 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Halogenases: a palette of emerging opportunities for synthetic biology–synthetic chemistry and C–H functionalisation
Charlotte
Crowe†‡
,
Samuel
Molyneux‡
,
Sunil V.
Sharma
,
Ying
Zhang
,
Danai S.
Gkotsi§
,
Helen
Connaris
and
Rebecca J. M.
Goss
 *
*
School of Chemistry, and BSRC, University of St Andrews, North Haugh, St Andrews KY16 9ST, UK. E-mail: rjmg@st-andrews.ac.uk
First published on 9th August 2021
Abstract
The enzymatic generation of carbon–halogen bonds is a powerful strategy used by both nature and synthetic chemists to tune the bioactivity, bioavailability and reactivity of compounds, opening up the opportunity for selective C–H functionalisation. Genes encoding halogenase enzymes have recently been shown to transcend all kingdoms of life. These enzymes install halogen atoms into aromatic and less activated aliphatic substrates, achieving selectivities that are often challenging to accomplish using synthetic methodologies. Significant advances in both halogenase discovery and engineering have provided a toolbox of enzymes, enabling the ready use of these catalysts in biotransformations, synthetic biology, and in combination with chemical catalysis to enable late stage C–H functionalisation. With a focus on substrate scope, this review outlines the mechanisms employed by the major classes of halogenases, while in parallel, it highlights key advances in the utilisation of the combination of enzymatic halogenation and chemical catalysis for C–H activation and diversification.
Introduction
Halogenated organic compounds play an important role in society. In the agrochemical sector, a staggering 96% of herbicides, fungicides, insecticides, acaricides and nematicides produced since 2010 contain halogen atoms.1 Such compounds are valuable commodities; for example, the top-selling insecticide imidacloprid (Fig. 1A) 1 had sales of 1.09 billion USD in 2011 alone,2 and in 2013, 50% of marketed top drugs contain halogen atoms.3,4 The fluorinated hepatitis C ledipasvir/sofosbuvir combination (Harvoni®) 6 was the second best-selling blockbuster pharmaceutical in 2016 with USA sales revenues of 4.9 billion USD (Fig. 1B),5 whilst, in the same year, US sales of the chlorinated antihistamine loratadine (Claritin®) 11 exceeded 2 billion USD.6 An economic report analysing the annual sales of the 100 top selling pharmaceuticals reveals that 12.9% of the final active pharmaceutical ingredient (API) is chlorinated or brominated, whilst a further 62.7% require chlorination at some stage in their manufacture.7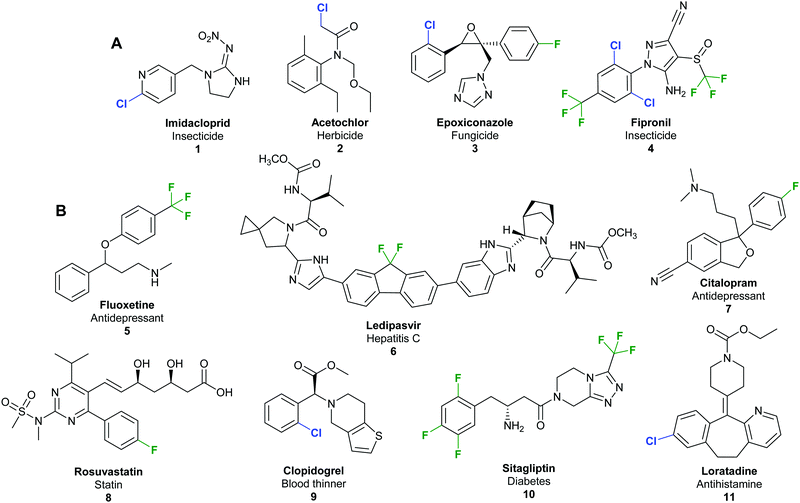 | ||
| Fig. 1 Examples of commercially important and structurally diverse organohalides. (A) Examples of top-selling halogenated agrochemicals include imidacloprid 1, biggest selling insecticide 2011 sales of 1.09 billion USD per annum;2 acetochlor 2, herbicide in the top 10 best-selling agrochemicals in 2003;16 epoxiconazole 3, a triazole based fungicide with sales of 215 million USD in 2001;17 and fipronil 4, a pyrazole based insecticide with sales of 270 million USD in 2001.17 (B) Examples of top-selling halogenated pharmaceuticals include antidepressants fluoxetine 5 (Prozac®)17 and citalopram 7 (Celexa®);18 anti-hepatitis C agent ledipasvir 6;5 statin rosuvastatin 8;19 the blood thinner clopidogrel 9;16 the type 2 diabetes treatment sitagliptin 10;20 and antihistamine loratadine 11.6 | ||
The incorporation of a carbon–halogen bond is a regularly employed strategy in medicinal and agro-chemistry. Due to bond polarisation and the ability to form highly directional halogen bonds to electron-rich oxygen, nitrogen, sulfur and aromatic rings, halogen incorporation may be used to enhance stability, and increase lipophilicity which in turn can influence bioavailability and modulate compound bioactivity.8 Beyond these applications, series of fluorinated and iodinated compounds are used in radiotherapy and biomedical imaging.9,10 Halogenation is also central to molecule construction and manufacture. The reactivity and chemical orthogonality of organohalides enables many selective transformations including, most notably, cross-coupling and substitution chemistries. Synthetically, the generation of organohalides is currently achieved through the addition of molecular halogens, hydrohalogenation, or the use of reagents enabling nucleophilic substitution and electrophilic aromatic substitution [EAS]. These reactions are generally accompanied by environmental and selectivity challenges due to the toxic, corrosive and reactive nature of the reagents used, and the propensity to generate complex product mixtures.11 Whilst recent advances in homogenous catalyst design has meant more selective catalytic approaches towards halogenation are known, such catalysts can be complex, costly and time consuming to synthesise.12
Though naturally occurring organohalides were initially speculated to be nothing more than a biological curiosity, artefact, or to be anthropogenic in origin, today, over 5000 halogenated natural products are known, with genes encoding halogenases identified across all kingdoms of life.13 These represent a resource for greener and often highly regioselective halogenation. An increased appreciation of mechanisms underpinning enzymatic halogenation has evolved over the past 25 years, and may broadly be classified as electrophilic, nucleophilic or radical halogenation. Whilst a good level of understanding of the enzymatic active sites and structural requirements of these enzymes exists, a number of fundamental details remain to be elucidated.
This review overviews the mechanisms of the different classes of halogenases, the natural context of the halogenases, their biocatalytic applications, and explores their powerful potential for use in the hyphenation of Synthetic Biology and Synthetic Chemistry (SynBio–SynChem) enabling C–H activation and diversification.
1. Mechanisms for biocatalytic halogenation
Enzymatic halogenation is classified mechanistically in accordance with whether the halogen is introduced by an electrophilic, nucleophilic, or radical halide species, with the vast majority of the enzymes that have been discovered so far, being electrophilic halogenases, mediating oxidation of the halogen to generate a reactive electrophilic species. There is further subdivision within the broader halogenase categories, based on the exact catalytic species and cofactor which enables halogen oxidation.1.1.1 Haloperoxidases. Haloperoxidases are defined as enzymes that utilise hydrogen peroxide and chloride, bromide or iodide to generate HOX. These enzymes are named in accordance with the most electronegative halogen for which they can affect oxidation. Haloperoxidases are generally the least regioselective of the enzymes enabling halogenation; catalysing the generation of free hypohalous acid, which in all but a few cases, is released by the enzyme. The earliest reported studies of halogenation enabled by an enzyme date back to 1959, when Lowell Hager discovered a haloperoxidase (HPO) from the fungus Calariomyces fumago.14 The enzyme that Hager had utilised as part of a lysate was named chloroperoxidase or (CPO) and was shown to be capable of affecting dichlorination of 1,3-cyclopentanedione 12. It has thus been postulated to be involved in the biosynthesis of the antibiotic caldariomycin 14 (Scheme 1C).15 CPO may be described more specifically as a heme iron-dependent haloperoxidase, a subset of haloperoxidases that contain a heme-iron cofactor coordinated to an axial cysteine ligand within the enzyme. Hydrogen peroxide binds axially to heme-Fe(III) in its resting state to form a ˙+heme-Fe(IV)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O intermediate, denoted compound I.23 Addition of halide to the ferryl oxygen produces a heme-Fe(III)–OX species (Scheme 1A, IV). Free hypohalous acid, HOX, is generated which reacts with an electron-rich substrate (Scheme 1A).24 Generation of electrophilic X+ allows halogenation of electron-rich compounds by electrophilic aromatic substitution, generally at the most reactive site(s) within the substrate. CPO has been used biocatalytically to mediate the chlorination, bromination or iodination of a variety of electron-rich substrates (see Section 2), however, as it lacks a site to position the substrate, no regiocontrol beyond that conferred by the electronics of the substrate is observed.
O intermediate, denoted compound I.23 Addition of halide to the ferryl oxygen produces a heme-Fe(III)–OX species (Scheme 1A, IV). Free hypohalous acid, HOX, is generated which reacts with an electron-rich substrate (Scheme 1A).24 Generation of electrophilic X+ allows halogenation of electron-rich compounds by electrophilic aromatic substitution, generally at the most reactive site(s) within the substrate. CPO has been used biocatalytically to mediate the chlorination, bromination or iodination of a variety of electron-rich substrates (see Section 2), however, as it lacks a site to position the substrate, no regiocontrol beyond that conferred by the electronics of the substrate is observed.
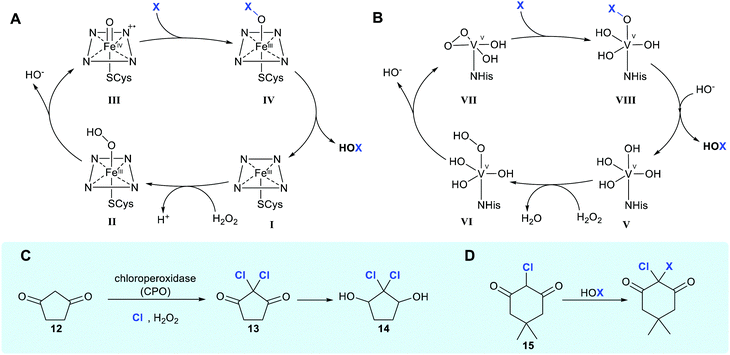 | ||
| Scheme 1 Haloperoxidase HOX generation and utilisation. (A and B) Biocatalytic cycle of heme iron-dependent and vanadium-dependent haloperoxidases, respectively, showing the release of freely diffusible hypohalous acid (NB the order of events in B remains unclear, with the protonation state of VII being ambiguous).21 (C) Chlorination of 1,3-cyclopentanedione 12 by HOCl in the presence of a heme iron-dependent chloroperoxidase (CPO) and hydrogen peroxide suggested to occur in caldariomycin 14 biosynthesis.15 (D) The reaction of monochlorodimedone (MCD) 15 with HOX, the benchmark assay for haloperoxidase activity.22 | ||
The vanadium-dependent haloperoxidases (V-HPOs) operate by an analogous mechanism, again generating and, mostly releasing, free hypohalous acid. This subcategory of haloperoxidase is widely found in algae, fungi and bacteria, and utilises a vanadium cofactor bound by an axial histidine ligand.25 In the resting state, the cofactor is in a trigonal bipyramidal configuration (Scheme 1B, V), a distal H2O2 displaces two hydroxyl ligands giving rise to formation of the known η2-peroxido complex (Scheme 1B, VII), with the distorted square pyramidal geometry. Though the exact order of events and level of protonation here is not known, a postulated catalytic cycle is shown. A lysine residue hydrogen bonded to an equatorial V–O oxygen activates the peroxide complex for X− attack and a return to the trigonal bipyramidal geometry (Scheme 1B, VIII) is observed. The resulting V–OX species undergoes hydrolysis to generate electrophilic X+ in the form of free HOX.26 Electrophilic aromatic substitution occurs, as in the case with heme-iron haloperoxidases.27 A notable difference with the heme iron-dependent biocatalytic cycle is that the vanadium centre maintains its oxidation state throughout the process.28
Haloperoxidases had initially been considered to lack significant enzyme-mediated selectivity due to the release of freely diffusible hypohalous acid. The free electrophilic species allows halogenation at the most electron-rich carbon, and a mixture of mono-, di- and tri-substituted products can often be observed. Detection of the free HOX through reaction with nucleophilic monochlorodimedone (MCD) 15 has previously been used to assay for haloperoxidase activity (Scheme 1D).29 However, for a number of vanadium-dependent haloperoxidases, substrate, regio- and even stereo-specific halogenation is evident, as noted in the biosynthesis of napyradiomycins 76,30 and merochlorins 77, 78.31 (Scheme 5). Currently, the exact mechanistic rationale for this specificity is not known. Here, free HOCl is notably not detected in assay with MCD.31 It is thus postulated that residues within the enzyme might transiently support the electrophilic HOCl, whilst the electron-rich substrate is appropriately and proximally positioned. Moore invokes the involvement of an active site lysine forming a haloamine, in a manner analogous to the FDHs.31,32 Though NapH1 from napyradiomycin biosynthesis is associated with the first chlorination event, this enzyme is also capable of mediating nonspecific bromination through generation and release of HOBr. Greater understanding of the mechanism enabling selective halogenation by these series of enzymes would be valuable, potentially enabling informed discovery of further enzymes with biotechnological potential.
1.1.2 Flavin-dependent halogenases. The haloperoxidases remained the only known halogenases for several decades until in 1995, the first of a second series of halogenases, the flavin-dependent halogenases (FDHs), Chl from 7-chlortetracycline 73 biosynthesis in Streptomyces aureofaciens was revealed in a study by Dairi.33 The enzyme was implicated as being involved in chlorination with knockouts of the gene encoding Chl lacking halogenated product. However, from these initial studies, it was not fully apparent that this new series of enzymes were flavin-dependent, only a partial protein sequence lacking 100 amino acids at the N-terminal end including the flavin binding site. In vitro analysis of the tryptophan 7-halogenase PrnA, by van Pée and co-workers, demonstrated the powerful regioselectivity of this new class of halogenase and the requirement for both flavin and molecular di-oxygen.34,35
The FDHs can be separated into those acting on free substrates (variant A), and those requiring their substrate to be covalently tethered to a carrier protein via a phosphopantatheine linker (variant B).36 The most extensively investigated and engineered FDHs are the tryptophan halogenases, variant A FDHs enabling regioselective halogenation of tryptophan at the 5, 6 or 7 positions. The first structural insights as to how FDHs mediate their exquisite regioselectivity were revealed by van Pée and Naismith.37 However, 15 years on and debate still remains as to the exact way in which these enzymes function. Details as to which residues within the active site participate may vary from one enzyme to another. The mechanisms proposed by Walsh38 and van Pée39 for the flavin dependent tryptophan halogenase are presented in Scheme 2B and C.
