Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tailoring copper(II) complexes with pyridine-4,5-dicarboxylate esters for anti-Candida activity†
Tina P.
Andrejević
 a,
Ivana
Aleksic
a,
Ivana
Aleksic
 b,
Marta
Počkaj
b,
Marta
Počkaj
 c,
Jakob
Kljun
c,
Jakob
Kljun
 c,
Dusan
Milivojevic
c,
Dusan
Milivojevic
 b,
Nevena Lj.
Stevanović
b,
Nevena Lj.
Stevanović
 a,
Jasmina
Nikodinovic-Runic
a,
Jasmina
Nikodinovic-Runic
 *b,
Iztok
Turel
*b,
Iztok
Turel
 *c,
Miloš I.
Djuran
*c,
Miloš I.
Djuran
 *d and
Biljana Đ.
Glišić
*d and
Biljana Đ.
Glišić
 *a
*a
aUniversity of Kragujevac, Faculty of Science, Department of Chemistry, R. Domanovića 12, 34000 Kragujevac, Serbia. E-mail: biljana.glisic@pmf.kg.ac.rs
bInstitute of Molecular Genetics and Genetic Engineering, University of Belgrade, Vojvode Stepe 444a, 11042 Belgrade, Serbia. E-mail: jasmina.nikodinovic@imgge.bg.ac.rs
cUniversity of Ljubljana, Faculty of Chemistry and Chemical Technology, Večna pot 113, SI-1000 Ljubljana, Slovenia. E-mail: Iztok.Turel@fkkt.uni-lj.si
dSerbian Academy of Sciences and Arts, Knez Mihailova 35, 11000 Belgrade, Serbia. E-mail: milos.djuran@pmf.kg.ac.rs
First published on 18th January 2021
Abstract
Five novel copper(II) complexes with pyridine-4,5-dicarboxylate esters as ligands, [Cu(NO3)(py-2tz)(H2O)3]NO3 (1), [Cu(NO3)2(py-2metz)(H2O)] (2), [Cu(NO3)2(py-2py)(H2O)]·H2O (3), [CuCl2(py-2tz)]2 (4) and [CuCl2(py-2metz)]n (5) (py-2tz is dimethyl 2-(thiazol-2-yl)pyridine-4,5-dicarboxylate, py-2metz is dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate and py-2py is dimethyl 2,2′-bipyridine-4,5-dicarboxylate), were synthesized and structurally characterized by different spectroscopic and electrochemical methods. The structure of these complexes was determined by single-crystal X-ray diffraction analysis, confirming the bidentate coordination mode of the corresponding pyridine-4,5-dicarboxylate ester to the Cu(II) ion through the nitrogen atoms. The antimicrobial potential of copper(II) complexes 1–5 was assessed against two bacterial and two Candida species. These complexes showed better growth inhibiting activity against Candida spp. with respect to the tested bacterial species, also being moderately toxic towards normal human lung fibroblast cells (MRC-5). Complexes 1 and 4 showed the greatest ability to inhibit the filamentation of C. albicans, which is an important process during fungal infection, and these two complexes efficiently inhibited the biofilm formation of C. albicans at subinhibitory concentrations. Complex 4 also successfully prevented the adhesion of C. albicans in an in vitro epithelial cell model. The mechanism of the antifungal activity of copper(II) complexes 1–5 was studied through their interaction with ct-DNA, as one of the possible target biomolecules, by fluorescence spectroscopy and gel electrophoresis. Finally, the ability of these complexes to bind to bovine serum albumin (BSA) was studied by fluorescence emission spectroscopy.
Introduction
In the last few decades, medicinal chemistry has seen an impressive advance, with special interest in the study of metal complexes, which have found a wide range of applications in medicine for the treatment of numerous diseases. In this context, medicinal inorganic chemistry has directed special attention to the synthesis of complexes of essential metal ions.1 One of the most important essential metals for living organisms is copper, which can be found in the active sites of different metalloproteins and enzymes, including superoxide dismutase, cytochrome c oxidase and tyrosinase.2 The significant role of copper in the human body is confirmed by the fact that the deficiency of this metal can result in anaemia, as a consequence of lower activity of copper ferroxidase.3 On the other hand, the reduced ability of the organism to excrete an excess of copper can result in Wilson's disease, an autosomal recessive genetic disorder.4Many copper(II) complexes have shown to be effective as superoxide scavenging5 and anti-inflammatory agents.6 For example, copper(II) salicylate was found to be approximately 30 times more effective in comparison with aspirin.7 In addition to the anti-inflammatory activity, copper(II) complexes have remarkable antibacterial,8 antifungal,9 antioxidant10 and anticancer11 activities, which takes precedence and makes copper(II) complexes attractive in metal-based drug design. In general, the biological activity of metal complexes depends on the oxidation state of metal ions, number and types of ligands, redox behaviour, hydro- and lipophilicity and coordination geometry.1 Moreover, a change of the substituent on the coordinated ligand can have an influence on the complex biological activity.12 The Cu(II) ion behaves as a Lewis acid intermediate and shows affinity toward nitrogen- and oxygen-donor atoms, forming complexes with coordination numbers 4, 5 and 6.13 The antimicrobial activity of copper(II) complexes with nitrogen-donor ligands has been found to be higher than that in the case of uncoordinated ligands.14 Considering this, pyridine-containing compounds represent a very attractive class of ligands for the synthesis of copper(II) complexes as potential antimicrobial agents. Copper(II) complexes of nicotinic acid with 2-hydroxypyridine, 2-aminopyridine and pyridine-2-carboxylic acid inhibited the growth of Bacillus subtilis ATCC 6633 and partially inhibited the growth of Candida albicans ATCC 90028,5 while [Cu(2,2′-bipy)Cl2(thiouracil)] (2,2′-bipy is 2,2′-bipyridine) showed fungicidal and fungistatic activities against 21 clinical isolates of Candida species.15 On the other hand, copper(II) complexes with the phosphate and hydroxymethyl derivatives of pyridine and benzimidazole did not inhibit completely the growth of the studied bacteria and fungi up to 200 μg mL−1.16
In the present study, pyridine-4,5-dicarboxylate esters, namely dimethyl 2-(thiazol-2-yl)pyridine-4,5-dicarboxylate (py-2tz), dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate (py-2metz) and dimethyl 2,2′-bipyridine-4,5-dicarboxylate (py-2py) (Scheme 1), were used as ligands for the synthesis of copper(II) complexes. These ligands have been previously used for the synthesis of silver(I) complexes as efficient agents for the control of cow mastitis associated pathogens17 and of ruthenium(II) complexes as inhibitors of aldo–keto reductases (AKRs)18 and 15-lipoxygenase-1.19 Given the versatile activities obtained upon coordination of these ligands to Ag(I) and Ru(II) ions, the aim of this study was to comprehensively evaluate, in terms of biological activity and interaction with biomolecules, the set of copper(II) complexes. Two copper(II) salts, Cu(NO3)2·3H2O and CuCl2·2H2O, were used in the synthesis, due to the fact that by altering the nature of the counter-anion of the starting inorganic salt, it is possible to tune the nuclearity of the corresponding copper(II) complexes.20–22 The synthesized copper(II) complexes, [Cu(NO3)(py-2tz)(H2O)3]NO3 (1), [Cu(NO3)2(py-2metz)(H2O)] (2), [Cu(NO3)2(py-2py)(H2O)]·H2O (3), [CuCl2(py-2tz)]2 (4) and [CuCl2(py-2metz)]n (5), were characterized by spectroscopy (IR and UV-Vis), mass spectrometry, cyclic voltammetry and single-crystal X-ray diffraction analysis. These complexes were further assessed for their in vitro antimicrobial potential, which was compared to their cytotoxicity to normal human lung fibroblast cells (MRC-5). In order to understand the mode of action of the copper(II) complexes, their interactions with possible biological targets, including DNA and proteins, were studied.
