Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19†
E. K. Hayes a,
C. L. Sweeney
a,
C. L. Sweeney a,
L. E. Anderson
a,
L. E. Anderson a,
B. Li
a,
B. Li a,
G. B. Erjaveca,
M. T. Gouthroa,
W. H. Krkosekb,
A. K. Stoddart
a,
G. B. Erjaveca,
M. T. Gouthroa,
W. H. Krkosekb,
A. K. Stoddart a and
G. A. Gagnon
a and
G. A. Gagnon *a
*a
aCentre for Water Resources Studies, Faculty of Engineering, Department of Civil and Resource Engineering, Dalhousie University, 1360 Barrington St. Halifax, Halifax, NS, Canada B3H 4R2. E-mail: graham.gagnon@dal.ca; Tel: +1 902 494 6070
bHalifax Water, 450 Cowie Hill Road, Halifax, Nova Scotia, Canada B3P 2V3
First published on 20th July 2021
Abstract
The overall objective of this work was to develop a simple and effective passive sampling protocol for the detection of SARS-CoV-2 in sewer catchments at targeted institutional-level sampling sites in a region of low COVID-19 prevalence. We developed a new 3D-printed sampling cage and assessed four commercially-available materials (cotton gauze, cotton cheesecloth, cellulose sponges, and electronegative filters) for RNA adsorption in the cage. We determined that cheesecloth and electronegative filters provided an effective approach for collecting and measuring SARS-CoV-2 in wastewater. We also compared the performance of three elution mixtures (a commercially-available lysis buffer, a Tween®20-based buffer, and a 1:1 acetonitrile:water mixture) for detection of heat-inactivated SARS-CoV-2 reference material (HI-SCV-2) spiked into municipal wastewater at 1.0 × 103 genomic units per millilitre (GU mL−1). The highest mean RNA concentrations were achieved using the cheesecloth (7.0 × 104 ± 3.7 × 104 GU per eluate) and electronegative filters (2.3 × 104 ± 2.5 × 104 GU per eluate) in combination with the Tween®20-based buffer with positive detections in all three biological replicates for both material types. We deployed passive samplers at two sewer catchments (locations A and B) to compare the performance of each passive sampler material type in the field. Over 15 sampling events at each site, we demonstrated that both cheesecloth (location A) and electronegative filters (location B) coupled with a Tween®20-based elution technique could be utilized for the reliable detection of SARS-CoV-2. These results have demonstrated a quick and effective passive sampling approach for SARS-CoV-2 detection in targeted locations in wastewater collection systems, which may have long-term applicability as global vaccination programs evolve.
Water impactThis study demonstrated the effectiveness of an innovative 3D-printed passive sampling device with new extraction techniques for SARS-CoV-2 detection in municipal wastewater in a region with low COVID-19 prevalence. When collected alongside grab samples, the passive sampling approach was more sensitive. Therefore, this approach has global implications as it can provide targeted monitoring as vaccination programs expand. |
1 Introduction
Since the onset of the coronavirus 2019 (COVID-19) pandemic, the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been identified in both respiratory and gastrointestinal tracts,1 with viral shedding reported in faeces and urine of both symptomatic and asymptomatic individuals through all stages of infection.2 The global spread of COVID-19 has led to the progression of wastewater-based epidemiology (WBE) as a macro-scale surveillance tool that is sought to aid in public health decision making.3–9 The WBE approach is relatively new and was developed based on the analysis of biomarkers and pollutants in wastewater to obtain both quantitative and qualitative data on the activity of individuals in a wastewater catchment area.10–14 For example, recent studies have shown that SARS-CoV-2 RNA was detected in sewage samples before any reported cases, suggesting that virus monitoring could be feasible before cases are documented through the health surveillance system.8,15–17 This approach also offers a broader viral surveillance method within the populous at a relatively low cost compared to individual clinical laboratory tests.Although the WBE approach has been applied for SARS-CoV-2 monitoring, much is yet to be understood surrounding sampling methods. Conventionally, samples for WBE have been taken through grab or 24 hour composite sampling techniques.18 Grab sampling, although simple and convenient, provides only a snapshot of representation for a population's wastewater system. Similarly, composite sampling offers a final volume that is more representative of a given population over time but can be time consuming and costly. Further, composite sampling may not be a reliable monitoring approach in areas of low COVID-19 prevalence as composite samples can dilute SARS-CoV-2 signals.19,20 Alternatively, sampling upstream of WWTPs at sewershed pump stations, manholes, or institution-level sites (e.g., commercial properties, airports, university campuses, etc.) can target specific areas of interest. Consideration of the challenges of each sampling approach is critical for developing effective wastewater monitoring programs, which could become increasingly more relevant as vaccination programs begin to take shape globally.21,22
A passive sampling approach provides a cost effective and relatively easy option to grab and composite sampling. This approach involves the use of a passive sampling device which is deployed at a location in the sewershed for a predetermined period, and contaminants in the wastewater are allowed to interact with an adsorbent material housed inside the device,23,24 which results in the concentration of the virus. A passive sampling technique commonly referred to as the “Moore swab” has been previously used to extract enteric pathogens from cotton gauze in water.25,26 Recently, the Moore swab approach was used for passive sampling of SARS-CoV-2 in wastewater and it was determined that passive samplers were at least as sensitive as conventional sampling for detecting SARS-CoV-2 in wastewater.24 Another benefit of the passive sampling approach for WBE is that these devices can be deployed at smaller scales at sewer locations or pump stations for targeted monitoring at a specific location or building where grab or composite sampling may not be feasible.
The Moore swab method involves the use of cotton gauze as a swab and has been used in the collection of microorganisms in water and wastewater, including poliovirus,27 and human norovirus.28 Sikorski and Levine (2020) noted that the type of materials that have been used range from cheesecloth to cotton gauze.25 Specific to SARS-CoV-2 monitoring, Schang et al. (2020) tested medical gauze, laboratory grade electronegative filter paper and cotton buds for passive sampling of viruses in wastewater.24 Additionally, Liu et al. (2020) have successfully used cotton gauze in the passive sampling of wastewater to measure SARS-CoV-2.29 A more recent study successfully implemented the use of tampon swabs in the detection SARS-CoV-2 RNA in wastewater.30 Although these materials have proven to be effective, there is opportunity to further optimize the selection of adsorbent material for passive sampling for SARS-CoV-2 in wastewater. Schang et al. (2020) also stated that further research is needed for laboratory methods, particularly with respect to elution and extraction of the virus from the various adsorbent materials.24 For example, a mixed elution buffer consisting of phosphate buffer solution, Tween 80 and antifoam emulsion was used for the recovery of SARS-CoV-2 RNA from adsorbent material, but it is recognized that there are a number of chemical agents that could be used to increase elution from specific swabbing materials. Thus, there is a need to comparatively evaluate elution efficacy of the various buffers or mixtures that have been considered in the literature.