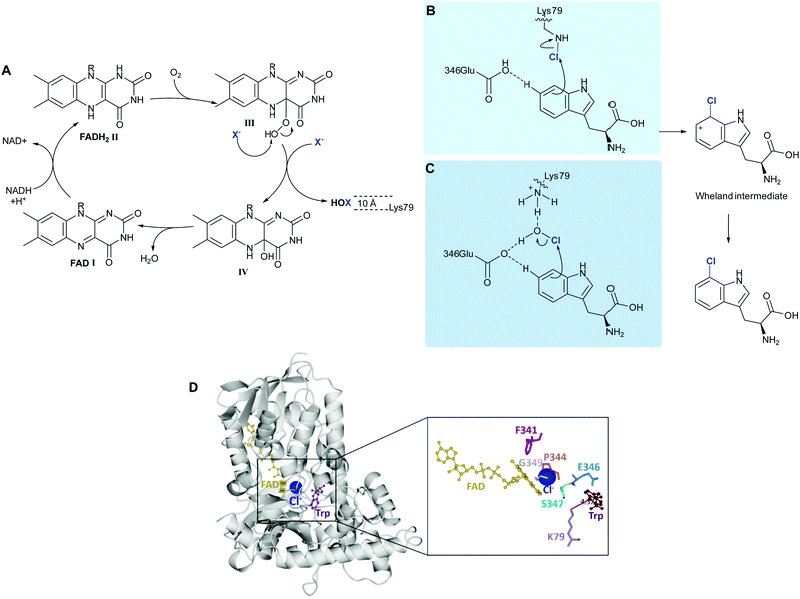 | ||
| Scheme 2 FDH-mediated electrophilic halogenation. (A) Biocatalytic cycle of a flavin-dependent halogenase, showing the release of hypohalous acid. HOX is channelled through a ∼10 Å tunnel. The regioselectivity of the electrophilic aromatic substitution is postulated to be achieved by: (B) a lysine residue, at the end of the tunnel that reacts with HOX to form a haloamine intermediate,43 and (C) lysine and glutamine, acting in concert to position HOX and further electronically activate the tryptophan.39 (D) Extrapolation of the crystal structure of PrnA showing how Lys79 is positioned over C7 of the indole ring mediating regioselective halogenation. | ||
Reduced flavin adenine dinucleotide (FADH2), (Scheme 2A, I) and dioxygen in the presence of chloride, bromide or iodide are utilised by these enzymes to catalyse C–H to C–X. Freely diffusible FAD binds to a conserved GxGxxG motif. The reduced cofactor, FADH2, (Scheme 2A, II) reacts with molecular oxygen to form FAD(C4α)–OOH (III), which subsequently reacts with a nucleophilic halide releasing HOX and FAD(C4α)–OH (Scheme 2A, IV).40 Unlike haloperoxidases, the HOX generation and electrophilic aromatic substitution (EAS) events are spatially separated by a ∼10 Å tunnel. At the end of this tunnel is a conserved lysine, proposed by Walsh to react with the HOX thereby forming a haloamine intermediate, guiding the regiochemistry of halogenation. (Scheme 2B).38 Mutation of this lysine residue to alanine, across a series of halogenases including RebH, PrnA, and Rdc2, results in loss of activity.41 In a subtly different mechanism, van Pée invokes a proximal glutamate residue, as well as the lysine residue binding to HOX, thereby activating it and positioning it (Scheme 2C).39 In both models, the electrophilic halogenating species is directed towards the substrate, promoting a regioselective EAS reaction. The exact mechanism and halogenating species remain elusive. Mutation of this active site glutamate to glutamine in PrnA results in a drop in rate by two orders of magnitude.37 This glutamate is not present in the more electron-rich phenol halogenases. A different proposed role for this active site glutamate, invoked in deprotonation of the Wheland intermediate indole benzo ring in the tryptophan halogenases, is that it perhaps plays a role in reducing the activation energy of halogenation of these less electron-rich species.41 Notably no kinetic isotope effect is observed for halogenation of tryptophan by RebH.42
These FDHs share a similar flavin binding domain to the flavin-dependent monoxygenases, and though most utilise free and diffusible FAD, which is converted to FADH2, variants exist where the flavin is covalently bound to the enzyme, such as in the case of CmlS involved in chloramphenicol biosynthesis.44 Naturally, most FDHs are usually provided the reduced flavin by a partner flavin reductase that utilises NADH (nicotinamide adenine dinucleotide) and FAD to generate reduced FADH2.35 The partner flavin reductase can be readily substituted for any other capable of producing FADH2. Single component FDHs, that have this reductase function present within the one protein, also exist; the first discovered example of such an enzyme was Bmp5.45 This enzyme shows other notable differences to all previously studied FDHs; in analogy to the decarboxylative hydroxylation observed for a single component monooxygenase, decarboxylative dibromination of 18 occurs (Fig. 2F).45 Other intriguing variations of the FDHs also exist, recently, the first of a new class of zinc-binding FDHs, MalA from the biosynthesis of the malbrancheamides 21 and 22 (Fig. 2G) was reported. This remarkable enzyme is capable of mediating the late stage dichlorination of 5, and 6 positions of the indole benzo ring of pre-malbrancheamide 20, a complex prenylated indole alkaloid. In the first step, positions 5 and 6 appear to be halogenated with comparable rates, perhaps due to increased electron density, the second halogenation even occurs far more quickly. Within the enzyme, the active site glutamate is seen proximal to the NH of the indole. It is clearly implicated in deprotonating this moiety, rendering the substrate more nucleophilic, and whilst mutation of this glutamate to aspartate results in low but observable activity, mutation to glutamine renders the enzyme inactive.46
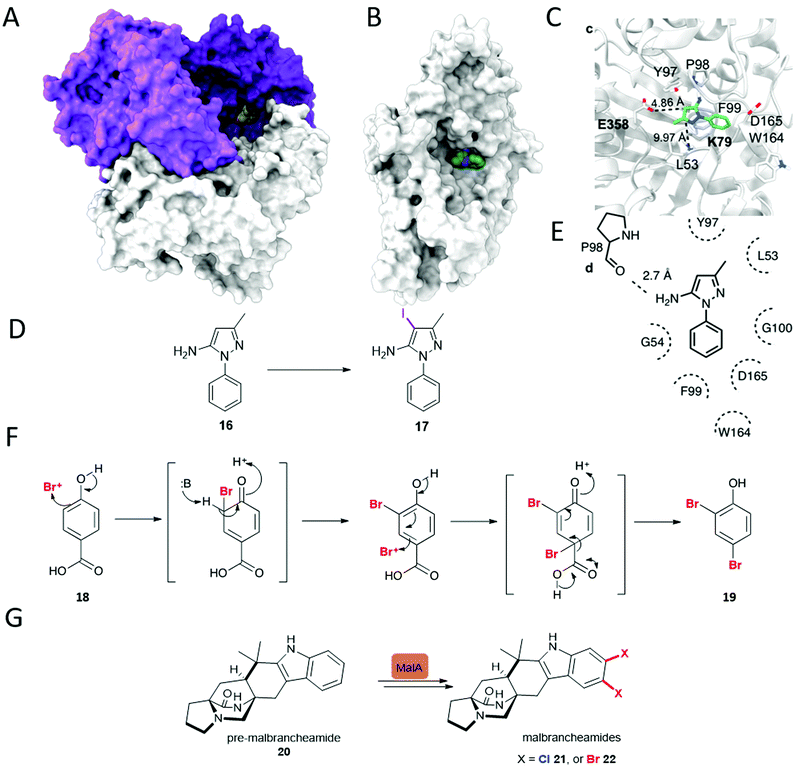 | ||
| Fig. 2 Unusual FDHs. (A) Trimer of VirX1, the first FDH with a preference for iodination, and the first halogenase from a virus showing the large and accessible active site, with a bulky substrate 16 modelled in the active site. (B) View of a subunit of VirX1, again with substrate 16 modelled into the active site, and embedded in a hydrophobic cleft. (C) Key hydrogen bonds between VirX1 and 16 as predicted by structural modelling. (D) Regiospecific iodination of 16 by VirX1.47 (E) Model of compound 16 in the active site of VirX1. (F) Decarboxylative bromination of 18 observed for Bmp5.48 (G) Iterative dihalogenation in malbrancheamides 21 and 22 biosynthesis, by an unusual zinc-containing FDH.46 | ||
FDHs contain multiple sequence motifs; a series of amino acid residues integral for enzyme activity or structure, that are generally conserved throughout all FDHs. These are a dinucleotide-binding motif GxGxxG, part of the Rossman fold allowing for binding of flavin and NAD(P)H,49 and a tryptophan motif WxWxIP, suggested to block binding of substrate near the bound cofactor, thus preventing a monooxygenation reaction instead of halogenation.37 It is notable though, that GxGxxG, the flavin binding motif is present in almost all flavoenzymes. The WxWxIP motif or variants of this motif, is known to be absent in a number of FDHs including the more unusual Bmp5.48 A new motif, Fx.Px.Sx.G was recently reported. This, or variants of this motif in which G is replaced with a charge bearing residue, line the tunnel between the flavin binding site and the site of halogenation, may be found in all active FDHs.47 As such, it can be used as a method for quickly identifying candidate FDHs from uncurated sequence data. A complementary and overlapping motif FX[DE]PX[EFL] was subsequently identified by Lewis and coworkers.157
Notably, almost all halogenases studied, until recently, have been identified from bacteria and fungi, most being found due to their implication in the biosynthesis of a specific natural product or series of natural products. Iodination had also been missing from the FDH portfolio of reactions, though postulated to be involved in the biosynthesis of iodinated natural products.50 Whilst efforts had been made to explore the utilisation of enzymes such as RebH for iodination, incubation of such enzymes with iodide had been unfruitful, and competition studies had revealed that the presence of such salts procluded conversion, perhaps due to formation and inability to release the bulky product.43 More recently the FDH, PltM, was shown to be able to process sodium iodide, though preference for bromination, then chlorination was apparent.51 VirX1, the first halogenase to be characterised from a virus, is the first FDH for a preference for halogenation, and may be seen to have a large and readily accessible active site (Fig. 2A and B). Hydrophobic residues anchor the substrate in position, enabling regiospecific halogenation as guided by the proximal lysine (K79). Glutamate enables further activation, of otherwise less electrophilic substrates. It is postulated that by virtue of openness of this active site not only can a large range of small to sterically bulky substrates be processed, but halogenation by the largest and most readily oxidised of the halides is enabled.47
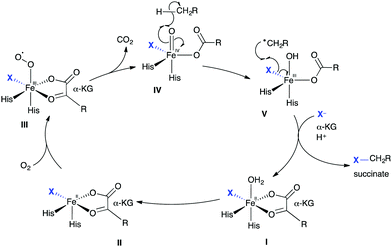 | ||
| Scheme 3 Radical halogenation. Biocatalytic cycle of a non-heme iron α-ketoglutarate-dependent halogenase, showing the release of halogenated product. | ||
Significant conformational changes are observed within these NHFe proteins. These changes are postulated to occur directly after oxygen binding and the subsequent decarboxylation event, orientating the substrate for the observed regioselective halogenation. Crystal structures of WelO5 with NO (an O2 analogue) reveal a significant change in conformation. Initially, NHFeHals appeared to be difficult to handle in vitro due to oxygen sensitivity of the Fe(II) core,58 however, recent advances suggest an iron-reconstitution step can be performed to activate enzymes purified aerobically.59 The majority of NHFeHals, such as SyrB1,58 BarB1 and BarB2,60 and CurA halogenase,61 like the variant B FDHs, all required the substrate to be presented on carrier proteins, tethered via a phosphopantatheine linker (Ppant). In 2014, WelO5 was discovered, becoming the first member of a new family of NHFeHals (that we will refer to as variant A NHFeHals), which act on freestanding substrates.62 Excitingly, these open up the possibility for using such proteins more readily in biotransformations.
The nucleophilic halogenases characterised so far all operate by holding the halide proximal to the electrophile, SAM. Crystallographic studies of the fluorinase reveal a small pocket, the fluoride binding site, giving insight into how displacement of the hydration sphere might be achieved. Whilst predominantly hydrophobic, this small pocket contains a serine.67 Dehydration is proposed to occur when SAM binds with the positive charge on the sulphur possibly assisting with stabilisation of the naked fluoride. The requirement for the fluoride ion to bind first in order for a productive complex to be formed, is argued based on observed substrate inhibition by SAM. Within the crystal structure, SAM may be seen bound with its ribose ring forced into a high energy, eclipsed conformation. The halide anion is positioned collinearly with the SAM C–S bond, thus promoting an SN2 backside attack (Scheme 4).67 When the enzyme is crystallised in the presence of both SAM and fluoride, the products L-methionine and 5′-fluoro-5′-deoxyadenosine (5′-FDA) with the ribose ring in the relaxed conformation may be observed.
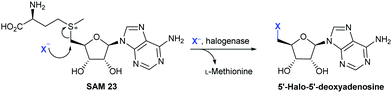 | ||
| Scheme 4 Nucleophilic halogenation. Halogenation of S-adenosyl-L-methionine 23 (SAM) by X− mediated by a nucleophilic halogenase, yielding L-methionine and 5′-halo-5′-deoxyadenosine.67 | ||
The fluorinase has a strong preference for fluoride, but can incorporate chloride to a low level.68 SalL, from Salinispora tropica bacterium has 54% sequence similarity to the fluorinase, and in the halide binding pocket, a glycine residue resides in place of a serine. This enzyme shows a strong preference for chlorination and, whilst capable of utilising bromide and iodide, it cannot process fluoride.64
Notable uses of the fluorinase include exploration of its application to 18F incorporation for positron emission tomography (PET),69 and it's the utilisation of the fluorinase in parallel with a polyketide synthase to enable synthetic biological access to simple fluorinated polyketides.70
| Enzyme | Substrate | Classification | Halide | k cat (min−1) | K M (μM) | k cat/KM (μM−1 min−1) | Ref. |
|---|---|---|---|---|---|---|---|
| VirX1 | 6-Azaindole | FDH | I | 5 ± 0.5 | 28 ± 2 | 0.179 ± 0.004 | 47 |
| Br | 2.4 ± 0.6 | 53 ± 3 | 0.044 ± 0.008 | ||||
| PltM | Resorcinol | FDH | Br | 1.9 ± 0.1 | 0.71 ± 0.3 | 2.7 ± 0.6 | 51 |
| Cl | 2.3 ± 0.1 | 0.076 ± 0.013 | 30 ± 5 | ||||
| PyrH | Tryptophan | FDH | Cl | 3.56 ± 1.1 | 109 ± 44 | 0.0325 ± 0.017 | 51 |
| KtzQ | 7-Cl-L-Tryptophan | FDH | Cl | 1.4 | 114 | 0.012 | 82 |
| MalA | Premalbrancheamide | FDH | Cl | 0.08 ± 0.05 | 7.0 ± 2.9 | 11.49 ± 0.02 | 46 |
| WelO5 | 12-Epi-fisherindole U | NHFeHal | Cl | 1.8 ± 0.2 | ND | ND | 83 |
| PfHalA | Lysine | NHFeHal | Cl | 14.7 ± 0.71 | 280 ± 60 | 0.0525 ± 0.0118 | 84 |
| AdeV | 2′-Deoxyadenosine monophosphate | NHFeHal | Cl | 0.6 | 2500 | 0.24 × 10−3 | 85 |
| CPO | Catechol | HPO | Cl | 33 ± 1 | 27 ± 8 | 1.22 ± 0.36 | 86 |
| V-BrHPO | 2-Chlorodimedone | V-HPO | Br | 5 | ND | ND | 26 |
| FlA | SAM | Nucleophilic | F | 0.07 ± 0.001 | 6.5 ± 0.3 | 0.0108 ± 0.0005 | 87 |
| Enzymes | No. residues | PDB code | Cofactors and ligands | Identification time | Relevant BGC | Oligomer | Reported substrates | Halogen | Halogenation stage | Source | Subgroup | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a No in vitro activity demonstrated to date. b Natural protein length from UniProt. c Not specifically mentioned, tested by standard substrate monochlorodimedone/TB iodoperoxidase assay. d FlA is not in a BGC, but participates in the synthesis of fluoroacetate and 4-fluorothreonine. A. Flavin dependent halogenases. TBP To be published. TBP: To be published (compound structures reported in the PDP but not yet formally published elsewhere). | ||||||||||||
| PrnA | 538 | 2AR8 | FAD, Cl−, 7-Cl-Trp | 2005 | Pyrrolnitrin | Dimer | Tryptophan 7C, indolic substrates | Cl, Br | Early stage | Pseudomonas fluorescens | A | 37 |
| 2APG | FAD, Cl−, | 37 | ||||||||||
| 2AQJ | FAD, Cl−, Trp | 37 | ||||||||||
| 2ARD | FDA | 37 | ||||||||||
| 4Z43 (E450K) | FAD, Cl− | 2015 | 79 | |||||||||
| 4Z44 (F454K) | FAD, Cl−, PO4 | 79 | ||||||||||
| 2JKC (E346D) | FAD, Cl−, Trp | 2008 | 39 | |||||||||
| RebH | 530 | 2OA1 | FAD, ADN, Trp, Cl− | 2008 | Rebeccamycin | Dimer | Tryptophan 7C, indolic substrates, aniline substrates | Cl, Br | Early stage | Lechevalieria aerocolonigenes | A | 88 |
| 2O9Z | PO4 | 88 | ||||||||||
| 2OAL | FAD, Cl− | 2007 | 43 | |||||||||
| 2OAM | Apo form | 43 | ||||||||||
| 2E4G | Trp | 43 | ||||||||||
| 4LU6 (thermostabilized) | PO4 | 2013 | 89 | |||||||||
| 6P00 (11 mutations) | FAD | 2020 | TBP | |||||||||
| 6P2V (6 mutations) | FAD | TBP | ||||||||||
| 7JU0 (1 mutation) | FAD, TSS | TBP | ||||||||||
| CndHa | 512 | 3E1T | FAD, Cl− | 2009 | Chondrochloren | Monomer | Aliphatic substrates | Cl | Early stage | Chondromyces crocatus | B | 36 |
| PyrH | 511 | 2WET | FAD, Trp, Cl− | 2010 | Pyrroindomycin | Dimer | Tryptophan 5C | Cl, Br | Early stage | Streptomyces rugosporus | A | 90 |
| 2WEU | Trp | 2009 | 90 | |||||||||
| 2WES (E46Q) | FAD, Cl− | 2009 | 90 | |||||||||
| CmlSa | 570 | 3I3L | FAD | 2010 | Chloramphenicol | Monomer | Aliphatic substrates | Cl, Br | Late stage | Streptomyces venezuelae | A | 44 |
| PltA | 455 | 5DBJ | FAD, Cl− | 2015 | Pyoluteorin | Dimer | Pyrrolic substrates | Cl | Early stage | Pseudomonas fluorescens | B | 91 |
| Mpy16 | 447 | 5BUK | FAD | 2016 | Pyrrolyl-S-Mpy15 | Monomer | Pyrrolic substrates | Cl | Late stage | Streptomyces sp. CNQ-418 | B | 48 |
| Th-Hal | 510 | 5LV9 | — | 2016 | — | Monomer | Tryptophan | Cl, Br | — | Streptomyces violaceusniger | A | 92 |
| 5LVA | FMN | 2016 | 92 | |||||||||
| SttH | 519 | 5HY5 | FAD, Cl− | 2016 | — | Dimer | Tryptophan 6C, N-methyltryptophan, kynurenine, anthranilamide | Cl, Br | — | Streptomyces toxytricini | A | 80 |
| Bmp2 | 409 | 5BVA | FAD, EDO | 2016 | Tetrabromo-pyrrole | Monomer | Pyrrolic substrates | Br, I | — | Pseudoalteromonas luteoviolacea | B | 48 |
| 5BUL (Y302S F306V A345W) | FAD | 48 | ||||||||||
| MalA | 667 | 5WGZ | FAD, IM7, Cl− | 2017 | Malbrancheamide | Monomer | Premalbrancheamide, malbrancheamide B, isomalbrancheamide B | Cl, Br | Late stage | Malbranchea aurantiaca | A | 46 |
| 5WGV (C112S C128S) | FAD, PM7, Cl− | 46 | ||||||||||
| 5WGU (E494D) | FAD, PM7, Cl− | 46 | ||||||||||
| 5WGX (H253A) | FAD, MB5, Cl− | 46 | ||||||||||
| 5WGY (C112S C128S) | FAD, MB5, Cl− | 46 | ||||||||||
| 5WGR | FAD, PM7, Cl− | 46 | ||||||||||
| 5WGS (H253F) | FAD, PM7, Cl− | 46 | ||||||||||
| 5WGT | FAD, PM7, Cl− | 46 | ||||||||||
| 5WGW | FAD, MB5, Cl− | 46 | ||||||||||
| BrvH | 502 | 6FRL | — | 2018 | — | Dimer | Indolic substrates | Br > Cl | — | Brevundimonas BAL3 | B | 93 |
| Tar14 | 532 | 6NSD | FAD | 2019 | Taromycin | Dimer | Tryptophan | Cl, Br | Early stage | Saccharomonospora sp. CNQ-490 | A | 94 |
| Thal | 534 | 6H43 | PO4 | 2019 | Thienodolin | Dimer | Tryptophan 6C, indolic substrates | Cl, Br | Early stage | Streptomyces albogriseolus | A | 81 |
| 6H44 | Trp, PO4 | 2019 | 81 | |||||||||
| 6IB5 (Thal-RebH5, 5 mutations) | PO4 | 2019 | 81 | |||||||||
| 6SLT | FAD, AMP, Trp, PO4 | 2019 | 95 | |||||||||
| 6SLS | FAD, PO4 | 95 | ||||||||||
| 7AQV (N-6His-Thal-RebH5, 5 mutations) | — | 2020 | TBP | |||||||||
| 7AQU (N-6His-Thal-RebH5, 5 mutations) | BCN, SER, ALA, GLY | 2020 | TBP | |||||||||
| 7CU0 | Trp | 2020 | 96 | |||||||||
| 7CU1 | FAD, AMP | 2020 | 96 | |||||||||
| 7CU2 | FDA | 2020 | 96 | |||||||||
| PltM | 522 | 6BZN | — | 2019 | Pyoluteorin | Monomer | Phenolic substrates, aniline substrates | Cl, Br, I | — | Pseudomonas protegens Pf-5 | A | 51 |
| 6BZZ | FAD | 2019 | 51 | |||||||||
| 6BZA | FAD, 13X, Cl− | 2019 | 51 | |||||||||
| 6BZQ | FAD, Br−, Cl− | 2019 | 51 | |||||||||
| 6BZT (L111Y) | FAD, Br−, Cl− | 2019 | 51 | |||||||||
| 6BZI | EMC, HG | 2019 | 51 | |||||||||
| Virx1 | 531 | 6QGM | Apo form | 2019 | — | Trimer | Aromatic substrates, heterocycles, azaspirocycles | I > Br > Cl | — | Cyanophage Syn10 | A | 47 |
