measures to assess the positive and negative predictive value of passive sampling and RT-LAMP.
Experimental
Primary influent and raw sewage samples
During the RT-LAMP validation experiments, 24-hour time-based composite samples of primary influent, referred to as “primary influent” throughout this text, were collected at eleven wastewater treatment plants (WWTPs) whose characteristics are summarized in Table S1.† In addition to primary influent, wastewater samples were collected from manholes, referred to as “raw sewage” throughout this text. The manholes served populations ranging from 94 to 299 people with an average population of 181 ± 61. Per sewer system maps, the diameter of the sewage pipes entering and exiting these manholes ranged from 8′′ to 24′′ with code-specified slopes ranging from 2% to 0.08%. Raw sewage samples were collected using two techniques: 24-hour time-based composite samples and tampon swab passive samplers (detailed further below). In all cases, immediately after collection, both primary influent and raw sewage samples were stored and transported on ice or at 4 °C until processed.Tampon swab samplers
During RT-LAMP validation experiments, tampons (OB Brand Organic Tampons Super & Tampax Pearl Super unscented) were used as low-cost and readily available swabs for passive sampling of raw sewage in the wastewater collection system. OB Organic and Tampax Pearl tampons are free of dyes, perfume, and chlorine. OB Organic tampons are made from 100% organic cotton while Tampax Pearl tampons are made from cotton and rayon. Prior to deployment in manholes, the tampons were removed from the applicator and tied to fishing line with a 20-pound tensile strength. The fishing line was secured to the ladder within each manhole. After recovery, swabs were placed in sterile WhirlPak bags (Nasco, Fort Atkinson, WI) and saturated with 20 mL of sterile PBS. Saturated swabs were then hand massaged (while wearing gloves) through the sealed WhirlPak bag for two minutes to elute viruses and then the sorbate was hand squeezed from the swab within the WhirlPak bag and transferred to a sterile 50 mL centrifuge tube for immediate extraction.During the longitudinal monitoring period at ND, with the assistance of utilities personnel, tampon swabs (Tampax Pearl Super unscented) were deployed into the wastewater collection system once per week for six weeks from approximately 8:00 am to 11:00 am (same day) at nine different locations selected to isolate individual residential halls (RH) (anonymized as RH 1 to 9). During the monitoring period, these RHs housed 1627 students accounting for 25% of the on-campus residents. Upon retrieval from manholes, swabs were placed into sterile WhirlPak bags and stored on ice. In the lab, swabs were hand squeezed while in the WhirlPak bag to remove most of the sorbate and then aseptically placed into a 60 mL Luer-lock syringe (ML60, Air-Tite Products Co, Virginia Beach, VA). The sorbate remaining in the WhirlPak bag was then poured into the syringe and pressed into a 50 mL centrifuge tube using the syringe plunger typically resulting in 25 to 35 mL of sorbate. After the first press, a volume of PBS/Tween20 solution (10 mM sodium phosphate, 0.15 M NaCl, 0.05% Tween 20) was pipetted into the syringe (typically 15 to 25 mL) such that the total volume of sorbate resulting from each swab was 50 mL and pressed through the swab into the centrifuge tube. The resulting 50 mL of sorbate was then immediately concentrated or extracted as described below.
To optimize the RT-LAMP and tampon swab/RT-LAMP workflow a variety of approaches were trialed. For a subset of raw sewage and primary influent samples, no concentration or fractionation was performed prior to extraction. For other subsets of wastewater samples, different forms of concentration (centrifugal ultrafilter concentration) and fractionation (swab sorbate solids fractionation) as fully described in the ESI† were trialed. The resulting sample types from these workflows included “concentrated swab sorbate” from ultrafilter retentate, “sorbate supernatant” from the sorbate supernatant after centrifugation, and “sorbate solids” the material pelleted after sorbate centrifugation. The resulting sample sizes for each of these approaches is reported in the Results and discussion section.
RNA extraction
For a subset of wastewater samples, RNA was extracted from 280 μL aliquots of unconcentrated tampon sorbate and primary influent (composite samples) using a QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany). Purified RNA was eluted in 60 μL of PCR-grade water. But for the majority of wastewater samples, DNA and RNA were extracted from tampon sorbate and primary influent (composite samples) using an AllPrep PowerViral DNA/RNA kit (Qiagen, Hilden, Germany). Prior to extraction, membrane filters, Amicon ultrafilter retentate, and raw sewage and sorbate solids were homogenized by adding 600 μL of PM1 and 6 μL of 2-mercaptoethanol (MP Biomedicals, Irvine, CA, USA) to the PowerBead tubes. These tubes were bead beat for four rounds of 20 seconds each at 4.5 M s−1 on a FastPrep 24 (MP Biomedicals, Irvine, CA, USA). The bead tubes were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 minute and 500 μL of the resulting supernatant was transferred into a clean 2 mL microcentrifuge tube and DNA/RNA was extracted per the Qiagen protocol. Purified nucleic acids were eluted in 100 μL of RNase-free water. In addition to kit-based extractions, RT-LAMP was also trialed using no extraction and heat extraction as detailed in the ESI.†
000 × g for 1 minute and 500 μL of the resulting supernatant was transferred into a clean 2 mL microcentrifuge tube and DNA/RNA was extracted per the Qiagen protocol. Purified nucleic acids were eluted in 100 μL of RNase-free water. In addition to kit-based extractions, RT-LAMP was also trialed using no extraction and heat extraction as detailed in the ESI.†
RT-ddPCR
To characterize the sensitivity, specificity, and limit of detection of RT-LAMP qualitative LAMP results were compared with quantitative SARS-CoV-2 RNA data produced using electronegative membrane filtration and RT-ddPCR as detailed in the ESI† and elsewhere (https://dx.doi.org/10.17504/protocols.io.bhiuj4ew).43RT-LAMP
