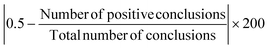Towards resolution of antibacterial mechanisms in metal and metal oxide nanomaterials: a meta-analysis of the influence of study design on mechanistic conclusions†
Eva
Albalghiti
 a,
Lisa M.
Stabryla
a,
Lisa M.
Stabryla
 b,
Leanne M.
Gilbertson
b,
Leanne M.
Gilbertson
 b and
Julie B.
Zimmerman
b and
Julie B.
Zimmerman
 *ac
*ac
aSchool of the Environment, Yale University, New Haven, Connecticut 06520, USA. E-mail: Julie.zimmerman@yale.edu; Tel: +1 203 432 9702
bDepartment of Civil and Environmental Engineering, University of Pittsburgh, Pittsburgh, Pennsylvania 15261, USA
cDepartment of Chemical and Environmental Engineering, Yale University, New Haven, Connecticut 06520, USA
First published on 24th November 2020
Abstract
While the antibacterial potency of metal and metal oxide engineered nanomaterials (MMO ENMs) has been well-established in the literature, the underlying mechanisms of antibacterial activity are regarded by many as uncertain, despite a considerable volume of publications on this subject. In order to illuminate sources of perceived uncertainty and disagreement, 318 articles pertaining to the mechanism of antimicrobial activity of Ag, Cu, CuO, TiO2 and ZnO ENMs were analyzed. The 318 studies all aimed to assess one or more of eight mechanistic questions, and both positive (i.e. affirmative) and negative conclusions were reported for each mechanistic question for each of the five core compositions. Differences in study design, including the exposure conditions and experimental methods used, were found to statistically significantly correlate with differences in reported mechanistic conclusions. Further analysis of studies which investigated two or more mechanisms revealed how assumptions about which mechanisms predominate for a given core composition may influence study design and, in turn, conclusions. Finally, 181 distinct experimental methods were identified, many of which are relatively untested and have not been evaluated in the published literature, while many frequently-used methods were found to have limitations that may obscure interpretation and mechanistic insight.
Environmental significanceThe antibacterial activity of metal and metal oxide nanomaterials is of great environmental relevance; while these materials have been touted as promising candidates to alleviate the mounting crisis of antibiotic resistance, concerns have also been raised that accidental release may have a detrimental impact on environmental systems. Designing nanomaterials to maximize desired functionality while minimizing undesired environmental consequences requires thorough understanding of their mechanisms of antibacterial activity. Despite widespread pursuit of mechanistic understanding through research efforts spanning more than a decade, there remains a significant amount of controversy and purported uncertainty in the published literature. This review elucidates the underlying sources contributing to these disagreements including experimental conditions to assess the mechanistic basis for metal and metal oxide engineered nanomaterial antibacterial activity. |
1. Introduction
Over the past two decades, the antibacterial properties of metal and metal oxide engineered nanomaterials (MMO ENMs) have garnered significant interest.1–3 While these materials may be promising alternatives to conventional antibiotics in some applications, with the potential to alleviate the antibiotic resistance crisis,4–7 there remain concerns of possible unintended consequences upon environmental release, including their potential adverse impacts on essential processes mediated by bacteria, such as nitrogen cycling8,9 and wastewater treatment.10,11 Many researchers, therefore, aim to design and select MMO ENMs with desirable functionality and minimal environmental hazard, a goal which requires an understanding of not only the magnitude of ENM antibacterial activity but also the underlying mechanisms.12–14 However, the significant complexities inherent to the study of MMO ENM antibacterial mechanisms may inhibit mechanism-informed design. Specifically, the selection and interpretation of experimental methods presents many opportunities for ambiguous, confounding, and even contradictory results in identifying likely mechanisms, even for a given core composition.The possible antibacterial mechanisms in various MMO ENM core chemistries were explored in a 2016 book chapter.15 Briefly, existing mechanistic literature encompasses investigations into four main areas: 1. the physicochemical processes determining ENM exposure and bioavailability, 2. the role of intact ENMs versus dissolved ions and/or reactive oxygen species (ROS), 3. the transport mechanisms and outcomes (i.e. where particles localize and/or accumulate) of ENMs and dissolved species within the cell, and 4. the physical and chemical effects on cellular components including membranes, DNA, enzymes, and others. Given the vast range of points of inquiry along a mechanistic pathway from initiation to outcome, the very framing and articulation of the research question(s) can, increasingly, be a source of complexity within the mechanistic literature.
As mechanistic knowledge has accumulated across different core chemistries, a divergence in research aims has emerged. Researchers interested in mechanism as it pertains to nano-design for specific applications often ask whether or not a given process, transport outcome, or cell effect contributes significantly to antibacterial activity for a given core composition.16–32 In contrast, biologists, nanotoxicologists, and ecosystem scientists often ask to what extent and in what conditions a given mechanistic pathway contributes to a toxicity outcome,33–38 inquiries with nuances that are not as readily captured within such a binary framework. These different framings of mechanistic questions may create apparent uncertainty in what is “known” and “unknown” about antibacterial mechanisms for a given core chemistry; while some studies report that long-established mechanistic questions have yet to be resolved in even well-studied core chemistries39–44 and others report seemingly definitive but contradictory conclusions,45–51 new questions continue to be raised about the conditions under which various mechanistic pathways may predominate.52,53
A 2014 study examined over 600 published articles to understand the disparities between ENM concentrations used in laboratory-scale ecotoxicity assessments and modeled or measured concentrations of ENMs in the environment.53 While a main finding was the dearth of ecotoxicity studies conducted at environmentally-relevant (e.g., sufficiently low dose) ENM concentrations, the review also identified numerous experimental conditions that could potentially impact the conclusions drawn from experimental work. The impact of study conditions on reported conclusions was investigated in more detail in a 2016 follow-up report, which determined that exposure conditions such as aqueous chemistry, temperature, pH, and exposure duration play a large role in elucidating the mechanistic basis of antibacterial activity,52 in addition to the known effects of ENM size, shape, and surface coating.17,54–59 These variables, which are often underreported in mechanistic studies, introduce additional complexity to the interpretation of mechanistic conclusions reported in the literature, as well as the design of new mechanistic studies. In response, efforts by communities of experts have aimed to harmonize and standardize both exposure conditions60 and ENM characterization61 in ecotoxicological studies of ENMs.
In addition to diverging research aims and dissimilar exposure conditions between studies, a third source of complexity is the variety and lack of standardization of experimental methods used to evaluate antibacterial mechanisms. While some methods are specifically tailored to provide mechanistic insight (e.g. those targeting the gene transcription level52), others are adaptations of existing cellular or acellular methods which are not necessarily representative of real exposure conditions (e.g., using a growth inhibition or CFU count assay to compare ENMs to scaled ion controls,62 or assessing ion release from ENMs in a cell-free environment63–65).
Just as mechanistically-relevant research questions may differ by disciplinary orientation, preferred experimental methods for mechanistic investigation may also be related to familiarity and acceptance within ones' research community. The limitations of different methods,62,66–71 as well as the nuances of what information they can and cannot provide, may not be obvious to those who are expert in nano-related topics but may not have sufficient depth, through training or collaboration, in microbiology. Proposed frameworks for producing high-quality studies on the magnitude of antibacterial activity for ENMs72–77 may prove to be invaluable guides for refining and standardizing mechanism-targeted experimental methods, but further work remains to adapt these frameworks for the complexity and variety of mechanistic investigations. Similarly, recent studies and expert reviews have provided new insights into accurate interpretation and artifact avoidance in ENM toxicity studies across a range of organisms,69–71 which presents an opportunity to connect what is known about the limitations of experimental methods with mechanistic conclusions reported in the literature thus far.
The factors discussed above introduce complexity into mechanistic research at three stages: articulating areas of inquiry, obtaining relevant empirical results, and interpreting results to yield conclusions. Accordingly, even when two studies ask the same question about a given core chemistry, seemingly contradictory conclusions may be reported for different reasons. In some cases, differences in study conditions or ENM properties between two studies (which may or may not have been reported) cause a different mechanism to predominate; the two sets of conclusions taken together thus represent valuable information for the community's understanding of that core chemistry. In other cases, rather than a “true” difference in mechanism between the two studies, the difference arises because the method(s) used could not elucidate a conclusive answer for the research question being investigated, contributing to a perception of disagreement or ambiguity in the literature and serving to confound mechanistic resolution. Understanding the prevalence and influence of these two sources of disagreement would enhance both the interpretation of existing studies and the design of new ones. Such understanding requires, however, a relatively comprehensive list of the many mechanism-targeted experimental methods currently in use, as well as an analysis of how method choice affects reported mechanistic conclusions, as compared to the known effects of ENM properties and exposure conditions.
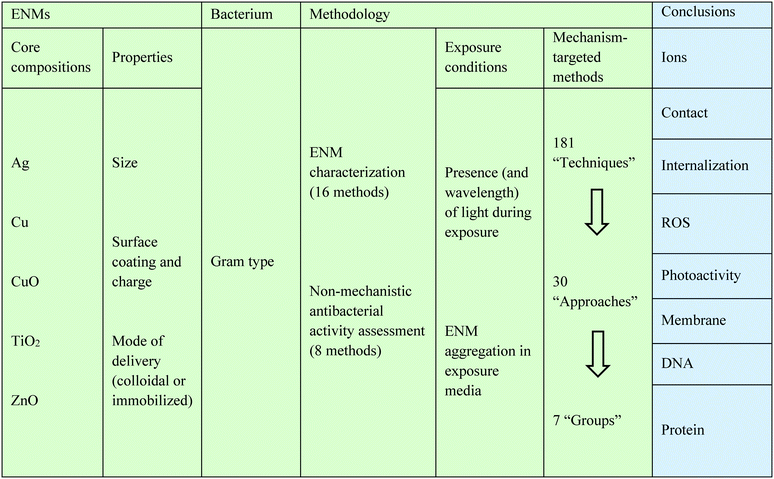
As such, this study applies statistical analyses to assess the potential influence of study design, including method choice, on reported mechanistic conclusions. To this end, a dataset of studies which reported conclusions on eight straightforward, frequently studied mechanistic questions (described in Table 1) is systematically retrieved. Five of the most heavily studied core compositions (Ag, Cu, CuO, TiO2, and ZnO) are selected to produce a robust dataset. The first phase of the analysis quantifies the frequency with which different exposure conditions were reported by studies in the dataset, as well as the impact of exposure conditions on reported mechanistic conclusions. Studies that reported conclusions to multiple mechanistic questions are also examined in order to capture the effect that presumed relationships between mechanistic investigations may have on the reported conclusions. Subsequently, the effects of experimental method choice on reported conclusions are quantified, and these findings are presented alongside a review of the advantages and limitations of methods currently documented in the literature. Finally, experimental methods are evaluated based on their tendency to produce consistent or inconsistent mechanistic conclusions for a given core composition, demonstrating how methods may differ in terms of their sensitivity, appropriateness, or both. Through these analyses, this study will facilitate method selection and interpretation of results, while also supporting the refinement and standardization of experimental methods aimed at elucidating antibacterial mechanisms for ENMs.
| Name | Question reviewed | Notes |
|---|---|---|
| Ion | “Are dissolved metal ions, released from particles into the exposure medium, necessary for antibacterial activity?” | • A related question not reviewed here is whether ions exert toxicity directly or indirectly through another antibacterial pathway (e.g., ROS production)9,78–81 |
| • Proposed mechanisms predicated on the local release of ions from an ENM in close proximity with the cell would instead constitute a positive conclusion to the “Contact” question described below; this is because the release of ions into the exposure medium alone may not be sufficient to cause antibacterial activity in these cases (i.e., a “nano-specific” effect can be said to exist) | ||
| Contact | “Is close association between the ENM and cell necessary for antibacterial activity?” | • Example mechanisms include mechanical damage to the cell envelope,82,83 internalization of particles,84–86 local ion release,87,88 and ROS production via direct electron transfer89 |
| Internalization | “Does internalization of intact ENMs across an intact cell membrane contribute significantly to antibacterial activity?” | • A related question not reviewed here is whether and under what conditions nano-sized particles can cross bacterial membranes without membrane permeabilization as a prerequisite.90,91 |
| • When intracellular objects are observed, it may be difficult to discern whether these truly represent intact ENMs internalized across the membrane, as some studies have reported the internalization of ions into the cytoplasm followed by oxidation to metal oxide19,92,93 or reduction to metal,47,94,95 giving the appearance of intracellular particles | ||
| ROS | “Does the production or accumulation of ROS, intracellularly or extracellularly, contribute significantly to antibacterial activity?” | • ROS may be generated directly from ENM surfaces, potentially through a light-mediated mechanism, and/or may accumulate naturally in cells as a result of other stresses exerted by ENMs.96 A related question, not reviewed here, is whether ROS generated endogenously is sufficient to cause cell death.96 |
| • If ROS accumulation is driven by endogenous production, ROS levels may continue to rise even after the stressor is removed, leading to delayed cell death97 that may not be captured depending on the timeframe of the study | ||
| • This question overlaps with the “Ion” and “Contact” questions as some proposed mechanisms involve the generation of ROS by dissolved ions9,78–81 or through direct electron transfer between ENMs and cells89 | ||
| Photoactivity | “Is the presence of light, of any wavelength, necessary for antibacterial activity?” | • While many studies treat “light-mediated mechanisms” as interchangeable with “photoactivated production of ROS,” ion-driven mechanisms involving photo-dissolution processes have also been proposed.98–100 |
| • Since photoactivated ROS production can occur at both visible and UV wavelengths, light-mediated mechanisms may be overlooked when ambient lighting is not considered.101 | ||
| Membrane | “Does permeabilization of the cell membrane contribute significantly to antibacterial activity?” | • Although membrane permeability has long been used as an indicator for cell death,102 membrane permeability may occur without antibacterial activity, and vice versa.103,104 |
| • Related questions not reviewed here include whether membrane permeability contributes to cell death per se105 or simply increases cell vulnerability to other injuries (e.g. internalization of toxic components and/or ROS accumulation),91,106 as well as the different responses of the outer membrane (OM) and inner membrane (IM) in Gram negative bacteria.107,108 | ||
| • This question overlaps with the “Contact” and “ROS” questions when the proposed pathway involves mechanical abrasion82,83 or lipid peroxidation,20,47 respectively | ||
| DNA | “Does damage to bacterial DNA contribute significantly to antibacterial activity?” | • Related questions not reviewed here include whether DNA damage occurs via a primary or secondary pathway (i.e., from interaction of ENMs themselves with DNA or from ion- or ROS-mediated damage)109 as well as which types of DNA damage contribute most significantly to antibacterial activity.96 |
| • This question overlaps with the “Protein” question, since DNA repair enzyme abundance and activity were found to be key determinants of antibacterial activity for some antibiotics,110,111 and ENMs may bind and deactivate cellular proteins, including enzymes,112,113 as discussed below | ||
| Protein | “Does the binding or inactivation of intra- or extracellular proteins by ENMs or their dissolved ions contribute significantly to antibacterial activity?” | • Many bacterial proteins have been shown to bind specifically to ENMs of one or more compositions,113,114 and the dissolved ions of both silver and copper have been shown to target thiol groups present in amino acids.88,115,116 |
| • Enzymes are of particular interest due to their importance in bacterial metabolic processes117 and may be inactivated by ENMs in several ways including direct blockage of the active site113 or alteration of enzyme structure.114 Additionally, the relationship between ENM concentration and enzyme activity may not be straightforward.114 | ||
| • Protein damage is often investigated alongside holistic metabolic effects using techniques such as transcriptome and proteome analysis,118 incorporating a variety of questions not captured within this binary framework |
2. Methods
2.1 Data retrieval
A literature search was conducted in the Web of Science database using search terms listed in Table S1,† and an initial screening of titles and abstracts was performed in order to identify experimental (rather than review) articles that pertained to antibacterial activity of the core compositions of interest (i.e., Ag, ZnO, TiO2,Cu, CuO). Articles were separated by core composition because this study aims to investigate reported conclusions within ENM “classes” as they are typically defined in the literature, and different core compositions are typically treated as different “classes” of ENMs for the purposes of mechanistic investigation. Studies on nanocomposites (i.e., materials in which the “nano” component of the system is deliberately engineered to be of heterogeneous core composition) could not be accommodated within this framework and were excluded. However, ENMs composed of a metal or metal oxide core shell with a capping agent or MMO ENMs embedded on or within a non-nanostructured surface or matrix were not excluded; this is because studies comparing capped and uncapped or colloidal and immobilized particles often do not treat these as separate classes of ENMs in discussions of antibacterial mechanism. For the same reason, metal ENMs which are known to form an outer layer of oxide were included.Since this analysis involves tabulating reported mechanistic conclusions and attributing differences in conclusions to ENM properties, exposure conditions, and experimental method choice, articles in the dataset must also meet three additional criteria: 1) one or more mechanistic conclusion(s) must be reported, 2) mechanistic conclusion(s) must be supported by at least one experiment, and 3) ENM properties must be documented. Therefore, only articles which specifically articulated a conclusion about the mechanism of antibacterial activity (as defined by the respective authors), supported this conclusion with at least one experimental method, and documented some type of ENM “characterization” (as defined by the respective authors) were included. No restrictions were imposed on definitions of “mechanism” and “characterization” because, in addition to affecting the ease of comparison across studies, it is possible that assumptions about which mechanistic pathways and ENM properties are relevant may influence or prematurely restrict the conclusions reached. Therefore, it was important to capture how discrepancies in the way these terms are defined by different authors may contribute to confusion or disagreement. As summarized in Table 2, the initial search returned more than 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 articles, but applying the screens described above yielded a final dataset of 318 articles.
