Open Access Article
Open Access ArticleDirected genome evolution driven by structural rearrangement techniques
Sijie
Zhou†
 ab,
Yi
Wu†
ab,
Yi
Wu†
 ab,
Ze-Xiong
Xie
ab,
Ze-Xiong
Xie
 ab,
Bin
Jia
ab,
Bin
Jia
 ab and
Ying-Jin
Yuan
ab and
Ying-Jin
Yuan
 *ab
*ab
aFrontier Science Center for Synthetic Biology (Ministry of Education), Tianjin University, Tianjin, 300072, China. E-mail: yjyuan@tju.edu.cn
bKey Laboratory of Systems Bioengineering (Ministry of Education), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300072, China
First published on 15th October 2021
Abstract
Directed genome evolution simulates the process of natural evolution at the genomic level in the laboratory to generate desired phenotypes. Here we review the applications of recent technological advances in genome writing and editing to directed genome evolution, with a focus on structural rearrangement techniques. We highlight how these techniques can be used to generate diverse genotypes, and to accelerate the evolution of phenotypic traits. We also discuss the perspectives of directed genome evolution.
1. Introduction to directed genome evolution
The concept of biological evolution refers to iterated mutations under natural selection, and provides solutions to challenges that individuals have faced to adapt to the natural world over billions of years.1 To accelerate evolution for phenotypes in our favor, humans have used artificial selection for centuries, beginning with the selective breeding of crops2 and domestication of animals.3 Directed evolution has proved to be a highly effective and broadly applicable framework to generate desired traits through iterative rounds of mutagenesis and library screening or selection.4 While many synthetic evolution strategies link a particular organismal phenotype to a single gene, more complex phenotypes often require coordinated and global changes in the whole-genome due to the complexity and interconnectivity of metabolic networks.5 Furthermore, even in the minimal bacterial genome, JCVI-syn3.0, many genes (∼30%) are of unknown function.6 Recent studies on the noncoding regions of the genome suggest these sequences may have important physiological functions.7 Taken together, techniques allowing quick introduction of global changes to the whole genome are particularly important as screening of favorable phenotypes can be conducted before the functions of all genomic loci are understood. In addition, systematic analyses of the observed phenotypes may in turn help us characterize the function(s) of an individual gene.Physical, chemical, and transposon mutagenesis techniques have been widely used to mutagenize target genomes and are usually coupled to a screen or selection for an altered phenotype. Recently, researchers have developed various mature and efficient directed genome evolution strategies to introduce small-scale variations such as single nucleotide polymorphisms (SNPs) and Indels throughout the genome (Table 1). Several recent reviews summarized and discussed the strategies and applications of genome-wide directed evolution driven by small-scale variations.33,34 Other methods developed by contrast introduce genome structural rearrangements (Table 1), which induce large-scale variations throughout the genome including gene deletion, gene fusion, changes in gene orders and copy-numbers of genes, as well as chromatin structure.
| Variation scale | Category | Method | Feasibility | Beneficial mutants |
|---|---|---|---|---|
| Small (SNP, Indel) | Oligo-mediated | Multiplex automated genome evolution. (MAGE,8 eMAGE,9 YOGE,10etc.) | Short oligonucleotides-mediated recombination; introduction of SNP and Indel | Higher yield of lycopene8 and carotene9 |
| CRISPR/Cas-mediated | Nicking Cas9 (nCas9)-mediated recruitment of error-prone DNA polymerases. (EvolvR,11 yEvolvR.12) | Continuous targeted mutagenesis on targeted regions | Antibiotic resistance11 | |
| dCas9-recruitment of base-editing proteins. (CRISPR-STOP,13 TAM,14 BARBEKO15) | Multiplexed, continuous mutation of targeted cytosines, guanines or potentially adenosines | Imatinib resistance14 | ||
| Homologous recombination mediated genome editing. (CHAnGE,16 MAGIC,17 CREATE,18 iCREATE,19 MAGESTIC20) | Homologous repair mediated by gRNA promotes the accuracy and effectiveness of genome editing | Tolerance to growth inhibitors,16 fast growth,17 stress tolerance18 | ||
| Large (SV) | Genome writing-mediated | Synthetic chromosome rearrangement and modification by LoxP-mediated evolution (ring chromosome SCRaMbLE,21 multiplex SCRaMbLE Iterative Cycling,22 heterozygous and interspecies SCRaMbLE,23in vitro SCRaMbLE,24 orthogonal SCRaMbLE25) | LoxPsym sites were inserted in genome in advance. Upon expression of Cre recombinase, a variety of rearrangements including deletions, inversions, duplications, and translocations can be generated | Tolerance to growth inhibitors,26,27 higher yield of carotene22 and violacein25 |
| Genome editing-mediated | CRISPR-Cas928–30 | CRISPR-Cas9 stimulates genome rearrangement by introducing double-strand breaks (DSBs), and then DSBs are repaired by NHEJ or HDR | Tolerance to environmental stress,26 higher yield of taxadiene28 | |
| TaqI31 and MseI32 | Activated TaqI or MseI targeted 4 base sequence to induce DSB | Tolerance to environmental stress31,32 |
We will review the quickly expanded toolbox of directed genome evolution with a focus on those new tools used to generate genome structural rearrangements (Table 2), including Synthetic Chromosome Rearrangement and Modification by LoxP-mediated Evolution (SCRaMbLE) and endonuclease-mediated rearrangement. The review highlights the mechanism by which they can be used to accelerate phenotypic evolution and the insights into the relationship between genomic structure and function. We also emphasize the challenges and future perspectives in directed genome evolution.
| Category | Technology | Principles | Advantages | Limitations | Applications |
|---|---|---|---|---|---|
| Recombinase-mediated site-specific recombination | SCRaMbLE | LoxPsym sites were inserted among synthetic yeast genome. A variety of rearrangements including deletions, inversions, duplications, and translocations can be generated upon expression of Cre recombinase | High controllable; generation of a large library of structural variations in a short time; rearrangement site determination35 | Genome sequences with multiple loxPsym sites must be constructed in advance; LoxPsym site left on genome | Strain development;21,22,26 genotype-phenotype correlations;23,27 genome minimization;36 genetic interaction;36 loss of heterozygosity;37 aneuploidy38 |
| Nuclease mediated DSB | CRISPR-Cas9 | CRISPR-Cas9 stimulates genome rearrangement by introducing DSBs, which are mainly repaired by NHEJ or HDR to generate numerous rearrangement libraries | Marker less integration;39 ease to multiplexing; scarless | Possible off-target; the size of the rearrangement library is limited by the target gRNA | Improving microbial phenotypes;29,30 loss of heterozygosity;40 aneuploidy;38 reproductive isolation;28 karyotype restructuring41,42 |
2. Genome writing-mediated directed genome evolution
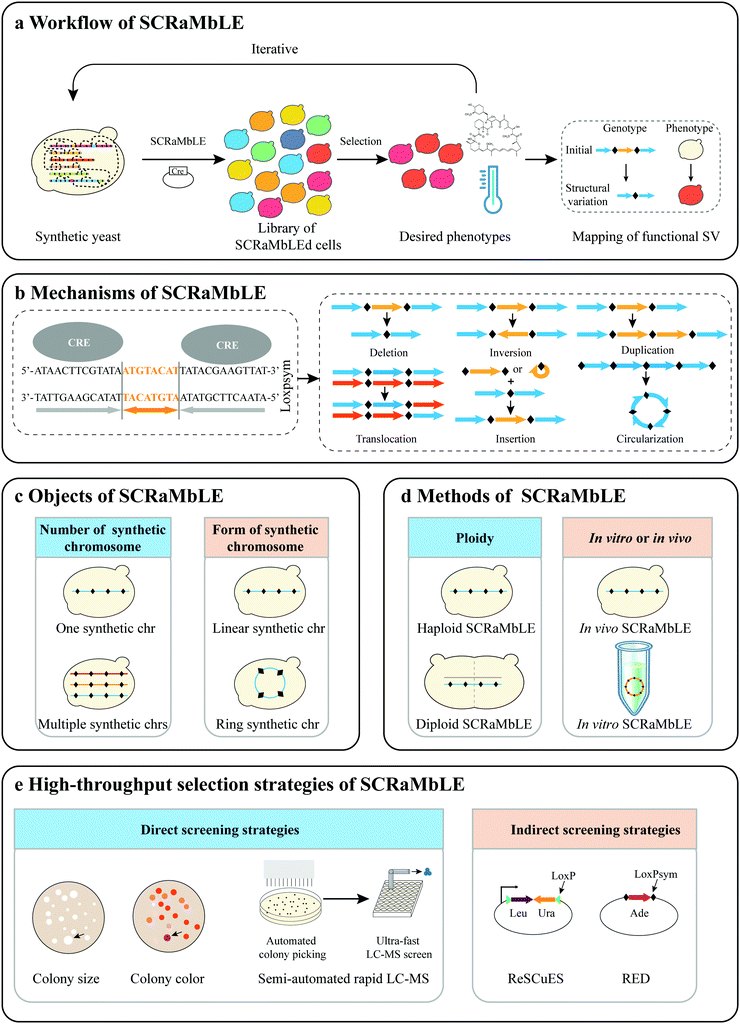
Genome structural variation proved challenging to study until the recent development of sequencing technology, in particular, the advent of long-read sequencing technology.43,44 Genome rearrangements naturally occur in all kingdoms of life45–50 and are considered an important driving factor in species evolution,50,51 crop improvement,46,52 and the emergence of human diseases.53 However, diverse structural rearrangements are difficult to induce experimentally, and this has impeded further investigation and exploitation of directed genome evolution. Recent progress in synthetic genomics allows de novo design and synthesis of large fragments of DNA or even genome, offering an opportunity to systematically introduce a site-specific recombination system at the genome-level.54,55 This introduction of a site-specific recombination system enables the induction of structural rearrangements at the genome level and facilitates the investigation of directed genome evolution.2.1 The technique of SCRaMbLE
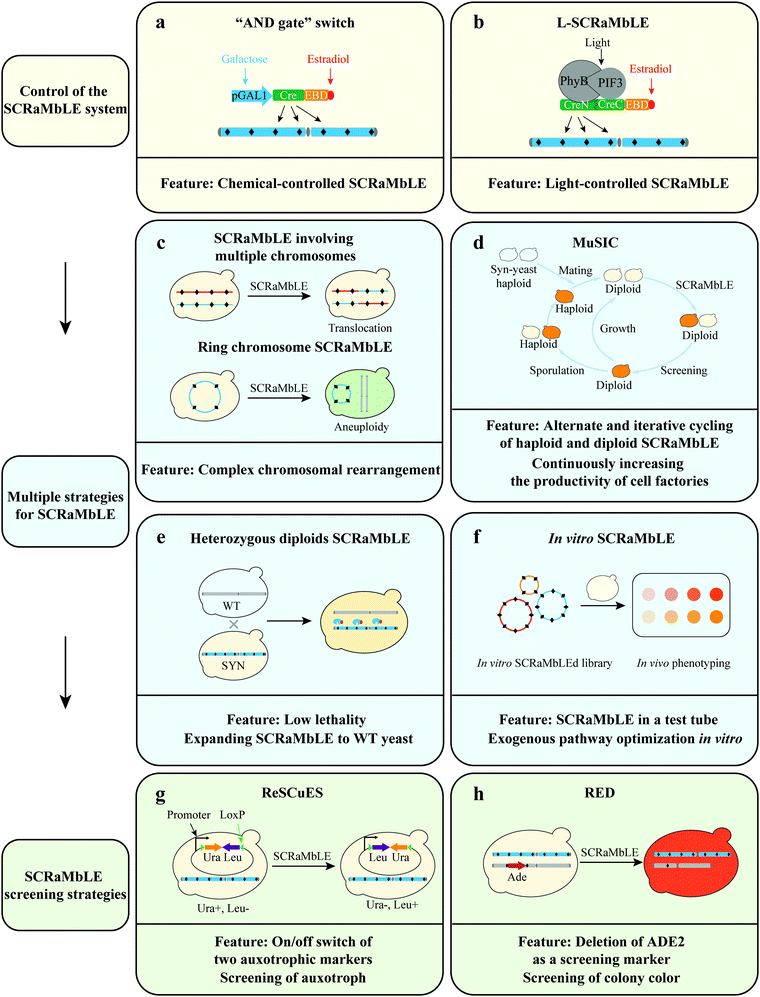
Control of the SCRaMbLE system. In previous SCRaMbLE studies, β-estradiol binding to Cre-EBD triggered the translocation of Cre recombinase into the nucleus where Cre recombinase executes its activity.55 However in the absence of β-estradiol residual Cre-recombinase activity was still observed, resulting in a lack of control of SCRaMbLE, and genomic instability,55,61,62 which is problematic in the screen for desired phenotypes. To address this bottleneck, Jia et al. constructed a genetic “AND gate” switch for SCRaMbLE, which is turned on only in the presence of both galactose and β-estradiol (Fig. 2a).22 The switch uses the pGAL1 promoter for transcriptional control of Cre-EBD, thereby controlling the subcellular localization of Cre-EBD enzyme activity.