The overall objective of this work was to develop a simple and effective passive sampling protocol for the detection of SARS-CoV-2 in sewer catchments at targeted institutional-level sampling sites in a region of low COVID-19 prevalence. Our specific aims were to 1) design and construct a 3-D-printed passive sampling device that protects the enclosed adsorbent material from being lost, torn, or obstructed by larger wastewater debris; 2) assess four materials for maximum recovery of heat-inactivated SARS-CoV-2 reference material from deionized water and wastewater; 3) compare the performance of three elution mixtures for maximum recovery of SARS-CoV-2 surrogate RNA (heat-inactivated SARS-CoV-2) from spiked wastewater; and 4) conduct field-scale testing at an institutional level using this passive sampling protocol to monitor SARS-CoV-2 in a region of low COVID-19 prevalence.
2 Materials and methods
2.1 Reagents and materials
Heat inactivated SARS-CoV-2 (HI-SCV-2) (ATCC® VR1986HK™) was sourced from American Type Culture Collection (Virginia, USA). DI water was obtained from a Milli-Q system (Reference A+, Millipore) and contained a total organic carbon (TOC) concentration <5 μg L−1 and a resistivity of 18.2 MΩ cm. Ethanol (EtOH) and acetonitrile (ACN) were purchased from Fisher Scientific (Ottawa, ON, CA). Cellulose sponges, cotton gauze, and cheesecloth were acquired from a local pharmacy, and electronegative filter membranes (4.7 cm, 0.1 μm or 9.0 cm, 0.22 μm cellulose nitrate membrane filters) were purchased from Fisher Scientific (Ottawa, ON, CA) and Sigma-Aldrich (St. Louis, MO, USA). For preliminary work, a premade elution buffer comprised of 0.075% Tween®20 + 25 mM Tris HCl was obtained from Innovaprep Technologies (Drexel, MO, USA). For subsequent experiments, this mixture was made using Tween®20 and Tris-HCl sourced from Sigma Aldrich (Ottawa, ON, CA); 75 μL of Tween®20 and 250 μL of a 0.1 M Tris-HCl intermediate was added to DI water for a total volume of 100 mL. Magnetic binding beads (20 g L−1), RNA extraction kits, and SARS-CoV-2 assay kits were obtained from LuminUltra Technologies Ltd (Fredericton, NB, CA). Samples were stirred on a Fisherbrand™ Isotemp™ magnetic plate stirrer (Fisher Scientific, Ottawa, ON, CA). Bovine serum albumin (BSA), used to reduce inhibition in RT-qPCR reactions, was purchased from Alfa Aesar by Thermo Fisher Scientific (Tewksbury, MA, US) to make a 1 mg mL−1 BSA solution (10 mg lyophilized BSA in 10 mL DI water).2.2 The passive sampling device: COVID-19 sewer cage (COSCa)
To build upon the Moore swab concept as a passive sampling approach for SARS-CoV-2 in low prevalence areas, the COVID-19 Sewer Cage (COSCa) (ESI† Fig. S1) was developed to minimize over-saturation of solids on the adsorbent material (e.g., swab) and to prevent loss or damage of the adsorbent material itself. The COSCa is a 10 cm diameter hollowed sphere with 26 holes, with each hole having a 1.5 cm diameter to foster non-restrictive flow. The COSCa was designed and exported from Fusion 360 software (2018) and sent to a material jetting 3D printer (AnyCubic Mega Zero 2.0) and was printed with acrylonitrile butadiene styrene (ABS) plastic, an engineered thermoplastic with a high melting point that can withstand high autoclave temperatures. The COSCa was printed with solid walls to provide sufficient mass for complete submersion in stagnant waters or in moderate flow catchments.2.3 Wastewater collection for method development
Four 1 L wastewater samples (24 h influent composite) were collected from a wastewater treatment facility (WWTF) in Moncton, New Brunswick, Canada on different calendar days in November and December 2020. Six 1 L wastewater samples (24 h influent composite) were collected from two WWTFs in Halifax and Dartmouth, Nova Scotia, Canada between March, and April 2021. Samples were transported to Dalhousie University on ice and kept at 4 °C for up to 24 h prior to initial RNA extraction to determine background levels of SARS-CoV-2. Remaining sample volumes were stored at −20 °C until used in passive sampling experiments.2.4 Bench-scale experimental set-up for HI-SCV-2 RNA recovery in DI water and wastewater
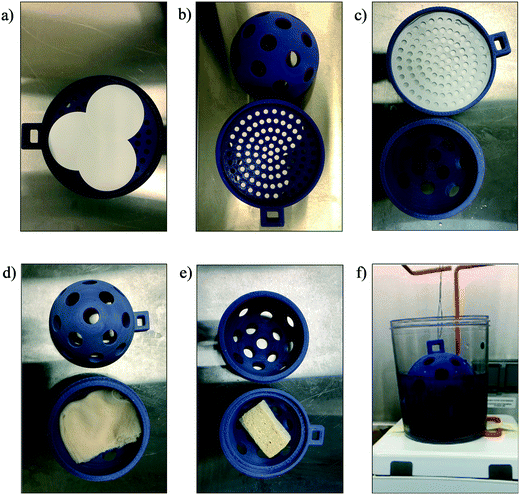
For each bench-scale experiment, 500 mL samples were prepared. DI water and wastewater samples were spiked with HI-SCV-2 (1 × 103 GU mL−1) in triplicate and left stirring continuously at 100 rpm at room temperature for 30 min to equilibrate before adsorbent material was added. To simulate deployment in sewer catchments, adsorbent material was placed inside each COSCa, which were then suspended in the prepared water samples from a biohazard waste bag holder and continuously stirred on a stir plate at 100 rpm for 24 h at room temperature. After 24 h, the material was extracted from the COSCa and placed into 50 mL falcon tubes for subsequent RNA extraction. For each sample batch, a single matrix sample was left unspiked to serve as a blank.Bulk samples of gauze and cheesecloth were cut to approximately 7.6 × 183 cm and