000 articles, but applying the screens described above yielded a final dataset of 318 articles.
| Core composition | Total returned in search | Passed initial screening | Reached mechanism conclusion | Performed mechanism-targeted experiment | Included ENM characterization |
|---|---|---|---|---|---|
| Cu/CuO | 1623 | 96 | 72 | 66 | 60 |
| TiO2 | 2434 | 81 | 61 | 54 | 44 |
| Ag | 4780 | 359 | 205 | 178 | 160 |
| ZnO | 2211 | 191 | 133 | 119 | 99 |
| Total |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 048 048
|
727 | 471 | 417 | 363 |
2.2 Correlating study design variables with mechanistic conclusions
The data collected from each article were represented as binary “conclusions” and “study design variables” (Table 3). For each mechanism question (Table 1) investigated in each study, a “positive” conclusion was recorded if the authors of the study reported an affirmative answer to the question and a “negative” conclusion was recorded if the authors of the study reported a negative answer. In order to accurately represent authors' interpretations of their results, no re-interpretation of experimental findings was performed. For example, if reported data appeared to support enhanced antibacterial activity in the presence of light, but the authors of the study did not explicitly discuss the mechanistic implications of this finding, no conclusion was recorded for “photoactivity.”The study design variables analyzed here are listed in full in Table S2.† Examples of study design variables include “ENM had at least one dimension smaller than 10 nm as measured by TEM” (part of the “ENMs… Properties… Size” set of variables), “Used a Gram-negative bacterium” (part of the “Bacterium… Gram type” set), and “Performed mechanism-targeted experiments under ambient (visible-spectrum) light” (part of the “Methodology… Exposure Conditions… Presence (and wavelength) of light” set). Within the “Mechanism-targeted experimental methods” category of study design variables, the 181 distinct methods identified (referred to as “Techniques” hereafter) were further sorted into 30 “Approaches” and seven “Groups.” This categorization aims to lend coherence to the long list of techniques by highlighting key similarities and differences. Different techniques within one approach utilize different instruments or reagents but purport to measure similar endpoints; different approaches within the same group aim to measure different endpoints, but researchers apply similar rationales to interpret results and draw mechanistic conclusions. Aggregation of individual techniques into approaches and groups also provided larger sample sizes for comparing methods via statistical tests, which was desirable because the majority of techniques were used in a small number of (i.e. fewer than five) studies.
For statistical analyses, a binomial distribution of conclusions was constructed for each of the eight mechanism questions (Table 1) within each core composition. The influence of each study design variable on reported conclusions was then assessed as follows. First, the distribution of conclusions reported in studies where a given study design variable was true was compared to the distribution of conclusions reported in studies where a given study design variable was false. If the difference between these distributions was found by Fisher's exact test to have a less than five percent probability of occurring by coincidence (i.e. p < 0.05), the study design variable was determined to affect conclusions to a statistically significant degree. Since core composition is expected to influence mechanism, this analysis was performed within core compositions only. To complement statistical analyses, a review of the available literature pertaining to the advantages and limitations of the reviewed techniques, approaches, and groups of methods was conducted.
Further analysis was directed at studies which investigated multiple mechanisms. In order to reveal correlations between reported conclusions for different mechanism questions as they exist in the literature, the “null hypothesis” adopted for this analysis was that the eight mechanism questions are independent of one another. When this “null hypothesis” is shown to be untrue, it may be because a relationship between mechanisms actually exists (e.g., an ENM internalization-driven mechanism is necessarily also a contact-mediated mechanism) or because a relationship is only assumed to exist (e.g. ion- and contact-mediated mechanisms are sometimes believed to be mutually exclusive). Metrics used include 1) the percent of studies which investigated two mechanisms simultaneously (of studies which investigated one or the other), and 2) the percent of studies which reported positive conclusions for two mechanisms (of studies which reported positive conclusions for one or the other).
2.3 Assessing the consistency of mechanism-targeted experimental methods
Subsequent analyses aimed to quantify the tendency of experimental methods to yield similar or different conclusions for a given mechanism question, within a given core composition. Such differences are a potentially important consideration when evaluating experimental methods, since they may stem from different degrees of sensitivity and/or ease of interpretation between methods. A new metric, termed the “Consistency Score,” was defined as:Since the absolute value of the average of a binomial distribution captures the “spread” of the distribution (similar to a standard deviation), the consistency score provides a metric for the degree of consensus that exists within a set of studies without regard for whether that consensus favors positive or negative conclusions. This allows the consistency of conclusions to be quantified even across sets of studies which are expected to differ in terms of the “true” conclusion. The highest possible consistency score is 100, which corresponds to unanimous agreement within a set of studies, while a score of zero corresponds to equal numbers of positive and negative conclusions within a set of studies.
For each of the eight mechanism questions, consistency scores were calculated for subsets of studies which used each group, approach, and technique to evaluate the relevant question. This was first done within each core composition. Weighted averages were then calculated across all core compositions to yield an overall consistency score for each group, approach, and technique as applied to each mechanism. As described above, the weighted average could be used because the consistency score depends only on the degree of spread within the distribution of conclusions, not on whether the consensus favors positive or negative conclusions, rendering the differences in conclusions that would be expected to arise between different core compositions irrelevant. A bootstrapping technique, in which the original dataset was randomly sampled with replacement to generate 200 “re-samples” of the same size as the original dataset and a consistency score was calculated for each of these re-samples, was used to obtain distributions of consistency scores. The mean and standard error of the resultant distributions were then used to obtain 95% confidence intervals for the overall consistency score of each group, approach, and technique.
3. Results and discussion
3.1 Summary of reported mechanistic conclusions and relationship with study design
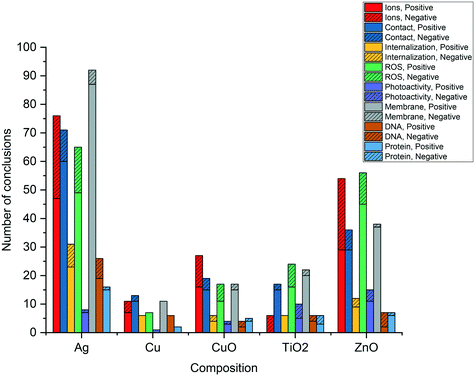
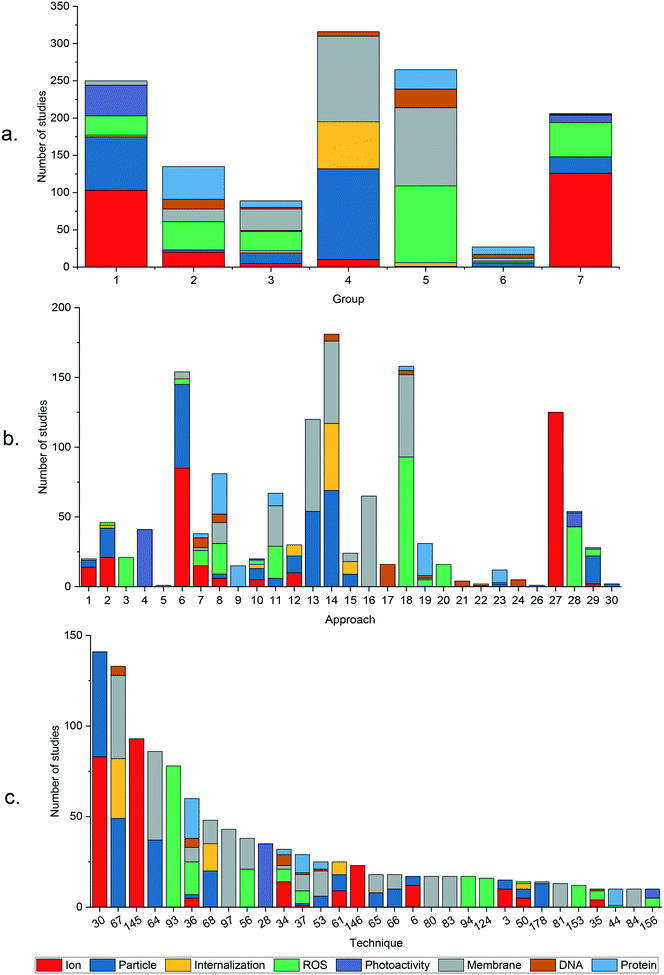
Fig. 1 summarizes the mechanistic conclusions reported by studies in the dataset. Within the body of literature analyzed, all eight mechanistic questions have yielded both positive and negative conclusions within all five core compositions. The number of studies for each core composition is unequally distributed, with nAg the subject of significantly more studies than other ENM core compositions. The prevalence of nAg studies is particularly clear for less-studied questions including internalization, DNA damage, and protein damage. The proportion of studies investigating each mechanistic question is roughly equal across core compositions, with the exception of photoactivity, where nTiO2 and nZnO account for the majority of studies.A majority of studies in the dataset investigated questions of ion-, contact-, ROS-, and membrane permeabilization-mediated toxicity, with each of these questions being the subject of more than 150 studies across the five core compositions. Questions of particle internalization, photoactivity, DNA damage, and protein damage have received less attention, with a total of between 30 and 60 studies each. This discrepancy could have several explanations. One reason may be that some questions are investigated less frequently because they are perceived to be “settled” already, that is, they are widely believed to either contribute or not contribute to the antibacterial activity of a given core composition. Alternatively, the complexity and expense of the experiments required to test for some mechanistic questions may cause these questions to be investigated less frequently.
The ratio of positive to negative conclusions for most mechanism questions across all core composition groups suggests considerable influence from ENM properties, exposure conditions, and/or experimental methods. Exceptions include membrane permeabilization and protein damage, for which there is near-unanimous agreement in conclusions within the more commonly studied core compositions, Ag and ZnO. For other questions, all cases in which conclusions appear to agree unanimously draw on fewer than 10 studies, indicating that the question may be settled or may not yet be sufficiently well-studied. Subsequent analysis aims to explore the causes of disagreement as they relate to study design variables outlined in Table 3.
Fig. 2 summarizes three important indicators of the relationship between study design and reported conclusions. First, the percent of conclusions for each question which were supported by experiment is given in the leftmost column, since studies may have performed an experiment for one question but based conclusions for other questions on assumptions and/or previous studies, which may have utilized dissimilar exposure conditions. Second, the percentage of studies in the dataset which reported any information for each category of study design variable is given, because not all studies specified all ENM properties or exposure conditions. Finally, for each of the eight mechanism questions, categories of study design variables containing at least one variable that generated statistically significant differences in conclusions (as defined in section 2.2, “Correlating Study Design Variables with Mechanistic Conclusions”) are highlighted.
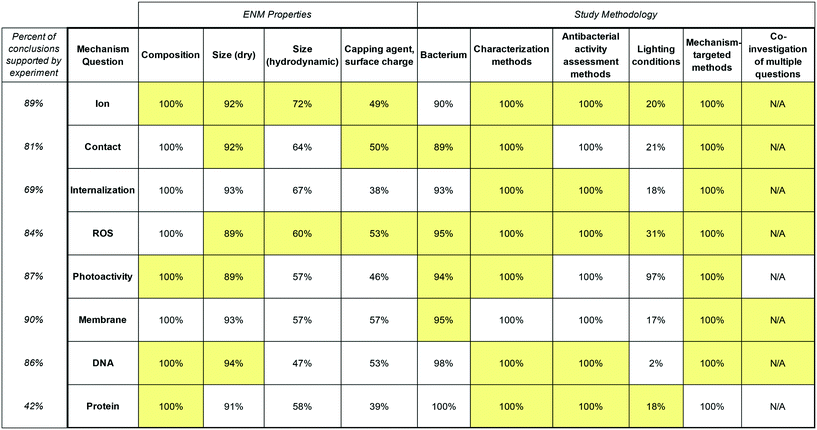 | ||
| Fig. 2 The leftmost column indicates the percent of reported conclusions for each mechanism question which were found to be supported by at least one experiment. The remaining columns show statistically significant differences in conclusions for each question based on variables related to ENM properties and study methodology, overlaid with the percent of studies that reported any information about each variable. Highlighted cells indicate that dividing the dataset based on the variable on the horizontal axis created a difference in conclusions about the mechanism question on the vertical axis that was large enough to be considered statistically significant (p < 0.05 according to Fisher's exact test). With the exception of core composition, all variables were examined within core composition groups; a highlighted cell therefore indicates that a statistically significant difference was identified within one or more compositions. No statistically significant differences in conclusions emerged for immobilized versus colloidal delivery methods, so this variable category is not displayed. In Fig. S1 and S2,† the analysis is disaggregated by core composition and whether the variable in question skewed conclusions towards positive or negative is indicated. | ||
Despite the criterion that included studies must include at least one experiment that targeted antibacterial mechanism, the number of studies that reported a conclusion for a given mechanism was generally found to be higher (usually by 10–20%, but more in some cases) than the number of studies which performed an experiment to test for that mechanism. This was found to result from the aforementioned practice of investigating some questions experimentally while drawing conclusions about others via “process of elimination” or a review of previous literature. These common practices may compound disagreement and perceived ambiguity in the literature by, for the former, prematurely assuming the mutual exclusivity of mechanisms and, for the latter, combining conclusions that were reached under different conditions. Conclusions reached through these means are therefore excluded from Fig. 1 and subsequent analyses.
Additionally, reporting of potentially relevant ENM properties and exposure conditions was found to be incomplete. Since comprehensive reporting of exposure conditions52 and characterization of ENMs73 have both been identified as important to the interpretation and comparison of results, this gap in reporting is a likely contributor to real or perceived mechanistic uncertainty. While the vast majority of studies specified ENM core composition and size as measured by TEM or equivalent method, as well as bacterium used, fewer studies characterized the aggregation state of ENMs in exposure media and the surface charge or presence of any capping agents. Furthermore, only a small minority of studies which did not specifically investigate light-mediated mechanisms indicated the lighting conditions used in toxicity or mechanistic experiments, suggesting that the importance of light in some mechanistic pathways may be underexplored.
Contrary to expectation, few statistically significant differences in conclusions were identified between core compositions, which may indicate that the five core compositions reviewed overlap more in antibacterial mechanism than their separate treatment in the literature may suggest. In contrast, other study design variables were found to yield many statistically significant differences in conclusions. Differences in conclusions which emerge based on ENM size, capping agent, bacterium, antibacterial activity assessment methods, and lighting conditions suggest that different mechanisms may predominate in different conditions (e.g., a separate mechanism actually exists for Gram positive versus Gram negative bacteria, or for small versus large ENMs, or for antibacterial activity in UV light versus in darkness). In contrast, differences in conclusions which emerge based on characterization methods suggest that researcher's subjective interpretations of results may influence conclusions (e.g., information gleaned from characterization may influence how the results of mechanism-targeted methods are interpreted by researchers to formulate conclusions).
Taken together, these results confirm that some perceived ambiguity surrounding mechanism is likely attributable to differences in study design that are not reported or not considered when researchers design experiments and interpret their results to yield conclusions. However, it is difficult to differentiate between cases of different conclusions representing real differences in mechanism (due to dissimilar ENM properties, model organisms, or exposure conditions) and cases of different conclusions resulting from choice of experimental methods and/or subsequent interpretation of results. Simple measures to promote clarity include more complete reporting of ENM properties and exposure conditions as previously suggested,52,73 as well as separating mechanistic conclusions reached through experiment from mechanistic conclusions inferred from previous literature.
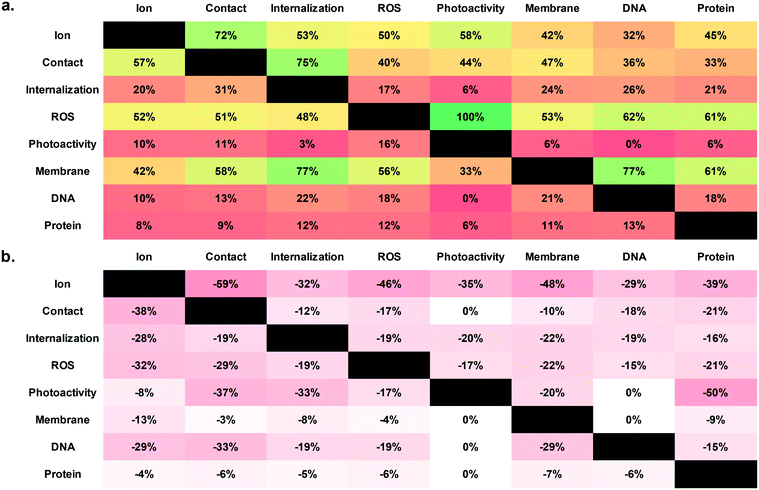
Among studies in the dataset, a tendency to frame possible mechanisms as mutually exclusive was noted. One common example is the perceived “ion” versus “particle” dichotomy also noted in previous reviews.62 In order to better understand whether and how this may affect conclusions, studies which investigated more than one mechanistic question within a given core composition were examined (Fig. 3). This investigation also sought to identify areas in which the relationships between mechanisms may warrant further exploration, a development which would complement moving away from “binary” definitions of questions to introduce greater nuance into mechanistic discussions.
The patterns that emerge highlight existing preconceptions of the relationships between mechanisms which may, in turn, shape study design. For example, ion- and particle-driven mechanisms are frequently investigated together, and photoactivity is always investigated alongside ROS, which aligns with the general consensus that these two pairs of mechanisms are related. On the other hand, mechanisms related to specific cellular targets (i.e., membrane damage, DNA damage, and protein damage) are usually not investigated alongside ion- or particle-driven antibacterial activity. These relationships could be explored more in future work in order to relate the “origins” of ENM antibacterial activity to specific cellular effects. Additionally, future membrane studies can focus on protein damage in order to obtain more detailed information about the nature and origins of membrane permeabilization, as well as the relationship between membrane effects and metabolic effects. Further, the link between ions and photoactivity remains underexplored, and may yield insights about proposed “photo-dissolution” processes in some core compositions.98–100 Finally, while studies on internalization often feature investigations of particle-driven antibacterial activity, other mechanisms can be explored more extensively in combination with internalization. For example, studies on DNA and protein damage can provide insights to the intracellular targets of ENMs following internalization.
Interestingly, it appears that studies which reached a positive conclusion for one mechanism were, to varying degrees, more likely to draw negative conclusions for any other mechanism. This is especially true for ion- and particle-mediated antibacterial activity. While one possible explanation is that this trend is reflective of reality (i.e. that one mechanism actually does dominate at a given time), a more plausible explanation is that the trend is not reflective of reality (i.e. that belief in a single, dominant mechanism continues to guide many investigations). In this case, reaching a positive conclusion about one mechanism would introduce bias that would lead to other mechanisms being prematurely ruled out. While it was not possible to test these hypotheses with greater statistical rigor using this dataset, the questions raised merit further investigation, which would be supported by more studies examining multiple mechanistic questions.