In addition to the aforementioned chemical-controlled Cre recombinase induction system, Hochrein et al. developed a light-controlled one (L-SCRaMbLE) (Fig. 2b).63 In L-SCRaMbLE the N- and C-terminal ends of Cre recombinase are fused to photoreceptor phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3), respectively. Exposure to red light induces interaction between the plant-originating proteins PhyB and PIF3 and brings together the two parts of Cre recombinase, reconstituting its enzyme activity. This system not only effectively overcomes the leakage problem but also allows induction of high levels of Cre activity in a short time frame. With this system, induction time, light dose, and sensor concentration all become means to regulate the Cre activity. Both approaches offer precise control of the SCRaMbLE system.
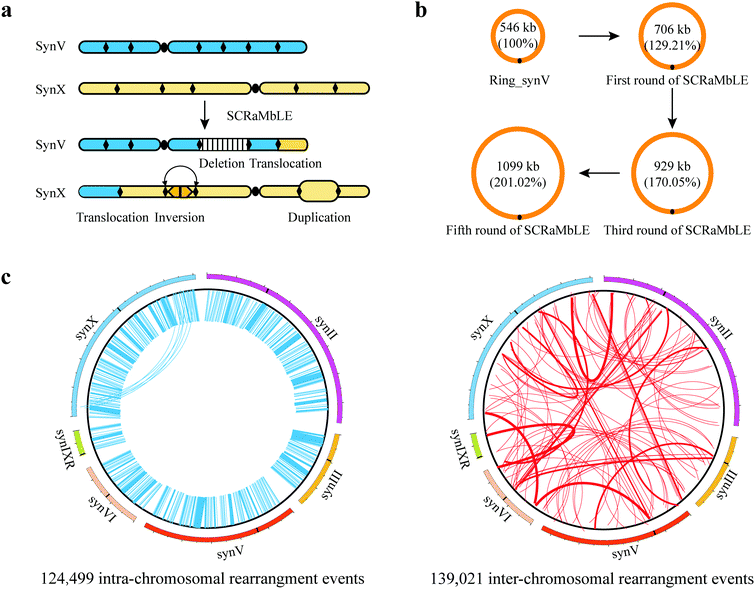
SCRaMbLE involving multiple chromosomes. SCRaMbLE of a strain harboring a single synthetic chromosome has validated the capability of SCRaMbLE to induce deletions, inversions, duplications, and additional complex rearrangements (Fig. 1c).62 To further explore the power of the SCRaMbLE recombination assay in generating translocation, Sarah et al. combined synthetic chromosomes with an “endoreduplication intercross” to generate Sc2.0 poly-synthetic chromosome strains.54 The obtained strain carrying synIII and synIXR was induced by SCRaMbLE and the not previously observed translocation event was detected (Fig. 2c).64 More recently, Sijie et al. consolidated 5.5 synthetic chromosomes (synII, synIII, synV, synVI, synIXR, and synX) into a single haploid strain based on Vika/vox.65 Enormous inter-chromosomal events were observed in this poly-synthetic chromosome strain following SCRaMbLE.
Ring chromosome SCRaMbLE. In addition to SCRaMbLEing linear chromosomes, Shen et al. SCRaMbLEd ring_synIXR strains and revealed that relatively complex rearrangement events occurred, including not only deletions and inversions but also a high frequency of duplications in 64 SCRaMbLEd colonies (Fig. 1c).62 Wang et al. SCRaMbLEd haploid yeast cells carrying a ring_synV and suggested that the topological structure of chromosomes exerts a functional impact on chromosome rearrangement.21 The total length increased by 552
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410 bp, accounting for ∼101.02% of ring_synV after five rounds of SCRaMbLE. Furthermore, aneuploidies of chromosomes I, III, VI, XII, XIII, and ring_synV were generated in SCRaMbLEd strains (Fig. 2c). Ring chromosome SCRaMbLE exhibited the capability to continuously produce complex genotypes and obtain corresponding phenotypes, thereby effectively expanding the scale and number of structural variations. Compared with SCRaMbLE of linear chromosomes, its use on ring chromosomes may result in increased duplication events or more complex chromosomal rearrangement events.
410 bp, accounting for ∼101.02% of ring_synV after five rounds of SCRaMbLE. Furthermore, aneuploidies of chromosomes I, III, VI, XII, XIII, and ring_synV were generated in SCRaMbLEd strains (Fig. 2c). Ring chromosome SCRaMbLE exhibited the capability to continuously produce complex genotypes and obtain corresponding phenotypes, thereby effectively expanding the scale and number of structural variations. Compared with SCRaMbLE of linear chromosomes, its use on ring chromosomes may result in increased duplication events or more complex chromosomal rearrangement events.
Multiplex SCRaMbLE iterative cycling (MuSIC). To generate genome diversity rapidly and continuously in a large population of synthetic strains, Jia et al. established Multiplex SCRaMbLE Iterative Cycling (MuSIC), which is used to accumulate preponderant rearrangements through multiple cycles of SCRaMbLE (Fig. 2d).22 High stability and reliability of the “AND gate” switch enables the iterative SCRaMbLEing of synthetic chromosomes without removing the Cre plasmid. Through this iterative genome rearrangement reaction strategy, the yield of strains is continuously and gradually increased. In addition, high-yield diploid strains can produce spores through meiosis, and the screened spores can be mated with a yeast strain harboring new synthetic chromosome to produce a new diploid strain, which can subsequently be used to perform the next round of MuSIC. The alternate and iterative cycling of the haploid and diploid SCRaMbLE strategy can continuously increase the productivity of cell factories, expanding the scope of genome rearrangement and screening of new targets on a larger scale.
Heterozygous diploid and interspecies SCRaMbLE system. SCRaMbLEing haploid synthetic yeast results in a high lethality rate due to the deletion of essential genes and S. cerevisiae laboratory strain background has limited its industrial applications (Fig. 1d). Consequently, Shen et al. extended SCRaMbLE to heterozygous diploids and cross-species diploids, and used the genome rearrangement system to rapidly drive industrial yeast evolution (Fig. 2e).23 Combining synthetic yeasts with flexible genotypes and wild-type (WT) yeasts with diversified phenotypes through yeast mating allows the genome rearrangement system to drive the genome rearrangement of heterozygous diploids and cross-species diploids. The authors mated a haploid strain harboring one synthetic chromosome (synX) with a Y12 sake-brewing S. cerevisiae strain or Saccharomyces paradoxus strain CBS5829 to generate heterozygous diploid strains. The results indicate that the heterozygous diploid SCRaMbLE system could reduce the lethality of the SCRaMbLE method. The development of heterozygous diploid (and interspecies) SCRaMbLE system expands the application of the SCRaMbLE system to native yeast strains and will accelerate their improvement of phenotypes.
In vitro and in vivo SCRaMbLE system. SCRaMbLE is compatible with cell-free systems.66 Direct addition of recombinase to the test tube and incubation with a plasmid harboring genes involved in a metabolic pathway has been proved to be a valid method in the optimization of this prototyping pathway. To enable rapid metabolic engineering through pathway prototyping and chassis optimization in yeast, Wu et al. developed an “in vitro SCRaMbLE system”, which uses Cre recombinase mixed in a test tube with purified DNA encoding multiple loxPsym sites (Fig. 2f).24
This method can generate a structural variant library containing gene deletions, duplications, inversions, or insertions (transpositions) and achieve efficient DNA rearrangement in test tubes. Using a β-carotene pathway designed for expression in yeast as an example, Wu et al. demonstrated top-down and bottom-up in vitro SCRaMbLE, which optimized the biosynthetic pathway flux through the rearrangement of relevant transcription units.24 The top-down method specifies the use of a single DNA construct encoding multiple loxPsym sites and generates a SCRaMbLEd DNA library. A total of 94 unique pathway structures were determined by PacBio sequencing of a SCRaMbLEd library. The bottom-up system consists of an acceptor vector and a pool of donor fragments flanked by loxPsym sites. Both two ways provide a straightforward method to optimize the exogenous pathway.
Exogenous pathway optimization and chassis engineering are two key methods to achieve high expression of heterologous pathways. Liu et al. developed an orthogonal SCRaMbLE system “SCRaMbLE-in” to tackle both challenges simultaneously.67 The researchers used VCre/VloxP, Cre/loxP, or Dre/rox to integrate a range of regulatory elements such as promoters into the pathways of interest and generate a pathway library in vitro. The library was then integrated into the synthetic yeast chromosomes through the SCRaMbLE-in system, which also causes large-scale genome rearrangements of chassis cells. As the three recombinases are orthogonal to each other, the pathway and chassis can be optimized either independently or in combination to enable rapid metabolic engineering, realizing the precise rearrangement of functional modules. In summary, the SCRaMbLE-in method was used to increase the production of violacein and β-carotene, and resulted in co-optimization of the pathway and the chassis.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) the SCRaMbLEd pool.65 In total, over 260
000×) the SCRaMbLEd pool.65 In total, over 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rearrangement events, including 124
000 rearrangement events, including 124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 499 intra-chromosomal events and 139
499 intra-chromosomal events and 139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021 inter-chromosomal events were detected (Fig. 3c). The intra-chromosomal rearrangement events consisted of 62
021 inter-chromosomal events were detected (Fig. 3c). The intra-chromosomal rearrangement events consisted of 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 inversions, 22
106 inversions, 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526 deletions and 39
526 deletions and 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 complex rearrangement events.65 In the future, all synthetic chromosomes will be consolidated into a synthetic yeast enabling massive genome structural diversity. Enormous numbers of rearrangements by SCRaMbLE provide a driving force to potentiate directed genome evolution.
867 complex rearrangement events.65 In the future, all synthetic chromosomes will be consolidated into a synthetic yeast enabling massive genome structural diversity. Enormous numbers of rearrangements by SCRaMbLE provide a driving force to potentiate directed genome evolution.
