Open Access Article
Open Access ArticleA dithiacyclam-coordinated silver(I) polymer with anti-cancer stem cell activity†
Alice
Johnson
 *a,
Linda
Iffland
b,
Kuldip
Singh
a,
Ulf-Peter
Apfel
*a,
Linda
Iffland
b,
Kuldip
Singh
a,
Ulf-Peter
Apfel
 *bc and
Kogularamanan
Suntharalingam
*bc and
Kogularamanan
Suntharalingam
 *a
*a
aSchool of Chemistry, University of Leicester, Leicester, LE1 7RH, UK. E-mail: k.suntharalingam@leicester.ac.uk; alice.johnson@leicester.ac.uk
bRuhr-Universität Bochum, Anorganische Chemie I, Universitätsstraße 150, 44801 Bochum, Germany. E-mail: ulf.apfel@rub.de
cFraunhofer UMSICHT, Osterfelder Str. 3, 46047 Oberhausen, Germany
First published on 14th April 2021
Abstract
A cancer stem cell (CSC) active, solution stable, silver(I) polymeric complex bearing a dithiacyclam ligand is reported. The complex displays similar potency towards CSCs to salinomycin in monolayer and three-dimensional cultures. Mechanistic studies suggest CSC death results from cytosol entry, an increase in intracellular reactive oxygen species, and caspase-dependent apoptosis.
Cancer stem cells (CSCs) are a small sub-population of highly resistant tumour cells found within solid and liquid tumours.1 Conventional cancer therapies target rapidly proliferating bulk cancer cells and consequently are often ineffective towards slow growing CSCs, which share many characteristics with normal stem cells.2,3 CSCs have the ability to self-renew, differentiate, and form secondary or tertiary tumours, leading to cancer relapse which is currently one of the leading causes of cancer-related deaths worldwide.4,5 An increasing number of drug development and oncology research programmes are focused on identifying chemical agents and biologics which can specifically target and remove CSCs.6 Despite these efforts, to date, no anti-CSC agent has been approved for clinical use. Many of the compounds undergoing clinical trials and preclinical development as anti-CSC agents are organic small molecules. We and others have shown that metal complexes also possess promising anti-CSC properties.7 Indeed, transition metal complexes offer distinct chemical and physical properties that can be exploited to develop effective anti-CSC agents.8
Silver plays no known biological role, however, the body can tolerate low doses of silver without any toxic side effects.9,10 Despite a long history in antibacterial research,11 the application of silver(I) complexes as anticancer agents is relatively underexplored (and the mechanism of action of several cytotoxic silver(I) complexes has not been fully elucidated).12 Nevertheless, a structurally diverse range of silver(I) compounds with carboxylic acids, amino acids, nitrogen, phosphorus and sulphur donor ligands have been studied as antitumour agents.12–15 Silver(I) complexes can suffer from poor light stability and aqueous solubility, hence the careful choice of ligands is mandatory to prepare complexes with biologically compatible properties. Of note, certain silver(I) complexes with diphosphine and N-heterocyclic carbene ligands have been reported to exhibit promising in vivo antitumour activity in mice bearing leukaemia, reticulum cell sarcoma, and ovarian cancer.16,17 Although initial reports suggested that strong σ-donor ligands were required to elicit a robust anticancer effect, silver(I) complexes featuring more weakly coordinating nitrogen and sulphur donor (mixed) ligands have subsequently been reported with reasonable in vitro bulk cancer cell potency.18–20 Within this sub-class of compounds, two water soluble silver(I) complexes containing 2,2′-bipyridine and 4,6-diamino-5-hydroxy-2-mercaptopyrimidine or 2-amino-4,5,6,7-tetrahydro-7-oxo-benzo[b]thiophene-3-carbonitrile are the only examples to have been tested in vivo.21,22 Administration of the complexes (0.01 mg per mice per day) to mice bearing Ehrlich ascites tumours resulted in a 21–24% increase in lifespan and a reduction in tumour size from 220.0 to 30.4–35.1 × 106 cells per cm3 compared with the untreated control group.21,22 Despite the existing body of work on the anticancer properties of silver(I) complexes, their impact on CSCs of any tissue type is untested. It should be noted that silver nanoparticles have been reported to show significant cytotoxic potential against ovarian and myeloma CSCs, as well as against the drug resistant breast cancer cell lines MCF7 and MDA-MB-231, which have appreciable breast CSC populations.23–25