folded four times, based on the approach described by Sikorski and Levine (2020).25 The sponge was cut into pieces of approximately 1 × 2.5 × 4 cm. Due to the fragility of electronegative filter papers, filter holders were designed and 3D-printed for each device. Filters were inserted between two attachments and placed inside the COSCas to allow contact with the wastewater while maintaining the integrity of the filter. For each laboratory-controlled sample, three filters were placed adjacent to each other within a 3D-printed electronegative filter holder inside a COSCa device. The placement of different adsorbent materials inside the COSCa passive sampler and experimental set-up are shown in Fig. 1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 acetonitrile
1 acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture. Following a 24 h stirring period, each sample (cheesecloth or filter) was immediately eluted by adding 2 mL of elution mixture, shaking vigorously by hand for 1 min, and incubating for 1 min at room temperature. Residual liquid was pressed out of the adsorbent material by kneading, and the eluate was transferred to a separate falcon tube. This process was repeated twice more for a total elution volume of 6 mL, 1 mL of which was used for RNA extraction using the magnetic bead-based method described in section “1.8 RNA extraction”. This 6 mL elution volume was determined using a scaled down approximation of elution buffer volume used in a Moore swab processing method described by Liu et al. (2020).29
water mixture. Following a 24 h stirring period, each sample (cheesecloth or filter) was immediately eluted by adding 2 mL of elution mixture, shaking vigorously by hand for 1 min, and incubating for 1 min at room temperature. Residual liquid was pressed out of the adsorbent material by kneading, and the eluate was transferred to a separate falcon tube. This process was repeated twice more for a total elution volume of 6 mL, 1 mL of which was used for RNA extraction using the magnetic bead-based method described in section “1.8 RNA extraction”. This 6 mL elution volume was determined using a scaled down approximation of elution buffer volume used in a Moore swab processing method described by Liu et al. (2020).292.5 Comparing the performance of cheesecloth and electronegative filters in COSCa passive samplers at two sewer catchments
Two sewer catchments at targeted institutional-level sampling sites were chosen for this study: location A, a university residence and location B, a business center comprised of five adjacent buildings (ESI† Fig. S2). These sites were selected after news releases indicated that there was at least one known case of COVID-19 at a university residence on the same campus as location A, as well as known cases amongst the five buildings that service location B. Multiple COSCas were not deployed at the same sewer location as we did not want to lose sampling equipment in the sewer system and harm sewer infrastructure. To compare the performance of cheesecloth (7.6 × 183 cm) and electronegative filters (9.0 cm diameter) in the detection of SARS-CoV-2 in wastewater using a passive sampling device, location A was regularly sampled using a COSCa containing cheesecloth swabs and location B was sampled using a COSCa containing filters. Fifteen sampling events were conducted at each location between January and May 2021.For each sampling event, a COSCa was deployed for 24, 48 or 72 h, depending on site accessibility. A paired grab sample (125 mL) was collected with most COSCa samples at location A; however, grab samples were not feasible at location B due to manhole depth restrictions. Following each deployment period, the COSCa was retrieved, and the swab or filter was immediately placed inside a sterile 50 mL Falcon tube and transported to the lab on ice for analysis. All COSCa samples obtained through field experiments were eluted with a Tween®20-based elution buffer. To conserve reagents, single aliquots for each field sample eluate were extracted for SARS-CoV-2 RNA and analyzed via RT-qPCR. In cases when inhibition was expected (e.g., sample eluate with high solids content), extracted RNA was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with BSA. For cheesecloth samples containing a large amount of solids, the eluate was diluted (up to 5-fold) to facilitate RNA extraction. Sample eluate and RNA dilutions for each sampling event are summarized in ESI† Tables S1 and S2. For redeployment, the COSCa was disinfected with EtOH, a new swab or filter was placed inside, and the COSCa was then lowered and placed directly into the wastewater flow.
1 with BSA. For cheesecloth samples containing a large amount of solids, the eluate was diluted (up to 5-fold) to facilitate RNA extraction. Sample eluate and RNA dilutions for each sampling event are summarized in ESI† Tables S1 and S2. For redeployment, the COSCa was disinfected with EtOH, a new swab or filter was placed inside, and the COSCa was then lowered and placed directly into the wastewater flow.