 | ||
| Scheme 1 Structural formulas of the pyridine-4,5-dicarboxylate esters used for the synthesis of copper(II) complexes. | ||
Results and discussion
Synthesis and structural characterization of complexes 1–5
Three pyridine-4,5-dicarboxylate esters, dimethyl 2-(thiazol-2-yl)pyridine-4,5-dicarboxylate (py-2tz), dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate (py-2metz) and dimethyl 2,2′-bipyridine-4,5-dicarboxylate (py-2py) (Scheme 1), which were synthesized in accordance with the previously published method,23 were used for the synthesis of copper(II) complexes.A copper(II) salt CuX2 (X = NO3− and Cl−) was reacted with an equimolar amount of the corresponding pyridine-4,5-dicarboxylate ester in ethanol at room temperature to yield [Cu(NO3)(py-2tz)(H2O)3]NO3 (1), [Cu(NO3)2(py-2metz)(H2O)] (2), [Cu(NO3)2(py-2py)(H2O)]·H2O (3), [CuCl2(py-2tz)]2 (4) and [CuCl2(py-2metz)]n (5) complexes (Scheme 2). The characterization of these complexes was done by the results of elemental analysis, UV-Vis and IR spectroscopy, mass spectrometry and cyclic voltammetry, while their structures were determined by single-crystal X-ray diffraction analysis.
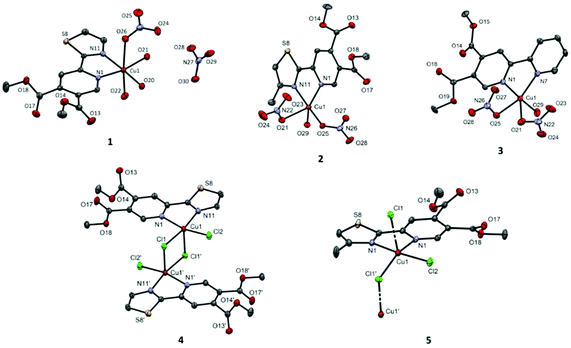
Complexes 1–3, which result from the reactions between Cu(NO3)2·3H2O and the pyridine-4,5-dicarboxylate esters, are mononuclear (Fig. 1). The nitrato ligand is monodentately coordinated to the Cu(II) ion in all three complexes. The orientation of the nitrato ligands in 2 and 3 is such that the distances from the Cu(II) ion to the uncoordinated nitrato oxygen atoms are relatively short, but still significantly longer than the length of the coordinated bonds (see the caption of Fig. 1).
On the other hand, compounds 4 and 5, prepared from CuCl2·2H2O, are either dinuclear (4) or polymeric (5) in the solid state. In both cases, the chlorido anion acts as a μ2-bridging ligand. In 4, two such bridges lead to the formation of a Cu2Cl2 motif, whereas in 5, only one bridging chlorido ligand on each side of the coordination center is present, forming zig–zag chains.
The wavelengths of the absorption maxima (λmax) for copper(II) complexes 1–5 and molar extinction coefficients (ε, M−1 cm−1) determined immediately after their dissolution in DMSO are in accordance with those reported for other copper(II) complexes with aromatic nitrogen-containing heterocycles.25 The presently reported complexes exhibit a single band in the expected region,28 attributed to the dz2, dxy, dxz, dyz → dx2−y2 transitions with a dx2−y2 ground state.29
The solution stability of complexes 1–5 was studied by recording the UV-Vis spectra in DMSO after dissolution, as well as after 24 and 48 h (Fig. S1† shows the time-dependant UV-Vis spectra of complex 1). No significant changes in the intensity and position of the absorption maxima with time were observed for 1–5, suggesting that the pyridine-4,5-dicarboxylate ester remains coordinated to the Cu(II) ion in solution.
From the molar conductivity measurements, and after comparison of the obtained data with those reported in the literature for different metal complexes,30,31 it can be concluded that complexes 1–3 behave as electrolytes, while complexes 4 and 5 act as nonelectrolytes in DMSO solution. As previously reported, the molar conductance ranges for the nonelectrolytes and electrolytes in DMSO are <50 and >50 Ω−1 cm2 mol−1, respectively.30,31
Bioactivity evaluation
The inhibitory concentrations of copper(II) complexes 1–5 were evaluated for two bacterial pathogenic strains and two common fungal pathogens (Table 1). As can be seen from Table 1, these complexes did not affect the bacterial growth under the tested conditions. On the other hand, a moderate antifungal activity was observed, especially in the case of complexes 2 and 5 with a minimal inhibitory concentration (MIC) of 31.25 μg mL−1 against C. albicans ATCC 10231. Notably, complex 5 was not highly toxic to healthy human fibroblasts MRC-5 (Table 1). As it was previously reported, none of the pyridine-4,5-dicarboxylate esters was active against the tested strains up to 500 μg mL−1,17 while the MIC values of Cu(NO3)2·3H2O were reported to be 500 and 250 μg mL−1 against the tested bacterial and fungal species, respectively.25| Test | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| P. aeruginosa NCTC 10332 | >500 | >500 | >500 | >500 | >500 |
| S. aureus ATCC 25923 | >500 | >500 | >500 | >500 | 500 |
| C. albicans ATCC 10231 | 62.5 | 31.25 | 62.5 | 125 | 31.25 |
| C. parapsilosis ATCC 22019 | 500 | >500 | >500 | 500 | 250 |
| MRC-5 | 75 ± 5 | 50 ± 2 | 60 ± 4 | 35 ± 2 | 70 ± 5 |
Previously it was shown that the copper(II) complexes of nicotinic acid with 2-hydroxypyridine, 2-aminopyridine and pyridine-2-carboxylic acid inhibited the growth of Gram-positive Bacillus subtilis ATCC 6633 (MIC = 256 μg mL−1), while these complexes partially inhibited the growth of C. albicans ATCC 90028 (MIC = 128–256 μg mL−1).5 The more potent fungicidal and fungistatic activities against 21 clinical isolates of Candida spp. (MICs = 31–125 μg mL−1) for the [Cu(2,2′-bipy)Cl2(thiouracil)] complex were also reported.15
The observed antifungal activity has triggered the evaluation of the effect of complexes 1–5 on the filamentation of C. albicans (Fig. 3). The morphological transformation from the yeast to hyphae form is characteristic of the majority of fungal species, and of Candida species, and it is the indicator of their pathogenic potential.35,36 Complexes 1, 2 and 4, although not causing the growth inhibition, almost completely inhibited the formation of hyphae even after 72 h of incubation, whereas 1 and 4 were even effective after 120 h. The latter two complexes were efficient at inhibiting the hyphae formation even in the liquid RPMI (Roswell Park Memorial Institute) medium (Fig. S3†).