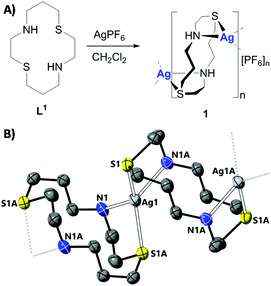
When coordinated to tetra-aza macrocycles, silver(I) is known to undergo rapid disproportionation.26,27 However when chelated to N2S2-donor macrocycles redox stable silver(I) complexes can be achieved, as a result of the strong affinity of the soft thioether donor for silver(I) ions.28–30 Despite their simple synthesis and high stability, no silver(I) complexes containing N2S2-donor macrocyclic ligands have been challenged with bulk cancer cells, let alone CSCs of any tissue type. Here we report an air, light, and solution stable silver(I) 1,8-dithia-4,11-diazacyclotetradecane polymeric complex, 1 (Fig. 1A) and its anti-breast CSC properties in monolayer and three-dimensional cell culture systems. Insight into the likely mechanism of action of 1 is also provided.
The silver(I) complex 1 was prepared as outlined in Fig. 1A. Reaction of 1,8-dithia-4,11-diazacyclotetradecane (dithiacyclam) L1,31 with a stoichiometric amount of AgPF6 in dichloromethane led to the formation of 1, which was isolated in a good yield (72%) as a pale-yellow solid (Fig. 1A). The silver(I) complex 1 was fully characterised by 1H, 13C, 31P{1H}, 19F{1H} NMR spectroscopy, high-resolution ESI-QTOF mass spectrometry, and elemental analysis (Fig. S1–S6 and see ESI†). Crystals of 1 suitable for X-ray diffraction were grown by the slow diffusion of pentane into a dichloromethane solution of 1 (CCDC 2053375, Fig. 1B, Tables S1 and S2†). The silver(I) complex 1 has a 1D polymeric structure with each silver(I) centre bridging two dithiacyclam ligands (L1) as depicted in Fig. 1B and S7.† The metal centre adopts a distorted tetrahedral coordination environment, with the silver(I) ion bound to one sulphur and one nitrogen atom from two separate L1 molecules. The Ag–N (2.394(5) Å) and Ag–S (2.5355(19) Å) bond lengths are consistent with bond parameters reported for a related silver(I) complex.29 Upon exposure of the solid form of 1 to air and light for 7 months, the 1H NMR spectrum of the solid (in DMSO-d6) remained unaltered. This suggests that 1 is stable in air and light, in the solid form, over long periods of time (Fig. S8†).
The lipophilicity of 1 was determined by the extent to which it partitioned between octanol and water, P. The experimentally determined log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value for 1 was −0.61 ± 0.03, indicative of amphiphilicity. This suggests that 1 should display reasonable water solubility and be readily internalised by cells. Time course 1H NMR spectroscopy and ESI mass spectrometry studies were carried out to assess the stability of 1 in solution. There was no observable change in the 1H NMR spectra of 1 in DMSO-d6 or D2O
P value for 1 was −0.61 ± 0.03, indicative of amphiphilicity. This suggests that 1 should display reasonable water solubility and be readily internalised by cells. Time course 1H NMR spectroscopy and ESI mass spectrometry studies were carried out to assess the stability of 1 in solution. There was no observable change in the 1H NMR spectra of 1 in DMSO-d6 or D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO-d6 (5
DMSO-d6 (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) over the course of 72 h at 37 °C, suggestive of solution stability (Fig. S9 and S10†). In H2O
1) over the course of 72 h at 37 °C, suggestive of solution stability (Fig. S9 and S10†). In H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (100
DMSO (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the ESI mass spectra (positive mode) of 1 (100 μM) exhibited a distinctive peak corresponding to the intact complex, with the expected isotopic pattern, throughout the course of 72 h at 37 °C (m/z = 343 a.m.u., [1–PF6]+) (Fig. S11–S14†), with no observable speciation. Taken together, the NMR spectroscopy and ESI mass spectrometry studies clearly show that 1 is stable in solution.
