Open Access Article
Open Access ArticleEnumeration and characterization of five pathogenic Legionella species from large research and educational buildings†
Alshae' R.
Logan-Jackson
 *a,
Matthew
Flood
b and
Joan B.
Rose
ab
*a,
Matthew
Flood
b and
Joan B.
Rose
ab
aDepartment of Microbiology and Molecular Genetics, Michigan State University, East Lansing, Michigan 48824, USA. E-mail: loganals@msu.edu
bDepartment of Fisheries and Wildlife, Michigan State University, East Lansing, Michigan 48824, USA
First published on 15th December 2020
Abstract
Legionella pneumophila is the species that is most often cultured from the natural environment, while disease-relevant Legionella species, such as Legionella micdadei, Legionella bozemanii, Legionella anisa, and Legionella longbeachae have yet to be extensively explored in premise plumbing systems. This study examined the concentrations of five pathogenic Legionella species (listed previously) in the influent and the taps of five different large buildings (BPS, ERC, F, FH, and M), undertaken during the start of two semesters (late summer/fall (August–September) and early winter/spring (January)). A total of 37 large-volume samples to examine building water quality (influents to the buildings and exposure sites (taps)) were collected and analyzed using droplet digital™ PCR. Legionella spp. (23S rRNA) were present in all water samples during both seasons. The majority (66%) of the exposure sites (bathroom taps) were positive for at least one target Legionella species (listed above). Results showed that pathogenic Legionella species were most often detected during the winter/spring sampling event – the percent positives for any one of the pathogenic Legionella species at the hot-water taps was 80% in building F and 40% in BPS, M, FH, and ERC. Legionella pneumophila and L. longbeachae were found in the highest concentrations (2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 gene copies (GC)/100 mL) at the hot-water taps in buildings F and ERC, respectively. No strong relationships were found with the physical–chemical parameters. Overall, general Legionella spp. concentrations increased in the winter/spring samples due possibly to lower water usage (lower occupancy and no use of cooling towers, which led to more water stagnation or time in the system).
log10 gene copies (GC)/100 mL) at the hot-water taps in buildings F and ERC, respectively. No strong relationships were found with the physical–chemical parameters. Overall, general Legionella spp. concentrations increased in the winter/spring samples due possibly to lower water usage (lower occupancy and no use of cooling towers, which led to more water stagnation or time in the system).
Water impactLegionnaires disease cases have been linked to non-pneumophila Legionella species. Pathogenic Legionella species were identified in 66% buildings' taps at concentrations that ranged from 1.4 to 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL compared to the influent which ranged from 1.5 to 1.8 log10 GC/100 mL compared to the influent which ranged from 1.5 to 1.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL (20%). Significant concentrations of pathogenic Legionella spp. are found in buildings and hot water taps with greater levels observed during lower water use. log10 GC/100 mL (20%). Significant concentrations of pathogenic Legionella spp. are found in buildings and hot water taps with greater levels observed during lower water use.
|
Introduction
Legionella spp. are Gram-negative, opportunistic waterborne pathogens that reside in premise plumbing (i.e., building) as well as other engineered water systems. Legionella pneumophila is the etiologic agent responsible for most Legionnaires disease (LD) with other species identified less frequently causing severe pneumonia and the less-studied Pontiac fever (an acute, but generally milder set of cold-like signs and symptoms).1Legionella naturally colonizes freshwater and groundwater environments, as well as engineered systems including cooling towers, air conditioners, hot tubs, taps, and showers.2,3Legionella infections are acquired via inhalation of aerosols and air droplets generated from these structures containing the bacteria.4 The first recognized outbreak of LD, caused by L. pneumophila, occurred in 1976.5 In the United States (US), LD prevalence has increased significantly since 2000, and in 2018 there were approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reported cases.6Legionella species are difficult to assess and control in the drinking water system because they survive in the biofilm on the surface of the pipes and within amoebae hosts.7,8 The difficulty in assessing and controlling Legionella species makes these bacteria and their associated diseases a paramount public health concern.
000 reported cases.6Legionella species are difficult to assess and control in the drinking water system because they survive in the biofilm on the surface of the pipes and within amoebae hosts.7,8 The difficulty in assessing and controlling Legionella species makes these bacteria and their associated diseases a paramount public health concern.
Many outbreaks of LD occur at the community level, as was the case in Flint, Michigan between 2014 and 2015 when Michigan saw a 375% increase in cases, most of which were part of the Flint outbreak.9,10 During the 2014–2015 Flint outbreak, it was suggested that there were multiple sources of exposure, including the hospital water system, water at home (showers or taps), and residential proximity to cooling towers.9 Although the LD outbreak during the water crisis in Flint, Michigan, was the largest in the state, there has been an increased number of cases statewide from 2000 to 2016.10
Legionella pneumophila, serogroup 1 is the most often diagnosed agent accounting for 90% of identified LD pneumonia cases, perhaps due to the restriction of the urinary antigen test.11–15 In recent years, other Legionella species found in drinking water have also been identified in about 10% of cases.16–23Legionella micdadei, L. bozemanii, L. longbeachae, and L. anisa have been isolated from human patients.24,25 There have been five drinking water outbreaks caused by L. micdadei,26–33 one by L. bozemanii34 and two by L. anisa26 in the US. To date, there have not been any reports of L. longbeachae related infections associated with building water systems in the US, but there have been outbreaks in Australia, and cases reported in New Zealand, and some parts of Asia [Thailand].35,36 In Australia, Legionella infections are commonly caused by L. longbeachae, and one of the exposure pathways was suggested to be potting mixes and compost.37
The majority of reported LD outbreaks have occurred in large complex plumbing systems, which are used in hospitals and healthcare facilities.38 However, 97% of LD cases are sporadic infections,39 for which the environmental source of exposure is usually unknown. The National Academies report on “Management of Legionella in Water Systems” (2019) stated that for every one outbreak case, there are nine more sporadic cases.40 Despite a substantial amount of research on the molecular virulence mechanisms and ecology of Legionella, annual incidence rates of the disease continue to rise along with great uncertainty on how to control the colonization of water systems.
Currently, only a few studies have simultaneously characterized multiple pathogenic Legionella species (L. anisa, L. micdadei, L. bozemanii, and L. longbeachae) in drinking water systems including the source, distribution system and tap18,41–45 although most have not evaluated the concentrations and only focused on presence absence. Legionella bozemanii, L. dumoffii, L. longbeachae, L. anisa, L. moravica, L. parisiensis, L. brunensis, L. londinensis, and L. hackeliae, among many others, have been detected in water samples collected from hospitals in Italy,44 warm water systems in Germany,18 and in utility drinking water systems in the Netherlands.41,42 In 2016, a research group in Germany, found L. pneumophila, L. longbeachae, L. worsleiensis, L. anisa, and L. dumoffii, (among many others) from source water to the cold- and hot-water taps in Germany using genus-specific PCR amplicons (16S rRNA) and single-strand conformation polymorphism fingerprint analyses.45Legionella anisa was detected in the Netherlands in three of four dental care units (75%) at a concentration of 1 × 102 CFU mL−1 using the Dutch Legionella standard culture technique, identified by whole-genome sequencing (MALDI-TOF).43
The goals of this study were to assess the concentrations of general Legionella spp., compared to pathogenic species L. pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae, to understand the microbial quality of the drinking water entering five large research, classroom, and office buildings (all utilizing the same water source) compared to the water quality of the buildings. Utilizing droplet digital PCR, this study addressed the following objectives (i) quantification of L. pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae in the influents and at the bathroom taps (points of use) of five large buildings using large volume composite sampling (ii) exploration of the associations of Legionella species with respect to temperature, chlorine, conductivity, pH, and heterotrophic plate count (HPC), and (iii) assessment of whether there were differences between two sampling periods of the year (August/September and January) for five pathogenic Legionella species. The quantitative data presented in this study should improve quantitative risk assessment of various specific pathogenic Legionella species within a drinking water system. This study gives an estimated concentration of Legionella species which may help improve exposure analysis.