SARS-CoV-2 RNA was detected by RT-LAMP using the SARS-CoV-2 rapid colorimetric LAMP assay kit (Cat no. E2019S) from New England BioLabs (NEB) (Ipswich, MA, USA), a 30-minute 65 °C colorimetric assay. The kit includes an internal inhibition control (LAMP primer mix targeting human RNA rActin) and a SARS-CoV-2 LAMP primer mix targeting the N and E genes (N2 and E1, respectively, Table S2†). NEB reports positive detections observable down to 50 copies per reaction (NEB product specification). Each sample was assayed in triplicate RT-LAMP reactions and in parallel with the previously mentioned inhibition control, and with positive controls and negative controls for each experiment. For each reaction, template RNA (4 μL) was mixed with WarmStart Colorimetric LAMP 2X master mix with UDG (12.5 μL), LAMP primer mix (2.5 μL), guanidine hydrochloride (2.5 μL), and PCR-grade water to a final reaction volume of 25 μL. The reaction was vortexed gently and briefly spun down prior to incubation at 65 °C for 30 minutes. Reactions were cooled at room temperature for 5 min before reading color change and interpreting the results per the NEB protocol. RT-LAMP results were acceptable if the inhibition control was successfully detected in each sample, the SARS-CoV-2 positive and negative controls (two each per experiment) were appropriately positive and negative, and the negative extraction controls were negative for both the inhibition control and SARS-CoV-2. When the inhibition control was not detected for a sample, the sample was interpreted as inhibited.COVID-19 clinical surveillance at ND
During the period of wastewater monitoring at ND, COVID-19 safety protocols were in place including universal masking, physical distancing, daily health checks, and weekly asymptomatic and symptomatic COVID-19 testing. COVID-19 testing methods included saliva-based PCR tests, primarily for asymptomatic surveillance, nasal swab PCR tests, and rapid antigen tests. All undergraduate and professional students participated in mandatory weekly surveillance testing. At the beginning of the semester, all students selected the day of the week they wished to participate in surveillance testing for the duration of the semester. Critically, the clinical testing was not informed by the wastewater testing, such that the results are independent of one another. Students testing positive for COVID-19 immediately entered isolation in residential facilities outside of their residence hall and their close contacts entered isolation as soon as they were identified by contact tracing. Close contacts were tested by nasal swab PCR test on day four of isolation and rapid antigen test on day seven of isolation. If both tests were negative, close contacts departed isolation on day 7. If either test was positive, close contacts began a new 10-day period of isolation. Students testing positive for COVID-19 completed isolation per United States Centers for Disease Control and Prevention protocols with at least 10 days from symptom onset for symptomatic cases or 10 days from positive test results for asymptomatic cases. Although visitation between residence halls was restricted, the possibility of a non-resident COVID-19 case or convalescent case shedding into the wastewater system of another residence hall cannot be precluded.Deidentified COVID-19 case data including the date of positive test, date of isolation start, and date of isolation end were acquired for the nine residence halls over the wastewater monitoring period. The research protocol was reviewed by the University of Notre Dame Institutional Review Board (21-04-6586). In addition to de-identification of the COVID-19 case data for the study, the residence halls have also been anonymized (RH1 to RH9), and the monitoring period has been anonymized by the use of elapsed days (0 to 73) rather than dates. The wastewater surveillance was performed in coordination with the ND Covid response unit.
Data analysis
The RT-LAMP 95% limit of detection (LOD) was estimated using N1 copy number data (RT-ddPCR) and the proportion of RT-LAMP reactions positive along an N1 concentration gradient. A cumulative Gaussian distribution was fit to the RT-LAMP proportion positive along the gradient and the 95th percentile estimated.44 The true negative rate (specificity) was estimated using RT-ddPCR non-detections and paired RT-LAMP results. The true positive rate (sensitivity) was estimated using RT-ddPCR detections and paired RT-LAMP results. The relationship between N1 copy number and RT-LAMP classification was modeled using a simple logistic regression45 with statistical significance determined by a likelihood ratio test46 and fit assessed using Tjur's R-squared.47 Comparisons between two groups (e.g., inhibition between sample types) were made using Mann–Whitney tests and between multiple groups (e.g., inhibition between extraction methods and positivity rate between sorbate fractions) using Kruskal–Wallis tests with Dunn's post test.48–50 The positive and negative predictive values (PPV, NPV) of wastewater testing by tampon swab and RT-LAMP for COVID-19 cases was estimated for incident COVID-19 cases in the residence hall from the day of wastewater sampling (day 0) out to six days after wastewater sampling (day 6). The rationale for this comparison is that wastewater sampling from 8 am to 11 am could detect shedding cases living in the residence hall and not yet in isolation before the case is identified by clinical testing on that same day. Additionally, students could be shedding into the wastewater prior to being identified as a case during clinical testing in the subsequent six days. In this case PPV is the probability of an incident COVID-19 case following a positive wastewater sample, and, conversely, NPV is the probability of no incident COVID-19 cases following a negative wastewater sample. PPV and NPV were estimated across all nine residence halls each week, among single residence halls across all weeks, and across all residence halls and all weeks.51 PPV and NPV were estimated using three different RT-LAMP positivity cutoff values – 1 of 3, 2 of 3, and 3 of 3 replicates positive. All graphing and statistical analyses associated with the described experiments were performed using GraphPad Prism version 9.0.0 (GraphPad software, La Jolla, CA, USA).Results and discussion