Direct screening strategies. Phenotypes that either affect growth or elicit a colorful phenotype can be directly selected by colony size or color (Fig. 1e). Tolerance to environmental stresses and drug resistance including high temperature, acetic acid, alkaline, and caffeine are all screened by colony size,23,26,67 while colorful phenotypes allowed the isolation of high-yielding strains by colony color in previous SCRaMbLE studies.21,22,67 For phenotypes that are not easily selectable, an automated high-throughput screening strategy may provide a solution. Gowers et al. employed semi-automated technologies, an ultra-fast 84 second liquid chromatography coupled to mass spectrometry (LC-MS) method to rapidly isolate the best performing strains.68 This SCRaMbLE screening system consists of automated colony picking and sample extraction, ultra-fast LC-MS measurement and multiplexed nanopore sequencing. The selected genotypes were quickly resolved using multiplex nanopore sequencing. This method has wide applicability for other industrially relevant metabolites and can rapidly isolate improved strain from a variant library, making it an important tool for directed genome evolution.
Indirect screening strategies. SCRaMbLE is a powerful system to generate massive strains with genotypic diversity. However, the strains with genotypic changes are difficult to screen efficiently. To address this bottleneck, Dymond et al. detected SCRaMbLEd cells via lys1 and/or met28 auxotroph in a synthetic strain harboring synIXR. Furthermore, Luo et al. developed a reporter SCRaMbLE system, termed ReSCuES (Fig. 2g), to efficiently select SCRaMbLEd cells.27 ReSCuES was designed based on an alternating on/off switch of two auxotrophic markers flanked by two opposite loxP sites. Upon induction of Cre, two auxotrophic markers between the two loxP sites are inverted, subsequently turning on the expression of one marker and turning off the other. In a recent study, Wightman et al. developed RED (rapid evolution detection) to identify SCRaMbLEd strains in a SCRaMbLEd population (Fig. 2h).69 This system disrupted the native ADE2 gene and reintroduced ADE2 flanked by two loxPsym sites into a synthetic chromosome. The visibly red colonies produced by the deletion of ADE2 as a screening marker determine whether rearrangements have occurred. The above SCRaMbLE screening strategies allow the efficient detection of the genomic rearrangements that occurred within a SCRaMbLEd population.
Most of the above works use relatively straightforward screening strategies which are performed on solid agar. Furthermore, like CRISPR-mediated approaches,17 liquid culture of pooled organisms is also feasible for SCRaMbLE when a screening protocol being used enriches the strain of interest (i.e., fitness based selection). In this case identification of each structural variations can be achieved by the sequencing read counts in a SCRaMbLEd pool.65
2.2 Applications of SCRaMbLE
Genome-wide loxPsym-mediated recombination can produce abundant genomic structural variations and accelerate the evolution of yeast strains with various genotypes and phenotypes. Here, we highlight a few examples of how SCRaMbLE drive directed genome evolution by improving the adaptability of growth and chemical production of cell factories (Table 3).| Category of phenotypes | Phenotypic changes | Associated structural variations | Objects of SCRaMbLE | Methods of SCRaMbLE | Selection strategies of SCRaMbLE | Beneficial strains | Ref. |
|---|---|---|---|---|---|---|---|
| Improving adaptability of growth | Thermotolerance | Deletion of the region spanning YJL154C-YJL140W | Single synthetic chrX | In vivo diploid SCRaMbLE | Colony size | Two strains | 23 |
| Alkali tolerance | Deletion of YER161C (SPT2) | Single synthetic chrV | In vivo haploid SCRaMbLE | Colony size | Seven strains | 26 | |
| Acetic acid tolerance | Unresolved | Single synthetic chrXII | In vivo haploid SCRaMbLE | ReSCuES and colony size | Three strains | 27 | |
| Ethanol tolerance | An inversion involves ZRT2 and ACE2 | Single synthetic chrXII | In vivo haploid SCRaMbLE | ReSCuES and colony size | Three strains | 27 | |
| Improved xylose utilization | An inversion of a 7 kb region encoding GCN4, YEL008W, MIT1, and YEA6 genes and deletion of MXR1 | Single synthetic chrV | In vivo haploid SCRaMbLE | Colony size | One strain | 67 | |
| Rapamycin tolerance | Deletion of gene GLN3, long-range LOH in the left arm of synX, whole chromosome LOH of synX. | Two synthetic chrs (chrV&X) | In vivo interspecies SCRaMbLE | Colony size | Seven strains | 37 | |
| Hygromycin B tolerance | Deletions of YBR219C and YBR220C | Single synthetic chrII | In vivo haploid SCRaMbLE | Colony size | Ten strains | 71 | |
| Caffeine tolerance | Duplication of POL32 | Single synthetic chrX | In vivo interspecies SCRaMbLE | Colony size | Ten strains | 23 | |
| Improving chemical production of cell factories | Production of carotenoid (increased to 1.5-fold) | Deletion of the YEL013W | Single synthetic chrV | In vivo haploid SCRaMbLE | Colony color | Five strains | 22 |
| Production of carotenoid (increased to 38.8-fold) | Unresolved | Multiple synthetic chrs | MuSIC | Colony color | One strain | 22 | |
| Production of β-carotene (increased to 5.1-fold) | Duplication and inversion of crtI gene | Exogenous pathway | In vitro SCRaMbLE | Colony color | Eleven strains | 24 | |
| Production of β-carotene (increased to 2-fold) | Unresolved | Single synthetic chrII | In vitro SCRaMbLE and in vivo haploid SCRaMbLE | Colony color | One strain | 25 | |
| Production of lycopene (increased to 129.5-fold) | Unresolved | Single synthetic chrII | In vivo diploid SCRaMbLE | Colony color | One strain | 72 | |
| Production of astaxanthin (increased to 2.7-fold) | An inversion (YJL052C-A-YJR071C) event and a translocation exchange between synV and synX | Two synthetic chrs (chrV&X) | In vivo haploid SCRaMbLE | Colony color | Two strains | 73 | |
| Production of violacein (increased to 17-fold) | Unresolved | Single synthetic chrII | In vitro SCRaMbLE and in vivo haploid SCRaMbLE | Colony color | Seven strains | 25 | |
| Production of PDV (increased to 3.48-fold) | Deletions of YEL017C-A, YEL017W, YER151C, and YER182W | Single ring synthetic chrV | In vivo haploid SCRaMbLE | Colony color | Fourteen strains | 21 | |
| Production of violacein (increased to 2.1-fold) | Deletion of UBP3 and an inversion of 5337 bp region containing SWI4 and LSM4 | Single synthetic chrV | In vivo haploid SCRaMbLE | Colony color | One strain | 67 | |
| Production of betulinic acid (increased to 3-fold) | Deletion of 723![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) bp of non-coding DNA bp of non-coding DNA |
Single synthetic chrV | In vivo haploid SCRaMbLE | Semi-automated rapid LC-MS screening | Twelve strains | 68 |
Thermotolerance is one of the major limitations in the industrial production of ethanol using yeast.70 Recent studies showed that both haploid and diploid SCRaMbLE can increase tolerance to high temperatures. Luo et al. generated three thermotolerant haploid strains using ReSCuES, which was 1.28 ± 0.03 times faster than the control.27 Another study employed heterozygous diploid SCRaMbLE by crossing a Y12 sake-brewing S. cerevisiae strain and a synX-bearing strain, and two SCRaMbLEd strains that displayed an improvement in growth at 42 °C were obtained.23 Whole-genome sequencing analysis showed that deletion of the region spanning YJL154C-YJL140W is responsible for the improvement. For increased or decreased pH tolerance, Ma et al. generated 7 strains with increased alkali tolerance at pH 8.0 through SCRaMbLEing haploid yeast strains harboring either one (synV) or two (synV and synX) synthetic chromosomes.26 They determined that the deletion of YER161C (SPT2) could increase alkali tolerance in yeast through comparative analysis of structural variations. Luo et al. generated three strains with increased tolerance to acetic acid and one of the SCRaMbLEd strains achieved nearly a 21-fold increase in growth capacity compared to the control.27 In addition to thermotolerance and pH, other chemical or environmental tolerances can also be enhanced through SCRaMbLE. Three strains with increased ethanol tolerances were generated using ReSCuES.27 Furthermore, the authors demonstrated that the disruption of ACE2 is responsible for the increased tolerance. Blount et al. introduced the heterologous pathway of xylose utilization into yeast with synV, and successfully obtained a strain with enhanced growth on a medium with xylose as the sole carbon source through SCRaMbLE.67
SCRaMbLE also can increase drug tolerance. Seven evolved strains with increased rapamycin resistance were generated via SCRaMbLE of heterozygous diploid yeast strains.37 The deletion of gene GLN3, long-range loss of heterozygosity (LOH) in the left arm of the synthetic chromosome X, whole chromosome LOH of the synthetic chromosome X, and duplication of chromosome VIII (trisomy) were each found to lead to increased rapamycin resistance in synthetic yeast. Similarly, ten strains exhibiting caffeine-tolerant phenotypes were obtained through SCRaMbLE.23 Whole genome sequencing analysis determined that the duplication of POL32 is responsible for the increase of caffeine tolerance. Recently, Ong et al. SCRaMbLEd a haploid yeast strain harboring synII resulting in increased hygromycin B resistance, showing that deletions of YBR219C and YBR220C contribute to hygromycin B resistance phenotypes.71
Carotenoids are important antioxidants and display colored phenotypes. Through one round of SCRaMbLE, five SCRaMbLEd strains exhibiting different colors were obtained.22 The maximum yield of carotenoids was increased to 1.5-fold and the deletion of YEL013W is responsible for the increase. Using the precise control switch of “AND gate” and the MuSIC strategy, production of carotenoids (37.39 mg L−1) was increased up to 38.8-fold through 5 iterative cycles of SCRaMbLE. Wu et al. used in vitro SCRaMbLE to increase the production of β-carotene.24 A total of 17 unique β-carotene pathway structures were identified and the highest yield of β-carotene was 5.1-fold greater than that of the original construct. Correlation analysis between the rearrangement structure of the pathway and the production of β-carotene revealed that crtI is the key gene in the metabolic pathway, and the duplication and inversion of this gene led to increased β-carotene production. Similarly, a 2-fold improvement in β-carotene production (500 μg L−1) was generated by the SCRaMbLE-in method.25 In combination with optimization of culturing conditions, Zhang et al. obtained a high-yield strain of lycopene (41.47 mg L−1) via SCRaMbLE.72 More recently, two cycles of SCRaMbLE in a synV&X bearing strain revealed that the combined effect of translocation and inversion increased the yield of astaxanthin by 2.7-fold. Violacein is another pathway which is easy to screen. Liu et al. used the SCRaMbLE-in method to achieve efficient combinatorial library construction, exhibiting a 17-fold improvement (16.8 mg L−1) in violacein production compared with the control strain.25 Wang et al. SCRaMbLEd ring_synV to generate complex genomic variations and determined that 11 of the 29 novel SVs prompted the pro-deoxy violacein (PDV) biosynthesis.21 The highest productions of PDV increased by approximately 3.48-fold compared with the control strain. The deletions of YEL017C-A, YEL017W, YER151C, and YER182W have an effect on the increase of PDV production. Through combining SCRaMbLE with long-read nanopore sequencing, Blount et al. quickly identified genomic rearrangements and obtained high-producing strains of violacein and penicillin through in vivo SCRaMbLE of synV.67
The above studies all had colorful phenotypes that allowed easy selection of improved phenotypes. However, most industrially relevant metabolites exhibit no easily selectable phenotype.74 To address this bottleneck, Gowers et al. introduced automated and high-throughput sample preparation to expand the screening capabilities of SCRaMbLEd strains.68 This semi-automated workflow screened 1000 colonies, and identified 12 strains with between 2- to 7-fold improvement in betulinic acid (BA) titer. The deletion of 723 bp of non-coding DNA led to approximately 3-fold increase in BA titer.