2.6 RNA extraction
RNA extraction for raw wastewater and passive sampler swabs was carried out using a magnetic bead-based RNA extraction procedure.31 A volume of 1 mL of sample (wastewater or COSCa eluate) was used to perform this extraction protocol, resulting in a total of 50 μL of eluted RNA for RT-qPCR analysis. This RNA extraction method was selected for several reasons: A. the LuminUltra GeneCount SARS-CoV-2 Wastewater RT-qPCR Assay Kit utilizes a commercially-available patent-pending method based on a simple and rapid extraction that produces results within a couple of hours; B. this magnetic bead-based extraction protocol was used in our regular wastewater surveillance program and offered direct comparison between sampling approaches; C. the passive sampling swabs required a direct RNA extraction method that could process low-volume particulate-laden samples, as other commonly used RNA extraction method involve large sample volumes and extensive sample pre-processing (i.e., preconcentration, filtration, etc.). The RNA extraction protocol was followed according to manufacturer's instruction and are summarized in the ESI.†2.7 RT-qPCR analysis
All RNA samples were processed by RT-qPCR on a GeneCount® Q-16 instrument (LuminUltra Technologies Ltd, Fredericton, CA). The primers and probes sequences used were published by the US CDC as shown in Table 1.32 For the analysis of SARS-CoV-2, 20-μL reactions were prepared using the GeneCount SARS- CoV-2 Screening kit (LuminUltra Technologies Ltd, Fredericton, CA), containing 15 μL of Master Mix and 5 μL of template RNA. When inhibition was expected, 2.5 μL template RNA was diluted with 2.5 μL BSA solution. Thermal cycling reactions were carried out as follows: a pre-denaturation step at 55 °C for 10 min followed by a second pre-denaturation step at 95 °C for 1 min. The two pre-denaturation steps were followed by 45 cycles of 95 °C for 10 s and 55 °C for 45 s, along with a final hold step at 50 °C for 1 min. Positive detections were indicated by cycle threshold (Ct) values under 40. The RT-qPCR upper Ct value detection threshold being 40 cycles, which corresponds to 1.4 copies per reaction.| Organism | Sequence type | Sequence (5′ – 3′) |
|---|---|---|
| SARS-CoV-2 N2 gene | N2 forward primer | TTACAAACATTGGCCGCAAA |
| N2 reverse primer | GCGCGACATTCCGAAGAA | |
| N2 probe | FAMACAATTTGCCCCCAGCGCTTCAG ZEN/3IABkFQ/ |
2.8 Quantitative analysis of SARS-CoV-2 from passive sampling material
In this work, quantitative analysis is carried out to assess the relative performance of each swab material type and elution mixture. RNA concentrations that reflect the amount of viral RNA eluted from the adsorbent material (total genomic units per 6 mL eluate) were calculated using eqn (1). Recovery of HI-SCV-2 was calculated using eqn (2). A flowchart showing both calculations is shown in ESI† Fig. S3.| RNA concentration (GU per eluate) ≈ sample concentration (GU mL−1) × 6 mL eluate | (1) |
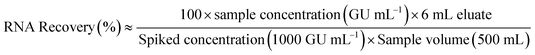 | (2) |
2.9 Quality control
All passive sampling experiments, and RNA extractions were performed in a Thermo Scientific 1300 Series A2 biosafety cabinet, with RT-qPCR assays being prepared in a separate laboratory to minimize contamination. All materials were sterilized in an autoclave to eliminate any pre-contamination, and blank samples were run with all RNA assays to ensure no contamination occurred during sample extractions and preparation. Extracted RNA was analyzed with the RT-qPCR immediately after extraction, with all experiments being performed in triplicate. Standards outlined in MIQE guidelines were consulted for evaluating qPCR-based experiments. Internal positive and negative controls were implemented in each RNA extraction and qPCR assay. The LuminUltra qPCR system utilizes a master standard curve incorporated into the software. Mean Ct values are provided for each assay. To assess sampling efficacy in bench-scale experiments, biological replicates were performed in triplicate while technical replicates were omitted to conserve reagents and materials.The method limit of detection (MLOD) determined by Parra et al. (2021) for the direct extraction of SARS-CoV-2 RNA from raw wastewater was experimentally determined as 40 GU mL−1, and the method recovery efficiency for Accuplex, a SARS-CoV-2 surrogate, from wastewater was reported as ∼12%.31 However, these values for MLOD and recovery efficiency are not directly applicable to this passive sampling approach for two reasons: A) a representative MLOD requires calculations of recovery efficiencies based on adsorption kinetics data, which are beyond the scope of this work; and B) a different surrogate was used to determine recovery efficiency and MLOD for the direct magnetic beads extraction method. To compare the performance of each method parameter, relative recovery was calculated for each adsorbent material type and elution mixture using the HI-SCV-2 surrogate, as shown in ESI† Fig. S3.
2.10 Statistical analysis
A Welch two-sample t-test (two-tailed, α = 0.05) was performed to evaluate the statistical significance between mean HI-SCV-2 RNA concentrations obtained from experiments carried out using different adsorbent materials and elution buffers in both DI water and wastewater. All experiments were performed using three biological replicates and standard deviation was used to determine error bars. All statistical analyses were performed using R (version 4.0.4) software.333 Results and discussion
3.1 Comparison of adsorbent materials for the detection and recovery of HI-SCV-2 from different water matrices
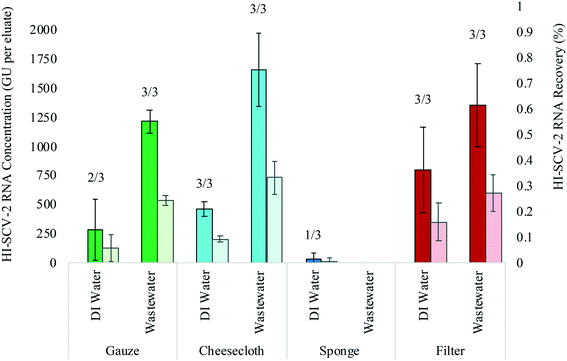
A controlled bench-scale experiment was conducted using four adsorbent materials and two different water matrices (DI water and municipal wastewater, both spiked with HI-SCV-2) to evaluate the detection and recovery of HI-SCV-2. RNA concentrations (GU per 6 mL eluate), recoveries (%), and percent positive detections are shown in Fig. 2. In the controlled experiment using DI water spiked to 1.0 × 103 GU mL−1, HI-SCV-2 RNA was recovered from all four materials. The electronegative filters resulted in the highest mean RNA concentration (8.0 × 102 ± 3.7 × 102 GU per eluate) followed by cheesecloth (4.6 × 102 ± 6.3 × 101 GU per eluate). The sponge resulted in the lowest mean RNA concentrations (3.2 × 101 ± 5.6 × 101 GU per eluate). Ct values for RNA extracted from cheesecloth, gauze, and electronegative filters in DI water ranged from 36.8 to 37.2, 36.8 to 37.5, and 35.6 to 37.3 respectively. The Ct value for the single positive detection in the sponge replicates was 39.5, which is approaching the qPCR limit of detection.To further explore the performance of each material in recovering HI-SCV-2, the experiment was carried out in municipal wastewater that previously tested negative for SARS-CoV-2 and was spiked to 1.0 × 103 GU mL−1 with HI-SCV-2. In this matrix, the highest mean HI-SCV-2 RNA concentration was recovered from the cheesecloth (1.7 × 103 ± 3.1 × 102 GU per eluate). The electronegative filters resulted in the second highest mean RNA concentrations (1.4 × 103 ± 3.6 × 102 GU per eluate), but there was no statistically significant difference (p = 0.331) in the recovered concentrations from the cheesecloth and filters. Although the use of a cellulose sponge was promising due to its absorbance capacity, the surrogate was not detected in any of the sponge replicates. Ct values for RNA extracted from cheesecloth, gauze, and electronegative filters in wastewater ranged from 34.6 to 35.2, 35.3 to 35.6, and 34.8 to 35.5 respectively. In these initial experiments, recoveries were below 1% for all passive sampling material types eluted with the lysis buffer in both matrices.
