Open Access Article
Open Access ArticleBacterial transmission and colonization in activated carbon block (ACB) point-of-use (PoU) filters†
Chia-Chen
Wu
 a,
Nancy G.
Love
a,
Nancy G.
Love
 b and
Terese M.
Olson
b and
Terese M.
Olson
 *b
*b
aInstitue of Environmental Health Sciences, Wayne State University, Detroit, MI, USA
bDepartment of Civil and Environmental Engineering, University of Michigan, Ann Arbor, MI, USA. E-mail: tmolson@umich.edu
First published on 4th May 2021
Abstract
Drinking water bacteria are known to colonize commercial activated carbon block (ACB) point-of-use (PoU) drinking water filters. The breakthrough pattern of drinking water bacteria through these filters, however, is unclear. A manifold system was constructed with two of the most common filter brands in the U.S. and operated to simulate normal household diurnal use. The filters were fed tap water inoculated with two fluorescent-tagged bacterial species that were acclimated to drinking water conditions (GFP-Pseudomonas aeruginosa and mCherry-Escherichia coli). Separate breakthrough tests with abiotic fluorescent microspheres (1 μm) were also conducted. The fluorescent P. aeruginosa and E. coli strains were detected in filter effluent within the first 6 and 28 L processed, respectively. The fluorescent microspheres were also detected in the effluent within the first 6 L processed, indicating that preferential flow pathways with diameters larger than 1 μm allow influent particles to transit the entire thickness of the carbon block. P. aeruginosa broke through the filters more quickly than E. coli, possibly due to its smaller cell size. P. aeruginosa also displayed greater colonization advantages over E. coli, and its effluent cell number plateaued at levels two times the influent level. This study provides evidence that some bacteria, including species known to include opportunistic and enteric pathogenic strains, can be readily transmitted through solid block activated carbon filters and that these filters can elevate the levels of effective colonizers in drinking water.
Water impactThe colonization of opportunistic pathogens in activated carbon block point-of-use drinking water filters can pose health effects to users who are immune-compromised and vulnerable to infection. Understanding the breakthrough pattern of drinking water bacteria through the filters is crucial to evaluate the risk of using this device as a sole unit to treat tap water. |
Introduction
Growing public concern for tap water safety has increased consumer demand for commercial point-of-use (PoU) filters, especially in the U.S. In a 2017 U.S. market survey, it was estimated that 72% of consumers own at least one filtration product.1 During several recent water quality contamination events in the U.S., in fact, governmental and charitable organizations have provided consumers with PoU filters to protect their health.2,3 These distributed PoU filters are faucet-mounted, activated carbon block (ACB) type filters that are commercially available and certified4–6 for the removal of disinfectant residual, chemicals of aesthetic and health concern, and particles including protozoan cysts. As the use of ACB PoU filters becomes an accepted practice to limit exposure to selected contaminants, the potential limitations of these devices warrant further study.While ACB PoU filters are effective for the aesthetic and health-related contaminants they are certified for, they are not designed or certified to remove bacteria. Studies of bacterial levels in ACB-type tap water filters have demonstrated this limitation. For example, multiple studies that rely on culture-based methods have shown that heterotrophic bacterial counts increase with time in the effluent of ACB PoU filters to levels that exceed those in the tap water influent, even when used within the manufacturers' recommended usage period.7–10 Other culture-based studies found that the composition of bacteria in the filter effluent was different from the tap water.10–12 Chaidez et al. (2004) found that indigenous Aeromonas hydrophila, Pseudomonas aeruginosa and total coliforms were more abundant in the effluent than in the influent using culture-based techniques.11 Using culture-independent techniques, studies have similarly found that ACB PoU filters significantly alter the bacterial abundance and composition in filter effluents.10,13
Despite evidence that some bacteria can pass through or grow in ACB PoU filters, it is unclear how efficiently pathogens or opportunistic pathogens of interest can move through them. ACB filters employ a solid block of compressed and polymer-fused powdered activated carbon as the filter media. The ACB contains extensive surface area for physical adsorption of chemical contaminants and has low porosity for mechanical filtration of some particulate contaminants.10 The porosity of the ACB is expected to be very small; for example, line pressures of at least 210 kPa (30 psi) are required for water to pass through the ACB cylinder radially and exit to the annular effluent channel.10 In addition, in an earlier study we estimated that the average micropore size of the activated carbon in one brand of ACB PoU filters is 22 Å.10 Influent microorganisms are unlikely to traverse the entire filter media thickness, therefore, unless there are flow pathways with diameters that are larger than the bacterial cell size. Even if preferential flow channels of bacterial size are present in the ACB, previously observed differences between the bacterial community structure of influent and effluent samples suggests that a fraction of influent bacterial species are able to remain viable11,12 and migrate through an ACB filter.10–12 Beyond questions about the transport efficiency, survival- and growth-related selection factors would be expected to control the changes in bacterial community structure within ACB filters.
The goal of this study was to assess the transmission and growth of two bacteria species with different physiological characteristics and public health significance in commonly used commercial faucet-mounted ACB PoU filters. Bacterial breakthrough experiments were conducted by simultaneously spiking fluorescent protein-tagged forms of P. aeruginosa and non-pathogenic Escherichia coli into tap water containing monochloramine as the residual disinfectant. An important distinction in the approach relative to other literature studies were the steps taken to ensure that the spiked strains were acclimated to the oligotrophic conditions of tap water by first exposing them to limited nutrient and carbon sources at concentrations typical of tap water. E. coli was selected because it is an indicator for fecal contamination and is known to be readily outcompeted by indigenous bacteria in low-nutrient environments.14 It is not commonly detected in drinking water unless there are deficiencies in the treatment process or in the distribution system. P. aeruginosa is not regulated for municipal drinking water and is an opportunistic pathogen that is significant for immunocompromised populations.15 Possibly due to its ability to form biofilms,16P. aeruginosa has been detected in chlorinated water and chloraminated water with a range from 2 to 28 cell equivalent per liter water.17,18 Exposure routes that pose the greatest health risk for P. aeruginosa are direct contact and inhalation,19 and E. coli is oral consumption. Studying the intra-filter migration of two species with different properties and exposure routes offers a better understanding of the bacterial exposure risks posed by ACB PoU filters.