Considering these findings, the effect of the subinhibitory concentrations of complexes 1 and 4 to prevent the C. albicans biofilm formation was also examined (Fig. 4). The concentration gradient was observed for both investigated complexes. Almost 70% of biofilm formation is inhibited at 25 μg mL−1 of complex 1 and 50% of complex 4, with the concentration being lower than their respective MIC and IC50 values (Table 1). These results are much better than the anti-biofilm activity of the [Cu(2,2′-bipy)Cl2(thiouracil)] complex, where the percentage inhibition of about 10% against C. krusei and C. glabrata biofilm formations was observed at concentrations higher than the MIC values against planktonic cells.15
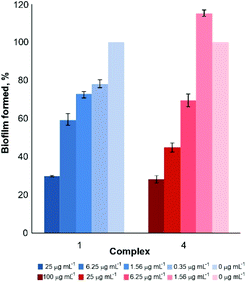 | ||
| Fig. 4 Effect of growth subinhibitory concentrations of complexes 1 and 4 on C. albicans ATCC 10231 biofilm formation. | ||
Protein binding study
In the present study, the emission spectra of BSA (5 μM) in the presence of increasing concentrations of complexes 1–5 were recorded in phosphate buffered saline (PBS; pH = 7.4) in the wavelengths range from 295 to 500 nm, by exciting the protein at 290 nm. As can be seen from Fig. 6, in the presence of complexes 1–5, an intense quenching of a fluorophore was observed, as a consequence of complex binding to BSA which resulted in the changes in the protein tertiary structure.42
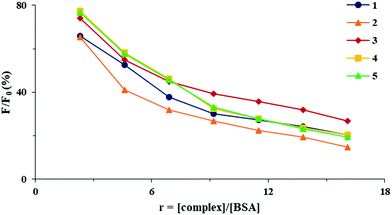 | ||
| Fig. 6 Plot of relative fluorescence intensity (F/F0, %) vs. r (r = [complex]/[BSA]) for complexes 1–5 in PBS (pH = 7.4). | ||
The data obtained using the Stern–Volmer and Scatchard equations (Stern–Volmer constant (Ksv), quenching rate constant (Kq), binding constant (KA) and the number of binding sites per BSA (n)) are given in Table 2. The values of quenching constant Kq are higher than 1010 M−1 s−1, indicating the static mechanism of quenching.43 The KA values for all complexes are relatively high, showing the ability of the complexes to bind to BSA and transport to their biological targets.42 Similar binding constants to those of BSA were previously reported for copper(II) complexes with non-steroidal anti-inflammatory agents, such as tolfenamic, meclofenamic, mefenamic, clonixic and niflumic acids, and different nitrogen-donor ligands.44–46 The number of binding sites per BSA molecule is similar for all complexes, which is in accordance with their tendency to bind to one site in the protein (Table 2).
| Complex | K sv (M−1) | Hypochromism (%) | K q (M−1 s−1) | K A (M−1) | n |
|---|---|---|---|---|---|
| 1 | (7.03 ± 0.10) × 104 | 82.2 | 7.03 × 1012 | 1.14 × 105 | 1.07 |
| 2 | (1.26 ± 0.02) × 105 | 85.3 | 1.26 × 1013 | 3.53 × 105 | 1.16 |
| 3 | (6.00 ± 0.80) × 104 | 85.7 | 6.00 × 1012 | 6.93 × 104 | 1.06 |
| 4 | (2.88 ± 0.02) × 105 | 86.9 | 2.88 × 1013 | 1.81 × 106 | 1.36 |
| 5 | (4.98 ± 0.02) × 105 | 86.3 | 4.98 × 1013 | 2.43 × 106 | 1.38 |
 | ||
| Fig. 7 Synchronous fluorescence spectra of BSA in the absence and presence of increasing concentrations of complex 5 at Δλ = 15 (A) and Δλ = 60 nm (B). | ||
| Δλ (nm) | K sv (M−1) | Hypochromism (%) | K q (M−1 s−1) | K A (M−1) | n |
|---|---|---|---|---|---|
| 15 | (3.08 ± 0.02) × 105 | 83.8 | 3.08 × 1013 | 2.87 × 106 | 1.36 |
| 60 | (1.16 ± 0.01) × 106 | 82.3 | 1.16 × 1014 | 1.48 × 108 | 1.77 |
Lipophilicity assay
Studies on the potential biological activity of the metal complexes have shown that the cellular uptake of these compounds increases with their lipophilicity.52 Therefore, this physicochemical property of a compound is critical for its potential application as a therapeutic agent.53,54 The partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) between the hydrophobic octanol phase and the hydrophilic water phase is a measure of lipophilicity and indicates the ability of a compound to be transported through the cell membrane.55 The values of log
P) between the hydrophobic octanol phase and the hydrophilic water phase is a measure of lipophilicity and indicates the ability of a compound to be transported through the cell membrane.55 The values of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for different clinically used therapeutic agents, including metal complexes, are found to be in the range from −0.4 to 5.6.56 All complexes 1–5 have the values in this ideal lipophilicity range, 0 ≤ log
P for different clinically used therapeutic agents, including metal complexes, are found to be in the range from −0.4 to 5.6.56 All complexes 1–5 have the values in this ideal lipophilicity range, 0 ≤ log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 2.30. Complexes 1, 4 and 5 have much greater log
P ≤ 2.30. Complexes 1, 4 and 5 have much greater log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values than the other two complexes, indicating that these three complexes are mostly distributed in the octanol phase and that their cellular uptake efficiency is higher.
P values than the other two complexes, indicating that these three complexes are mostly distributed in the octanol phase and that their cellular uptake efficiency is higher.