In total, 147 wastewater samples were tested via RT-LAMP. To characterize the sensitivity, specificity, and analytical sensitivity of RT-LAMP, we used split extracts derived from 24-hour composite samples of primary influent (n = 42) and raw sewage samples collected via tampon swabs (n = 7). To analyze RT-LAMP performance with various extraction and processing methods, we also leveraged split extracts of samples from primary influent composites (n = 42) and raw sewage from tampon swabs (n = 68). Lastly, during a prospective wastewater surveillance campaign at ND, we used RT-LAMP to test raw sewage (n = 53) samples collected via tampon swabs. One tampon swab could not be recovered because it broke free while deployed in a manhole. Although, the pipe exiting the manhole was 8′′ in diameter and the swab was unlikely to clog such a pipe, the potential for passive sampler break offs should be considered when selecting sampling sites. The types and number of samples are summarized in Table S3.†RT-LAMP specificity and sensitivity
Compared to RT-ddPCR non-detections (n = 13), RT-LAMP demonstrated an overall specificity (true negative rate) of 100% among both primary influent composites and raw sewage samples. We estimated the sensitivity (true positive rate) using RT-ddPCR (n = 36) quantifications (N1 target in triplicate) compared to positivity among all RT-LAMP reactions. Across all samples positive for SARS-CoV-2 RNA by RT-ddPCR (n = 36), the RT-LAMP positivity was 57% (Fig. S1A†). A logistic regression model (Fig. S1B†) fit to the data indicated that increasing N1 GC/reaction was associated with increasing probability of detection by RT-LAMP performed in triplicate (likelihood ratio test, p = 0.0034). However, the model fit was poor (Tjur's R-squared = 0.24). Nonetheless, the logistic model indicates that at 18 N1 GC/reaction there is a 50% probability of detection via RT-LAMP reactions in triplicate, while at the NEB-reported “limit of detection” (50 copies) there is an 83% probability of detection by RT-LAMP triplicates.Analytical sensitivity
Using paired RT-LAMP positivity and RT-ddPCR N1 copy number data, we estimated the RT-LAMP 95% LOD to be 76 N1 gene copies (GC) for a single reaction (95% CI: 67–87) using a fitted cumulative Gaussian distribution (Fig. S2;†R2 = 0.997). The RT-LAMP 95% LOD is approximately 20 times our previous estimate of the N1 RT-ddPCR 95% LOD.44 NEB reports “positive detection observable down to 50 copies”, which is comparable to our estimated 67% LOD (51 N1 GC/reaction).Viral RNA mini versus PowerViral DNA/RNA inhibition rate
We assessed the rate of RT-LAMP inhibition (using the previously described rActin inhibition control) for samples extracted using the viral RNA mini kit (n = 14) and PowerViral DNA/RNA kit (n = 96). For 24-hour primary influent composite samples (Fig. S3,†n = 9), no inhibition was observed following extraction with the viral RNA mini kit. But we observed a significantly higher inhibition rate (60%, p = 0.0275) for raw sewage sorbate from swabs extracted with the same kit (n = 5). Among primary influent composite samples extracted with the PowerViral kit (n = 33), 18% were inhibited. While for raw sewage sorbate, sorbate solid fraction, and sorbate liquid fraction samples (n = 63) from swabs, the PowerViral kit produced an significantly lower (p = 0.0317) inhibition rate of 4% (Fig. S3†). As shown in Fig. S4,† the difference in inhibition rates between the viral RNA mini kit and PowerViral DNA/RNA kit was not statistically significant for primary influent composite samples (panel A) or all wastewater samples (panel C). However, we did observe a significantly lower rate of inhibition for swab samples extracted via PowerViral compared to viral RNA mini (Fig. S4B;†p = 0.0030). For no extraction and heat extraction, inhibition rates, indicated by the non-detection of the inhibition control human rActin, of RT-LAMP were prohibitively high for reliable use (details in ESI†).Tampon swab sorbate processing
To optimize the workflow for SARS-CoV-2 RNA detection in raw sewage via tampon swabs and RT-LAMP, we assessed the rates of inhibition and positivity between Amicon-concentrated swab sorbate, the solid fraction of swab sorbate, and the liquid fraction of swab sorbate during two wastewater surveillance experiments. Amicon-concentrated sorbate (detailed in ESI†) extracted via PowerViral produced no inhibited RT-LAMP reactions and an overall SARS-CoV-2 RNA positivity of 40% (11 of 27 RT-LAMP replicates) in samples collected from nine RHs. However, filtering the swab sorbate through the Amicon ultrafilters required several hours of centrifugation. Given our interest in a rapid testing procedure, we abandoned centrifugal ultrafiltration. During the next wastewater surveillance experiment, the swab sorbate was centrifuged (detailed in ESI†), then the resulting supernatant was concentrated via Amicon and extracted with PowerViral. The solid fraction pellet was also extracted via PowerViral. The rate of RT-LAMP inhibition among the sorbate supernatant samples was 38% and SARS-CoV-2 RNA was not detected in any of 24 RT-LAMP replicates. For the extracted solid fractions, there was no inhibition observed and the SARS-CoV-2 RNA positivity was 33% among 30 RT-LAMP replicates. Both the Amicon-concentrated and sorbate solids exhibited lower rates of inhibition (Fig. S5A†) and higher rates of SARS-CoV-2 positivity (Fig. S5B†) than the sorbate supernatant. Since inhibition rates (p > 0.9999) and SARS-CoV-2 RNA positivity rates (p > 0.9999) were comparable between Amicon-concentrate and the sorbate solids fraction, we elected to continue wastewater surveillance at ND using only the swab sorbate solid fraction to allow for faster processing.COVID-19 clinical data
During the entire 72-day period, 143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 884 COVID-19 clinical tests (symptomatic and asymptomatic) were performed at ND. During the wastewater surveillance (day 31 to 66), an average of 13
884 COVID-19 clinical tests (symptomatic and asymptomatic) were performed at ND. During the wastewater surveillance (day 31 to 66), an average of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 748 clinical tests were performed each week. The COVID-19 positivity and case number trends among the subpopulation accounted for in wastewater surveillance (Fig. S6†) are similar to the trends for the entire campus. The proportion of wastewater RT-LAMP tests that were positive decreased abruptly from 30% to 0 from week 3 to week 4, and then increased slightly in the following two weeks.
748 clinical tests were performed each week. The COVID-19 positivity and case number trends among the subpopulation accounted for in wastewater surveillance (Fig. S6†) are similar to the trends for the entire campus. The proportion of wastewater RT-LAMP tests that were positive decreased abruptly from 30% to 0 from week 3 to week 4, and then increased slightly in the following two weeks.