| Classification | Enzymes | No. residuesb | PDB code | Cofactors and ligands | Idt. time | Relevant BGC | Oligomer | Reported substrates | Halogen | Source | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vanadium-dependent haloperoxidases | Ci-VCPO | 609 | 1VNC | VO4, AZI | 1996 | — | Monomer | Chlorinated 1-(4-ethoxy-3-methoxyphenyl)-2-(2-methoxyphenoxy)-1,3-dihydroxypropane | Cl, Br | Curvularia inaequalis | 97 |
| 1VNS | SO4 | 1999 | 98 | ||||||||
| 1VNI | Apo form | 98 | |||||||||
| 1VNH (H496A) | VO4 | 98 | |||||||||
| 1VNG (H404A) | VO4 | 98 | |||||||||
| 1VNF (R360A) | VO4 | — | 98 | ||||||||
| 1VNE (D292A) | VO4 | 98 | |||||||||
| 1IDQ | VO4 | 1997 | — | 99 | |||||||
| 1IDU | VO4 | — | 99 | ||||||||
| 3BB0 | PO3 | 2008 | 100 | ||||||||
| An-VBPO1 | 556 | 1QI9 | VO4, I− | 1999 | — | Dimer | 2-Methylindole, 2-phenylindole, phenolsulfonephthalein, derivatives of O-methyl pyrrole-2-carboxylate | Br, I | Ascophyllum nodosum | 101–103 | |
| An-VBPO2 | 597 | 5AA6 | VO4 | 2015 | — | Hexamer | Refer to An-VBPO1 | Br, I | Ascophyllum nodosum | TBP | |
| Co-VBPO | 596 | 1QHB | PO4 | 1999 | — | Dodecamer | (E)-(+)-Nerolidol | Br | Corallina officinalis | 104 | |
| Cp-VBPO | 598 | 1UP8 | PO4 | 2005 | — | Dodecamer | Br | Corallina pilulifera | 105 | ||
| NapH1 | 531 | 3W35 (Apo form) | — | 2012 | Napyradiomycin | Dimer | Polyketide-terpenoid substrates | Cl, Br | Streptomyces sp. CNQ525 | TBP | |
| 3W36 | VO4 | 2012 | TBP | ||||||||
| Zg-VIPO1 | 458 | 4CIT | VO4 | 2014 | — | Monomer | TB iodoperoxidase assay | I | Zobellia galactanivorans | 106 | |
| Zg-VIPO2 | 458 | 4USZ | VO4 | 2014 | — | Monomer | TB iodoperoxidase assay | I | Zobellia galactanivorans | 106 | |
| AmVHPO | 639 | 5LPC | PO4 | 2016 | — | Dodecamer | Aromatic substrates | Cl, Br | Acaryochloris marina | 107 | |
| Heme-dependent haloperoxidases | HI-HPO | 373 | 1CPO | HEM | 1995 | Caldariomycin | Monomer | 1,3-Cyclopentanedione, electron-rich aromatic substrates | Cl, Br | Leptoxyphium fumago | 108 |
| 2CPO | HEM | 108 | |||||||||
| 2CJ0 | HEM | 2006 | 109 | ||||||||
| 2CIW | HEM, I− | 109 | |||||||||
| 2CIV | HEM, Br− | 109 | |||||||||
| 2CJ2 | HEM, FMT | 109 | |||||||||
| 2CJ1 | HEM, FMT | 109 | |||||||||
| 2CIZ | HEM, Br− | 109 | |||||||||
| 2CIY | HEM, Br−, CYN, DMSO | 109 | |||||||||
| 2CIX | HEM, CEJ, Br−, EDO | 109 | |||||||||
| NHFe dependent halogenases (NHFeHal) | CytC3 | 319 | 3GJA | ACT | 2009 | γ,γ-Dichloroaminobutyrate | Monomer | L-2-Aminobutyric acid | Cl | Streptomyces sp. | 110 |
| 3GJB | AKG, ACT, Fe2+ | 110 | |||||||||
| SyrB2 | 310 | 2FCT | DSU, AKG, Cl−, Fe2+ | 2006 | Syringomycin E | Monomer | L-Threonine | Cl, Br | Pseudomonas syringae | 111 | |
| 2FCV | DSU, AKG, Br−, Fe2+ | 111 | |||||||||
| 2FCU | DSU, AKG | 111 | |||||||||
| CurA-Hal | 2311 | 2LIU | — | 2011 | Curacin A | Domain (NMR structure) | 3-Hydroxyl-3-methyl-glutaryl | Cl | Lyngbya majuscula | 111 | |
| 2LIW | PNS, MAH | 2011 | Domain (NMR structure) | 111 | |||||||
| WelO5 | 315 | 5IQS | AKG, Fe2+, Cl− | 2016 | Welwitindoline | Monomer | 12-Epi-fischerindole U | Cl, Br | Hapalosiphon welwitschii | 111 | |
| 5IQT | 6CU, AKG, Fe2+, Cl− | 111 | |||||||||
| 5IQU (G166D) | 6CU, AKG, Fe2+ | 111 | |||||||||
| 5IQV | 6CU, AKG, Fe2+, Cl−, NO | 111 | |||||||||
| 5J4R | AKG, Ni2+ | 2016 | TBP | ||||||||
| 5T22 | AKG, Ni2+ | 2016 | TBP | ||||||||
| 5TRQ | SIN, ACT, Ni2+ | 2016 | TBP | ||||||||
| SAM (S-adenosyl-L-methionine)-dependent halogenases | FIA | 299 | 1RQP | SAM | 2004 | Hexamer | SAM | F | Streptomyces cattleya | 67 | |
| 1RQR | 5FD, MET | 67 | |||||||||
| 2C2W | 5CD, Cl− | 2005 | 68 | ||||||||
| 2C4T | SA8, Cl− | 2007 | TBP | ||||||||
| 2C4U | — | 2006 | 112 | ||||||||
| 2C5B | 5F1, MET | 112 | |||||||||
| 2CBX | CC5 | 112 | |||||||||
| 2CC2 | — | 112 | |||||||||
| 2C5H | 3D1, MET, Cl− | 2006 | TBP | ||||||||
| 2V7T (S158G) | SAH, Cl− | 2008 | 87 | ||||||||
| 2V7U (S158G) | SAM, Cl− | 87 | |||||||||
| 2V7V | 5FD | 87 | |||||||||
| 2V7W (S158G) | 5FD | 87 | |||||||||
| 2V7X (S158G) | 5FD, MET | 87 | |||||||||
| 4CQJ | EFA | 2014 | TBP | ||||||||
| 5FIU | Y3J, TLA | 2015 | 113 | ||||||||
| 5LMZ | 1DA, Cl− | 2016 | 114 |
2. Natural and unnatural aromatic substrate scope for biocatalytic halogenation
2.1.1 Early stage halogenation in biosynthetic pathways. Indoles are commonly utilised motifs in medicine and agrochemistry, and new tools that enable their chemical diversification are desirable. There are a series of FDHs that allow for this diversification, via halogenation. Many FDHs mediating the regiospecific halogenation of the indole moiety of tryptophan have been found so far. Most of these enzymes have been found to be involved in the early stages of bioactive natural product synthesis. Such enzymes include PrnA and RebH, tryptophan 7-halogenases, from the first step of the biosynthesis of the antibiotic pyrrolnitrin 24 and the indolocarbazole anticancer agent rebeccamycin 36, respectively.35,43 Tryptophan 6-halogenases include ThdH (alternatively known as ThaI) from the first step of thienodolin 25 biosynthesis, a plant growth promoting compound,71 SttH identified in silico adjacent to an NRPS cluster,72 SatH from Streptomyces albus,73 BorH, a thermophilic halogenase from borregomycin 33–35 biosynthesis,74 and AfnX responsible for generating the 6-chlorotryptophan precursor of alboflavusin A 26. Alboflavusin is an analogue of the dimeric non-ribosomal peptide (NRP) anti-tumor agent and apoptosis inducing agents, himastatin and chloptosin.75
Though less common so far, 5-chlorotryptophans may also be seen as motifs in natural products. Examples include the antibiotic pyrroindomycin B 27; the first step in the biosynthesis of this compound is the generation of 5-chlorotryptophan by PyrH.76 Sequence analysis of Xenorhabdus szentirmaii revealed a further flavin dependent tryptophan 5-halogenase XszenFHal. Though the natural function of this halogenase remains unknown, it was demonstrated to be capable of regioselectively halogenating a series of indolic and anthranilic substrates.77
The first successful demonstration of the ability to modify the regioselectivity of a FDH was via site directed mutagenesis of tryptophan 7-halogenase PrnA. The modification of a large phenylalanine residue in the active site to a smaller alanine allowed for a different binding orientation of tryptophan, leading to halogenation at the 5 position, though it must be noted that halogenation at the 7 position still dominated (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of 7-/5-bromination).78 Despite useful changes in selectivity being rare, there are a number of further successes.42,79 Positioning of the lysine residue over the substrate was thought to be critical to site of halogenation. Various studies toward gaining deeper understanding of subtle factors beyond the positioning of the lysine residue in the active site have been carried out, including structural comparison of PyrH, SttH and PrnA, and, specifically, a loop proximal to the active site, informing the design of mutants with modified regiochemistries.80 Modifications to ThaI, to promote chlorination at the 7 position, rather than the 6 position have also been reported.81
1 ratio of 7-/5-bromination).78 Despite useful changes in selectivity being rare, there are a number of further successes.42,79 Positioning of the lysine residue over the substrate was thought to be critical to site of halogenation. Various studies toward gaining deeper understanding of subtle factors beyond the positioning of the lysine residue in the active site have been carried out, including structural comparison of PyrH, SttH and PrnA, and, specifically, a loop proximal to the active site, informing the design of mutants with modified regiochemistries.80 Modifications to ThaI, to promote chlorination at the 7 position, rather than the 6 position have also been reported.81
Biosynthesis of 4-chlorotryptophan remains a mystery. Although it was isolated from immature pea seedlings in 1970, and postulated to be a precursor to a plant growth hormone,115 the halogenase has not yet been found, hinting that it may be strikingly different to the other, well known FDHs. There have also been fewer reports of natural products containing 2-halotryptophans, perhaps due to their inherent instability. The keramamides 30116 and linked family of jaspamides, chondramides 28 are two exceptions, with CmdE implicated in the generation of 2-chlorotryptophan,117 though for this enzyme, no activity with free tryptophan can be shown. It is not clear as to whether lack of observation of production of this species by CmdE is due to the enzyme requiring a different substrate or due to the instability of the product. 2-Chlorotryptophans are very unstable and both the free amine and carboxy groups might be considered to promote dechlorination.
Tryptophan halogenases capable of halogenating substituted tryptophans are also known. These include the tryptophan 6-halogenase KtzR, which acts in tandem with KtzQ in the first steps of kutzneride 29 assembly, installing a second chlorine into the 7-chlorotryptophan that KtzQ generates118 and KrmI, another tryptophan-6-halogenase, which processes 4-hydroxy-tryptophan as its natural substrate, though the wild type enzyme shows ability to process a fairly broad range of other substrates. KrmI is an unusual halogenase, involved in the biosynthesis of the keramamides 30, this FDH is translationally fused to a ThiF protein119 (Fig. 3).
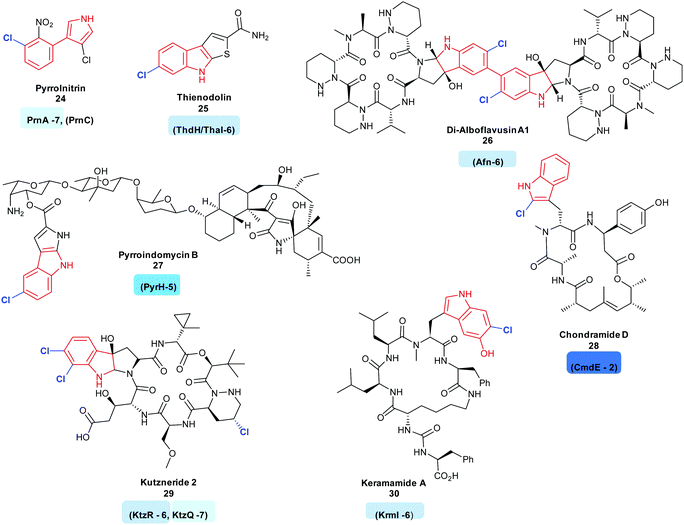 | ||
| Fig. 3 Halogenated natural products arising from the halogenation of indolic substrates: all include regioselective halogenation of tryptophan as a first step in their biosynthesis (tryptophan halogenase name and regiochemistry noted below each corresponding natural product). Examples include (C7) pyrrolnitrin 24,120 (C6) thienodolin 25,71 Di-alboflavusin A1 26,75 (C5) pyrroindomycin B 27,76 (C2) chondramide D 28117 (C6, C7) kutzneride 29,121 keramamide A 30116 Tryptophan halogenases involved in the biosyntheses of these natural products are depicted, and are colour coded in accordance with the regiochemistry of the halogenation that they mediate. Chlorination is indicated in blue. Different regio-chemistries for chlorination are represented by different shades of blue. | ||
A large variety of symmetrically and asymmetrically halogenated indolocarbazole compounds are known, including the cladoniamides 31, 32,122 borregomycins 33–35,123 and rebeccamycins 36, 37.40 From genomic analysis of the producing organisms, the first step in the biosynthesis of these compounds also involved the generation of 5, 6 or 7-halotryptophan. This biosynthetic logic has been utilised to enable the generation of combinatorial libraries of known and novel compounds in which halogenation and glycosylation patterns are modulated.124 Similar to these compounds are the bisindole alkaloids indimicins and related spiroindimicins 38–40 (Fig. 4).125
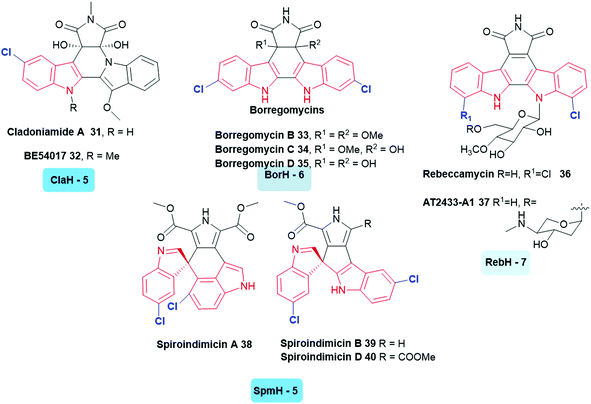 | ||
| Fig. 4 Naturally occurring halogenated bisindole alkaloids. These compounds incorporate a halogen into tryptophan in the first stage of their biosynthesis. (The tryptophan halogenase responsible and its regioselectivity that it confers, are noted below each natural product.) Examples include indolocarbazoles (C5) cladoniamide A 31,122 (C5) BE-54017 32,128 (C6) borregomycins B 33, C 34 and D 35123 (C7) rebeccamycin 36,40 (C7) AT2433-A1 37,129 (C5) spiroindimicins 38–40.125 Non-halogenated analogues of the latter have been accessed through deactivation of SpmH.125 Tryptophan halogenases involved in the biosyntheses of these natural products are depicted, and are colour coded in accordance with the regiochemistry of the halogenation that they mediate. Chlorination is indicated in blue. Different regio-chemistries for chlorination are represented by different shades of blue. | ||
The tryptophan halogenases have been previously thought to possess only modest innate substrate flexibility. RebH is capable of processing tryptolines, whilst KrmI will halogenate unsubstituted indoles and fluoro-tryptophan.42,119 The tryptophan halogenases continue to be subject to extensive rational reengineering and directed evolution due to their potential as tools for biocatalysis. In addition to studies that explore and modulate factors governing regiocontrol, considerable effort continues to be invested in extending substrate scope.41 Wild-type RebH (the tryptophan 7-halogenase from rebeccamycin 36 biosynthesis) has been demonstrated to be capable of regioselectively halogenating a series of indolic (Fig. 5) and arene substrates (Fig. 9).126
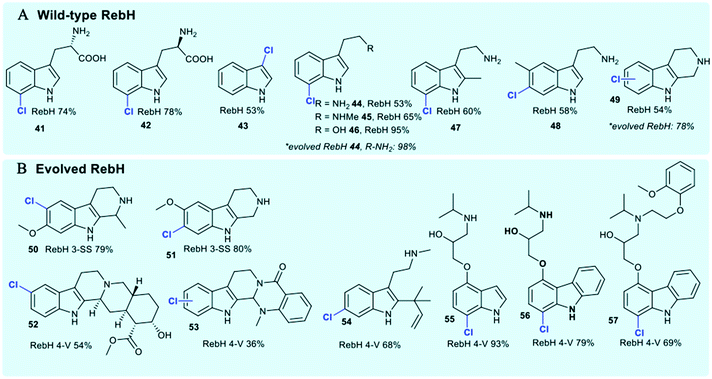 | ||
| Fig. 5 Representation of some of the diverse substrate scope of tryptophan halogenases. Enzymes include (A) wild-type RebH chlorinating indoles;42,126 (B) evolved RebH 3-SS and RebH 4-V which accept tricyclic tryptoline and large indole carbazoles.127 | ||
Natural substrate scope for the FDHs RebH, GsfI, ThaI and Rdc2 is seen to be a little broader, and these enzymes have capability to process a range of indolic substrates.42 Directed evolution of RebH has been employed by the Lewis group to tune the enzyme for use in 7- 44, 6- and 5-chlototryptamine generation enabling quantitative conversions in some instances, and high selectivities.42 Evolved RebH variants have been employed, achieving selective late stage functionalisation of chlorination of sterically bulky substrates, including tricyclic tryptolines 50–53 and large indole carbazoles 56, 57, improving conversions of these unnatural substrates as well considerably extending substrate scope from what was previously known (Fig. 4).126,127
Production of halogenated indolic substrates by flavin-dependent halogenases is commonly reported at small scale.47,126,127 The use of Cross-Linked Enzyme Aggregates (CLEAs), in which the physical aggregation of enzymes with a cross-linking agent is mediated, can enable enzyme stabilisation and enhanced yields. By crosslinking RebH and PrnF (the PrnA-related reductase) with glutaraldehyde, Sewald was able to achieve yields on a gram scale with CLEAs for the halogenation of L-tryptophan, D-tryptophan and L-5-hydroxytryptophan, establishing the foundations for halogenase biocatalysis on a preparative scale130 (see Section 3).
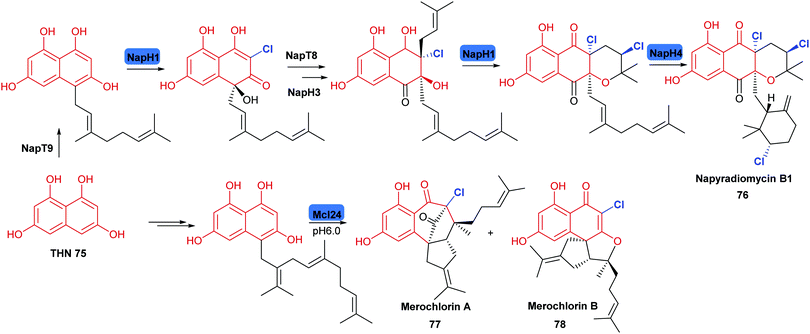 | ||
| Scheme 5 Halogenation in the biosynthesis of the phenolic napyradiomycin 76 and merochlorins 77, 78: these natural products are synthesised from polyketide derived tetrahydroxynaphthalene (THN), the biosynthesis of these suites of metabolites involve versatile VHPOs and employ a chemical logic of chloronium formation promoting enantioselective intramolecular cyclisation.153,155 Chlorination is indicated in blue. | ||
2.2.1 Early stage halogenation by FDH variant B enzymes. Whilst there has been considerable and detailed investigations of a number of the variant A halogenases that work on substrates that are not covalently tethered to a carrier enzyme (in particular the tryptophan halogenases), fewer in vitro investigations of variant B halogenases have been published. However, a diverse range of bioactive natural products that contain halogenated phenols have been isolated. Here, the operation of variant B FDHs may be seen to be prevalent, and investigations indeed indicate many of the substrates to be enzyme tethered. Halophenol-containing natural products include the actinobacteria-produced aminocoumarins clorobiocin 58 and simocyclinone D8 59. The initial steps of the biosyntheses of these metabolites are postulated to include halogenation of enzyme-tethered tyrosine (mediated by Clo-hal and SimX1, respectively).133,134 Evidence points to a similar halogenation strategy also being utilised in the biosynthesis of the peptidic cyanobacterial products aeruginosin (AerJ), cyanopeptolin (McnD), and cryptophycin A (CrpH) 60 of mixed PKS NRPS origin.135,140 A similar biosynthetic logic is also predicted for the glycopeptide antibiotics (GPAs). Vancomycin 62 (VhaA) and teicoplanin 61 (Tcp21), are two potent GPAs; again, these compounds contain chlorinated tyrosine residues. In the case of the GPA teicoplanin 61, careful in vitro investigations have shown that Tcp21 accepts the peptidyl carrier protein (PCP) supported aminoacyl substrate (tethered tyrosine), but will not accept the dipeptide or extended peptides as substrate.136 In teicoplanin biosynthesis, this amino acid residue is likely to be β hydroxylated at a later stage by a non-heme iron oxygenase, whilst in balhimycin biosynthesis OH-Tyr, and not tyrosine, is incorporated by the NRPS machinery.141 Analogues of tyrosine may also be accepted by halogenases. SgcC3 accepts β-tyrosine, probably also in carrier protein tethered form, in the biosynthesis of the enediyne C-1027 64.137 In a similar manner, enzyme tethered 3-hydroxy-4-methoxy-L-phenylalanine is seen to be processed to 2-chloro-3-hydroxy-4-methoxy-L-phenylalanine by Pep1 in the first steps of the biosynthesis of pepticinnamin G.142 In chondrochloren 65, 66 biosynthesis, CndH is indicated to process a different, perhaps decarboxylated yet enzyme tethered analogue of tyrosine.36 Whilst many of the halogenases identified so far are seen to accept electron-rich aromatic amino acids in the biosynthesis of chlorothricin 63, ChlB4 is postulated to act on the small polyketide subunit, 6-methyl salicylic acid139 (Fig. 6).