3.2 Summary and assessment of mechanism-targeted methods
The remainder of the analysis aims to further illuminate the role that method choice may play in sowing perceived uncertainty. 181 distinct experimental “Techniques” of mechanistic investigation, further categorized into 30 “Approaches” and seven “Groups,” are summarized in Table 4. The “Uses” column lists the full set of mechanistic questions to which each technique was applied in studies in the dataset, which demonstrates how a single method is often applied to multiple questions. While some methods aim to provide binary information about the presence or absence of a given mechanism, others are tailored towards more detailed insight. For example, the propidium iodide (PI) stain (group 5, approach 18, technique 97) is commonly used to establish loss of membrane integrity, but Fourier-transform infrared spectroscopy (FTIR) of the cell mass (group 3, approach 11, technique 53) and scanning electron microscopy (SEM) (group 4, approach 13, technique 64) can be used to understand chemical changes in the cell membrane and the extent and character of membrane permeabilization, respectively. All methods are expected to have advantages and limitations, although these may not be documented in the literature; those which were found to be documented in the literature are summarized in Table S5.†| Group | Approach | Technique | Endpoint | Type of assay | Uses | Ref. |
|---|---|---|---|---|---|---|
| 1 “Elimination” methods: modifications of antibacterial activity or other assays designed to isolate the contribution from a single mechanism (i.e., by blocking the action of that mechanism or alternative mechanisms) | 1 Isolate dissolved ion effect | 2 Insoluble salt-forming agent addition, including orthophosphate, sulfide, sodium thiosulfate, sodium chloride | Difference in cell death relative to control | (Dependent on antibacterial activity assay) | Ion, contact | [119–122] |
| 3 Chelator addition, including NAC, EDTA, bathocuproine, neocuproine | Ion, contact | [45, 78, 119, 123–129] | ||||
| 2 Isolate particle effect | 5 Compare colloidal to immobilized particles | Ion, contact, internalization | [122, 130, 131] | |||
| 6 Compare to ENM-free filtrate | Ion, contact | [18, 20, 51, 54, 120, 132–138] | ||||
| 7 Membrane barrier | Ion, contact | [87, 139–141] | ||||
| 8 Induce aggregation, including extracellular polymeric substance (EPS) addition | Ion, contact | [49, 51, 141–143] | ||||
| 9 Compare to inert ENMs of same size and morphology | Contact | [81, 88, 144–146] | ||||
| 10 Compare antibacterial activity in aerobic and anaerobic environment | Ion, contact, ROS | [28, 119, 147] | ||||
| 3 Isolate ROS effect | 11 Cysteine (e.g. NAC) addition | ROS | [20, 28, 81, 135, 148–151] | |||
| 12 Ascorbic acid addition | ROS | [149, 152, 153] | ||||
| 16 GSH addition | ROS | [121, 139, 152] | ||||
| 17 SOD addition | ROS | [126, 135, 151, 154] | ||||
| 18 CAT addition | ROS | [126, 151, 155, 156] | ||||
| 4 Isolate light exposure effect | 27 NP pre-irradiation | Photoactivity | [9, 99, 157–159] | |||
| 28 Compare antibacterial activity in light vs. dark conditions | Photoactivity | [85, 129, 154, 160–183] | ||||
| 6 Compare to positive control | 30 Soluble salt (e.g. AgNO3, CuSO4, ZnCl2) | Ion, contact | [1, 2, 23, 25, 45, 50, 55, 56, 64, 78, 81, 83, 87, 88, 99, 112, 123, 125, 129, 132, 139–142, 145, 158, 163, 165, 174, 180, 184–219] | |||
| 32 H2O2 | ROS | [20, 129, 141, 216] | ||||
| 33 Detergent or bacteriolytic agent | Membrane | [17, 191, 220, 221] | ||||
| 2 “Genomic” methods: using information about bacterial genomes to infer the mechanism of antibacterial activity based on cellular responses to ENMs | 7 Cellular methods: bioreporters and knockout strains | 34 Recombinant bioluminescent reporter strain | Various (incl. intracellular ROS species, bioavailable metal, DNA damage, membrane damage) | Chemiluminometric | Ion, ROS, membrane, DNA, protein | [1, 55, 56, 64, 78, 126, 138, 148, 175, 184, 215, 222–225] |
| 35 Single-gene deletion (“knockout”) strain | Difference in antibacterial activity | (Dependent on antibacterial activity assay) | Ion, ROS, DNA | [55, 56, 64, 225, 226] | ||
| 8 High-throughput methods: transcriptome and proteome analysis | 36 Transcriptome analysis | Quantity of RNA produced relative to control (indicator for gene up- and down-regulation) | Various, usually microarray or high-throughput sequencing | Ion, contact, ROS, membrane, DNA, protein | [8, 20, 88, 137, 148, 175, 178, 192, 209, 214, 226–238] | |
| 37 Proteome analysis | Quantity of protein produced relative to control (indicator for gene up- and down-regulation) | Various, usually mass spectrometry | Ion, contact, ROS, membrane, DNA, protein | [127, 152, 174, 191, 194, 195, 219, 232, 234, 235, 239–242] | ||
| 9 Enzyme activity | 39 Ammonium molybdate assay | CAT activity | Colorimetric | ROS, protein | [45, 127, 135, 152, 221, 243] | |
| 43 DTNB assay | GR activity | ROS, protein | [129, 135, 244] | |||
| 44 NADH assay (with INT or resorufin) | Dehydrogenase, including LDH, activity | Colorimetric or fluorometric | Protein, ROS | [32, 127, 198, 208, 217, 239, 243–246] | ||
| 3 “Chemical analysis” methods: using information about the chemical and electronic state of metals or biomolecules following ENM exposure to bacteria in order to pinpoint the origin of antibacterial activity and relevant chemical processes | 10 Chemical analysis of metals | 50 XAS analysis of metal in cell mass or supernatant | Local geometric and electronic structure of metal atoms | XAS | Ion, contact, internalization, ROS | [19, 92, 93, 247, 248] |
| 11 Chemical analysis of cellular components | 53 FTIR of cellular fraction or extracellular polymeric substances (EPS) | Chemical changes in cellular components | FTIR | Contact, membrane, protein, DNA | [26, 85, 93, 155, 174, 185, 206, 249–256] | |
| 54 Raman spectroscopy of cellular fraction | Raman spectroscopy | Protein, DNA | [85, 136, 257] | |||
| 56 TBA/MDA assay | Degree of lipid peroxidation | Colorimetric or fluorometric | ROS, membrane | [20, 47, 121, 135, 151, 161, 162, 169, 184, 202, 243, 254, 258–267] | ||
| 4 “ENM localization or visualization” methods: probing the localization of ENMs and/or ions and cell morphology changes after cell exposure to ENMs | 12 Metal partitioning study | 61 Metal concentration in cellular fraction, including sucrose gradient centrifugation assay | Quantity of metal associated with cells | ICP-MS | Ion, contact, internalization | [11, 24, 50, 112, 219, 238, 247, 268, 269] |
| 13 High-resolution imaging of cell surfaces | 64 SEM | Qualitative attributes of cell/particle interaction | Image of cell exterior | Contact, membrane | [26, 27, 32, 84, 121, 129, 136, 141, 143, 150, 151, 155, 166, 173, 198, 204, 238, 262, 264, 265, 270–296] | |
| 65 SEM with elemental mapping (EDX or synchrotron XFM) | Image of cell exterior with elemental mapping | Contact, membrane | [16, 217, 218, 253, 297–300] | |||
| 66 AFM | Image of cell surface | Contact, membrane | [27, 55, 56, 125, 131, 224, 252, 255, 301–303] | |||
| 14 High-resolution imaging of cell interiors | 67 TEM | Image of cell interior | Membrane, contact, internalization, DNA | [17, 32, 47, 55, 56, 82, 86, 92, 128, 129, 141, 152, 165, 167, 171, 185, 204, 207, 210, 229, 237–239, 249, 254, 265, 288, 304–318] | ||
| 68 TEM with elemental mapping (EDX or synchrotron XFM) | Image of cell interior with elemental mapping | Contact, internalization, membrane | [29, 31, 50, 54, 126, 184, 193, 195, 237, 250, 253, 260, 297–299, 319–321] | |||
| 15 Light microscopy and light scattering techniques | 69 CLSM | Qualitative attributes of cell/ENM interaction (for fluorescently labeled or intrinsically fluorescent ENMs) | Fluorescence microscopy | Contact, internalization, membrane, DNA | [183, 309, 322–325] | |
| 72 Dark-field microscopy (may be equipped with HSI) | Relative strength of interactions between ENMs and cell surfaces | Light microscopy | Contact, membrane | [50, 192, 269, 326] | ||
| 74 ENM tracking with intrinsic fluorescence or fluorescent label (e.g. rhodamine B) | Localization of NPs within cells | Fluorescence microscopy | Contact, internalization | [196, 242, 281] | ||
| 75 Light scattering method for particle internalization | Ratio of forward- to side-scattered light (varies with cell granularity) | Light microscopy | Internalization | [86, 129, 276, 327] | ||
| 5 “Cell effects” methods: assays which gauge the degree of disruption caused to various cellular systems, thereby elucidating the specific effects that ENMs have on the cell | 16 Membrane permeability via release of cellular components into extracellular space | 76 ONPG hydrolysis assay | GAL leakage | Colorimetric | Membrane | [153, 242, 251, 265, 271, 283, 328] |
| 77 K+ and/or Mg2+ leakage | K+ and/or Mg+ in supernatant | AAS/AES or selective electrode | Membrane | [129, 139, 197, 213, 232, 249, 265, 295, 316] | ||
| 80 Nucleic acid leakage | Nucleic acids in supernatant | UV-vis | Membrane | [16, 112, 124, 151, 153, 221, 242, 251, 270, 280, 287, 311, 323, 329–331] | ||
| 81 Sodium pyruvate assay | LDH in supernatant | Membrane | [11, 23, 49, 195, 253, 256, 260, 324, 332, 333] | |||
| 82 Lowry method | Protein in supernatant | Colorimetric | Membrane | [153, 245, 276, 325, 328] | ||
| 83 Bradford method | Membrane | [32, 155, 208, 221, 243, 246, 251, 290, 294, 295, 300, 312, 330, 332, 334–336] | ||||
| 84 Miller method | Reducing sugar in supernatant | Membrane | [208, 217, 243, 245, 246, 312, 330, 334–336] | |||
| 85 DNA release, including diphenylamine and PicoGreen assays | DNA in supernatant | Fluorometric | Membrane | [112, 124, 143, 293, 326] | ||
| 17 Genomic DNA extraction and analysis | 89 DNA ladder assay | Degree of gDNA fragmentation | Gel electrophoresis | DNA, protein | [47, 92, 153, 198, 258–261, 291, 295, 333, 337, 338] | |
| 18 Intracellular probes for stress markers | 93 DCFH-DA (including variants such as CM-H2DCFDA, ab113851-DCFDA, H2DCFDA, DCF-DA) | Intracellular ROS | Fluorometric | ROS | [8, 9, 16–18, 20, 21, 23, 24, 28, 50, 84, 86, 121, 135, 143, 150–152, 166, 167, 170–172, 178–180, 184, 188, 211, 219, 236, 238, 243, 245, 253–256, 258, 260–264, 267–272, 276, 281, 282, 287, 288, 292, 310, 320, 324, 325, 327, 333, 339–343] | |
| 94 NBT assay | Usually superoxide anion concentration; inhibition of stain is also used as a measure for SOD activity | Colorimetric | ROS | [16, 99, 112, 127, 135, 152, 154, 253, 256, 259, 324, 330, 340, 344] | ||
| 95 Rhodamine dyes, including DHR6G and dihydrorhodamine 123 | Intracellular ROS | Fluorometric | ROS | [26, 262, 345, 346] | ||
| 97 Propidium iodide (PI) stain | Membrane permeability (enters cells with compromised membranes) | Membrane | [16, 18, 19, 23, 24, 50, 84–86, 92, 93, 124, 128, 129, 132, 143, 151, 171, 180, 199, 204, 205, 217, 226, 238, 250, 253, 255, 264, 272, 274, 280, 288, 292, 331, 337, 341, 342, 347, 348] | |||
| 99 DiBAC4 stain | Membrane potential | Membrane | [205, 217, 236] | |||
| 100 DiSC3(5) assay for membrane potential | Membrane | [47, 191, 276, 284, 326] | ||||
| 101 1-NPN | Outer membrane permeability | Membrane | [124, 197, 242, 264, 270, 287] | |||
| 102 DPH membrane fluidity assay | Membrane fluidity | Membrane | [17, 276, 295, 341] | |||
| 104 TUNEL assay | Degree of apoptosis | DNA | [236, 276, 292] | |||
| 107 Annexin V | Protein | [152, 236, 349] | ||||
| 109 RedoxSensor assay | Reductase activity | ROS | [19, 92, 93] | |||
| 19 Other assays targeting general cell respiration or disturbance to cellular processes | 110 DTNB assay for cellular GSH | Quantity of disulfide-containing molecules | Colorimetric | ROS, protein | [129, 243, 255, 260, 262] | |
| 115 Luciferin/trichloroacetic (TCA) assay for ATP content | Cell respiration | Chemiluminometric | Protein | [47, 51, 82, 151, 184, 191, 217, 243, 295] | ||
| 118 Intracellular K+ or Ca2+ | Cell respiration (as a reflection of membrane damage and metabolic disruption) | Flame AES | Protein | [47, 191, 236] | ||
| 119 EPS quantification | EPS production (as a measure of cell viability or metabolic activity) | Colorimetric | Protein | [11, 16, 93] | ||
| 121 Phag-GFP expression assay | Degree of Phag-GFP expression relative to control | Chemiluminometric | Protein | [58, 92, 93] | ||
| 20 Other ROS quantification methods | 124 EPR/ESR | ROS quantity (species depends on spin trap used) | ESR | ROS | [30, 154, 170, 174, 181, 184, 192, 235, 238, 254, 280, 292, 343, 350] | |
| 21 Reverse mutation assay | 125 Ames test | Number of mutations caused by material | Number of colonies | DNA | [270, 287, 351, 352] | |
| 6 “Acellular” methods: acellular techniques which examine interactions between ENMs and biomolecules | 23 Acellular ENM-biomolecule interaction study | 132 In vitro enzyme activity assay | Inhibition of enzyme activity in vitro | Various, usually colorimetric | Protein | [23, 47, 49, 195] |
| 24 Acellular DNA damage assays | 136 In vitro DNA fragmentation assay | Degree of plasmid fragmentation | Gel electrophoresis | DNA | [24, 259, 334] | |
| 7 “Correlation” methods: establishing correlations between magnitude of antibacterial activity and other measured variables to identify what attributes of ENMs are relevant to antibacterial activity | 27 Correlate antibacterial activity with ion release | 145 ICP-MS, -OES, -AES | Concentration of ions in exposure media | ICP-MS, -OES, -AES | Ion | [45, 49, 54, 56, 123, 125, 130, 132, 145, 161, 162, 185, 247, 259] [2, 8, 17, 24–26, 29, 30, 50, 55, 64, 88, 127–129, 147, 150, 151, 164, 165, 167, 174, 175, 180, 182, 191, 192, 194, 199, 201–203, 205, 207, 211–213, 216, 218, 219, 254–256, 285, 286, 292, 293, 298, 320, 327, 329, 340, 346, 347, 350, 353–364] |
| 146 AAS | Concentration of ions in exposure media | AAS | Ion | [11, 19, 55, 56, 87, 99, 126, 138, 140, 143, 154, 155, 204, 206, 215, 223, 226, 274, 275, 280, 365, 366] | ||
| 150 Silver/sulfide electrode | Concentration of ions in exposure media | Selective electrode | Ion | [58, 87, 140, 151, 188, 345] | ||
| 28 Correlate antibacterial activity with in vitro redox assay | 152 GSH oxidation (DTNB) | ROS quantity | Colorimetric | ROS, protein | [54, 161, 162, 256, 274, 282, 364] | |
| 153 XTT | Superoxide radical quantity | Colorimetric | ROS | [84, 95, 163–165, 182, 270, 274, 287, 364] | ||
| 155 Methyl orange | ROS quantity | Colorimetric | Photoactivity, ROS | [254, 357, 358] | ||
| 156 Methylene blue | ROS quantity | Colorimetric | Photoactivity, ROS | [157, 171, 172, 233, 329] | ||
| 157 3′-(p-aminophenyl) fluorescein (APF) | Hydroxyl radical quantity | Fluorometric | [20, 188, 233] | |||
| 166 Terepthalic acid (or Phth) | Hydroxyl radical quantity | Fluorometric | ROS | [154, 294, 344, 363] | ||
| 167 Luminol | Superoxide radical quantity | Chemiluminometric | ROS | [184, 363] | ||
| 169 p-Chlorobenzoic acid (pCBA) | ROS | [163–165, 182, 362] | ||||
| 170 FFA | ROS | [163–165, 182] | ||||
| 29 Correlate antibacterial activity with ENM property | 176 PL spectroscopy for defect sites/oxygen vacancies | Difference in antibacterial activity | (Dependent on antibacterial activity assay) | ROS | [233, 344, 361, 367, 368] | |
| 177 Morphology | Ion, contact | [337, 356, 364] | ||||
| 178 Zeta potential | Contact, membrane | [2, 125, 149, 168, 223, 224, 253, 255, 256, 268, 320, 358] | ||||
| 179 Surface coating | Ion, contact | [46, 144, 189, 252, 369] |
Statistically significant differences in conclusions were identified between studies which used certain groups, approaches, and techniques as compared to studies which did not; these groups, approaches, and techniques are represented in bold in Table 4. These differences were sometimes, but not always, identified in methods for which advantages or limitations were reported in the literature (Table S5†). Some methods which produced differences in conclusions were the same methods for which multiple limitations were reported (e.g., group 1 “elimination” methods and group 4 “ENM localization or visualization” methods) while others were methods for which multiple advantages were reported (e.g. some group 2 “genomic” and group 3 “chemical analysis” methods).