These and other studies provide a roadmap for current and future applications of the SCRaMbLE system, allowing creation of industrially-relevant phenotypes, and deepening understanding of the relationship between genotype and phenotype. Tremendous pools of genetic diversity differing in gene orders and genome structures can be obtained in just a matter of days, and thus diverse traits can be evolved. Overall, SCRaMbLE is a promising way to generate superior strains for industrial applications.75
Genotype-phenotype correlations. Genotype to phenotype correlations have been difficult to establish due to the complexity of both phenotypes and variations. SCRaMbLE is a useful strategy to efficiently link genotype to phenotype, and can frequently obtain the desired phenotype by changing copy-numbers of genes and gene orders often with surprising results (Table 3).76 For example, deletion of YEL013W caused a global transcriptome change and increased the flux of the mevalonate (MVA) pathway, but had not been reported before.22 Deletion of 723 bp of non-coding DNA led to the deletion of the 3′ untranslated region of TIR1 and an approximately 3-fold increase in BA titer, but had not been previously reported to affect the BA pathway. POL32, a nonessential subunit of DNA polymerase δ, was previously unlinked to caffeine resistance.23 A recent study demonstrated that duplication of POL32 is responsible for an increase in caffeine tolerance. In terms of gene order changes, inversion resulted in loss of the 3′UTR of ACE2 and disruption of ACE2, which is responsible for increased ethanol tolerance.27 More recently, the combined effects of translocation and inversion increased the yield of astaxanthin, and combined structural variations were identified that led increased mRNA levels of STE18, RPS5 and MCM22, which contribute to astaxanthin biosynthesis.73 None of them were previously reported to directly affect astaxanthin production.
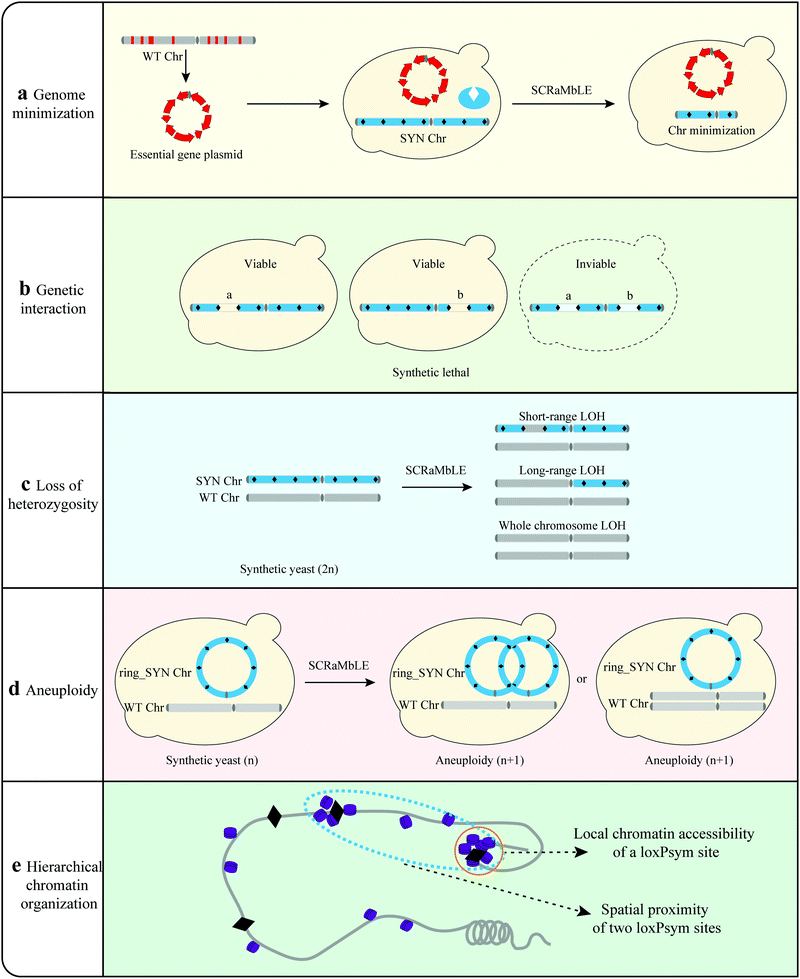
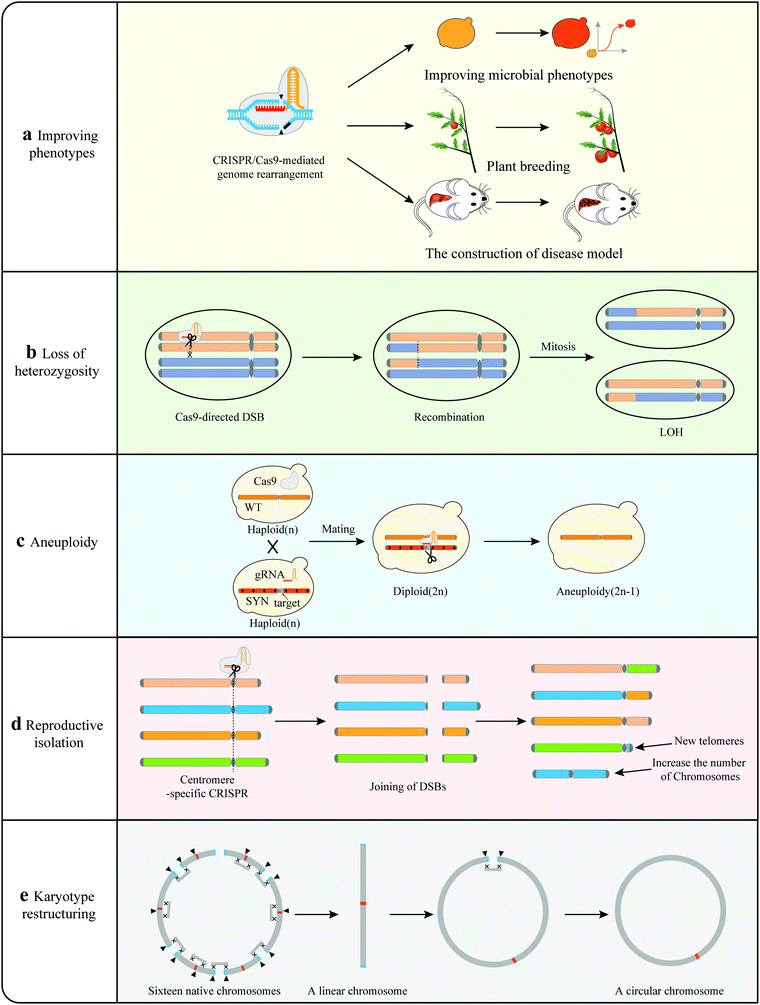
Genome minimization. A fundamental question in biology regards the minimal set or number of genes required to support cellular life in the absence of environmental stress.77 Hutchison III et al. designed and synthesized a minimal bacterial genome using a top-down approach.78 However, most eukaryotic organisms have more complicated genomes, and the construction of a minimal genome would be extremely difficult.79 The SCRaMbLE method has the potential to create reduced genome sets by inducing random deletions to eliminate non-essential genomic regions over time.55 Wang et al. SCRaMbLEd a haploid yeast strains harboring synIII with all essential genes of yeast chromosome III (chrIII) clustered in a centromeric plasmid to simplify the yeast genome (Fig. 4).36 Investigation of all SCRaMbLEd strains revealed a total deletion of 97 kb in synIII, which is 35.7% of the length of synIII, guiding the construction of the minimal gene set. Luo et al. revealed that 39 out of 65 nonessential genes in synXIIL could be removed collectively without affecting cell viability at 30 °C in a rich medium with clustered essential genes.80 This indicated that SCRaMbLE with clustered essential genes is an effective method to streamline the synthetic yeast genome. Through multiple rounds of SCRaMbLE, it is possible to continuously simplify the synthetic yeast genome. In the future, all essential genes of the yeast genome can be clustered in a fully synthetic yeast to facilitate the study of simplified yeast genomes.36
Genetic interaction. Single-, double- and triple-gene disruptions of the yeast genome have been constructed to explore the resulting genetic interactions.81,82 They offer a fundamental way to understand global genetic interactions, although they are relatively time-consuming and laborious. SCRaMbLE provides a platform to investigate complex genetic interactions by randomly deleting different regions of the genome (Fig. 4). A synIII strain with clustered essential genes was used as an example to explore the interactions.36 A total of 141 SCRaMbLEd strains with various deletions were detected and the deletion frequencies were statistically analyzed. For regions with no deletion event, a synthetic lethal interaction was detected in the region of synIII 82–88 kb, providing a strategy to study complex genetic interactions within the genome.
Loss of heterozygosity (LOH). SCRaMbLE has also been able to induce different levels of loss of heterozygosity (LOH) events. Li et al. utilized the SCRaMbLE system to comparatively analyze the mechanisms underlying the phenotypic adaption.37 Different levels of LOH events including short-range LOH, long-range LOH and whole chromosome LOH were detected in SCRaMbLEd strains. By independently constructing corresponding strains with different levels of LOH, Li et al. revealed that different levels of LOH enhanced rapamycin resistance in synthetic yeast. The underlying genetic variations ranged from genes to the whole chromosome, showing the diverse processes of species evolution and providing a valuable insight into yeast genome architecture and function.37
Aneuploidy. Aneuploidy is the condition of having missing or extra chromosomes compared with the wild-type organism, resulting from chromosome segregation and replication errors. Aneuploidy may drive adaptive evolution in yeast, and can contribute to resistance to environmental or other stressors.83,84 SCRaMbLE has been verified to generate aneuploid chromosomes and provide a platform for the investigation of aneuploidy (Fig. 4).21 Researchers SCRaMbLEd a synthetic strain harboring ring_synV and generated aneuploidies of chromosomes I, III, VI, XII, XIII, and ring_synV in SCRaMbLEd strains. The ring synthetic chromosome tended to generate neochromosomes with complex structure variations. Rearrangements can produce relatively large interference to strains, resulting in aneuploidy via rearrangement directly or generating aneuploidy to adapt to the interference of the genome.
Hierarchical chromatin organization. SCRaMbLE also provides a unique model to systematically interrogate the dynamic process of eukaryotic genome evolution (Fig. 4). Sijie et al. found a specific landscape of rearrangement frequency by investigating 260
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rearrangement events.65 To explore the mechanisms underlying the rearrangement patterns, the rearrangement map was compared with local chromatin structures and three-dimensional architecture. These analyses revealed that the rearrangement pattern was affected by chromatin accessibility and spatial contact probability, and that the rearrangements tended to occur in three-dimensional spatially proximal and chromatin-accessible regions. This investigation of numerous rearrangement events indicated that hierarchical chromatin organization play an important role in structural rearrangement-mediated directed genome evolution.
000 rearrangement events.65 To explore the mechanisms underlying the rearrangement patterns, the rearrangement map was compared with local chromatin structures and three-dimensional architecture. These analyses revealed that the rearrangement pattern was affected by chromatin accessibility and spatial contact probability, and that the rearrangements tended to occur in three-dimensional spatially proximal and chromatin-accessible regions. This investigation of numerous rearrangement events indicated that hierarchical chromatin organization play an important role in structural rearrangement-mediated directed genome evolution.