To learn more about the particle physical transport pathways in the filters, separate breakthrough studies were conducted with bacteria-sized microspheres. Negatively charged fluorescent microspheres were selected to simulate “abiotic” bacteria in these studies. A previous study by Paramonova et al.20 demonstrated that latex microspheres serve as reasonable surrogates for Gram negative bacteria in granular activated carbon beds. Microsphere breakthrough, however, has not been studied in an ACB PoU filter configuration with intermittent (on/off) usage patterns. By tracking the breakthrough of these abiotic particles under conditions of household filter use it was possible to characterize the physical filtration efficiency of the ACB filters for bacteria surrogates.
Materials and methods
Filter manifold system
A manifold system with the two most commonly used commercial brands of ACB PoU filters in the U.S. was constructed for the breakthrough tests of fluorescent bacteria and microspheres (Fig. 1a). Both filter brands have similar construction. Each filter contains a synthetic fabric mesh that is wrapped around the ACB and serves as a particle pre-filter. During operation, the influent water flows radially from the outside surface through the carbon block pore network, and then exits the filter from a center effluent channel of the ACB (Fig. 1b). Duplicate filters of each brand (brand A and B) were tested and connected in parallel, with two brands in alternating manifold positions (A – B – A – B). To simulate household filter usage, influent was mixed with known amounts of fluorescent-tagged bacteria or microspheres and intermittently fed to the PoU filters under the control of a LabVIEW program. Details of the program are given in the ESI,† Operational Program of the filter manifold system. Approximately 500 mL influent was pumped (Brass Rotary Vane Pump, Procon) to individual filters each hour for 11 h (bacteria experiment) or 16 h (microsphere experiment), followed by an overnight stagnation period of 13 h (bacteria) or 8 h (microspheres). Influent pumping during each hourly cycle lasted 5 to 7 s, depending upon the life stage of the filter (pumping times increased slightly as the filters aged and headloss increased). The filtration order was rotated every day so that each filter received the first influent water sample every fourth day. For the bacteria breakthrough experiment, the influent was tap water mixed with mCherry-tagged E. coli and GFP-tagged P. aeruginosa to obtain final concentrations of 5 × 102 cell per mL. The system was operated for a total of 29 days, comprising an application phase of 23 days during which fluorescent bacteria were mixed with the tap water influent, and a washout phase of 6 days with no supplemental fluorescent bacteria. For the microsphere experiment, the operation was similarly prepared with a few modifications. The influent was deionized water mixed with fluorescent microspheres to generate final concentrations of 5 × 105 particle per mL. The run period was ended at day 13, when the processed water volume reached approximately 100 L, because the four filters were impacted by clogging and one of the filters (B-4) developed a crack due to backpressure.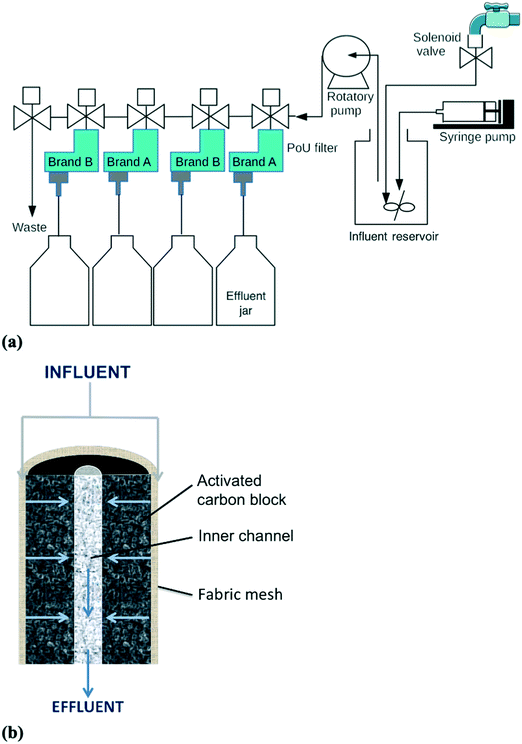 | ||
| Fig. 1 Schematics of the (a) PoU filter manifold system and (b) cross sectional profile of the ACB filter cartridge (image modified from Wu et al.10). | ||
Cultures and reagents
The two strains were acclimated to low organic carbon growth conditions by growing them for 24 hours each in 1X, 10X, 50X, then 250X diluted R2A medium.25 The total organic carbon (TOC) level of 250X diluted R2A medium was measured as 9.25 ± 0.60 mg L−1. Then, 5 mL of 250X-diluted R2A bacterial solution was transferred to 45 mL of dechloraminated, 0.22 μm-filtered tap water for 24 h prior to its use as a syringe stock. The concentration of syringe stock was approximately 3–5 × 105 cell per mL. The additional cumulative background total organic carbon (TOC) mass contributed by the R2A medium in the acclimated syringe stock cultures was estimated to be 206 μg TOC per filter throughout the application phase. From an earlier study in our laboratory, the influent tap water contained 2.3 ± 0.4 mg L−1 TOC over a two month period.10 Thus the additional TOC contributed by the R2A medium was too small to affect TOC levels in the influent water. The tap water for acclimation was dechloraminated with sodium thiosulfate (Na2S2O3); however, the tap water used to dilute the culture stocks in the influent reservoir was not dechloraminated to simulate the actual conditions expected with tap water filtration. The mean influent total chlorine concentration over the period of operation was 0.86 ± 0.12 mg L−1 as Cl2. The fluorescence of E. coli and P. aeruginosa determined by flow cytometry was similar before and after mixing with tap water for 2 min. The total chloramine contact time with the spiked bacteria was calculated to be 2.12 min, including mixing and pumping times.