In some cases, relatively untested methods for which no advantages or limitations are yet reported yielded a difference in conclusions. Examples include the membrane barrier and filtration methods for ion isolation (group 1, approach 2, techniques 6 and 7). A possible explanation may be “bio-dissolution” processes (Table 4) in which cells mediate the release of ions from ENMs, which would not be captured in these techniques.63–65 Additionally, some methods were reported to have strong advantages but did not yield any statistically significant difference in conclusions from other methods; an example of this phenomenon is electron paramagnetic resonance (EPR) spectroscopy, group 5, approach 20, technique 119, as compared to other group 5 “cell effects” methods. It is possible that this is due to small sample size, since statistically significant differences tended to emerge only for the most commonly used methods. While it is not possible to identify with certainty the source of differing conclusions between methods, these instances confirm that the experimental methods used may influence the conclusions reached, and that further exploration by microbiologists of the advantages and limitations of commonly used methods would be beneficial.
Fig. 4 demonstrates that a small set of methods, many of which are facile and yield binary yes/no answers about the presence or absence of mechanisms, dominate mechanistic investigations. Several of the most-used assays, including comparison to soluble salts (group 1, approach 5, technique 30), correlation with ion release (group 7, approach 27, techniques 145–150), electron microscopy (group 4, approaches 13 and 14, techniques 64 and 67), and DCFH-DA (group 5, approach 18, technique 93), have significant, documented limitations (Table S5†) that may impede resolving the intended mechanistic question(s). Additionally, nearly 150 of 181 techniques are used in fewer than 10 studies; statistical analysis of these methods is difficult given the small sample size, and no attention has been devoted to their advantages and limitations in the literature. Taken together, these observations suggest a need to critically evaluate the wide breadth of mechanism-targeted methods currently in use and promote a smaller set of methods that are known to be reliable and tailored to provide the insight needed drive forward understanding. This shift may entail moving away from long-standing and commonly used assays in favor of those which may yield more information or be subject to fewer confounding variables.
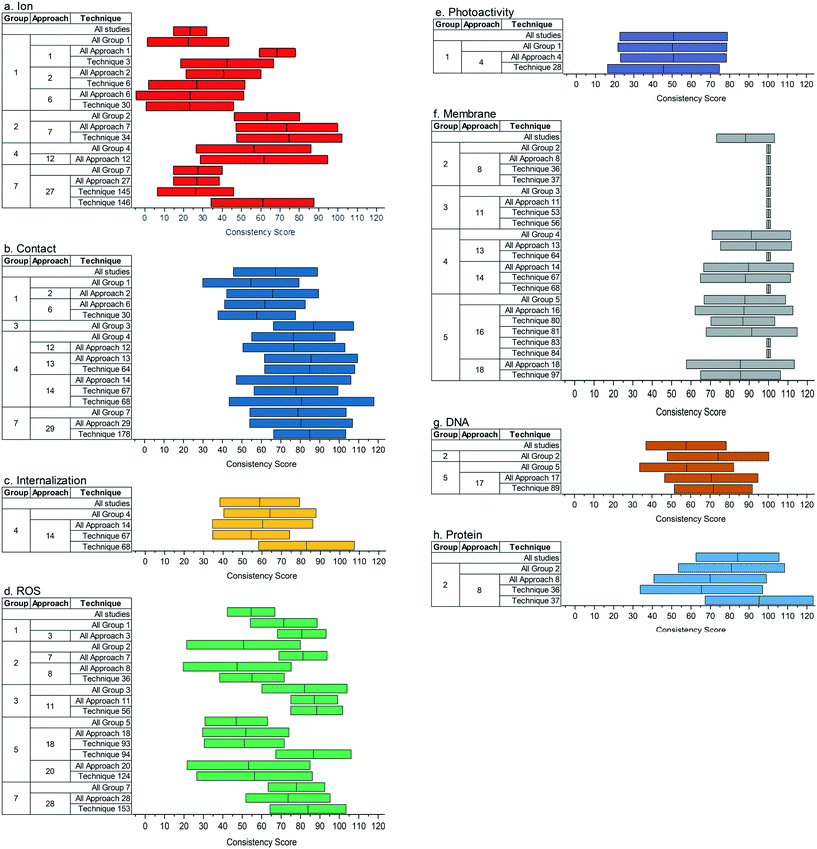
Fig. 5 compares the consistency score (weighted average across all core compositions, with 95% bootstrap confidence intervals) of method groups, approaches, and techniques used in 10 or more studies. A high consistency score for a method indicates that studies which used that method tended to draw the same conclusion about a given mechanism within a given core composition, regardless of whether that conclusion was positive or negative. A high consistency score does not, however, indicate that the method tends to produce the “correct” conclusion.
As previously noted, there are several possible explanations for the observed differences in consistency score between methods. The first explanation is that disagreement between studies is reflective of real differences in mechanism that may arise from, for example, differing ENM properties or experimental conditions; this would imply that methods with a low consistency score are relatively sensitive (i.e. precise and accurate). Another explanation is that disagreement between studies is not reflective of real differences in mechanism and arises instead from differing interpretations of a method's output; this would imply that methods with a lower consistency score are relatively difficult to interpret. Finally, a third explanation is that some methods are uninformative (i.e. precise but not necessarily accurate), consistently yielding the same, seemingly unambiguous result even when it does not reflect the “true” antibacterial mechanism for the core composition and exposure conditions of interest.
Identifying the underlying reasons for varying consistency scores will require further investigation. Possible avenues might include examining the statistically significant differences (as indicated by non-overlapping confidence intervals) between methods applied to the ions (Fig. 5a) and ROS (Fig. 5d) questions, as well as why the same method may have different consistency scores when applied to different questions, as in the case of group 4 methods for particle (Fig. 5b) versus internalization (Fig. 5c) questions (although these differences are not statistically significant). Additionally, since some methods applied to the membrane permeabilization question (Fig. 5f) had the highest possible consistency score of 100, corresponding to unanimous agreement among the studies reviewed within every core composition, it may be beneficial to examine more closely those methods for which disagreement was present. If this disagreement is found to be attributable to real mechanistic differences, in line with the first explanation described above, this may reveal facets of the membrane permeabilization question which remain to be resolved despite relative consensus about its general role in antibacterial activity (as discussed in section 3.1, “Summary of Reported Mechanistic Conclusions and Relationship with Study Design”).
4. Implications
The purpose of this study was two-fold: to synthesize the mechanism(s) of antibacterial activity that have been previously reported for five core compositions, and to investigate the underlying reasons that ENM antibacterial mechanisms continue to appear ambiguous. All of the mechanisms considered here were reported to contribute, to some extent and in some conditions, to the antibacterial activity of all of the core compositions reviewed. Contrary to expectation, differences in conclusions between core compositions were found to be minimal, suggesting that considering these core compositions together may yield useful insights that may also be transferable to other MMO ENMs. Taken together, these findings indicate a need to move beyond models which assume that mechanisms are mutually exclusive and re-focus on the conditions under which each mechanism might predominate, as these have so far eluded succinct articulation.While the contributions of ions, ENM contact, ROS, and membrane permeabilization have been the subject of hundreds of studies, questions of internalization, photo-activation, DNA damage, and protein damage remain relatively under-studied, which may stem partly from assumptions about which mechanisms apply to which core compositions. Additionally, even seemingly well-studied questions (e.g. membrane permeabilization) are often framed in binary terms (i.e., yes or no), to the detriment of nuanced understanding. Rather than continuing to conduct new research according to the status quo, a more fundamental shift in the framing and methodology of studies may be necessary to advance understanding of ENM antibacterial mechanisms and inform advanced ENM design.
With regard to the impact of the selected study design parameters (which included ENM properties, experimental conditions, choice of experimental methods, and investigation of multiple mechanisms), almost all of the parameters investigated were shown to influence conclusions to a statistically significant degree. It is not, however, possible to discern the origin of these differences based on this analysis alone. A closer investigation of experimental method choice reveals that there is much variety in methods used to study mechanism, some of which have documented advantages and limitations, but the majority of which have yet to be evaluated. Additionally, many methods are applied to the study of multiple questions, for which they may not be equally suited. Both the conclusions reached and the level of disagreement between studies were found to vary based on the methods chosen, occasionally to a statistically significant degree, although the reasons for this cannot yet be explained.
Ultimately, this analysis highlights the need for the experimental methods used to support mechanistic investigation to match the growing level of nuance in mechanistic discussions, as binary questions of “does X mechanism contribute or not to Y core composition's antibacterial activity?” are being gradually replaced with lines of inquiry that recognize the importance of ENM properties, study conditions, and possible interactions between multiple mechanisms of antibacterial activity. It is only with this level of knowledge that the full functional potential of ENMs can be realized while minimizing unintended, adverse impacts.
Conflicts of interest
There are no conflicts to declare.References
- M. Heinlaan, et al., Toxicity of nanosized and bulk ZnO, CuO and TiO2 to bacteria Vibrio fischeri and crustaceans Daphnia magna and Thamnocephalus platyurus, Chemosphere, 2008, 71(7), 1308–1316 CrossRef CAS.
- W. Jiang, H. Mashayekhi and B. S. Xing, Bacterial toxicity comparison between nano- and micro-scaled oxide particles, Environ. Pollut., 2009, 157(5), 1619–1625 CrossRef CAS.
- X. K. Hu, et al., In vitro evaluation of cytotoxicity of engineered metal oxide nanoparticles, Sci. Total Environ., 2009, 407(8), 3070–3072 CrossRef CAS.
- E. Hoseinzadeh, et al., A Review on Nano-Antimicrobials: Metal Nanoparticles, Methods and Mechanisms, Curr. Drug Metab., 2017, 18(2), 120–128 CrossRef CAS.
- A. Raghunath and E. Perumal, Metal oxide nanoparticles as antimicrobial agents: a promise for the future, Int. J. Antimicrob. Agents, 2017, 49(2), 137–152 CrossRef CAS.
- N. Beyth, et al., Alternative antimicrobial approach: nano-antimicrobial materials, J. Evidence-Based Complementary Altern. Med., 2015, 2015, 246012 Search PubMed.
- J. T. Seil and T. J. Webster, Antimicrobial applications of nanotechnology: methods and literature, Int. J. Nanomed., 2012, 7, 2767–2781 CAS.
- Y. Yang, et al., Impacts of silver nanoparticles on cellular and transcriptional activity of nitrogen-cycling bacteria, Environ. Toxicol. Chem., 2013, 32(7), 1488–1494 CAS.
- O. Choi and Z. Hu, Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria, Environ. Sci. Technol., 2008, 42(12), 4583–4588 CrossRef CAS.
- T. Wang, et al., Effects of Metal Nanoparticles on Methane Production from Waste-Activated Sludge and Microorganism Community Shift in Anaerobic Granular Sludge, Sci. Rep., 2016, 6, 25857 CrossRef CAS.
- Z. Z. Zhang, et al., Short-term impacts of Cu, CuO, ZnO and Ag nanoparticles (NPs) on anammox sludge: CuNPs make a difference, Bioresour. Technol., 2017, 235, 281–291 CrossRef CAS.
- L. M. Gilbertson, et al., Designing nanomaterials to maximize performance and minimize undesirable implications guided by the Principles of Green Chemistry, Chem. Soc. Rev., 2015, 44(16), 5758–5777 RSC.
- M. M. Falinski, et al., A framework for sustainable nanomaterial selection and design based on performance, hazard, and economic considerations, Nat. Nanotechnol., 2018, 13(8), 708–714 CrossRef CAS.
- A. M. Voutchkova, T. G. Osimitz and P. T. Anastas, Toward a Comprehensive Molecular Design Framework for Reduced Hazard, Chem. Rev., 2010, 110(10), 5845–5882 CrossRef CAS.
- Y. H. Ge, A. M. Horst, J. Kim, J. Priester, Z. S. Welch and P. Holden, Toxicity of Manufactured Nanomaterials to Microorganisms, In Engineered Nanoparticles and the Environment: Biophysicochemical Processes and Toxicity, ed. B. Xing, C. D. Vecitis and N. Senesi, 2016, pp. 320–346 Search PubMed.
- B. Ahmed, et al., ROS mediated destruction of cell membrane, growth and biofilms of human bacterial pathogens by stable metallic AgNPs functionalized from bell pepper extract and quercetin, Adv. Powder Technol., 2018, 29(7), 1601–1616 CrossRef CAS.
- J. C. Jin, et al., One-step synthesis of silver nanoparticles using carbon dots as reducing and stabilizing agents and their antibacterial mechanisms, Carbon, 2015, 94, 129–141 CrossRef CAS.
- X. M. Liu, et al., Mechanisms of oxidative stress caused by CuO nanoparticles to membranes of the bacterium Streptomyces coelicolor M145, Ecotoxicol. Environ. Saf., 2018, 158, 123–130 CrossRef CAS.
- Y. H. Hsueh, P. H. Tsai and K. S. Lin, pH-Dependent Antimicrobial Properties of Copper Oxide Nanoparticles in Staphylococcus aureus, Int. J. Mol. Sci., 2017, 18(4), 793 CrossRef.
- U. Kadiyala, et al., Unexpected insights into antibacterial activity of zinc oxide nanoparticles against methicillin resistant Staphylococcus aureus (MRSA), Nanoscale, 2018, 10(10), 4927–4939 RSC.
- S. Baek, et al., Antibacterial effect and toxicity pathways of industrial and sunscreen ZnO nanoparticles on Escherichia coli, J. Environ. Chem. Eng., 2017, 5(3), 3024–3032 CrossRef CAS.
- M. Seong and D. G. Lee, Silver Nanoparticles Against Salmonella enterica Serotype Typhimurium: Role of Inner Membrane Dysfunction, Curr. Microbiol., 2017, 74(6), 661–670 CrossRef CAS.
- Y. G. Chen, et al., The impacts of silver nanoparticles and silver ions on wastewater biological phosphorous removal and the mechanisms, J. Hazard. Mater., 2012, 239, 88–94 CrossRef.
- C. Kaweeteerawat, et al., Cu Nanoparticles Have Different Impacts in Escherichia coli and Lactobacillus brevis than Their Microsized and Ionic Analogues, ACS Nano, 2015, 9(7), 7215–7225 CrossRef CAS.
- J. Rousk, et al., Comparative Toxicity of Nanoparticulate CuO and ZnO to Soil Bacterial Communities, PLoS One, 2012, 7(3), e34197 CrossRef CAS.
- I. Rago, et al., Zinc oxide microrods and nanorods: different antibacterial activity and their mode of action against Gram-positive bacteria, RSC Adv., 2014, 4(99), 56031–56040 RSC.
- D. Laha, et al., Shape-dependent bactericidal activity of copper oxide nanoparticle mediated by DNA and membrane damage, Mater. Res. Bull., 2014, 59, 185–191 CrossRef CAS.
- H. Y. Xu, et al., Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7, BioMetals, 2012, 25(1), 45–53 CrossRef CAS.
- S. W. Kim, Y. W. Baek and Y. J. An, Assay-dependent effect of silver nanoparticles to Escherichia coli and Bacillus subtilis, Appl. Microbiol. Biotechnol., 2011, 92(5), 1045–1052 CrossRef CAS.
- I. Perelshtein, et al., The influence of the crystalline nature of nano-metal oxides on their antibacterial and toxicity properties, Nano Res., 2015, 8(2), 695–707 CrossRef CAS.
- B. L. Espana-Sanchez, et al., Early Stages of Antibacterial Damage of Metallic Nanoparticles by TEM and STEM-HAADF, Curr. Nanosci., 2018, 14(1), 54–61 CAS.
- W. R. Li, et al., Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli, Appl. Microbiol. Biotechnol., 2010, 85(4), 1115–1122 CrossRef CAS.
- C. Kaweeteerawat, et al., Toxicity of metal oxide nanoparticles in Escherichia coli correlates with conduction band and hydration energies, Environ. Sci. Technol., 2015, 49(2), 1105–1112 CrossRef CAS.
- T. Klanjscek, et al., Dynamic energy budget approach to modeling mechanisms of CdSe quantum dot toxicity, Ecotoxicology, 2013, 22(2), 319–330 CrossRef CAS.
- M. Mortimer, et al., Physical Properties of Carbon Nanomaterials and Nanoceria Affect Pathways Important to the Nodulation Competitiveness of the Symbiotic N2-Fixing Bacterium Bradyrhizobium diazoefficiens, Small, 2020, 1906055 CrossRef CAS.
- J. H. Priester, et al., Integrated approach to evaluating the toxicity of novel cysteine-capped silver nanoparticles to Escherichia coli and Pseudomonas aeruginosa, Analyst, 2014, 139(5), 954–963 RSC.
- J. H. Priester, et al., Effects of soluble cadmium salts versus CdSe quantum dots on the growth of planktonic Pseudomonas aeruginosa, Environ. Sci. Technol., 2009, 43(7), 2589–2594 CrossRef CAS.
- J. H. Priester, et al., Effects of TiO2 and Ag nanoparticles on polyhydroxybutyrate biosynthesis by activated sludge bacteria, Environ. Sci. Technol., 2014, 48(24), 14712–14720 CrossRef CAS.
- S. Akhtar, et al., Antibacterial and antiviral potential of colloidal Titanium dioxide (TiO2) nanoparticles suitable for biological applications, Mater. Res. Express, 2019, 6(10), 105409 CrossRef CAS.
- C. A. P. Bastos, et al., Copper nanoparticles have negligible direct antibacterial impact, NanoImpact, 2020, 17, 100192 CrossRef.
- R. Dadi, et al., Antibacterial activity of ZnO and CuO nanoparticles against gram positive and gram negative strains, Mater. Sci. Eng., C, 2019, 104, 109968 CrossRef CAS.
- Y. Dong and X. Sun, Antibacterial Mechanism of Nanosilvers, Curr. Pharmacol. Rep., 2019, 5(6), 401–409 CrossRef CAS.