The recovery of SARS-CoV-2 may be impacted by the presence of solids in wastewater. Retention of solids can improve RNA recovery, as SARS-CoV-2 partitions to solids,34–36 which can act as a vehicle for viral transport. In contrast, excessive retention of solids can impede recovery by inhibiting the RNA extraction process. The optimal passive sampling material should balance the benefit of retaining virus-laden particles and the negative impact of solids on efficient RNA extraction. The cheesecloth and gauze samples retained more solids from wastewater than did the filters. The concentration of HI-SCV-2 RNA recovered using cheese cloth and gauze was not significantly different in DI water (p = 0.373) nor in the wastewater matrix (p = 0.091). The comparable performance of these two readily-available materials suggest that either may be used as an alternative when laboratory-grade materials (e.g., electronegative filters) are not available, which may be relevant for wastewater surveillance programs in remote areas. In some sampling locations where solids content is expected to be high, a filter may be a preferred choice for passive sampling to allow efficient RNA extraction. As such, the solids content of wastewater should be considered when selecting passive sampling material.
The affinity of the solids in the wastewater to the passive sampling material also plays a role in the adsorption of the virus. While COVID-19 virion sizes range from 0.07 to 0.09 μm,37 researchers have found that cellulose nitrate membranes were capable of recovering viruses despite pore sizes exceeding that of the viruses.38 This phenomenon is understood to be the result of multiple reactive sites covering the filters causing viral adhesion or the adsorption of the virus-laden particles that may adhere to the membrane's surface. Consequently, due to the nature of wastewater matrices, high solid content is often observed, which may result in preferential adsorption of organics to the filter membrane thus reducing viral adsorption efficiencies. However, the quantity of solids retained over a 24 h period did not impact RNA extraction processes in bench-scale experiments.
The results of these experiments indicate that electronegative filters and cheesecloth resulted in the highest mean HI-SCV-2 RNA concentrations in spiked DI water and wastewater, respectively. Although cheesecloth may be used as a quick and effective passive sampling material for SARS-CoV-2 detection in municipal wastewater, particularly when electronegative filters may not be readily available, the use of laboratory grade materials provides reproducibility and consistency in results that household materials may not. The cheesecloth and the electronegative filters were selected for further investigation in subsequent bench-scale experiments and field studies because of the comparable performance of these materials in both matrices.
3.2 Comparison of three elution techniques in the analysis of SARS-CoV-2
To optimize laboratory methods for this passive sampling approach, two other elution mixtures in addition to lysis buffer were tested, including a Tween®20-based elution buffer and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 acetonitrile
1 acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
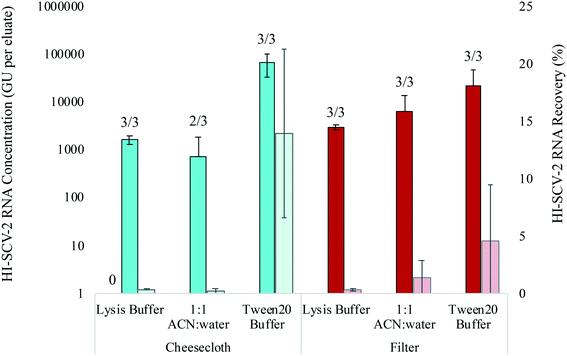
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture. All samples were run in biological triplicates for cheesecloth swabs and electronegative filters in municipal wastewater spiked to 1.0 × 103 GU mL−1 with HI-SCV-2 (Fig. 3). All elution mixtures resulted in positive detections for both material types. The Tween®20-based elution buffer resulted in highest mean RNA concentrations for both materials with positive detections in all three replicates. The cheesecloth resulted in a mean RNA concentration of 7.0 × 104 ± 3.7 × 104 GU per eluate, while a mean concentration of 2.3 × 104 ± 2.5 × 104 GU per eluate was obtained with the filters. There was no statistical difference in mean RNA concentrations between the cheesecloth and filters eluted with the Tween®20-based buffer (p = 0.149). In wastewater, mean recoveries of HI-SCV-2 increased from 0.3 to 13.9% and from 0.3 to 4.5% using the Tween20®-based elution buffer to elute cheesecloth and filters, respectively. All other recoveries in this series of experiments were below 2%. For wastewater, Ct values for RNA extracted from cheesecloth and electronegative filters using the Tween20®-based elution buffer ranged from 28.1 to 29.6 and 29.4 to 34.1 respectively. By comparison, Ct values for all other elution mixtures were less reliable and ranged between 31.3 (one filter eluted with 1
water mixture. All samples were run in biological triplicates for cheesecloth swabs and electronegative filters in municipal wastewater spiked to 1.0 × 103 GU mL−1 with HI-SCV-2 (Fig. 3). All elution mixtures resulted in positive detections for both material types. The Tween®20-based elution buffer resulted in highest mean RNA concentrations for both materials with positive detections in all three replicates. The cheesecloth resulted in a mean RNA concentration of 7.0 × 104 ± 3.7 × 104 GU per eluate, while a mean concentration of 2.3 × 104 ± 2.5 × 104 GU per eluate was obtained with the filters. There was no statistical difference in mean RNA concentrations between the cheesecloth and filters eluted with the Tween®20-based buffer (p = 0.149). In wastewater, mean recoveries of HI-SCV-2 increased from 0.3 to 13.9% and from 0.3 to 4.5% using the Tween20®-based elution buffer to elute cheesecloth and filters, respectively. All other recoveries in this series of experiments were below 2%. For wastewater, Ct values for RNA extracted from cheesecloth and electronegative filters using the Tween20®-based elution buffer ranged from 28.1 to 29.6 and 29.4 to 34.1 respectively. By comparison, Ct values for all other elution mixtures were less reliable and ranged between 31.3 (one filter eluted with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 acetonitrile
1 acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture) and 37.1. In addition to the material type and characteristics, the relative recovery of viral RNA is highly dependent on the performance of the elution buffer.
water mixture) and 37.1. In addition to the material type and characteristics, the relative recovery of viral RNA is highly dependent on the performance of the elution buffer.