1), the ESI mass spectra (positive mode) of 1 (100 μM) exhibited a distinctive peak corresponding to the intact complex, with the expected isotopic pattern, throughout the course of 72 h at 37 °C (m/z = 343 a.m.u., [1–PF6]+) (Fig. S11–S14†), with no observable speciation. Taken together, the NMR spectroscopy and ESI mass spectrometry studies clearly show that 1 is stable in solution.
The monolayer cytotoxicity of 1 against bulk breast cancer cells (HMLER) and breast CSC-enriched cells (HMLER-shEcad) was determined using the colorimetric MTT assay. The corresponding IC50 values, determined by plotting dose–response curves (Fig. S15 and S16†), are shown in Table 1. The silver(I) complex 1 displayed similar potency towards HMLER and HMLER-shEcad cells, within the micromolar range (Table 1). It is worth noting that the cytotoxicity of 1 towards breast CSC-enriched HMLER-shEcad cells is comparable to, or greater than, salinomycin (an established breast CSC-potent agent) and cisplatin (a clinically approved anticancer drug).32,33 Furthermore the potency of 1 towards bulk breast cancer cells (HMLER) is comparable to the potency of other silver(I) complexes towards related breast cancer cell lines (such as MCF7, MDA-MB-231, T-47D).12,15,17,34 Control studies showed that L1 is non-toxic towards HMLER or HMLER-shEcad cells (IC50 value >100 μM), while AgPF6 displayed ≈4-fold lower potency compared to 1 against HMLER or HMLER-shEcad cells (Fig. S17–S20† and Table 1). When dosed as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture, the combined treatment of L1 and AgPF6 showed a >4-fold reduction in potency towards breast CSC-enriched HMLER-shEcad cells compared to 1 (Fig. S21† and Table 1). This demonstrates that the preformed complex 1 is significantly (p < 0.05) better at killing breast CSCs than a mixture of its individual components.
1 mixture, the combined treatment of L1 and AgPF6 showed a >4-fold reduction in potency towards breast CSC-enriched HMLER-shEcad cells compared to 1 (Fig. S21† and Table 1). This demonstrates that the preformed complex 1 is significantly (p < 0.05) better at killing breast CSCs than a mixture of its individual components.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of L1 and AgPF6, cisplatin, and salinomycin against HMLER and HMLER-shEcad cells determined after 72 h incubation (mean of three independent experiments ± SD). n.d. not determined
1 mixture of L1 and AgPF6, cisplatin, and salinomycin against HMLER and HMLER-shEcad cells determined after 72 h incubation (mean of three independent experiments ± SD). n.d. not determined
As a measure of therapeutic potential, the cytotoxicity of 1 towards a panel non-cancerous cell lines; BEAS-2B (bronchial epithelium), MCF10A (epithelial breast), and HEK 293 (embryonic kidney) cells was determined. The complex, 1 was less potent toward BEAS-2B (IC50 value = 8.66 ± 0.48 μM, p < 0.05, Fig. S22†), MCF10A (IC50 value = 10.12 ± 0.74 μM, p < 0.05, Fig. S23†), and HEK 293 (IC50 value = 34.31 ± 0.10 μM, p < 0.05, Fig. S24†) cells than HMLER and HMLER-shEcad cells, therefore 1 has the potential to potently kill breast CSCs and bulk breast cancer cells over non-cancerous cells derived from various tissue types.