- S. Liao, et al., Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa, Int. J. Nanomed., 2019, 14, 1469–1487 CrossRef CAS.
- S. Rajeshkumar, et al., Antibacterial and antioxidant potential of biosynthesized copper nanoparticles mediated through Cissus arnotiana plant extract, J. Photochem. Photobiol., B, 2019, 197, 111531 CrossRef CAS.
- Y. Choi, et al., Comparative toxicity of silver nanoparticles and silver ions to Escherichia coli, J. Environ. Sci., 2018, 66, 50–60 CrossRef.
- C. C. S. Batista, et al., Antimicrobial activity of nano-sized silver colloids stabilized by nitrogen-containing polymers: the key influence of the polymer capping, RSC Adv., 2018, 8(20), 10873–10882 RSC.
- Q. Lv, et al., Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: Novel approach and mechanisms investigation, J. Hazard. Mater., 2018, 347, 141–149 CrossRef CAS.
- D. L. Wang, et al., Where does the toxicity of metal oxide nanoparticles come from: The nanoparticles, the ions, or a combination of both?, J. Hazard. Mater., 2016, 308, 328–334 CrossRef CAS.
- J. Hou, et al., Ecotoxicological effects and mechanism of CuO nanoparticles to individual organisms, Environ. Pollut., 2017, 221, 209–217 CrossRef CAS.
- A. Joe, et al., Antibacterial mechanism of ZnO nanoparticles under dark conditions, J. Ind. Eng. Chem., 2017, 45, 430–439 CrossRef CAS.
- K. Ouyang, et al., Effects of humic acid on the interactions between zinc oxide nanoparticles and bacterial biofilms, Environ. Pollut., 2017, 231, 1104–1111 CrossRef CAS.
- P. A. Holden, et al., Considerations of environmentally relevant test conditions for improved evaluation of ecological hazards of engineered nanomaterials, Environ. Sci. Technol., 2016, 50(12), 6124–6145 CrossRef CAS.
- P. A. Holden, et al., Evaluation of exposure concentrations used in assessing manufactured nanomaterial environmental hazards: are they relevant?, Environ. Sci. Technol., 2014, 48(18), 10541–10551 CrossRef CAS.
- L. M. Gilbertson, et al., Shape-Dependent Surface Reactivity and Antimicrobial Activity of Nano-Cupric Oxide, Environ. Sci. Technol., 2016, 50(7), 3975–3984 CrossRef CAS.
- A. Ivask, et al., Toxicity Mechanisms in Escherichia coli Vary for Silver Nanoparticles and Differ from Ionic Silver, ACS Nano, 2014, 8(1), 374–386 CrossRef CAS.
- A. Ivask, et al., Size-Dependent Toxicity of Silver Nanoparticles to Bacteria, Yeast, Algae, Crustaceans and Mammalian Cells In Vitro, PLoS One, 2014, 9(7), e102108 CrossRef.
- A. J. Kora, R. Manjusha and J. Arunachalam, Superior bactericidal activity of SDS capped silver nanoparticles: Synthesis and characterization, Mater. Sci. Eng., C, 2009, 29(7), 2104–2109 CrossRef CAS.
- G. A. Sotiriou and S. E. Pratsinis, Antibacterial activity of nanosilver ions and particles, Environ. Sci. Technol., 2010, 44(14), 5649–5654 CrossRef CAS.
- L. Xiong, et al., Morphology-dependent antimicrobial activity of Cu/CuxO nanoparticles, Ecotoxicology, 2015, 24(10), 2067–2072 CrossRef CAS.
- N. K. Geitner, et al., Harmonizing across environmental nanomaterial testing media for increased comparability of nanomaterial datasets, Environ. Sci.: Nano, 2020, 7(1), 13–36 RSC.
- G. Hunt, et al., Towards a Consensus View on Understanding Nanomaterials Hazards and Managing Exposure: Knowledge Gaps and Recommendations, Materials, 2013, 6(3), 1090–1117 CrossRef CAS.
- L. M. Stabryla, et al., Emerging investigator series: it's not all about the ion: support for particle-specific contributions to silver nanoparticle antimicrobial activity, Environ. Sci.: Nano, 2018, 5(9), 2047–2068 RSC.
- D. N. Freitas, et al., Beyond the passive interactions at the nano-bio interface: evidence of Cu metalloprotein-driven oxidative dissolution of silver nanoparticles, J. Nanobiotechnol., 2016, 14, 7 CrossRef.
- N. Joshi, et al., Use of bioreporters and deletion mutants reveals ionic silver and ROS to be equally important in silver nanotoxicity, J. Hazard. Mater., 2015, 287, 51–58 CrossRef CAS.
- M. A. Maurer-Jones, et al., Characterization of silver ion dissolution from silver nanoparticles using fluorous-phase ion-selective electrodes and assessment of resultant toxicity to Shewanella oneidensis, Chem. Sci., 2013, 4(6), 2564–2572 RSC.
- A. M. Horst, et al., An Assessment of Fluorescence- and Absorbance-Based Assays to Study Metal-Oxide Nanoparticle ROS Production and Effects on Bacterial Membranes, Small, 2013, 9(9–10), 1753–1764 CrossRef CAS.
- A. B. Djurisic, et al., Toxicity of metal oxide nanoparticles: mechanisms, characterization, and avoiding experimental artefacts, Small, 2015, 11(1), 26–44 CrossRef CAS.
- A. Ivask, et al., Methodologies and approaches for the analysis of cell-nanoparticle interactions, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10(3), e1486 Search PubMed.
- E. J. Petersen, et al., Identification and Avoidance of Potential Artifacts and Misinterpretations in Nanomaterial Ecotoxicity Measurements, Environ. Sci. Technol., 2014, 48(8), 4226–4246 CrossRef CAS.
- E. J. Petersen, et al., Determining what really counts: modeling and measuring nanoparticle number concentrations, Environ. Sci.: Nano, 2019, 6(9), 2876–2896 RSC.
- E. J. Petersen, et al., Strategies for robust and accurate experimental approaches to quantify nanomaterial bioaccumulation across a broad range of organisms, Environ. Sci.: Nano, 2019, 6(6), 1619–1656 RSC.
- R. Handy, Practical considerations for conducting ecotoxicity test methods with manufactured nanomaterials: what have we learnt so far?, Ecotoxicology, 2012, 21(4), 933–972 CrossRef CAS.
- M. E. Pettitt and J. R. Lead, Minimum physicochemical characterisation requirements for nanomaterial regulation, Environ. Int., 2013, 52, 41–50 CrossRef.
- J. W. Card and B. A. Magnuson, A method to assess the quality of studies that examine the toxicity of engineered nanomaterials, Internet J. Toxicol., 2010, 29(4), 402–410 CrossRef CAS.
- S. L. Harper, et al., Systematic Evaluation of Nanomaterial Toxicity: Utility of Standardized Materials and Rapid Assays, ACS Nano, 2011, 5(6), 4688–4697 CrossRef CAS.
- D. G. Thomas, et al., ISA-TAB-Nano: a specification for sharing nanomaterial research data in spreadsheet-based format, BMC Biotechnol., 2013, 13, 2 CrossRef.
- R. Damoiseaux, et al., No time to lose—high throughput screening to assess nanomaterial safety, Nanoscale, 2011, 3(4), 1345–1360 RSC.
- O. Bondarenko, et al., Sub-toxic effects of CuO nanoparticles on bacteria: Kinetics, role of Cu ions and possible mechanisms of action, Environ. Pollut., 2012, 169, 81–89 CrossRef CAS.
- N. Durán, et al., Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity, Nanomed.: Nanotechnol., Biol. Med., 2016, 12(3), 789–799 CrossRef.
- N. Hoshino, et al., Damage to the cytoplasmic membrane of Escherichia Coli by catechin-copper (II) complexes, Free Radical Biol. Med., 1999, 27(11), 1245–1250 CrossRef CAS.
- J. S. Kim, et al., Antimicrobial effects of silver nanoparticles, Nanomedicine, 2007, 3(1), 95–101 CrossRef CAS.
- J. N. Chen, et al., Various antibacterial mechanisms of biosynthesized copper oxide nanoparticles against soilborne Ralstonia solanacearum, RSC Adv., 2019, 9(7), 3788–3799 RSC.
- N. Padmavathy and R. Vijayaraghavan, Enhanced bioactivity of ZnO nanoparticles—an antimicrobial study, Sci. Technol. Adv. Mater., 2008, 9(3), 035004 CrossRef.
- S. Chakraborti, et al., The Molecular Basis of Inactivation of Metronidazole-Resistant Helicobacter pylori Using Polyethyleneimine Functionalized Zinc Oxide Nanoparticles, PLoS One, 2013, 8(8), e70776 CrossRef CAS.
- S. Dalai, et al., Studies on interfacial interactions of TiO2 nanoparticles with bacterial cells under light and dark conditions, Bull. Mater. Sci., 2014, 37(3), 371–381 CrossRef CAS.
- M. Kumari, et al., Enhanced Cellular Internalization: A Bactericidal Mechanism More Relative to Biogenic Nanoparticles than Chemical Counterparts, ACS Appl. Mater. Interfaces, 2017, 9(5), 4519–4533 CrossRef CAS.
- O. Bondarenko, et al., Particle-Cell Contact Enhances Antibacterial Activity of Silver Nanoparticles, PLoS One, 2013, 8(5), e64060 CrossRef CAS.
- J. S. McQuillan, et al., Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12, Nanotoxicology, 2012, 6(8), 857–866 CrossRef CAS.
- E. Prokhorov, et al., Chitosan/copper nanocomposites: Correlation between electrical and antibacterial properties, Colloids Surf., B, 2019, 180, 186–192 CrossRef CAS.
- A. Tiwari, et al., Passive membrane penetration by ZnO nanoparticles is driven by the interplay of electrostatic and phase boundary conditions, Nanoscale, 2018, 10(7), 3369–3384 RSC.
- Y. N. Slavin, et al., Metal nanoparticles: understanding the mechanisms behind antibacterial activity, J. Nanobiotechnol., 2017, 15(1), 65 CrossRef.
- Y. H. Hsueh, et al., The Antimicrobial Properties of Silver Nanoparticles in Bacillus subtilis Are Mediated by Released Ag+ Ions, PLoS One, 2015, 10(12), e0144306 CrossRef.
- Y.-H. Hsueh, et al., ZnO nanoparticles affect Bacillus subtilis cell growth and biofilm formation, PLoS One, 2015, 10(6), e0128457 CrossRef.
- R. K. Gupta, et al., Biosynthesis of silver nanoparticles from the novel strain of Streptomyces Sp. BHUMBU-80 with highly efficient electroanalytical detection of hydrogen peroxide and antibacterial activity, J. Environ. Chem. Eng., 2017, 5(6), 5624–5635 CrossRef CAS.
- K. J. P. Anthony, M. Murugan and S. Gurunathan, Biosynthesis of silver nanoparticles from the culture supernatant of Bacillus marisflavi and their potential antibacterial activity, J. Ind. Eng. Chem., 2014, 20(4), 1505–1510 CrossRef CAS.
- J. A. Imlay, Diagnosing oxidative stress in bacteria: not as easy as you might think, Curr. Opin. Microbiol., 2015, 24, 124–131 CrossRef CAS.
- Y. Hong, et al., Post-stress bacterial cell death mediated by reactive oxygen species, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(20), 10064–10071 CrossRef CAS.
- J. Han, W. Qiu and W. Gao, Potential dissolution and photo-dissolution of ZnO thin films, J. Hazard. Mater., 2010, 178(1), 115–122 CrossRef CAS.
- S. W. Kim and Y.-J. An, Effect of ZnO and TiO 2 nanoparticles preilluminated with UVA and UVB light on Escherichia coli and Bacillus subtilis, Appl. Microbiol. Biotechnol., 2012, 95(1), 243–253 CrossRef CAS.
- A. M. Mittelman, J. D. Fortner and K. D. Pennell, Effects of ultraviolet light on silver nanoparticle mobility and dissolution, Environ. Sci.: Nano, 2015, 2(6), 683–691 RSC.
- E. J. Petersen, et al., DNA Damaging Potential of Photoactivated P25 Titanium Dioxide Nanoparticles, Chem. Res. Toxicol., 2014, 27(10), 1877–1884 Search PubMed.
- K. H. Jones and J. A. Senft, An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide, J. Histochem. Cytochem., 1985, 33(1), 77–79 CrossRef CAS.
- L. Shi, et al., Limits of propidium iodide as a cell viability indicator for environmental bacteria, Cytometry, Part A, 2007, 71(8), 592–598 CrossRef.
- J. Trevors, Can dead bacterial cells be defined and are genes expressed after cell death?, J. Microbiol. Methods, 2012, 90(1), 25–28 CrossRef CAS.
- R. Pagán and B. Mackey, Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains, Appl. Environ. Microbiol., 2000, 66(7), 2829–2834 CrossRef.
- I. P. Mukha, et al., Antimicrobial activity of stable silver nanoparticles of a certain size, Appl. Biochem. Microbiol., 2013, 49(2), 199–206 CrossRef CAS.
- S. I. Miller, Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules, MBio, 2016, 7(5), e01541-16 CrossRef.
- K. L. May and M. Grabowicz, The bacterial outer membrane is an evolving antibiotic barrier, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(36), 8852–8854 CrossRef CAS.
- S. Charles, et al., Assessment of the in vitro genotoxicity of TiO2 nanoparticles in a regulatory context, Nanotoxicology, 2018, 12(4), 357–374 CrossRef CAS.
- X. Giroux, et al., Maladaptive DNA repair is the ultimate contributor to the death of trimethoprim-treated cells under aerobic and anaerobic conditions, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(43), 11512–11517 CrossRef CAS.
- E. Guillemet, et al., The bacterial DNA repair protein Mfd confers resistance to the host nitrogen immune response, Sci. Rep., 2016, 6(1), 29349 CrossRef CAS.
- S. Meghana, et al., Understanding the pathway of antibacterial activity of copper oxide nanoparticles, RSC Adv., 2015, 5(16), 12293–12299 RSC.
- N. S. Wigginton, et al., Binding of Silver Nanoparticles to Bacterial Proteins Depends on Surface Modifications and Inhibits Enzymatic Activity, Environ. Sci. Technol., 2010, 44(6), 2163–2168 CrossRef CAS.
- H. M. Zhang, et al., Effect of TiO2 nanoparticles on the structure and activity of catalase, Chem.-Biol. Interact., 2014, 219, 168–174 CrossRef CAS.
- M. E. Letelier, et al., Mechanisms underlying iron and copper ions toxicity in biological systems: Pro-oxidant activity and protein-binding effects, Chem.-Biol. Interact., 2010, 188(1), 220–227 CrossRef CAS.
- X. Zhao, T. Toyooka and Y. Ibuki, Synergistic bactericidal effect by combined exposure to Ag nanoparticles and UVA, Sci. Total Environ., 2013, 458–460, 54–62 CrossRef CAS.
- P. Belenky, et al., Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage, Cell Rep., 2015, 13(5), 968–980 CrossRef CAS.
- J. Zhang, et al., The effects and the potential mechanism of environmental transformation of metal nanoparticles on their toxicity in organisms, Environ. Sci.: Nano, 2018, 5(11), 2482–2499 RSC.
- Z. M. Xiu, J. Ma and P. J. J. Alvarez, Differential Effect of Common Ligands and Molecular Oxygen on Antimicrobial Activity of Silver Nanoparticles versus Silver Ions, Environ. Sci. Technol., 2011, 45(20), 9003–9008 CrossRef CAS.
- A. B. Smetana, et al., Biocidal activity of nanocrystalline silver powders and particles, Langmuir, 2008, 24(14), 7457–7464 CrossRef CAS.
- Y. M. Long, et al., Surface ligand controls silver ion release of nanosilver and its antibacterial activity against Escherichia coli, Int. J. Nanomed., 2017, 12, 3193–3206 CrossRef CAS.
- S. Agnihotri, S. Mukherji and S. Mukherji, Impact of background water quality on disinfection performance and silver release of immobilized silver nanoparticles: Modeling disinfection kinetics, bactericidal mechanism and aggregation behavior, Chem. Eng. J., 2019, 372, 684–696 CrossRef CAS.
- A. J. Calder, et al., Soil components mitigate the antimicrobial effects of silver nanoparticles towards a beneficial soil bacterium, Pseudomonas chlororaphis O6, Sci. Total Environ., 2012, 429, 215–222 CrossRef CAS.
- L. R. Christena, et al., Copper nanoparticles as an efflux pump inhibitor to tackle drug resistant bacteria, RSC Adv., 2015, 5(17), 12899–12909 RSC.
- C. O. Dimkpa, et al., Interaction of silver nanoparticles with an environmentally beneficial bacterium, Pseudomonas chlororaphis, J. Hazard. Mater., 2011, 188(1–3), 428–435 CrossRef CAS.
- F. F. Li, et al., Analysis of copper nanoparticles toxicity based on a stress-responsive bacterial biosensor array, Nanoscale, 2013, 5(2), 653–662 RSC.
- D. Barros, et al., Proteomics and antioxidant enzymes reveal different mechanisms of toxicity induced by ionic and nanoparticulate silver in bacteria, Environ. Sci.: Nano, 2019, 6(4), 1207–1218 RSC.
- C. L. Arnaout and C. K. Gunsch, Impacts of Silver Nanoparticle Coating on the Nitrification Potential of Nitrosomonas europaea, Environ. Sci. Technol., 2012, 46(10), 5387–5395 CrossRef CAS.
- J. Zhao, et al., Mitigation of CuO nanoparticle-induced bacterial membrane damage by dissolved organic matter, Water Res., 2013, 47(12), 4169–4178 CrossRef CAS.
- S. Agnihotri, S. Mukherji and S. Mukherji, Immobilized silver nanoparticles enhance contact killing and show highest efficacy: elucidation of the mechanism of bactericidal action of silver, Nanoscale, 2013, 5(16), 7328–7340 RSC.
- A. Taglietti, et al., Antibacterial Activity of Glutathione-Coated Silver Nanoparticles against Gram Positive and Gram Negative Bacteria, Langmuir, 2012, 28(21), 8140–8148 CrossRef CAS.