Experimental
Site location and sampling
Water samples were collected during the beginning of two semesters (fall 2018 and spring 2019) from five buildings (F, BPS, M, FH, and ERC) on a large research institution of higher education. This was a large-scale spatial study; samples collected the beginning of the fall semester will be referred to the summer sampling and the beginning of the spring semester will be referred to as the winter sampling. Sample collection was conducted on August 13th and 27th, September 4th, 2018, and January 7th–9th, 14th, and 15th, 2019. This included research buildings F, ERC, and BPS, as well as buildings FH and M containing offices and classrooms. Building age, water use, and distance from the reservoir are shown in Table 1. The buildings are listed based on its pipe mileage from the effluent reservoir. Each building was assessed at an influent point with the sample collected at the most accessible sampling port on each building's influent pipe with the exception of ERC. The ERC influent sampling port was inaccessible; thus, it was decided to sample the nearest valve to the influent pipe, which was an eye-wash station in the mechanical room where the influent pipe entered the building. The building water quality was assessed by composite sampling point of use locations for each building included cold- and hot-water taps (sink faucets and showerheads) located in bathrooms, locker rooms, and breakrooms. All sinks described below were used for sample collection. Building F had two floors, with two sinks on the first floor and three sinks on the second floor; BPS had six floors, with 20 sinks on the first floor and four sinks on the sixth floor; M had two floors, with four sinks on the first floor and two sinks on the second floor; FH had two floors, with 17 sinks on the first floor and ten sinks on the second floor; ERC had one floor, with 11 sinks and two showers.| Buildinga (construction year) and pipe material | Volume of water used geomean per month (Aug/Sept) [January] consumption (kGAL) | Distance from reservoir (km) | Building size (m2) | Floors sampled (# of floors) | # of taps | Volumeb of water collected from each tap (L) (summer) winterc |
|---|---|---|---|---|---|---|
| a 10 L composite sample were collected from all buildings; the volume of water from each tap was dependent upon the number of taps per floor per building. b For the summer event, hot and cold composite samples were collected as one 10 L sample from each building per floor. c Single faucet fixtures with two taps (1/2 were cold and the other 1/2 were hot water pipes). For the winter event, hot and cold composite samples were separated so that two 10 L samples were collected per floor. d The numbers of showerheads samples as part of the composite for ERC was two compared to 24 sink faucets. | ||||||
| F (1948) | (94.9) | 4.7 | 7118 | First | 4 | (2.5) 5 |
| 75% galvanized 25% copper | [90.9] | Second | 6 | (1.67) 3.33 | ||
| BPS (2001) | (4245) | 6.8 | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 045 045 |
First | 40 | (0.5) 0.25 |
| 50% galvanized 50% copper | [382] | Sixth | 8 | (2.5) 1.2 | ||
| M (1940) | (11.6) | 9.6 | 5926 | First | 8 | (2.5) 1.25 |
| 90% galvanized 10% copper | [60.9] | Second | 4 | (5) 2.5 | ||
| FH (1964) | (N/A) | 10.2 | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 057 057 |
First | 34 | (0.5) 0.29 |
| 50% galvanized 50% copper | [100] | Second | 20 | (1) 0.5 | ||
| ERC (1986) | (200.8) | 19.4 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 896 896 |
First | 26d | (0.77) 0.38 |
| 50% galvanized 50% copper | [289.8] | |||||
For influent samples, 10 L were collected from each building's influent sampling location. For tap samples, a large-volume (10 L) composite sample was collected to evaluate the water quality of each building's taps rather than the quality of individual taps. The first flush with equal total volumes from each tap was collected and composited into 10 L for the first floor and top floor, respectively. For the summer approach, a cold- and hot-water composite sample was collected to evaluate and compare the water quality on the first and top floors, separately. During the summer sampling, a total of three 10 L samples were collected from each building (influent, first floor taps, and top floor taps). For the winter approach, the cold- and hot-water taps were collected as separate samples to evaluate and compare the water quality of the cold-water taps and the hot-water taps on the first and top floor, respectively. For the winter sampling event, a total of five 10 L samples were collected per building, one influent, one cold- and one hot-water sample from the first and top floor, respectively. The one exception was building ERC, which only had one floor, where three 10 L samples were collected. The volume collected from each tap that was composited was determined by the number of taps on each floor. Table 1 shows the number of taps and the volume collected to construct the composite samples during the summer and winter. The goal was to assess the building water quality via composite sampling. Different sampling approaches for the two seasons were chosen initially to evaluate the water quality by floor (summer where cold and hot water were combined) after initial analysis, the decision was made to separate out the samples by hot and cold (winter). The difference between summer and winter at the tap was the volume of water collected (10 L for cold, 10 L for hot, and 10 L for combined); thus, this may have affected the detection limit as one more sample was collected from the buildings in the winter. All samples were collected in carboys (influent and tap samples) with 10% sodium thiosulfate to neutralize residual chlorine. Temperature and chlorine were recorded from each tap to examine the variation by tap and by floor.
Chemical–physical analysis
A 100 mL sample was collected for conductivity, pH, and turbidity analyses. Temperature and residual chlorine (total and free) were measured onsite. Chlorine was measured using the Test Kit Pocket Colorimeter II (HACH®, CO, USA) according to the manufacturer's instructions. Conductivity, pH, and turbidity were measured offsite at the laboratory according to the manufacturers' instructions using a Russell RL060C Portable Conductivity Meter (Thermo Scientific, MA, USA), UltraBasic pH meter (Denver Instrument, NY, USA), and a Turbidity Meter code 1970-EPA (LaMotte Company, MD, USA).Microbiological analysis
All samples were transported on ice to the laboratory and preserved at 4 °C until processed. While the on-campus water utility tests for coliform bacteria on a routine basis, all samples collected for this study were tested according to the standard methods for coliform bacteria and E. coli using Colilert (IDEXX Laboratories, ME, USA) as well as with heterotrophic plate count (HPC) analyses using membrane filters (47 mm diameter, 0.45 μm pore size) (PALL Corporation, NY, USA) on m-HPC agar (Becton, Dickinson and Company, Difco™, MI, USA), incubated for 48 ± 2 h at 35–37 °C, then enumerated for colony-forming units (CFU).46 Total coliforms were assayed for the summer and winter sampling events, while the HPC analyses were performed only for the winter.Water sample processing and DNA extraction
The 10 L water samples were processed using a single-use Asahi REXEED-25S dialysis filter (Dial Medical Supply, PA, USA), which was pretreated with 0.01% of sodium hexametaphosphate (used to trap microbial material onto each ultrafilter) and utilized in a dead-end mode. A high-pressure single-use elution fluid canister (Innovaprep LLC, MO, USA) was used to concentrate the 10 L to ∼50 mL.Molecular analysis
Each ultrafiltration concentrate was split into 10 mL subsamples. One 10 mL subsample was further filtered through a 47 mm, 0.45 μm polycarbonate filter (Whatman, Kent, UK) for DNA extraction and analyzed by ddPCR. The remaining subsamples were stored at −80 °C.DNA extraction and quantitative detection of Legionella by droplet digital PCR
DNA was extracted using QIAamp DNA Mini Kit (Qiagen, CA, USA). Each 10 mL subsample was filtered on a polycarbonate filter (described above) using a sterilized 0.47 mm magnetic filter funnel (PALL Corporation, NY, USA). Immediately afterward, the polycarbonate filter was folded into a 1/8 shape with contents of filter folded to the inside. The filter was then transferred to a 2.0 mL polypropylene screw cap tube (VWR, PA, USA) containing 0.3 g of 212–300 μm acid-washed glass beads (Sigma, MO, USA). DNA extraction was performed by adding 590 μL of AE buffer (Qiagen, CA, USA) to the samples then bead milling using a FastPrep-24™ 5G Instrument MP Biomedicals (VWR, PA, USA). Samples were milled at 6000 rpm for one minute, followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for one minute. The supernatant (∼400 μL) was transferred to a new clean microcentrifuge tube and centrifuged at 12
000 × g for one minute. The supernatant (∼400 μL) was transferred to a new clean microcentrifuge tube and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for an additional three minutes to pellet any remaining debris. Extracted nucleic acid was eluted (∼350 μL) into a final clean microcentrifuge tube. The eluted volume was then aliquoted (∼60 μL) into several microcentrifuge tubes (∼five extraction replicates per sample) for storage at −80 °C – to reduce the need for several freeze/thaw cycles. One aliquot per water sample was later used for PCR analysis (samples were held in −80 °C for up to 30 days before analysis).
000 × g for an additional three minutes to pellet any remaining debris. Extracted nucleic acid was eluted (∼350 μL) into a final clean microcentrifuge tube. The eluted volume was then aliquoted (∼60 μL) into several microcentrifuge tubes (∼five extraction replicates per sample) for storage at −80 °C – to reduce the need for several freeze/thaw cycles. One aliquot per water sample was later used for PCR analysis (samples were held in −80 °C for up to 30 days before analysis).
Droplet digital PCR (Bio-Rad Laboratories, CA, USA) technology was performed according to the manufacturer's instructions to analyze each sample for general Legionella spp. (23S rRNA), L. pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae. The primers and probes used in this study are listed in Table 2. Duplex reactions were performed for three separate assays: the first assay consisted of Legionella spp. and L. pneumophila, the second assay comprised of L. micdadei, and L. anisa, and the third assay consisted of L. bozemanii, and L. longbeachae (Table 2). All primers and probes were ordered from Eurofins (KY, USA).
| Target species | Primer/probe name | Primer/probe sequencea | Accession number | Amplicon length (bp) | Ref. |
|---|---|---|---|---|---|
a Hexachlorofluorescein (HEX), and fluorescein amidites (FAM), reporter dyes that are added to the 5′ end of an oligonucleotide.
b Black Hole Quencher (BHQ1), dark quenchers that does not absorb or emit light and are added to the 3′ end of an oligonucleotide. The temperature for each primer and probe set was 57.1 °C. The sample DNA did not need to be diluted before performing digital PCR as ddPCR is less sensitive to inhibitors. However, the positive control DNA was diluted to minimize the over-saturation of DNA to polymerase and this was identified by performing a dilution series with each positive control sample. The lowest dilution factor used for positive control DNA that yielded consistent quantification was 10−5. The limit of detection for 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log per 100 mL. log per 100 mL.