Sampling protocols and parameters
Sampling protocols were slightly different between the biotic (fluorescent bacteria) and abiotic (microsphere) breakthrough tests. Water sample types collected for the fluorescent bacteria breakthrough test included: 500 mL grabs of filter influent and effluent, and 11 h composite filter effluent samples. Effluent samples were collected every two days and measured for culturability and fluorescent bacteria concentration. Additional effluent samples were collected twice a week for bacterial measurements of total and membrane-intact cell counts. Grab and composite effluent samples were compared to evaluate the stability of fluorescent and total bacteria concentrations in the effluent container. After the washout phase, the four filters were sacrificed to collect biofilm samples from pieces of the fabric mesh in three locations (top, middle, and bottom) and from swabs of the interior core effluent channel walls at the top, middle, and bottom. During the microsphere breakthrough test, the influent and effluent grab samples from the 1st, 9th, and 15th hours were collected every day for analysis.Details of analytical methods are given in the ESI,† Fig. S1–S3, and Table S1. Briefly, the counts of fluorescent-tagged bacteria and fluorescent microspheres were analyzed by ZE5 Cell Analyzer (Bio-Rad, CA). The detection limit of fluorescent-tagged bacteria is 50 cell per mL. Total bacterial measurements were conducted using a BD Accuri C6 Plus flow cytometer (BD Accuri cytometers, Belgium). Bacterial cells were stained by SYTO Green and propidium iodide (PI). The membrane intact cells are determined by subtracting the membrane-compromised cells (PI stained cells) from the total cell counts (SYTO Green stained cells). For the fluorescent-tagged bacteria breakthrough test, biofilm DNA from ACB fabric mesh and the swab of the filter inner channel walls was extracted using the DNA IQTM Reference Sample Kit for MaxWell® 16 (Promega, Madison, WI, USA) with bead beating described in Wu et al.10 The extracted DNA was analyzed by qPCR to quantify GFP and mCherry gene counts. The culturability of fluorescent-tagged bacteria were evaluated by the membrane filtration method,31 or spreading one fabric piece or a biofilm swab on Pseudomonas isolation agar (Fisher Scientific, Hampton, NH) for P. aeruginosa32 and M Endo LES agar (Fisher Scientific, Hampton, NH) for E. coli.31 The size and cell hydrophobicity of fluorescent-tagged bacteria before and after acclimation were characterized by microscopic analysis and a bacterial-adhesion-to-hydrocarbons (BATH) assay,33 respectively.
Results and discussion
Breakthrough of fluorescent bacteria and 1 μm microspheres provides evidence of preferential pathways
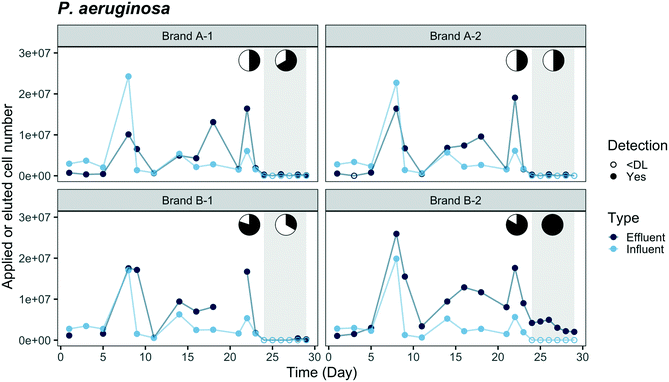
Breakthrough profiles are presented in Fig. 2 and 3 for the two fluorescent-tagged bacterial species, which were spiked into the tap water influent of two ACB PoU filter brands. The effluent daily cell numbers in Fig. 2 and 3 were estimated as the product of the measured bacterial concentration in 11 h composite samples times the daily processed water volume. We found that the concentrations of fluorescent bacteria from the grab samples of the 1st, 5th, and 11th hours of the day were similar to the daily composite samples (p > 0.05, one-sided t test, data not shown). Therefore, the composite samples can be used to represent the daily average of hourly effluent samples and the agreement between the grab and composite samples suggests that the fluorescent bacteria did not proliferate or die in the effluent jar. Both species, P. aeruginosa and E. coli, were detected in the effluents of brand A and B filters. In fact, P. aeruginosa was detected within the first day, indicating that there must be flow pathways of sufficient size to allow some organisms to traverse the entire distance from the influent to the effluent of the ACB.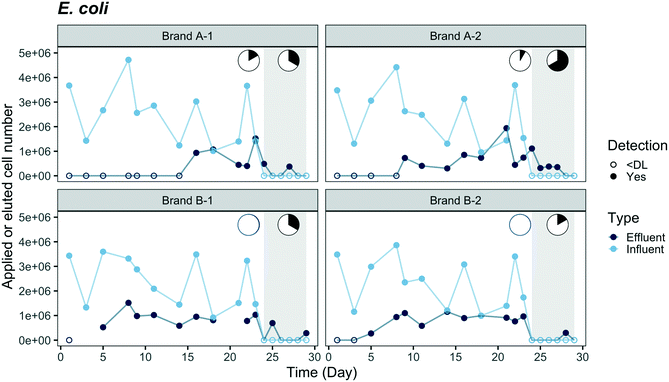 | ||
| Fig. 3 Breakthrough of mCherry-tagged E. coli through duplicate ACB PoU filters of brands A and B. Fluorescent-tagged bacteria were applied from day 0 to day 23 followed by a washout phase (shaded region) through day 29. Y-Axis coordinates were estimated by the same method described in Fig. 2. Cell numbers were plotted as “zero” with open circles when bacterial concentrations were below the detection limit (DL, 50 cell per mL). Average influent concentration was 4.2 × 102 ± 1.8 × 102 cell per mL (N = 12). Effluent data for filter B-1 was not available on days 3 and 21. The pie charts show the fraction of time during application and washout phases when effluent concentrations were less than or equal to influent (white) or exceeded the influent concentration (black). | ||
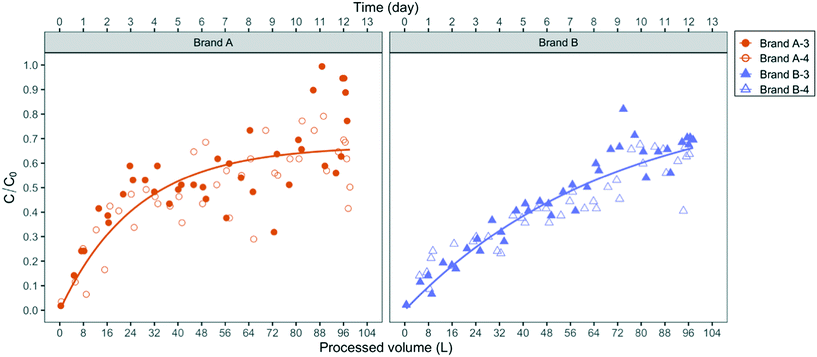
The existence of such preferential flow pathways is also supported by breakthrough experiments with the abiotic fluorescent microspheres (1 μm diameter). As shown in Fig. 4, the microspheres were detected in the effluent of both filter brands immediately after the first half-liter of influent was applied. The breakthrough of microspheres, which are not subject to growth and decay, demonstrates the capability of negatively charged micron-sized particles to migrate from the influent to the effluent at process volumes that represent 0.1% of the filter manufacturer's design process volume (100 gal or 379 L).