- C. Gunawan, et al., Cytotoxic origin of copper (II) oxide nanoparticles: comparative studies with micron-sized particles, leachate, and metal salts, ACS Nano, 2011, 5(9), 7214–7225 CrossRef CAS.
- J. Pasquet, et al., The contribution of zinc ions to the antimicrobial activity of zinc oxide, Colloids Surf., A, 2014, 457, 263–274 CrossRef CAS.
- J. Pasquet, et al., Antimicrobial activity of zinc oxide particles on five micro-organisms of the Challenge Tests related to their physicochemical properties, Int. J. Pharm., 2014, 460(1–2), 92–100 CrossRef CAS.
- S. S. Khan, S. S. Ghouse and P. Chandran, Toxic effect of environmentally relevant concentration of silver nanoparticles on environmentally beneficial bacterium Pseudomonas putida, Bioprocess Biosyst. Eng., 2015, 38(7), 1243–1249 CrossRef.
- Y. Liu, et al., Antibacterial activities of zinc oxide nanoparticles against Escherichia coli O157:H7, J. Appl. Microbiol., 2009, 107(4), 1193–1201 CrossRef CAS.
- A. Nagy, et al., Silver nanoparticles embedded in zeolite membranes: release of silver ions and mechanism of antibacterial action, Int. J. Nanomed., 2011, 6, 1833–1852 CAS.
- M. Visnapuu, et al., Dissolution of Silver Nanowires and Nanospheres Dictates Their Toxicity to Escherichia coli, BioMed Res. Int., 2013, 819252 Search PubMed.
- Y. H. Jiang, et al., Role of physical and chemical interactions in the antibacterial behavior of ZnO nanoparticles against E. coli, Mater. Sci. Eng., C, 2016, 69, 1361–1366 CrossRef CAS.
- O. Bondarenko, et al., Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review, Arch. Toxicol., 2013, 87(7), 1181–1200 CrossRef CAS.
- L. L. Zhang, et al., Mechanistic investigation into antibacterial behaviour of suspensions of ZnO nanoparticles against E. coli, J. Nanopart. Res., 2010, 12(5), 1625–1636 CrossRef CAS.
- E. Bae, et al., Bacterial cytotoxicity of the silver nanoparticle related to physicochemical metrics and agglomeration properties, Environ. Toxicol. Chem., 2010, 29(10), 2154–2160 CrossRef CAS.
- A. L. Ulloa-Ogaz, et al., Oxidative damage to Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 induced by CuO-NPs, Environ. Sci. Pollut. Res., 2017, 24(27), 22048–22060 CrossRef CAS.
- A. Dror-Ehre, et al., Silver nanoparticle-E. coli colloidal interaction in water and effect on E-coli survival, J. Colloid Interface Sci., 2009, 339(2), 521–526 CrossRef CAS.
- J. Fabrega, et al., Silver nanoparticle impact on bacterial growth: effect of pH, concentration, and organic matter, Environ. Sci. Technol., 2009, 43(19), 7285–7290 CrossRef CAS.
- M. I. Hossain, et al., Antimicrobial properties of nanorods: killing bacteria via impalement, IET Nanobiotechnol., 2017, 11(5), 501–505 CrossRef.
- Z. M. Xiu, et al., Negligible Particle-Specific Antibacterial Activity of Silver Nanoparticles, Nano Lett., 2012, 12(8), 4271–4275 CrossRef CAS.
- E. T. Hwang, et al., Analysis of the toxic mode of action of silver nanoparticles using stress-specific bioluminescent bacteria, Small, 2008, 4(6), 746–750 CrossRef CAS.
- M. R. Ajdari, et al., Phytosynthesis of Silver Nanoparticles Using Myrtus communis L. Leaf Extract and Investigation of Bactericidal Activity, J. Electron. Mater., 2017, 46(12), 6930–6935 CrossRef CAS.
- S. Baek, S. H. Joo and M. Toborek, Treatment of antibiotic-resistant bacteria by encapsulation of ZnO nanoparticles in an alginate biopolymer: Insights into treatment mechanisms, J. Hazard. Mater., 2019, 373, 122–130 CrossRef CAS.
- S. L. Loo, et al., Bactericidal Mechanisms Revealed for Rapid Water Disinfection by Superabsorbent Cryogels Decorated with Silver Nanoparticles, Environ. Sci. Technol., 2015, 49(4), 2310–2318 CrossRef CAS.
- S. J. Lia, et al., Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa, Int. J. Nanomed., 2019, 14, 1469–1487 CrossRef.
- M. Singh, Elucidation of biogenic silver nanoparticles susceptibility towards Escherichia coli: an investigation on the antimicrobial mechanism, IET Nanobiotechnol., 2016, 10(5), 276–280 CrossRef.
- V. L. Prasanna and R. Vijayaraghavan, Insight into the Mechanism of Antibacterial Activity of ZnO: Surface Defects Mediated Reactive Oxygen Species Even in the Dark, Langmuir, 2015, 31(33), 9155–9162 CrossRef.
- J. A. Quek, et al., Visible light responsive flower-like ZnO in photocatalytic antibacterial mechanism towards Enterococcus faecalis and Micrococcus luteus, J. Photochem. Photobiol., B, 2018, 187, 66–75 CrossRef CAS.
- L. L. Zhang, et al., The properties of ZnO nanofluids and the role of H2O2 in the disinfection activity against Escherichia coli, Water Res., 2013, 47(12), 4013–4021 CrossRef CAS.
- R. J. Barnes, et al., Comparison of TiO 2 and ZnO nanoparticles for photocatalytic degradation of methylene blue and the correlated inactivation of gram-positive and gram-negative bacteria, J. Nanopart. Res., 2013, 15(2), 1432 CrossRef.
- M. Veerapandian, et al., Glucosamine functionalized copper nanoparticles: Preparation, characterization and enhancement of anti-bacterial activity by ultraviolet irradiation, Chem. Eng. J., 2012, 209, 558–567 CrossRef CAS.
- L. L. Zhang, et al., ZnO nanofluids - A potential antibacterial agent, Prog. Nat. Sci.: Mater. Int., 2008, 18(8), 939–944 CrossRef CAS.
- O. Akhavan and E. Ghaderi, Cu and CuO nanoparticles immobilized by silica thin films as antibacterial materials and photocatalysts, Surf. Coat. Technol., 2010, 205(1), 219–223 CrossRef CAS.
- T. P. Dasari and H. M. Hwang, Effect of humic acids and sunlight on the cytotoxicity of engineered zinc oxide and titanium dioxide nanoparticles to a river bacterial assemblage, J. Environ. Sci., 2013, 25(9), 1925–1935 CrossRef CAS.
- T. P. Dasari, K. Pathakoti and H. M. Hwang, Determination of the mechanism of photoinduced toxicity of selected metal oxide nanoparticles (ZnO, CuO, Co3O4 and TiO2) to E-coli bacteria, J. Environ. Sci., 2013, 25(5), 882–888 CrossRef CAS.
- Y. Li, et al., Mechanism of Photogenerated Reactive Oxygen Species and Correlation with the Antibacterial Properties of Engineered Metal-Oxide Nanoparticles, ACS Nano, 2012, 6(6), 5164–5173 CrossRef CAS.
- Y. Li, et al., Influence of aqueous media on the ROS-mediated toxicity of ZnO nanoparticles toward green fluorescent protein-expressing Escherichia coli under UV-365 irradiation, Langmuir, 2014, 30(10), 2852–2862 CrossRef CAS.
- E. X. Shang, et al., Effect of aqueous media on the copper-ion-mediated phototoxicity of CuO nanoparticles toward green fluorescent protein-expressing Escherichia coli, Ecotoxicol. Environ. Saf., 2015, 122, 238–244 CrossRef CAS.
- A. Al-Sharqi, et al., Enhancement of the Antibacterial Efficiency of Silver Nanoparticles against Gram-Positive and Gram-Negative Bacteria Using Blue Laser Light, Int. J. Photoenergy, 2019, 2528490 CAS.
- T. P. Dasari and H. M. Hwang, The effect of humic acids on the cytotoxicity of silver nanoparticles to a natural aquatic bacterial assemblage, Sci. Total Environ., 2010, 408(23), 5817–5823 CrossRef CAS.
- A. Erdem, et al., Inhibition of bacteria by photocatalytic nano-TiO2 particles in the absence of light, Int. J. Environ. Sci. Technol., 2015, 12(9), 2987–2996 CrossRef CAS.
- A. Erdem, et al., The short-term toxic effects of TiO2 nanoparticles toward bacteria through viability, cellular respiration, and lipid peroxidation, Environ. Sci. Pollut. Res., 2015, 22(22), 17917–17924 CrossRef CAS.
- G. Gedik, et al., Production of Metal Oxide Containing Antibacterial Coated Textile Material and Investigation of the Mechanism of Action, Fibers Polym., 2018, 19(12), 2548–2563 CrossRef CAS.
- B. Jalvo, et al., Antimicrobial and antibiofilm efficacy of self-cleaning surfaces functionalized by TiO2 photocatalytic nanoparticles against Staphylococcus aureus and Pseudomonas putida, J. Hazard. Mater., 2017, 340, 160–170 CrossRef CAS.
- A. Joe, et al., Antimicrobial activity of ZnO nanoplates and its Ag nanocomposites: Insight into an ROS-mediated antibacterial mechanism under UV light, J. Solid State Chem., 2018, 267, 124–133 CrossRef CAS.
- U. Joost, et al., Photocatalytic antibacterial activity of nano-TiO2 (anatase)-based thin films: Effects on Escherichia coli cells and fatty acids, J. Photochem. Photobiol., B, 2015, 142, 178–185 CrossRef CAS.
- Y. H. Leung, et al., Toxicity of ZnO and TiO2 to Escherichia coli cells, Sci. Rep., 2016, 6, 35243 CrossRef CAS.
- J. S. McQuillan and A. M. Shaw, Whole-cell Escherichia coli-based bio-sensor assay for dual zinc oxide nanoparticle toxicity mechanisms, Biosens. Bioelectron., 2014, 51, 274–279 CrossRef CAS.
- J. Nesic, et al., New evidence for TiO2 uniform surfaces leading to complete bacterial reduction in the dark: Critical issues, Colloids Surf., B, 2014, 123, 593–599 CrossRef CAS.
- K. Pathakoti, et al., Using experimental data of Escherichia coli to develop a QSAR model for predicting the photo-induced cytotoxicity of metal oxide nanoparticles, J. Photochem. Photobiol., B, 2014, 130, 234–240 CrossRef CAS.
- T. A. Qiu, et al., A mechanistic study of TiO2 nanoparticle toxicity on Shewanella oneidensis MR-1 with UV-containing simulated solar irradiation: Bacterial growth, riboflavin secretion, and gene expression, Chemosphere, 2017, 168, 1158–1168 CrossRef CAS.
- D. M. Rocca, et al., Biocompatibility and photo-induced antibacterial activity of lignin-stabilized noble metal nanoparticles, RSC Adv., 2018, 8(70), 40454–40463 RSC.
- L. Valenzuela, et al., Antimicrobial surfaces with self-cleaning properties functionalized by photocatalytic ZnO electrosprayed coatings, J. Hazard. Mater., 2019, 369, 665–673 CrossRef CAS.
- D. Visinescu, et al., Zinc Oxide Spherical-Shaped Nanostructures: Investigation of Surface Reactivity and Interactions with Microbial and Mammalian Cells, Langmuir, 2018, 34(45), 13638–13651 CrossRef CAS.
- W. Zhang, et al., Photogeneration of Reactive Oxygen Species on Uncoated Silver, Gold, Nickel, and Silicon Nanoparticles and Their Antibacterial Effects, Langmuir, 2013, 29(15), 4647–4651 CrossRef CAS.
- L. V. Zhukova, Evidence for Compression of Escherichia coli K12 Cells under the Effect of TiO2 Nanoparticles, ACS Appl. Mater. Interfaces, 2015, 7(49), 27197–27205 CrossRef CAS.
- G. Applerot, et al., Understanding the Antibacterial Mechanism of CuO Nanoparticles: Revealing the Route of Induced Oxidative Stress, Small, 2012, 8(21), 3326–3337 CrossRef CAS.
- S. Bandyopadhyay, et al., Comparative toxicity assessment of CeO2 and ZnO nanoparticles towards Sinorhizobium meliloti, a symbiotic alfalfa associated bacterium: use of advanced microscopic and spectroscopic techniques, J. Hazard. Mater., 2012, 241, 379–386 CrossRef.
- F. Benakashani, A. R. Allafchian and S. A. H. Jalali, Biosynthesis of silver nanoparticles using Capparis spinosa L. leaf extract and their antibacterial activity, Karbala International Journal of Modern Science, 2016, 2(4), 251–258 CrossRef.
- C. R. Bowman, et al., Effects of silver nanoparticles on zebrafish (Danio rerio) and Escherichia coli (ATCC 25922): A comparison of toxicity based on total surface area versus mass concentration of particles in a model eukaryotic and prokaryotic system, Environ. Toxicol. Chem., 2012, 31(8), 1793–1800 CrossRef CAS.
- A. O. Choi, et al., Quantum dot-induced cell death involves Fas upregulation and lipid peroxidation in human neuroblastoma cells, J. Nanobiotechnol., 2007, 5, 1 CrossRef.
- A. M. El Badawy, et al., Surface Charge-Dependent Toxicity of Silver Nanoparticles, Environ. Sci. Technol., 2011, 45(1), 283–287 CrossRef CAS.
- A. Ivask, et al., Profiling of the reactive oxygen species-related ecotoxicity of CuO, ZnO, TiO2, silver and fullerene nanoparticles using a set of recombinant luminescent Escherichia coli strains: differentiating the impact of particles and solubilised metals, Anal. Bioanal. Chem., 2010, 398(2), 701–716 CrossRef CAS.
- C.-N. Lok, et al., Proteomic analysis of the mode of antibacterial action of silver nanoparticles, J. Proteome Res., 2006, 5(4), 916–924 CrossRef CAS.
- J. D. Moore, et al., Time-dependent bacterial transcriptional response to CuO nanoparticles differs from that of Cu2+ and provides insights into CuO nanoparticle toxicity mechanisms, Environ. Sci.: Nano, 2017, 4(12), 2321–2335 RSC.
- J. R. Morones, et al., The bactericidal effect of silver nanoparticles, Nanotechnology, 2005, 16(10), 2346 CrossRef CAS.
- A. L. Neal, et al., Can the soil bacterium Cupriavidus necator sense ZnO nanomaterials and aqueous Zn2+ differentially?, Nanotoxicology, 2012, 6(4), 371–380 CrossRef CAS.
- Y. L. Su, et al., Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles, Sci. Rep., 2015, 5, 15824 CrossRef CAS.
- M. Alqahtany, et al., Nanoscale reorganizations of histone-like nucleoid structuring proteins in Escherichia coli are caused by silver nanoparticles, Nanotechnology, 2019, 30(38), 385101 CrossRef CAS.
- O. M. Bondarenko, et al., Plasma membrane is the target of rapid antibacterial action of silver nanoparticles in Escherichia coil and Pseudomonas aeruginosa, Int. J. Nanomed., 2018, 13, 6779–6790 CrossRef CAS.
- A. Q. Carlos, et al., Toxicity Effects in Pathogen Microorganisms Exposed to Silver Nanoparticles, Nanosci. Nanotechnol. Lett., 2017, 9(2), 165–173 CrossRef.
- J. N. Chen, et al., Enhancement of the Antibacterial Activity of Silver Nanoparticles against Phytopathogenic Bacterium Ralstonia solanacearum by Stabilization, J. Nanomater., 2016, 7135852 Search PubMed.
- C. O. Dimkpa, et al., CuO and ZnO nanoparticles differently affect the secretion of fluorescent siderophores in the beneficial root colonizer, Pseudomonas chlororaphis O6, Nanotoxicology, 2012, 6(6), 635–642 CrossRef CAS.
- M. A. Doody, et al., Differential antimicrobial activity of silver nanoparticles to bacteria Bacillus subtilis and Escherichia coli, and toxicity to crop plant Zea mays and beneficial B. subtilis-inoculated Z. mays, J. Nanopart. Res., 2016, 18(10), 290 CrossRef.
- R. K. Dutta, et al., Antibacterial effect of chronic exposure of low concentration ZnO nanoparticles on E. coli, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2013, 48(8), 871–878 CrossRef CAS.
- A. Gelabert, et al., Testing nanoeffect onto model bacteria: Impact of speciation and genotypes, Nanotoxicology, 2016, 10(2), 216–225 CrossRef CAS.
- N. T. Giao, et al., Influence of silver nanoparticles and liberated silver ions on nitrifying sludge: ammonia oxidation inhibitory kinetics and mechanism, Environ. Sci. Pollut. Res., 2017, 24(10), 9229–9240 CrossRef CAS.
- Y. T. Guo, et al., Heterogenic response of prokaryotes toward silver nanoparticles and ions is facilitated by phenotypes and attachment of silver aggregates to cell surfaces, Cytometry, Part A, 2017, 91(8), 775–784 CrossRef CAS.
- H. Y. Li, et al., A comparative study of the antibacterial mechanisms of silver ion and silver nanoparticles by Fourier transform infrared spectroscopy, Vib. Spectrosc., 2016, 85, 112–121 CrossRef CAS.
- M. Li, L. Z. Zhu and D. H. Lin, Toxicity of ZnO Nanoparticles to Escherichia coli: Mechanism and the Influence of Medium Components, Environ. Sci. Technol., 2011, 45(5), 1977–1983 CrossRef CAS.
- N. Mapara, et al., Antimicrobial potentials of Helicteres isora silver nanoparticles against extensively drug-resistant (XDR) clinical isolates of Pseudomonas aeruginosa, Appl. Microbiol. Biotechnol., 2015, 99(24), 10655–10667 CrossRef CAS.
- J. S. McQuillan and A. M. Shaw, Differential gene regulation in the Ag nanoparticle and Ag+-induced silver stress response in Escherichia coli: A full transcriptomic profile, Nanotoxicology, 2014, 8, 177–184 CrossRef CAS.