RT-LAMP PPV and NPV for COVID-19
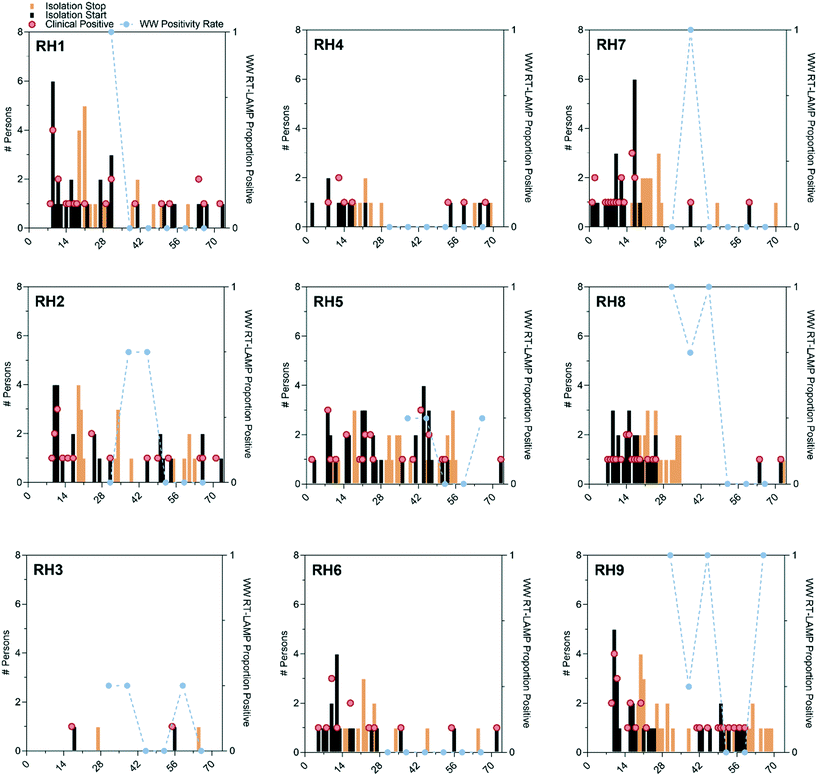
RT-LAMP wastewater testing results (proportion of positive RT-LAMP replicates), COVID-19 clinical positives, residents exiting the RH for isolation, and residents returning from isolation are shown for each RH in Fig. 1. RT-LAMP positives in wastewater were coincident with COVID-19 cases on the same day on four occasions (RH1, RH2, RH7, RH9). For two residence halls (RH4, RH6) RT-LAMP results were negative across the entire sampling period with one occurring on the same day as a positive COVID-19 clinical test in RH4. There were also RT-LAMP positives during periods without incident COVID-19 cases in RH2, RH3, RH8, and RH9.Although the ND COVID-19 response unit was informed of the wastewater sampling results, the clinical surveillance testing was performed independently and thus allows for an estimation of the tampon swab and RT-LAMP wastewater testing PPV and NPV. PPV and NPV were calculated for each day from the day of wastewater testing (day 0) out to six days after. The PPVs displayed a wider range across residence halls (0 to 100%; Fig. S8A†) than weeks (0 to 75%; Fig. S8C†). In general, PPV increased from the day of wastewater monitoring to three days after as incident COVID-19 cases increased in the days following. PPV could not be estimated for RH4, RH6, or week 4 monitoring since there were no positive wastewater results. NPV displayed a similar pattern of variation with the range observed between residence halls (0 to 100%) being greater than the range between weeks of monitoring (22 to 100%). NPV decreased from the day of wastewater monitoring out to three days as incident COVID-19 cases increased.
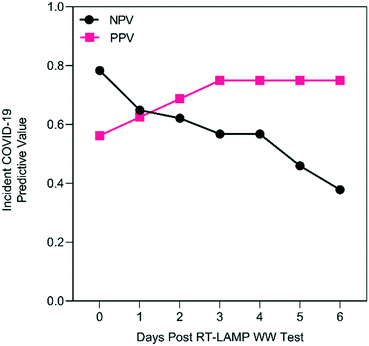
Across all residence halls and weeks, tampon swab and RT-LAMP wastewater monitoring, with any replicate positive classified as a positive wastewater result, displayed a PPV of 19 to 38% for clinically detected COVID-19 without accounting for convalescent cases during the six days following wastewater testing (Fig. S8A†). As shown in Fig. S8B,† NPV was greater with a maximum of 78% on the day of wastewater testing to a day six minimum of 38%. The PPV of wastewater testing could be adversely affected by positive RT-LAMP results attributable to convalescent COVID-19 cases returning to residence halls after isolation. As shown in Fig. S9,† there were six instances where RT-LAMP replicates were positive despite no incident COVID-19 cases, but with returning convalescent cases in the prior seven days. In these six instances, it required four or more convalescent cases before 2 of 3 RT-LAMP replicates were positive, suggesting that a cutoff value of 67% positivity (2 of 3 replicates) could increase the PPV of the wastewater method. If the detection of convalescent COVID-19 cases by wastewater surveillance is accounted for (e.g., the true detection of SARS-CoV-2 RNA shed into the wastewater system), then the PPV improves to 56% on day 0 up to 75% by day three after wastewater sampling (Fig. 2) while the NPV remains unchanged.