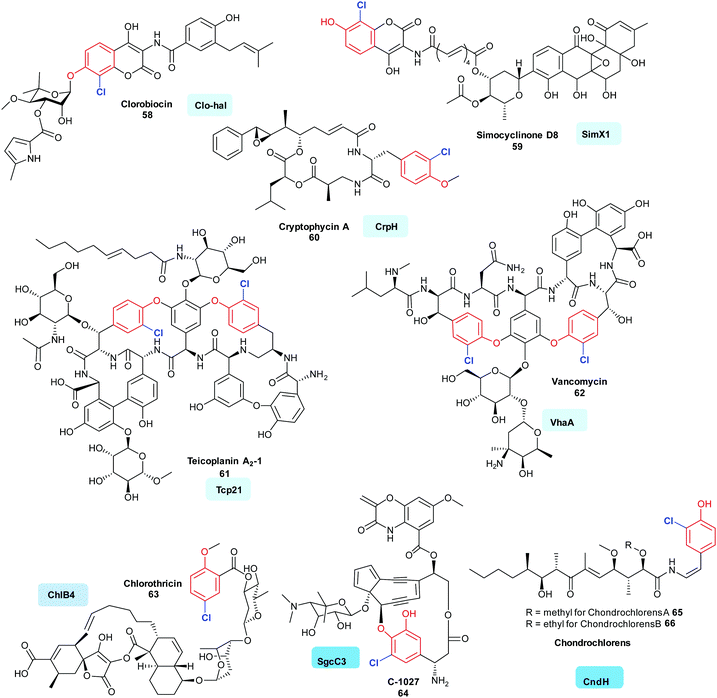 | ||
| Fig. 6 Representative bioactive natural products containing halophenol motifs, where the halogen is installed by a variant B FDH. Clorobiocin 58,133 simocyclinone D8 59,134 cryptophycin A 60,135 are all postulated to be generated from the halogenation of an enzyme tethered tyrosine residue, in the first step (halogen added ortho in EAS fashion as mediated by Clo-hal, SimX1 and CrpH respectively). In vitro evidence supporting the early stage halogenation of an enzyme tethered tyrosine exists for teicoplanin 61 and vancomycin 62 (mediated by Tcp21136 and VhaA).136 C-1027 64137 and the chondroclorens 65, 66 are both postulated to arise from the halogenation of an enzyme tethered analogue of tyrosine, in the first stages of the biosynthesis,36,138 whereas a first step of chlorothricin 63 assembly is thought to be the halogenation of enzyme tethered salicylate (catalysed by ChlB4).139 Chlorination is indicated in blue. Different regio-chemistries for chlorination are represented by different shades of blue. | ||
2.2.2 Late stage halogenation by variant A FDHs. Whilst the variant B FDHs are not easily amenable to in vitro investigations, and are indicated to have a very limited substrate scope, the variant A FDHs are less limited by these problems and offer great opportunity for harnessing as biocatalysts. Amongst the most exciting of these, are enzymes that are reported to act at a late stage of the biosynthesis of structurally complex and sterically bulky phenolic/quinolic natural products. Rdc2143 and RadH144 share 87% sequence similarity; these two enzymes are identified to act in the final steps of biosynthesis of the fungal polyketide natural product radicicol 67, an Hsp90 inhibitor, within the fungi Pochonia chlamydosporia and Floropilus chiversii respectively. GsfI operates in the final stages of the assembly of griseofulvin 68, a clinically utilised antifungal agent (Fig. 7).145 TiaM from tiacumicin B biosynthesis operates in the final step of the biosynthetic pathway, functionalising a complex and glycosylated polyketide generating both the mono and dihalogenated products.226
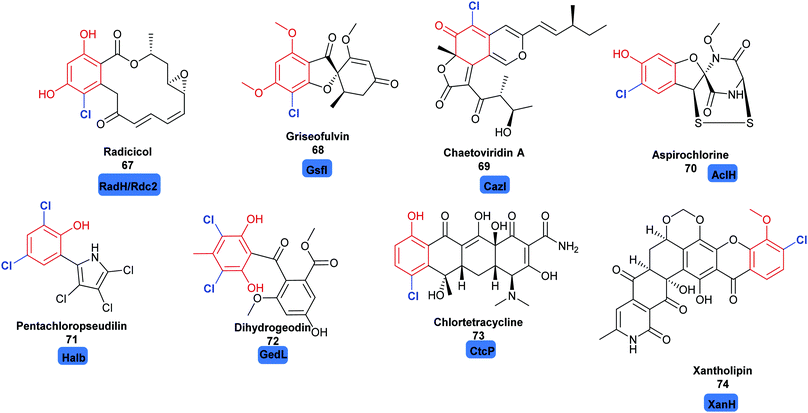 | ||
| Fig. 7 Halogenated natural products where installation of halogen on a structurally complex non-tethered freestanding phenol is performed selectively by a halogenase: including radicicol 67,143 griseofulvin 68,145 chaetoviridin A 69,146 aspirochlorine 70,149 pentachloropseudilin 71151 dihydrogeodin 72,148 chlortetracycline 73147 and xantholipin 74.150 Chlorination is indicated in blue. | ||
CazI, from chaetoviridin A 68 biosynthesis, is postulated to operate on a complex phenolic intermediate, in the final stages of this cytotoxic polyketide.146 In a similar manner, CtcP is seen to halogenate an advanced type II, aromatic polyketides core, in the biosynthesis of chlortetracycline 73.147 Heterologous pathway reconstruction studies indicate that in geodin biosynthesis GedL dichlorinates the advanced PKS metabolite sulochrin, directly affording dihydrogeodin 72.148 Aspirochlorine 70 is an epidithiodiketepiperazine (ETP) toxin, produced by Aspergullus orizae, a mold used for millennia in Asian cuisine. The first stages of the biosynthesis of aspirochlorine involve the incorporation of two phenylalanines into a symmetric diketopiperazine (DKP). Intriguingly, one phenylalanine undergoes a C–C cleavage to convert it into glycine. The other phenylalanine is hydroxylated. AclH, operates in the final step of the biosynthesis, chlorinating the hydroxylated phenylalanine residue and affording the toxin.149 XanH, a bifunctional FDH fused to its concomitant flavin reductase, has been shown capable of regioselectively chlorinating a complex late stage xanthone intermediate on the path to xantholipin 74.150 Pentachloropseudilin 71 has a series of interesting biological activities including inhibiting TGF-beta signalling, and impairing angiogenesis; yet its exact biosynthesis remains unclear. HalB was found in a cosmid library of the producing organism; the first halogenase identified from an actinomycetes. Notably, HalB shows 55% sequence identity to the pyrrole FDH PrnC. HalB has been shown to be capable, in vitro, of installing a single halogen into 2-(3,5-dibromophenyl pyrrole), but, whether or not it acts iteratively to install all 5 halogens in both the phenol and pyrrole ring in the natural system remains unclear.151
2.2.3 Late stage halogenation by haloperoxidases. Vanadium-dependent haloperoxidases (V-HPOs) have been identified that are involved in the biogenesis of halophenol containing natural products. The napyradiomycins (including napradiomycin B1 76) are polyketide-terpenoid natural products identified in marine streptomycetes, over 50 members of this suite of natural products have been identified so far. V-HPOs identified within the biosynthetic gene cluster include NapH1, and NapH4. NapH1 is particularly notable, this versatile enzyme acts upon geranylated tetrahydroxynapthalene (THN) derivative, stereoselectively hydroxylating it whilst mediating regioselective chlorination (Scheme 5). NapT8 and NapH3 operate successively to catalyse prenylation and then an α-hydroxy ketone rearrangement. NapH1 then operates once more to halogenate the newly introduced prenyl unit. The reaction is most likely to proceed via a halonium species, which is then opened by the hydroxyl that NapH1 previously introduced, thereby enabling enantioselective cyclisation. In a similar manner, NapH4 mediates halogenation of the geranyl unit, again most likely proceeding via the halonium species, and promoting intramolecular cyclisation.152 The complex cyclic scaffolds of the merochlorins 77, 78 are formed using similar chemical logic. Enzymatic total syntheses of napyradiomycin B1 76 has been enabled in 18% yield.153
2.2.4 Substrate scope: phenols. Smaller, freestanding, non-native phenolic substrates have been demonstrated to be processed by variant A FDHs and HPOs (Fig. 8), revealing these enzymes to be useful biocatalysts. The employment of the HPOs, in this context, whilst enabling halogenation of sterically bulky substrates, results in mixtures of regioisomers and polyhalogenated compounds. This is a direct result of most HPOs releasing free hypohalous acid; in this context, the enzyme has little control over the regiochemistry of the transformation, and it is simply the most electron-rich region(s) of the substrate that are halogenated. In contrast, the FDHs usually accept much smaller substrates but are generally highly regioselective. PltM has been shown capable of mono 117 or di-halogenation 118 of phloroglucinol (Fig. 8J), as well as being able to accept resveratrol as a substrate.51 Wild type RebH, GsfI, and ThaI have been shown capable of processing a broad range of non-natural substrates containing phenolic moieties.154 Exceptions may be seen with RadH/Rdc2 and VirX1, FDHs with much more open and accessible active sites, capable of regioselectively monohalogenating much larger phenolic substrates.47,143,144
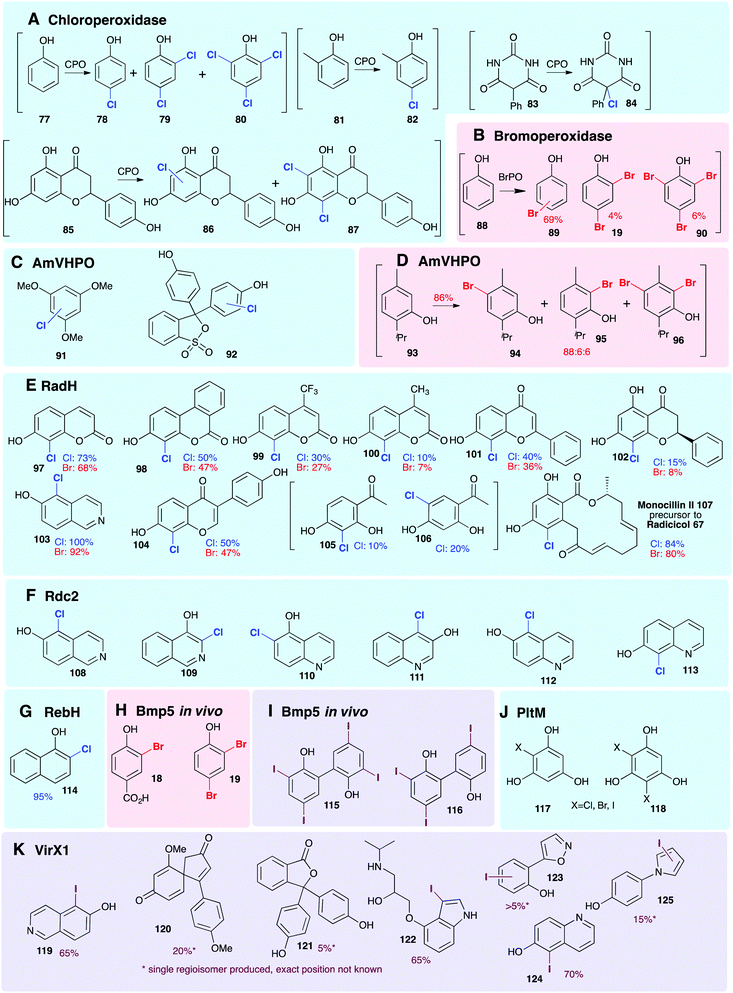 | ||
| Fig. 8 Representation of some of the diverse phenols that are accepted as non-natural substrates by different wild-type halogenases. All experiments are in vitro, unless specified otherwise, and % conversions reported as determined by LC/LC-MS. Enzymes include (A) chloroperoxidase (heme iron-dependent),156–158 (B) bromoperoxidase (vanadium-dependent)159 (C and D) AmVHPO (vanadium-dependent)107 (E) RadH (FDH) chlorination;144 (F) Rdc2 (FDH) chlorination and bromination;143,160 (G) RebH (FDH) chlorination126 (H and I) Bmp5 brominase (FDH) in vivo.45 (J) PltM (FDH) associated with transcriptional regulation of pyoluteorin 43 biosynthesis,51,161 (K) VirX1 (FDH from a virus, the first FDH with a preference for iodination).47 Chlorination, bromination and iodination are colour coded blue, red and purple respectively. | ||
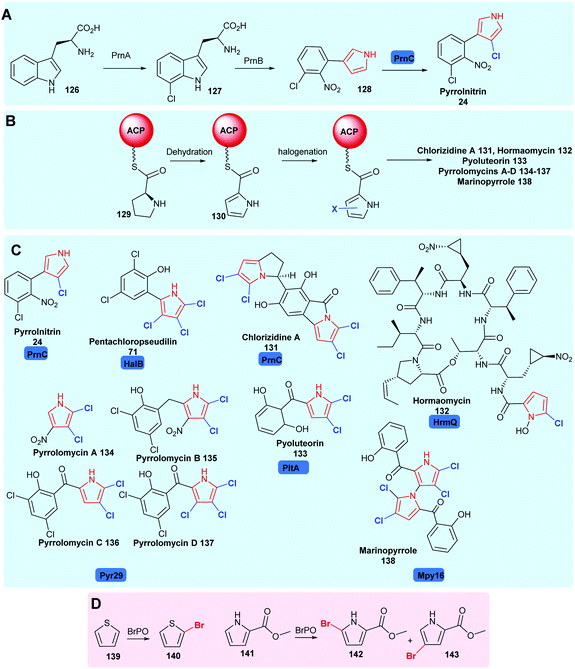 | ||
| Scheme 6 Naturally occurring halopyrrole containing compounds. (A) The biogenesis of pyrrolnitrin 24 involving a variant A FDH PrnC. (B) The enzyme tethered biosynthesis of halopyroles from proline, employing variant B FDHs. (C) Bioactive compounds containing halopyrroles including pyrrolnitrin 24,34 pentachloropseudilin 71,151 chlorizidine A 131,162 hormaomycin 132,163 pyoluteorin 133,164 pyrrolomycins 134–137165 and marinopyrrole 138,166 noting the halogenases utilised in their construction. (D) Biocatalytically vanadium-dependent bromoperoxidases may be utilised to monobrominate thiophene and pyrroles.167 Chlorination is colour coded blue. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14% para
14% para![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ortho mixture. Mutations could be effectively employed to modulate ortho:para selectivities and improve yields for these unnatural substrates.79 Chlorination of naphthalen-2-amine to give 165 by RebH ortho to –NH2 is achieved in a high yield of 93%.126
ortho mixture. Mutations could be effectively employed to modulate ortho:para selectivities and improve yields for these unnatural substrates.79 Chlorination of naphthalen-2-amine to give 165 by RebH ortho to –NH2 is achieved in a high yield of 93%.126
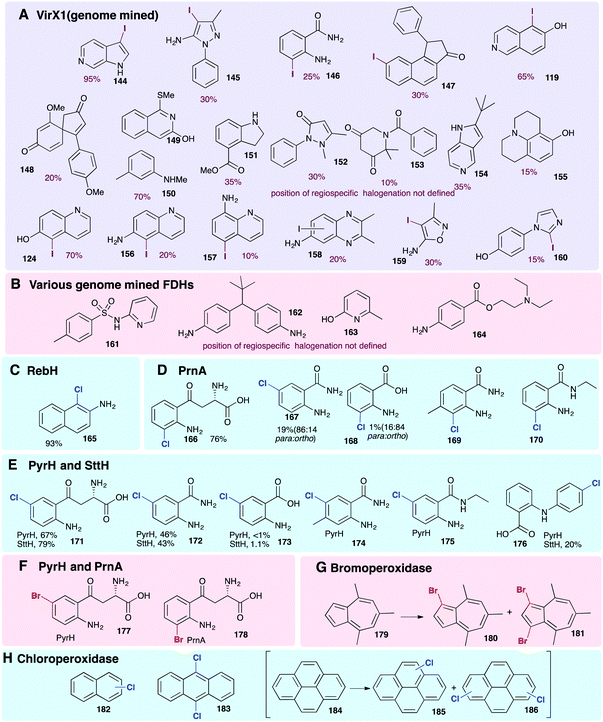 | ||
| Fig. 9 Representative selection of aromatic moieties (beyond indole, pyrrole and phenol) that may be accepted as substrates for enzymatic halogenation. Regiochemistry and conversions are given where reported in the primary literature. Enzymes include (A) genome mined VirX1, a variant A FDH with a very broad substrate scope, the first halogenase to be isolated from a virus and the first FDH to show preference for iodination.47 Notably, the regioselective halogenation of a diverse range of both small and large substrates may be seen including several less electron-rich and less activated substrates (bromination of all substrates also possible). (B) A series of genome mined FDHs, again, halogenation of sterically more bulky substrates may be seen (bromination of all substrates also possible).154 (C–F) Anilines and anthranilates processed by variant A FDHs RebH, PrnA, PyrH and SttH, respectively.79,80,126 (G) Vanadium-dependent bromoperoxidase capable of brominating bulky substrates.168 (H) Heme-iron dependent chloroperoxidases shown to be capable of processing bulky, planar modestly activated compounds such as pyrenes, mixtures of regiochemistries and levels of substitution are observed.169 Chlorination, bromination and iodination are colour coded blue, red and purple respectively. | ||
The halogenation of sterically bulky substrates can be enabled using haloperoxidases. As there is generally no substrate binding site, and free HOX is released, the substrates that can be halogenated using these systems are not limited by size. However, as is usually expected for an HPO that releases HOX, series of regioisomers, as well as mono, di, and tri halogenated compounds result (Fig. 9G).169 For example, heme iron-dependent chloroperoxidase (CPO) from C. fumago has been used to give chlorinated analogues of naphthalene 182, anthracene 183 and pyrenes 185 and 186. A vanadium-dependent bromoperoxidase from the marine algae Ascophyllum nodosum has also been employed mono and dibromination 4,6,8-trimethylazulene 179 (Fig. 9G), though the instability of the resultant product hindered purification.168
To achieve regioselective electrophilic halogenation, generally FDHs are needed, however, historically these have shown fairly narrow substrate scope. In 2019, new FDHs were revealed that had been found using in silico genome mining approaches (Fig. 9A and B). These halogenases were able to accept much larger substrates and showed a much broader substrate scope than any FDHs examined previously. VirX1, with a preference for iodination, was demonstrated to be capable of regiospecifically iodinating or brominating a diverse portfolio of substrates with a variety of steric and electronic demands, and some of which might be considered to be only weakly activated. Good conversions of a wide range of unnatural substrates are possible with this unusual enzyme from a virus.47 Similar infomatics-led approaches have been utilised by the Sewald and Lewis groups, further demonstrating that natural FDHs do exist with broader substrate scope.170
 | ||
| Fig. 10 Selected water-soluble ligands used for aqueous Suzuki–Miyaura cross-coupling reactions in the presence of halogenase enzymes or even living systems. | ||
Like the FDHs, the NHFeHals may be subdivided into those that operate only on protein tethered substrates (which we call here the variant B NHFeHals, to be consistent with the variant B FDHs that also operate only on protein tethered substrates), and the perhaps more biotechnologically useful, and recently discovered variant A NHFeHals that process free substrates (see Section 1.2). The variant B type NHFeHals have proved challenging to handle requiring anaerobic conditions, nevertheless they catalyse series of intriguing chemistries, including cryptic halogenation en route to the installation of cyclopropyl or alkynyl motifs, notable examples may be seen in jamaicamide 204,61 curacin A 208,174 coronatine 213175 and kutzneride 29 biosynthesis (Scheme 8).118 The need for covalent substrate tethering prior to halogenation means that variant B halogenases have less biotechnological potential at the current time, and will be discussed just briefly in the following section.