|
|||||
| Legionella species | 23SF | 5′-CCCATGAAGCCCGTTGAA-3′ | Not available | 92 | 80 |
| 23SR | 5′-ACAATCAGCCAATTAGTACGAG TTAGC-3′ | ||||
| 23SP probe | 5′-HEXa-TCCACACCTCGCCTATCAACGTCGTAGT-BHQ1b-3′ | ||||
| L. pneumophila (mip gene) | mipF | 5′-AAAGGCATGCAAGACGCTATG-3′ | S42595 | 78 | |
| mipR | 5′-GAAACTTGTTAAGAACGTCTTTCATTTG-3′ | ||||
| LmipP | 5′-FAMa-TGGCGCTCAATTGGCTTTAACCGA-BHQ1b-3′ | ||||
| L. micdadei | Pan-Legionella F | 5′-GTACTAATTGGCTGATTGTCTTG-3′ | Z30460 | Not available | 81 |
| L. anisa | Pan-Legionella R | 5′-TTCACTTCTGAGTTCGAGATGG-3′ | Z30535 | ||
| L. bozemanii | LmicdadeiP | 5′-FAMa-AGCTGATTGGTTAATAGCCCAATCGG-BHQ1b-3′ | AY883058 | ||
| LanisaP | 5′-HEXa-CTCAACCTACGCAGAACTACTTGAGG-BHQ1b-3′ | ||||
| L. longbeachae | Lbozemanii P | 5′-FAMa-TACGCCCATTCATCATGCAAACCAGnT-BHQ1b-3′ | Z30456 | ||
| LlongbeachaeP | 5′-HEXa-CTGAGTATCATGCCAATAATGCGCGC-BHQ1b-3′ | ||||
For each reaction mixture, 2X supermix (no dUTP) (Bio-Rad Laboratories CA, USA) was mixed with a final concentration of 900 nM of forward and reverse primers, 250 nM for each probe (Eurofins Genomics Co., AL, USA), and DNA template (up to 330 ng) to a final volume of 22.0 μL (10% excess), as recommended by Bio-Rad. Exactly 20 μL of each of the samples' reaction mixtures were loaded into a DG8 cartridge (Bio-Rad Laboratories, CA, USA), followed by 70 μl of droplet generator oil (Bio-Rad Laboratories, CA, USA). The samples were then loaded into the QX200 Droplet Generator, and droplets were generated. The droplet emulsion (∼40 μl) was then transferred into a 96-well plate using a multichannel pipet. The plate was then heat sealed with pierceable foil heat seals using a PX1™ PCR Plate Sealer (Bio-Rad, Laboratories, CA USA). The sample reaction mixture was amplified using a Benchmark TC9639 thermal cycler (Benchmark Scientific Inc, NJ, USA) with the following thermocycling parameters: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 57 °C for 1 min, with a final 10 min cycle at 98 °C for 10 min. Droplets were then read using a QX200 droplet reader (Bio-Rad QX200™ Droplet Digital™ PCR System, CA, USA).
The strains of these five species were obtained from American Type Culture Collection (ATCC®) and are listed below in parenthesis. Two negative controls, a filtration blank (phosphate-buffered water) and a non-template control (molecular grade water) were run with each ddPCR plate. Positive controls using DNA from L. pneumophila (ATCC No. 33152), L. micdadei (ATCC No. 33218), L. anisa (ATCC No. 35292), L. bozemanii, (ATCC No. 33217) and L. longbeachae (ATCC No. 33462) for each assay target were run with each ddPCR plate. Sample results were only considered for analysis when the reader accepted 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 or more droplets as part of the quality control. Sample reactions with three or more positive droplets per well were identified as positive for their assay target. Three technical replicates were run for each sample to determine the reproducibility of the assay results. Further detailed ddPCR experimental information is in Table 1 Supplementary material.
000 or more droplets as part of the quality control. Sample reactions with three or more positive droplets per well were identified as positive for their assay target. Three technical replicates were run for each sample to determine the reproducibility of the assay results. Further detailed ddPCR experimental information is in Table 1 Supplementary material.
Statistical analysis
Descriptive statistics were conducted in GraphPad Prism 8 software (GraphPad Software, CA, USA). Sample concentrations were transformed from gene copies (GC)/100 mL into log10 GC/100 mL for statistical analysis. A geometric mean for each sample was calculated using only the technical replicates that had ≥ three positive droplets. If one technical replicate was positive, only that value was used. The biological data were expressed as geometric means with standard deviations (SD). Correlation analysis was performed between the concentrations of Legionella species (23S rRNA) present in samples and water quality parameters tested (temperature, chlorine, turbidity, pH, and conductivity). One-way analysis of variance (ANOVA) was also performed to compare each variable (building influents, taps on the first and top floor (if any), cold- and hot-water taps, and among both sampling events). Statistical results were interpreted at the level of significance p < 0.05.Results
Characterization and concentrations of Legionella 23S rRNA and five pathogenic Legionella species
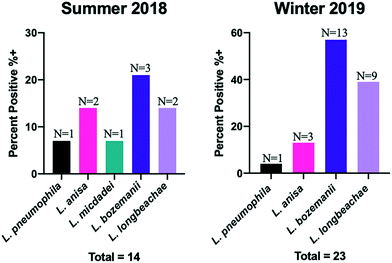
Overall, a total of 37 large volume composite samples from five buildings were analyzed during this study: 14 from the beginning of the summer sampling event (fall term) and 23 from the beginning of the winter term. Legionella species (23S rRNA) were found in all water samples at concentrations ranging from 1.4 to 4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL, and 54% of the samples were positive for at least one of the target species: L. pneumophila (2/37), L. anisa (5/37), L. micdadei, (1/37), L. bozemanii, (16/37), and L. longbeachae (11/37) at average geomean concentrations of 1.7, 1.6, 1.7, 1.6, and 1.6
log10 GC/100 mL, and 54% of the samples were positive for at least one of the target species: L. pneumophila (2/37), L. anisa (5/37), L. micdadei, (1/37), L. bozemanii, (16/37), and L. longbeachae (11/37) at average geomean concentrations of 1.7, 1.6, 1.7, 1.6, and 1.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL, respectively (Fig. 1).
log10 GC/100 mL, respectively (Fig. 1).
Five Legionella species detected in the influent and tap water samples in five different buildings
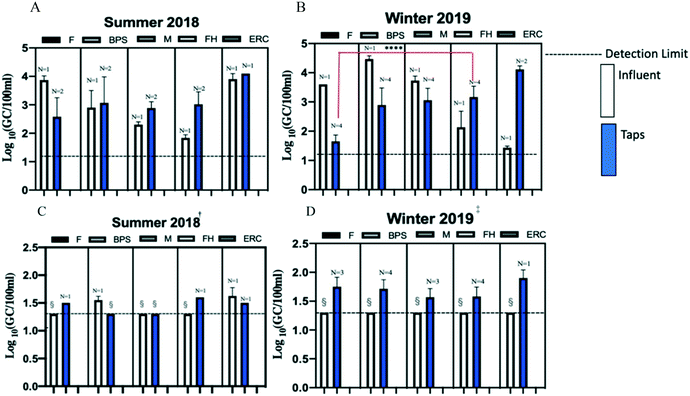
Two of the five influent samples were positive for at least one of the target pathogenic Legionella species in the summer sampling and no pathogenic species were detected in the influents to the buildings during the winter sampling event. Legionella bozemanii and L. longbeachae were detected in the influent of the BPS building at concentrations of 1.6 and 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL, respectively. Legionella pneumophila, L. micdadei, L. bozemanii, and L. longbeachae were detected in the influent (eyewash site) of the ERC building at concentrations of 1.5, 1.7, 1.8, 1.5
log10 GC/100 mL, respectively. Legionella pneumophila, L. micdadei, L. bozemanii, and L. longbeachae were detected in the influent (eyewash site) of the ERC building at concentrations of 1.5, 1.7, 1.8, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL, respectively (Fig. 2).
log10 GC/100 mL, respectively (Fig. 2).
In the taps of the buildings, L. pneumophila was detected in 3.7% of the composite samples (1 of 27; January 7th, 2019). Legionella anisa, L. bozemanii and L. longbeachae were detected in 18.5, 51.8, and 33% respectively (during both the summer and winter sampling events). Legionella micdadei was not detected in the composite tap water samples (Fig. 2).
Potential amplification of general (23S rRNA) and five pathogenic Legionella species between the influent and the taps
Fig. 2 compares the influent concentrations to the taps for buildings F, BPS, M, FH, and ERC and demonstrates the potential for amplification of general (23S rRNA) and pathogenic Legionella species in the premise plumbing. In the summer sampling event, 80% of the buildings (BPS, M, FH, and ERC) had higher Legionella (23S rRNA) concentrations at the exposure sites (taps) compared to influent water samples (Fig. 2A). In the winter sampling event, 40% of the buildings (FH, and ERC) had higher Legionella (23S rRNA) concentrations observed at the taps compared to influent water samples (Fig. 2B). The concentration of general Legionella spp. (23S rRNA) in building FH was significantly higher than the concentration detected in building F (Fig. 2B). Two buildings (F and FH) showed suspected amplification for L. anisa in the summer samples (Fig. 2C). All buildings in the winter showed potential amplification, L. pneumophila, L. anisa, L. bozemanii, and L. longbeachae (Fig. 2D). Overall, there were higher Legionella concentrations (23S rRNA and target pathogenic species) seen at these exposure sites (taps) compared to influent samples (Fig. 2).Comparison of five targeted Legionella species in summer and winter semesters
In the summer sampling event, all five specific Legionella spp. (L. pneumophila, L. anisa, L. micdadei, L. bozemanii and L. longbeachae) were detected. In the winter, four species were detected as L. micdadei was not found. Overall, L. bozemanii (43%, 16/37) and L. longbeachae (29.7%, 11/37) accounted for the majority of the Legionella positive samples detected in both sampling events. In the summer for the composite samples buildings F and FH only had one species detected (L. anisa) and building M had no detects. Building BPS was positive for both L. bozemanii, and L. longbeachae and ERC was positive for L. pneumophila, L. bozemanii, L. longbeachae and L. micdadei. In the winter, hot water taps were positive in F, BPS, M, FH, and ERC for L. pneumophila (detected once), L. anisa, L. bozemanii, and L. longbeachae. The cold-water taps were positive in F, BPS, M, and FH for L. anisa, L. bozemanii, and L. longbeachae species (see Fig. 1 in ESI†).