When grown under serum-free, low-attachment conditions, HMLER-shEcad cells can form three-dimensional spheroids called mammospheres, providing a more representative model of tumours than monolayer cell cultures.35 The inhibitory effect of 1 on the formation of HMLER-shEcad mammospheres was probed using an inverted microscope. Addition of 1 at 2 μM (for 5 days) to single cell suspensions of HMLER-shEcad cells did not significantly affect the number of mammospheres formed compared to the untreated control, while addition of 1 at 8 μM led to a 27% reduction in the number of mammospheres formed (Fig. 2A). Addition of salinomycin at 2 μM (for 5 days) led to an 82% decrease in the number of mammospheres formed (Fig. 2A). Despite 1 displaying a lower mammosphere inhibitory effect than salinomycin, 1 (at the IC20 value for 5 days) did reduce the size of mammospheres formed to a similar extent as salinomycin when compared to the untreated control (Fig. 2B). To determine the ability of 1 to reduce mammosphere viability, the resazurin-based TOX8 colorimetric reagent was used. The IC50 value of 1, the concentration required to reduce mammosphere viability by 50%, was determined from a dose–response curve (Fig. S25†). The mammosphere potency of 1 (IC50 value = 12.95 ± 1.35 μM) was greater than salinomycin (IC50 value = 18.50 ± 1.50 μM) and comparable to cisplatin (IC50 value = 13.50 ± 2.34 μM) under identical conditions.33,36 Unsurprisingly, L1 was non-toxic towards mammospheres (IC50 > 133 μM, Fig. S26†). This shows that the silver(I) ion in 1 is a major determinant of mammosphere toxicity.
To shed light on the mechanism of toxicity of 1 further cell-based studies were conducted. Cellular uptake studies were carried out to determine the breast CSC permeability of 1. HMLER-shEcad cells were incubated with 1 (5 μM for 24 h) and the intracellular silver content was determined by inductively coupled plasma mass spectrometry (ICP-MS) (Fig. 3). Identical studies were also carried out with AgPF6 (5 μM for 24 h). The results showed that the silver content in HMLER-shEcad cells incubated with 1 was 364.7 ng per 106 cells, approximately 5 times higher than the silver content in HMLER-shEcad cells incubated with AgPF6 (75.6 ng per 106 cells) (Fig. 3). This shows that complexation of silver to L1 improves breast CSC internalisation. Fractionation studies were carried out with HMLER-shEcad cells incubated with 1 (5 μM, 24 h) to determine the localisation of 1 within breast CSCs. Significant amounts of internalised silver were detected in the cytoplasm (55%) and cell membrane (38%), with minimal quantities detected in the nucleus (Fig. 3). This implies that the intracellular target for 1 in breast CSCs is unlikely to be genomic DNA, which is found in the nucleus, and more likely to be biomolecules within the cytoplasm.
 | ||
| Fig. 3 Silver content (ng of Ag per 106 cells) in various cellular components upon treatment of HMLER-shEcad cells with 1 or AgPF6 (5 μM for 24 h). | ||
Given that the biological activity of many silver(I) complexes is associated to their interaction with thiol (sulfhydryl) groups in proteins,37 the reaction of 1 with thiol-containing biomolecules was probed using 1H NMR and high-resolution ESI-QTOF-MS studies (over 24 h at 37 °C). In DMSO-d6 or DMSO-d6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D2O (1
D2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the addition of 1 (10 mM) to a stoichiometric amount of cysteine (Cys), N-acetylcysteine (NAC), or glutathione (GSH) led to the rapid precipitation of a white solid in all cases. The 1H NMR spectrum of the resultant solution in each case revealed the presence of the mono-protonated analogue of L1, L1–H+ only (Fig. S27–S29,† see Fig. S30† for chemical structure of L1–H+). L1–H+ was independently prepared in situ by reacting L1 (10 mM) with one equivalent of HCl and characterised by 1H NMR spectroscopy (in DMSO-d6 and DMSO-d6
1), the addition of 1 (10 mM) to a stoichiometric amount of cysteine (Cys), N-acetylcysteine (NAC), or glutathione (GSH) led to the rapid precipitation of a white solid in all cases. The 1H NMR spectrum of the resultant solution in each case revealed the presence of the mono-protonated analogue of L1, L1–H+ only (Fig. S27–S29,† see Fig. S30† for chemical structure of L1–H+). L1–H+ was independently prepared in situ by reacting L1 (10 mM) with one equivalent of HCl and characterised by 1H NMR spectroscopy (in DMSO-d6 and DMSO-d6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D2O (1