2.5.1 Tethered substrates with unactivated sp3 carbon centres. NHFeHals capable of processing a variety of very different tethered substrates are known and have been shown to be capable of selectively functionalising terminal methyl groups and internal methylenes. Tri-chlorination of the pro-R methyl of peptidyl carrier protein (BarA) tethered leucine 187 by tandem action of BarB1 and BarB2,60 and di-chlorination and mono-chlorination of the γ-methyl of peptidyl carrier protein tethered L-2-aminobutyric acid 193 and L-threonine 190 by CytC3 and SyrB2 has been demonstrated through in vitro reconstitution experiments by Walsh and co-workers (Scheme 7).58,171 CytC3 and SyrB2, have a level of similarity (58% identity, 71%) similarity and process these subtly different substrates. Beyond amino acids, NHFeHals are known that can process tethered piperazines, fatty acids and tethered polyketide intermediates. Examples include, KthP, which mediates the generation of the 5-chloropiperazyl motif in kutzneride 29 biosynthesis,118 and HctB responsible for chlorination at C5, of an acyl-carrier protein tethered hexanoate 199, in the biosynthesis of the antifungal hectochlorin by cyanobacterium Lyngbya majuscula (Scheme 7).172
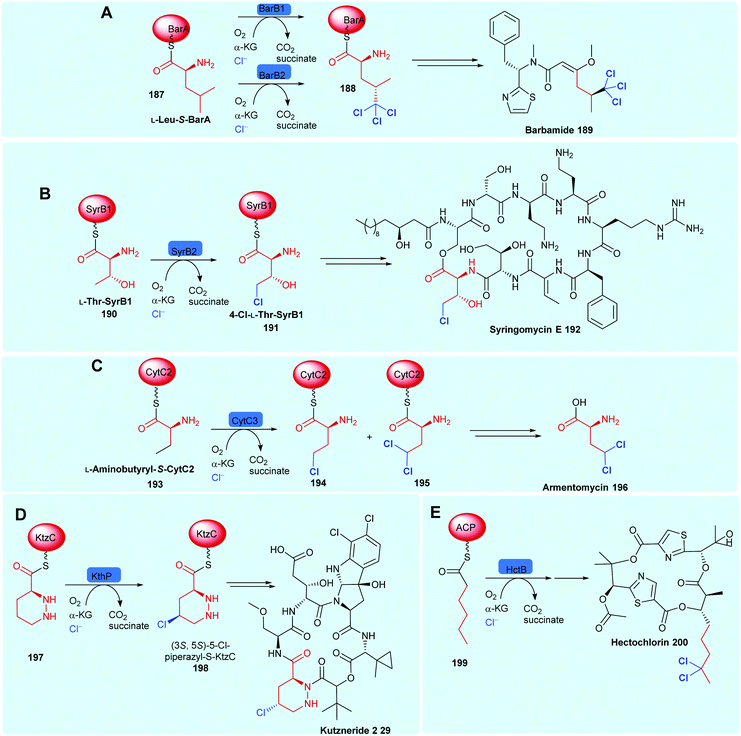 | ||
| Scheme 7 Bioactive compounds where installation of halogen on a protein-tethered aliphatic substrate is performed selectively by a NHFeHal, including (A) barbamide 189,60 (B) syringomycin E 192,58 (C) armentomycin 196,171 (D) kutzneride 2 29118 and (E) hectochlorin 200.172 | ||
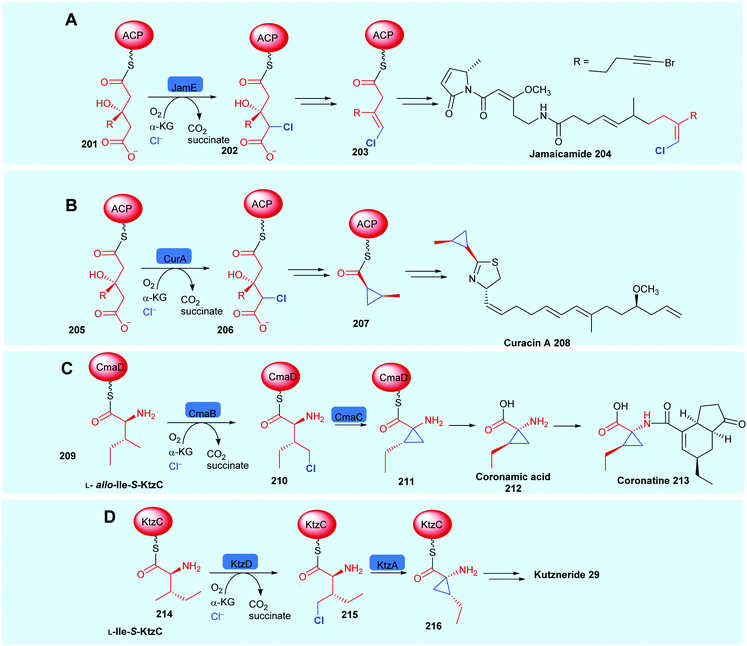 | ||
| Scheme 8 Installation of halogen on protein-tethered aliphatic substrate performed selectively by a NHFeHals, in the biosynthesis of natural products, enabling alkene generation and cyclopropane generation. (A) Jamaicamide 204,61 (B) curacin A 208,174 (C) coronatine 213,175 and (D) kutzneride 29.118 Notable similarities may be observed between the first steps in jamaicamide, vinylchloride formation and curacin cyclopropyl biosynthesis. Coronamic acid and kutzneride 29 biosynthesis are initiated by the halogenation of the γ-methyl of different diastereoisomers of L-isoleucine. | ||
2.5.2 Unactivated sp3 carbon centres, processed as free, non-tethered substrates: enzymatic generation of alkyl halides. The discovery of WelO5 from Hapalosiphon welwitschii UTEX B1830, the welwintindolinone A 219 producer, by Liu and coworkers was ground-breaking; for the first time, a NHFeHal could be seen to process non-enzyme tethered substrates. Its regioselective and stereoselective halogenation of a complex carbocycle, renders it an exciting tool for biotechnological applications. WelO5 monochlorinates 12-epi-fischerindole U 217 and 12-epi-hapalindole C 220 (Scheme 12),62 and has been demonstrated to enable bromination of the same substrates, leading to their enhanced antibacterial activity.176 Sharing 79% sequence identity with WelO5, AmbO5 was subsequently discovered through the analysis of the ambiguine 53 biosynthetic gene cluster.83,177 AmbO5 accepts a wider scope of substrates than WelO5 and selectively chlorinates a range of fischerindole (52, 53), hapalindole (222) and ambiguine alkaloids (56) (Scheme 9A).83 By generating hybrid enzymes consisting of the N-terminus of WelO5 and C-terminus of AmbO5 (and vice-versa), some success of halogenating a slightly wider scope of hapalindole type molecules was achieved.83
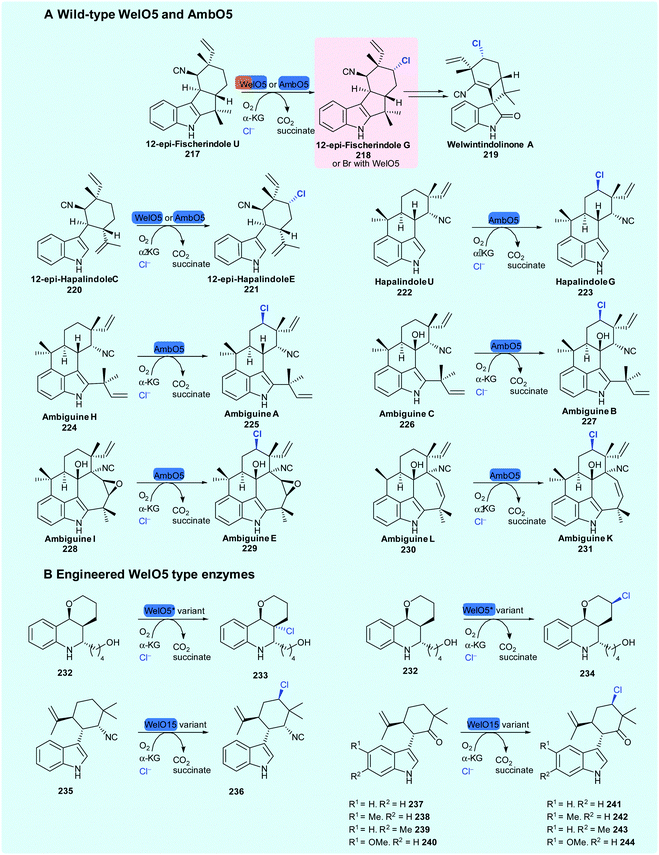 | ||
| Scheme 9 Known substrate scope of wild-type NHFeHals WelO562 and AmbO583 and engineered varieties of WelO15178 and WelO5*.179 | ||
Initial sequence and mutational analysis of WelO5 and AmbO5 pointed to eleven C-terminal residues, likely influencing substrate utilisation.180 The potential to develop WelO5 through directed evolution to improve the biocatalytic capabilities and extend substrate scope has been attempted by a number of groups. Hoebenreich and coworkers used structure-guided direct evolution to develop variants of WelO5 homologue Wi-WelO15 from Westiella intricata HT-29-1 capable of selectively chlorinating non-natural hapalindole 235, and hapalindoles 237–240 containing a ketone moiety at the position of the natural isonitrile.178 Buller and coworkers engineered variants of another homologue WelO5* for selective halogenation at two separate positions on a martinelline analogue 232,179 representing the first successful biocatalytic installation of a halogen by WelO5 type enzymes on a substrate that is notably different to their natural hapalindole targets. Both Buller and Hoebenreich identified amino acid positions, which seem to play an important role in the regioselectivity of these enzymes towards non-natural substrates, opening up possible opportunities for rational re-engineering of these catalysts.
The co-crystal structure of WelO5 and native substrate was used in a structural search for other enzymes capable of alkyl halogenation of different substrates. In this manner, the hydroxylase SadA was identified. Minor modification to the active site and coordination of the FeII (D157GSadA) resulted in an enzyme capable of chlorinating as well as hydroxylating its natural substrate.182
A second series of variant A NHFHals that operate on substrates that are not covalently tethered to a carrier protein (free substrate NHFe halogenases) have recently been discovered, and named the BesD family. Like CurA, CmaB, KtzD (Scheme 8) and JamE, BesD is a cryptic halogenase, and was discovered in the biosynthetic pathway of the amino acid β-ethynylserine 247 (βes) from S. cattleya, after knockouts of non-essential amino acid desaturases pointing towards a novel enzymatic production of the alkyne moiety.183 Purified BesD was shown to selectively chlorinate free lysine 245, producing 4-Cl lysine 246.84 Subsequent C–C cleavage and elimination of the installed chlorine was shown to lead to the terminal alkyne (Scheme 10A). BesD has low sequence identity to both substrate bound SyrB2 (7%) and variant B prototype WelO5 (11%), instead having a much higher identity to predicted hydroxylases (>46%). Rather than being a standalone enzyme, BesD was shown to be part of a variant A NHFeHals.84 Homology networks were produced from hits arising from a sequence-based homology search of BesD. This approach resulted in the discovery of 20 more halogenases that act on free amino acid substrates. The range of substrates was diverse, with BesD and other enzymes accepting lysine but also ornithine, while the hydrophobic amino acids leucine, isoleucine, and norleucine, were shown to be substrates for PrHalE (Scheme 10A). A selection of these halogenases was shown to perform alongside amino acid metabolising enzymes to produce chlorinated heterocycles, diamines, and α-keto acids from lysine (SwHalB), ornithine (PkHalD), and norleucine (PrHalE). Remarkably, all enzymes retained fidelity for halogenation over hydroxylation, while the radical halogenase HalB (different to the FDH HalB) from Streptomyces wuyuanensis (SwHalB) was able to accept bromine and azide anions to produce bromo-lysine and azido-lysine. Chlorolysine, generated by SwHalB and PfHalA, could also be incorporated into a 9 amino acid peptide using an in vitro transcription and translation system-suggesting a potential use for these enzymes in production of more complex natural product analogues.84
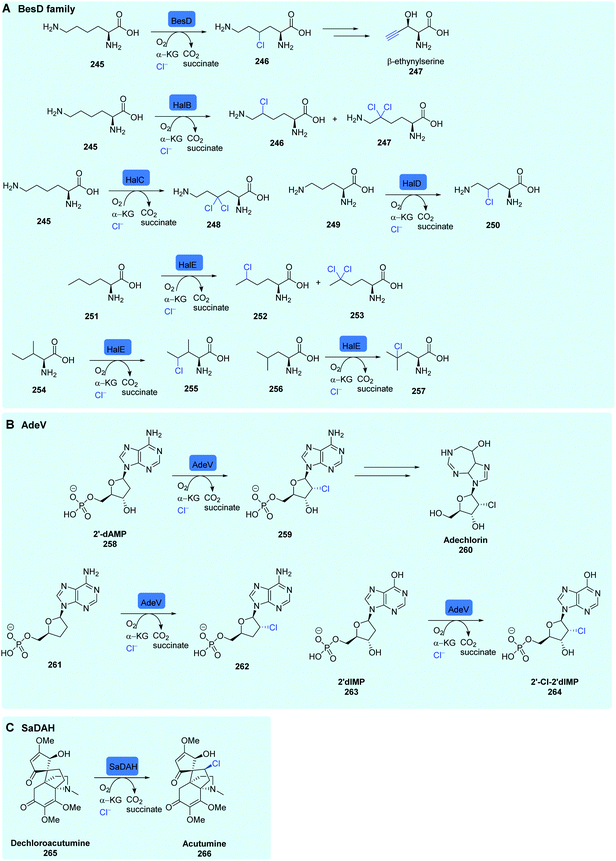 | ||
| Scheme 10 NHFe halogenases. (A) Known substrate scope of the NHFe halogense BesD and enzymes from the BesD family,84 (B) AdeV chlorination of 2′-dAMP in adechlorin biosynthesis and other known substrates of AdeV,85 (C) SaDAH chlorination in acutumine biosynthesis.181 | ||
Recently, variant A NHFHal capable of halogenating nucleosides, named AdeV, has been discovered in the biosynthesis of the chlorinated natural product adechlorin 260 in Actinomadura sp. ATCC 39365, opening up the path to access valuable halonucleosides85 (Scheme 10B). AdeV was shown to have 15% similarity to WelO5. Gene knockout experiments confirmed its role in adechlorin biosynthesis. AdeV acts in the early stages of biosynthesis on free nucleoside 2′-deoxyadenosine-5′-monophosphate (2′-dAMP) 258 to generate 2′-Cl-2′-dAMP. In vitro assays revealed that 2′-deoxyadenosine (2′-dA), identical to 2′-dAMP apart from the 5′- phosphate moiety, was not accepted as a substrate, indicating that the presence of this phosphate is essential for substrate binding and halogenation activity. Consistent with this logic, two other phosphorylated nucleosides 2′,3′-dideoxyadenosine monophosphate 261 and 2′deoxyinosine-5′-monophosphate (2′-dIMP) 263, were accepted by AdeV and converted to their chlorinated counterparts indicating a level of enzyme promiscuity. However, the natural substrate 2′-dAMP showed the highest levels of conversion.
Several plant species are known to generate halogenated metabolites. The toxin fluoroacetate is produced in many plants including Camellia sinensis (from which tea is generated). Fatty acid derivatives of fluoroacetate such as ω-fluorooleic acid accumulating in the seeds of Dichapetalum toxicarium.181 An exciting new NHFe halogenase DAH that performs the stereoselective late stage chlorination of the complex alkaloid dechloroacutimine 265, (Scheme 10C) produced by menispermaceae plants, has been found and its activity demonstrated in vitro. This represents the very first NHFeHal found in plants.181 Phylogenetic analysis indicates that DAH, which has a variant in Menispermaceae canadense (McDAH) and Sinomenium acutum (SaDAH), evolved independently from the previously discussed bacterial NHFeHAls, potentially being an example of parallel evolution in halogenated natural product metabolism. In vitro assays against a wide range of alkaloids indicated that DAH was highly selective towards its natural substrate, with no other small molecules being accepted for halogenation. As with some other halogenases, DAH was shown to accept azide anions as well as halides, and convert (−)-dechloroacutumine to 11-azido-dechloroacutumine.
Beyond the NHFe halogenases, alkyl halides can be generated by a series of vanadium-dependent haloperoxidases. An elegant cyclisation cascade is initiated by the selective bromination of a single alkene within a terpenoid; the resultant bromonium undergoes ring opening affording the alkyl bromide. For example, the vanadium bromoperoxidase from C. officinalis brominates the terpenoid precursor (E)-(+)nerolidol 267, yielding cyclised snyderols 268-230.184 For β- and γ-snyderol, a single diastereoisomer is produced. This provides an exciting biocatalytic opportunity as synthetic methods have succeeded only in forming a mixture of two diastereoisomers of each product (Scheme 11). In a similar manner, chlorination, mediated by haloperoxidases, may be seen to generate chloronium species, initiating cyclisation cascades. Installation of chlorine onto the aliphatic moiety of SF2415B1 by NapH1 allows cyclisation of the structure to afford a cyclic ether, offering potential for synthesis of napyradiomycins 76 (Scheme 5) with cytotoxic and antibacterial properties.155
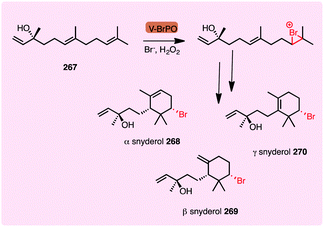 | ||
| Scheme 11 V-BrHPO catalysed terpene bromination and cyclisation event in snyderol biosynthesis.184 | ||
Notably, a small number of FDHs process aliphatic compounds. One such example is CmlS, an FDH from chloramphenicol biosynthesis implicated, through in vivo studies, in the generation of a dichloroacetyl moiety.185 In this unusual FDH, the flavin cofactor is covalently linked to the enzyme. The malonyl-CoA substrate is proposed to form an enolate, stabilised through hydrogen bonding to tyrosine, this species may then react with a proximal chloramine.44 Beyond the halogenation of tethered malonyl-CoA by CmlS, other exciting observations of the applications of these enzymes are noted. Intriguingly, an FDH from a fungus, AoiQ, has been demonstrated to regioselectively chlorinate an unactivated terminal sp3 carbon of a freestanding molecule 271, in the biosynthesis of orthosporin (Scheme 12). So far, AoiQ is unique amongst other flavin-dependent halogenases in its ability to perform this task. Although the mechanism for this reaction is unknown, it has been postulated by Hertweck and coworkers that, though it is a FDH, it may proceed via a radical reaction.186 Notably, like KrmI, it is one of the few fused halogenases to be explored. In the case of AoiQ, it contains a functional O-methylation domain. The mystery as to how it operates remains to be revealed.
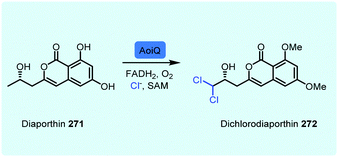 | ||
| Scheme 12 FDH AoiQ dichlorination and dimethylation of diaporthin 271.186 | ||
3. Approaches toward improved biocatalytic halogenation, and steps toward upscaling
Considerable effort has been invested into engineering and evolving the biocatalytic activity of halogenases. Studies have predominantly focussed upon the FDHs in order to alter substrate profiles, modify regiochemistries of halogenation and enhance biocatalytic stability. Greater structural understanding of these enzymes has enabled progress to be made in their rational redesign.187,188 Such approaches combined with substrate walking methods189 have enabled the utilisation of halogenases in the selective decoration of increasingly more complex substrates, as well as stabilising enzymes to operate at higher temperatures.190 Engineering of the NHFeHals has also enabled their application to a broader series of substrates (see Section 2.2.4).178,179 Though the evolution of a small number of tryptophan halogenases for utilisation for halogenation of a broader series of substrates has proved useful, other approaches have considered the natural diversity of the halogenases. The broad range of halogenated structures that have been noted, over 5000 to date,13 together with the variety of organisms from which they are produced, indicates that many diverse halogenases remain to be discovered. In silico discovery approaches have been utilised to mine for various halogenases within uncurated genomic deposits, leading to the discovery of more substrate flexible halogenases from diverse sources including viruses.47,170 Discovery of novel halogenases remains at the forefront of biocatalytic halogenation research. For effective biocatalysis, good amounts of soluble protein are required. The Lewis group have effectively improved heterologous production of RebF through including a cleavable maltose binding protein (MBP) tag, to enhance solubility of the enzyme, and co-expressing both the halogenase and the concomitant reductase, RebF with the chaperonins GroEL/ES, in addition to developing a cofactor recycling system.126 Advances on FADH2 regeneration have recently been achieved by replacing NADH with synthetic mimics.191 A photocatalytic approach has been successfully applied to the VHPOs. The Gulder group cleverly demonstrated that by illuminating their biocatalytic system with 455 nm light, and including sacrificial electron donors, cofactor recycling could be addressed, and a ready supply of reduced cofactor afforded in a relatively inexpensive manner.192,193 An approach that has made a significant difference to the utilisation of FDHs is the application of cross-linked enzyme aggregates (CLEA). FDHs generally have poor stability, impacting their catalytic longevity and utility in scaled up reactions. The robust nature and solvent tolerance of Cross-Linked Enzyme Crystals was first reported in the early 1990s, but the technique required the enzymes of interest to first be crystalised.194 The application of CLEA was first demonstrated for PenAcylase, an aminoacylase utilised in the generation of ampicillin, where CLEA were generated from precipitating, rather than crystallising, the enzyme prior to cross-linking.195 By using a CombiCLEAS approach of 1 g of L-tryptophan to 1.8 g of L-7-Br-tryptophan could be achieved with complete conversion within an 8 days period.130 To achieve this, the lysate from Escherichia coli (E. coli) (6 L), that had been grown with a RebH expression vector, was combined with PrnF, the flavin reductase from Pseudomonas fluorescens, and an alcohol dehydrogenase from Rhodococcus sp., that had been precipitated with ammonium sulfate, was then cross-linked with 0.5%/wt glutaraldehyde to form the CombiCLEAS.4. Site selective C–H activation through hyphenation of enzymatic halogenation and synthetic diversification
The opportunity to combine enzymatic halogenation with chemical modification, in particular catalytic cross-coupling, is exciting and provides a much-needed tool for molecule constructions. This concept, first reported in 2010, with the tryptophan halogenase PrnA being utilised to give access to a new to nature haloindolic natural product, which was subsequently functionalised through Suzuki–Miyaura cross-coupling,196 has been gaining momentum. This section comprehensively reviews the current state-of-the-art and reflects on technologies enabling the hyphenation.Specifically, with the goal of enabling C–H activation through enzymatic halogenation and chemical cross-coupling, our team has developed series of aqueous cross-coupling procedures, enabling the cross-coupling to be compatible with molecules decorated with sensitive functional groups, as well as with enzymes and even in live cell environments. We have initiated our development of these catalytic conditions by focussing on challenging free and unprotected halotryptophans. Our choice has been directed by three factors:
– The availability of a well-characterised series of tryptophan halogenases that may be utilised in standalone format in conjunction with a biosynthetic pathway to access halotryptophans and synchronously new-to-nature halotryptophan containing natural products;
– The utility of tryptophan as a fluorescence tag and a moiety that governs protein folding, and the attraction to being able to tune these properties;
– And the challenge that free tryptophan confers, rendering it a suitably stimulating test bed. We reasoned that by developing conditions that were suitable to address the cross-coupling of free halotryptophan, a small molecule with the propensity to coordinate to the catalyst and render it inactive, such conditions could be readily applied to other more tractable moieties. To this end, we developed a range of aqueous conditions enabling the Suzuki–Miyaura,209 Heck,210 Sonogashira,211 keto-arylation212 and Buchwald–Hartwig213 diversification of free unprotected tryptophan, peptides, and natural products (Scheme 13).