Reliable RT-LAMP workflow and analytical performance
To develop more accessible wastewater surveillance methods, we characterized the performance of tampon swabs and RT-LAMP to detect SARS-CoV-2 RNA in building-level wastewater and subsequently COVID-19 cases among residents. The 95% LOD for a single RT-LAMP reaction was 23 times higher than RT-ddPCR. Several studies have found that SARS-CoV-2 RNA shedding in feces can outlast nasopharyngeal shedding in up to 50% of COVID-19 patients.52–54 In such cases, the higher RT-LAMP LOD could be advantageous by allowing for convalescent cases to go undetected, while newly incident COVID-19 cases could still be detected. RT-LAMP demonstrated an overall sensitivity of 57% and specificity of 100% compared to RT-ddPCR. Unfortunately, we were not able to replicate the findings of an earlier preprint as all of our attempts to test wastewater without extraction were inhibited.42 Our attempts at heat extraction were also consistently inhibited despite the success with saliva and other clinical samples.55 We found that regardless of the wastewater type (primary influent composite or raw sewage sorbate) the use of an extraction kit for testing by RT-LAMP was important to produce uninhibited RT-LAMP reactions. When paired with tampon swab sorbate, the Qiagen AllPrep PowerViral DNA/RNA kit yielded a 4% inhibition rate among all samples. Concentrating sorbate with Amicon ultrafilters proved burdensome due to clogging. Since wastewater solids have been proposed as an efficient and sensitive partition for SARS-CoV-2 RNA detection,14,56 we opted to abandon Amicon concentration in favor of testing the sorbate solids fraction. We found that the solids fraction yielded a comparable SARS-CoV-2 positivity and inhibition rate to ultrafilter concentrate.RT-LAMP predictive capability compared to RT-qPCR
The optimized tampon swab and RT-LAMP workflow yielded a three-day PPV of 75% and a same-day NPV of 80% in six weeks of wastewater surveillance. The PPV and NPV we observed were lower than the 82% and 88.9%, respectively, reported during another study leveraging PEG precipitation and RT-qPCR.21 Nonetheless, the tampon swab and RT-LAMP approach may offer a reasonable PPV and NPV without requiring the complex equipment and lab infrastructure of more sophisticated monitoring methods. Several epidemiological modeling studies have suggested that an optimal strategy for managing COVID-19 on college campuses should include high-frequency screening tests that are highly specific.57,58Rapidity of RT-LAMP results
These models have also consistently emphasized rapid results reporting over sensitivity as a critical feature of effective screening. Wong et al. found that wastewater monitoring with one day to results and four days or less to follow up clinical testing could keep infection rates within 5% of those achieved by clinical testing of individuals.59 Following extraction, the RT-ddPCR workflows used in the study required 7 hours to produce results. Whereas, the RT-LAMP workflow required only 1.5 hours (45 minute preparation, 30 minute incubation, 15 minutes to read results). Typical RT-qPCR workflows require approximately 2 hours to generate results. Additional time is required for tampon swab deployment, collection, sorbate harvesting, and extraction. At ND, tampon swabs were deployed at 8:00 am, retrieved at 11:00 am, and results were transmitted to the COVID response unit by 3:00 pm each surveillance day. Though we only conducted the wastewater monitoring weekly, the workflow could easily be modified to achieve results daily by noon. For example, a tampon swab could be deployed in the sewer for 24 hours, retrieved at 8:00 am, at which time another could be deployed, and results could be reported by noon at which time clinical testing could be mobilized in response. Based on a 5-day incubation and 1.2 day medical seeking period,60 Zhu et al. have suggested a 6.2-day window to efficiently interrupt transmission chains.61 The tampon swab and RT-LAMP method described in this study is capable of producing wastewater results well within this window. Efficient transmission control through timely wastewater results is even more important on college campuses since asymptomatic infections are more prevalent among younger populations.26Wastewater monitoring scalability and accessibility
In addition to rapid results, the tampon swab and RT-LAMP method could also improve accessibility to wastewater surveillance in low-resource settings. Many of the COVID-19 wastewater surveillance efforts to date, including those on college campuses, have made use of composite samplers and RT-qPCR techniques to detect and quantify SARS-CoV-2 RNA.20,62 While these techniques have proven useful for tracking COVID-19 in some communities, the expense of composite samplers and the apparatus required to perform RT-qPCR greatly limits the accessibility and scalability of wastewater monitoring for SARS-CoV-2. The World Health Organization has identified wastewater surveillance approaches for pooled testing of high-risk lower-resource settings as a critical need to expand the application of the tool.63 While we could not avoid using a kit-based RNA extraction, the method does not require a composite sampler or thermal cycler for RT-qPCR, relying instead on tampons for wastewater sampling and basic lab equipment including centrifuges, microcentrifuges, vortexes, and single temperature incubators for swab processing and RT-LAMP testing. The per sample analytical cost was comparable between RT-ddPCR ($35) and the NEB RT-LAMP kit ($31); however, we estimate that a self-assembled RT-LAMP kit using the same primers could halve the per-sample cost once optimized. Even with the off-the-shelf RT-LAMP kit, the per sample consumables cost for the entire workflow was approximately $43 USD and could be driven as low as $27 USD. The average per capita wastewater surveillance cost using tampon swabs and RT-LAMP during this study was $0.24 USD per week.Limitations