DNA binding study
Interactions of metal ions with DNA, its constituents and derivatives, are of great importance for the design of novel metal-based antimicrobial agents.57 Firstly, the interaction of the copper(II) complexes with λDNA was investigated by gel electrophoresis. After 2 h of incubation, complexes 3 and 4 prevented the ethidium bromide (EthBr) binding, which was reflected as a lower intensity of DNA bands on the agarose gel in comparison with the DMSO control (Fig. 8A). This activity increased with the incubation time (complex 2 was the least effective one after 4 h of incubation), and upon 24 h of incubation all complexes were able to prevent the EthBr intercalation with almost 100% efficiency.The binding affinity of the metal complexes to ct-DNA has also been investigated by monitoring the emission intensity of EthBr bound to ct-DNA.41 Ethidium bromide is the most studied fluorescent probe that intercalates to ct-DNA via its planar phenanthroline ring between the adjacent base pairs of the double helix, resulting in the intensive fluorescence emission of the EthBr-DNA conjugate.39,58 The addition of the metal complex, which may substitute EthBr in the EthBr-DNA conjugate or bind to this conjugate, results in a decrease of the emission.59 The extent of the decrease in the fluorescence quenching of the EthBr-DNA system indicates the interaction mode between the studied complex and ct-DNA.60
Within this context, the emission spectra of the EthBr-DNA system ([ct-DNA]/[EthBr] = 10) were recorded in the absence and presence of an increasing amount of the investigated complexes 1–5 (0–175 μM) (Fig. 8B). In all cases, upon addition of the copper(II) complex, a decrease in the fluorescence intensity was observed, indicating its interaction with ct-DNA. However, the values of the binding constants (KA, Table S1†) of the investigated complexes are much lower than that of EthBr itself (KA = 2 × 106 M−1),43 and those of previously reported copper(II) complexes with fenamates and isonicotinamide, for which an intercalation mode of interaction with ct-DNA was proposed.44 Considering this, a more reasonable explanation for reduction in the emission intensity of the EthBr-DNA system in the presence of 1–5 could be the binding of the complexes to the EthBr-DNA conjugate, leading to the formation of a new non-fluorescent EthBr-DNA-complex system.
Experimental
Materials and measurements
The copper(II) salts (Cu(NO3)2·3H2O and CuCl2·2H2O), ethanol, methanol, acetonitrile, dimethyl sulfoxide (DMSO), phosphate-buffered saline (PBS), bovine serum albumin (BSA), calf thymus DNA (ct-DNA) and ethidium bromide (EthBr) were obtained from Sigma-Aldrich. All chemicals and solvents were of analytical grade and used without further purification.Elemental microanalyses of the synthesized copper(II) complexes for carbon, hydrogen and nitrogen were performed using a PerkinElmer 2400 Series II instrument (CHN). The ESI-HRMS spectra in the positive mode were recorded after dissolving complexes 1–5 in CH3CN with an Agilent 62224 accurate mass spectrometer, using time of flight liquid chromatography/mass spectrometry. The IR spectra were recorded as KBr pellets on a PerkinElmer Spectrum 100 spectrometer over the wavenumber range of 4000–450 cm−1. The UV-Vis spectra were recorded on a Shimadzu double-beam spectrophotometer after dissolving copper(II) complexes 1–5 in DMSO, as well as after 24 and 48 h of standing at ambient temperature, over the wavelength range of 1100–200 nm. The concentration of the solutions used for these measurements was 1 × 10−2 M. Molar conductance data were measured at room temperature using a digital conductivity-meter Crison Multimeter MM 41. The concentration of the solutions of complexes 1–5 in DMSO used for these measurements was 1 × 10−3 M. The cyclic voltammetry (CV) measurements were performed using a potentiostat/galvanostat AutoLab PGSTAT204. The cell (5.0 mL) consisted of a three electrode system, a glassy carbon (GC) electrode as a working electrode, Ag/AgCl (saturated KCl) as a reference electrode and platinum wire as a counter electrode. All reported potentials are referred versus the Ag/AgCl (saturated KCl) reference electrode. The electrode surface was renewed before every measurement by polishing with Al2O3 micro-powder and with a piece of cotton due to the strong adsorption of the complexes. The concentration of the solutions of complexes 1–5 in DMSO used for the electrochemical measurements was 1 × 10−3 M. The emission spectra for the ct-DNA interactions of the complexes were recorded using a Jasco FP-6600 spectrophotometer.
Synthesis of pyridine-4,5-dicarboxylate esters
The pyridine-4,5-dicarboxylate esters, namely dimethyl 2-(thiazol-2-yl)pyridine-4,5-dicarboxylate (py-2tz), dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate (py-2metz) and dimethyl 2,2′-bipyridine-4,5-dicarboxylate (py-2py) used for the synthesis of copper(II) complexes were prepared by a previously described method starting from different methyl ketones.23 All pyridine-4,5-dicarboxylate esters were pure based on elemental microanalysis and 1H NMR spectroscopy.Synthesis of copper(II) complexes 1–5
Copper(II) complexes, [Cu(NO3)(py-2tz)(H2O)3]NO3 (1), [Cu(NO3)2(py-2metz)(H2O)] (2), [Cu(NO3)2(py-2py)(H2O)]·H2O (3), [CuCl2(py-2tz)]2 (4) and [CuCl2(py-2metz)]n (5), were synthesized according to the modified procedure for the preparation of silver(I) complexes with pyridine-4,5-dicarboxylate esters.17 A solution of 1.0 mmol of the corresponding pyridine-4,5-dicarboxylate ester (278.3 mg of py-2tz for 1 and 4, 292.3 mg of py-2metz for 2 and 5 and 272.3 mg of py-2py for 3) in 10.0 mL of ethanol was added slowly under stirring to the solution containing an equimolar amount of the copper(II) salt (241.6 mg of Cu(NO3)2·3H2O for 1–3 and 170.5 mg of CuCl2·2H2O for 4 and 5) in 5.0 mL of ethanol. The reaction mixture was stirred for 3 h on a magnetic stirrer at room temperature. Complex 5 crystallized from the mother ethanol solution after evaporation at ambient temperature for 4 days, while other copper(II) complexes were obtained after recrystallization of the obtained powder in methanol for complex 1 and acetonitrile for 2, 3 and 4. The crystals of complexes 1–5 suitable for single-crystal X-ray crystallography were filtered off and dried at room temperature. Yield (calculated on the basis of pyridine-4,5-dicarboxylate ester): 447.1 mg (86%) for 1, 318.6 mg (64%) for 2, 317.3 mg (64%) for 3, 321.9 mg (78%) for 4 and 102.4 mg (24%) for 5.Anal. calcd for 1 = C12H16CuN4O13S (MW = 519.89): C, 27.73; H, 3.10; N, 10.78. Found: C, 27.84; H, 2.77; N, 10.96%. IR (KBr, ν, cm−1): 3421br (ν(O–H)), 3112w (ν(Car–H)), 2956w (ν(C–H)), 1734vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1616m, 1556m, 1501w, 1488w, 1438m (ν(Car
O)), 1616m, 1556m, 1501w, 1488w, 1438m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(Car
Car) and ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1384s, 1330s, 1299s (νas(NO3)), 1280m (ν(C–O)), 795m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 836 (ε = 37.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 381.9920 (381.9917). ΛM (DMSO): 69.5 Ω−1 cm2 mol−1.