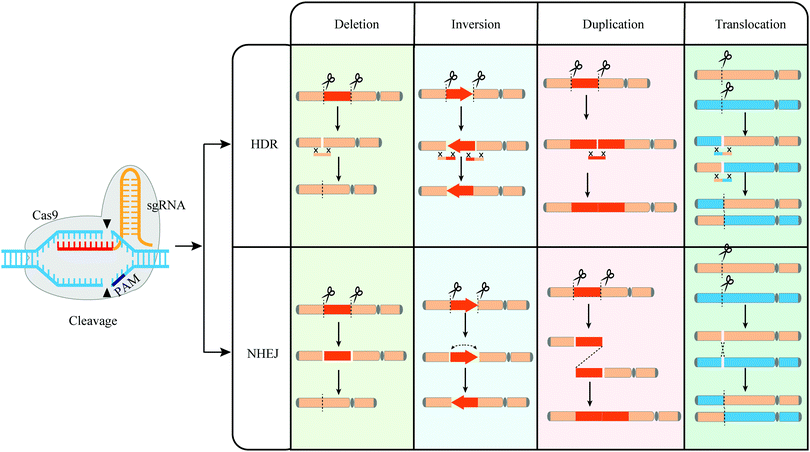
3 Genome editing-mediated directed genome evolution
SCRaMbLE requires synthetic genome and genome writing techniques, which are not essential in endonuclease-induced genome rearrangements for directed genome evolution. Endonucleases used to induce genome rearrangements include natural enzymes, TaqI,31 MseI32 and meganuclease (I-SceI, I-CreI, I-DmoI),85 and artificial nucleases such as CRISPR-Cas9, ZFNs, and TALENs.86,87 Endonuclease-induced genome rearrangements are initiated by forming DSBs at specific loci, followed by homologous recombination (HR) or non-homologous end joining (NHEJ) repairing. Among all endonucleases, CRISPR-Cas9, TaqI, and MseI are more applicable in directed genome evolution as they can readily cleave a large number of loci in the genome.3.1 The technique of CRISPR-Cas9 mediated genome rearrangement
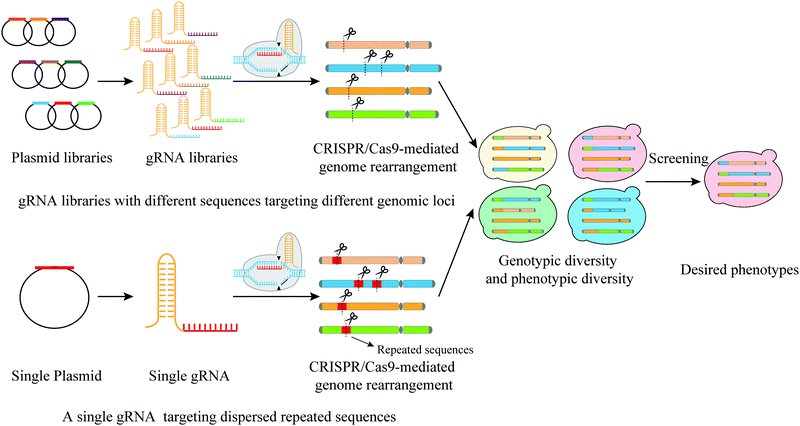 | ||
| Fig. 6 Two main CRISPR-Cas9 mediated strategies to generate rearrangement libraries: targeting different genomic loci by gRNA libraries and targeting dispersed repeated sequences by a single gRNA. | ||
The first strategy requires the simultaneous introduction of many gRNAs with different sequences targeting different genomic loci. Sadhu et al. designed 95 gRNAs with all targeting sequences at different loci on the left arm of yeast chromosome 7.40 With this design, the authors used CRISPR-Cas9 to generate DSBs and introduced local recombination events, which allowed fine mapping of trait variants. A single nucleotide polymorphism located in the ORF encoding the transporter Pmr1 was found responsible for varied degrees of manganese sensitivities in yeast. Apel et al. used this strategy, but with gRNAs targeting 23 sequences on 16 chromosomes to address chromosomal integration locus of heterologous gene.30 The authors successfully obtained an evolved yeast strain with a high yield of taxadiene.
The second approach reshuffles the genome through targeting dispersed repeated sequences. For instance, Fleiss et al. employed a single gRNA targeting 5 different Ty3 Long Terminal Repeats (LTRs) that are located on 4 different chromosomes.29 In this study, they were able to induce chromosomal rearrangements yielding 22 yeast strains with different karyotypes. The growth phenotypes of these strains under different conditions were carefully recorded. These growth conditions include various types of stresses with some caused by the addition of different drugs that interfere with replication, transcription, translation or affect subcellular structures. These strains were found even more competent in growth than the WT type in the presence of caffeine, cycloheximide and fluconazole.29 Similarly, Mitsui et al. targeted the repeated δ-sequences in order to shuffle the genome.101 The S. cerevisiae genome harbors at least 100 copies of δ sequence that are distributed throughout the haploid genome.102 A heat-resistant strain was obtained, which also exhibited resistance to the growth inhibition of high ethanol and acid. As genome structural rearrangements can be used to generate strains with increased tolerance to stress, Mitsui et al. further combined CRISPR-δ integration aiming to obtain acid tolerance strain with rational metabolic engineering that integrates a series of genes from Leuconostoc mesenteroides into the yeast genome to enable the yeast strain to produce LA.103 This combined method resulted in a high yield of D-LA.
Loss of heterozygosity. Like SCRaMbLE, CRISPR also generates LOH events but through a different mechanism (Fig. 7). CRISPR-mediated LOH events require the induction of DSBs in heterozygous diploids. In the aforementioned study by Sadhu et al., the gRNAs were designed to target only one chromosome in a heterozygous diploid, using PAMs polymorphic between the two homologous chromosomes.40 A specific DSB could induce different LOH events at different crossover sites, usually within 20 kb of the DSB, thus generating a large number of LOH lines. Using 95 gRNAs targeting the left arm of one of the two yeast chromosome VII, 384 LOH lines were obtained and assayed for growth rates under 12 different conditions. Specific genetic variants can be quickly linked to specific growth phenotypes.40 However, this approach may not be applicable to the organisms in which mitotic recombination events occur at a relatively low frequency.
Aneuploidy. Like SCRaMbLE, CRISPR-Cas9 can eliminate an entire chromosome resulting in aneuploidy (Fig. 7). Xu et al. designed a specific gRNA targeting the neighborhood of the centromere of one chromosome in a yeast diploid (2n),38 and they found that only one DSB around the centromere is sufficient to cause the elimination of the entire chromosome to form aneuploidy (2n − 1). Similar observations have also been made in mammalian cells; the authors have revealed that the CRISPR-Cas9 system can enable complete elimination of the targeted chromosomes by multiple CRISPR-Cas9 mediated DNA cleavages.104,105 Chromosome elimination via CRISPR-Cas9 offers a powerful tool to generate aneuploidy in different species that could be used as models for human disease.
Karyotype restructuring and reproductive isolation. Apart from generating mutations and gene disruptions, chromosome translocations can also result in reproductive isolation during meiosis.106 In a recent study, Yadav et al. applied CRISPR-Cas9 to drive reproductive isolation in C. neoformans genome, in which the centromeres of 9 chromosomes harbor repeated retrotransposons (Tcn2) (Fig. 7).107 The authors induced multiple DSBs at the centromeric retrotransposons via the CRISPR-Cas9 system and shuffled the genome through chromosomal translocations.28 Only two rearranged strains survived, with both containing intensive interchromosomal rearrangements and increased numbers of chromosomes. They also failed to undergo normal meiosis, resulting in postzygotic reproductive isolation. Yadav's observation is consistent with the results of comparative genomic studies which concluded that inter-centromeric translocations could cause reproductive isolation and promote incipient speciation.108,109 Shao et al. and Luo et al. extended the applications of CRISPR-Cas9 to karyotype restructuring in yeast (Fig. 7).41,42 These researchers were able to reduce the number of yeast chromosomes from 16 to two and even one by CRISPR-mediated chromosome end-to-end fusions. Circularization was shown by our group possible for synthetic yeast chromosome.21 Similarly, the karyotype of that single-chromosome yeast was further altered, generating a single circular chromosome yeast strain via CRISPR-Cas9.110
In summary, karyotype restructuring results in changes of chromosome, centromere and telomere numbers, leading to reproductive isolation.28,41,42
3.2 TaqI and MseI mediated genome rearrangement
In regard to natural enzymes, Muramoto et al. developed a genome-restructuring system, namely the TAQing system, to induce large-scale genome rearrangements through introducing the heat-activated endonuclease TaqI into S. cerevisiae and A. thaliana cells.31 TaqI is an endonuclease that recognizes the TCGA palindrome and can tolerate base analogue and chemically modified substrates. The TaqI-mediated genome rearrangements were assayed for their link to various phenotypes. New strains with improved ethanol-production at high temperature and increased biomass of A. thaliana were obtained. The TAQing system was then developed into the Ex-TAQing system, in which MseI capable of efficient cleavage of the genome at room temperature was used instead of TaqI.32 This modified system also resulted in several strains with beneficial phenotypes that tolerate environmental stress, such as high salinity stress tolerance and hypersensitivity to abscisic acid.4. Conclusions and future perspectives
Synthetic biology has been greatly developed recently. According to a recent report by Reportlinker in 2021, the current global synthetic biology market size is USD 9.5 billion, which has doubled from that in 2018 and is expected to continuously grow and to reach USD 30.7 billion in 2026 (https://www.reportlinker.com/p05246309/Synthetic-Biology-Market-by-Tool-Technology-Application-Industrial-Global-Forecast-to.html). Directed genome evolution has recently drawn great attention, as it has contributed to the growth of the synthetic biology market. With the development of novel tools for directed genome evolution, many industrially-relevant phenotypes of microbial cells have been altered in our favor. One would anticipate that through directed genome evolution, microbial chassis suited for high yields of different types of molecules can be broadly applied to industry.The techniques used to induce structural rearrangements provide useful tools for the investigation of the roles of genome rearrangements in natural evolution and tumorigenesis. Genome structural rearrangements are known to result in fusion genes that cause human cancers. For example, chromosomal translocation t(15;17) results in a chimera of the PML gene and the retinoic acid receptor alpha (RAR alpha) gene and acute promyelocytic leukemia (APL).111 Such newly developed techniques render possibilities to study early cellular and molecular events in the context of an ongoing structural rearrangement.
Both CRISPR-Cas9 and SCRaMbLE techniques used in directed genome evolution have their own advantages and limits. CRISPR-Cas9 requires the use of specific gRNA and the presence of PAM sites in the target; therefore the editing capabilities need to be expanded.112,113 Future efforts include exploring a PAM-free system.114 In comparison with the CRISPR-Cas9 method, SCRaMbLE can induce a much larger number of rearrangement events at a much higher frequency. The high-throughput, genome-scale or randomized creation of structural variations usually depend on the formation and the follow-up repair of DSBs, failure of which can cause lethality and thus are less efficient in generation of a large pool of structural variations. Whereas, SCRaMbLE system is recombinase-mediated and DSB-independent methods. Consolidating 5.5 synthetic chromosomes in S. cerevisiae, followed by a short-time induction of SCRaMbLE generates genomic libraries that contain up to 105 rearrangement events,65 which are expected to be increased tremendously if all 16 yeast chromosomes are replaced by synthetic chromosomes. For SCRaMbLE in Sc2.0, loxPsym sites were inserted into downstream of the 3′UTR of all non-essential genes through synthetic genomes, and most of the insertions have no effects on the growth fitness. The strategy can be extended to other annotated genomes. However, introduction of targeted loci to generate structural variants into the genome is still challenging, as there are many regions with limited or unknown function in the genome, especially for higher organisms. The insertion of recombination sites may influence the function of particular gene. For instance, the insertion of a loxPsym site in synX disrupts transcription of neighboring gene ATP2, leading to a growth defect.115 Nevertheless, the process of debugging can be used to explore the regions with limited or unknown function, and then we can redesign the inserted position based on the feedback.