Many factors of an elution mixture can impact the elution efficiency of viruses from materials, including difference in pH, salinity, or the use of a surfactant.39 Tween®20 is a non-ionic polysorbate surfactant widely used in biochemical applications and is known for being a gentle surfactant at lower concentrations to prevent premature cell lysis.40 In other work, Tween®20 has been successfully implemented for ultrafiltration techniques, significantly increasing microbial recovery efficiencies.41 Tween®80, has been utilized for its capability to elute viruses from filtration media,42 and most recently, SARS-CoV-2 from passive sampling material.24,29 The use of either Tween®20 or Tween®80 are often interchangeable, with the main difference of the two being the composition of fatty acids.43 Liu et al., (2020) and Schang et al., (2020) used 0.05% Tween®80 mixed with sterile phosphate buffer solution and 0.001% Y-30 antifoam emulsion for elution of SARS-CoV-2 from cotton gauze collected from passive samplers.24,29
In this study, quantitative analysis of viral RNA was carried out to assess the performance (i.e., relative recovery efficiency) of each adsorbent material type and elution mixture. Although recovery assessment for other quantitative viral RNA extraction methods is often carried out using the spike-and-recovery approach with a surrogate, this may not accurately represent recovery efficiency, as many surrogates used to assess SARS-CoV-2 extraction efficiency do not appreciably partition to the solids fraction of wastewater when seeded as SARS-CoV-2 does naturally.44 This difference in behaviour between SARS-CoV-2 and its surrogates may introduce variability in, and impact interpretability of, results for not only controlled recovery studies, but for our assessment of relative recovery as well. As such, field studies to assess the optimized passive sampling approach (i.e., combination of materials and elution buffers) were a critical next step from bench-scale experiments to optimize the detection and quantitation of SARS-CoV-2. Based on the performance of both the electronegative filters and cheesecloth from our laboratory-controlled experiments, these two material types in combination with the Tween®20-based elution buffer were used in subsequent field experiments.
3.3 Detection of SARS-CoV-2 in two sewer catchments using cheesecloth and electronegative filters in COSCa passive samplers
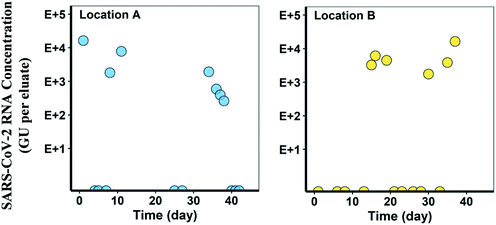
Over 15 sampling events at each location, SARS-CoV-2 was detected on seven separate occasions at location A (COSCa with cheesecloth swabs) and five sampling events at location B (COSCa with electronegative filters) (Fig. 4). Based on results from bench-scale experiments, the Tween®20-based buffer was used for viral elution from cheesecloth swabs and filters in all field experiments. Detection levels varied at location A, ranging from 2.6 × 102 to 1.6 × 104 GU per eluate and from 1.8 × 103 to 6.1 × 103 GU per eluate at location B. SARS-CoV-2 was not detected in any of the grab samples paired with the passive samples collected from location A.At location A, there were three positive SARS-CoV-2 RNA detections in the first six sampling events. The viral signal was not detected in the next two sampling events but reappeared in the following four consecutive sampling events. For these positive detections, there was a decline in the eluate viral RNA concentration from 1.9 × 103 to 2.6 × 102 GU mL−1, and the signal was eventually no longer detected in the last three sampling events. At location B, three consecutive positive detections were observed following four non-detects. The signal appeared again in two of the four remaining sampling events. SARS-CoV-2 RNA concentrations ranged from 1.8 × 103 to 6.1 × 103 GU mL−1 at location B. Although the sampling sites in this study were targeted based on known cases in the areas at the time, the actual number of cases in each location, and the contributing population of each catchment, were unknown.
The results of passive sampling at these two sewer catchments demonstrate that both cheesecloth (location A) and electronegative filters (location B) are effective materials for detecting SARS-CoV-2 in wastewater when eluted with a Tween®20-based buffer. Furthermore, this COSCa sampling approach successfully detected changes in viral presence in two small contributing populations, with distinct resolution in viral RNA concentrations observed across two orders of magnitude. However, it is possible that the adsorption capacity of the passive sampling materials used in this study has been exceeded, as maximum mean RNA concentrations do not exceed 7 × 104 GU per eluate in bench-scale and field experiments. To determine the maximum absorbance capacity of these passive sampling materials, further research investigating the adsorption kinetics of the COSCa passive sampling materials is required.
Our results also demonstrated the lack of sensitivity of grab sampling when paired with passive sampling. However, the suitability of this passive sampling approach may ultimately depend on specific site characteristics and water quality parameters. The deployment of these devices is ideal for low flow locations, such as manholes and thus, are best suited to target specific buildings, designated catchment areas within the sewershed, or remote communities.
Conclusions
In assessing the performance of four adsorption materials and three elution mixtures for the analysis of SARS-CoV-2 in municipal wastewater using our 3D-printed passive sampling devices, the results of this study show that cheesecloth and electronegative filters in combination with the Tween®20-based elution buffer resulted in the highest mean concentrations of a SARS-CoV-2 surrogate in bench-scale studies. When deployed at two targeted locations within the sewer catchment, both cheesecloth (location A) and electronegative filters (location B) allowed the reliable detection of SARS-CoV-2 in wastewater. Furthermore, this passive sampling approach revealed fluctuations in viral presence in the two small contributing populations at these locations.This work demonstrates the effectiveness of passive sampling to detect SARS-CoV-2 in wastewater, and the lack of sensitivity of grab sampling in low prevalence areas when grab samples were collected along with COSCa samples. During prolonged periods of low COVID-19 prevalence, detection in wastewater using grab and composite sampling strategies can be inconsistent and ineffective. To overcome these challenges, the COSCa provides a solution that can foster more direct and targeted analysis when the number of COVID-19 cases are low, which may have increased relevance as vaccination programs expand. The potential use of the described passive method provides added sensitivity and straightforward approach to concentrating samples during collection. The passive sampling approach outlined offers a quick and effective wastewater monitoring tool for SARS-CoV-2 detection in targeted locations that may provide an early warning signal during low COVID-19 prevalence.