- D. A. Mosselhy, et al., Comparative synthesis and antimicrobial action of silver nanoparticles and silver nitrate, J. Nanopart. Res., 2015, 17(12), 473 CrossRef.
- H. Mu and Y. G. Chen, Long-term effect of ZnO nanoparticles on waste activated sludge anaerobic digestion, Water Res., 2011, 45(17), 5612–5620 CrossRef CAS.
- Q. Qin, J. Y. Li and J. B. Wang, Antibacterial Activity Comparison of Three Metal Oxide Nanoparticles and their Dissolved Metal Ions, Water Environ. Res., 2017, 89(4), 378–383 CrossRef CAS.
- T. S. Radniecki, et al., Influence of liberated silver from silver nanoparticles on nitrification inhibition of Nitrosomonas europaea, Chemosphere, 2011, 85(1), 43–49 CrossRef CAS.
- N. Singh, K. M. Paknikar and J. Rajwade, Gene expression is influenced due to 'nano' and 'ionic' copper in pre-formed Pseudomonas aeruginosa biofilms, Environ. Res., 2019, 175, 367–375 CrossRef CAS.
- G. Y. Su, et al., Comparison on the molecular response profiles between nano zinc oxide (ZnO) particles and free zinc ion using a genome-wide toxicogenomics approach, Environ. Sci. Pollut. Res., 2015, 22(22), 17434–17442 CrossRef CAS.
- V. Svetlichnyi, et al., ZnO nanoparticles obtained by pulsed laser ablation and their composite with cotton fabric: Preparation and study of antibacterial activity, Appl. Surf. Sci., 2016, 372, 20–29 CrossRef CAS.
- H. Wang, et al., Silver nanoparticles: A novel antibacterial agent for control of Cronobacter sakazakii, J. Dairy Sci., 2018, 101(12), 10775–10791 CrossRef CAS.
- J. A. Wang, et al., Effects of Silver Nanoparticles on Soil Microbial Communities and Bacterial Nitrification in Suburban Vegetable Soils, Pedosphere, 2017, 27(3), 482–490 CrossRef.
- X. T. Yan, et al., Antibacterial mechanism of silver nanoparticles in Pseudomonas aeruginosa: proteomics approach, Metallomics, 2018, 10(4), 557–564 RSC.
- I.-S. Hwang, et al., Synergistic effects between silver nanoparticles and antibiotics and the mechanisms involved, J. Med. Microbiol., 2012, 61(12), 1719–1726 CrossRef CAS.
- L. S. Reddy, et al., Antimicrobial activity of zinc oxide (ZnO) nanoparticle against Klebsiella pneumoniae, Pharm. Biol., 2014, 52(11), 1388–1397 CrossRef CAS.
- S. Kim and Y. Yoon, Assessing bioavailability and genotoxicity of heavy metals and metallic nanoparticles simultaneously using dual-sensing Escherichia coli whole-cell bioreporters, Appl. Biol. Chem., 2016, 59(4), 661–668 CrossRef CAS.
- A. L. Kubo, et al., Antimicrobial potency of differently coated 10 and 50 nm silver nanoparticles against clinically relevant bacteria Escherichia coli and Staphylococcus aureus, Colloids Surf., B, 2018, 170, 401–410 CrossRef CAS.
- D. Deryabin, et al., Investigation of copper nanoparticles antibacterial mechanisms tested by luminescent Escherichia coli strains, Nanotechnol. Russ., 2013, 8(5–6), 402–408 CrossRef.
- M. A. Radzig, et al., Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action, Colloids Surf., B, 2013, 102, 300–306 CrossRef CAS.
- K. R. Raghupathi, R. T. Koodali and A. C. Manna, Size-Dependent Bacterial Growth Inhibition and Mechanism of Antibacterial Activity of Zinc Oxide Nanoparticles, Langmuir, 2011, 27(7), 4020–4028 CrossRef CAS.
- N. Singh, K. M. Paknikar and J. Rajwade, RNA-sequencing reveals a multitude of effects of silver nanoparticles on Pseudomonas aeruginosa biofilms, Environ. Sci.: Nano, 2019, 6(6), 1812–1828 RSC.
- B. A. Chambers, et al., Effects of chloride and ionic strength on physical morphology, dissolution, and bacterial toxicity of silver nanoparticles, Environ. Sci. Technol., 2013, 48(1), 761–769 CrossRef.
- C. Fajardo, et al., Impact of Ag and Al2O3 nanoparticles on soil organisms: In vitro and soil experiments, Sci. Total Environ., 2014, 473–474, 254–261 CrossRef CAS.
- N. Gou and A. Z. Gu, A New Transcriptional Effect Level Index (TELI) for Toxicogenomics-based Toxicity Assessment, Environ. Sci. Technol., 2011, 45(12), 5410–5417 CrossRef CAS.
- N. Gou, A. Onnis-Hayden and A. Z. Gu, Mechanistic Toxicity Assessment of Nanomaterials by Whole-Cell-Array Stress Genes Expression Analysis, Environ. Sci. Technol., 2010, 44(15), 5964–5970 CrossRef CAS.
- B. Sohm, et al., Insight into the primary mode of action of TiO2 nanoparticles on Escherichia coli in the dark, Proteomics, 2015, 15(1), 98–113 CrossRef CAS.
- Y. K. Qin, et al., Cytotoxicity of TiO2 nanoparticles toward Escherichia coli in an aquatic environment: effects of nanoparticle structural oxygen deficiency and aqueous salinity, Environ. Sci.: Nano, 2017, 4(5), 1178–1188 RSC.
- D. Soni, et al., Stress response of pseudomonas species to silver nanoparticles at the molecular level, Environ. Toxicol. Chem., 2014, 33(9), 2126–2132 CrossRef CAS.
- A. Kubacka, et al., Understanding the antimicrobial mechanism of TiO2-based nanocomposite films in a pathogenic bacterium, Sci. Rep., 2014, 4, 4134 CrossRef.
- W. Lee, K. J. Kim and D. G. Lee, A novel mechanism for the antibacterial effect of silver nanoparticles on Escherichia coli, BioMetals, 2014, 27(6), 1191–1201 CrossRef CAS.
- R. Yu, et al., Short-term effects of TiO2, CeO2, and ZnO nanoparticles on metabolic activities and gene expression of Nitrosomonas europaea, Chemosphere, 2015, 128, 207–215 CrossRef CAS.
- L. Zhang, et al., Size-dependent cytotoxicity of silver nanoparticles to Azotobacter vinelandii: Growth inhibition, cell injury, oxidative stress and internalization, PLoS One, 2018, 13(12), e0209020 CrossRef CAS.
- W. R. Li, et al., Antibacterial effect of silver nanoparticles on Staphylococcus aureus, BioMetals, 2011, 24(1), 135–141 CrossRef CAS.
- F. Mirzajani, et al., Proteomics study of silver nanoparticles toxicity on Bacillus thuringiensis, Ecotoxicol. Environ. Saf., 2014, 100, 122–130 CrossRef CAS.
- S. Vidovic, et al., ZnO Nanoparticles Impose a Panmetabolic Toxic Effect Along with Strong Necrosis, Inducing Activation of the Envelope Stress Response in Salmonella enterica Serovar Enteritidis, Antimicrob. Agents Chemother., 2015, 59(6), 3317–3328 CrossRef CAS.
- P. Patra, et al., Damage of lipopolysaccharides in outer cell membrane and production of ROS-mediated stress within bacteria makes nano zinc oxide a bactericidal agent, Appl. Nanosci., 2015, 5(7), 857–866 CrossRef CAS.
- Y. G. Yuan, Q. L. Peng and S. Gurunathan, Effects of Silver Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus aureus and Pseudomonas aeruginosa from Mastitis-Infected Goats: An Alternative Approach for Antimicrobial Therapy, Int. J. Mol. Sci., 2017, 18(3), 569 CrossRef.
- A. Alum, A. Alboloushi and M. Abbaszadegan, Copper nanoparticles toxicity: Laboratory strains verses environmental bacterial isolates, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2018, 53(7), 643–650 CrossRef CAS.
- E. Z. Gomaa, Silver nanoparticles as an antimicrobial agent: A case study on Staphylococcus aureus and Escherichia coli as models for Gram-positive and Gram-negative bacteria, J. Gen. Appl. Microbiol., 2017, 63(1), 36–43 CrossRef CAS.
- R. S. Thombre, et al., Antimicrobial Activity and Mechanism of Inhibition of Silver Nanoparticles against Extreme Halophilic Archaea, Front. Microbiol., 2016, 7, 1424 Search PubMed.
- J. G. Clar, et al., Copper nanoparticle induced cytotoxicity to nitrifying bacteria in wastewater treatment: a mechanistic copper speciation study by X-ray absorption spectroscopy, Environ. Sci. Technol., 2016, 50(17), 9105–9113 CrossRef CAS.
- A. Gitipour, et al., Anaerobic toxicity of cationic silver nanoparticles, Sci. Total Environ., 2016, 557, 363–368 CrossRef.
- R. Aminedi, et al., Shape-dependent bactericidal activity of TiO2 for the killing of Gram-negative bacteria Agrobacterium tumefaciens under UV torch irradiation, Environ. Sci. Pollut. Res., 2013, 20(9), 6521–6530 CrossRef CAS.
- C. M. Hessler, et al., The influence of capsular extracellular polymeric substances on the interaction between TiO2 nanoparticles and planktonic bacteria, Water Res., 2012, 46(15), 4687–4696 CrossRef CAS.
- I. M. Sadiq, N. Chandrasekaran and A. Mukherjee, Studies on Effect of TiO2 Nanoparticles on Growth and Membrane Permeability of Escherichia coli, Pseudomonas aeruginosa, and Bacillus subtilis, Curr. Nanosci., 2010, 6(4), 381–387 CrossRef CAS.
- P. Bhadra, et al., Interaction of chitosan capped ZnO nanorods with Escherichia coli, Mater. Sci. Eng., C, 2011, 31(5), 929–937 CrossRef CAS.
- D. Kumar, et al., Qualitative toxicity assessment of silver nanoparticles on the fresh water bacterial isolates and consortium at low level of exposure concentration, Ecotoxicol. Environ. Saf., 2014, 108, 152–160 CrossRef CAS.
- Y. H. Leung, et al., Antibacterial activity of ZnO nanoparticles with a modified surface under ambient illumination, Nanotechnology, 2012, 23(47), 475703 CrossRef CAS.
- B. Ramalingam, T. Parandhaman and S. K. Das, Antibacterial Effects of Biosynthesized Silver Nanoparticles on Surface Ultrastructure and Nanomechanical Properties of Gram-Negative Bacteria viz. Escherichia coli and Pseudomonas aeruginosa, ACS Appl. Mater. Interfaces, 2016, 8(7), 4963–4976 CrossRef CAS.
- S. Dalai, et al., A comparative cytotoxicity study of TiO2 nanoparticles under light and dark conditions at low exposure concentrations, Toxicol. Res., 2012, 1(2), 116–130 CrossRef CAS.
- L. Cui, et al., In Situ Study of the Antibacterial Activity and Mechanism of Action of Silver Nanoparticles by Surface-Enhanced Raman Spectroscopy, Anal. Chem., 2013, 85(11), 5436–5443 CrossRef CAS.
- A. K. Chatterjee, R. Chakraborty and T. Basu, Mechanism of antibacterial activity of copper nanoparticles, Nanotechnology, 2014, 25(13), 135101 CrossRef.
- K. Giannousi, et al., Hydrothermal synthesis of copper based nanoparticles: Antimicrobial screening and interaction with DNA, J. Inorg. Biochem., 2014, 133, 24–32 CrossRef CAS.
- A. Kumar, et al., Engineered ZnO and TiO2 nanoparticles induce oxidative stress and DNA damage leading to reduced viability of Escherichia coli, Free Radical Biol. Med., 2011, 51(10), 1872–1881 CrossRef CAS.
- R. Chakraborty, et al., A simple, fast and cost-effective method of synthesis of cupric oxide nanoparticle with promising antibacterial potency: Unraveling the biological and chemical modes of action, Biochim. Biophys. Acta, Gen. Subj., 2015, 1850(4), 845–856 CrossRef CAS.
- N. Liu, et al., {101}-{001} Surface Heterojunction-Enhanced Antibacterial Activity of Titanium Dioxide Nanocrystals Under Sunlight Irradiation, ACS Appl. Mater. Interfaces, 2017, 9(7), 5907–5915 CrossRef CAS.
- M. A. Quinteros, et al., Oxidative stress generation of silver nanoparticles in three bacterial genera and its relationship with the antimicrobial activity, Toxicol. In Vitro, 2016, 36, 216–223 CrossRef CAS.
- S. B. Subramaniyan, et al., Evaluation of the toxicities of silver and silver sulfide nanoparticles against Gram-positive and Gram-negative bacteria, IET Nanobiotechnol., 2019, 13(3), 326–331 CrossRef.
- S. Swetha, et al., Elucidation of Cell Killing Mechanism by Comparative Analysis of Photoreactions on Different Types of Bacteria, Photochem. Photobiol., 2012, 88(2), 414–422 CrossRef CAS.
- R. K. Dutta, B. P. Nenavathu and M. K. Gangishetty, Correlation between defects in capped ZnO nanoparticles and their antibacterial activity, J. Photochem. Photobiol., B, 2013, 126, 105–111 CrossRef CAS.
- X. C. Lin, et al., Toxicity of TiO2 Nanoparticles to Escherichia coli: Effects of Particle Size, Crystal Phase and Water Chemistry, PLoS One, 2014, 9(10), e110247 CrossRef.
- D. Mandal, et al., Bio-fabricated silver nanoparticles preferentially targets Gram positive depending on cell surface charge, Biomed. Pharmacother., 2016, 83, 548–558 CrossRef CAS.
- W. Liu, et al., TiO2 nanoparticles alter iron homeostasis in Pseudomonas brassicacearum as revealed by PrrF sRNA modulation, Environ. Sci.: Nano, 2016, 3(6), 1473–1482 RSC.
- N. Dasgupta and C. Ramalingam, Silver nanoparticle antimicrobial activity explained by membrane rupture and reactive oxygen generation, Environ. Chem. Lett., 2016, 14(4), 477–485 CrossRef CAS.
- N. Bala, et al., Phenolic compound-mediated single-step fabrication of copper oxide nanoparticles for elucidating their influence on anti-bacterial and catalytic activity, New J. Chem., 2017, 41(11), 4458–4467 RSC.
- V. Gopinath, et al., In vitro toxicity, apoptosis and antimicrobial effects of phyto-mediated copper oxide nanoparticles, RSC Adv., 2016, 6(112), 110986–110995 RSC.
- S. Nair, et al., Role of size scale of ZnO nanoparticles and microparticles on toxicity toward bacteria and osteoblast cancer cells, J. Mater. Sci.: Mater. Med., 2009, 20, 235–241 CrossRef.
- T. O. Okyay, et al., Antibacterial properties and mechanisms of toxicity of sonochemically grown ZnO nanorods, RSC Adv., 2015, 5(4), 2568–2575 RSC.
- M. Raffi, et al., Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli, Ann. Microbiol., 2010, 60(1), 75–80 CrossRef CAS.
- S. Sarwar, et al., The antimicrobial activity of ZnO nanoparticles against Vibrio cholerae: Variation in response depends on biotype, Nanomedicine, 2016, 12(6), 1499–1509 CrossRef CAS.
- S. Sonia, et al., Synthesis of hierarchical CuO nanostructures: Biocompatible antibacterial agents for Gram-positive and Gram-negative bacteria, Curr. Appl. Phys., 2016, 16(8), 914–921 CrossRef.
- C. Wang, et al., Antibacterial effects of zinc oxide nanoparticles on Escherichia coli K88, Afr. J. Biotechnol., 2012, 11(44), 10248–10254 CAS.
- Y. P. Xie, et al., Antibacterial Activity and Mechanism of Action of Zinc Oxide Nanoparticles against Campylobacter jejuni, Appl. Environ. Microbiol., 2011, 77(7), 2325–2331 CrossRef CAS.
- J. Du, et al., Antibacterial activity of a novel Forsythia suspensa fruit mediated green silver nanoparticles against food-borne pathogens and mechanisms investigation, Mater. Sci. Eng., C, 2019, 102, 247–253 CrossRef CAS.
- B. Das, et al., Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage, Arabian J. Chem., 2017, 10(6), 862–876 CrossRef CAS.
- S. Khan, et al., Bacterial cellulose-titanium dioxide nanocomposites: nanostructural characteristics, antibacterial mechanism, and biocompatibility, Cellulose, 2015, 22(1), 565–579 CrossRef CAS.
- N. Kumar, et al., Photocatalytic and antibacterial biomimetic ZnO nanoparticles, Anal. Methods, 2017, 9(33), 4776–4782 RSC.
- R. Lotha, et al., Plant nutraceuticals (Quercetrin and Afzelin) capped silver nanoparticles exert potent antibiofilm effect against food borne pathogen Salmonella enterica serovar Typhi and curtail planktonic growth in zebrafish infection model, Microb. Pathog., 2018, 120, 109–118 CrossRef CAS.
- S. Oliver, et al., Enhancing the antimicrobial and antibiofilm effectiveness of silver nanoparticles prepared by green synthesis, J. Mater. Chem. B, 2018, 6(24), 4124–4138 RSC.
- S. F. Pan, et al., Synthesis of Silver Nanoparticles Embedded Electrospun PAN Nanofiber Thin-Film Composite Forward Osmosis Membrane to Enhance Performance and Antimicrobial Activity, Ind. Eng. Chem. Res., 2019, 58(2), 984–993 CrossRef CAS.
- S. Ranjan and C. Ramalingam, Titanium dioxide nanoparticles induce bacterial membrane rupture by reactive oxygen species generation, Environ. Chem. Lett., 2016, 14(4), 487–494 CrossRef CAS.
- M. A. Rauf, et al., Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals, RSC Adv., 2017, 7(58), 36361–36373 RSC.