N)), 1384s, 1330s, 1299s (νas(NO3)), 1280m (ν(C–O)), 795m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 836 (ε = 37.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 381.9920 (381.9917). ΛM (DMSO): 69.5 Ω−1 cm2 mol−1.
Anal. calcd for 2 = C13H14CuN4O11S (MW = 497.88): C, 31.37; H, 2.84; N, 11.26. Found: C, 31.22; H, 3.08; N, 11.68%. IR (KBr, ν, cm−1): 3368br (ν(O–H)), 3151w, 3026w (ν(Car–H)), 2963w (ν(C–H)), 1743s, 1730s (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1650w, 1614m, 1553w, 1528w, 1486m, 1457m, 1439m (ν(Car
O)), 1650w, 1614m, 1553w, 1528w, 1486m, 1457m, 1439m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(Car
Car) and ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1384vs, 1342s (νas(NO3)), 1289s, 1268s (ν(C–O)), 768w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 842 (ε = 33.0 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 396.0068 (396.0074). ΛM (DMSO): 55.4 Ω−1 cm2 mol−1.
N)), 1384vs, 1342s (νas(NO3)), 1289s, 1268s (ν(C–O)), 768w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 842 (ε = 33.0 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 396.0068 (396.0074). ΛM (DMSO): 55.4 Ω−1 cm2 mol−1.
Anal. calcd for 3 = C14H16CuN4O12 (MW = 495.85): C, 33.92; H, 3.25; N, 11.30. Found: C, 33.53; H, 3.32; N, 11.08%. IR (KBr, ν, cm−1): 3454br (ν(O–H)), 3044w (ν(Car–H)), 2956w, 2924w, 2855w (ν(C–H)), 1751s, 1738vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1616m, 1565w, 1492m, 1436m (ν(Car
O)), 1616m, 1565w, 1492m, 1436m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(Car
Car) and ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1385vs, 1317s (νas(NO3)), 1278s, 1256s (ν(C–O)), 781w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 728 (ε = 41.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 376.0363 (376.0353). ΛM (DMSO): 54.6 Ω−1 cm2 mol−1.
N)), 1385vs, 1317s (νas(NO3)), 1278s, 1256s (ν(C–O)), 781w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 728 (ε = 41.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 376.0363 (376.0353). ΛM (DMSO): 54.6 Ω−1 cm2 mol−1.
Anal. calcd for 4 = C24H20Cl4Cu2N4O8S2 (MW = 825.47): C, 34.92; H, 2.44; N, 6.79. Found: C, 35.17; H, 2.12; N, 6.55%. IR (KBr, ν, cm−1): 3108w, 3024w (ν(Car–H)), 2940w (ν(C–H)), 1737vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1615m, 1554w, 1443m, 1428m (ν(Car=Car) and ν(Car=N)), 1296vs, 1280vs (ν(C–O)), 794m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 919 (ε = 85.3 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 381.991 (381.9917). ΛM (DMSO): 23.3 Ω−1 cm2 mol−1.
O)), 1615m, 1554w, 1443m, 1428m (ν(Car=Car) and ν(Car=N)), 1296vs, 1280vs (ν(C–O)), 794m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 919 (ε = 85.3 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 381.991 (381.9917). ΛM (DMSO): 23.3 Ω−1 cm2 mol−1.
Anal. calcd for 5 = C13H12Cl2CuN2O4S (MW = 426.76): C, 36.59; H, 2.84; N, 6.57. Found: C, 36.22; H, 3.11; N, 6.84%. IR (KBr, ν, cm−1): 3098w, 3076w (ν(Car–H)), 2951w, 2925w (ν(C–H)), 1729vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1614m, 1552m, 1524w, 1486w, 1452m, 1433m (ν(Car
O)), 1614m, 1552m, 1524w, 1486w, 1452m, 1433m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(Car
Car) and ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1285vs (ν(C–O)), 792w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 935 (ε = 87.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 396.0073 (396.0079); (found for ([Cu + 2L]+) (calcd)): 647.0326 (647.0331). ΛM (DMSO): 31.8 Ω−1 cm2 mol−1.
N)), 1285vs (ν(C–O)), 792w (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 935 (ε = 87.9 M−1 cm−1). ESI-HRMS (CH3CN) m/z (found for ([Cu + L + CH3CN]+ (calcd)): 396.0073 (396.0079); (found for ([Cu + 2L]+) (calcd)): 647.0326 (647.0331). ΛM (DMSO): 31.8 Ω−1 cm2 mol−1.
Crystallographic data collection and refinement of the structures
For X-ray structural analysis, single crystals of compounds 1–5 were coated with silicon grease, mounted onto the tip of glass fibre and transferred to the goniometer head in the liquid nitrogen cryostream (150(2) K). The data were collected using a SuperNova diffractometer equipped with an Atlas detector using the CrysAlis software either with monochromated Mo Kα (0.71073 Å) or Cu Kα radiation (1.54184 Å).61 The initial structural models were obtained via direct methods using the Olex2 graphical user interface62 implemented in SHELXT. A full-matrix least-squares refinement method on F2 magnitudes with anisotropic displacement parameters for all nonhydrogen atoms using Olex2 or SHELXL-2018/3 was employed.62,63 All non-hydrogen atoms were refined anisotropically, while the hydrogen atoms were placed at the calculated positions and further treated as riding models on their parent atoms. The coordinates of the water hydrogen atoms were obtained from difference Fourier maps and were further refined using appropriate distance restraints.Additional details of the crystal data, data collection and refinement are given in the ESI (Table S2†). Figures depicting the structures were obtained with Mercury.64
Determination of minimum inhibitory concentrations (MICs)
The MIC values of complexes 1–5 were determined according to the standard broth microdilution assays, recommended by the National Committee for Clinical Laboratory Standards (M07-A8) for bacteria and Standards of European Committee on Antimicrobial Susceptibility Testing (v 7.3.1: the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts) for Candida species. The tested compounds were dissolved in DMSO at a concentration of 50 mg mL−1. The highest used concentration was 500 μg mL−1. Two bacterial test organisms, Pseudomonas aeruginosa NCTC 10332 and Staphylococcus aureus ATCC 25923, and two Candida strains, C. albicans ATCC 10231 and C. parapsilosis ATCC 22019, were included in the study. The inoculums were 5 × 105 colony forming units, cfu mL−1, for bacteria and 1 × 105 cfu mL−1 for Candida species. The MIC value was recorded as the lowest concentration that inhibited the growth after 24 h at 37 °C, using a Tecan Infinite 200 Pro multiplate reader (Tecan Group Ltd, Männedorf, Switzerland).Cytotoxicity
The antiproliferative activities of complexes 1–5 were examined using the standard colorimetric MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay.65 MRC-5 cells (human lung fibroblasts obtained from ATCC, American Type Culture Collection) were plated in a 96-well flat-bottom plate at a concentration of 1 × 104 cells per well, grown in a humidified atmosphere of 95% air and 5% CO2 at 37 °C and maintained as monolayer cultures in RPMI 1640 medium (Gibco). Each tested compound was added to the cells at a concentration ranging from 5 to 250 μg mL−1 and the treatment lasted for 48 h. The MTT assay was performed two times in four replicates and the extent of MTT reduction was measured spectrophotometrically at 540 nm using a Tecan Infinite 200 Pro multiplate reader (Tecan Group Ltd, Männedorf, Switzerland). Cytotoxicity was expressed as the concentration of the compound inhibiting the cell growth by 50% (IC50) in comparison with the control (DMSO-treated cells).Filamentation and anti-biofilm activity
Candida albicans ATCC 10231 was grown in Sabouraud dextrose broth medium overnight at 30 °C with shaking at 180 rpm on a rotary shaker. The overnight culture was washed with PBS and diluted in RPMI 1640 medium with 2% (v/w) glucose to the final concentration of 1 × 106 cells per mL. The cell suspension was treated with 0.5 × MIC concentrations of complexes 1–5 for 3 h at 37 °C with shaking at 180 rpm on a rotary shaker. The cells treated with DMSO were used as the negative control. Finally, the cells were pelleted at 3000g, and concentrated 10 times in fresh PBS and the hyphae formation was observed using bright field microscopy (Olympus BX51, Applied Imaging Corp., San Jose, CA, United States) under 20× magnification.