There are limitations that should be considered in generalizing the findings of this study. First, our comparison of RT-LAMP and RT-ddPCR used samples from a limited number of WWTPs and sewer systems. Although we made use of raw sewage and primary influent from diverse sources, wastewater and therefore RT-LAMP performance could be variable among sites. We did not assess the process recovery for the passive samplers via an exogenous control. Nor did we assess the mechanistic basis for sorption of SARS-CoV-2 from wastewater. Interestingly, we are not aware of a mechanistic characterization of the Moore swab, despite their use since the 1940s. We also only used two brands of tampon during the current study. Since material and method of fabrication varies by brand, the performance of tampons as passive samplers is also likely to vary by brand. Each of these should be investigated for further development of passive sampling methods. For comparison with clinical surveillance, we monitored wastewater at nine ND residence halls. We note that while COVID-19 protocols during the sampling period did not allow guests into the residence halls, it is not possible to completely exclude the possible shedding of SARS-CoV-2 RNA into the residence hall wastewater by non-residents. In settings without strict COVID-19 protocols, the movement of people into and through various residential buildings could greatly complicate the interpretation of positive wastewater results from individual facilities. The predictive performance was variable between halls and weeks and the study was not designed to further investigate these differences. The tampon swabs were only deployed for a three-hour interval between 8:00 am and 11:00 am. This period accounted for roughly 20% of daily domestic water use, but the performance of the workflow could potentially be improved with longer deployments of the tampon swabs, assuming this does not lead to increased rates of inhibition. We independently monitored the wastewater from residence halls during a large and robust clinical surveillance program that featured weekly testing of every single student. In the midst of such a large clinical surveillance effort, the predictive performance of wastewater surveillance is likely to be conservative compared to a typical application.Conclusions
If wastewater surveillance is to play a meaningful role in controlling infectious disease, and in particular COVID-19, methods which are broadly applicable and widely scalable must be developed. While less sensitive for SARS-CoV-2 RNA than more sophisticated PCR-based methods, the tampon swab and RT-LAMP protocol yielded a PPV and NPV for incident COVID-19 that were reasonable for interrupting transmission chains. Importantly, it does so without the need of expensive composite samplers or thermal cycling platforms and at a low per capita cost. Our experience suggests that tampon swabs in combination with RT-LAMP could afford a specific, rapid, cost-effective, and accessible screening method for building-level wastewater surveillance. As vaccination efforts continue to progress and COVID-19 incidence decreases, swabs and RT-LAMP may offer a scalable platform for non-intrusive screening of at-risk populations, even in low-resource settings.Author contributions
Conceptualization – KB, AB, EL, ML; data curation – ML, AB; formal analysis – ML, AB; investigation and methodology – AB, ML, MS, ZW, DN; supervision – EL, KB; validation – AB, ML; visualization – AB; writing – original draft – AB, ML; writing – review and editing – AB, KB, EL, ML, MS, ZW, DN.Conflicts of interest
The authors declare no competing financial or non-financial interests.Acknowledgements
This work was funded in part by the UGA College of Public Health and Athens-Clarke County, the University of Notre Dame COVID response unit, and NSF grant 2027752. We would like to thank ND utilities, the ND Covid response unit, our local WWTP partners, Athens-Clarke County, and the public utilities department for their logistical support of the work described herein.References
- M. Cevik, M. Tate, O. Lloyd, A. E. Maraolo, J. Schafers and A. Ho, SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis, Lancet Microbe, 2021, 2(1), e13–e22 CrossRef CAS PubMed.
- A. L. Wyllie, J. Fournier, A. Casanovas-Massana, M. Campbell, M. Tokuyama and P. Vijayakumar, et al., Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2, N. Engl. J. Med., 2020, 383(13), 1283–1286, DOI:10.1056/NEJMc2016359.
- A. H. Kashi, J. Rosette, E. Amini, H. Abdi, M. Fallah-karkan and M. Vaezjalali, Urinary Viral Shedding of COVID-19 and its Clinical Associations: A Systematic Review and Meta-analysis of Observational Studies, Urol. J., 2020, 17(05), 433–441, DOI:10.22037/uj.v16i7.6248.
- A. Bivins, D. North, A. Ahmad, W. Ahmed, E. Alm and F. Been, et al. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight Against COVID-19, Environ. Sci. Technol., 2020, 54(13), 7754–7757, DOI:10.1021/acs.est.0c02388.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins and J. W. O'Brien, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community, Sci. Total Environ., 2020, 138764, DOI:10.1016/j.scitotenv.2020.138764.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands, Environ. Sci. Technol. Lett., 2020, 7(7), 511–516, DOI:10.1021/acs.estlett.0c00357.
- R. Johnson, C. J. F. Muller, S. Ghoor, J. Louw, E. Archer and S. Surujlal-Naicker, et al. Qualitative and quantitative detection of SARS-CoV-2 RNA in untreated wastewater in Western Cape Province, South Africa, S. Afr. Med. J., 2021, 111(3), 198 CrossRef CAS PubMed.
- G. Fongaro, P. H. Stoco, D. S. M. Souza, E. C. Grisard, M. E. Magri and P. Rogovski, et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019, Sci. Total Environ., 2021, 778, 146198 CrossRef CAS PubMed.
- F. Wu, J. Zhang, A. Xiao, X. Gu, W. L. Lee and F. Armas, et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases, mSystems, 2020, 5(4), e00614-20 Search PubMed.
- J. Carrillo-Reyes, M. Barragán-Trinidad and G. Buitrón, Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico, J. Water Process. Eng., 2021, 40, 101815 CrossRef.
- R. Gonzalez, K. Curtis, A. Bivins, K. Bibby, M. H. Weir and K. Yetka, et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology, Water Res., 2020, 186, 116296 CrossRef CAS PubMed.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan and A. Casanovas-Massana, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics, Nat. Biotechnol., 2020, 38(10), 1164–1167 CrossRef CAS PubMed.
- S. Feng, A. Roguet, J. S. McClary-Gutierrez, R. J. Newton, N. Kloczko and J. G. Meiman, et al. Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities, ACS ES&T Water, 2021, 1(8), 1955–1965, DOI:10.1021/acsestwater.1c00160 , preprint:acsestwater.1c00160.
- P. M. D'Aoust, E. Mercier, D. Montpetit, J.-J. Jia, I. Alexandrov and N. Neault, et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence, Water Res., 2021, 188, 116560 CrossRef PubMed.
- P. M. D'Aoust, T. E. Graber, E. Mercier, D. Montpetit, I. Alexandrov and N. Neault, et al. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations, Sci. Total Environ., 2021, 770, 145319 CrossRef PubMed.
- A. Nemudryi, A. Nemudraia, T. Wiegand, K. Surya, M. Buyukyoruk and C. Cicha, et al. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater, Cell. Rep. Med., 2020, 1(6), 100098 CrossRef PubMed.
- F. Saguti, E. Magnil, L. Enache, M. P. Churqui, A. Johansson and D. Lumley, et al. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19, Water Res., 2021, 189, 116620 CrossRef CAS PubMed.