Since the influent fluorescent microspheres were suspended in slightly buffered deionized water with calculated near-neutral pH, their breakthrough pattern cannot be compared directly to the breakthrough of bacteria suspensions in pH 8.3 Ann Arbor tap water. The experimental design of the microsphere experiment, however, unequivocally demonstrates that flow pathways larger in diameter than bacteria-sized particles exist in both PoU filter brands. As the microspheres deposit in those channels, their net deposition rate decreases over the first approximately 50 L such that breakthrough concentrations increase and approach a steady-state C/C0 value in the brand A filter. This steady-state condition represents a net balance of particle attachment and detachment rates. In the brand B filters, steady-state microsphere breakthrough was more slowly attained but the breakthrough patterns were otherwise similar to the brand A filter.
P. aeruginosa and E. coli exhibit different breakthrough and washout patterns
Comparisons of the breakthrough of both fluorescent-tagged bacteria reveal that P. aeruginosa was detected earlier and reached a higher effluent level than E. coli during the application phase (Fig. 2 and 3). The effluent P. aeruginosa cell number plateaued at levels up to 1.8 times greater than influent counts after 21% of the filter manufacturer's design life water volume was processed (around two weeks). Effluent E. coli concentrations, on the other hand, were below the detection limit (50 cell per mL) until day 5 for brand B filters and day 9 for brand A filters, then the cell number plateaued at 35% of the influent level. During the plateau periods for both bacteria, we observed a wide variation in effluent level. The effluent P. aeruginosa cell number varied between 30% to 400% of the influent level, and E. coli varied between 24% to 60% of the influent cell number (Fig. S4†). These variations are likely caused by clogging, the subsequent buildup of hydraulic back pressure, followed by bacterial release as flow pathways became unclogged. During the release or sloughing phases of these cycles, effluent cell numbers of P. aeruginosa exceeded the influent level while E. coli approached the influent level. Since the flow cytometry method used in this study only accounted for single cell counts, it should be noted that some underestimation of the true sloughing concentrations is possible, as the method does not detect large clumps of fluorescent particles.The later breakthrough of E. coli relative to P. aeruginosa indicates that E. coli was either more readily strained by the ACB PoU filters, more favorably attached to the ACB pore walls than P. aeruginosa, or more vulnerable to die-off during filtration. The transport of bacteria through porous media is greatly affected by bacterial variables such as size, shape, hydrophobicity, and surface charge.34,35 The migration of bacteria through the flow pathways in the ACB PoU filters is likely to be affected by similar factors. To examine the role these variables might play, we measured the cell size and hydrophobicity of the two species after growing the bacteria with the acclimation procedure. The average size of P. aeruginosa (length × width = 1.3 ± 0.2 × 0.6 ± 0.1 μm) was smaller than E. coli (3.8 ± 0.8 × 0.8 ± 0.1 μm) (Fig. S5†), whereas the cell hydrophobicities as measured by the BATH method were similar between the two species (Table S2†). The later breakthrough of E. coli relative to P. aeruginosa may be due to the fact that E. coli cells have longer length and larger width than P. aeruginosa. Thus, E. coli was more readily strained by the ACB pore network. These findings differ from Geldreich et al.'s study, which introduced P. aeruginosa, E. coli and other organisms in a single spike into dechlorinated tap water fed to ACB PoU filters.36 Although there were differences in how bacteria were introduced between this study (continuous spike) and Geldreich et al.'s (one-time spike), they found that the spiked E. coli passed through the ACB PoU filter immediately but P. aeruginosa broke through later. The outcome differences between the two studies could be due to differences in the preparation of the spiked species and the use of different strains of the same species. The spiked E. coli used in Geldreich et al.'s study was prepared in a fresh rich medium that was then added to the dechlorinated tap water without any acclimation. In our study, the bacteria were acclimated by serial dilution of the media to obtain a TOC concentration similar to drinking water. The nutritional state of microorganisms is known to impact the surface chemistry and charge of cells,37 as well as their size.38 We found that the two test species after acclimation were significantly (p < 0.05) smaller than those that were grown in fresh rich medium (P. aeruginosa = 1.6 ± 0.4 × 0.5 ± 0.1 μm, E. coli = 4.2 ± 1.2 × 0.8 ± 0.2 μm) (Fig. S5†). In addition, Bolster et al. have shown that different E. coli strains have significantly different filtration rates in sand media under the same chemical conditions.39 Since the E. coli strain was not specified in the Geldreich et al. study, it is not possible to assess whether strain differences contribute to the bacterial breakthrough differences in our experiment.