 | ||
Scheme 13 Mild aqueous conditions enabling the cross-coupling of free unprotected halotryptophan and halotryptophans embedded into natural products, biomolecules and bioactive compounds. Colour code: Blue background: conditions enable processing of aryl-chlorides in addition to more reactive aryl-bromides, Red background: least activated aryl halide processed is Ar–Br under these conditions (broad substrate scope and extension of chemistry to other systems not shown here). (A) Suzuki–Miyaura cross-coupling on bromotryptophans: phenylboronic acid (1.1 equiv.), Na2PdCl4 (2.5 mol%), TXPTS (25 mol%), K2CO3 (5 equiv.), water, 40–80 °C, 70–90% isolated yields.209 (B) Suzuki–Miyaura cross-coupling on chlorotryptohans, and 7-chlorotryptophan pacidamycin: phenylboronic acid (1.5 equiv.), Na2PdCl4 (5 mol%), SPhos (10 mol%), K2CO3 (5 equiv.), water:acetonitrile 80 °C, on purified material and crude extract 60–70% isolated yields.217 (C) Suzuki–Miyaura cross-coupling of bromotryptohans, p-tolylboronic acid (3 equiv.), Pd(OAc)2-[2-amino-pyrimidine-4,6-diol]2 (5 mol%), K2CO3 (6 equiv.), 45 °C, 54–85% isolated yields and 7-bromotryptophan pacidamycin in living cultures: reaction carried out in optimised culture medium at 37 °C.206 (D) Ketoarylation of N-Boc-bromotryptohans, and small water-soluble chloro and bromo pharmacophores: ketone (4 equiv.), [Pd(DtBPF)Cl2] (DtBPF = 1,1′-bis(di-tert-butylphosphino)ferrocene) (2 mol%), NaOH (4 equiv.), dioxane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water 1 water 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 60 °C, 45–94% isolated yields.212 (E) Buchwald–Hartwig diversification of chloro and bromotryptophans, peptides containing bromotryptophan and the bromotryptophan containing natural product barettin: aniline (2 equiv.), [Pd(tBu-XPhos)G1] (5 mol%), KOH (4 equiv.), dioxane 1, 60 °C, 45–94% isolated yields.212 (E) Buchwald–Hartwig diversification of chloro and bromotryptophans, peptides containing bromotryptophan and the bromotryptophan containing natural product barettin: aniline (2 equiv.), [Pd(tBu-XPhos)G1] (5 mol%), KOH (4 equiv.), dioxane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water 1 water 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100 °C, 8–48 min, 31–75% isolated yields.213 (F) Sonogashira diversification of bromotryptophans, peptides containing bromotryptophan and the new to nature bromotryptophan containing natural product cystargamide: alkyne (1 equiv.), [PdCl2(CH3CN)2] (5 mol%), sXPhos (15 mol%), Cs2CO3 (2.5 equiv.), acetonitrile 1, 100 °C, 8–48 min, 31–75% isolated yields.213 (F) Sonogashira diversification of bromotryptophans, peptides containing bromotryptophan and the new to nature bromotryptophan containing natural product cystargamide: alkyne (1 equiv.), [PdCl2(CH3CN)2] (5 mol%), sXPhos (15 mol%), Cs2CO3 (2.5 equiv.), acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water 1 water 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100 °C, 25–97% isolated yields.211 (G) Heck diversification of bromotryptophans, peptides containing bromotryptophan 7-bromotryptophan containing pacidamycin: alkene (1.5 equiv.), Na2PdCl4 (10 mol%), TXPTS (23 mol%), Na2CO3 (4 equiv.), acetonitrile 1, 100 °C, 25–97% isolated yields.211 (G) Heck diversification of bromotryptophans, peptides containing bromotryptophan 7-bromotryptophan containing pacidamycin: alkene (1.5 equiv.), Na2PdCl4 (10 mol%), TXPTS (23 mol%), Na2CO3 (4 equiv.), acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water 1 water 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 90 °C, 51–95% isolated yields.210 3, 90 °C, 51–95% isolated yields.210 | ||
The development of mild aqueous chemistries has facilitated chemo-enzymatic and GenoChemetic approaches. Site selective C–H activation is enabled by a halogenase and synthetic diversification carried out, in some cases synchronously, through aqueous compatible Pd mediated catalysis. More reactive aryl iodides and bromides can be coupled under much milder conditions than their less activated chloro counterparts.
 | ||
Scheme 14 Hyphenation of halogenation and cross-coupling to achieve C–H activation, (A) general concept in which the halogenase operates first, and then the cross-coupling is carried out on the halogenated material as a component of the crude extract. This is carried out stepwise in (B–F), enabling access to a broad series of compounds, and synchronously in G where the halogenation and cross-coupling events are spatially separated through application of a membrane. (B) Bromination of L-tryptophan by Thal, followed by Suzuki–Miyaura cross-coupling of the brominated product as a component of the crude lysate.215 The product of cross-coupling (λex 300 nm/λem 430 nm) is used in screening the directed evolution of RebH, resulting in a 2.5-fold increase in enzymatic activity. Cross-coupling conditions: 3-aminophenylboronic acid (10 equiv.), Na2PdCl4 (50 mol%), SSphos (150 mol%), K3PO4 (15 equiv.), 95 °C.215 (C) One-pot bromination of L-tryptophan by RebH-CLEA, followed by Mizoroki–Heck cross-coupling, in a stepwise fashion to afford C-7 substituted 7-(4-carboxystyryl)tryptophan with a λem of 485 nm (λex 360 nm). Cross-coupling conditions: 4-carboxystyrene (5 equiv.), PdOAc2 (0.1–0.2 equiv.), TPPTS (0.3–0.6 equiv.), K2CO3 (5 equiv.), water, degassed under argon, 100 °C.189 (D–F) Sequential enzymatic bromination by evolved RebH variants, followed by (D) Suzuki–Miyaura, (E) Buchwald–Hartwig, (F) alkoxylation of tryptophan as a component of a crude extract. Conditions D: ArB(OH)2 (1.5 equiv.), PdOAc2 (0.05 equiv.), SSPhos (0.05 equiv.), iPrOH: phosphate buffer (170 mM, pH 8.5) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 90 °C. (E) ArNH2 (3 equiv.), PdOAc2 (0.03 equiv.), BrettPhos (0.03 equiv.), NaOt-Bu (6 equiv.), dioxane, 100 °C. (F) CF3CH2OH (2 equiv.), [(allyl)PdCl]2(0.005 equiv.), RockPhos (0.015 equiv.), Cs2CO3 (2 equiv.), toluene, 90 °C.189 (G) One-pot, synchronous halogenation and Suzuki–Miyaura cross-coupling by PDMS membrane separated RebH-CLEA and Pd catalyst. Cross-coupling conditions: PdOAc2 (10 mol%), 2-(dimethylamino)-pyrimidine-4,6-diol (20 mol%), aryl boronic acid (5 equiv.) and CsF (10 equiv.), r.t. overnight then 80 °C.211 1, 90 °C. (E) ArNH2 (3 equiv.), PdOAc2 (0.03 equiv.), BrettPhos (0.03 equiv.), NaOt-Bu (6 equiv.), dioxane, 100 °C. (F) CF3CH2OH (2 equiv.), [(allyl)PdCl]2(0.005 equiv.), RockPhos (0.015 equiv.), Cs2CO3 (2 equiv.), toluene, 90 °C.189 (G) One-pot, synchronous halogenation and Suzuki–Miyaura cross-coupling by PDMS membrane separated RebH-CLEA and Pd catalyst. Cross-coupling conditions: PdOAc2 (10 mol%), 2-(dimethylamino)-pyrimidine-4,6-diol (20 mol%), aryl boronic acid (5 equiv.) and CsF (10 equiv.), r.t. overnight then 80 °C.211 | ||
This larger Stokes’ shift renders this technology potentially very useful for molecule tagging and enzyme assay, as well as valuable for molecule diversification. Even when stabilised as CLEA, enzymes and Pd catalysts can have poor compatibility. To achieve a synchronous one-pot enzymatic halogenation and chemical cross-coupling, and to avoid the additional filtration step, Micklefield and co-workers elegantly partitioned CLEA stabilised PyrH, RebH, RadH and SttH from the Pd catalyst through use of a polydimethylsiloxane (PDMS) membrane. Whilst the brominated enzymatic product could pass through the PMDS, it was reported to be impervious to the enzymes and cofactors. The cross-coupled product, once formed, remained in the chemo-catalytic chamber (Scheme 14G).218
Though the majority of chemo-enzymatic approaches published so far have focussed on tryptophans and other indolic systems, this is predominantly a reflection of the most extensively studied FDHs. Through substrate walking methodology, Jared Lewis and co-workers have impressively evolved RebH to work on substrates that are chemically distinct from its natural tryptophan substrate. Utilisation of evolved halogenases enabled bromination of a diverse series of substrates including tryptolines, sterically bulky substituted anilines as well as carvediol and pindolol, two clinically utilised beta-blockers. Conditions to enable Suzuki–Miyaura, Buchwald–Hartwig amination and alkoxylation were developed and applied mostly to the chlorinated and brominated intermediates as a component of a crude extract, resulting in high yields of the respective products 284–289 (Scheme 13D–F).189 With the discovery of more substrate diverse halogenases, engineering of known halogenases, enzyme stabilisation and partitioning strategies and the development of milder reaction conditions, the portfolio of functionalisable systems is set to expand significantly.
FDHs are not the only halogenating enzymes to be merged with palladium catalysed cross-coupling. V-HPOs and HPOs have been successfully used in various chemoenzymatic halocyclization reactions,219,220 and recently this has been merged with homogenous metal catalysis for further diversification. Deska and coworkers enabled the one-pot combination of allenol halocylization using CPO from Caldariomyces fumago and glucose oxidase from Aspergillus niger (GOx) with SPhos catalysed Suzuki–Miyaura and Sonogashira coupling to diversify a range of dihydrofurans. Excitingly, a nanobiohybrid was also generated using GOx as a support for palladium nanoparticles, which led to good yields of stepwise halocyclization and Suzuki coupling.221
The halogenated metabolites have been subjected to cross-coupling chemistries. In the first example, the tryptophan halogenase PrnA was installed, through use of an integrative vector, into the genome of Streptomyces coeruleorubidis. It was demonstrated that there was no need to introduce PrnA's concomitant flavin reductase, as the organism's own reductases sufficed. A chlorinated analogue of pacidamycin, a uridyl peptide antibiotic, was produces at a titre of ∼1 mg L−1. In spite of bearing series of sensitive functional groups, this metabolite could be readily cross-coupled using the moderately mild Suzuki-Miyaura conditions that were developed. The cross-coupling was carried out both on the purified material and on material as a component of a crude extract (Scheme 15). The latter was preferable, as the more lipophilic product that resulted was much more readily amenable to purification.217 The generation of brominated analogues would, of course, facilitate cross-coupling under even milder conditions. However, Streptomyces coeruleorubidis was found to be sensitive to bromide salts, and the brominated analogue cannot readily be accessed through use of this strain. Streptomyces coelicolor showed good tolerance to bromide salts in a screen of series of potential heterologous expression hosts, and so the pacidamycin biosynthetic gene cluster, identified previously, was ported into this strain conferring it with the ability to produce pacidamycins. The gene encoding the halogenase PrnA was engineered into the strain and ability to produce bromo-pacidamycin was confirmed. In order to enable bromometabolite production and cross-coupling synchronously, mild cross-coupling reactions that enabled derivatisation of 7-Br-pacidamycin D 305 in the presence of the living culture were carried out. In a similar manner, E. coli was engineered with the ability to make bromo-tryptophan and its cross-coupling to 306 affected in the living fermentative culture.222 A similar system was designed in hairy root cultures of Catharanthus roseus in which they were engineered to include RebH, PyrH and their concomitant reductases. This led to the production of a range of chlorinated and brominated analogues of the plant's monoterpene indole alkaloid natural products 299–301, which were successfully extracted and coupled in non-aqueous Suzuki–Miyaura couplings, either as crude extracts or purified compounds (Scheme 15A).223,224
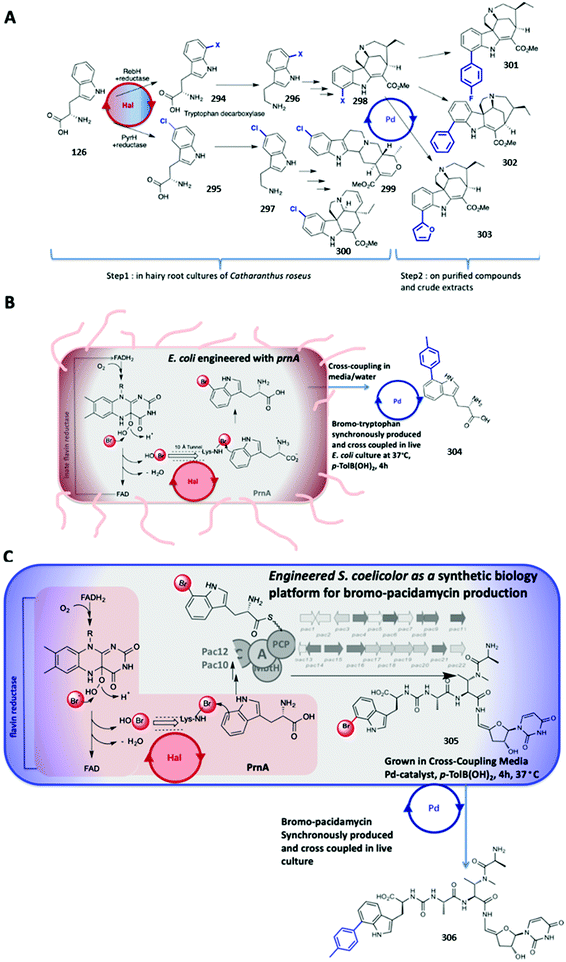 | ||
| Scheme 15 GenoChemetic approaches to C–H functionalisation of small metabolites and complex natural products. Here, a halogenase is installed to complement an existing metabolic/biosynthetic pathway, the halogenated natural product is diversified through application of cross-coupling chemistries. (A) Two step approach: hairy root cultures of Catharanthus roseus are engineered to include RebH, PyrH and their concomitant reductases. A series of chlorinated and brominated analogues are produced, and cross coupling has been carried out on purified compounds and on the metabolites as components of crude extracts, enabling access to a wide portfolio of functionalised molecules.224 Cross-coupling conditions: arylboronic acid (3 equiv.), Pd(OAc)2 (5 mol%), Sphos (13 mol%), K3PO4, n-butanol, 100 °C, 10–60 min. (B and C) One pot approach, pathways constructed in heterologous hosts, with halogenation (mediated by PrnA)225 and cross-coupling occurring synchronously in the presence of the living E. coli and Streptomyces coelicolor,222 respectively. | ||
Conclusions
Though enzyme mediated halogenation was initially considered to be nothing more than an artefact or a rare event, today over 30 enzymes capable of mediating such chemistry have been structurally and biochemically characterised (Table 2), and an even greater number subject to in vitro and in vivo investigation. Halogenated compounds play a major role in the construction of molecules via cross-coupling methodologies, as well as making up a large percentage of medicinally and agrochemically relevant molecules. In a world that seeks more selective, greener and more sustainable solutions to molecule manufacture, halogenases hold an opportunity. They also offer potential for the vectorisation of fragments for SAR. Whilst the kinetic analyses of the halogenases is rather potted, drawing this data together is useful in identifying potential halogenases for development and use in biotransformations. Whilst the HPOs are capable of mediating halogenation of large and sterically demanding substrates, as most neither bind the substrate nor the electrophilic species, such systems generally lack regiocontrol. A few intriguing departures from this trend have been identified from marine systems, indicating that there is much more to learn about these enzymes, their operation and evolution. New variant A FDHs with reasonable catalytic efficiency and broad substrate scopes have been revealed in the last two years, enabling regioselective chlorination, bromination and even iodination on sterically and electronically demanding aromatic systems, even prior to rational redesign or evolution. Excitingly, in recent years the range of variant A NHFeHals that enable the controlled halogenation, including stereoselective halogenation, have been identified; clearly, there are many more such systems awaiting discovery. Inspired by such systems, it may be possible to generate artificial architectures that, perhaps via haloamines, position and deliver electrophilic halogenating agents to substrates held in the required position to affect desired regiochemistries.Halogenation introduces a chemically reactive, chemically orthogonal moiety into a compound. This single atom modification, that can be genetically/enzymatically installed has the potential to act as a superior tag to azide or alkyne modifications. In parallel with halogenase discovery, a variety of aqueous cross-coupling reactions compatible with sensitive functionalities in natural products and biomolecules have been developed. Some of these reactions are even biorthogonal. The opportunities for building pathways and circuits of SynBio-SynChem present themselves.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank ERC GenoChemetics (FP7/2007-2013/ERC consolidator grant GCGXC grant agreement no. 614779 RJMG) for funding, BBSRC Follow on Funding, Synthetic Biology Highlight award. RSE Enterprise Fellowship (SVS), China Scholarship Council and EPSRC CRITICAT EP/L016419/1 for studentship support (Y. Z. & S. M. respectively).References
- P. Jeschke, Pest Manage. Sci., 2017, 73, 1053–1066 CrossRef CAS.
- P. Jeschke, R. Nauen, M. Schindler and A. Elbert, J. Agric. Food Chem., 2011, 59, 2897–2908 CrossRef CAS PubMed.
- Z. Xu, Z. Yang, Y. Liu, Y. Lu, K. Chen and W. Zhu, J. Chem. Inf. Model., 2014, 54, 69–78 CrossRef CAS PubMed.
- M. C. Ford and P. S. Ho, J. Med. Chem., 2016, 59, 1655–1670 CrossRef CAS PubMed.
- C. W. Lindsley, ACS Chem. Neurosci., 2017, 8, 1635–1636 CrossRef CAS PubMed.
- B. E. Blass, Basic Principles of Drug Discovery and Development, 2015, pp. 499–529 Search PubMed.
- R. Whitfield and F. Brown, The Benefits of Chlorine Chemistry in Pharmaceuticals in the United States and Canada, 2016 Search PubMed.
- E. Parisini, P. Metrangolo, T. Pilati, G. Resnati and G. Terraneo, Chem. Soc. Rev., 2011, 40, 2267–2278 RSC.
- J. Ruiz-Cabello, B. P. Barnett, P. A. Bottomley and J. W. M. Bulte, NMR Biomed., 2011, 24, 114–129 CrossRef CAS.
- J. M. Widmark, Bayl. Univ. Med. Cent. Proc., 2007, 20, 408–417 CrossRef PubMed.
- J. G. Speight, Environmental Organic Chemistry for Engineers, 2017, pp. 87–151 Search PubMed.
- D. A. Petrone, J. Ye and M. Lautens, Chem. Rev., 2016, 116, 8003–8104 CrossRef CAS PubMed.
- G. W. Gribble, Environ. Chem., 2015, 12, 396–405 CrossRef CAS.
- P. D. Shaw and L. P. Hager, J. Am. Chem. Soc., 1959, 81, 6527–6528 CrossRef CAS.
- J. R. Beckwith, R. Clark and L. P. Hager, J. Biol. Chem., 1963, 9, 3086–3090 CrossRef.
- J. Delaney, E. Clarke, D. Hughes and M. Rice, Drug Discovery Today, 2006, 11, 839–845 CrossRef PubMed.