D2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)) to confirm the abovementioned assignment (Fig. S31 and S32†). The ESI-QTOF-MS spectra of the 1
1)) to confirm the abovementioned assignment (Fig. S31 and S32†). The ESI-QTOF-MS spectra of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys/NAC/GSH reaction solutions (after 24 h at 37 °C) all displayed a single peak corresponding to L1–H+ (m/z = 235.1299 a.m.u. for the Cys reaction, 235.1303 a.m.u. for the NAC reaction, and 235.1303 a.m.u. for the GSH reaction) (Fig. S33–S35†), confirming that the major component in solution was L1–H+. Due to the insolubility of the precipitate formed during the reactions, it could not be directly characterised by 1H NMR or ESI-QTOF-MS. Instead, ICP-MS studies were performed to determine the proportion of silver present in the precipitate (and reaction solution). ICP-MS analysis indicated that after the reaction of 1 with Cys, NAC, or GSH (24 h at 37 °C) the majority of the silver content was contained in the precipitate, with only small amounts of silver detected in solution (<1% total silver in solution for the reactions with Cys and GSH, and 17% total silver in solution for the reaction with NAC) (Tables S3–S5†). This is consistent with previous reports that show silver(I) salts tend to react with thiol-containing biomolecules to form (poorly soluble) extended polymeric networks.38 Taken together, the 1H NMR, ESI-QTOF-MS, and ICP-MS studies suggest that 1 reacts with Cys, NAC, and GSH to give L1–H+ which remains in solution, and an insoluble silver-rich precipitate which is most likely a silver-biomolecule polymer (Scheme S1†).
Cys/NAC/GSH reaction solutions (after 24 h at 37 °C) all displayed a single peak corresponding to L1–H+ (m/z = 235.1299 a.m.u. for the Cys reaction, 235.1303 a.m.u. for the NAC reaction, and 235.1303 a.m.u. for the GSH reaction) (Fig. S33–S35†), confirming that the major component in solution was L1–H+. Due to the insolubility of the precipitate formed during the reactions, it could not be directly characterised by 1H NMR or ESI-QTOF-MS. Instead, ICP-MS studies were performed to determine the proportion of silver present in the precipitate (and reaction solution). ICP-MS analysis indicated that after the reaction of 1 with Cys, NAC, or GSH (24 h at 37 °C) the majority of the silver content was contained in the precipitate, with only small amounts of silver detected in solution (<1% total silver in solution for the reactions with Cys and GSH, and 17% total silver in solution for the reaction with NAC) (Tables S3–S5†). This is consistent with previous reports that show silver(I) salts tend to react with thiol-containing biomolecules to form (poorly soluble) extended polymeric networks.38 Taken together, the 1H NMR, ESI-QTOF-MS, and ICP-MS studies suggest that 1 reacts with Cys, NAC, and GSH to give L1–H+ which remains in solution, and an insoluble silver-rich precipitate which is most likely a silver-biomolecule polymer (Scheme S1†).
As 1 readily reacts with GSH (Fig. S29, S35 and Table S5†) and enters the cytoplasm of breast CSCs (Fig. 3) where GSH is predominately localised, 1 could potentially perturb the GSH redox buffering system in breast CSCs and induce intracellular reactive oxygen species (ROS) elevation.39 The ability of 1 to increase ROS levels in HMLER-shEcad cells over a 24 h period was determined using 2′,7′-dichlorodihydro-fluorescein diacetate (DCFH-DA), a well-established ROS indicator. HMLER-shEcad cells treated with 1 (4 μM) exhibited a substantial increase in intracellular ROS levels after 6 h exposure (81% increase, p < 0.05) (Fig. 4A). Such an increase in intracellular ROS levels can induce apoptosis.40 Immunoblotting studies showed that HMLER-shEcad cells treated with 1 (4–8 μM) for 72 h displayed a marked increase in expression of cleaved caspase-3 and -7, and PARP-1, compared to untreated cells (Fig. 4B). This suggests that 1 induces caspase-dependent apoptosis in breast CSC-enriched HMLER-shEcad cells. To further corroborate this, cytotoxicity studies were carried out in the presence of z-VAD-FMK (5 μM), a peptide-based caspase-dependent apoptosis inhibitor. The IC50 value of 1 towards HMLER-shEcad cells increased significantly in the presence of z-VAD-FMK (IC50 value = 7.45 ± 0.74 μM, p < 0.05, Fig. S36†) further confirming that 1 induces caspase-dependent apoptosis in breast CSCs.