4 Research ethics statement for wastewater surveillance studies
In consultation with the Research Ethics Board (REB) at Dalhousie University, it was determined that REB review was not required for research that involves analysis of anonymous human biological materials (such as municipal waste) without generating identifiable information. This research complies with Article 2.4 described in the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans (TCPS 2, 2018).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded through support from a Research Nova Scotia Special Initiatives Grant [grant number #1367], an NSERC Collaborative Research and Development Grant in partnership with Halifax Water [grant number CRDPJ 539387-19] and the NSERC/Halifax Water Industrial Research Chair program [grant number IRCPJ: 349838-16]. The authors would like to extend thanks to municipal staff at institutions A and B and Halifax Water for sample collection. As well, the authors would like to extend thanks to researchers Drs. Ana Parra and Yannan Huang from the Centre for Water Resources Studies at Dalhousie University for providing technical reviews, sample collection and laboratory support during the study, and to Dr. Jason LeBlanc from Dalhousie University and Nova Scotia Health for technical support. The graphical abstract was adapted from http://BioRender.com (2021); retrieved from https://app.biorender.com.References
- Y. Tian, L. Rong, W. Nian and Y. He, Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission, Aliment. Pharmacol. Ther., 2020, 51(9), 843–851 CrossRef CAS PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins and J. W. O'Brien, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS PubMed.
- O. E. Hart and R. U. Halden, Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges, Sci. Total Environ., 2020, 730, 138875 CrossRef CAS PubMed.
- A. Carducci, I. Federigi, D. Liu, J. R. Thompson and M. Verani, Making Waves: Coronavirus detection, presence and persistence in the water environment: State of the art and knowledge needs for public health, Water Res., 2020, 179, 115907 CrossRef PubMed.
- P. Foladori, F. Cutrupi, N. Segata, S. Manara, F. Pinto and F. Malpei, et al., SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review, Sci. Total Environ., 2020, 743, 140444 CrossRef CAS PubMed.
- T. Prado, T. M. Fumian, C. F. Mannarino, P. C. Resende, F. C. Motta and A. L. F. Eppinghaus, et al., Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil, Water Res., 2021, 191, 116810 CrossRef CAS PubMed.
- W. Q. Betancourt, B. W. Schmitz, G. K. Innes, S. M. Prasek, K. M. Pogreba Brown and E. R. Stark, et al., COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention, Sci. Total Environ., 2021, 779, 146408 CrossRef CAS PubMed.
- Y. Zhu, W. Oishi, C. Maruo, M. Saito, R. Chen and M. Kitajima, et al., Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks, Sci. Total Environ., 2021, 767, 145124 CrossRef CAS PubMed.
- K. Mao, H. Zhang, Y. Pan and Z. Yang, Biosensors for wastewater-based epidemiology for monitoring public health, Water Res., 2021, 191, 116787 CrossRef CAS PubMed.
- F. Y. Lai, K. Lympousi, F. Been, L. Benaglia, R. Udrisard and O. Delémont, et al., Levels of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in raw wastewater as an innovative perspective for investigating population-wide exposure to third-hand smoke, Sci. Rep., 2018, 8(1), 13254 CrossRef PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6125383/.
- F. Been, M. Bastiaensen, F. Y. Lai, A. L. N. van Nuijs and A. Covaci, Liquid Chromatography–Tandem Mass Spectrometry Analysis of Biomarkers of Exposure to Phosphorus Flame Retardants in Wastewater to Monitor Community-Wide Exposure, Anal. Chem., 2017, 89(18), 10045–10053 CrossRef CAS PubMed.
- P. M. Choi, B. Tscharke, S. Samanipour, W. D. Hall, C. E. Gartner and J. F. Mueller, et al., Social, demographic, and economic correlates of food and chemical consumption measured by wastewater-based epidemiology, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(43), 21864–21873 CrossRef CAS PubMed.
- L. Lopardo, D. Adams, A. Cummins and B. Kasprzyk-Hordern, Verifying community-wide exposure to endocrine disruptors in personal care products – In quest for metabolic biomarkers of exposure via in vitro studies and wastewater-based epidemiology, Water Res., 2018, 143, 117–126 CrossRef CAS PubMed.
- N. I. Rousis, E. Gracia-Lor, E. Zuccato, R. Bade, J. A. Baz-Lomba and E. Castrignanò, et al., Wastewater-based epidemiology to assess pan-European pesticide exposure, Water Res., 2017, 121, 270–279 CrossRef CAS PubMed.
- G. Orive, U. Lertxundi and D. Barcelo, Early SARS-CoV-2 outbreak detection by sewage-based epidemiology, Sci. Total Environ., 2020, 732, 139298 CrossRef CAS PubMed.
- W. Randazzo, P. Truchado, E. Cuevas-Ferrando, P. Simón, A. Allende and G. Sánchez, SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area, Water Res., 2020, 181, 115942 CrossRef CAS PubMed.
- W. Ahmed, B. Tscharke, P. M. Bertsch, K. Bibby, A. Bivins and P. Choi, et al., SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study, Sci. Total Environ., 2021, 761, 144216 CrossRef CAS PubMed.
- K. Curtis, D. Keeling, K. Yetka, A. Larson and R. Gonzalez, Wastewater SARS-CoV-2 Concentration and Loading Variability from Grab and 24-Hour Composite Samples, medRxiv, 2020, DOI:10.1101/2020.07.10.20150607.
- S. Harris-Lovett, K. Nelson, P. Beamer., H. N. Bischel, A. Bivins and A. Bruder, et al. Wastewater surveillance for SARS-CoV-2 on college campuses: Initial efforts, lessons learned and research needs, medRxiv, 2021, DOI:10.1101/2021.02.01.21250952.
- W. Ahmed, S. Simpson, P. Bertsch, K. Bibby, A. Bivins and L. Blackall, et al., Minimizing Errors in RT-PCR Detection and Quantification of SARS-CoV-2 RNA for Wastewater Surveillance, 2021 Apr 19 [cited 2021 May 18]; DOI:10.20944/preprints202104.0481.v1, Available from: https://www.preprints.org/manuscript/202104.0481/v1.
- M. Levine-Tiefenbrun, I. Yelin, R. Katz, E. Herzel, Z. Golan and L. Schreiber, et al., Decreased SARS-CoV-2 viral load following vaccination, medRxiv, 2021, DOI:10.1101/2021.02.06.21251283.