- P. Maienfisch and R. G. Hall, Chimia, 2004, 58, 93–99 CrossRef CAS.
- R. J. Milne and K. L. Goa, Drugs, 1991, 41, 450–477 CrossRef CAS PubMed.
- M. Baumann and I. R. Baxendale, Beilstein J. Org. Chem., 2013, 9, 2265–2319 CrossRef PubMed.
- P. Oakley, J. Pharm. Res. Int., 2018, 21, 1–9 CrossRef.
- A. Timmins and S. P. de Visser, Catalysts, 2018, 8, 314 CrossRef.
- J. Reedijk and E. Bouwman, Bioinorganic Catalysis, CRC Press, Boca Raton, 2nd edn, 1999 Search PubMed.
- H. B. Dunford, A. M. Lambeir, M. A. Kashem and M. Pickard, Arch. Biochem. Biophys., 1987, 252, 292–302 CrossRef CAS.
- L. C. Blasiak and C. L. Drennan, Acc. Chem. Res., 2009, 42, 147–155 CrossRef CAS PubMed.
- M. Fritzsche, V. Vergopoulos and D. Rehder, Inorg. Chim. Acta, 1993, 211, 11–16 CrossRef CAS.
- E. de Boer and R. Wever, J. Biol. Chem., 1988, 263, 12326–12332 CrossRef CAS.
- G. J. Colpas, B. J. Hamstra, J. W. Kampf and V. L. Pecoraro, J. Am. Chem. Soc., 1996, 118, 3469–3478 CrossRef CAS.
- C. C. McLauchlan, H. A. Murakami, C. A. Wallace and D. C. Crans, J. Inorg. Biochem., 2018, 186, 267–279 CrossRef CAS PubMed.
- R. Morris and L. P. Hager, J. Biol. Chem., 1966, 8, 1763–1768 CrossRef.
- P. Bernhardt, T. Okino, J. M. Winter, A. Miyanaga and B. S. Moore, J. Am. Chem. Soc., 2011, 133, 4268–4270 CrossRef CAS PubMed.
- S. Diethelm, R. Teufel, L. Kaysser and B. S. Moore, Angew. Chem., Int. Ed., 2014, 53, 11023–11026 CrossRef CAS PubMed.
- B. S. Moore, Synlett, 2018, 401–409 CAS.
- T. Nakano, K. Aisaka, R. Katsumata, M. Hasegawa and T. Dairi, Biosci., Biotechnol., Biochem., 1995, 59, 1099–1106 CrossRef PubMed.
- S. Kirner, P. E. Hammer, D. S. Hill, A. Altmann, I. Fischer, L. J. Weislo, M. Lanahan and J. M. Ligon, J. Bacteriol., 1998, 180, 1939–1943 CrossRef CAS PubMed.
- S. Keller, T. Wage, K. Hohaus, M. Hölzer, E. Eichhorn and P. Kh, Angew. Chem., Int. Ed., 2000, 39, 2300–2302 CrossRef CAS PubMed.
- S. Buedenbender, S. Rachid, R. Müller and G. E. Schulz, J. Mol. Biol., 2009, 385, 520–530 CrossRef CAS PubMed.
- C. Dong, S. Flecks, S. Unversucht, C. Haupt, K. H. Van Pée and J. H. Naismith, Science, 2005, 309, 2216–2219 CrossRef CAS PubMed.
- E. Yeh, L. C. Blasiak, A. Koglin, C. L. Drennan and C. T. Walsh, Biochemistry, 2007, 46, 1284–1292 CrossRef CAS PubMed.
- S. Flecks, E. P. Patallo, X. Zhu, A. J. Ernyei, G. Seifert, A. Schneider, C. Dong, J. H. Naismith and K. H. Van Pée, Angew. Chem., Int. Ed., 2008, 47, 9533–9536 CrossRef CAS PubMed.
- E. Yeh, L. J. Cole, E. W. Barr, J. M. Bollinger, D. P. Ballou and C. T. Walsh, Biochemistry, 2006, 45, 7904–7912 CrossRef CAS PubMed.
- M. C. Andorfer and J. C. Lewis, Annu. Rev. Biochem., 2018, 87, 159–185 CrossRef CAS PubMed.
- M. C. Andorfer, H. J. Park, J. Vergara-Coll and J. C. Lewis, Chem. Sci., 2016, 7, 3720–3729 RSC.
- E. Yeh, L. C. Blasiak, A. Koglin, C. L. Drennan and C. T. Walsh, Biochemistry, 2007, 46, 1284–1292 CrossRef CAS PubMed.
- K. Podzelinska, R. Latimer, A. Bhattacharya, L. C. Vining, D. L. Zechel and Z. Jia, J. Mol. Biol., 2010, 397, 316–331 CrossRef CAS.
- V. Agarwal, A. A. El Gamal, K. Yamanaka, D. Poth, R. D. Kersten, M. Schorn, E. E. Allen and B. S. Moore, Nat. Chem. Biol., 2014, 10, 640–647 CrossRef CAS PubMed.
- A. E. Fraley, M. Garcia-Borràs, A. Tripathi, D. Khare, E. V. Mercado-Marin, H. Tran, Q. Dan, G. P. Webb, K. R. Watts, P. Crews, R. Sarpong, R. M. Williams, J. L. Smith, K. N. Houk and D. H. Sherman, J. Am. Chem. Soc., 2017, 139, 12060–12068 CrossRef CAS PubMed.
- D. S. Gkotsi, H. Ludewig, S. V. Sharma, J. A. Connolly, J. Dhaliwal, Y. Wang, W. P. Unsworth, R. J. K. Taylor, M. M. W. McLachlan, S. Shanahan, J. H. Naismith and R. J. M. Goss, Nat. Chem., 2019, 11, 1091–1097 CrossRef CAS PubMed.
- A. El Gamal, V. Agarwal, S. Diethelm, I. Rahman, M. A. Schorn, J. M. Sneed, G. V. Louie, K. E. Whalen, T. J. Mincer, J. P. Noel, V. J. Paul and B. S. Moore, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3797–3802 CrossRef CAS PubMed.
- O. Dym and D. Eisenberg, Protein Sci., 2001, 10, 1712–1728 CrossRef CAS PubMed.
- J. Ahlert, E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet and J. S. Thorson, Science, 2002, 297, 1173–1176 CrossRef CAS PubMed.
- S. Mori, A. H. Pang, N. T. Chandrika, S. Garneau-tsodikova and O. V. Tsodikov, Nat. Commun., 2019, 1–11 Search PubMed.
- S. S. Gao, N. Naowarojna, R. Cheng, X. Liu and P. Liu, Nat. Prod. Rep., 2018, 35, 792–837 RSC.
- W. S. Aik, M. A. McDonough, A. Thalhammer, R. Chowdhury and C. J. Schofield, Curr. Opin. Struct. Biol., 2012, 22, 691–700 CrossRef CAS.
- M. L. Matthews, C. M. Krest, E. W. Barr, F. H. Vaillancourt, C. T. Walsh, M. T. Green, C. Krebs and J. M. Bollinger, Biochemistry, 2009, 48, 4331–4343 CrossRef CAS PubMed.
- D. P. Galonić, E. W. Barr, C. T. Walsh, J. M. Bollinger and C. Krebs, Nat. Chem. Biol., 2007, 3, 113–116 CrossRef PubMed.
- M. L. Matthews, C. S. Neumann, L. A. Miles, T. L. Grove, S. J. Booker, C. Krebs, C. T. Walsh and J. M. Bollinger, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17723–17728 CrossRef CAS.
- S. M. Pratter, K. M. Light, E. I. Solomon and G. D. Straganz, J. Am. Chem. Soc., 2014, 136, 9385–9395 CrossRef CAS PubMed.
- F. H. Vaillancourt, J. Yin and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10111–10116 CrossRef CAS.
- X. Liu, In Vitro Analysis of Cyanobacterial Nonheme Iron-Dependent Aliphatic Halogenases WelO5 and AmbO5, Elsevier Inc., 1st edn, 2018, vol. 604 Search PubMed.
- D. P. Galonić, F. H. Vaillancourt and C. T. Walsh, J. Am. Chem. Soc., 2006, 128, 3900–3901 CrossRef.
- L. Gu, B. Wang, A. Kulkarni, T. W. Geders, R. V. Grindberg, L. Gerwick, K. Hkansson, P. Wipf, J. L. Smith, W. H. Gerwick and D. H. Sherman, Nature, 2009, 459, 731–735 CrossRef CAS PubMed.
- M. L. Hillwig and X. Liu, Nat. Chem. Biol., 2014, 10, 921–923 CrossRef CAS.
- D. O’Hagan, C. Schaffrath, S. L. Cobb, J. T. G. Hamilton and C. D. Murphy, Nature, 2002, 416, 279 CrossRef PubMed.
- A. S. Eustáquio, F. Pojer, J. P. Noel and B. S. Moore, Nat. Chem. Biol., 2008, 4, 69–74 CrossRef PubMed.
- D. O’Hagan and H. Deng, Chem. Rev., 2015, 115, 634–649 CrossRef.
- H. M. Senn, D. O’Hagan and W. Thiel, J. Am. Chem. Soc., 2005, 127, 13643–13655 CrossRef CAS PubMed.
- C. Dong, F. Huang, H. Deng, C. Schaffrath, J. B. Spencer, D. O’Hagan and J. H. Naismith, Nature, 2004, 427, 561–565 CrossRef CAS PubMed.
- H. Deng, S. L. Cobb, A. R. McEwan, R. P. McGlinchey, J. H. Naismith, D. O’Hagan, D. A. Robinson and J. B. Spencer, Angew. Chem., Int. Ed., 2006, 45, 759–762 CrossRef CAS PubMed.
- S. Thompson, M. Onega, S. Ashworth, I. N. Fleming, J. Passchier and D. O’Hagan, Chem. Commun., 2015, 51, 13542–13545 RSC.
- M. C. Walker, B. W. Thuronyi, L. K. Charkoudian, B. Lowry, C. Khosla and M. C. Y. Chang, Science, 2013, 341, 1089–1094 CrossRef CAS PubMed.
- D. Milbredt, E. P. Patallo and K. H. Van Pée, ChemBioChem, 2014, 15, 1011–1020 CrossRef CAS PubMed.
- J. Zeng and J. Zhan, Biotechnol. Lett., 2011, 33, 1607–1613 CrossRef CAS PubMed.
- J. Lee, J. Kim, H. Kim, E. J. Kim, H. J. Jeong, K. Y. Choi and B. G. Kim, ChemBioChem, 2020, 21, 1446–1452 CrossRef CAS PubMed.
- K. Lingkon and J. J. Bellizzi, ChemBioChem, 2020, 21, 1121–1128 CrossRef CAS PubMed.
- Z. Guo, P. Li, G. Chen, C. Li, Z. Cao, Y. Zhang, J. Ren, H. Xiang, S. Lin, J. Ju and Y. Chen, J. Am. Chem. Soc., 2018, 140, 18009–18015 CrossRef CAS.
- S. Zehner, A. Kotzsch, B. Bister, R. D. Süssmuth, C. Méndez, J. A. Salas and K. H. Van Pée, Chem. Biol., 2005, 12, 445–452 CrossRef CAS PubMed.
- J. Domergue, D. Erdmann, A. Fossey-Jouenne, J.-L. Petit, A. Debard, V. de Berardinis, C. Vergne-Vaxelaire and A. Zaparucha, AMB Express, 2019, 9, 175 CrossRef PubMed.
- A. Lang, S. Polnick, T. Nicke, P. William, E. P. Patallo, J. H. Naismith and K. H. Van Pée, Angew. Chem., Int. Ed., 2011, 50, 2951–2953 CrossRef CAS PubMed.
- S. A. Shepherd, C. Karthikeyan, J. Latham, A. W. Struck, M. L. Thompson, B. R. K. Menon, M. Q. Styles, C. Levy, D. Leys and J. Micklefield, Chem. Sci., 2015, 6, 3454–3460 RSC.
- S. A. Shepherd, B. R. K. Menon, H. Fisk, A. W. Struck, C. Levy, D. Leys and J. Micklefield, ChemBioChem, 2016, 17, 821–824 CrossRef CAS PubMed.
- A. C. Moritzer, H. Minges, T. Prior, M. Frese, N. Sewald and H. H. Niemann, J. Biol. Chem., 2019, 294, 2529–2542 CrossRef CAS PubMed.
- J. R. Heemstra and C. T. Walsh, J. Am. Chem. Soc., 2008, 130, 14024–14025 CrossRef CAS PubMed.
- M. L. Hillwig, Q. Zhu, K. Ittiamornkul and X. Liu, Angew. Chem., Int. Ed., 2016, 55, 5780–5784 CrossRef CAS PubMed.
- M. E. Neugebauer, K. H. Sumida, J. G. Pelton, J. L. McMurry, J. A. Marchand and M. C. Y. Chang, Nat. Chem. Biol., 2019, 15, 1009–1016 CrossRef CAS PubMed.
- C. Zhao, S. Yan, Q. Li, H. Zhu, Z. Zhong, Y. Ye, Z. Deng and Y. Zhang, Angew. Chem., Int. Ed., 2020, 59, 9478–9484 CrossRef CAS PubMed.
- R. D. Libby, N. S. Rotberg, T. Emerson, T. C. White, G. M. Yen, S. H. Friedman, N. S. Sun and R. Goldowski, J. Biol. Chem., 1989, 264, 15284–15292 CrossRef CAS.
- X. Zhu, D. A. Robinson, A. R. McEwan, D. O’Hagan and J. H. Naismith, J. Am. Chem. Soc., 2007, 129, 14597–14604 CrossRef CAS PubMed.
- E. Bitto, Y. Huang, C. A. Bingman, S. Singh, J. S. Thorson and G. N. Phillips, Proteins: Struct., Funct., Bioinf., 2007, 70, 289–293 CrossRef PubMed.
- C. B. Poor, M. C. Andorfer and J. C. Lewis, ChemBioChem, 2014, 15, 1286–1289 CrossRef CAS PubMed.
- X. Zhu, W. De Laurentis, K. Leang, J. Herrmann, K. Ihlefeld, K. H. van Pée and J. H. Naismith, J. Mol. Biol., 2009, 391, 74–85 CrossRef CAS PubMed.
- A. H. Pang, S. Garneau-Tsodikova and O. V. Tsodikov, J. Struct. Biol., 2015, 192, 349–357 CrossRef CAS PubMed.
- B. R. K. Menon, J. Latham, M. S. Dunstan, E. Brandenburger, U. Klemstein, D. Leys, C. Karthikeyan, M. F. Greaney, S. A. Shepherd and J. Micklefield, Org. Biomol. Chem., 2016, 14, 9354–9361 RSC.
- P. R. Neubauer, C. Widmann, D. Wibberg, L. Schröder, M. Frese, T. Kottke, J. Kalinowski, H. H. Niemann and N. Sewald, PLoS One, 2018, 13, 1–21 Search PubMed.
- H. Luhavaya, R. Sigrist, J. R. Chekan, S. M. K. McKinnie and B. S. Moore, Angew. Chem., Int. Ed., 2019, 58, 8394–8399 CrossRef CAS PubMed.
- A. C. Moritzer and H. H. Niemann, Protein Sci., 2019, 28, 2112–2118 CrossRef CAS PubMed.
- A. Phintha, K. Prakinee, A. Jaruwat, N. Lawan, S. Visitsatthawong, C. Kantiwiriyawanitch, W. Songsungthong, D. Trisrivirat, P. Chenprakhon, A. Mulholland, K.-H. van Pée, P. Chitnumsub and P. Chaiyen, J. Biol. Chem., 2021, 296, 100068 CrossRef CAS PubMed.
- A. Messerschmidt and R. Wever, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 392–396 CrossRef CAS PubMed.
- S. Macedo-Ribeiro, W. Hemrika, R. Renirie, R. Wever and A. Messerschmidt, J. Biol. Inorg. Chem., 1999, 4, 209–219 CrossRef CAS PubMed.
- A. Messerschmidt, L. Prade and R. Wever, Biol. Chem., 1997, 378, 309–316 CAS.
- S. De Macedo-Ribeiro, R. Renirie, R. Wever and A. Messerschmidt, Biochemistry, 2008, 47, 929–934 CrossRef CAS PubMed.
- M. Weyand, H. J. Hecht, M. Kieß, M. F. Liaud, H. Vilter and D. Schomburg, J. Mol. Biol., 1999, 293, 595–611 CrossRef CAS PubMed.
- J. N. Carter-Franklin, J. D. Parrish, R. A. Tschirret-Guth, R. D. Little and A. Butler, J. Am. Chem. Soc., 2003, 125, 3688–3689 CrossRef CAS PubMed.
- D. Wischang, M. Radlow and J. Hartung, Dalton Trans., 2013, 42, 11926–11940 RSC.
- M. N. Isupov, A. R. Dalby, A. A. Brindley, Y. Izumi, T. Tanabe, G. N. Murshudov and J. A. Littlechild, J. Mol. Biol., 2000, 299, 1035–1049 CrossRef CAS PubMed.
- E. Garcia-Rodriguez, T. Ohshiro, T. Aibara, Y. Izumi and J. Littlechild, J. Biol. Inorg. Chem., 2005, 10, 275–282 CrossRef CAS PubMed.
- J. B. Fournier, E. Rebuffet, L. Delage, R. Grijol, L. Meslet-Cladière, J. Rzonca, P. Potin, G. Michel, M. Czjzek and C. Leblanc, Appl. Environ. Microbiol., 2014, 80, 7561–7573 CrossRef PubMed.
- A. Frank, C. J. Seel, M. Groll and T. Gulder, ChemBioChem, 2016, 17, 2028–2032 CrossRef CAS PubMed.
- M. Sundaramoorthy, J. Terner and T. L. Poulos, Structure, 1995, 3, 1367–1377 CrossRef CAS PubMed.
- K. Ku, W. Blankenfeldt, J. Terner and I. Schlichting, J. Biol. Chem., 2006, 281, 23990–23998 CrossRef PubMed.
- C. Wong, D. G. Fujimori, C. T. Walsh and C. L. Drennan, J. Am. Chem. Soc., 2009, 131, 4872–4879 CrossRef CAS PubMed.
- L. C. Blasiak, F. H. Vaillancourt, C. T. Walsh and C. L. Drennan, Nature, 2006, 440, 368–371 CrossRef CAS PubMed.
- S. L. Cobb, H. Deng, A. R. McEwan, J. H. Naismith, D. O’Hagan and D. A. Robinson, Org. Biomol. Chem., 2006, 4, 1458–1460 RSC.
- S. Thompson, S. A. McMahon, J. H. Naismith and D. O’Hagan, Bioorg. Chem., 2016, 64, 37–41 CrossRef CAS PubMed.
- H. Deng, L. Ma, N. Bandaranayaka, Z. Qin, G. Mann, K. Kyeremeh, Y. Yu, T. Shepherd, J. H. Naismith and D. O’Hagan, ChemBioChem, 2014, 15, 364–368 CrossRef CAS PubMed.
- S. Marumo and H. Hattori, Planta, 1970, 90, 208–211 CrossRef CAS PubMed.
- M. Ishibashi, Y. Li, M. Sato and J. Kobayashi, Nat. Prod. Lett., 1994, 4, 293–296 CrossRef CAS.
- S. Rachid, D. Krug, B. Kunze, I. Kochems, M. Scharfe, T. M. Zabriskie, H. Blöcker and R. Müller, Chem. Biol., 2006, 13, 667–681 CrossRef CAS PubMed.
- W. Jiang, J. R. Heemstra, R. R. Forseth, C. S. Neumann, S. Manaviazar, F. C. Schroeder, K. J. Hale and C. T. Walsh, Biochemistry, 2011, 50, 6063–6072 CrossRef CAS.
- D. R. M. Smith, A. R. Uria, E. J. N. Helfrich, D. Milbredt, K. H. Van Pée, J. Piel and R. J. M. Goss, ACS Chem. Biol., 2017, 12, 1281–1287 CrossRef CAS PubMed.
- K. Hohaus, A. Altmann, W. Burd, I. Fischer, P. E. Hammer, D. S. Hill, J. M. Ligon and K. Van Pce, Angew. Chem., Int. Ed., 2012, 36, 2012–2013 CrossRef.
- D. G. Fujimori, S. Hrvatin, C. S. Neumann, M. Strieker, M. A. Marahiel and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16498–16503 CrossRef PubMed.
- D. E. Williams, J. Davies, B. O. Patrick, H. Bottriell, T. Tarling, M. Roberge and R. J. Andersen, Org. Lett., 2008, 10, 3501–3504 CrossRef CAS PubMed.