In summary, we report an air, light, and solution stable macrocyclic silver(I) complex with a polymeric structure, 1. To the best of our knowledge complex 1 is the first silver(I) complex to be investigated for its anti-CSC properties. The silver(I) complex 1 displays greater or comparable breast CSC potency to salinomycin and cisplatin in monolayer breast CSC and three-dimensional mammosphere cultures. Biophysical studies suggest that 1 rapidly reacts with thiol-containing biomolecules. Cell-based mechanistic studies indicate that 1 readily enters breast CSCs, localises in the cytoplasm (and cell membrane), increases intracellular ROS levels, and induces caspase-dependent apoptosis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
K. S. is supported by an EPSRC New Investigator Award (EP/S005544/1). U.-P. A. is supported by the DFG (AP242/5-1) and Fraunhofer Internal Programs (Attract 097-602175).Notes and references
- L. V. Nguyen, R. Vanner, P. Dirks and C. J. Eaves, Nat. Rev. Cancer, 2012, 12, 133–143 CrossRef CAS PubMed.
- L. N. Abdullah and E. K. Chow, Clin. Transl. Med., 2013, 2, 3 Search PubMed.
- K. Rycaj and D. G. Tang, Int. J. Radiat. Biol., 2014, 90, 615–621 CrossRef CAS PubMed.
- J. Marx, Science, 2007, 317, 1029–1031 CrossRef CAS PubMed.
- D. R. Pattabiraman and R. A. Weinberg, Nat. Rev. Drug Discovery, 2014, 13, 497–512 CrossRef CAS PubMed.
- J. Kaiser, Science, 2015, 347, 226–229 CrossRef CAS PubMed.
- K. Laws and K. Suntharalingam, ChemBioChem, 2018, 19, 2246–2253 CrossRef CAS PubMed.
- A. Johnson, J. Northcote-Smith and K. Suntharalingam, Trends Chem., 2021, 3, 47–58 CrossRef CAS.
- G. Drasch, H. J. Gath, E. Heissler, I. Schupp and G. Roider, J. Trace Elem. Med. Biol., 1995, 9, 82–87 CrossRef CAS.
- N. Hadrup and H. R. Lam, Regul. Toxicol. Pharmacol., 2014, 68, 1–7 CrossRef CAS PubMed.
- J. S. Mohler, W. Sim, M. A. T. Blaskovich, M. A. Cooper and Z. M. Ziora, Biotechnol. Adv., 2018, 36, 1391–1411 CrossRef PubMed.
- C. N. Banti and S. K. Hadjikakou, Metallomics, 2013, 5, 569–596 CrossRef CAS PubMed.
- W. Liu and R. Gust, Chem. Soc. Rev., 2013, 42, 755–773 RSC.
- A. Gautier and F. Cisnetti, Metallomics, 2012, 4, 23–32 CrossRef CAS PubMed.
- X. Liang, S. Luan, Z. Yin, M. He, C. He, L. Yin, Y. Zou, Z. Yuan, L. Li, X. Song, C. Lv and W. Zhang, Eur. J. Med. Chem., 2018, 157, 62–80 CrossRef CAS PubMed.
- S. J. Berners-Price, R. K. Johnson, A. J. Giovenella, L. F. Faucette, C. K. Mirabelli and P. J. Sadler, J. Inorg. Biochem., 1988, 33, 285–295 CrossRef CAS PubMed.
- D. A. Medvetz, K. M. Hindi, M. J. Panzner, A. J. Ditto, Y. H. Yun and W. J. Youngs, Met.-Based Drugs, 2008, 2008, 384010 Search PubMed.
- M. Gottschaldt, A. Pfeifer, D. Koth, H. Görls, H.-M. Dahse, U. Möllmann, M. Obata and S. Yano, Tetrahedron, 2006, 62, 11073–11080 CrossRef CAS.