Washout tests were conducted from day 24 to 29 after the effluent cell number had plateaued to examine if the fluorescent-tagged bacteria within the filter continued to seed the effluent after they were no longer introduced to the influent. During this period, the effluent concentrations of E. coli and P. aeruginosa decreased to near the detection limit from all filters except for P. aeruginosa in the B-2 filter, which had a stable P. aeruginosa level (3.5 × 103 ± 1.3 × 103 cell per day) in the effluent throughout the washout phase. The fluorescent bacteria detected in the effluent suggest they accumulated and survived inside the filters. It is not clear why only one of the brand B filters established a steady-state level of P. aeruginosa during the washout phase; however, it may be related to hydraulic characteristic differences between the filters. The permeability of the B-2 filter was the least among all tested filters, based on the hydraulic volumes processed over a five minute period before the experiment started, whereas the B-1 filter was the most permeable (Table S3†). In our previous work and other studies, ACB filter units of the same design model were found to vary in permeability, porosity, and surface area, and these filter-to-filter differences were hypothesized to account for different microbiological outcomes.10,36 Given the lower hydraulic conductivity of the B-2 filter, we suspect that greater bacterial accumulation and biofilm shear conditions may have occurred in this filter, resulting in higher levels of effluent seeding by P. aeruginosa. Higher rates of hydraulic shear can cause greater detachment of biofilm colonists.40 It is also noteworthy, that a rapid decrease in E. coli effluent cell number was observed even in the B-2 filter, perhaps because it was less able to adapt to the filter environment and compete with other microorganisms.
Evidence of fluorescent-tagged bacterial growth in ACB PoU filters
Cell balance evaluations were performed to test for evidence that fluorescent-tagged bacteria grew in the ACB filters. The cumulative cell counts of fluorescent-tagged bacteria in the filter influent and effluent over the application period are presented in Table S4.† The average integrated P. aeruginosa counts leaving the brand A and B filters were 1.1 times (1.2 × 108) and 2.1 times (2.0 × 108 cells), respectively, more than the average influent loading (9.7 × 107 cells). The effluent P. aeruginosa of brand A and B were also culturable over the course of application period (Fig. S6†). The cell balance confirms that net P. aeruginosa growth must occur in the brand B filters, allowing the daily effluent cell numbers to exceed influent levels. Since the cumulative influent and effluent cell counts of P. aeruginosa in the brand A filters were nearly equal, more limited growth was observed in these filters. In contrast to P. aeruginosa, E. coli levels in the effluent were consistently lower than the influent in both brands; a cumulative 25% of the applied E. coli were detected in the effluent during the period that fluorescent-tagged bacteria were applied to the filters.The growth evidence for P. aeruginosa suggests it may adapt to the ACB PoU filter environment better than E. coli. P. aeruginosa is known to utilize various carbon sources and can adapt to low-nutrient environments.41,42 Indigenous P. aeruginosa is frequently detected in ACB filter effluents when tap water is fed continuously, which suggests that the species' surface and growth characteristics allow it to pass through and colonize these filters.9,11 Conversely, some E. coli strains are known to have inferior kinetic properties for competing at low carbon concentrations with indigenous bacteria.43–45
Total membrane-intact cell counts in the effluent of ACB PoU filters did not exceed the influent level
In addition to the growth evidence of fluorescent bacteria in the ACB PoU filters, we also tracked the fate of indigenous bacteria in the influent that originated from the tap water using flow cytometry to count total membrane-intact cells (Fig. S7†). The effluent total cell counts slowly approached the influent during the experiment but did not exceed the influent level. Cell counts of approximately 40% of the influent level, however, were observed in the first sampling period after processing 5 L. The effluent bacterial counts ranged from 4.8 × 104 to 1.8 × 105 cells per day and increased over time to 83% of the influent level (2.0 × 105 ± 0.5 × 105). We only processed 44% of the water volume that the PoU filters were rated to process, so it is not unexpected that effluent total membrane-intact cell counts did not yet exceed the influent counts, as is typically found when counts are determined by heterotrophic plate counts.7–10 Furthermore, membrane-intact counts do not necessarily correspond to heterotrophic plate counts,46 making a direct comparison to past studies difficult.P. aeruginosa colonized entire ACB while E. coli was absent near filter exit port
We sought to determine whether the fluorescent bacteria could colonize surfaces across the entire depth of the ACB by evaluating their abundance at the entrance and exit surfaces of the PoU filters at the end of the breakthrough experiment. Samples tested on the upstream side of the ACB included the fabric mesh that wraps around the influent side ACB surface, and swabs of the activated carbon surface of the interior core of the effluent channel at the downstream side of the ACB. In an earlier study, the fabric mesh in the brand B filter was shown to consist of silicic fibers.10 Since the biofilm samples were collected after the washout phase, the amount of biomass collected might have been larger if it had been collected immediately after the application phase. Some bacterial cells that were loosely attached to the filter surfaces during the application period may have sloughed out to the effluent during the washout phase. Cells that were found on the fabric and effluent channel surfaces after the washout phase may be those that were irreversibly attached.The abundance patterns of the two types of fluorescent bacteria in upstream and downstream biofilms of the ACB, as shown in Table 1, are consistent with greater mobility and survivability of P. aeruginosa relative to E. coli in ACB PoU filters. Bacterial abundance was characterized in terms of the number of fluorescent gene counts of both bacteria and the number of culturable P. aeruginosa cells. We were not able to enumerate culturable E. coli from the M Endo LES agar, presumably due to the competition from other non-coliform bacteria.47 The GFP genes and culturable P. aeruginosa cells were detected from both the fabric biofilm and the surface of the interior core channel. Therefore, it is likely that P. aeruginosa cells colonized the surface of flow pathways within the ACB.