Adherence assay
The ability of C. albicans SC5314 cells to infect adenocarcinomic human alveolar basal epithelial cells A549 (obtained from ATCC) was studied by adherence assay, as described previously.68 A549 cells were grown on 22 mm glass coverslips in RPMI 1640 medium for two days. A549 monolayers on the glass coverslips were inoculated with C. albicans cells in a log growth phase (without centrifugation), and the complexes were added at MIC and incubated for 1 h in RPMI 1640 without FBS (fetal bovine serum) at 37 °C and 5% CO2. After co-incubation, the non-adherent cells were removed by extensively rinsing three times with PBS and samples fixed with 4% paraformaldehyde. Next, the A549 cells were stained with 1 μg mL−1 of 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, Sigma-Aldrich, Munich, Germany) for 10 min in the dark. Both A549 and C. albicans cells were visualized using a fluorescent microscope (Olympus BX51, Applied Imaging Corp., San Jose, USA), at 60× magnification.Protein binding study by fluorescence emission spectroscopy
The protein binding study was performed by carrying out tryptophan fluorescence quenching experiments using BSA (5 μM) in phosphate buffer solution (pH = 7.4). The quenching of the emission intensity of the tryptophan residues of BSA at 365 nm was monitored at increasing concentrations of complexes 1–5 (up to 130 μM). The fluorescence spectra were recorded in the range of 295–500 nm at an excitation wavelength of 290 nm. The corresponding binding constants of the complexes (KA) and apparent binding sites (n) were calculated as previously explained.41,69 For the synchronous fluorescence spectroscopy (SFS) studies, the same concentration of BSA and complexes 1–5 was used, and the spectra were measured at two different Δλ values (Δλ = λem − λex), 15 and 60 nm, which are characteristic of the tyrosine (Tyr) and tryptophan (Trp) residues, respectively.Lipophilicity assay
The lipophilicity of the copper(II) complexes was determined by the flask-shaking method.53 Complexes 1–5 were dissolved in DMSO and added to the water/n-octanol system. The mixture was vortexed for 1 h at room temperature to allow partitioning. After this time, the solutions were allowed to stand for 24 h until the two phases were separated. The concentration of the complexes in both phases was determined by measuring the absorbance values using the previously determined calibration curves. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values were calculated according to the following equation:
P values were calculated according to the following equation:log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = log(C0/Cw) P = log(C0/Cw) |
DNA binding study
In vitro DNA binding by the electrophoresis assay
Commercially available λDNA (Thermo Scientific, 0.3 μg μL−1) was used for studying the effect of complexes 1–5 on Lambda DNA (λDNA) (Thermo Scientific). The final concentration in the water solution used was 20 μg μL−1 of λDNA and 500 μg μL−1 of the compounds (100 μL final volume). The ability of complexes 1–5 to bind λDNA was examined by using agarose gel electrophoresis, a methodology published earlier,71 with some modifications. The mixtures (DNA and complexes) were incubated for 2, 4 and 24 h at 37 °C, and then subjected to gel electrophoresis on 1% (w/v) agarose gel containing 0.5 μg μL−1 of EthBr in TAE buffer (40 mM Tris acetate, 1 mM EDTA, pH = 7.4) at 70 V for 2 h. Control was the DNA solution treated with DMSO and incubated for the same time periods. The gels were visualized and analyzed using the Gel Doc EZ system (Bio-Rad, Life Sciences, Hercules, USA), with the Image Lab™ Software.| F0/F = 1 + Kqτ0[complex] = 1 + Ksv[complex] |
log(F0 − F)/F = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KA + n KA + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[complex] log[complex] |
Conclusions
In this study, five new copper(II) complexes with pyridine-4,5-dicarboxylate esters as ligands, [Cu(NO3)(py-2tz)(H2O)3]NO3 (1), [Cu(NO3)2(py-2metz)(H2O)] (2), [Cu(NO3)2(py-2py)(H2O)]·H2O (3), [CuCl2(py-2tz)]2 (4) and [CuCl2(py-2metz)]n (5), were synthesized, structurally characterized and biologically evaluated. Reactions of copper(II) nitrate with pyridine-4,5-dicarboxylate esters result in the formation of mononuclear complexes 1–3, while copper(II) chloride readily forms dinuclear (4) and polynuclear (5) species. Moreover, complexes 1–3, along with the corresponding pyridine-4,5-dicarboxylate ester and nitrato ligands, contain water molecules in the coordination sphere of the Cu(II) ion, while no coordination of this ligand is observed in the cases of 4 and 5. Only complex 1 shows an elongated octahedral geometry, while the geometry of the remaining complexes is distorted square pyramidal. The structural varieties of these complexes were reflected as slight differences in their biological activities. All complexes showed moderate to good activity against Candida albicans, and the ability to inhibit the hyphae formation of this strain, while being non-toxic to healthy human fibroblasts. Two complexes, 1 and 4, efficiently inhibited the biofilm formation of C. albicans at the subinhibitory concentrations, which is a highly desirable property of the novel antifungal agent. Importantly, dinuclear complex 4 efficiently prevented the adhesion of C. albicans in an in vitro epithelial cell model, which suggests that this complex could be brought further for structural optimizations and the assessment of efficiency to prevent candidiasis. All synthesized complexes showed affinity toward protein and DNA, providing possible insights into the mode of their antimicrobial activity.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research was financially supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (agreement no. 451-03-68/2020-14/200042 and 451-03-68/2020-14/200122) and by the Slovenian Research Agency (grant P1-0175; funding in 2019-20). The EN → FIST Centre of Excellence, Trg OF 13, SI-1000 Ljubljana, Slovenia, is acknowledged for the use of the SuperNova diffractometer. This research also received funding from the Serbian Academy of Sciences and Arts under strategic projects programme - grant agreement no. 01-2019-F65 and project of this institution no. F128.Notes and references
- G. Gasser and N. Metzler-Nolte, Curr. Opin. Chem. Biol., 2012, 16, 84 CrossRef CAS.