Fig. 1 compares the presence and absence of the pathogens in tap water samples from the summer and winter seasons. Legionella pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae were present in low quantities (near the detection limit, 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL) throughout the buildings drinking water system. The concentrations of these species ranged from 1.5 to 1.8
log10 GC/100 mL) throughout the buildings drinking water system. The concentrations of these species ranged from 1.5 to 1.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL in the summer samples, and from 1.4 to 2.0
log10 GC/100 mL in the summer samples, and from 1.4 to 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL in the winter samples (Fig. 2). More specifically, during the summer, 7% (1/14) of the composite samples were positive for L. pneumophila and L. micdadei, 14% (2/14) for L. anisa, and L. longbeachae, and 21% (3/14) for L. bozemanii (Fig. 1). Collectively, five target Legionella species were detected in 36% (5/14) of the summer samples. During the winter, 65% (15/23) samples were positive with one or more of the target Legionella species. Legionella bozemanii had the highest occurrence at 57% (13/23), followed by L. longbeachae at 39% (9/23), L. anisa at 13% (3/23), and L. pneumophila at 4% (1/23) (Fig. 1). Legionella micdadei was not detected in any of the winter samples (Fig. 1).
log10 GC/100 mL in the winter samples (Fig. 2). More specifically, during the summer, 7% (1/14) of the composite samples were positive for L. pneumophila and L. micdadei, 14% (2/14) for L. anisa, and L. longbeachae, and 21% (3/14) for L. bozemanii (Fig. 1). Collectively, five target Legionella species were detected in 36% (5/14) of the summer samples. During the winter, 65% (15/23) samples were positive with one or more of the target Legionella species. Legionella bozemanii had the highest occurrence at 57% (13/23), followed by L. longbeachae at 39% (9/23), L. anisa at 13% (3/23), and L. pneumophila at 4% (1/23) (Fig. 1). Legionella micdadei was not detected in any of the winter samples (Fig. 1).
Legionella species in cold compared to hot taps
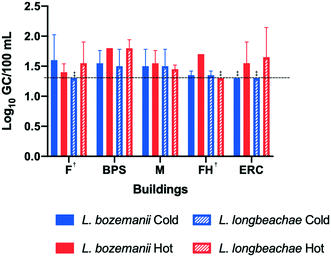
Fig. 3 compares the log10 gene copies of the most prevalent species, L. bozemanii and L. longbeachae in the cold- and hot-water taps. Legionella bozemanii concentrations were higher in the hot-water samples (geomean of 1.7) than in the cold-water samples (geomean of 1.6) in BPS, M, FH, and ERC. Legionella longbeachae concentrations were also higher in the hot-water samples (geomean of 1.8) compared to the cold-water samples (geomean of 1.6) in F, BPS, and ERC buildings. Concentrations of L. bozemanii and L. longbeachae in the cold-water taps were significantly different (unpaired t-test, p = 0.03) than the hot-water taps. Overall, the five target Legionella species were more prevalent in hot-water samples (39% positive, 9/23) compared to the cold-water samples (26% positive, 6/23). Within the hot-water samples, there appeared to be more diversity of the target Legionella species (L. pneumophila, L. anisa, L. bozemanii, or L. longbeachae) present, compared to the cold-water tap samples, where L. pneumophila was not detected (Fig. 3).Chemical–physical water quality
For the summer sampling event, the cold- and hot-water taps were composite samples; thus, the chemical–physical parameters are reflective of this as the interest was determining the difference of water quality by floors. In August and September, water temperatures in the influent of all buildings ranged from 12.6 to 20.2 °C, with an average of 16.5 °C (Table 3). The temperatures of the composite cold- and hot-water samples were similar in range among both floors but slightly different across buildings. The water temperature across the buildings ranged from 25.8 to 34.2 °C and 27.1 to 36.7 °C on the first floor and top floor, respectively. Free chlorine ranged from 0.04 mg L−1 to 0.52 mg L−1, with an average of 0.3 mg L−1 in the influents. The buildings' average free chlorine on the first floor was 0.09 mg L−1 and increased on the top floor to 0.21 mg/ (Table 3). This may be due to a higher occupancy on the top floor and greater use of the bathrooms due to research labs and office space being active at the start of the semester, compared to the first floor with more classrooms which were not in use at the time of sampling. The conductivity ranged from 750 to 867 μS cm−1, with an average of 802 μS cm−1 in the buildings' influent. The buildings' average conductivity was 915.8 μS cm−1 on the first floor and then decreased on the top floor (827.8 μS cm−1). Turbidity ranged from 1.3 to 66.2 NTU (this groundwater sources being notorious for high iron content with many “red water” alerts), with an average of 19.5 NTU in the buildings' influent. The buildings' average turbidity dropped to 3.2 NTU on the first floor and slightly increased on the top floor (5.9 NTU). The mean pH was 7.4 in the influents, first floors, and the top floors.| Composite cold and hot-water sample | ||||||
|---|---|---|---|---|---|---|
| F | BPS | M | FH | ERCa | Building average | |
| a ERC, has one floor. b Composite cold and hot taps. | ||||||
| Influent | ||||||
| Temperature (°C) | 15.7 | 12.6 | 18.3 | 15.9 | 20.2 | 16.5 |
| Conductivity (μS) | 750 | 867 | 755 | 780 | 858 | 802 |
| Turbidity (NTU) | 24.5 | 3.58 | 1.3 | 1.89 | 66.2 | 19.5 |
| pH | 7.3 | 7.5 | 7.5 | 7.5 | 7.6 | 7.48 |
| Total (free) chlorine residual (mg L−1) | 0.58 (0.52) | 0.52 (0.32) | 0.39 (0.3) | 0.53 (0.43) | 0.04 (0.04) | 0.4 (0.3) |
| Coliforms (MPN/100 mL) | <1 | <1 | <1 | <1 | <1 | <1 |
| E. coli (MPN/100 ml) | <1 | <1 | <1 | <1 | <1 | <1 |
| 1st floor composite samplesb | ||||||
| Temperatureb (°C) | 33.4 | 29.2 | 34.2 | 31.9 | 25.8 | 30.9 |
| Conductivity (μS) | 814 | 857 | 807 | 1256 | 845 | 915.8 |
| Turbidity (NTU) | 0.55 | 0.98 | 6.36 | 3.44 | 4.43 | 3.2 |
| pH | 7.3 | 7.4 | 7.5 | 7.2 | 7.6 | 7.4 |
| Total (free) chlorine residualb (mg L−1) | 0.16 (0.13) | 0.16 (0.02) | 0.12 (0.03) | 0.24 (0.17) | 0.17 (0.1) | 0.17 (0.09) |
| Coliforms (MPN/100 mL) | <1 | <1 | <1 | <1 | <1 | <1 |
| E. coli (MPN/100 mL) | <1 | <1 | <1 | <1 | <1 | <1 |
| Top floorb | ||||||
| Temperatureb (°C) | 32 | 27.1 | 36.7 | 35 | N/A | 32.7 |
| Conductivity (μS) | 794 | 904 | 794 | 819 | N/A | 827.8 |
| Turbidity (NTU) | 2.06 | 5.07 | 13.1 | 3.46 | N/A | 5.9 |
| pH | 7.3 | 7.5 | 7.5 | 7.4 | N/A | 7.4 |
| Total (free) chlorine residualb (mg L−1) | 0.29 (0.22) | 0.33 (0.21) | 0.1 (0.05) | 0.1 (0.04) | N/A | 0.2 (0.21) |
| Coliforms (MPN/100 mL) | <1 | <1 | <1 | <1 | N/A | <1 |
| E. coli (MPN/100 mL) | <1 | <1 | <1 | <1 | N/A | <1 |
For the winter sampling event, the cold- and hot-water taps were collected as separate composite samples; thus, the chemical–physical parameters are reflective of this. In the winter, building influent water temperature ranged from 11.2 to 26.9 °C, with an average of 17.9 °C (Table 4). The average (21.4 °C for the first floor and 22.4 °C on the top floor) cold-water temperature for the buildings did not differ between floors; however, the buildings' hot-water was slightly warmer, on average, on the top floor (36.1 °C) compared to the first floor (31.6 °C). Free chlorine ranged from 0.17 to 1.46 mg L−1 (influent of FH to influent of BPS) with an average of 0.6 mg L−1. The buildings' mean for free chlorine (first and top floors) differed between the cold- (0.07 mg L−1) and hot-water taps (0.04 mg L−1) (Table 4). The buildings' average conductivity in winter ranged from 794 to 931 μS cm−1 in the influent with an average of 847 μS cm−1. The conductivity of the cold- (947 μS cm−1) and hot-water taps (931 μS cm−1) on the first floor of the buildings were only slightly different. The conductivity on the top floors varied more between the cold- (890 μS cm−1) and hot-water taps (918 μS cm−1). Turbidity ranged from 4.6 to 155 NTU (influent of FH Hall to influent of BPS) with an average of 58.3 NTU (may be due to high iron content). The mean turbidity for the cold-water taps was 7.6, and 2.4 for the hot-water taps on the buildings' first floors. However, the mean turbidity slightly increased on the top floor for both taps, 20.6 NTU (cold-water tap), and 2.81 NTU (hot-water tap). The pH was approximately the same as the summer sampling, ranging from 7.3 (influent) to 7.6 (first-floor hot-water tap) and 7.5 (top-floor hot-water taps).