- F. Hassard, L. Lundy, A. C. Singer, J. Grimsley and M. Di Cesare, Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools, Lancet Microbe, 2021, 2(1), e4–e5 CrossRef PubMed.
- R. Spurbeck, A. T. Minard-Smith and L. A. Catlin, Applicability of Neighborhood and Building Scale Wastewater-Based Genomic Epidemiology to Track the SARS-CoV-2 Pandemic and other Pathogens, medRxiv, 2021, DOI:10.1101/2021.02.18.21251939v1.
- S. Harris-Lovett, K. L. Nelson, P. Beamer, H. N. Bischel, A. Bivins and A. Bruder, et al. Wastewater Surveillance for SARS-CoV-2 on College Campuses: Initial Efforts, Lessons Learned, and Research Needs, Int. J. Environ. Res. Public Health, 2021, 18(9), 4455 CrossRef CAS PubMed.
- W. Q. Betancourt, B. W. Schmitz, G. K. Innes, S. M. Prasek, K. M. Pogreba Brown and E. R. Stark, et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention, Sci. Total Environ., 2021, 779, 146408 CrossRef CAS PubMed.
- S. Karthikeyan, N. Ronquillo, P. Belda-Ferre, D. Alvarado, T. Javidi and C. A. Longhurst, et al. High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County, mSystems, 2021, 6(2), e00045-21 CrossRef PubMed.
- C. Gibas, K. Lambirth, N. Mittal, M. A. I. Juel, V. B. Barua and L. Roppolo Brazell, et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus, Sci. Total Environ., 2021, 782, 146749 CrossRef CAS PubMed.
- M. Monod, A. Blenkinsop, X. Xi, D. Hebert, S. Bershan and S. Tietze, et al. Age groups that sustain resurging COVID-19 epidemics in the United States, Science, 2021, 371(6536), eabe8372, DOI:10.1126/science.abe8372.
- D. F. M. Reukers, M. van Boven, A. Meijer, N. Rots, C. Reusken and I. Roof, et al. High infection secondary attack rates of SARS-CoV-2 in Dutch households revealed by dense sampling, Clin. Infect. Dis., 2021, ciab237, DOI:10.1093/cid/ciab237/6209401.
- K. K. Bjorkman, T. K. Saldi, E. Lasda, L. C. Bauer, J. Kovarik, P. K. Gonzales, M. R. Fink, K. L. Tat, C. R. Hager, J. C. Davis and C. D. Ozeroff, Higher viral load drives infrequent severe acute respiratory syndrome coronavirus 2 transmission between asymptomatic residence hall roommates, J. Infect. Dis., 2021, 224(8), 1316–1324 CrossRef CAS PubMed.
- E. Lavezzo, E. Franchin, C. Ciavarella, G. Cuomo-Dannenburg, L. Barzon and C. Del Vecchio, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’, Nature, 2020, 584(7821), 425–429 CrossRef CAS PubMed.
- M. M. Arons, K. M. Hatfield, S. C. Reddy, A. Kimball, A. James and J. R. Jacobs, et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility, N. Engl. J. Med., 2020, 382(22), 2081–2090, DOI:10.1056/NEJMoa2008457.
- K. A. Walsh, K. Jordan, B. Clyne, D. Rohde, L. Drummond and P. Byrne, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection, J. Infect., 2020, 81(3), 357–371 CrossRef CAS PubMed.
- S. K. Park, C. W. Lee, D. I. Park, H. Y. Woo, H. S. Cheong, H. C. Shin, K. Ahn, M. J. Kwon and E. J. Joo, Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea, Clin. Gastroenterol. Hepatol., 2021, 19(7), 1387–1394 CrossRef CAS PubMed.
- D. P. Oran and E. J. Topol, Prevalence of Asymptomatic SARS-CoV-2 Infection, Ann. Intern. Med., 2020, 173(5), 362–367, DOI:10.7326/M20-3012.
- B. Moore, The Detection of Paratyphoid Carriers in Towns by means of Sewage Examination, Mon. Bull. Minist. Health Public Health Lab. Serv., 1948, 7, 241–248 Search PubMed.
- T. J. Barrett, P. A. Blake, G. K. Morris, N. D. Puhr, H. B. Bradford and J. G. Wells, Use of Moore swabs for isolating Vibrio cholerae from sewage, J. Clin. Microbiol., 1980, 11(4), 385–388 CrossRef CAS PubMed.
- P. Tian, D. Yang, L. Shan, D. Wang, Q. Li and L. Gorski, et al. Concurrent Detection of Human Norovirus and Bacterial Pathogens in Water Samples from an Agricultural Region in Central California Coast, Front. Microbiol., 2017, 8, DOI:10.3389/fmicb.2017.01560/full.
- P. Liu, M. Ibaraki, J. Van Tassell, K. Geith, M. Cavallo and R. Kann, et al. A Novel COVID-19 Early Warning Tool: Moore Swab Method for Wastewater Surveillance at an Institutional Level, medRxiv, 2020, DOI:10.1101/2020.12.01.20238006v1.
- C. Schang, N. D. Crosbie, M. Nolan, R. Poon, M. Wang and A. Jex, et al. Passive Sampling of SARS-CoV-2 for Wastewater Surveillance, Environ. Sci. Technol., 2021, 55(15), 10432–10441, DOI:10.1021/acs.est.1c01530.
- T. Notomi, Loop-mediated isothermal amplification of DNA, Nucleic Acids Res., 2000, 28(12), e63, DOI:10.1093/nar/28.12.e63.
- B. Schermer, F. Fabretti, M. Damagnez, V. Di Cristanziano, E. Heger and S. Arjune, et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay, PLoS One, 2020, 15(11), e0238612, DOI:10.1371/journal.pone.0238612.
- A. Ganguli, A. Mostafa, J. Berger, M. Y. Aydin, F. Sun and S. A. S. Ramirez, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(37), 22727–22735, DOI:10.1073/pnas.2014739117.