- C. Gibas, K. Lambirth, N. Mittal, M. A. I. Juel, V. B. Barua and L. Roppolo Brazell, et al., Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus, Sci. Total Environ., 2021, 146749, 782 Search PubMed.
- M. I. G. S. Almeida, A. M. L. Silva, R. A. Coleman, V. J. Pettigrove, R. W. Cattrall and S. D. Kolev, Development of a passive sampler based on a polymer inclusion membrane for total ammonia monitoring in freshwaters, Anal. Bioanal. Chem., 2016, 408(12), 3213–3222 CrossRef CAS PubMed.
- C. Schang, N. Crosbie, M. Nolan, R. Poon, M. Wang and A. Jex, et al., Passive sampling of viruses for wastewater-based epidemiology: a case-study of SARS-CoV-2, 2020 Dec 2 Search PubMed.
- M. J. Sikorski and M. M. Levine, Reviving the “Moore Swab”: a Classic Environmental Surveillance Tool Involving Filtration of Flowing Surface Water and Sewage Water To Recover Typhoidal Salmonella Bacteria, Appl. Environ. Microbiol., 2020, 86(13), e00060-20 CrossRef PubMed , Available from: https://aem.asm.org/content/86/13/e00060-20.
- B. Moore, The Detection of Enteric Carriers in Towns By Means of Sewage Examination, R. Sanit. Inst. J., 1951, 71(1), 57–60 CrossRef CAS PubMed.
- G. Matrajt, B. Naughton, A. S. Bandyopadhyay and J. S. Meschke, A Review of the Most Commonly Used Methods for Sample Collection in Environmental Surveillance of Poliovirus, Clin. Infect. Dis., 2018, 67(suppl_1), S90–S97 CrossRef CAS PubMed.
- P. Tian, D. Yang, L. Shan, D. Wang, Q. Li and L. Gorski, et al., Concurrent Detection of Human Norovirus and Bacterial Pathogens in Water Samples from an Agricultural Region in Central California Coast, Front. Microbiol., 2017, 8 DOI:10.3389/fmicb.2017.01560/full.
- P. Liu, M. Ibaraki, J. VanTassell, K. Geith, M. Cavallo and R. Kann, et al., A Novel COVID-19 Early Warning Tool: Moore Swab Method for Wastewater Surveillance at an Institutional Level, medRxiv, 2020, DOI:10.1101/2020.12.01.20238006.
- A. Bivins, M. Lott, M. Shaffer, Z. Wu, D. North and E. Lipp, et al., Building-Level Wastewater Monitoring for COVID-19 Using Tampon Swabs and RT-LAMP for Rapid SARS-Cov-2 RNA Detection, 2021 May 17 [cited 2021 May 18], Available from: https://www.preprints.org/manuscript/202105.0381/v1.
- A. L. Parra Guardado, C. L. Sweeney, E. K. Hayes, B. F. Trueman, Y. Huang and R. C. Jamieson, et al., Development and optimization of a new method for direct extraction of SARS-CoV-2 RNA from municipal wastewater using magnetic beads, medRxiv, 2020 Jan 1, DOI:10.1101/2020.12.04.20237230.
- CDC, CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel: CDC-006-00019, Revision: 05, CDC/DDID/NCIRD/Division of Viral Disease, 2020, [cited 2020 Sep 29], Available from:https://www.fda.gov/media/134922/download Search PubMed.
- R Core Team, R: A language and environment for statistical computing, Vienna, Austria; 2020, Available from: https://www.r-project.org/ Search PubMed.
- Y. Ye, R. M. Ellenberg, K. E. Graham and K. R. Wigginton, Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater, Environ. Sci. Technol., 2016, 50(10), 5077–5085 CrossRef CAS PubMed.
- K. E. Graham, S. K. Loeb, M. K. Wolfe, D. Catoe, N. Sinnott-Armstrong and S. Kim, et al., SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed, Environ. Sci. Technol., 2020, 55(1), 448–498 Search PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7737534/.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan and A. Casanovas-Massana, et al., Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics, Nat. Biotechnol., 2020, 38(10), 1164–1167 CrossRef CAS PubMed.
- S. Kumar, R. Nyodu, V. K. Maurya and S. K. Saxena, Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), in Coronavirus Disease 2019 (COVID-19): Epidemiology, Pathogenesis, Diagnosis, and Therapeutics, Medical Virology: From Pathogenesis to Disease Control, ed. Saxena S. K., Springer, Singapore, 2020 [cited 2021 May 18]. pp. 23–31, DOI:10.1007/978-981-15-4814-7_3.
- L. A. Ikner, C. P. Gerba and K. R. Bright, Concentration and Recovery of Viruses from Water: A Comprehensive Review, Food Environ. Virol., 2012, 4(2), 41–67 CrossRef PubMed.
- N. L. Turnage and K. E. Gibson, Sampling methods for recovery of human enteric viruses from environmental surfaces, J. Virol. Methods, 2017, 248, 31–38 CrossRef CAS PubMed.
- S. Srivatsan, P. D. Han, K. van Raay, C. R. Wolf, D. J. McCulloch and A. E. Kim, et al., Preliminary support for a “dry swab, extraction free” protocol for SARS-CoV-2 testing via RT-qPCR, bioRxiv, 2020 Jan 1, DOI:10.1101/2020.04.22.056283.
- E. Forés, S. Bofill-Mas, M. Itarte, S. Martínez-Puchol, A. Hundesa and M. Calvo, et al., Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater, Sci. Total Environ., 2021, 768, 144786 CrossRef PubMed.
- S. R. Farrah, Chemical Factors Influencing Adsorption of Bacteriophage MS2 to Membrane Filterst, Appl. Environ. Microbiol., 1982, 43, 5 CrossRef PubMed.
- F. O. Ayorinde, S. V. Gelain, J. H. Johnson and L. W. Wan, Analysis of some commercial polysorbate formulations using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, Rapid Commun. Mass Spectrom., 2000, 14(22), 2116–2124 CrossRef CAS PubMed.
- Canadian Water Network, Phase I Inter-Laboratory Study: Comparison of approaches to quantify SARS-CoV-2 RNA in wastewater, 2020, [cited 2021 Mar 2]. Available from: https://cwn-rce.ca/covid-19-wastewater-coalition/phase-1-inter-laboratory-study Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ew00207d |
| This journal is © The Royal Society of Chemistry 2021 |