- F. Y. Chang and S. F. Brady, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2478–2483 CrossRef CAS.
- C. Sánchez, L. Zhu, A. F. Braña, A. P. Salas, J. Rohr, C. Méndez and J. A. Salas, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 461–466 CrossRef PubMed.
- Z. Liu, L. Ma, L. Zhang, W. Zhang, Y. Zhu, Y. Chen, W. Zhangb and C. Zhang, Org. Biomol. Chem., 2019, 17, 1053–1057 RSC.
- J. T. Payne, M. C. Andorfer and J. C. Lewis, Angew. Chem., Int. Ed., 2013, 52, 5271–5274 CrossRef CAS PubMed.
- J. T. Payne, C. B. Poor and J. C. Lewis, Angew. Chem., Int. Ed., 2015, 54, 4226–4230 CrossRef CAS PubMed.
- F. Y. Chang and S. F. Brady, ChemBioChem, 2014, 15, 815–821 CrossRef CAS PubMed.
- C. Carrasco, M. Facompré, J. D. Chisholm, D. L. Van Vranken, W. D. Wilson and C. Bailly, Nucleic Acids Res., 2002, 30, 1774–1781 CrossRef CAS.
- M. Frese and N. Sewald, Angew. Chem., Int. Ed., 2015, 54, 298–301 CrossRef CAS.
- X. M. Chu, C. Wang, W. Liu, L. L. Liang, K. K. Gong, C. Y. Zhao and K. L. Sun, Eur. J. Med. Chem., 2019, 161, 101–117 CrossRef CAS PubMed.
- F. Prati, C. Bergamini, R. Fato, O. Soukup, J. Korabecny, V. Andrisano, M. Bartolini and M. L. Bolognesi, ChemMedChem, 2016, 1284–1295 CrossRef CAS PubMed.
- G. A. Castillon, N. R. Adames, C. H. Rosello, H. S. Seidel, M. S. Longtine, J. A. Cooper and R. A. Heil-Chapdelaine, Curr. Biol., 2003, 13, 654–658 CrossRef CAS PubMed.
- O. Bilyk, E. Brötz, B. Tokovenko, A. Bechthold, T. Paululat and A. Luzhetskyy, ACS Chem. Biol., 2016, 11, 241–250 CrossRef CAS PubMed.
- N. A. Magarvey, Z. Q. Beck, T. Golakoti, Y. Ding, U. Huber, T. K. Hemscheidt, D. Abelson, R. E. Moore and D. H. Sherman, ACS Chem. Biol., 2006, 1, 766–779 CrossRef CAS PubMed.
- T. Kittila, C. Kittel, J. Tailhades, D. Butz, M. Schoppet, A. Buttner, R. J. A. Goode, R. B. Schittenhelm, K.-H. van Pee, R. D. Sussmuth, W. Wohlleben, M. J. Cryle and E. Stegmann, Chem. Sci., 2017, 8, 5992–6004 RSC.
- S. Lin, S. G. Van Lanen and B. Shen, J. Am. Chem. Soc., 2007, 129, 12432–12438 CrossRef CAS PubMed.
- S. Rachid, M. Scharfe, H. Blöcker, K. J. Weissman and R. Müller, Chem. Biol., 2009, 16, 70–81 CrossRef CAS PubMed.
- X. Y. Jia, Z. H. Tian, L. Shao, X. D. Qu, Q. F. Zhao, J. Tang, G. L. Tang and W. Liu, Chem. Biol., 2006, 13, 575–585 CrossRef CAS PubMed.
- S. Cadel-Six, C. Dauga, A. M. Castets, R. Rippka, C. Bouchier, N. Tandeau de Marsac and M. Welker, Mol. Biol. Evol., 2008, 25, 2031–2041 CrossRef CAS PubMed.
- S. Stinchi, L. Carrano, A. Lazzarini, M. Feroggio, A. Grigoletto, M. Sosio and S. Donadio, FEMS Microbiol. Lett., 2006, 256, 229–235 CrossRef CAS PubMed.
- Y. Ge, G. Wang, J. Jin, T. Liu, X. Ma, Z. Zhang, Y. Ge, G. Wang, J. Jin, T. Liu, X. Ma, Z. Zhang and T. Geng, J. Org. Chem., 2020, 85, 8673–8682 CrossRef CAS PubMed.
- J. Zeng, A. K. Lytle, D. Gage, S. J. Johnson and J. Zhan, Bioorg. Med. Chem. Lett., 2013, 23, 1001–1003 CrossRef CAS PubMed.
- B. R. K. Menon, E. Brandenburger, H. H. Sharif, U. Klemstein, S. A. Shepherd, M. F. Greaney and J. Micklefield, Angew. Chem., Int. Ed., 2017, 56, 11841–11845 CrossRef CAS PubMed.
- Y. H. Chooi, R. Cacho and Y. Tang, Chem. Biol., 2010, 17, 483–494 CrossRef CAS PubMed.
- J. M. Winter, M. Sato, S. Sugimoto, G. Chiou, N. K. Garg, Y. Tang and K. Watanabe, J. Am. Chem. Soc., 2012, 134, 17900–17903 CrossRef CAS PubMed.
- T. Zhu, X. Cheng, Y. Liu, Z. Deng and D. You, Metab. Eng., 2013, 19, 69–78 CrossRef CAS PubMed.
- M. T. Nielsen, J. B. Nielsen, D. C. Anyaogu, D. K. Holm, K. F. Nielsen, T. O. Larsen and U. H. Mortensen, PLoS One, 2013, 8, e72871 CrossRef CAS PubMed.
- P. Chankhamjon, D. Boettger-Schmidt, K. Scherlach, B. Urbansky, G. Lackner, D. Kalb, H. M. Dahse, D. Hoffmeister and C. Hertweck, Angew. Chem., Int. Ed., 2014, 53, 13409–13413 CrossRef CAS PubMed.
- L. Kong, Q. Wang, Z. Deng and D. You, Appl. Environ. Microbiol., 2020, 86, 1–14 Search PubMed.
- I. Wynands and K. H. Van Pée, FEMS Microbiol. Lett., 2004, 237, 363–367 CAS.
- S. M. K. McKinnie, Z. D. Miles and B. S. Moore, Methods in Enzymology, Elsevier Inc., 1st edn, 2018, vol. 604, pp. 405–424 Search PubMed.
- S. M. K. McKinnie, Z. D. Miles, P. A. Jordan, T. Awakawa, H. P. Pepper, L. A. M. Murray, J. H. George and B. S. Moore, J. Am. Chem. Soc., 2018, 140, 17840–17845 CrossRef CAS PubMed.
- M. C. Andorfer, J. E. Grob, C. E. Hajdin, J. R. Chael, P. Siuti, J. Lilly, K. L. Tan and J. C. Lewis, ACS Catal., 2017, 7, 1897–1904 CrossRef CAS PubMed.
- Z. D. Miles, S. Diethelm, H. P. Pepper, D. M. Huang, J. H. George and B. S. Moore, Nat. Chem., 2017, 9, 1235–1242 CrossRef CAS PubMed.
- C. Wannstedt, D. Rotella and J. F. Siuda, Bull. Environ. Contam. Toxicol., 1990, 282–287 CrossRef CAS PubMed.
- M. C. R. Franssen and H. C. van der Plas, Bioorg. Chem., 1987, 15, 59–70 CrossRef CAS.
- P. Yaipakdee and L. W. Robertson, Phytochemistry, 2001, 57, 341–347 CrossRef CAS PubMed.
- D. Wischang and J. Hartung, Tetrahedron, 2012, 68, 9456–9463 CrossRef CAS.
- F. Xu, A. Merkley, D. Yu and J. Zhan, Tetrahedron Lett., 2016, 57, 5262–5265 CrossRef CAS.
- Q. Yan, B. Philmus, J. H. Chang and J. E. Loper, eLife, 2017, 6, 1–24 CrossRef PubMed.
- S. M. Mantovani and B. S. Moore, J. Am. Chem. Soc., 2013, 135, 18032–18035 CrossRef CAS PubMed.
- I. Höfer, M. Crüsemann, M. Radzom, B. Geers, D. Flachshaar, X. Cai, A. Zeeck and J. Piel, Chem. Biol., 2011, 18, 381–391 CrossRef PubMed.
- P. C. Dorrestein, E. Yeh, S. Garneau-Tsodikova, N. L. Kelleher and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13843–13848 CrossRef CAS.
- X. Zhang and R. J. Parry, Antimicrob. Agents Chemother., 2007, 51, 946–957 CrossRef CAS PubMed.
- K. Yamanaka, K. S. Ryan, T. A. M. Gulder, C. C. Hughes and B. S. Moore, J. Am. Chem. Soc., 2012, 134, 12434–12437 CrossRef CAS PubMed.
- N. Itoh, A. K. M. Q. Hasan, Y. Izumi and H. Yamada, Eur. J. Biochem., 1988, 172, 477–484 CrossRef CAS PubMed.
- J. Hartung, Y. Dumont, M. Greb, D. Hach, F. Köhler, H. Schulz, M. Casný, D. Rehder and H. Vilter, Pure Appl. Chem., 2009, 81, 1251–1264 CAS.
- R. Vázquez-Duhalt, M. Ayala and F. J. Márquez-Rocha, Phytochemistry, 2001, 58, 929–933 CrossRef.
- B. F. Fisher, H. M. Snodgrass, K. A. Jones, M. C. Andorfer and J. C. Lewis, ACS Cent. Sci., 2019, 5, 1844–1856 CrossRef CAS PubMed.
- M. Ueki, D. P. Galonić, F. H. Vaillancourt, S. Garneau-Tsodikova, E. Yeh, D. A. Vosburg, F. C. Schroeder, H. Osada and C. T. Walsh, Chem. Biol., 2006, 13, 1183–1191 CrossRef CAS PubMed.
- S. M. Pratter, J. Ivkovic, R. Birner-Gruenberger, R. Breinbauer, K. Zangger and G. D. Straganz, ChemBioChem, 2014, 15, 567–574 CrossRef CAS PubMed.
- N. Sitachitta, J. Rossi, M. A. Roberts, W. H. Gerwick, M. D. Fletcher and C. L. Willis, J. Am. Chem. Soc., 1998, 120, 7131–7132 CrossRef CAS.
- Z. Chang, N. Sitachitta, J. V. Rossi, M. A. Roberts, P. M. Flatt, J. Jia, D. H. Sherman and W. H. Gerwick, J. Nat. Prod., 2004, 67, 1356–1367 CrossRef CAS PubMed.
- F. H. Vaillancourt, E. Yeh, D. A. Vosburg, S. E. O’Connor and C. T. Walsh, Nature, 2005, 436, 1191–1194 CrossRef CAS PubMed.
- Q. Zhu, M. L. Hillwig, Y. Doi and X. Liu, ChemBioChem, 2016, 17, 466–470 CrossRef CAS PubMed.
- M. L. Hillwig, Q. Zhu and X. Liu, ACS Chem. Biol., 2014, 9, 372–377 CrossRef CAS PubMed.
- S. Duewel, L. Schmermund, T. Faber, K. Harms, V. Srinivasan, E. Meg-gers and S. Hoebenreich, ACS Catal., 2019, 10, 1272–1277 CrossRef.
- T. Hayashi, M. Ligibel, E. Sager, M. Voss, J. Hunziker, K. Schroer, R. Snajdrova and R. Buller, Angew. Chem., Int. Ed., 2019, 18535–18539 CrossRef CAS PubMed.
- Q. Zhu and X. Liu, Beilstein J. Org. Chem., 2017, 13, 1168–1173 CrossRef CAS PubMed.
- C. Y. Kim, A. J. Mitchell, C. M. Glinkerman, F. S. Li, T. Pluskal and J. K. Weng, Nat. Commun., 2020, 11, 1–7 Search PubMed.
- A. J. Mitchell, N. P. Dunham, J. A. Bergman, B. Wang, Q. Zhu, W. C. Chang, X. Liu and A. K. Boal, Biochemistry, 2017, 56, 441–444 CrossRef CAS PubMed.
- J. A. Marchand, M. E. Neugebauer, M. C. Ing, C. I. Lin, J. G. Pelton and M. C. Y. Chang, Nature, 2019, 567, 420–424 CrossRef CAS PubMed.
- J. N. Carter-Franklin and A. Butler, J. Am. Chem. Soc., 2004, 126, 15060–15066 CrossRef CAS PubMed.
- M. Piraee, R. L. White and L. C. Vining, Microbiology, 2004, 150, 85–94 CrossRef CAS PubMed.
- P. Chankhamjon, Y. Tsunematsu, M. Ishida-Ito, Y. Sasa, F. Meyer, D. Boettger-Schmidt, B. Urbansky, K. D. Menzel, K. Scherlach, K. Watanabe and C. Hertweck, Angew. Chem., Int. Ed., 2016, 55, 11955–11959 CrossRef CAS PubMed.
- M. C. Andorfer and J. C. Lewis, Annu. Rev. Biochem., 2018, 87, 159–185 CrossRef CAS PubMed.
- H. Ludewig, S. Molyneux, S. Ferrinho, K. Guo, R. Lynch, D. S. Gkotsi and R. J. M. Goss, Curr. Opin. Struct. Biol., 2020, 65, 51–60 CrossRef CAS.
- L. J. Durak, J. T. Payne and J. C. Lewis, ACS Catal., 2016, 6, 1451–1454 CrossRef CAS PubMed.
- H. Minges, C. Schnepel, D. Böttcher, M. S. Weiß, J. Sproß, U. T. Bornscheuer and N. Sewald, ChemCatChem, 2020, 12, 818–831 CrossRef CAS.
- M. Ismail, L. Schroeder, M. Frese, T. Kottke, F. Hollmann, C. E. Paul and N. Sewald, ACS Catal., 2019, 9, 1389–1395 CrossRef CAS PubMed.
- C. J. Seel, A. Králík, M. Hacker, A. Frank, B. König and T. Gulder, ChemCatChem, 2018, 10, 3960–3963 CrossRef CAS.
- C. J. Seel and T. Gulder, ChemBioChem, 2019, 20, 1871–1897 CrossRef CAS.
- N. L. S. Clair and M. A. Navia, J. Am. Chem. Soc., 1992, 114, 7314–7316 CrossRef.
- L. Cao, F. Van Rantwijk and R. A. Sheldon, Org. Lett., 2000, 2, 1361–1364 CrossRef CAS PubMed.
- A. D. Roy, S. Grüschow, N. Cairns and R. J. M. Goss, J. Am. Chem. Soc., 2010, 132, 12243–12245 CrossRef CAS PubMed.
- C. C. C. J. Seechurn, M. O. Kitching, T. J. Colacot and V. Snieckus, Angew. Chem., Int. Ed., 2012, 5062–5085 CrossRef PubMed.
- A. L. Casalnuovo and J. C. Calabrese, J. Am. Chem. Soc., 1990, 112, 4324–4330 CrossRef CAS.
- E. C. Western, J. R. Daft and K. H. Shaughnessy, J. Org. Chem., 2003, 68, 6767–6774 CrossRef CAS PubMed.
- T. E. Schmid, D. C. Jones, O. Songis, O. Diebolt, M. R. L. Furst, A. M. Z. Slawin and C. S. J. Cazin, Dalton Trans., 2013, 42, 7345–7353 RSC.
- G. Hervé, G. Sartori, G. Enderlin, G. MacKenzie and C. Len, RSC Adv., 2014, 4, 18558–18594 RSC.
- B. H. Lipshutz, T. B. Petersen and A. R. Abela, Org. Lett., 2008, 10, 1333–1336 CrossRef CAS PubMed.
- A. Fihri, M. Bouhrara, B. Nekoueishahraki, J. M. Basset and V. Polshettiwar, Chem. Soc. Rev., 2011, 40, 5181–5203 RSC.
- K. W. Anderson and S. L. Buchwald, Angew. Chem., Int. Ed., 2005, 44, 6173–6177 CrossRef CAS PubMed.
- J. M. Chalker, C. S. C. Wood and B. G. Davis, J. Am. Chem. Soc., 2009, 16346–16347 CrossRef CAS PubMed.
- T. Willemse, K. Van Imp, R. J. M. Goss, H. W. T. Van Vlijmen, W. Schepens, B. U. W. Maes and S. Ballet, ChemCatChem, 2015, 2055–2070 CrossRef CAS.
- J. H. Li, X. D. Zhang and Y. X. Xie, Eur. J. Org. Chem., 2005, 4256–4259 CrossRef CAS.
- Z. Gao, V. Gouverneur and B. G. Davis, J. Am. Chem. Soc., 2013, 135, 13612–13615 CrossRef CAS PubMed.
- A. D. Roy, R. J. M. Goss, G. K. Wagner and M. Winn, Chem. Commun., 2008, 4831–4833 RSC.
- R. J. M. Goss, C. Pubill-Ulldemolins, S. Sharma, C. Cartmell, J. Zhao and P. Cárdenas, Chem. – Eur. J., 2019, 25, 10866–10875 CrossRef PubMed.
- M. J. Corr, S. V. Sharma, C. Pubill-Ulldemolins, R. T. Bown, P. Poirot, D. R. M. Smith, C. Cartmell, A. Abou Fayad and R. J. M. Goss, Chem. Sci., 2017, 8, 2039–2046 RSC.
- E. Marelli, Y. Renault, S. V. Sharma, S. P. Nolan and R. J. M. Goss, Chem. – Eur. J., 2017, 23, 3832–3836 CrossRef CAS PubMed.
- Y. J. G. Renault, R. Lynch, E. Marelli, S. V. Sharma, C. Pubill-Ulldemolins, J. A. Sharp, C. Cartmell, P. Cárdenas and R. J. M. Goss, Chem. Commun., 2019, 55, 13653–13656 RSC.
- M. Frese, C. Schnepel, H. Minges, H. Voß, R. Feiner and N. Sewald, ChemCatChem, 2016, 8, 1799–1803 CrossRef CAS.
- C. Schnepel, H. Minges, M. Frese and N. Sewald, Angew. Chem., Int. Ed., 2016, 55, 14159–14163 CrossRef CAS PubMed.
- H. Gruß, C. Belu, L. M. Bernhard, A. Merschel and N. Sewald, Chem. – Eur. J., 2019, 25, 5880–5883 CrossRef PubMed.
- A. D. Roy, S. Grüschow, N. Cairns and R. J. M. Goss, J. Am. Chem. Soc., 2010, 132, 12243–12245 CrossRef CAS PubMed.
- M. F. Greaney, J. Micklefield, B. R. K. Menon, S. A. Shepherd, H. H. Sharif, J. Latham and J.-M. Henry, Nat. Commun., 2016, 7, 1–8 Search PubMed.
- S. H. H. Younes, F. Tieves, D. Lan, Y. Wang, P. Süss, H. Brundiek, R. Wever and F. Hollmann, ChemSusChem, 2019, 97–101 Search PubMed.
- J. Naapuri, J. D. Rolfes, J. Keil, C. Manzuna Sapu and J. Deska, Green Chem., 2017, 19, 447–452 RSC.
- J. M. Naapuri, G. A. Åberg, J. M. Palomo and J. Deska, ChemCatChem, 2021, 13, 763–769 CrossRef CAS.
- S. V. Sharma, X. Tong, C. Pubill-Ulldemolins, C. Cartmell, E. J. A. Bogosyan, E. J. Rackham, E. Marelli, R. B. Hamed and R. J. M. Goss, Nat. Commun., 2017, 8, 1–9 CrossRef CAS PubMed.
- W. Runguphan, X. Qu and S. E. O’Connor, Nature, 2010, 468, 461–467 CrossRef CAS.
- W. Runguphan and S. E. O’Connor, Org. Lett., 2013, 15, 2850–2853 CrossRef CAS PubMed.
- E. J. Rackham, S. Grüschow, A. E. Ragab, S. Dickens and R. J. M. Goss, ChemBioChem, 2010, 11, 1700–1709 CrossRef CAS PubMed.
- Y. Xiao, S. Li, S. Niu, L. Ma, G. Zhang, H. Zhang, G. Zhang, J. Ju and C. Zhang, J. Am. Chem. Soc., 2011, 133, 1092–1105 CrossRef CAS PubMed.
Footnotes |
| † Now studying at: Division of Biological Chemistry and Drug Discovery, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, UK. |
| ‡ These authors contributed equally. |
| § Now carrying out postdoctoral research at: School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, West Yorkshire, LS2 9JT, UK. |
| This journal is © The Royal Society of Chemistry 2021 |