- M. A. Lalli, J. S. Langmade, X. Chen, C. C. Fronick, C. S. Sawyer and L. C. Burcea, et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification, Clin. Chem., 2021, 67(2), 415–424 CrossRef PubMed.
- Y. Zhang, G. Ren, J. Buss, A. J. Barry, G. C. Patton and N. A. Tanner, Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride, BioTechniques, 2020, 69(3), 178–185, DOI:10.2144/btn-2020-0078.
- J. E. Ongerth and R. E. Danielson, RT qLAMP--Direct Detection of SARS-CoV-2 in Raw Sewageitle, medRxiv, 2020, DOI:10.1101/2020.10.01.20205492v1.
- A. Bivins, D. North, Z. Wu, M. Shaffer, W. Ahmed and K. Bibby, Within- and between-Day Variability of SARS-CoV-2 RNA in Municipal Wastewater during Periods of Varying COVID-19 Prevalence and Positivity, ACS ES&T Water, 2021, 1(9), 2097–2108, DOI:10.1021/acsestwater.1c00178.
- A. Bivins, D. North, Z. Wu, M. Shaffer, W. Ahmed and K. Bibby, Within-Day Variability of SARS-CoV-2 RNA in Municipal Wastewater Influent During Periods of Varying COVID-19 Prevalence and Positivity, medRxiv, 2021, DOI:10.1101/2021.03.16.21253652v1.
- J. H. McDonald, Handbook of Biological Statistics, Sparky House Publishing, Baltimore, Maryland, 3rd edn, 2015, pp. 238–246 Search PubMed.
- J. Fox, Applied regression analysis, linear models, and related methods, Sage Publications, Inc., 1997 Search PubMed.
- T. Tjur, Coefficients of Determination in Logistic Regression Models—A New Proposal: The Coefficient of Discrimination, Am. Stat., 2009, 63(4), 366–372, DOI:10.1198/tast.2009.08210.
- O. J. Dunn, Multiple Comparisons Using Rank Sums, Technometrics, 1964, 6(3), 241–252, DOI:10.1080/00401706.1964.10490181.
- W. H. Kruskal and W. A. Wallis, Use of Ranks in One-Criterion Variance Analysis, J. Am. Stat. Assoc., 1952, 47(260), 583–621, DOI:10.1080/01621459.1952.10483441.
- H. B. Mann and D. R. Whitney, On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other, Ann. Math. Stat., 1947, 18(1), 50–60 CrossRef.
- R. Parikh, A. Mathai, S. Parikh, G. Chandra Sekhar and R. Thomas, Understanding and using sensitivity, specificity and predictive values, Indian J. Ophthalmol., 2008, 56(1), 45 CrossRef PubMed.
- R. Elbeblaw, Gastrointestinal SARS CoV-2 Infection and The Dynamic of Its Detection in Stool, J. Respir. Infect., 2020, 4(1), 61 Search PubMed.
- D. L. Jones, M. Q. Baluja, D. W. Graham, A. Corbishley, J. E. McDonald and S. K. Malham, et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19, Sci. Total Environ., 2020, 749, 141364 CrossRef PubMed.
- X. Wang, J. Zheng, L. Guo, H. Yao, L. Wang and X. Xia, et al. Fecal viral shedding in COVID-19 patients: Clinical significance, viral load dynamics and survival analysis, Virus Res., 2020, 289, 198147 CrossRef PubMed.
- S. A. Mahmoud, E. Ibrahim, S. Ganesan, B. Thakre, J. G. Teddy and R. Preeti, Evaluation of RNA extraction free method for detection of SARS-COV-2 in salivary samples for mass screening for COVID-19, medRxiv, 2021, DOI:10.1101/2021.03.15.21253570v1.
- K. Kitamura, K. Sadamasu, M. Muramatsu and H. Yoshida, Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater, Sci. Total Environ., 2021, 763, 144587 CrossRef PubMed.
- A. D. Paltiel, A. Zheng and R. P. Walensky, Assessment of SARS-CoV-2 Screening Strategies to Permit the Safe Reopening of College Campuses in the United States, JAMA Netw. Open, 2020, 3(7), e2016818 CrossRef PubMed.
- B. Lopman, C. Y. Liu, A. Le Guillou, A. Handel, T. L. Lash and A. P. Isakov, et al. A modeling study to inform screening and testing interventions for the control of SARS-CoV-2 on university campuses, Sci. Rep., 2021, 11(1), 5900 CrossRef PubMed.
- T. E. Wong, G. M. Thurston, N. Barlow, N. Cahill, L. Carichino and K. Maki, et al. Evaluating the Sensitivity of SARS-CoV-2 Infection Rates on College Campuses to Wastewater Surveillance, medRxiv, 2020, DOI:10.1101/2020.10.09.20210245v1.
- S. A. Lauer, K. H. Grantz, Q. Bi, F. K. Jones, Q. Zheng and H. R. Meredith, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application, Ann. Intern. Med., 2020, 172(9), 577–582, DOI:10.7326/M20-0504.
- Y. Zhu, W. Oishi, C. Maruo, M. Saito, R. Chen and M. Kitajima, et al. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks, Sci. Total Environ., 2021, 767, 145124 CrossRef CAS PubMed.
- W. Ahmed, A. Bivins, P. M. Bertsch, K. Bibby, P. M. Choi and K. Farkas, et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information, Curr. Opin. Environ. Sci. Health, 2020, 17, 82–93 CrossRef PubMed.
- World Health Organization, Status of environmental surveillance for SARS-CoV-2 virus, Geneva, Switzerland, 2020 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ew00496d |
| ‡ The datasets analyzed during the current study, excluding clinical data, are available in the OSF.IO repository, https://osf.io/2jdbs/ doi: http://10.17605/OSF.IO/2JDBS. |
| This journal is © The Royal Society of Chemistry 2022 |