The synthetic chromosomes after SCRaMbLE can be also transferred to wild-type strains via CRISPR-Cas9, which can transmit rearrangement related traits and expand the application of SCRaMbLE.38 SCRaMbLE needs the insertion of loxPsym sites in the genome in advance and the rates of rearrangements are dependent on the numbers of inserted loxPsym sites. Synthetic DNA has become much less expensive, providing different choices of methods used to insert loxPsym sites. The Cre/loxP system has been used in various organisms.116–118 Thus the use of SCRaMbLE can be expanded to other synthetic organisms. In addition, SCRaMbLE is expected to be extrapolated to natural genomes, such as industrially relevant microbes,75via genome editing tools for multiplex genomic insertion of recombination sites. SCRaMbLE is supposed to be compatible with many methods that have been used to create small scale variations (SNP and InDel). A combination of multiple technologies may further increase genome diversity and pave new avenues in directed genome evolution.
Synthetic biology has been successfully used in the production of highly valuable compounds in engineered microbes with the introduction of exogenous DNA. With the development of large DNA writing technology (i.e. design–assembly–delivery of large DNA), a fit of SCRaMbLE involving writing non-native DNA has great potential to further improve the scales of variation and diversity of SCRaMbLE. Moreover, synthetic biology has become an attractive media for the development of data storage.119,120
Directed genome evolution involves the changes of many loci throughout the genome producing numerous intermediate strains and thus requires complex genetic manipulations and culturing procedures, which are heavily labor- and time-consuming. Nevertheless, introducing automation technology that offers a high-throughput platform would facilitate the development of directed genome evolution.68 The Biofoundries Alliance has endeavored to provide automation to support the engineering of biological systems and its participation will be important for the future perspectives of directed genome evolution.121,122 Resulting giant datasets (genomics, transcriptomics, proteomics, metabolomics, protein interaction maps, etc.) require sophisticated analysis and conversion to comprehensible outputs.123 Therefore, the involvement of artificial intelligence (AI)/machine learning (ML) is a future direction.123,124 These datasets can train and refine machine learning models that can effectively predict beneficial variations and guide the design of directed genome evolution.34 Data-driven directed genome evolution aided by automation and AI may become a powerful and routine methodology for the construction of the next-generation microbial cell factories (MCFs) that supports the growth of synthetic biology market.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was funded by the National Key Research and Development Program of China (2021YFC2100800), the National Natural Science Foundation of China (21621004, 31861143017, 31971351, 31800719, 21907074), and the Natural Science Foundation of Tianjin (20JCQNJC02090). We thank Yan Zhang from Tianjin University for fruitful discussions and advices.References
- M. S. Packer and D. R. Liu, Nat. Rev. Genet., 2015, 16, 379–394 CrossRef CAS PubMed.
- S. I. Wright, I. V. Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley, M. D. McMullen and B. S. Gaut, Science, 2005, 308, 1310–1314 CrossRef CAS PubMed.
- C. A. Driscoll, D. W. Macdonald and S. J. O'Brien, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9971–9978 CrossRef CAS PubMed.
- R. E. Cobb, R. Chao and H. Zhao, AIChE J., 2013, 59, 1432–1440 CrossRef CAS PubMed.
- M. Cao, V. G. Tran and H. Zhao, Curr. Opin. Biotechnol., 2020, 66, 95–104 CrossRef CAS PubMed.
- C. A. Hutchison, R. Y. Chuang, V. N. Noskov, N. Assad-Garcia, T. J. Deerinck, M. H. Ellisman, J. Gill, K. Kannan, B. J. Karas, L. Ma, J. F. Pelletier, Z. Q. Qi, R. A. Richter, E. A. Strychalski, L. Sun, Y. Suzuki, B. Tsvetanova, K. S. Wise, H. O. Smith, J. I. Glass, C. Merryman, D. G. Gibson and J. C. Venter, Science, 2016, 351, d6253 CrossRef PubMed.
- R. P. Alexander, G. Fang, J. Rozowsky, M. Snyder and M. B. Gerstein, Nat. Rev. Genet., 2010, 11, 559–571 CrossRef CAS PubMed.
- H. H. Wang, F. J. Isaacs, P. A. Carr, Z. Z. Sun, G. Xu, C. R. Forest and G. M. Church, Nature, 2009, 460, 894–898 CrossRef CAS PubMed.
- E. M. Barbieri, P. Muir, B. O. Akhuetie-Oni, C. M. Yellman and F. J. Isaacs, Cell, 2017, 171, 1453–1467 CrossRef CAS PubMed.
- J. E. DiCarlo, A. J. Conley, M. Penttilä, J. Jäntti, H. H. Wang and G. M. Church, ACS Synth. Biol., 2013, 2, 741–749 CrossRef CAS PubMed.
- S. O. Halperin, C. J. Tou, E. B. Wong, C. Modavi, D. V. Schaffer and J. E. Dueber, Nature, 2018, 560, 248–252 CrossRef CAS PubMed.
- C. J. Tou, D. V. Schaffer and J. E. Dueber, ACS Synth. Biol., 2020, 9, 1911–1916 CrossRef CAS PubMed.
- C. Kuscu, M. Parlak, T. Tufan, J. Yang, K. Szlachta, X. Wei, R. Mammadov and M. Adli, Nat. Methods, 2017, 14, 710–712 CrossRef CAS PubMed.
- Y. Ma, J. Zhang, W. Yin, Z. Zhang, Y. Song and X. Chang, Nat. Methods, 2016, 13, 1029–1035 CrossRef CAS PubMed.
- P. Xu, Z. Liu, Y. Liu, H. Ma, Y. Xu, Y. Bao, S. Zhu, Z. Cao, Z. Wu, Z. Zhou and W. Wei, Nat. Biotechnol., 2021, 1–11 Search PubMed.
- Z. Bao, M. HamediRad, P. Xue, H. Xiao, I. Tasan, R. Chao, J. Liang and H. Zhao, Nat. Biotechnol., 2018, 36, 505–508 CrossRef CAS PubMed.
- J. Lian, C. Schultz, M. Cao, M. HamediRad and H. Zhao, Nat. Commun., 2019, 10, 1–10 CrossRef PubMed.
- A. D. Garst, M. C. Bassalo, G. Pines, S. A. Lynch, A. L. Halweg-Edwards, R. Liu, L. Liang, Z. Wang, R. Zeitoun, W. G. Alexander and R. T. Gill, Nat. Biotechnol., 2017, 35, 48–55 CrossRef CAS PubMed.
- R. Liu, L. Liang, A. Choudhury, M. C. Bassalo, A. D. Garst, K. Tarasava and R. T. Gill, Metab. Eng., 2018, 47, 303–313 CrossRef CAS PubMed.
- K. R. Roy, J. D. Smith, S. C. Vonesch, G. Lin, C. S. Tu, A. R. Lederer, A. Chu, S. Suresh, M. Nguyen, J. Horecka, A. Tripathi, W. T. Burnett, M. A. Morgan, J. Schulz, K. M. Orsley, W. Wei, R. S. Aiyar, R. W. Davis, V. A. Bankaitis, J. E. Haber, M. L. Salit, R. P. St. Onge and L. M. Steinmetz, Nat. Biotechnol., 2018, 36, 512–520 CrossRef CAS PubMed.
- J. Wang, Z. X. Xie, Y. Ma, X. R. Chen, Y. Q. Huang, B. He, J. Bin, B. Z. Li and Y. J. Yuan, Nat. Commun., 2018, 9, 3783 CrossRef PubMed.
- B. Jia, Y. Wu, B. Li, L. A. Mitchell, H. Liu, S. Pan, J. Wang, H. Zhang, N. Jia, B. Li, M. Shen, Z. Xie, D. Liu, Y. Cao, X. Li, X. Zhou, H. Qi, J. D. Boeke and Y. Yuan, Nat. Commun., 2018, 9, 1–13 CrossRef CAS PubMed.
- M. J. Shen, Y. Wu, K. Yang, Y. Li, H. Xu, H. Zhang, B. Li, X. Li, W. Xiao, X. Zhou, L. A. Mitchell, J. S. Bader, Y. Yuan and J. D. Boeke, Nat. Commun., 2018, 9, 1–8 CrossRef CAS PubMed.
- Y. Wu, R. Zhu, L. A. Mitchell, L. Ma, R. Liu, M. Zhao, B. Jia, H. Xu, Y. Li, Z. Yang, Y. Ma, X. Li, H. Liu, D. Liu, W. Xiao, X. Zhou, B. Li, Y. Yuan and J. D. Boeke, Nat. Commun., 2018, 9, 1935–1939 CrossRef PubMed.
- W. Liu, Z. Luo, Y. Wang, N. T. Pham, L. Tuck, I. Pérez-Pi, L. Liu, Y. Shen, C. French, M. Auer, J. Marles-Wright, J. Dai and Y. Cai, Nat. Commun., 2018, 9, 1–10 CrossRef PubMed.
- L. Ma, Y. Li, X. Chen, M. Ding, Y. Wu and Y. Yuan, Microb. Cell Fact., 2019, 18, 1–11 CrossRef PubMed.
- Z. Luo, L. Wang, Y. Wang, W. Zhang, Y. Guo, Y. Shen, L. Jiang, Q. Wu, C. Zhang, Y. Cai and J. Dai, Nat. Commun., 2018, 9, 1–10 CrossRef PubMed.
- V. Yadav, S. Sun, M. A. Coelho and J. Heitman, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 7917–7928 CrossRef CAS PubMed.
- A. Fleiss, S. O'Donnell, T. Fournier, W. Lu, N. Agier, S. Delmas, J. Schacherer and G. Fischer, PLoS Genet., 2019, 15, e1008332 CrossRef CAS PubMed.
- A. Reider Apel, L. D'Espaux, M. Wehrs, D. Sachs, R. A. Li, G. J. Tong, M. Garber, O. Nnadi, W. Zhuang, N. J. Hillson, J. D. Keasling and A. Mukhopadhyay, Nucleic Acids Res., 2017, 45, 496–508 CrossRef PubMed.
- N. Muramoto, A. Oda, H. Tanaka, T. Nakamura, K. Kugou, K. Suda, A. Kobayashi, S. Yoneda, A. Ikeuchi, H. Sugimoto, S. Kondo, C. Ohto, T. Shibata, N. Mitsukawa and K. Ohta, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- H. Tanaka, N. Muramoto, H. Sugimoto, A. H. Oda and K. Ohta, Plant J., 2020, 103, 2139–2150 CrossRef CAS PubMed.
- A. J. Simon, S. D. Oelsnitz and A. D. Ellington, Nat. Biotechnol., 2019, 37, 730–743 CrossRef CAS PubMed.
- Y. Wang, P. Xue, M. Cao, T. Yu, S. T. Lane and H. Zhao, Chem. Rev., 2021 DOI:10.1021/acs.chemrev.1c00260.
- J. Jin, B. Jia and Y. Yuan, Curr. Opin. Biotechnol., 2020, 66, 165–170 CrossRef CAS PubMed.
- P. Wang, H. Xu, H. Li, H. Chen, S. Zhou, F. Tian, B. Li, X. Bo, Y. Wu and Y. Yuan, ACS Synth. Biol., 2020, 9, 1181–1189 CrossRef CAS PubMed.