| Composite cold-water | ||||||
|---|---|---|---|---|---|---|
| Composite hot-water | ||||||
| F | BPS | M | FH | ERCa | Building average | |
| a ERC, only has one floor. | ||||||
| Influent | ||||||
| Temperature °C | 14.5 | 23.6 | 13.3 | 11.2 | 26.9 | 17.9 |
| Conductivity μS | 914 | 794 | 931 | 799 | 797 | 847 |
| Turbidity NTU | 18.6 | 155 | 6.33 | 4.6 | 106.9 | 58.3 |
| pH | 7.3 | 7.5 | 7.3 | 7.3 | 7.3 | 7.3 |
| Total (free) chlorine residual mg L−1 | 0.24(0.23) | 1.16(1.46) | 0.09(0.18) | 0.05(0.17) | 1.19(1.33) | 0.5(0.6) |
| Coliforms MPN/100 ml | <1 | <1 | <1 | <1 | <1 | <1 |
| E. coli MPN/100 ml | <1 | <1 | <1 | <1 | <1 | <1 |
| HPCs (CFU/100 mL) | 8.8 × 101 | 7.40 × 103 | 2.70 × 102 | 3.00 × 101 | 4.00 × 105 | 8.16 × 104 |
| 1st floor from composite samples | ||||||
| Temperature °C (cold and hot taps) | 17.8 | 22.8 | 23.3 | 21.8 | 21.1 | 21.4 |
| 37.6 | 31.3 | 36.3 | 28.2 | 24.7 | 31.6 | |
| Conductivity μS (cold and hot taps) | 793 | 1161 | 913 | 865 | 924 | 931 |
| 801 | 1220 | 904 | 895 | 914 | 947 | |
| Turbidity NTU (cold and hot taps) | 2.4 | 6.44 | 16.4 | 4.22 | 8.55 | 7.6 |
| 0.1 | 1.44 | 2.39 | 4.36 | 3.78 | 2.4 | |
| pH (cold and hot taps) | 7.4 | 7.3 | 7.6 | 7.6 | 7.7 | 7.5 |
| 7.7 | 7.4 | 7.7 | 7.8 | 7.6 | 7.6 | |
| Total (free) chlorine residual mg L−1 (cold and hot taps) | 0.04(0.03) | 0.14(0.1) | 0.14(0.13) | 0.05(0.16) | 0.07(0.09) | 0.09(0.1) |
| 0.03(0) | 0.03(0.05) | 0.03(0.02) | 0.08(0.12) | 0.09(0.11) | 0.05(0.06) | |
| Coliforms MPN/100 ml | <1 | <1 | <1 | <1 | <1 | <1 |
| E. coli MPN/100 ml | <1 | <1 | <1 | <1 | <1 | <1 |
| HPCs (CFU/100 mL) | 2.6 × 101 | 1.02 × 105 | 3.53 × 103 | 1.37 × 103 | 8.1 × 103 | 2.3 × 104 |
| 1.31 × 103 | 3.0 × 104 | 6.8 × 102 | 2.8 × 103 | 9.20 × 104 | 2.54 × 104 | |
| Top floor | ||||||
| Temperature (°C) (cold and hot taps) | 19.9 | 22 | 24.2 | 23.3 | N/A | 22.4 |
| 38.9 | 31.1 | 44.9 | 29.6 | 36.1 | ||
| Conductivity μS (cold and hot taps) | 786 | 977 | 882 | 916 | N/A | 890 |
| 802 | 1062 | 895 | 914 | 918 | ||
| Turbidity NTU (cold and hot taps) | 4.04 | 3.73 | 71 | 3.64 | N/A | 20.6 |
| 0.32 | 1.62 | 7.14 | 2.16 | 2.81 | ||
| pH (cold and hot taps) | 7.5 | 7.3 | 7.4 | 7.5 | N/A | 7.4 |
| 7.6 | 7.2 | 7.8 | 7.6 | 7.5 | ||
| Total (free) chlorine residual mg L−1 (cold and hot taps) | 0.05(0.07) | 0.05(0.05) | 0.4(0.4) | 0.03(0.05) | N/A | 0.13(0.14) |
| 0.07(0.03) | 0.02(0.01) | 0.09(0.09) | 0.02(0.04) | 0.05(0.04) | ||
| Coliforms MPN/100 ml | <1 | <1 | <1 | <1 | N/A | <1 |
| E. coli MPN/100 ml | <1 | <1 | <1 | <1 | N/A | <1 |
| HPCs (CFU/100 mL) | 7.00 × 101 | 1.14 × 104 | 5.3 × 102 | 2.4 × 103 | N/A | 3.6 × 103 |
| 1.31 × 103 | 1.56 × 103 | 1.54 × 102 | 1.04 × 103 | 9.72 × 102 | ||
Relationship between the presence of Legionella and water quality parameters
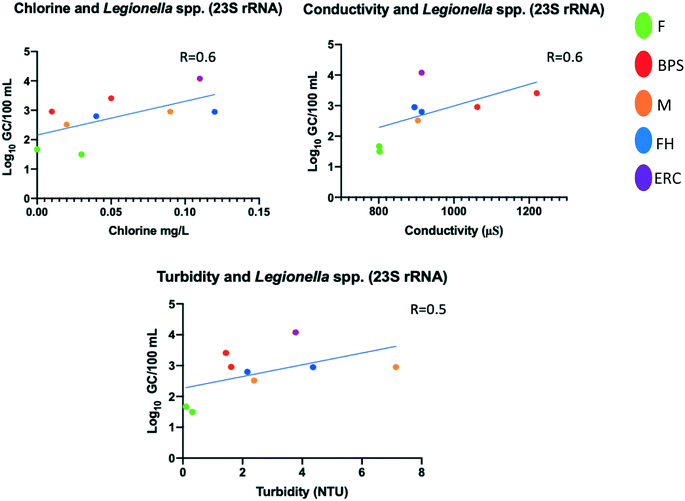
During the summer sampling event, there was not a relationship (positive or negative) between temperature, HPCs, chlorine, turbidity, pH, or conductivity with respect to general Legionella spp. (23S rRNA). Fig. 4 shows the correlation (R = 0.5 to 0.6) between Legionella spp. 23S rRNA and three water quality parameters (free residual chlorine concentration, conductivity, and turbidity) in the hot water taps during the winter semester events. While not statistically significant the positive trend between turbidity, pH, and HPCs, was driven more by the characteristics of the building with F being low and ERC being high with the other buildings in between.Chlorine residuals were very low in the building taps and no chlorine residual was detected when L. pneumophila and L. anisa were both detected in the hot-water taps on the first floor of building F. For the L. anisa positive sample (in the hot-water tap on the second floor on building FH), the residual chlorine was 0.04 mg L−1. Legionella bozemanii, and L. longbeachae occurred in hot-water taps on both floors (except ERC) of all five buildings; in the positive samples, the free residual chlorine ranged from 0 to 0.32 mg L−1. Free residual chlorine concentrations were below the US Centers for Disease Control and Prevention (US CDC) (under the safe water system) minimal 0.2 mg L−1 threshold47 in all Legionella-positive samples, except the BPS influent sample, which was 0.32 mg L−1. This sampling port (BPS influent) is located at the point water that enters into the building where turbidity levels were 3.58 NTU and water temperature was 12.6 °C. There was no significant correlation between pathogenic Legionella species and any water quality parameter (water temperature, residual chlorine, turbidity, pH, HPCs, or conductivity) tested; thus, this suggests the need for further investigation with larger data sets.
Discussion
This study revealed new quantitative information about the distribution of general Legionella and five pathogenic species in a complex of five buildings on the same community drinking water system. Legionella spp. (23S rRNA), L. pneumophila, L. micdadei, L. bozemanii, and L. longbeachae were found in the influent water pipes and four target species (excluding L. micdadei) were found at distal points of use. Legionella and pathogenic species are part of the water microbiome as reported previously.48,49 However, these studies did not provide concentrations on the pathogenic species other than L. pneumophila and the hospital systems are often the primary focus. In Schwake et al. (2016)49 20 single-story buildings were examined, and no pathogenic Legionella spp. were detected. This work presented herein examined large volume samples of the influent to buildings compared to composite large volume tap water samples and suggests amplification occurs of pathogenic species of Legionella in the system which is observable at the taps (Fig. 2). In addition, it was found that possible amplification can occur immediately at locations where the water enters the building as shown by the eye wash station at the ERC building. The reasons for this possible amplification are not completely clear however residence time, stagnation, water age could all be involved as the ERC building is furthest away from the water source (discussed below). However, all five buildings showed evidence of colonization with pathogenic Legionella species at the taps in the winter sampling event.The concentrations of the specific Legionella pathogenic species ranged 1.4 to 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL (average: 1.6
log10 GC/100 mL (average: 1.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL) at the exposure taps. The maximum contaminant level goal (MCLG) for Legionella is zero as established in the 1989 Surface Water Treatment Rule; however, this target is not federally regulated and thus no maximum contaminant level has been established.50 Moreover, the CDC provides a toolkit, which is a step-by-step guidance from ASHRAE Standard 188; this helps building owners evaluate the water system in their buildings to determine if a water management program is required51 however no numerical level for PCR exists at which action should be taken to remediate the building's water. Previous risk assessments suggest that the level which equates to the 10−4 annual infection risk target for drinking water safety (analysis of acceptable risk levels used a 1 in 10
log10 GC/100 mL) at the exposure taps. The maximum contaminant level goal (MCLG) for Legionella is zero as established in the 1989 Surface Water Treatment Rule; however, this target is not federally regulated and thus no maximum contaminant level has been established.50 Moreover, the CDC provides a toolkit, which is a step-by-step guidance from ASHRAE Standard 188; this helps building owners evaluate the water system in their buildings to determine if a water management program is required51 however no numerical level for PCR exists at which action should be taken to remediate the building's water. Previous risk assessments suggest that the level which equates to the 10−4 annual infection risk target for drinking water safety (analysis of acceptable risk levels used a 1 in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000/person-year as a target) is around 103 CFU L−1 (102 CFU/100 mL) for faucets.52 While the referenced acceptable risk level (described previously) was evaluated using CFU (culture method) and early studies suggested a ratio of around 7 CFU to 3 GC (PCR method) this assumed that all cells were culturable.53 There is a need for a greater comparison examining live and dead as well as viable- but- non-culturable cells to ultimately address risk via molecular tools. These concentrations presented herein at the taps are nearing the level of the acceptable annual risk – assuming that these species are 100% cultivatable and have similar dose-response characteristics (for example, the expression of virulence factors) as L. pneumophila.