| Biofilm location | Parameters | Brand A | Brand B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-1 | A-2 | B-1 | B-2 | ||||||||||
| Top | Center | Bottom | Top | Center | Bottom | Top | Center | Bottom | Top | Center | Bottom | ||
| +: detected 1–10 colonies. ++: detected 10–300 colonies. +++: detected >300 colonies. a E. coli could not be cultured because of competition with the indigenous bacteria from the background. | |||||||||||||
| Fabric | Culturabilitya | ||||||||||||
| P. aeruginosa | + | + | + | + | + | + | ++ | + | ++ | ++ | ++ | +++ | |
| DNA (copy gene number/cm2) | |||||||||||||
| GFP gene | 3.3 × 10 | 5.2 × 10 | 5.5 × 10 | 3.1 × 10 | 2.9 × 10 | 1.9 × 10 | 1.7 × 102 | 9.6 × 10 | 3.1 × 102 | 8.1 × 102 | 6.4 × 102 | 6.7 × 102 | |
| mCherry gene | 5.4 × 102 | 5.5 × 102 | 4.0 × 102 | 8.3 × 102 | 7.7 × 102 | 4.5 × 102 | 6.0 × 103 | 2.0 × 103 | 2.0 × 103 | 2.1 × 103 | 2.2 × 103 | 1.4 × 103 | |
| Inner effluent channel | Culturabilitya | ||||||||||||
| P. aeruginosa | + | + | + | + | + | + | ++ | + | ++ | + | + | +++ | |
| DNA (copy gene number) | |||||||||||||
| GFP gene | ND | ND | 4 | ND | ND | 2 | 3 | ND | 1.5 × 10 | 2.3 × 102 | 4.2 × 10 | 7.1 × 102 | |
| mCherry gene | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
Unlike P. aeruginosa, mCherry genes were detected from the fabric biofilm but not the effluent channel swabs. The mean estimated E. coli cell counts measured on the fabric (1.5 × 105 cell per total fabric area, Table S4†) was 0.3% of the E. coli applied to the filter over the whole application period (5.5 × 107 cell counts, Table S4†). The number of mCherry genes was also uniformly distributed between the top, center and bottom of the fabric (Table 1). Since mCherry genes were not detected in swabs of the effluent channel surface, E. coli was either less able to colonize and survive in downstream locations of the filter or it was more effectively filtered at upstream locations within the ACB, relative to P. aeruginosa. During the washout phase of the experiment, low but detectable levels of E. coli cells were still occasionally sloughed to the effluent. The accumulation of E. coli within upstream regions of the filter could serve as a reservoir for E. coli to seed the filter effluent at low cell numbers, even when no inoculation was spiked into the influent.
Two brands of ACB PoU filters exhibit different performance
Bacterial migration patterns between brand A and B filters differed in several respects. E. coli, for example, was detected later in the effluent of brand A filters than brand B, as shown in Fig. 3, while similar breakthrough times were observed between the two brands for P. aeruginosa. Over the course of the application period, the integrated effluent cell counts of P. aeruginosa from the brand B filter were two times higher than brand A and approximately two times the number of influent cells added, suggesting that brand B filters allowed slightly greater net growth of P. aeruginosa than brand A (Table S4†). In addition, the number of P. aeruginosa and E. coli cells on the brand B fabric biofilm were eleven and four times higher than brand A, respectively (Table S4†). These differences suggest that certain design features of the brand B filters favor greater colonization and accumulation of P. aeruginosa, as well as greater filtration of E. coli, which as we demonstrated, is the larger of the two fluorescent bacteria. The empirical exponential models of microsphere breakthrough concentration in Fig. 4 exhibit similar patterns between the two filter brands. The breakthrough concentration data for brand A, however, was noticeably more variable than brand B, as demonstrated by its greater deviation from the empirical pattern (residual sum of squares (RSS) = 0.36) relative to brand B (RSS = 0.09). Microsphere breakthrough concentration variability in brand A is likely caused by a somewhat greater susceptibility to hydraulic plugging. We note that the empirical model did not include the data greater than 100 L processed volume since all four filters were impacted by clogging (Fig. S8†) and filter B-4 developed a crack due to excessive back pressure.Diameter differences in the hydraulic flow paths of the two filter brands may explain the observed breakthrough differences if the brand B filter had larger diameter flow paths than brand A. Larger diameter paths would better allow passage of the larger fluorescent E. coli strain and be less susceptible to plugging. Larger diameter paths would also allow thicker biofilms (and hence greater biomass) to develop at the same volumetric flow rate, due to the resulting smaller shear rates.
The microsphere and bacterial breakthrough trends of the PoU filters differ from the general behavior of continuous flow filtration of colloidal particles in granular beds under strongly repulsive interaction conditions.48 For these filter conditions, normalized effluent concentrations typically attain a near steady-state concentration after just a few pore volumes. In our system, stagnation periods allow particle deposition due to diffusion and some colonization of pore surfaces by bacteria. In addition, hydraulic pulses likely cause shearing of deposited particles and bacteria after each stagnation event. The impacts caused by stagnation and bacterial growth could lead to a gradual increase in the normalized effluent concentration (C/C0) compared to the steady-state breakthrough patterns of abiotic particles in continuous process flow granular filters.
Conclusions
• Breakthrough of two tap-water spiked fluorescently-tagged bacteria, an opportunistic pathogen, P. aeruginosa, and an enteric indicator, E. coli, was detected early in the effluent of two popular US brands of faucet-mounted ACB PoU filters under simulated daily use patterns. Detection of the bacteria occurred at average process volumes representing 1.5% (P. aeruginosa) and 7% (E. coli), of the manufacturer's design life volume throughput of the filters. P. aeruginosa attained effluent concentrations that periodically exceeded influent levels, as a result of filter plugging and back pressure release patterns and its ability to grow within the filter media. E. coli was more effectively filtered than P. aeruginosa possibly because it was larger in size, more hydrophobic, and less able to grow within the filter. These bacterial variables determine the bacterial transport through the filter and abundance in the filter effluent.• The use of both culture and culture-independent methods (flow cytometry) in the case of P. aeruginosa, demonstrated that viable and intact cells, respectively, were present in the PoU filter effluent and that viable opportunistic pathogens can pass through ACB filters.
• Breakthrough of 1 μm diameter negatively charged fluorescent microspheres in the ACB PoU filters demonstrated that pore networks with diameters larger than 1 μm exist in the filters.
• The spiked bacteria in our PoU filter breakthrough experiments were first pre-acclimated to the oligotrophic conditions of drinking water, a step that has not been used in other studies. After the acclimation step, both of the fluorescent strains became more hydrophobic and the mean cell size of P. aeruginosa decreased. Given the physiological and metabolic changes that cells undergo during acclimation and the influence these changes can have on bacteria transport and growth, pre-acclimation is recommended in order to understand bacterial breakthrough patterns under field drinking water conditions.
Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
The authors would like to thank Dr. Josha Shrout at the University of Notre Dame for providing GFP-tagged P. aeruginosa isolates and Dr. Mattheos Koffas at the Rensselaer Polytechnic Institute for providing mCherry-tagged E. coli isolates. We also thank Dr. Shawn McElmurry at the Wayne State University for sharing equipment and techniques for flow cytometric total bacterial measurements. We are also grateful for Nicholas Lowe and Katie Stroh for their contributions through laboratory work associated with this study. This work was supported by a Rackham Pre-Doctoral Fellowship, a Rackham Graduate Student Research Grant, the Borchardt and Glysson Collegiate Professorship (NGL), and the College of Engineering at the University of Michigan. Any opinions, findings, and conclusions are solely the responsibility of the authors and do not necessarily represent the official view of the University of Michigan.References
- R. Cullen, Ownership and Interest in Water Filtration Products, in Water Filtration - US - October 2017, Mintel Report, 2017 Search PubMed.
- Assoc. Press, Michigan distribution water filters to residents of Flint, 2015 Search PubMed.
- A. Biolchini, A testing prompts wolverine to offer water filters to more homes, MLive Media Gr. Grant Rapids Press, 2017 Search PubMed.
- NSF International/ANSI, NSF Standard 42: Drinking Water Treatment Units—Aesthetic Effects, Ann Arbor, 2015 Search PubMed.
- NSF International/ANSI, NSF Standard 53 Drinking Water Treatment Units—Health Effects, Ann Arbor, 2015 Search PubMed.
- NSF International, NSF Protocol P473: Drinking Water Treatment Units—PFOA and PFOS, Ann Arbor, 2016 Search PubMed.
- S. L. Molloy, R. Ives, A. Hoyt, R. Taylor and J. B. Rose, The use of copper and silver in carbon point-of-use filters for the suppression of Legionella throughput in domestic water systems, J. Appl. Microbiol., 2008, 104, 998–1007 CrossRef CAS PubMed.
- D. J. Reasoner, J. C. Blannon and E. E. Geldreich, Microbiological characteristics of third-faucet point-of-use devices, J. - Am. Water Works Assoc., 1987, 79, 60–66 CrossRef CAS.
- R. S. Tobin, D. K. Smith and J. A. Lindsay, Effects of activated carbon and bacteriostatic filters on microbiological quality of drinking water, Appl. Environ. Microbiol., 1981, 41, 646–651 CrossRef CAS PubMed.
- C.-C. Wu, S. Ghosh, K. J. Martin, A. J. Pinto, V. J. Denef, T. M. Olson and N. G. Love, The microbial colonization of activated carbon block point-of-use (PoU) filters with and without chlorinated phenol disinfection by-products, Environ. Sci.: Water Res. Technol., 2017, 3, 830–843 RSC.
- C. Chaidez and C. P. Gerba, Comparison of the microbiologic quality of point-of-use (POU)-treated water and tap water, Int. J. Environ. Health Res., 2004, 14, 253–260 CrossRef PubMed.
- F. Su, M. Luo, F. Zhang, P. Li, K. Lou and X. Xing, Performance of microbiological control by a point-of-use filter system for drinking water purification, J. Environ. Sci., 2009, 21, 1237–1246 CrossRef CAS.
- D. Stalter, E. O'Malley, U. Von Gunten and B. I. Escher, Point-of-use water filters can effectively remove disinfection by-products and toxicity from chlorinated and chloraminated tap water, Environ. Sci.: Water Res. Technol., 2016, 2, 875–883 RSC.
- M. J. Lehtola, E. Torvinen, J. Kusnetsov, T. Pitkänen, L. Maunula, C.-H. Von Bonsdorff, P. J. Martikainen, S. A. Wilks, C. W. Keevil and I. T. Miettinen, Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow, Appl. Environ. Microbiol., 2007, 73, 2854–2859 CrossRef CAS PubMed.
- C. Hardalo and S. C. Edberg, Pseudomonas aeruginosa: assessment of risk from drinking water, Crit. Rev. Microbiol., 1997, 23, 47–75 CrossRef CAS PubMed.
- M. M. Moritz, H.-C. Flemming and J. Wingender, Integration of Pseudomonas aeruginosa and Legionella pneumophila in drinking water biofilms grown on domestic plumbing materials, Int. J. Hyg. Environ. Health, 2010, 213, 190–197 CrossRef CAS PubMed.
- H. Wang, M. Edwards, J. O. Falkinham and A. Pruden, Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems, Appl. Environ. Microbiol., 2012, 78, 6285–6294 CrossRef CAS PubMed.
- J. Lu, I. Struewing, E. Vereen, A. E. Kirby, K. Levy, C. Moe and N. Ashbolt, Molecular Detection of Legionella spp. and their associations with Mycobacterium spp., Pseudomonas aeruginosa and amoeba hosts in a drinking water distribution system, J. Appl. Microbiol., 2015, 120, 509–521 CrossRef CAS PubMed.
- K. D. Mena and C. P. Gerba, Risk assessment of Pseudomonas aeruginosa in water, Rev. Environ. Contam. Toxicol., 2009, 201, 71–115 CrossRef CAS PubMed.
- E. Paramonova, E. L. Zerfoss and B. E. Logan, Measurement of biocolloid collision efficiencies for granular activated carbon by use of a two-layer filtration model, Appl. Environ. Microbiol., 2006, 72, 5190–5196 CrossRef CAS PubMed.
- K. Choi, J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-schweizer and H. P. Schweizer, A Tn 7-based broad-range bacterial cloning and expression system, Nat. Methods, 2005, 2, 443–448 CrossRef CAS PubMed.
- J. A. Englaender, J. A. Jones, B. F. Cress, T. E. Kuhlman, R. J. Linhardt and M. A. G. Koffas, Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli, ACS Synth. Biol., 2017, 6, 710–720 CrossRef CAS PubMed.
- L. Eberl, R. Schulze, A. Ammendola, O. Geisenberger, R. Erhart, C. Sternberg, S. Molin and R. Amann, Use of green fluorescent protein as a marker for ecological studies of activated sludge communities, FEMS Microbiol. Lett., 1997, 149, 77–83 CrossRef CAS.