- Y. Shao, et al., Green synthesis of sodium alginate-silver nanoparticles and their antibacterial activity, Int. J. Biol. Macromol., 2018, 111, 1281–1292 CrossRef CAS.
- B. Senthil, A. Rajasekar and T. Devasena, Mechanism of Bactericidal Action of Biosynthesized Silver Nanoparticles, Res. J. Biotechnol., 2018, 13(1), 72–78 CAS.
- D. P. Tamboli and D. S. Lee, Mechanistic antimicrobial approach of extracellularly synthesized silver nanoparticles against gram positive and gram negative bacteria, J. Hazard. Mater., 2013, 260, 878–884 CrossRef CAS.
- X. Tian, et al., Bactericidal Effects of Silver Nanoparticles on Lactobacilli and the Underlying Mechanism, ACS Appl. Mater. Interfaces, 2018, 10(10), 8443–8450 CrossRef CAS.
- Z. P. Wang, et al., Anti-microbial activities of aerosolized transition metal oxide nanoparticles, Chemosphere, 2010, 80(5), 525–529 CrossRef CAS.
- K. A. Wong, S. M. Lam and J. C. Sin, Wet chemically synthesized ZnO structures for photodegradation of pre-treated palm oil mill effluent and antibacterial activity, Ceram. Int., 2019, 45(2), 1868–1880 CrossRef CAS.
- J. L. Yi, et al., Effect of polyhexamethylene biguanide functionalized silver nanoparticles on the growth of Staphylococcus aureus, FEMS Microbiol. Lett., 2019, 366(4), fnz036 CrossRef CAS.
- L. L. Zhang, et al., Investigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids), J. Nanopart. Res., 2007, 9(3), 479–489 CrossRef CAS.
- R. Sinha, et al., Interaction and nanotoxic effect of ZnO and Ag nanoparticles on mesophilic and halophilic bacterial cells, Bioresour. Technol., 2011, 102(2), 1516–1520 CrossRef CAS.
- H. T. Li, et al., Transport, fate, and long-term impacts of metal oxide nanoparticles on the stability of an anaerobic methanogenic system with anaerobic granular sludge, Bioresour. Technol., 2017, 234, 448–455 CrossRef CAS.
- P. Cervantes-Aviles and G. Cuevas-Rodriguez, Changes in nutrient removal and flocs characteristics generated by presence of ZnO nanoparticles in activated sludge process, Chemosphere, 2017, 182, 672–680 CrossRef CAS.
- A. Parveen, et al., Emphasized Mechanistic Antimicrobial Study of Biofunctionalized Silver Nanoparticles on Model Proteus mirabilis, J. Drug Delivery, 2018, 3850139 Search PubMed.
- B. Alfaro-Gonzalez, et al., Chitosan-silver nanoparticles as an approach to control bacterial proliferation, spores and antibiotic-resistant bacteria, Biomed. Phys. Eng. Express, 2018, 4(3), 035011 CrossRef.
- C. Z. Wang, C. J. Ehrhardt and V. K. Yadavalli, Nanoscale imaging and hydrophobicity mapping of the antimicrobial effect of copper on bacterial surfaces, Micron, 2016, 88, 16–23 CrossRef CAS.
- A. Maillard, et al., Studies on interaction of green silver nanoparticles with whole bacteria by surface characterization techniques, Biochim. Biophys. Acta, Biomembr., 2019, 1861(6), 1086–1092 CrossRef.
- A. Grigor'eva, et al., Fine mechanisms of the interaction of silver nanoparticles with the cells of Salmonella typhimurium and Staphylococcus aureus, BioMetals, 2013, 26(3), 479–488 CrossRef.
- R. Brayner, et al., Toxicological impact studies based on Escherichia coli bacteria in ultrafine ZnO nanoparticles colloidal medium, Nano Lett., 2006, 6(4), 866–870 CrossRef CAS.
- A. T. Le, et al., Synthesis of oleic acid-stabilized silver nanoparticles and analysis of their antibacterial activity, Mater. Sci. Eng., C, 2010, 30(6), 910–916 CrossRef CAS.
- K. H. Tam, et al., Antibacterial activity of ZnO nanorods prepared by a hydrothermal method, Thin Solid Films, 2008, 516(18), 6167–6174 CrossRef CAS.
- I. Sondi and B. Salopek-Sondi, Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria, J. Colloid Interface Sci., 2004, 275(1), 177–182 CrossRef CAS.
- F. Baldi, et al., Polysaccharide-based silver nanoparticles synthesized by Klebsiella oxytoca DSM 29614 cause DNA fragmentation in E-coli cells, BioMetals, 2016, 29(2), 321–331 CrossRef CAS.
- S. M. N. Gallon, et al., Characterization and study of the antibacterial mechanisms of silver nanoparticles prepared with microalgal exopolysaccharides, Mater. Sci. Eng., C, 2019, 99, 685–695 CrossRef.
- S. N. Hazarika, et al., One-pot facile green synthesis of biocidal silver nanoparticles, Mater. Res. Express, 2016, 3(7), 075401 CrossRef.
- F. Mirzajani, et al., Effect of silver nanoparticles on Oryza sativa L. and its rhizosphere bacteria, Ecotoxicol. Environ. Saf., 2013, 88, 48–54 CrossRef CAS.
- F. Mirzajani, et al., Antibacterial effect of silver nanoparticles on Staphylococcus aureus, Res. Microbiol., 2011, 162(5), 542–549 CrossRef CAS.
- Z. H. Ni, et al., Synthesis of poly acrylic acid modified silver nanoparticles and their antimicrobial activities, Mater. Sci. Eng., C, 2014, 41, 249–254 CrossRef CAS.
- S. F. C. Orou, et al., Antibacterial activity by ZnO nanorods and ZnO nanodisks: A model used to illustrate "Nanotoxicity Threshold", J. Ind. Eng. Chem., 2018, 62, 333–340 CrossRef CAS.
- L. Rastogi and J. Arunachalam, Synthesis and characterization of bovine serum albumin-copper nanocomposites for antibacterial applications, Colloids Surf., B, 2013, 108, 134–141 CrossRef CAS.
- S. Soman and J. G. Ray, Silver nanoparticles synthesized using aqueous leaf extract of Ziziphus oenoplia (L.) Mill: Characterization and assessment of antibacterial activity, J. Photochem. Photobiol., B, 2016, 163, 391–402 CrossRef CAS.
- R. Thomas, et al., Microbially and phytofabricated AgNPs with different mode of bactericidal action were identified to have comparable potential for surface fabrication of central venous catheters to combat Staphylococcus aureus biofilm, J. Photochem. Photobiol., B, 2017, 171, 96–103 CrossRef CAS.
- D. G. Romero-Urbina, et al., Ultrastructural changes in methicillin-resistant Staphylococcus aureus induced by positively charged silver nanoparticles, Beilstein J. Nanotechnol., 2015, 6, 2396–2405 CrossRef CAS.
- P. Siritongsuk, et al., Two-Phase Bactericidal Mechanism of Silver Nanoparticles against Burkholderia pseudomallei, PLoS One, 2016, 11(12), e0168098 CrossRef.
- U. K. Parashar, et al., Study of mechanism of enhanced antibacterial activity by green synthesis of silver nanoparticles, Nanotechnology, 2011, 22(41), 415104 CrossRef.
- F. Fang, et al., A detailed approach to study the antibacterial mechanisms of nanostructure, Appl. Surf. Sci., 2012, 258(10), 4397–4401 CrossRef CAS.
- R. K. Bera, S. M. Mandal and C. R. Raj, Antimicrobial activity of fluorescent Ag nanoparticles, Lett. Appl. Microbiol., 2014, 58(6), 520–526 CrossRef CAS.
- A. Mathur, et al., Decreased Phototoxic Effects of TiO2 Nanoparticles in Consortium of Bacterial Isolates from Domestic Waste Water, PLoS One, 2015, 10(10), e0141301 CrossRef.
- A. Ravichandran, et al., Phyto-mediated synthesis of silver nanoparticles using fucoidan isolated from Spatoglossum asperum and assessment of antibacterial activities, J. Photochem. Photobiol., B, 2018, 185, 117–125 CrossRef CAS.
- E. McGivney, et al., Disruption of Autolysis in Bacillus subtilis using TiO2 Nanoparticles, Sci. Rep., 2017, 7, 44308 CrossRef CAS.
- M. Ramani, et al., Amino acid-mediated synthesis of zinc oxide nanostructures and evaluation of their facet-dependent antimicrobial activity, Colloids Surf., B, 2014, 117, 233–239 CrossRef CAS.
- M. Singh, et al., Loss of outer membrane integrity in Gram-negative bacteria by silver nanoparticles loaded with Camellia sinensis leaf phytochemicals: plausible mechanism of bacterial cell disintegration, Bull. Mater. Sci., 2016, 39(7), 1871–1878 CrossRef CAS.
- A. U. Khan, et al., Enzymatic browning reduction in white cabbage, potent antibacterial and antioxidant activities of biogenic silver nanoparticles, J. Mol. Liq., 2016, 215, 39–46 CrossRef CAS.
- V. Tiwari, et al., Mechanism of Anti-bacterial Activity of Zinc Oxide Nanoparticle Against Carbapenem-Resistant Acinetobacter baumannii, Front. Microbiol., 2018, 9, 1218 CrossRef.
- J. Du, et al., Biosynthesis of large-sized silver nanoparticles using Angelica keiskei extract and its antibacterial activity and mechanisms investigation, Microchem. J., 2019, 147, 333–338 CrossRef CAS.
- R. Varghese, et al., Silver nanopaticles synthesized using the seed extract of Trigonella foenum-graecum L. and their antimicrobial mechanism and anticancer properties, Saudi J. Biol. Sci., 2019, 26(1), 148–154 CrossRef CAS.
- G. D. Venkatasubbu, et al., Toxicity mechanism of titanium dioxide and zinc oxide nanoparticles against food pathogens, Colloids Surf., B, 2016, 148, 600–606 CrossRef CAS.
- S. Qayyum and A. U. Khan, Biofabrication of broad range antibacterial and antibiofilm silver nanoparticles, IET Nanobiotechnol., 2016, 10(5), 349–357 CrossRef.
- S. Rajesh, V. Dharanishanthi and A. V. Kanna, Antibacterial mechanism of biogenic silver nanoparticles of Lactobacillus acidophilus, J. Exp. Nanosci., 2015, 10(15), 1143–1152 CrossRef CAS.
- A. Rautela, J. Rani and M. Debnath, Green synthesis of silver nanoparticles from Tectona grandis seeds extract: characterization and mechanism of antimicrobial action on different microorganisms, J. Anal. Sci. Technol., 2019, 10, 5 CrossRef.
- Q. Cai, et al., Insight into Biological Effects of Zinc Oxide Nanoflowers on Bacteria: Why Morphology Matters, ACS Appl. Mater. Interfaces, 2016, 8(16), 10109–10120 CrossRef CAS.
- S. T. Khan, et al., Synthesis and characterization of some abundant nanoparticles, their antimicrobial and enzyme inhibition activity, Acta Microbiol. Immunol. Hung., 2017, 64(2), 203–216 CrossRef CAS.
- S. Baek, et al., Effects of coating materials on antibacterial properties of industrial and sunscreen-derived titanium-dioxide nanoparticles on Escherichia coli, Chemosphere, 2018, 208, 196–206 CrossRef CAS.
- N. Jain, et al., Removal of Protein Capping Enhances the Antibacterial Efficiency of Biosynthesized Silver Nanoparticles, PLoS One, 2015, 10(7), e0134337 CrossRef.
- J. C. Jin, et al., Ultrasmall silver nanoclusters: Highly efficient antibacterial activity and their mechanisms, Biomater. Sci., 2017, 5(2), 247–257 RSC.
- X. Z. Li, et al., Riboflavin-protected ultrasmall silver nanoclusters with enhanced antibacterial activity and the mechanisms, RSC Adv., 2019, 9(23), 13275–13282 RSC.
- N. Tripathy, et al., Outstanding Antibiofilm Features of Quanta-CuO Film on Glass Surface, ACS Appl. Mater. Interfaces, 2016, 8(24), 15128–15137 CrossRef CAS.
- V. L. Prasanna and R. Vijayaraghavan, Chemical manipulation of oxygen vacancy and antibacterial activity in ZnO, Mater. Sci. Eng., C, 2017, 77, 1027–1034 CrossRef.
- E. K. Fauss, et al., Disinfection action of electrostatic versus steric-stabilized silver nanoparticles on E. coli under different water chemistries, Colloids Surf., B, 2014, 113, 77–84 CrossRef CAS.
- S. P. Liu, et al., The influence of H2O2 on the antibacterial activity of ZnO, Mater. Res. Express, 2019, 6(8), 0850c6 CrossRef CAS.
- X. Jin, et al., High-Throughput Screening of Silver Nanoparticle Stability and Bacterial Inactivation in Aquatic Media: Influence of Specific Ions, Environ. Sci. Technol., 2010, 44(19), 7321–7328 CrossRef CAS.
- S. S. D. Kumar, et al., Cellular imaging and bactericidal mechanism of green-synthesized silver nanoparticles against human pathogenic bacteria, J. Photochem. Photobiol., B, 2018, 178, 259–269 CrossRef CAS.
- H. J. Bao, et al., New Toxicity Mechanism of Silver Nanoparticles: Promoting Apoptosis and Inhibiting Proliferation, PLoS One, 2015, 10(3), e0122535 CrossRef.
- Y. H. Ng, et al., Antibacterial activity of ZnO nanoparticles under ambient illumination - The effect of nanoparticle properties, Thin Solid Films, 2013, 542, 368–372 CrossRef CAS.
- I. Lopes, et al., Toxicity and genotoxicity of organic and inorganic nanoparticles to the bacteria Vibrio fischeri and Salmonella typhimurium, Ecotoxicology, 2012, 21(3), 637–648 CrossRef CAS.
- K. S. Butler, et al., Silver nanoparticles: correlating nanoparticle size and cellular uptake with genotoxicity, Mutagenesis, 2015, 30(4), 577–591 CrossRef CAS.
- L. A. Tamayo, et al., Release of silver and copper nanoparticles from polyethylene nanocomposites and their penetration into Listeria monocytogenes, Mater. Sci. Eng., C, 2014, 40, 24–31 CrossRef CAS.
- E. Amato, et al., Synthesis, Characterization and Antibacterial Activity against Gram Positive and Gram Negative Bacteria of Biomimetically Coated Silver Nanoparticles, Langmuir, 2011, 27(15), 9165–9173 CrossRef CAS.
- A. Ananth, et al., Soft jet plasma-assisted synthesis of Zinc oxide nanomaterials: Morphology controls and antibacterial activity of ZnO, Chem. Eng. J., 2017, 322, 742–751 CrossRef CAS.
- J. Y. Cheon, et al., Shape-dependent antimicrobial activities of silver nanoparticles, Int. J. Nanomed., 2019, 14, 2773–2780 CrossRef CAS.
- E. de Lucas-Gil, et al., The fight against multidrug-resistant organisms: The role of ZnO crystalline defects, Mater. Sci. Eng., C, 2019, 99, 575–581 CrossRef CAS.
- E. de Lucas-Gil, et al., ZnO Nanoporous Spheres with Broad-Spectrum Antimicrobial Activity by Physicochemical Interactions, ACS Appl. Nano Mater., 2018, 1(7), 3214–3225 CrossRef CAS.
- H. R. Kim, et al., Direct synthesis of magnetron sputtered nanostructured Cu films with desired properties via plasma chemistry for their efficient antibacterial application, Plasma Processes Polym., 2018, 15(9), 1800009 CrossRef.
- M. H. Li, et al., Stability, Bioavailability, and Bacterial Toxicity of ZnO and Iron-Doped ZnO Nanoparticles in Aquatic Media, Environ. Sci. Technol., 2011, 45(2), 755–761 CrossRef CAS.
- A. Kanwal, et al., Size-dependent inhibition of bacterial growth by chemically engineered spherical ZnO nanoparticles, J. Biol. Phys., 2019, 45(2), 147–159 CrossRef.
- A. Ronen, R. Semiat and C. G. Dosoretz, Antibacterial efficiency of composite nano-ZnO in biofilm development in flow-through systems, Desalin. Water Treat., 2013, 51(4–6), 988–996 CrossRef CAS.
- D. Wang, et al., Quantitative Analysis of Reactive Oxygen Species Photogenerated on Metal Oxide Nanoparticles and Their Bacteria Toxicity: The Role of Superoxide Radicals, Environ. Sci. Technol., 2017, 51(17), 10137–10145 CrossRef CAS.
- T. Yang, et al., Tuning crystallization and morphology of zinc oxide with polyvinylpyrrolidone: Formation mechanisms and antimicrobial activity, J. Colloid Interface Sci., 2019, 546, 43–52 CrossRef CAS.
- R. K. Sharma, M. Agarwal and K. Balani, Effect of ZnO morphology on affecting bactericidal property of ultra high molecular weight polyethylene biocomposite, Mater. Sci. Eng., C, 2016, 62, 843–851 CrossRef CAS.
- D. C. Tien, et al., Colloidal silver fabrication using the spark discharge system and its antimicrobial effect on Staphylococcus aureus, Med. Eng. Phys., 2008, 30(8), 948–952 CrossRef.
- M. Ramani, S. Ponnusamy and C. Muthamizhchelvan, Preliminary investigations on the antibacterial activity of zinc oxide nanostructures, J. Nanopart. Res., 2013, 15(4), 1557 CrossRef.
- L. C. Ann, et al., Antibacterial responses of zinc oxide structures against Staphylococcus aureus, Pseudomonas aeruginosa and Streptococcus pyogenes, Ceram. Int., 2014, 40(2), 2993–3001 CrossRef CAS.
- A. J. Kora, R. B. Sashidhar and J. Arunachalam, Gum kondagogu (Cochlospermum gossypium): A template for the green synthesis and stabilization of silver nanoparticles with antibacterial application, Carbohydr. Polym., 2010, 82(3), 670–679 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0en00949k |
| This journal is © The Royal Society of Chemistry 2021 |