000/person-year as a target) is around 103 CFU L−1 (102 CFU/100 mL) for faucets.52 While the referenced acceptable risk level (described previously) was evaluated using CFU (culture method) and early studies suggested a ratio of around 7 CFU to 3 GC (PCR method) this assumed that all cells were culturable.53 There is a need for a greater comparison examining live and dead as well as viable- but- non-culturable cells to ultimately address risk via molecular tools. These concentrations presented herein at the taps are nearing the level of the acceptable annual risk – assuming that these species are 100% cultivatable and have similar dose-response characteristics (for example, the expression of virulence factors) as L. pneumophila.
Both hot- and cold-water taps can be colonized, and Legionella species can survive in cold-water taps,3 but hot-water taps are known to be a major source for their amplification.54 Many studies report prevalence but not concentrations. Legionella pneumophila was detected in 82% of samples from a hot-water system at a university hospital located in Sherbrooke, Canada, by culture.55 In 2019, Bédard and colleagues found L. pneumophila SG1 positive in 41%, and L. pneumophila serogroups 4 and 10 in 91% of the water samples in hot-water taps and connecting pipes in an undisclosed Canadian hospital by culture followed by sequence-based typing.56
This study detected the presence of specific Legionella species in both cold-water and hot-water taps with slightly higher concentrations seen in hot-water; this is in agreement with previous studies3,57–61 but these were not statistically significant. Recently, Donohue and colleagues found a difference in concentrations for L. pneumophila in hot-water taps compared to cold-water taps collected from 46 states across the US served by public water utilities.62 The concentrations were higher (median concentrations: 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL) for L. pneumophila in the hot-water taps than in the cold-water taps (median concentrations: 1.5
log10 GC/100 mL) for L. pneumophila in the hot-water taps than in the cold-water taps (median concentrations: 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL).62 In this study, L. pneumophila in the hot-water tap at a concentration of 2.0
log10 GC/100 mL).62 In this study, L. pneumophila in the hot-water tap at a concentration of 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 GC/100 mL was similar to these studies. These results suggest that when examining the water quality of the building, hot water taps can be composited particularly from floors that are in less use and this will provide information on Legionella colonization.
log10 GC/100 mL was similar to these studies. These results suggest that when examining the water quality of the building, hot water taps can be composited particularly from floors that are in less use and this will provide information on Legionella colonization.
Increased water age in the distribution system has been suggested to have an adverse downstream effect within the building water system.63 The impacts of increased water age are increased water temperature and a loss of chemical residual in the building water system.64 These changes in combination influenced the occurrence (presence/absence) of Legionella species.57,64 In addition, several studies have shown that the concentrations of general Legionella spp. (23S rRNA) and L. pneumophila increased during the summer season65–67 which was presumed to be due to water temperature.68–72 However, in this study, L. pneumophila, L. anisa, and L. longbeachae increased in concentration during the winter sampling event. All three species (listed above) increased in the hot-water taps in buildings F, FH, and ERC. Hot-water taps are a source for Legionella amplification;58 thus, there is likely a need for a building monitoring approach that includes sampling the hot water taps separately from the cold-water taps.
Our study examined these buildings at the beginning of two semesters (summer/fall Aug/Sept compared to winter/January). Each building except M had flow meters and water usage could be evaluated. The data on water use showed key buildings in the complex use less water in January compared to August due to cooling towers (BPS and ERC) which are used in the summer. Prior to classes beginning the bottom floor bathrooms of some of the buildings which housed the classrooms and not the research laboratories showed less use. Seasonal effects in this study on water temperature were not observed. The water temperature averages on the first and top floors for all the buildings were close to the Legionella's optimal growth temperatures (25 to 45 °C).71 Turbidity values were high in BPS, M, and ERC building samples. There was a moderate correlation between turbidity and Legionella colonization in hot water tap samples—this relationship may be a direct result of iron.73 The water quality on the various floors looked only slightly different with respect to higher chlorine residuals on the top floors perhaps due to greater water usage as the research laboratories and offices were consistently occupied and water was used and replaced at the taps. However, buildings were different in their physical chemical quality. Similar to previous studies, higher chlorine residuals observed in the BPS building (0.32 mg L−1) showed little to no effect on disinfecting Legionella in the influent sample.74 Interestingly, the influent of the ERC building had the greatest variety of Legionella species (L. pneumophila, L. micdadei, L. bozemanii, and L. longbeachae) and the detection of these species appears to be related to the lower levels of residual chlorine (0.04 mg L−1) at this site. This could be due to the ERC building having increased water age as its influent water pipe is the furthest away from the water source (reservoir) at 19.4 km. These data suggest that the influent water is seeding the system and water quality of the building including microbial aspects (Legionella) was affected more by water stagnation (low water use) and that understanding water age as it plays a role in the occurrence of Legionella species warrants further exploration as monitoring moves forward.
Health departments should consider the role of other Legionella species (L. anisa, L. micdadei, L. bozemanii, and L. longbeachae) in the presentation of pneumonia as they may pose a equal to or greater risk than L. pneumophila as they are widely distributed in the environment.18,23,41–45,75,76 The detection method used for routine sampling of water in hospitals and cooling towers is the culture-dependent method, however, there are some limitations to this practice: the inability to rapidly and precisely identify specific Legionella species and the presence of viable-but- non-culturable species, however PCR methods are incapable of distinguishing live and dead cells.77,78 Yet quantitative assessment using PCR can provide important information on concentrations and the approach used in this study to assay composites means that the “building” water quality can be examined and not just individual taps. Digital droplet PCR was very useful and could be run with duplex assays for the various species in this study, overcoming time delay from sampling to quantitative results as reported by others79 compared to the “gold standard” culture method for Legionella detection and LD diagnosis.77 These types of data provide information for building operations that could improve water quality such as more flushing and increasing flows through the system.
Conclusions
Legionella are a part of the water microbiome and were found 100% of the time. Yet pathogenic species beyond L. pneumophila, including L. anisa, L. micdadei, L. bozemanii, and L. longbeachae are also important in describing “building” water quality in a community drinking water system in the US. Our results provide evidence that pathogenic species in addition to L. pneumophila are increasing and potentially amplifying when comparing presence and concentrations between the influent and the points of use (taps) in various large educational buildings. More Legionella species were found under conditions where water stagnation (water age) or longer retention times in the pipes (low water use) were observed. By monitoring pathogenic species L. pneumophila, L. anisa, L. bozemanii, L. longbeachae and L. micdadei changes to building management can be made to address potentially different risks at different times of the year. This supports the need for a water management plan for various building types to reach optimization which includes monitoring.The examination of large volume (10 L) water samples using ultrafiltration increased the detection limits for specific Legionella species. A monitoring scheme that includes composite, large-volume sampling, and rapid assessment by ddPCR could lead to better control of Legionella in building drinking water systems.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the U.S. EPA Grant No. (FAIN): 83689001 and U.S. EPA agreement number R836890. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the EPA. AL-J thanks MF, Parker Kelly, and Rebecca Ives for their assistance on sampling. AL-J thanks the Infrastructure Plant Facility Staff at MSU for their assistance sampling the buildings.References
- D. M. Pierre, J. Baron, V. L. Yu and J. E. Stout, Diagnostic testing for Legionnaires’ disease, Ann. Clin. Microbiol. Antimicrob., 2017, 16(1), 59–62 CrossRef.
- M. Farhat, M. Moletta-Denat, J. Frère, S. Onillon, M.-C. Trouilhé and E. Robine, Effects of Disinfection on Legionella spp., Eukarya, and Biofilms in a Hot Water System, Appl. Environ. Microbiol., 2012, 78(19), 6850–6858 CrossRef CAS.
- M. J. Donohue, K. O'Connell, S. J. Vesper, J. H. Mistry, D. King and M. Kostich, et al., Widespread Molecular Detection of Legionella pneumophila Serogroup 1 in Cold Water Taps across the United States, Environ. Sci. Technol., 2014, 48(6), 3145–3152 CrossRef CAS.
- A. J. Prussin, D. O. Schwake and L. C. Marr, Ten Questions Concerning the Aerosolization and Transmission of Legionella in the Built Environment, Build Environ, 2017, 123, 684–695 CrossRef.
- D. W. Fraser, T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham and R. G. Sharrar, et al., Legionnaires' disease: description of an epidemic of pneumonia, N. Engl. J. Med., 1977, 297(22), 1189–1197 CrossRef CAS.
- Legionnaires Disease and Pontiac Fever | For Media | CDC, 2020, [cited 2020 Dec 1], Available from: https://www.cdc.gov/legionella/qa-media.html.
- T. Nishida, N. Nakagawa, K. Watanabe, T. Shimizu and M. Watarai, Attenuated Legionella pneumophila Survives for a Long Period in an Environmental Water Site, BioMed Res. Int., 2019, 2019, 8601346 Search PubMed.
- T. S. Gomes, L. Vaccaro, A. Magnet, F. Izquierdo, D. Ollero and C. Martínez-Fernández, et al., Presence and interaction of free-living amoebae and amoeba-resisting bacteria in water from drinking water treatment plants, Sci. Total Environ., 2020, 719, 137080 CrossRef CAS.
- A. F. Smith, A. Huss, S. Dorevitch, L. Heijnen, V. H. Arntzen and M. Davies, et al., Multiple Sources of the Outbreak of Legionnaires' Disease in Genesee County, Michigan, in 2014 and 2015, Environ. Health Perspect., 2019, 127(12), 127001 CrossRef.