- L. H. Hansen, S. Knudsen and S. J. Sørensen, The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens, Curr. Microbiol., 1998, 36, 341–347 CrossRef CAS PubMed.
- D. J. Reasoner and E. E. Geldreich, A new medium for the enumeration and subculture of bacteria from potable water, Appl. Environ. Microbiol., 1985, 49, 1–7 CrossRef CAS PubMed.
- M. T. Madigan, J. M. Martinko, D. A. Stahl and D. P. Clark, Brock Biology of Microorganisms, Pearson Education, San Francisco, 13th edn, 2010 Search PubMed.
- C. L. Lewis, C. C. Craig and A. G. Senecal, Mass and density measurements of live and dead Gram-negative and Gram-positive bacterial populations, Appl. Environ. Microbiol., 2014, 80, 3622–3631 CrossRef CAS PubMed.
- G. M. Litton and T. M. Olson, Colloid deposition rates on silica bed media and artifacts related to collector surface preparation methods, Environ. Sci. Technol., 1993, 27, 185–193 CrossRef CAS.
- C. Huang, J. R. Pan and S. Huang, Collision efficiencies of algae and kaolin in depth filter: the effect of surface properties of particles, Water Res., 1999, 33, 1278–1286 CrossRef CAS.
- J. Bergendahl and D. Grasso, Prediction of colloid detachment in a model porous media: Thermodynamics, AIChE J., 1999, 45, 475–484 CrossRef CAS.
- APHA, AWWA and WEF, Standard methods for the examination of water and wastewater, American Public Health Association, Washinton, DC, 21st edn, 2005 Search PubMed.
- V. I. Brown and E. J. L. Lowbury, Use of an improved cetrimide agar medium and other culture methods for Pseudomonas aeruginosa, J. Clin. Pathol., 1965, 18, 752–756 CrossRef CAS PubMed.
- N.-Y. Choi, Y.-M. Bae and S.-Y. Lee, Cell surface properties and biofilm formation of pathogenic bacteria, Food Sci. Biotechnol., 2015, 24, 2257–2264 CrossRef CAS.
- H. Zhong, G. Liu, Y. Jiang, J. Yang, Y. Liu, X. Yang, Z. Liu and G. Zeng, Transport of bacteria in porous media and its enhancement by surfactants for bioaugmentation: a review, Biotechnol. Adv., 2017, 35, 490–504 CrossRef CAS PubMed.
- Y. Wang, F. Hammes, M. Düggelin and T. Egli, Influence of size, shape, and flexibility on bacterial passage through micropore membrane filters, Environ. Sci. Technol., 2008, 42, 6749–6754 CrossRef CAS PubMed.
- E. E. Geldreich, R. H. Taylor, J. C. Blannon and D. J. Reasoner, Bacterial colonization of point-of-use water treatment devices, J. - Am. Water Works Assoc., 1985, 77, 72–80 CrossRef.
- B. Z. Haznedaroglu, C. H. Bolster and S. L. Walker, The role of starvation on Escherichia coli adhesion and transport in saturated porous media, Water Res., 2008, 42, 1547–1554 CrossRef CAS PubMed.
- R. Y. Morita, Bioavailability of energy and its relationship to growth and starvation survival in nature, Can. J. Microbiol., 1988, 34, 436–441 CrossRef CAS.
- C. H. Bolster, B. Z. Haznedaroglu and S. L. Walker, Diversity in cell properties and transport behavior among 12 different environmental Escherichia coli isolates, J. Environ. Qual., 2009, 38, 465–472 CrossRef CAS PubMed.
- Y. Shen, G. L. Monroy, N. Derlon, D. Janjaroen, C. Huang, E. Morgenroth, S. A. Boppart, N. J. Ashbolt, W. Liu and T. H. Nguyen, Role of biofilm roughness and hydrodynamic conditions in Legionella pneumophila adhesion to and detachment from simulated drinking water biofilms, Environ. Sci. Technol., 2015, 49, 4274–4282 CrossRef CAS PubMed.
- D. Van der Kooij, A. Visser and J. P. Oranje, Multiplication of fluorescent pseudomonads at low substrate concentrations in tap water, Antonie van Leeuwenhoek, 1982, 48, 229–243 CrossRef CAS PubMed.
- D. Van der Kooij, J. P. Oranje and W. A. M. Hijnen, Growth of Pseudomonas aeruginosa in tap water in relation to utilization of substrates at concentrations of a few micrograms per liter, Appl. Environ. Microbiol., 1982, 44, 1086–1095 CrossRef CAS PubMed.
- M. Vital, F. Hammes and T. Egli, Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations, Water Res., 2012, 46, 6279–6290 CrossRef CAS PubMed.
- A. K. Camper, G. A. McFeters, W. G. Characklis and W. L. Jones, Growth kinetics of coliform bacteria under conditions relevant to drinking water distribution systems, Appl. Environ. Microbiol., 1991, 57, 2233–2239 CrossRef CAS PubMed.
- Y. Rollinger and W. Dott, Survival of selected bacterial species in sterilized activated carbon filters and biological activated carbon filters, Appl. Environ. Microbiol., 1987, 53, 777–781 CrossRef CAS PubMed.
- M. Berney, M. Vital, I. Hülshoff, H.-U. Weilenmann, T. Egli and F. Hammes, Rapid, cultivation-independent assessment of microbial viability in drinking water, Water Res., 2008, 42, 4010–4018 CrossRef CAS PubMed.
- I. D. Gaudet, L. Z. Florence and R. N. Coleman, Evaluation of test media for routine monitoring of Escherichia coli in nonpotable waters, Appl. Environ. Microbiol., 1996, 62, 4032–4035 CrossRef CAS PubMed.
- M. Elimelech and C. R. O'Melia, Kinetics of deposition of colloidal particles in porous media, Environ. Sci. Technol., 1990, 24, 1528–1536 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: More details on analytical methods, including figures showing the flow cytometry dot plots of fluorescent-tagged bacteria, fluorescent microspheres, and results of fluorescent-tagged bacteria and microsphere breakthrough tests are shown in the ESI. See DOI: 10.1039/d0ew00982b |
| This journal is © The Royal Society of Chemistry 2021 |