- M.-X. Li, D. Zhang, L.-Z. Zhang and J.-Y. Niu, Inorg. Chem. Commun., 2010, 13, 1268–1271 CrossRef CAS.
- P. C. Zachariadis, S. K. Hadjikakou, N. Hadjiliadis, S. Skoulika, A. Michaelides, J. Balzarini and E. De Clercq, Eur. J. Inorg. Chem., 2004, 2004, 1420–1426 CrossRef.
- S. I. Mostafa and F. A. Badria, Met.-Based Drugs, 2008, 2008, 723634 Search PubMed.
- R. R. Zaky and A. M. Abdelghay, Res. J. Pharm., Biol. Chem. Sci., 2011, 2, 757–764 CAS.
- Y.-J. Choi, J.-H. Park, J. W. Han, E. Kim, O. Jae-Wook, S. Y. Lee, J.-H. Kim and S. Gurunathan, Int. J. Mol. Sci., 2016, 17, 2077 CrossRef PubMed.
- J. Dou, X. He, Y. Liu, Z. Huang, C. Yang, F. Shi, D. Chen and N. Gu, J. Nanopart. Res., 2013, 15, 2127 CrossRef.
- A. Bandyopadhyay, B. Roy, P. Shaw, P. Mondal, M. K. Mondal, P. Chowdhury, S. Bhattacharya and A. Chattopadhyay, Nucleus, 2020, 63, 191–202 CrossRef.
- L. U. Tolentino and H. N. Po, J. Coord. Chem., 1984, 13, 341–344 CrossRef CAS.
- E. Suet, A. Laouenan, H. Handel and R. Guglielmetti, Helv. Chim. Acta, 1984, 67, 441–449 CrossRef CAS.
- P. C. Riesen and T. A. Kaden, Helv. Chim. Acta, 1995, 78, 1325–1333 CrossRef CAS.
- J. D. Chartres, M. S. Davies, L. F. Lindoy, G. V. Meehan and G. Wei, Inorg. Chem. Commun., 2006, 9, 751–754 CrossRef CAS.
- A. J. Blake and M. Schröder, in Adv. Inorg. Chem, ed. A. G. Sykes, Academic Press, 1990, vol. 35, pp. 1–80 Search PubMed.
- P. Gerschel, K. Warm, E. R. Farquhar, U. Englert, M. L. Reback, D. Siegmund, K. Ray and U. P. Apfel, Dalton Trans., 2019, 48, 5923–5932 RSC.
- J. N. Boodram, I. J. McGregor, P. M. Bruno, P. B. Cressey, M. T. Hemann and K. Suntharalingam, Angew. Chem., Int. Ed., 2016, 55, 2845–2850 CrossRef CAS PubMed.
- A. Eskandari, A. Kundu, S. Ghosh and K. Suntharalingam, Angew. Chem., Int. Ed., 2019, 58, 12059–12064 CrossRef CAS PubMed.
- D. C. F. Monteiro, R. M. Phillips, B. D. Crossley, J. Fielden and C. E. Willans, Dalton Trans., 2012, 41, 3720–3725 RSC.
- G. Dontu, W. M. Abdallah, J. M. Foley, K. W. Jackson, M. F. Clarke, M. J. Kawamura and M. S. Wicha, Genes Dev., 2003, 17, 1253–1270 CrossRef CAS PubMed.
- C. Lu, K. Laws, A. Eskandari and K. Suntharalingam, Dalton Trans., 2017, 46, 12785–12789 RSC.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS PubMed.
- B. O. Leung, F. Jalilehvand, V. Mah, M. Parvez and Q. Wu, Inorg. Chem., 2013, 52, 4593–4602 CrossRef CAS PubMed.
- F. Q. Schafer and G. R. Buettner, Free Radicals Biol. Med., 2001, 30, 1191–1212 CrossRef CAS PubMed.
- M. B. Hampton and S. Orrenius, FEBS Lett., 1997, 414, 552–556 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2053375. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1dt01155c |
| This journal is © The Royal Society of Chemistry 2021 |