- E. Alessio, Bioinorganic Medicinal Chemistry, Wiley-VCH, Weinheim, 2011 Search PubMed.
- R. M. Llanos and J. F. B. Mercer, DNA Cell Biol., 2002, 21, 259 CrossRef CAS.
- G. J. Brewer, Curr. Opin. Chem. Biol., 2003, 7, 207 CrossRef CAS.
- T. Suksrichavalit, S. Prachayasittikul, C. Nantasenamat, C. Isarankura-Na-Ayudhya and V. Prachayasittikul, Eur. J. Med. Chem., 2009, 44, 3259 CrossRef CAS.
- J. E. Weder, C. T. Dillon, T. V. Hambley, B. J. Kennedy, P. A. Lay, J. R. Biffin, H. L. Regtop and N. M. Davies, Coord. Chem. Rev., 2002, 232, 95 CrossRef CAS.
- J. R. J. Sorenson, J. Med. Chem., 1976, 19, 135 CrossRef CAS.
- M. L. Beeton, J. R. Aldrich-Wright and A. Bolhuis, J. Inorg. Biochem., 2014, 140, 167 CrossRef CAS.
- K. Alomar, A. Landreau, M. Allain, G. Bouet and G. Larcher, J. Inorg. Biochem., 2013, 126, 76 CrossRef CAS.
- C. Tolia, A. N. Papadopoulos, C. P. Raptopoulou, V. Psycharis, C. Garino, L. Salassa and G. Psomas, J. Inorg. Biochem., 2013, 123, 53 CrossRef CAS.
- W. C. Zhang, X. Tang and X. Lu, J. Inorg. Biochem., 2016, 156, 105 CrossRef CAS.
- M. Iqbal, A. Karim, S. Ali, M. N. Tahir and M. Sohail, Polyhedron, 2020, 178, 114310 CrossRef CAS.
- R. R. Conry , Copper: Inorganic & Coordination Chemistry, in Encyclopedia of Inorganic Chemistry, John Wiley & Sons, 2011 Search PubMed.
- B. M. Paterson and P. S. Donnelly, Chem. Soc. Rev., 2011, 40, 3005 RSC.
- F. G. da Silva Dantas, A. A. de Almeida-Apolonio, R. P. de Araújo, L. R. V. Favarin, P. F. de Castilho, F. de Oliveira Galvão, T. I. E. Svidzinski, G. A. Casagrande and K. M. P. de Oliveira, Molecules, 2018, 23, 1856 CrossRef.
- U. Kalinowska-Lis, I. Szabłowska-Gadomska, K. Lisowska, J. Ochocki, M. Małecki and A. Felczak, Z. Anorg. Allg. Chem., 2017, 643, 993 CrossRef CAS.
- T. P. Andrejević, D. Milivojevic, B. Đ. Glišić, J. Kljun, N. Lj. Stevanović, S. Vojnovic, S. Medic, J. Nikodinovic-Runic, I. Turel and M. I. Djuran, Dalton Trans., 2020, 49, 6084 RSC.
- K. Traven, M. Sinreih, J. Stojan, S. Seršen, J. Kljun, J. Bezenšek, B. Stanovnik, I. Turel and T. L. Rižner, Chem.-Biol. Interact., 2015, 234, 349 CrossRef CAS.
- K. Traven, N. Eleftheriadis, S. Seršen, J. Kljun, J. Bezenšek, B. Stanovnik, I. Turel and F. J. Dekker, Polyhedron, 2015, 101, 306 CrossRef CAS.
- P. Seppälä, R. Sillanpää and A. Lehtonen, Coord. Chem. Rev., 2017, 347, 98 CrossRef.
- V. Marvaud, C. Decroix, A. Scuiller, F. Tuyèras, C. Guyard-Duhayon, J. Vaissermann, J. Marrot, F. Gonnet and M. Verdaguer, Chem. – Eur. J., 2003, 9, 1692 CrossRef CAS.
- P. Mahapatra, S. Giri, M. G. B. Drew and A. Ghosh, Dalton Trans., 2018, 47, 3568 RSC.
- J. Bezenšek, B. Prek, U. Grošelj, M. Kasunič, J. Svete and B. Stanovnik, Tetrahedron, 2012, 68, 4719 CrossRef.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, Dalton Trans., 1984, 1349 RSC.
- B. Đ. Glišić, I. Aleksic, P. Comba, H. Wadepohl, T. Ilic-Tomic, J. Nikodinovic-Runic and M. I. Djuran, RSC Adv., 2016, 6, 86695 RSC.
- A. S. Potapov, E. A. Nudnova, A. I. Khlebnikov, V. D. Ogorodnikov and T. V. Petrenko, Inorg. Chem. Commun., 2015, 53, 72 CrossRef CAS.
- A. B. P. Lever, E. Mantovani and B. S. Ramaswamy, Can. J. Chem., 1971, 49, 1957 CrossRef CAS.
- B. J. Hathaway, in Comprehensive Coordination Chemistry, ed. G. Wilkinson, R. D. Gillard and J. A. McCleverty, vol. 5, Pergamon, Oxford, 1987 Search PubMed.
- N. Lj. Stevanović, T. P. Andrejević, A. Crochet, T. Ilic-Tomic, N. S. Drašković, J. Nikodinovic-Runic, K. M. Fromm, M. I. Djuran and B. Đ. Glišić, Polyhedron, 2019, 173, 114112 CrossRef.
- I. Ali, W. A. Wani and K. Saleem, Synth. React. Inorg., Met., 2013, 43, 1162 CrossRef CAS.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81 CrossRef CAS.
- J. M. S. Cardoso, I. Correia, A. M. Galvão, F. Marques and M. F. N. N. Carvalho, J. Inorg. Biochem., 2017, 166, 55 CrossRef CAS.
- A. P. Sandoval-Rojas, L. Ibarraa, M. T. Cortés, M. A. Macías, L. Suescun and J. Hurtado, J. Electroanal. Chem., 2017, 805, 60 CrossRef CAS.
- E. Franco, E. Lopez-Torres, A. Mendiola and T. Sevilla, Polyhedron, 2000, 19, 441 CrossRef CAS.
- J. A. Romo, C. G. Pierce, A. K. Chaturvedi, A. L. Lazzell, S. F. McHardy, S. P. Saville and J. L. Lopez-Ribot, mBio, 2017, 8, e01991 CrossRef CAS.
- L. Sun, K. Liao, C. Hang and D. Wang, PLoS One, 2017, 12, e0172228 CrossRef.
- B. Wächtler, F. Citiulo, N. Jablonowski, S. Förster, F. Dalle, M. Schaller, D. Wilson and B. Hube, PLoS One, 2012, 7, e36952 CrossRef.