- MDHHS - Michigan Urges Continued Legionella Precaution, [cited 2020 Dec 1]. Available from: https://www.michigan.gov/mdhhs/0,5885,7-339--385676--,00.html.
- K. D. Beer, J. W. Gargano, V. A. Roberts, V. R. Hill, L. E. Garrison and P. K. Kutty, et al., Surveillance for Waterborne Disease Outbreaks Associated with Drinking Water — United States, 2011–2012, Morb. Mortal. Wkly. Rep., 2015, 64(31), 842–848 CrossRef.
- Y. Shachor-Meyouhas, I. Kassis, E. Bamberger, T. Nativ, H. Sprecher and I. Levy, et al., Fatal hospital-acquired Legionella pneumonia in a neonate, Pediatr. Infect. Dis. J., 2010, 29(3), 280–281 CrossRef.
- V. C. Cheng, S. S. Wong, J. H. Chen, J. F. Chan, K. K. To and R. W. Poon, et al., An unprecedented outbreak investigation for nosocomial and community-acquired legionellosis in Hong Kong, Chin. Med. J., 2012, 125(23), 4283 CAS.
- T. E. Haupt, R. T. Heffernan, J. J. Kazmierczak, H. Nehls-Lowe, B. Rheineck and C. Powell, et al., An Outbreak of Legionnaires Disease Associated with a Decorative Water Wall Fountain in a Hospital, Infect. Control Hosp. Epidemiol., 2012, 33(2), 185–191 CrossRef.
- S. Jarraud, G. Descours, C. Ginevra, G. Lina and J. Etienne, Identification of Legionella in clinical samples, Methods Mol. Biol., 2013, 954, 27–56 CrossRef CAS.
- M. Foissac, L. Bergon, J. Vidal, P. Cauquil, A. Mainar and M. Mourguet, Pneumonia and pulmonary abscess due to Legionella micdadei in an immunocompromised patient, GERMS, 2019, 9(2), 89–94 CrossRef.
- A. Chakeri, F. Allerberger, M. Kundi, A. Stöger, S. Rehak and W. Ruppitsch, et al., Draft Genome Sequences of Legionella taurinensis Recovered from a Hot Water System in Austria, 2018, Microbiol. Resour. Announce., 2019, 8(3), e01478-18 CrossRef.
- T. Dilger, H. Melzl and A. Gessner, Legionella contamination in warm water systems: A species-level survey, Int. J. Hyg. Environ. Health, 2017, 1, 221 Search PubMed.
- C. Stallworth, L. Steed, M. A. Fisher and F. S. Nolte, Legionnaires' Disease Caused by Legionella londiniensis, J. Clin. Microbiol., 2012, 50(12), 4178–4179 CrossRef.
- C. W. Svarrer and S. A. Uldum, The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry, Clin. Microbiol. Infect., 2012, 18(10), 1004–1009 CrossRef CAS.
- R. L. Cameron, K. G. J. Pollock, D. S. J. Lindsay and E. Anderson, Comparison of Legionella longbeachae and Legionella pneumophila cases in Scotland; implications for diagnosis, treatment and public health response, J. Med. Microbiol., 2016, 65(2), 142–146 CrossRef CAS.
- H. L. Isenman, S. T. Chambers, A. D. Pithie, S. L. S. MacDonald, J. M. Hegarty and J. L. Fenwick, et al., Legionnaires' disease caused by Legionella longbeachae: Clinical features and outcomes of 107 cases from an endemic area, Respirology, 2016, 21(7), 1292–1299 CrossRef.
- L. Vaccaro, F. Izquierdo, A. Magnet, C. Hurtado, M. A. Salinas and T. S. Gomes, et al., First Case of Legionnaire's Disease Caused by Legionella anisa in Spain and the Limitations on the Diagnosis of Legionella non-pneumophila Infections, PLoS One, 2016, 11(7), e0162934 CrossRef.
- V. L. Yu, J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe and A. Widmer, et al., Distribution of Legionella Species and Serogroups Isolated by Culture in Patients with Sporadic Community-Acquired Legionellosis: An International Collaborative Survey, J. Infect. Dis., 2002, 186(1), 127–128 CrossRef.
- J. Beauté, E. Robesyn and B. Jong, Legionnaires' disease in Europe: all quiet on the eastern front?, Eur. Respir. J., 2013, 42(6), 1454–1458 CrossRef.
- G. F. Craun, J. M. Brunkard, J. S. Yoder, V. A. Roberts, J. Carpenter and T. Wade, et al., Causes of Outbreaks Associated with Drinking Water in the United States from 1971 to 2006, Clin. Microbiol. Rev., 2010, 23(3), 507–528 CrossRef CAS.
- M. Best, J. Stout, R. R. Muder, V. L. Yu, A. Goetz and F. Taylor, Legionellaceae In The Hospital Water-Supply: Epidemiological Link with Disease and Evaluation of a Method for Control of Nosocomial Legionnaires' Disease and Pittsburgh Pneumonia, Lancet, 1983, 322(8345), 307–310 CrossRef.
- C. A. Knirsch, K. Jakob, D. Schoonmaker, J. A. Kiehlbauch, S. J. Wong and P. Della-Latta, et al., An outbreak of Legionella micdadei pneumonia in transplant patients: evaluation, molecular epidemiology, and control, Am. J. Med., 2000, 108(4), 290–295 CrossRef CAS.
- B. Doebbeling, M. A. Ishak, B. H. Wade, M. A. Pasquale, R. E. Gerszten and D. H. M. Gröschel, et al., Nosocomial Legionella micdadei pneumonia: 10 years experience and a case-control study, J. Hosp. Infect., 1989, 13(3), 289–298 CrossRef CAS.
- R. D. Harrington, A. E. Woolfrey, R. Bowden, M. G. McDowell and R. C. Hackman, Legionellosis in a bone marrow transplant center, Bone Marrow Transplant., 1996, 18(2), 361–368 CAS.
- A. W. Pasculle, J. C. Feeley, R. J. Gibson, L. G. Cordes, R. L. Myerowitz and C. M. Patton, et al., Pittsburgh pneumonia agent: direct isolation from human lung tissue, J. Infect. Dis., 1980, 141(6), 727–732 CrossRef CAS.
- R. L. Myerowitz, A. W. Pasculle, J. N. Dowling, G. J. Pazin, M. Puerzer and R. B. Yee, et al., Opportunistic Lung Infection Due to Pittsburgh Pneumonia Agent, N. Engl. J. Med., 1979, 301(18), 953–958 CrossRef CAS.
- B. H. Rogers, G. R. Donowitz, G. K. Walker, S. A. Harding and M. A. Sande, Opportunistic Pneumonia, N. Engl. J. Med., 1979, 301(18), 959–961 CrossRef CAS.
- M. F. Parry, Waterborne Legionella bozemanii and Nosocomial Pneumonia in Immunosuppressed Patients, Ann. Intern. Med., 1985, 103(2), 205 CrossRef CAS.
- D. I. Grove, P. J. Lawson, J. S. Burgess, J. L. Moran, M. S. O'Fathartaigh and W. E. Winslow, An outbreak of Legionella longbeachae infection in an intensive care unit?, J. Hosp. Infect., 2002, 52(4), 250–258 CrossRef CAS.
- M. R. Amodeo, D. R. Murdoch and A. D. Pithie, Legionnaires' disease caused by Legionella longbeachae and Legionella pneumophila: comparison of clinical features, host-related risk factors, and outcomes, Clin. Microbiol. Infect., 2010, 16(9), 1405–1407 CrossRef CAS.
- T. W. Steele, C. V. Moore and N. Sangster, Distribution of Legionella longbeachae serogroup 1 and other legionellae in potting soils in Australia, Appl. Environ. Microbiol., 1990, 56(10), 2984–2988 CrossRef CAS.
- L. E. Garrison, Vital Signs: Deficiencies in Environmental Control Identified in Outbreaks of Legionnaires' Disease — North America, 2000–2014, Morb. Mortal. Wkly. Rep., 2016, 65(22), 576–584 CrossRef.
- E. D. Hilborn, T. J. Wade, L. Hicks, J. Garrison, E. Carpenter and E. Adam, Surveillance for waterborne disease outbreaks associated with drinking water and other nonrecreational water—United States, 2009-2010, Morb. Mortal. Wkly. Rep., 2016, 65(35), 714–720 Search PubMed.
- J. B. Rose, N. J. Ashbolt, R. L. Berkelman, B. J. Gutelius, C. N. Haas, M. W. Le Chevallier, J. T. Letson, S. A. Pergam, M. Prevost, A. Pruden, M. S. Swanson, P. W. J. J. van der Wielen and L. C. N. Weekes, Management of Legionella in Water Systems, National Academies of Sciences E, 2019 Search PubMed.
- B. A. Wullings and D. van der Kooij, Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15 degrees C, Appl. Environ. Microbiol., 2006, 72(1), 157–166 CrossRef CAS.
- B. A. Wullings, G. Bakker and D. van der Kooij, Concentration and Diversity of Uncultured Legionella spp. in Two Unchlorinated Drinking Water Supplies with Different Concentrations of Natural Organic Matter, Appl. Environ. Microbiol., 2011, 77(2), 634–641 CrossRef CAS.
- G. Fleres, N. Couto, M. Lokate, L. W. M. van der Sluis, C. Ginevra and S. Jarraud, Detection of Legionella anisa in Water from Hospital Dental Chair Units and Molecular Characterization by Whole-Genome Sequencing, Microorganisms, 2018, 6(3), 71–85 CrossRef CAS.
- L. Fiume, M. A. B. Sabattini and G. Poda, Detection of Legionella pneumophila in water samples by species-specific real-time and nested PCR assays, Lett. Appl. Microbiol., 2005, 41(6), 470–475 CrossRef CAS.