- Y. Li, Y. Wu, L. Ma, Z. Guo, W. Xiao and Y. Yuan, Sci. China Life Sci., 2019, 62, 381–393 CrossRef CAS PubMed.
- H. Xu, M. Han, S. Zhou, B. Li, Y. Wu and Y. Yuan, Nat. Commun., 2020, 11, 1–9 CAS.
- Z. Yang and M. Blenner, Curr. Opin. Biotechnol., 2020, 66, 255–266 CrossRef CAS PubMed.
- M. J. Sadhu, J. S. Bloom, L. Day and L. Kruglyak, Science, 2016, 352, 1113–1116 CrossRef CAS PubMed.
- Y. Shao, N. Lu, Z. Wu, C. Cai, S. Wang, L. Zhang, F. Zhou, S. Xiao, L. Liu, X. Zeng, H. Zheng, C. Yang, Z. Zhao, G. Zhao, J. Zhou, X. Xue and Z. Qin, Nature, 2018, 560, 331–335 CrossRef CAS PubMed.
- J. Luo, X. Sun, B. P. Cormack and J. D. Boeke, Nature, 2018, 560, 392–396 CrossRef CAS PubMed.
- W. De Coster, M. H. Weissensteiner and F. J. Sedlazeck, Nat. Rev. Genet., 2021, 1–16 Search PubMed.
- S. S. Ho, A. E. Urban and R. E. Mills, Nat. Rev. Genet., 2020, 21, 171–189 CrossRef CAS PubMed.
- J. Xia, Z. Guo, Z. Yang, H. Han, S. Wang, H. Xu, X. Yang, F. Yang, Q. Wu, W. Xie, X. Zhou, W. Dermauw, T. C. J. Turlings and Y. Zhang, Cell, 2021, 184, 1693–1705 CrossRef CAS PubMed.
- M. Alonge, X. Wang, M. Benoit, S. Soyk, L. Pereira, L. Zhang, H. Suresh, S. Ramakrishnan, F. Maumus, D. Ciren, Y. Levy, T. H. Harel, G. Shalev-Schlosser, Z. Amsellem, H. Razifard, A. L. Caicedo, D. M. Tieman, H. Klee, M. Kirsche, S. Aganezov, T. R. Ranallo-Benavidez, Z. H. Lemmon, J. Kim, G. Robitaille, M. Kramer, S. Goodwin, W. R. McCombie, S. Hutton, J. Van Eck, J. Gillis, Y. Eshed, F. J. Sedlazeck, E. van der Knaap, M. C. Schatz and Z. B. Lippman, Cell, 2020, 182, 145–161 CrossRef CAS PubMed.
- J. Svedberg, S. Hosseini, J. Chen, A. A. Vogan, I. Mozgova, L. Hennig, P. Manitchotpisit, A. Abusharekh, T. M. Hammond, M. Lascoux and H. Johannesson, Nat. Commun., 2018, 9, 1–13 CrossRef CAS PubMed.
- M. Cossu, C. Badel, R. Catchpole, D. Gadelle, E. Marguet, V. Barbe, P. Forterre and J. Oberto, PLoS Genet., 2017, 13, e1006847 CrossRef PubMed.
- J. O. Korbel, A. E. Urban, J. P. Affourtit, B. Godwin, F. Grubert, J. F. Simons, P. M. Kim, D. Palejev, N. J. Carriero and L. Du, Science, 2007, 318, 420–426 CrossRef CAS PubMed.
- J. Peter, M. De Chiara, A. Friedrich, J. Yue, D. Pflieger, A. Bergström, A. Sigwalt, B. Barre, K. Freel, A. Llored, C. Cruaud, K. Labadie, J. Aury, B. Istace, K. Lebrigand, P. Barbry, S. Engelen, A. Lemainque, P. Wincker, G. Liti and J. Schacherer, Nature, 2018, 556, 339–344 CrossRef CAS PubMed.
- J. Yue, J. Li, L. Aigrain, J. Hallin, K. Persson, K. Oliver, A. Bergström, P. Coupland, J. Warringer, M. C. Lagomarsino, G. Fischer, R. Durbin and G. Liti, Nat. Genet., 2017, 49, 913–924 CrossRef CAS PubMed.
- J. Kreplak, M. Madoui, P. Cápal, P. Novák, K. Labadie, G. Aubert, P. E. Bayer, K. K. Gali, R. A. Syme, D. Main, A. Klein, A. Bérard, I. Vrbová, C. Fournier, L. D. Agata, C. Belser, W. Berrabah, H. Toegelová, Z. Milec, J. Vrána, H. Lee, A. Kougbeadjo, M. Térézol, C. Huneau, C. J. Turo, N. Mohellibi, P. Neumann, M. Falque, K. Gallardo, R. McGee, B. Tar An, A. Bendahmane, J. Aury, J. Batley, M. Le Paslier, N. Ellis, T. D. Warkentin, C. J. Coyne, J. Salse, D. Edwards, J. Lichtenzveig, J. Macas, J. Doležel, P. Wincker and J. Burstin, Nat. Genet., 2019, 51, 1411–1422 CrossRef CAS PubMed.
- C. M. B. Carvalho and J. R. Lupski, Nat. Rev. Genet., 2016, 17, 224–238 CrossRef CAS PubMed.
- S. M. Richardson, L. A. Mitchell, G. Stracquadanio, K. Yang, J. S. Dymond, J. E. DiCarlo, D. Lee, C. L. Huang, S. Chandrasegaran, Y. Cai, J. D. Boeke and J. S. Bader, Science, 2017, 355, 1040–1044 CrossRef CAS PubMed.
- J. S. Dymond, S. M. Richardson, C. E. Coombes, T. Babatz, H. Muller, N. Annaluru, W. J. Blake, J. W. Schwerzmann, J. Dai, D. L. Lindstrom, A. C. Boeke, D. E. Gottschling, S. Chandrasegaran, J. S. Bader and J. D. Boeke, Nature, 2011, 477, 471–476 CrossRef CAS PubMed.
- W. Zhang, L. A. Mitchell, J. S. Bader and J. D. Boeke, Annu. Rev. Biochem., 2020, 89, 77–101 CrossRef CAS PubMed.
- Z. X. Xie, D. Liu, B. Z. Li, M. Zhao, B. X. Zeng, Y. Wu, Y. Shen, T. Lin, P. Yang, J. Dai, Y. Cai, H. Yang and Y. J. Yuan, Chem. Soc. Rev., 2017, 46, 7191–7207 RSC.
- Z. Xie, J. Zhou, J. Fu and Y. Yuan, Chem. Sci., 2021, 12, 5381–5389 RSC.
- N. J. Kilby, M. R. Snaith and J. A. Murray, Trends Genet., 1993, 9, 413–421 CrossRef CAS PubMed.
- R. H. Hoess, A. Wierzbicki and K. Abremski, Nucleic Acids Res., 1986, 14, 2287–2300 CrossRef CAS PubMed.
- N. Annaluru, H. Muller, L. A. Mitchell, S. Ramalingam, G. Stracquadanio, S. M. Richardson, J. S. Dymond, Z. Kuang, L. Z. Scheifele, E. M. Cooper, Y. Cai, K. Zeller, N. Agmon, J. S. Han, M. Hadjithomas, J. Tullman, K. Caravelli, K. Cirelli, Z. Guo, V. London, A. Yeluru, S. Murugan, K. Kandavelou, N. Agier, G. Fischer, K. Yang, J. A. Martin, M. Bilgel, P. Bohutskyi, K. M. Boulier, B. J. Capaldo, J. Chang, K. Charoen, W. J. Choi, P. Deng, J. E. DiCarlo, J. Doong, J. Dunn, J. I. Feinberg, C. Fernandez, C. E. Floria, D. Gladowski, P. Hadidi, I. Ishizuka, J. Jabbari, C. Y. L. Lau, P. A. Lee, S. Li, D. Lin, M. E. Linder, J. Ling, J. Liu, J. Liu, M. London, H. Ma, J. Mao, J. E. McDade, A. McMillan, A. M. Moore, W. C. Oh, Y. Ouyang, R. Patel, M. Paul, L. C. Paulsen, J. Qiu, A. Rhee, M. G. Rubashkin, I. Y. Soh, N. E. Sotuyo, V. Srinivas, A. Suarez, A. Wong, R. Wong, W. R. Xie, Y. Xu, A. T. Yu, R. Koszul, J. S. Bader, J. D. Boeke and S. Chandrasegaran, Science, 2014, 344, 55–58 CrossRef CAS PubMed.
- Y. Shen, G. Stracquadanio, Y. Wang, K. Yang, L. A. Mitchell, Y. Xue, Y. Cai, T. Chen, J. S. Dymond, K. Kang, J. Gong, X. Zeng, Y. Zhang, Y. Li, Q. Feng, X. Xu, J. Wang, J. Wang, H. Yang, J. D. Boeke and J. S. Bader, Genome Res., 2016, 26, 36–49 CrossRef CAS PubMed.
- L. Hochrein, L. A. Mitchell, K. Schulz, K. Messerschmidt and B. Mueller-Roeber, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- G. Mercy, J. Mozziconacci, V. F. Scolari, K. Yang, G. Zhao, A. Thierry, Y. Luo, L. A. Mitchell, M. Shen, Y. Shen, R. Walker, W. Zhang, Y. Wu, Z. Xie, Z. Luo, Y. Cai, J. Dai, H. Yang, Y. Yuan, J. D. Boeke, J. S. Bader, H. Muller and R. Koszul, Science, 2017, 355, f4597 CrossRef PubMed.
- S. Zhou, Y. Wu, Z. Zhang, L. Jiang, L. Liu, Y. Zhang, J. Tang and Y. Yuan, bioRxiv, 2021, 2021–2027 Search PubMed.
- A. D. Silverman, A. S. Karim and M. C. Jewett, Nat. Rev. Genet., 2020, 21, 151–170 CrossRef CAS PubMed.
- B. A. Blount, G. F. Gowers, J. C. H. Ho, R. Ledesma-Amaro, D. Jovicevic, R. M. McKiernan, Z. X. Xie, B. Z. Li, Y. J. Yuan and T. Ellis, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- G. O. F. Gowers, S. M. Chee, D. Bell, L. Suckling, M. Kern, D. Tew, D. W. McClymont and T. Ellis, Nat. Commun., 2020, 11, 1–7 Search PubMed.
- E. L. I. Wightman, H. Kroukamp, I. S. Pretorius, I. T. Paulsen and H. K. M. Nevalainen, Microorganisms, 2020, 8, 1914 CrossRef CAS PubMed.
- L. Caspeta, Y. Chen, P. Ghiaci, A. Feizi, S. Buskov, B. M. Hallstrom, D. Petranovic and J. Nielsen, Science, 2014, 346, 75–78 CrossRef CAS PubMed.
- J. Y. Ong, R. Swidah, M. Monti, D. Schindler, J. Dai and Y. Cai, Bioengineering, 2021, 8, 42 CrossRef CAS PubMed.
- Y. Zhang, T. Chiu, J. Zhang, S. Wang, S. Wang, L. Liu, Z. Ping, Y. Wang, A. Chen, W. Zhang, T. Chen, Y. Wang and Y. Shen, Bioengineering, 2021, 8, 14 CrossRef CAS PubMed.
- B. Jia, J. Jin, M. Han and Y. Yuan, bioRxiv, 2021, 2021–2027 Search PubMed.
- W. Liu and R. Jiang, Appl. Microbiol. Biot., 2015, 99, 2093–2104 CrossRef CAS PubMed.
- J. Steensels, A. Gorkovskiy and K. J. Verstrepen, Nat. Commun., 2018, 9, 1–3 CrossRef CAS PubMed.