- N. D. Savić, S. Vojnovic, B. Đ. Glišić, A. Crochet, A. Pavic, G. V. Janjić, M. Pekmezović, I. M. Opsenica, K. M. Fromm, J. Nikodinovic-Runic and M. I. Djuran, Eur. J. Med. Chem., 2018, 156, 760 CrossRef.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Plenum Press, New York, 3rd edn, 2006 Search PubMed.
- Y.-Q. Wang, H.-M. Zhang, G.-C. Zhang, W.-H. Tao and S.-H. Tang, J. Lumin., 2007, 126, 211 CrossRef CAS.
- P. Smoleński, C. Pettinari, F. Marchetti, M. F. C. Guedes da Silva, G. Lupidi, G. V. B. Patzmay, D. Petrelli, L. A. Vitali and A. J. L. Pomberio, Inorg. Chem., 2015, 54, 434 CrossRef.
- V. Rajendiran, R. Karthik, M. Palaniandavar, H. Stoeckli-Evans, V. S. Periasamy, M. A. Akbarsha, B. S. Srinag and H. Krishnamurthy, Inorg. Chem., 2007, 46, 8208 CrossRef CAS.
- Y. Shi, C. Guo, Y. Sun, Z. Liu, F. Xu, Y. Zhang, Z. Wen and Z. Li, Biomacromolecules, 2011, 12, 797 CrossRef CAS.
- F. Jozefíková, S. Perontsis, M. Šimunková, Z. Barbieriková, Ľ. Švorc, M. Valko, G. Psomas and J. Moncol’, New J. Chem., 2020, 44, 12827 RSC.
- R. P. Sharma, S. Kumar, P. Venugopalan, V. Ferretti, A. Tarushi, G. Psomas and M. Witwicki, RSC Adv., 2016, 6, 88546 RSC.
- F. Dimiza, S. Fountoulaki, A. N. Papadopoulos, C. A. Kontogiorgis, V. Tangoulis, C. P. Raptopoulou, V. Psycharis, A. Terzis, D. P. Kessissoglou and G. Psomas, Dalton Trans., 2011, 40, 8555 RSC.
- D. Li, M. Zhu, C. Xu and B. Ji, Eur. J. Med. Chem., 2011, 46, 588 CrossRef CAS.
- B. Hemmateenejad, M. Shamsipur, F. Samari, T. Khayamian, M. Ebrahimi and Z. Rezaei, J. Pharm. Biomed. Anal., 2012, 67, 201 CrossRef.
- C.-L. Zhang, Y.-X. Liu, X.-M. Zhang, S. Chen, F. Shen, Y.-H. Xiong, W. Liu, Z.-W. Mao and X.-Y. Le, Mater. Sci. Eng., C, 2018, 91, 414 CrossRef CAS.
- J. G. Da Silva, A. A. Recio Despaigne, S. R. W. Louro, C. C. Bandeira, E. M. Souza-Fagundes and H. Beraldo, Eur. J. Med. Chem., 2013, 65, 415 CrossRef CAS.
- D. İnci, R. Aydın, Ö. Vatan, T. Sevgi, D. Yılmaz, Y. Zorlu, Y. Yerli, B. Çoşut, E. Demirkan and N. Çinkılıç, J. Biol. Inorg. Chem., 2017, 22, 61 CrossRef.
- S. Mehanna, N. Mansour, H. Audi, K. Bodman-Smith, M. A. Mroueh, R. I. Taleb, C. F. Daher and R. S. Khnayzer, RSC Adv., 2019, 9, 17254 RSC.
- C. A. Puckett and J. K. Barton, J. Am. Chem. Soc., 2007, 129, 46 CrossRef CAS.
- A. Ghezzi, M. Aceto, C. Cassino, E. Gabano and D. Osella, J. Inorg. Biochem., 2004, 98, 73 CrossRef CAS.
- L. Fetzer, B. Boff, M. Ali, M. Xiangjun, J.-P. Collin, C. Sirlin, C. Gaiddon and M. Pfeffer, Dalton Trans., 2011, 40, 8869 RSC.
- A. K. Ghose, V. N. Viswanadhan and J. J. Wendoloski, J. Comb. Chem., 1999, 1, 55 CrossRef CAS.
- I. Turel and J. Kljun, Curr. Top. Med. Chem., 2011, 11, 2661 CrossRef CAS.
- A. M. Godin, W. C. Ferreira, L. T. S. Rocha, J. G. T. Seniuk, A. L. L. Paiva, L. A. Merlo, E. B. Nascimento Jr., L. F. S. Bastos and M. M. Coelho, Pharmacol., Biochem. Behav., 2011, 99, 782 CrossRef CAS.
- R. F. Pasternack, M. Cacca, B. Keogh, T. A. Stephenson, A. P. Williams and F. J. Gibbs, J. Am. Chem. Soc., 1991, 113, 6835 CrossRef CAS.
- J.-B. Lepecq and C. Paoletti, J. Mol. Biol., 1967, 27, 87 CrossRef CAS.
- CrysAlis PRO, Oxford Diffraction Ltd, Yarnton, Oxfordshire, England, 2011 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339 CrossRef CAS.
- G. M. Sheldrick, SHELXL2018/3, University of Göttingen, Germany, 2018 Search PubMed.
- C. F. Macrae, P. R. Edgington, P. McCabe, E. Pidcock, G. P. Shields, R. Taylor, M. Towler and J. van de Streek, J. Appl. Crystallogr., 2006, 39, 453 CrossRef CAS.
- M. B. Hansen, S. E. Nielsen and K. Berg, J. Immunol. Methods, 1989, 119, 203 CrossRef CAS.
- B. S. R. Mohamed, M. Subramanian and K. P. Shunmugiah, Appl. Microbiol. Biotechnol., 2014, 98, 6775 CrossRef.
- C. G. Pierce, P. Uppuluri, A. R. Tristan, F. L. Wormley Jr., E. Mowat, G. Ramage and J. L. Lopez-Ribot, Nat. Protoc., 2008, 3, 1494 CrossRef CAS.
- Á. Jakab, S. Mogavero, T. M. Forster, M. Pekmezovic, N. Jablonowski, V. Dombrádi, I. Pócsi and B. Hube, Microbiology, 2016, 162, 2116 CrossRef.
- A. Wolfe, G. H. Shimer Jr. and T. Meehan, Biochemistry, 1987, 26, 6392 CrossRef CAS.
- R. Bera, B. K. Sahoo, K. S. Ghosh and S. Dasgupta, Int. J. Biol. Macromol., 2008, 42, 14 CrossRef CAS.
- V. T. Yilmaz, E. Gocmen, C. Icsel, M. Cengiz, S. Y. Susluer and O. Buyukgungor, J. Photochem. Photobiol., B, 2014, 131, 31 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S3 and Tables S1–S2. CCDC 2041819–2041823. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0dt04061d |
| This journal is © The Royal Society of Chemistry 2021 |