- R. Lesnik, I. Brettar and M. G. Höfle, Legionella species diversity and dynamics from surface reservoir to tap water: from cold adaptation to thermophily, ISME J., 2016, 10(5), 1064–1080 CrossRef CAS.
- L. S. Clescerl, A. E. Greenberg and A. D. Eaton, Standard Methods for the examination of water and wastewater, United book press, 20th edn, 1998 Search PubMed.
- Chlorine Residual Testing | The Safe Water System | CDC, 2020, [cited 2020 Dec 1]. Available from: https://www.cdc.gov/safewater/chlorine-residual-testing.html.
- P. Ji, W. J. Rhoads, M. A. Edwards and A. Pruden, Impact of water heater temperature setting and water use frequency on the building plumbing microbiome, ISME J., 2017, 11(6), 1318–1330 CrossRef.
- D. O. Schwake, E. Garner, O. R. Strom, A. Pruden and M. A. Edwards, Legionella DNA Markers in Tap Water Coincident with a Spike in Legionnaires' Disease in Flint, MI, Environ. Sci. Technol. Lett., 2016, 3(9), 311–315 CrossRef.
- US EPA O, National Primary Drinking Water Regulations, US EPA, 2015, [cited 2020 Dec 1], Available from: https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations.
- Preventing occupational exposure to Legionella, U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institute for Occupational Safety and Health, 2019 Sep [cited 2020 Dec 8], Available from: https://www.cdc.gov/niosh/docs/wp-solutions/2019-131/.
- K. A. Hamilton, M. T. Hamilton, W. Johnson, P. Jjemba, Z. Bukhari and M. LeChevallier, et al., Risk-Based Critical Concentrations of Legionella pneumophila for Indoor Residential Water Uses, Environ. Sci. Technol., 2019, 53(8), 4528–4541 CrossRef CAS.
- G. Yang, R. Benson, T. Pelish, E. Brown, J. M. Winchell and B. Fields, Dual detection of Legionella pneumophila and Legionella species by real-time PCR targeting the 23S-5S rRNA gene spacer region, Clin. Microbiol. Infect., 2010, 16(3), 255–261 CrossRef CAS.
- J. Lu, I. Struewing, E. Vereen, A. E. Kirby, K. Levy and C. Moe, et al., Molecular Detection of Legionella spp. and their associations with Mycobacterium spp., Pseudomonas aeruginosa and amoeba hosts in a drinking water distribution system, J. Appl. Microbiol., 2016, 120(2), 509–521 CrossRef CAS.
- E. Bédard, I. Boppe, S. Kouamé, P. Martin, L. Pinsonneault and L. Valiquette, et al., Combination of Heat Shock and Enhanced Thermal Regime to Control the Growth of a Persistent Legionella pneumophila Strain, Pathogens, 2016, 5(2), 35–50 CrossRef.
- E. Bédard, K. Paranjape, C. Lalancette, M. Villion, C. Quach and C. Laferrière, et al., Legionella pneumophila levels and sequence-type distribution in hospital hot water samples from faucets to connecting pipes, Water Res., 2019, 156, 277–286 CrossRef.
- H. Wang, S. Masters, M. A. Edwards, J. O. Falkinham and A. Pruden, Effect of Disinfectant, Water Age, and Pipe Materials on Bacterial and Eukaryotic Community Structure in Drinking Water Biofilm, Environ. Sci. Technol., 2014, 48(3), 1426–1435 CrossRef CAS.
- G. E. Bollin, J. F. Plouffe, M. F. Para and B. Hackman, Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets, Appl. Environ. Microbiol., 1985, 50(5), 1128–1131 CrossRef CAS.
- A. Peter and E. Routledge, Present-day monitoring underestimates the risk of exposure to pathogenic bacteria from cold water storage tanks, PLoS One, 2018, 13(4), e0195635 CrossRef.
- M. Totaro, P. Valentini, A. L. Costa, L. Frendo, A. Cappello and B. Casini, et al., Presence of Legionella spp. in Hot Water Networks of Different Italian Residential Buildings: A Three-Year Survey, Int. J. Environ. Res. Public Health, 2017, 14(11), 1296–1304 CrossRef.
- K. Toyosada, T. Otani, Y. Shimizu and S. Managi, Water Quality Study on the Hot and Cold Water Supply Systems at Vietnamese Hotels, Water, 2017, 9(4), 251 CrossRef.
- M. J. Donohue, S. Vesper, J. Mistry and J. M. Donohue, Impact of Chlorine and Chloramine on the Detection and Quantification of Legionella pneumophila and Mycobacterium Species, Appl. Environ. Microbiol., 2019, 85(24) CrossRef CAS Available from: https://aem.asm.org/content/85/24/e01942-19.
- S. Masters, J. Parks, A. Atassi and M. A. Edwards, Distribution system water age can create premise plumbing corrosion hotspots, Environ. Monit. Assess., 2015, 187(9), 559–576 CrossRef.
- M. Ambrose, S. M. Kralovic, G. A. Roselle, O. Kowalskyj, V. Rizzo, D. L. Wainwright and S. D. Gamage, Implementation of Legionella Prevention Policy in Health Care Facilities: The United States Veterans Health Administration Experience, J. Public Health Manag. Pract., 2020, 26(2), E1–E11 CrossRef.
- W. J. Rhoads, A. Pruden and A. M. Edwards, Survey of green building water systems reveals elevated water age and water quality concerns, Environ. Sci.: Water Res. Technol., 2016, 2(1), 164–173 RSC.
- Y. Sharaby, S. Rodríguez-Martínez, M. G. Höfle, I. Brettar and M. Halpern, Quantitative microbial risk assessment of Legionella pneumophila in a drinking water supply system in Israel, Sci. Total Environ., 2019, 671, 404–410 CrossRef CAS.
- C. R. Proctor, D. Dai, M. A. Edwards and A. Pruden, Interactive effects of temperature, organic carbon, and pipe material on microbiota composition and Legionella pneumophila in hot water plumbing systems, Microbiome, 2017, 5(1), 130 CrossRef.
- P.-M. Kao, B.-M. Hsu, T.-Y. Chang, T.-K. Hsu, K.-J. Tzeng and Y.-L. Huang, Seasonal variation of Legionella in Taiwan's reservoir and its relationships with environmental factors, Environ. Sci. Pollut. Res., 2015, 22(8), 6104–6111 CrossRef CAS.
- H. Whiley, A. Keegan, H. Fallowfield and R. Bentham, The presence of opportunistic pathogens, Legionella spp., L. pneumophila and Mycobacterium avium complex, in South Australian reuse water distribution pipelines, J. Water Health, 2015, 13(2), 553–561 CrossRef CAS.
- L. Liu, X. Xing, C. Hu and H. Wang, One-year survey of opportunistic premise plumbing pathogens and free-living amoebae in the tap-water of one northern city of China, J Environ Sci, 2019, 77, 20–31 CrossRef.
- S. M. Katz and J. M. Hammel, The effect of drying, heat, and pH on the survival of Legionella pneumophila, Ann. Clin. Lab. Sci., 1987, 17(3), 150–156 CAS.
- A. Ohno, N. Kato, K. Yamada and K. Yamaguchi, Factors influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water, Appl. Environ. Microbiol., 2003, 69(5), 2540–2547 CrossRef CAS.
- R. Amfo-Otu, J. B. Agyenim and G. B. Nimba-Bumah, Correlation Analysis of Groundwater Colouration from Mountainous Areas, Ghana, Environ. Res. Eng. Manag., 2014, 67(1), 16–24 Search PubMed.
- M. Rafiee, A. Mesdaghinia, H. Hajjaran, M. Hajaghazadeh, A. Miahipour and M. Jahangiri-Rad, The Efficacy of Residual Chlorine Content on the Control of Legionella Spp. In Hospital Water Systems, Iran. J. Public Health, 2014, 43(5), 637–644 Search PubMed.
- N. Mee-Marquet, A.-S. Domelier, L. Arnault, D. Bloc, P. Laudat and P. Hartemann, et al., Legionella anisa, a Possible Indicator of Water Contamination by Legionella pneumophila, J. Clin. Microbiol., 2006, 44(1), 56–59 CrossRef.
- R. R. Muder and L. Y. Victor, Infection Due to Legionella Species Other Than L. pneumophila, Clin. Infect. Dis., 2002, 35(8), 990–998 CrossRef.
- J. W. Mercante and J. M. Winchell, Current and Emerging Legionella Diagnostics for Laboratory and Outbreak Investigations, Clin. Microbiol. Rev., 2015, 28(1), 95–133 CrossRef CAS.
- H. Whiley and M. Taylor, Legionella detection by culture and qPCR: Comparing apples and oranges, Crit. Rev. Microbiol., 2016, 42(1), 65–74 CrossRef CAS.
- M. Baume, A. Cariou, A. Leveau, N. Fessy, F. Pastori and S. Jarraud, et al., Quantification of Legionella DNA certified reference material by digital droplet PCR, J. Microbiol. Methods, 2019, 157, 50–53 CrossRef CAS.
- E. J. Nazarian, D. J. Bopp, A. Saylors, R. J. Limberger and K. A. Musser, Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples, Diagn. Microbiol. Infect. Dis., 2008, 62(2), 125–132 CrossRef CAS.
- K. E. Cross, J. W. Mercante, A. J. Benitez, E. W. Brown, M. H. Diaz and J. M. Winchell, Simultaneous detection of Legionella species and L. anisa, L. bozemanii, L. longbeachae and L. micdadei using conserved primers and multiple probes in a multiplex real-time PCR assay, Diagn. Microbiol. Infect. Dis., 2016, 85(3), 295–301 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ew00893a |
| This journal is © The Royal Society of Chemistry 2021 |