- S. Chen, Z. Xie and Y. Yuan, FEMS Yeast Res., 2020, 20, a12 CrossRef PubMed.
- J. I. Glass, N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison, H. O. Smith and J. C. Venter, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 425–430 CrossRef CAS PubMed.
- C. A. Hutchison, R. Y. Chuang, V. N. Noskov, N. Assad-Garcia, T. J. Deerinck, M. H. Ellisman, J. Gill, K. Kannan, B. J. Karas, L. Ma, J. F. Pelletier, Z. Q. Qi, R. A. Richter, E. A. Strychalski, L. Sun, Y. Suzuki, B. Tsvetanova, K. S. Wise, H. O. Smith, J. I. Glass, C. Merryman, D. G. Gibson and J. C. Venter, Science, 2016, 351, d6253 CrossRef PubMed.
- A. L. V. Coradini, C. B. Hull and I. M. Ehrenreich, Nat. Commun., 2020, 11, 1–11 Search PubMed.
- Z. Luo, K. Yu, S. Xie, M. Monti, D. Schindler, Y. Fang, S. Zhao, Z. Liang, S. Jiang, M. Luan, C. Xiao, Y. Cai and J. Dai, Genome Biol., 2021, 22, 1–18 CrossRef PubMed.
- E. Kuzmin, B. VanderSluis, W. Wang, G. Tan, R. Deshpande, Y. Chen, M. Usaj, A. Balint, M. M. Usaj and J. Van Leeuwen, Science, 2018, 360 DOI:10.1126/science.aao1729.
- A. H. Y. Tong, G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost and M. Chang, Science, 2004, 303, 808–813 CrossRef CAS PubMed.
- N. Pavelka, G. Rancati, J. Zhu, W. D. Bradford, A. Saraf, L. Florens, B. W. Sanderson, G. L. Hattem and R. Li, Nature, 2010, 468, 321–325 CrossRef CAS PubMed.
- G. Rancati, N. Pavelka, B. Fleharty, A. Noll, R. Trimble, K. Walton, A. Perera, K. Staehling-Hampton, C. W. Seidel and R. Li, Cell, 2008, 135, 879–893 CrossRef CAS PubMed.
- T. Si, H. Xiao and H. Zhao, Biotechnol. Adv., 2015, 33, 1420–1432 CrossRef CAS PubMed.
- R. Gabriel, A. Lombardo, A. Arens, J. C. Miller, P. Genovese, C. Kaeppel, A. Nowrouzi, C. C. Bartholomae, J. Wang, G. Friedman, M. C. Holmes, P. D. Gregory, H. Glimm, M. Schmidt, L. Naldini and C. von Kalle, Nat. Biotechnol., 2011, 29, 816–872 CrossRef CAS PubMed.
- T. Gaj, C. A. Gersbach and C. F. I. Barbas, Trends Biotechnol., 2013, 31, 397–405 CrossRef CAS PubMed.
- D. Maddalo, E. Manchado, C. P. Concepcion, C. Bonetti, J. A. Vidigal, Y. Han, P. Ogrodowski, A. Crippa, N. Rekhtman, E. de Stanchina, S. W. Lowe and A. Ventura, Nature, 2014, 516, 423–427 CrossRef CAS PubMed.
- R. J. Platt, S. Chen, Y. Zhou, M. J. Yim, L. Swiech, H. R. Kempton, J. E. Dahlman, O. Parnas, T. M. Eisenhaure, M. Jovanovic, D. B. Graham, S. Jhunjhunwala, M. Heidenreich, R. J. Xavier, R. Langer, D. G. Anderson, N. Hacohen, A. Regev, G. Feng, P. A. Sharp and F. Zhang, Cell, 2014, 159, 440–455 CrossRef CAS PubMed.
- R. Wu, M. Lucke, Y. Jang, W. Zhu, E. Symeonidi, C. Wang, J. Fitz, W. Xi, R. Schwab and D. Weigel, Plant Methods, 2018, 14, 1–9 CrossRef PubMed.
- N. Beying, C. Schmidt, M. Pacher, A. Houben and H. Puchta, Nat. Plants, 2020, 6, 638–645 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- P. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed.
- N. Hug, D. Longman and J. F. Cáceres, Nucleic Acids Res., 2016, 44, 1483–1495 CrossRef PubMed.
- K. Kraft, S. Geuer, A. J. Will, W. L. Chan, C. Paliou, M. Borschiwer, I. Harabula, L. Wittler, M. Franke, D. M. Ibrahim, B. K. Kragesteen, M. Spielmann, S. Mundlos, D. G. Lupiáñez and G. Andrey, Cell Rep., 2015, 10, 833–839 CrossRef CAS PubMed.
- N. R. Pannunzio, G. Watanabe and M. R. Lieber, J. Biol. Chem., 2018, 293, 10512–10523 CrossRef CAS PubMed.
- Y. Li, A. I. Park, H. Mou, C. Colpan, A. Bizhanova, E. Akama-Garren, N. Joshi, E. A. Hendrickson, D. Feldser, H. Yin, D. G. Anderson, T. Jacks, Z. Weng and W. Xue, Genome Biol., 2015, 16, 1–11 CrossRef PubMed.
- P. S. Choi and M. Meyerson, Nat. Commun., 2014, 5, 1–6 Search PubMed.
- M. R. Lieber, Annu. Rev. Biochem., 2010, 79, 181–211 CrossRef CAS PubMed.
- H. H. Y. Chang, N. R. Pannunzio, N. Adachi and M. R. Lieber, Nat. Rev. Mol. Cell Biol., 2017, 18, 495–506 CrossRef CAS PubMed.
- R. Mitsui, R. Yamada and H. Ogino, Appl. Biochem. Biotechnol., 2019, 189, 810–821 CrossRef CAS PubMed.
- J. R. Cameron, E. Y. Loh and R. W. Davis, Cell, 1979, 16, 739–751 CrossRef CAS PubMed.
- R. Mitsui, R. Yamada, T. Matsumoto, S. Yoshihara, H. Tokumoto and H. Ogino, Appl. Microbiol. Biotechnol., 2020, 104, 9147–9158 CrossRef CAS PubMed.
- E. Zuo, X. Huo, X. Yao, X. Hu, Y. Sun, J. Yin, B. He, X. Wang, L. Shi, J. Ping, Y. Wei, W. Ying, W. Wei, W. Liu, C. Tang, Y. Li, J. Hu and H. Yang, Genome Biol., 2017, 18, 1–18 CrossRef PubMed.
- F. Adikusuma, N. Williams, F. Grutzner, J. Hughes and P. Thomas, Mol. Ther., 2017, 25, 1736–1738 CrossRef CAS PubMed.
- L. H. Rieseberg, Trends Ecol. Evol., 2001, 16, 351–358 CrossRef PubMed.
- B. J. Loftus, E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson and J. A. Fraser, Science, 2005, 307, 1321–1324 CrossRef PubMed.
- K. Guin, Y. Chen, R. Mishra, S. R. B. Muzaki, B. C. Thimmappa, C. E. O’Brien, G. Butler, A. Sanyal and K. Sanyal, eLife, 2020, 9, e58556 CrossRef CAS PubMed.
- S. R. Sankaranarayanan, G. Ianiri, M. A. Coelho, M. H. Reza, B. C. Thimmappa, P. Ganguly, R. N. Vadnala, S. Sun, R. Siddharthan, C. Tellgren-Roth, T. L. J. Dawson, J. Heitman and K. Sanyal, eLife, 2020, 9, e53944 CrossRef PubMed.
- Y. Shao, N. Lu, C. Cai, F. Zhou, S. Wang, Z. Zhao, G. Zhao, J. Zhou, X. Xue and Z. Qin, Cell Res., 2019, 29, 87–89 CrossRef CAS PubMed.
- A. Kakizuka, W. H. Miller Jr, K. Umesono, R. P. Warrell Jr, S. R. Frankel, V. Murty, E. Dmitrovsky and R. M. Evans, Cell, 1991, 66, 663–674 CrossRef CAS.
- M. A. Moreno-Mateos, C. E. Vejnar, J. Beaudoin, J. P. Fernandez, E. K. Mis, M. K. Khokha and A. J. Giraldez, Nat. Methods, 2015, 12, 982–988 CrossRef CAS PubMed.
- X. Xiang, G. I. Corsi, C. Anthon, K. Qu, X. Pan, X. Liang, P. Han, Z. Dong, L. Liu, J. Zhong, T. Ma, J. Wang, X. Zhang, H. Jiang, F. Xu, X. Liu, X. Xu, J. Wang, H. Yang, L. Bolund, G. M. Church, L. Lin, J. Gorodkin and Y. Luo, Nat. Commun., 2021, 12, 1–9 CrossRef PubMed.
- D. Collias and C. L. Beisel, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- Y. Wu, B. Li, M. Zhao, L. A. Mitchell, Z. Xie, Q. Lin, X. Wang, W. Xiao, Y. Wang, X. Zhou, H. Liu, X. Li, M. Ding, D. Liu, L. Zhang, B. Liu, X. Wu, F. Li, X. Dong, B. Jia, W. Zhang, G. Jiang, Y. Liu, X. Bai, T. Song, Y. Chen, S. Zhou, R. Zhu, F. Gao, Z. Kuang, X. Wang, M. Shen, K. Yang, G. Stracquadanio, S. M. Richardson, Y. Lin, L. Wang, R. Walker, Y. Luo, P. Ma, H. Yang, Y. Cai, J. Dai, J. S. Bader, J. D. Boeke and Y. Yuan, Science, 2017, 355 DOI:10.1126/science.aaf4706.
- B. J. Yu, B. H. Sung, M. D. Koob, C. H. Lee, J. H. Lee, W. S. Lee, M. S. Kim and S. C. Kim, Nat. Biotechnol., 2002, 20, 1018–1023 CrossRef CAS PubMed.
- A. J. Smith, M. A. De Sousa, B. Kwabi-Addo, A. Heppell-Parton, H. Impey and P. Rabbitts, Nat. Genet., 1995, 9, 376–385 CrossRef CAS PubMed.
- H. Gu, Y. Zou and K. Rajewsky, Cell, 1993, 73, 1155–1164 CrossRef CAS PubMed.
- W. Chen, M. Han, J. Zhou, Q. Ge, P. Wang, X. Zhang, S. Zhu, L. Song and Y. Yuan, Natl. Sci. Rev., 2021, 8, b28 Search PubMed.
- X. Lu and T. Ellis, Natl. Sci. Rev., 2021 DOI:10.1093/nsr/nwab086.
- N. Hillson, M. Caddick, Y. Cai, J. A. Carrasco, M. W. Chang, N. C. Curach, D. J. Bell, R. Le Feuvre, D. C. Friedman and X. Fu, Nat. Commun., 2019, 10, 1–4 CrossRef PubMed.
- R. Chao, S. Mishra, T. Si and H. Zhao, Metab. Eng., 2017, 42, 98–108 CrossRef CAS PubMed.
- C. E. Lawson, J. M. Martí, T. Radivojevic, S. V. R. Jonnalagadda, R. Gentz, N. J. Hillson, S. Peisert, J. Kim, B. A. Simmons, C. J. Petzold, S. W. Singer, A. Mukhopadhyay, D. Tanjore, J. G. Dunn and H. Garcia Martin, Metab. Eng., 2021, 63, 34–60 CrossRef CAS PubMed.
- T. Oyetunde, F. S. Bao, J. Chen, H. G. Martin and Y. J. Tang, Biotechnol. Adv., 2018, 36, 1308–1315 CrossRef CAS PubMed.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |