Open Access Article
Open Access ArticleDetection of coronavirus in environmental surveillance and risk monitoring for pandemic control†
Linlin
Yao‡
a,
Wenting
Zhu‡
b,
Jianbo
Shi
 acde,
Tailin
Xu
acde,
Tailin
Xu
 f,
Guangbo
Qu
*acde,
Wenhua
Zhou
f,
Guangbo
Qu
*acde,
Wenhua
Zhou
 *b,
Xue-Feng
Yu
*b,
Xue-Feng
Yu
 b,
Xueji
Zhang
f and
Guibin
Jiang
acde
b,
Xueji
Zhang
f and
Guibin
Jiang
acde
aState Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, P. R. China. E-mail: gbqu@rcees.ac.cn
bMaterials Interfaces Center, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, P. R. China. E-mail: wh.zhou@siat.ac.cn
cSchool of Environment, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou 310000, P. R. China
dInstitute of Environment and Health, Jianghan University, Wuhan 430056, P. R. China
eUniversity of Chinese Academy of Sciences, Beijing 100049, P. R. China
fSchool of Biomedical Engineering, Health Science Center, Shenzhen University, Shenzhen 518060, P. R. China
First published on 12th March 2021
Abstract
The novel human infectious coronaviruses (CoVs) responsible for severe respiratory syndromes have raised concerns owing to the global public health emergencies they have caused repeatedly over the past two decades. However, the ongoing coronavirus disease 2019 (COVID-19) pandemic induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has received unprecedented attention internationally. Monitoring pathogenic CoVs in environmental compartments has been proposed as a promising strategy in preventing the environmental spread and tracing of infectious diseases, but a lack of reliable and efficient detection techniques is still a significant challenge. Moreover, the lack of information regarding the monitoring methodology may pose a barrier to primary researchers. Here, we provide a systematic introduction focused on the detection of CoVs in various environmental matrices, comprehensively involving methods and techniques of sampling, pretreatment, and analysis. Furthermore, the review addresses the challenges and potential improvements in virus detection techniques for environmental surveillance.
Key learning points(1) The strategy of sampling and sample preparation aiming at the CoVs in different environmental matrices.(2) Basic principles and mechanisms of promising detection methods aimed at CoVs. (3) The advantages and limitations of current detection techniques in the context of practical applications. (4) The challenges and potential improvements of detection techniques applied to the environmental surveillance of pathogenic CoVs. |
1. Introduction
1.1 Infection of CoVs in humans
Because of the jumping of some CoVs to humans, CoV infections have resulted in several large-scale epidemics. As the first epidemic of the twenty-first century, the outbreak of severe acute respiratory syndrome (SARS) caused by SARS-CoV-1 lasted for less than one year, from November 2002 to June 2003, and resulted in 8422 infected cases and 916 fatalities, reported by the World Health Organization (WHO) (http://www.emro.who.int/health-topics/severe-acute-respiratory-syndrome/). The Middle East respiratory syndrome (MERS) due to MERS-CoV infection was first detected in 2012. Since then, 2519 infected individuals and 866 associated deaths have been confirmed, and the WHO still keeps updating the infectious cases every year (http://www.emro.who.int/pandemic-epidemic-diseases/mers-cov/mers-situation-update-january-2020.html). The ongoing pandemic of COVID-19 due to the infection of SARS-CoV-2 has resulted in over 70 million infections and over 1.6 million deaths globally (updated on December 16, 2020, WHO). Unfortunately, unlike the previous CoV-induced epidemics, these numbers keep growing each day, and COVID-19 has spread to almost every country and territory worldwide at an unprecedented transmission rate. A unique feature of infection by asymptomatic individuals further promotes the prevalence of COVID-19. The transmission and infection characteristics of SARS-CoV-2 are not significantly changing along with the natural change of climatic factors, and the duration of the ongoing COVID-19 pandemic is unpredictable.CoVs are a group of enveloped viruses with a positive-sense single-stranded ribonucleic acid (RNA) genome, named for their characteristic crown-like spikes projecting from the virion surface.1 There are four genera of CoVs, including Alphacoronavirus and Betacoronavirus, which can infect mammals, and Gammacoronavirus and DeltacoronavirusI, which primarily infect birds.2 Until now, seven CoVs have been confirmed to infect humans, namely HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, SARS-CoV-1, SARS-CoV-2, and MERS-CoV.1 SARS-CoV-1, SARS-CoV-2, and MERS-CoV, which belong to the genus of Betacoronavirus, can cause severe respiratory syndrome and serious complications or death.
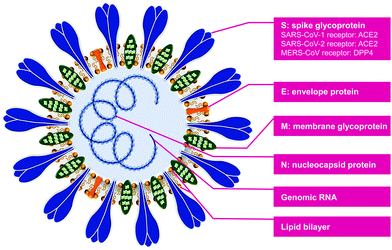
The unique features of these CoV genomic sequences and protein structures together contribute to their high infectivity, pathogenicity, and fatality, and they have led to multiple global public health emergencies over the past two decades. The genomic materials of the three CoVs (SARS-CoV-1, SARS-CoV-2, and MERS-CoV) encode four structural proteins, including membrane glycoprotein (M), envelope protein (E), spike glycoprotein (S), and nucleocapsid protein (N) (Fig. 1). The lipid bilayer is embedded with M, E, and S proteins, forming the outer envelope. The crown-like spikes on the virion surface are homotrimers of S proteins projecting outside the envelope. The genomic RNA is protected within the N protein, forming a helical nucleocapsid structure inside the virus envelope. The structural differences of S proteins determine the different mechanisms of viral binding to host cells. The S protein of SARS-CoV-1 and SARS-CoV-2 bind to the angiotensin-converting enzyme 2 (ACE2) and result in the entry of the virion into the host. In contrast, the target binding receptor of the MERS-CoV S protein is dipeptidyl-peptidase 4 (DPP4).
1.2 Environmental transmission of CoVs
The positive detection of viable CoVs in human respiratory specimens and fecal samples raised concerns for the possible presence and transmission of CoVs in the environment. Based on environmental sampling and CoV analysis, it was concluded that CoVs may spread across different environmental matrices and result in a viral infection through multiple pathways (Fig. 2). The respiratory droplets containing infective CoVs are expelled into the air in various sizes when infected individuals talk, cough, and sneeze.3 The virus-laden large droplets undergo gravitational settling and contaminate inanimate surfaces. Simultaneously, viruses in small aerosolized-droplets are readily carried by the air currents, leading to the contamination of the air, the environment, and surfaces at long distances from infected patients.4 The viable SARS-CoV-2 can be emitted by the host through defecation, leading to the entrance in sewage and wastewater treatment system.5 The CoVs in wastewater may contaminate the sewer plumbing, centralized water treatment facilities, and the recipient natural water bodies. Furthermore, CoVs in sewage-borne aerosols may result in air contamination through defected sewer plumbing and ventilation systems.As the dominant infection route, healthy individuals can be directly infected by inhaling and mucosa-contacting respiratory droplets/aerosols emitted from infected individuals. However, the contaminated environmental matrices may also play a pivotal role in the indirect transmission routes of CoVs. Contacting contaminated inanimate surfaces is considered a high risk for CoV transmission and infection, resulting in hand-to-mouth, hand-to-mucosa, and food-to-mouth routes. CoV contaminated sewage may drive the fecal–oral transmission and infection through the urban water cycle system. Although several transmission routes were proposed, the airborne transmission of CoVs may pose greater threat to humans compared to other routes. Relying on epidemiologic analysis and airflow-dynamics modeling, Yu et al. proposed transmission routes of SARS-CoV-1-laden aerosols through air shafts within buildings and airflow between buildings. They suggested that airborne transmission of SARS-CoV-1 contributed to a massive community outbreak of SARS in Hong Kong in 2003.6 The airborne transmission of SARS-CoV-2 could lead to the long-distance prevalence of COVID-19, which may make a contribution to exacerbate the current COVID-19 pandemic.
1.3 Environmental surveillance for the prevention and control of epidemics
The environmental surveillance mentioned herein refers to the analysis and monitoring of any possible aspect of the pathogenic virus in different environmental matrices. As a complementary strategy to clinical diagnosis in identifying viral infection and spread, the environmental surveillance of CoVs can be a promising strategy to prevent and early-warn the prevalence of infectious diseases. The information related to the presence, viability, infectivity, and environmental variation could be obtained through environmental surveillance. Environmental surveillance also facilitates the localization of the occurrence region and helps predict the risk and pattern of its onward transmission. On the one hand, environmental surveillance is a good early-warning tool buying time for the public health system to respond to the epidemic; on the other hand, it could serve to monitor the efficiency of outbreak control. Therefore, despite the epidemiological pattern that varies with different novel CoVs, reliable and rapid environmental surveillance of these viruses plays a crucial role in preventing and controlling viral epidemics.Based on the presence and potential transmission patterns of CoVs, the following practical scenarios can be emphatically considered during environmental surveillance: (1) interior space, especially with dense crowds and poor ventilation, e.g., hospital, airport, marketplace, restaurant, and entertainment venue; (2) logistic chain, especially cold food-related environment due to the low-temperature resistance of CoVs; and (3) sewage, wastewater treatment plant, and the recipient water body, which are also included in wastewater-based epidemiology.
1.4 Detection techniques for the environmental surveillance of CoVs
Various urgent objectives during the environmental surveillance of CoVs rely on dedicated detection techniques. These include the qualitative detection to confirm the presence of certain pathogens, a quantitative method for epidemiological modeling, viability and feasibility analysis for environmental transmission confirmation and risk evaluation, and environmental virology studies to characterize the properties of the virus. Furthermore, for long-term and large-scale environmental surveillance, detection techniques supporting portable, onsite, in-time, and online data transfer are urgently needed.Currently, the primary methods being applied to the environmental surveillance of CoVs include nucleic acid-based detection, intact virus characterization, and cell culture or plaque-forming units (PFU) for viability evaluation. After the positive detection of nucleic acids, the viability and infectivity of the virus still needs to be evaluated. The nucleic acid-based technique is considered to be the gold standard in the qualitative and quantitative analysis of CoVs RNA in any environmental matrix, similar to the clinical diagnosis. However, the conventional methods of computed tomography (CT) chest scan and serological testing frequently used for COVID-19 clinical diagnosis are, in principle, not suitable for the environmental surveillance of CoVs.
The primary challenges related to the detection of environmental CoVs include low viral load, matrix interference, and integrity damage. Therefore, to overcome the inherent limitation and accomplish the urgent objectives of environmental surveillance of CoVs, several guidelines were proposed for the sampling and pretreatment methods and analysis techniques, including (1) elimination or reduction of matrix-effects and impurities; (2) elevation of virus recovery, including viral nucleic acids, proteins, and intact virions; (3) maintaining structural integrity and viability of target virus; and (4) elevation of sensitivity and specificity of the detection technique.
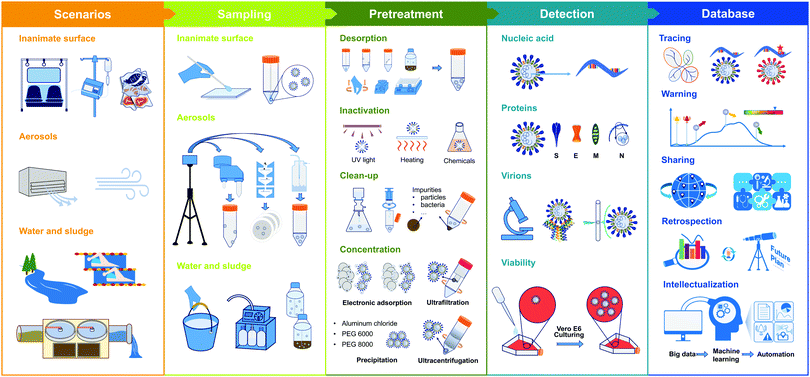
In this review, we introduce a major scheme of environmental detection of infectious CoVs and other emerging CoVs (as shown in Fig. 3), including (1) matrix-specific sampling and sample preparation methods; (2) advanced detection techniques that are target-specific, including nucleic acids, proteins, virions, and viability; and (3) the feasibility and necessity of these techniques in environmental surveillance. We also discuss the challenges and potential improvements of detection techniques for the environmental surveillance of pathogenic CoVs. We hope that this review will inspire researchers and foster relevant scientific studies.
2. Sampling and pretreatment
To understand the environmental spread of CoVs, representative and interpretive environmental samples should be obtained. The systematic and comprehensive design of the sample type, sampling site, and time interval between sample collection is essential when conducting wide-range and long-term environmental surveillance. Aiming at three CoVs (SARS-CoV-1, SARS-CoV-2, and MERS-CoV), the sampling and pretreatment methods in different environmental matrices are reviewed in Table 1 and Table S1 (in ESI†).| Sample Type | Virus | Sampling site | Sampling method | Pretreatment method | Analysis |
|---|---|---|---|---|---|
| NIOSH: the National Institute for Occupational Safety and Health, VIVAS: viable virus aerosol sampler, PTFE: polytetrafluoroethylene, WWTP: wastewater treatment plant, PEG: polyethylene glycol, RNA: ribonucleic acid, RT-PCR: reverse transcription-polymerase chain reaction, RT-qPCR: RT-quantitative real-time PCR, ddPCR: droplet digital PCR, SERS: surface enhanced Raman scattering. More information was provided in Table S1, ESI. | |||||
| Inanimate surface | SARS-CoV-1 SARS-CoV-2 MERS-CoV | Viral epidemic designated hospitals public places | Position: surfaces in patient wards, medical functional area, medical staff area, public area, etc. | Desorption: manually gently shake preservation liquid containing swab or pad | RT-PCR/RT-qPCR: viral RNA |
| Time: before or after disinfection | Clean-up: filtration of preservation liquid RNA extraction | Cell culture: viability | |||
| Equipment: pre-moistened swab or wipe pad | |||||
| Wiping within area or in a certain pattern | |||||
| Aerosol | SARS-CoV-2 MERS-CoV | Epidemic designated hospitals Public places | Position: indoor environment: patient wards, medical functional area, medical staff area, public area, etc. Outdoor environment | Desorption: dissolution of soluble filter, vortex of solid–liquid mixture | RT-PCR/RT-qPCR/ddPCR: Viral RNA |
| Time: before or after disinfection | Inactivation: addition of chemicals | Cell culture: viability (the distribution data of viral RNA in different sizes of aerosols were provided) | |||
| Equipment: automatic samplers, e.g., NIOSH cyclone sampler, SASS 2300 wetted wall cyclone sampler, cascade impactor, liquid impinger, VIVAS, filter cassette, etc. Filter type: gelatin, PTFE, quartz fiber, and glass microfiber. | Clean-up: centrifugation, filtration | ||||
| Concentration: ultracentrifugation RNA extraction | |||||
| Water | SARS-CoV-1 SARS-CoV-2 | Wastewater treatment plants Epidemic designated hospitals Water pipeline | Samples: pre-treated wastewater (also known as influent water or raw wastewater) treated wastewater (e.g., disinfected water, and effluent water from WWTPs) river (receiving treated wastewater) pipe water | Inactivation: heat treatment | RT-PCR/RT-qPCR: Viral RNA |
| Equipment: automatic sampler | Clean-up: centrifugation, filtration | SERS: Viral spike proteins | |||
| Sampling: grab sample (that is, instantaneously collected sample at a particular time or site) composite sample (that is, mixture of several samples collected from different time points and sites | Concentration: electrostatic adsorption, ultrafiltration, ultracentrifugation, and PEG or aluminum driven precipitation RNA extraction | Cell culture: Viability | |||
| Sludge | SARS-CoV-2 | Wastewater treatment plants | Samples: sludge from different treatment stage | Desorption: shake solid–liquid mixture | RT-qPCR: Viral RNA |
| Sampling: collected as wastewater–sludge mixture with solid content ranging between 1% and 5% | Clean-up: centrifugation, filtration | ||||
| Concentration: PEG driven precipitation RNA extraction | |||||
2.1 Sampling
Swabs or wipe pads are normally pre-moistened using a virus-specific preservation fluid, such as viral transport medium, universal transport medium, and phosphate buffer saline. Wiping with an “S” pattern in different directions and overall wiping within a certain area were used for collecting CoVs on the inanimate surface, which suggested a flexible strategy for selecting the shape and area of the sampling surface (Table S1, ESI†). After sampling, swabs or wipe pads potentially containing viruses are immediately packaged into a sterile container with or without preservation fluid.
van Doremalen et al. investigated the behavior of CoVs on various material surfaces that are commonly involved in the household and hospital environments.7 Based on the Bayesian regression model and virus stability experiments under laboratory circumstances, they reported that the viability and infectivity of SARS-CoV-1 and SARS-CoV-2 remained for hours to days and exhibited longer persistence on stainless steel and plastic surfaces, which positively suggested the transmission of CoVs through these contaminated surfaces. In order to evaluate the contamination of SARS-CoV-2 on inanimate surfaces, Ong et al. collected the inanimate surface samples from a dedicated isolation room for patients with COVID-19 either before or after the routine disinfection; in addition, the surface samples of personal protective equipment (PPE) of physicians entering the ward were collected as well.8 In this study, compared with the positive detection of SARS-CoV-2 RNA on inanimate surfaces before routine cleaning, the negative results from post-cleaning of the surface samples indicated that the routine disinfection measures in the hospital were effective and imperative during the pandemic; the positive results on doctor's PPE evidenced the necessity to protect medical staff. Relying on surface sample collection and nucleic acid detection, this study highlighted the transmission potential through inanimate surfaces and the importance of environment and hand disinfection.
Overall, the CoV nucleic acid presence on actual environmental surfaces as well as viability confirmation of CoVs under experimental conditions emphasized the infection risk of spreading infectious pathogens by contacting CoV contaminated inanimate surfaces. From the perspective of prevention and control of a pandemic, the environmental surveillance of CoVs on inanimate surfaces is supposed to facilitate the guidance and inspection of disinfection work in densely populated locations, e.g., a hospital. Furthermore, routine and random environmental surveillance throughout the cold food supply chain could minimize the contact-mediated infection risk.
Physical separation usually involves the aerodynamic diameter of aerosols and their inertial movement. The cascade impactor and the National Institute of Occupational Safety and Health (NIOSH) cyclone sampler were applied to aerosol sampling in an aerodynamic size-segregated manner. Liu et al. conducted aerosol sampling in two designated hospitals for patients with COVID-19, using a four-stage impactor at an airflow rate of 9 L min−1, which separately collected particles on a gelatin filter in five size ranges: >2.5 μm, 1.0 to 2.5 μm, 0.5 to 1.0 μm, 0.25 to 0.50 μm and <0.25 μm.9 Relying on this aerodynamic size-segregated sampling technique, the distribution and the presence of SARS-CoV-2 RNA on aerosols of different sizes could be determined. Similarly, using the NIOSH cyclone sampler with an in-flow rate of 3.5 L min−1, Chia et al. collected three sizes of aerosols in SARS-CoV-2 infection isolation wards (aerosols >4.0 μm and 1.0–4.0 μm in diameter were separately collected in empty centrifuge tubes, and <1.0 μm aerosols were collected on a polytetrafluoroethylene (PTFE) filter).10 However, Chia et al. reported the positive detection of SARS-CoV-2 RNA only in aerosols over 1.0 μm in diameter. The conflicting results of the above two studies need to be further demonstrated by thoroughly considering the reliability and efficiency of different sampling techniques. Overall, relying on the size-distribution based aerosol-sampling method, both studies showed SARS-CoV-2 RNA in submicrometer aerosols, suggesting the high possibility of long-distance spread and infection of CoVs through aerosol dispersion, especially considering that the smaller virus-laden particles may linger longer in the air and be more readily inhaled into the respiratory system. Lednicky et al. applied the VIVAS sampler to sample aerosols of CoVs from a respiratory infection area in a designated healthcare center for COVID-19.11 Using a laminar-flow water vapor condensation strategy to enlarge the small particles and collect enlarged aerosols more efficiently, this method enables the resistance of desiccation and maintenance of viability during sampling. However, no viable SARS-CoV-2 was detected in aerosol samples, and the efficiency of the sampling method in maintaining the viability of CoVs was not evaluated in this study. Nevertheless, it should be emphasized that maintaining viral stability during the technique selection in environmental sampling is essential, especially for viability and infectivity. Sufficient information about the viral load and viability is further needed to support the transmission and infectivity of CoVs through aerosols in a realistic environment. However, laboratory work with SARS-CoV-1 and SARS-CoV-2 as CoV model, based on aerosol sampling through a gelatin filter at designed time points, van Doremalen et al. experimentally demonstrated a similar stability of SARS-CoV-1 and SARS-CoV-2 in aerosols (half-life of approximately one hour), which suggested the infection potential of airborne CoVs.7
Based on the negative surface charge of virions and the positive potential magnetic beads in sampling solution of impingement air sampler, CoVs in aerosols could be specifically enriched from the air stream (Table S1, ESI†). More sampling methods and samplers aiming at CoVs in aerosols were listed in Table 1 and Table S1 (ESI†), such as a cascade impactor.
Unlike indoor aerosol sampling, in outdoor sampling, one should consider the sampling capacity in a more comprehensive dimensional range and natural environmental factors. Setti et al. collected PM10 samples in the Bergamo area, Italy, using a quartz fiber filter on a low-volume gravimetric air sampler.12 In this study, the sampling duration was 24 h with an airflow rate at 38.3 L min−1, which enabled an adequate sampling of air volume that reached 55 m3 per day. The study reported the presence of SARS-CoV-2 RNA in PM10 samples, suggesting the airborne transmission risk of SARS-CoV-2.
Environmental surveillance of CoV-laden aerosols enables an in-depth understanding of the aerodynamic characteristics in a realistic environment. Thus, it is beneficial to clarify the use of countermeasures during the epidemic. For instance, the presence of CoVs RNA in submicrometer-level aerosols indicated the necessity of respiratory protective measures in the confined space of dense crowds. The ventilation and sterilization of public places are imperative to prevent infection by reducing the concentration of virions in air environments.
Rimoldi et al. collected grab samples, including influent wastewater, treated wastewater of wastewater treatment plants (WWTPs), and treated water-receiving river water, demonstrating the presence of SARS-CoV-2 RNA in influent wastewater and receiving river water.13 The positive detection of SARS-CoV-2 RNA in river water samples in this work was attributed to the contamination of untreated wastewater, considering the negative results in treated water samples. Medema et al. collected a series of 24 h-based composite wastewater samples in WWTPs at different time points before and after the onset of the COVID-19 epidemic.14 They reported the positive correlation between SARS-CoV-2 RNA concentrations in wastewater samples and an increase in COVID-19 prevalence; what's more, SARS-CoV-2 RNA in wastewater was detected at several days ahead of the first case reported clinically. These studies thus demonstrated the timeliness and reliability of wastewater surveillance in epidemic prevention and epidemiology prediction, especially under a purposeful sampling strategy. Regarding the environmental surveillance in WWTPs, Bogler et al. reviewed the possible transmission and contamination of SARS-CoVs in the sewer system and natural environments (such as surface water and groundwater) as well as the potential daily occupational exposure (such as agricultural worker).15 Based on the comprehensive awareness of high risk mediated by the SARS-CoVs contained wastewater, they underscored the urgent need for environmental surveillance and risk assessment of SARS-CoV-2 in wastewater.
Considering the transmission potential through the water cycle system, the environmental surveillance of CoVs in different types of wastewater and natural water will be capable of explaining the environmental fate and evolution of CoVs and the corresponding health risk. Unlike the hard recognition of a specifically infected individual, the localization of the outbreak region or community can be easily identified through wastewater surveillance before identifying a large-scale outbreak. Considering the overwhelming burden of the individual test in the medical system, wastewater surveillance greatly helps identify the hotspot region of infection and alert the resurgence of an epidemic.
From the perspective of sludge treatment and the following agricultural usage, reliable data of abundance and survivability of CoVs in sludge will explain the environmental behaviors of CoVs across different environmental compartments. For instance, the CoVs contained in sludge may enter the air through aerosolization or contaminate water during the irrigation of sludge–fertilizer. Furthermore, the sludge surveillance of CoVs will contribute to monitoring the inactivation efficiency in WWTPs and the evaluation of infection risks through occupational exposure.
2.2 Sample pretreatment
The natural matrices in the environmental samples usually interfere with the virus detection. The pretreatment of initial environmental samples is imperative (workflow showed in Fig. 3). After sampling, the environmental samples containing CoVs should be transported to the biosafety laboratory under cold conditions, normally on ice, and immediately transferred to 4 °C to temporarily maintain viral stability. For long-term storage, laboratory pretreated samples and extracted nucleic acid samples should be transferred to −80 °C. Depending on the analysis purposed, at least one of the following steps is needed: inactivation, clean-up, and concentration. Of note, for nucleic acid testing, RNA extraction is included in the pretreatment.The selection of viral desorption depends on the sample type and filter material. For the inanimate surface sample, after manually inverting tubes containing swabs and the preservation liquid several times, the solution can be used directly for the RNA extraction (Fig. 3).12 For soluble gelatin filter used in aerosol sampling, the virions collected on it could be extracted by heating and dissolving the membrane (Table S1, ESI†). As for other insoluble membranes, a gentle vortex will facilitate the desorption of the virus from a soaked filter into solution, and then centrifugation and resuspension are needed to collect the virus pellet. Since sludge samples are collected as a solid–water mixture, the mixture can be shaken to adequately recover viruses in the liquid phase, collecting virus-containing supernatant after centrifugation for the subsequent steps (Table S1, ESI†).
Methods for concentration include electrostatic adsorption, ultrafiltration, precipitation, and ultracentrifugation (details in Table 1 and Table S1, ESI†). (1) Electrostatic adsorption is based on the principle of electrostatic interactions between filtration materials and virions. By decreasing the water sample's pH value, positive charges on the viral surface will be generated, and thus the electronegative filter can be used to concentrate CoVs.17 Alternatively, by using an electropositive material, the innate negatively charged virus can be adsorbed without preconditioning the water sample.18 (2) Ultrafiltration is a size-exclusion based method. Based on previous studies, three molecular weight cut-offs of 10, 30, and 100 kDa were used for ultrafiltration to concentrate CoVs in water samples (Table S1, ESI†). To minimize the virus loss during ultrafiltration, the backwash of ultrafilter was additionally employed to collect any filter-trapped virus.17 (3) The reported precipitation of CoVs in water samples include aluminum chloride-driven precipitation, and polyethylene glycol (PEG) (with a molecular weight of 6000 or 8000) driven precipitation due to its high hydrophilic properties (Table S1, ESI†). Of note, the PEG-driven precipitation technique normally requires an overnight incubation (or 12 h). Using surrogate viruses, the recovery of aluminum-driven precipitation method was around 10% and 5% for influent and effluent wastewater, respectively.19 As a reference, after conducting PEG precipitation and RNA isolation, Alpaslan Kocamemi et al. detected a 1–1.5 log titer loss of surrogate virus considering the initial amount of 300 μL of 105 copies per mL.20 (4) The ultracentrifugation method has also been used for the concentration of viruses. By spiking inactivated SARS-CoV-2 in wastewater samples, Green et al. estimated the ultracentrifugation method's recovery as approximately 12%.21 For aerosol samples collected in sampling solutions, the ultracentrifugation method was used to concentrate CoVs initially dispersed in the liquid phase.22 Considering multiple pretreatment procedures and the matrix effect of the environmental sample, the reported recoveries of the methods mentioned above may be acceptable for amplification-based nucleic acid detection. However, for highly sensitive environmental surveillance, the recovery rate warrant further optimization. Given the complexity of the wastewater sample, the above-introduced methods are also applied for battery pretreatment.18
2.3 Procedure and quality control
To prevent personnel infection and sample contamination, wearing PPE and changing gloves between each sample collection is highly suggested. Any equipment, material, solution, and container involved in the sampling and pretreatment of the virus sample must be sterilized. When equipment needs to be repeatedly used, such as autosamplers of aerosols and water, the sterilization of the sampler should be conducted between sample collections to avoid cross-contamination.To obtain a high-quality and convincing analysis result, the assessment of contamination, matrix effect, and virus recovery is crucial during sampling and preparation. When conducting the sampling campaign, the field blank is collected to control potential contamination during sampling. In addition, duplicate environmental samples are collected for the matrix effect evaluation and parallel sample treatment during quality control. During sample preparation, field blanks and laboratory blanks (e.g., molecular biology grade water) are treated as sampling and treatment procedure controls for contamination monitoring. By processing parallel samples, the relative standard deviation of the pretreatment method can be evaluated. Before the sample pretreatment, the surrogate virus or inactive target virus with a known quantity can be spiked as an internal standard for virus recovery assessment. The recovery control is highly required in any independent research, enabling the precise quantitation and evaluation of the virus presented in original environmental matrices. Given that the contamination during RNA extraction and polymerase chain reaction (PCR) can generate great bias or opposing results, an additional reagent blank or extraction blank is processed to monitor and minimize any potential contamination. Furthermore, for preventing cross-contamination, the RNA extraction and PCR setup are normally performed in separate rooms.
3. Advances in detection techniques of novel CoVs
Effective laboratory techniques enabling accurate, quick, and widely available testing of pathogenic viruses play critical roles in disease control and prevention, which is particularly evident during the ongoing COVID-19.23 The characterization and detection with high sensitivity and specificity for detecting viruses in different environmental conditions will also be crucial for researching the environmental behavior of CoVs. This section review a package of advanced methods listed in Table 2 for the comprehensive analysis of CoVs, including qualitative and quantitative detection, and the identification of virus variants. In addition, viability or infectivity testing is also critical for the risk assessment of contaminated environmental compartments by CoVs. Classical techniques are applied as the dominant methods and still play an irreplaceable role in the ongoing COVID-19 pandemic, especially considering the abundant commercial kits based on the detection of nucleic acid and antibody were developed and produced at an unprecedented rate since the outbreak of COVID-19.24 However, novel techniques developed from or integrated with traditional methods are urgently needed to satisfy the requirements of large-scale environmental surveillance. The innovative methods exhibit enhanced performance or simplified workflow, even though their actual sensitivity and reliability remain to be tested in practical applications.| Method | Feature | Advantage | Disadvantage | Example |
|---|---|---|---|---|
| PCR: polymerase chain reaction, RT-qPCR: reverse transcription quantitative real-time PCR, ddPCR: digital droplet PCR, RCA: rolling-circle amplification, NASBA: nucleic acid sequence-based amplification, LAMP: loop-mediated isothermal amplification, RPA: recombinase polymerase amplification, SHERLOCK: specific high-sensitivity enzymatic reporter unlocking, DETECR: DNA endonuclease-targeted CRISPR trans reporter, HOLMES: one-hour-low cost multipurpose highly efficient system, CRISDA: CRISPR-Cas9-triggered nicking endonuclease-mediated strand displacement amplification method, NGS: next-generation sequencing, RIA: radioimmunoassay, EIA: enzyme immunoassay, ELISA: enzyme linked immunosorbent assay, FPIA: fluorescence polarization immunoassay, MEIA: microparticle enzyme immunoassay, CLIA: chemiluminescent immunoassay, LFA: lateral flow assay, MALDI-TOF MS: matrix-assisted laser desorption/ionization-time of flight mass spectrometry, EM: electron microscopy, IEM: immune electron microscopy, Cyro-EM: cryo-electron microscopy, AFM: atomic force microscopy, COVID-19 FET sensor: graphene-based field-effect transistor, namely, the COVID-19 FET sensor, QCM: quartz crystal microbalance, SPR-based biosensor: surface plasmon resonance-based biosensor. The detailed information about examples was listed in Table S2, ESI. | ||||
| Nucleic acids-based analysis | According to base pairing, nucleic acids analysis can provide amounts of accurate information for virus diagnosis and virology research | High sensitivity and specificity, early detection of low viral titers, easily operated on a large scale | Requiring nucleic acids isolation process, time-consuming, and labor-intensive | Direct hybridization (dot-blot, or Southern blotting and northern blotting, dual-functional plasmonic biosensor, colorimetric assay based on gold nanoparticles) |
| PCR-based (PCR, RT-qPCR, ddPCR) | ||||
| Traditional isothermal amplification-based (RCA, NASBA, LAMP, RPA) | ||||
| CRISPR-based isothermal amplification-based (SHERLOCK, DETECR, HOLMES, CRISPR-chip, CRISDA) | ||||
| NGS (nanopore sequencing, Helicos sequencing and Real-time single-molecule sequencing with polymerase) | ||||
| Protein-based analysis | The analysis of antigens from viruses and antibodies, chemokines and interferons from human immune response | Determining the immune status of asymptomatic patients, easy and quick operation | Unlikely to play any role in screening or for the diagnosis of early infections, antigen detection may miss cases due to low infectious burden or sampling variability | Antigen or antibody immunoassay (RIA, EIA, ELISA, FPIA, MEIA, CLIA, LFA) |
| Mass spectrometry (MALDI-TOF MS) | ||||
| Virion detection | Analysis of intact virion without disrupting the integrity | Meaningful in virus classification | Time-consuming, technically high demanding, high cost involved in purchasing, and maintaining the facility | EM, IEM, Cyro-EM, AFM, COVID-19 FET sensor, QCM, flow cytometry technique |
| Viability and infectivity evaluation | Isolating viruses from samples and culturing in cells or tissues | Meaningful in virology research, e.g., viability and infectivity | Many viruses will not grow in cell culture at all, time-consuming, technically high demanding, and requiring well-controlled laboratory environments | Cell culture |
| Integrated methods | Combining advanced biomarker detection methods (e.g., nucleic acid-based or immunology-based techniques) with microfluidics | Highly sensitive, fast, cost-effective, multiplexing, portable, and suitable for field diagnosis | Newly developed and requiring various technologies | Microfluidics (a portable microfluidic immunoassay system) |
| Lab-on-a-chip (SPR-based biosensor) | ||||
3.1 Nucleic acid-based analysis
Nucleic acids serve as basic genetic materials for all living organisms and have routinely and widely been targeted for virus identification. The PCR-mediated amplification under variable temperatures and newly developed isothermal amplification revolutionized nucleic acids-based virus detection. The controllable amplification of virus-specific genomic or messenger ribonucleic acid (mRNA) sequences greatly improves the detection sensitivity and enables detection in a matrix of low virus concentration, such as in the environment.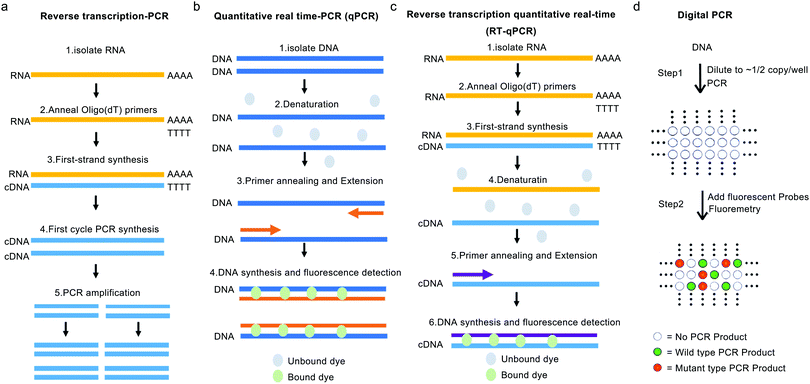 | ||
| Fig. 4 Schematic diagram of RT-PCR, qPCR, RT-qPCR and ddPCR. (a) After RNA isolation, using the reverse transcriptase enzyme and oligo (dT) primers, a single-stranded copy of cDNA is generated. Then single-stranded cDNA can be amplified by a DNA polymerase, generating double-stranded cDNA, feeding into a standard PCR-based amplification process. (b) Quantitative real-time PCR, by taking advantage of fluorescent dye or specific fluorescent labeled probes, has created the potential to monitor the DNA amplification process in “real-time” on an elegant equipment that is able to collect the fluorescent data from every PCR cycle. (c) After RNA isolation, the integrity analysis prior to cDNA generation and the qPCR assay is performed using intercalating dyes or hydrolysis probes. Fluorescence detection is performed throughout the PCR cycles, thus obtaining an amplification curve which is used to quantitate the target sample during the analysis of data. (a–c) were reproduced from ref. 25 with permission from the Portland Press, copyright 2020. (d) ddPCR is composed of two steps: following DNA sample dilution, PCR is carried out using fluorescent probes that distinguish wild type (WT) from mutant alleles by measuring fluorescence signals from specific molecular beacons. Reproduced from ref. 26 with permission from the National Academy of Sciences, USA, copyright 1999. | ||
RT-qPCR using fluorescent dyes or specific fluorophore-labeled probes can monitor the DNA amplification process in “real-time” on dedicated instruments capable of collecting the fluorescence data after every PCR cycle (Fig. 4b and c). In addition, these RT-qPCR assays can be designed to contain primer sets targeting multiple nucleic acid targets or different regions in a single target simultaneously, which is beneficial to improve the sensitivity, specificity, and throughput. For example, Yip et al. designed an RT-qPCR assay targeting two genomic targets of CoVs using a TaqMan probe pair, instead of one probe.27 Although other detection technologies are being developed rapidly over the last decade, RT-PCR and RT-qPCR are still the most commonly used techniques for detecting novel CoVs in environmental samples, even in the ongoing COVID-19 pandemic (Table 1, and detection results of CoVs shown in Table S1, ESI†). By using RT-PCR and RT-qPCR technique, different amounts of viral genomic RNA have been reported in different environmental samples. However, since different sample processing methods and viral nucleic acid extraction methods were employed, data obtained from different research studies and laboratories were not comparable to each other. In addition, the RT-PCR technique can only analyze the number of certain genomic fragments from virus rather than the number of viruses with viability.
Amplification techniques significantly elevate nucleic acid detection sensitivity and specificity, enabling the detection of CoVs at low concentrations in different environmental matrices. Among different amplification techniques, RT-PCR and RT-qPCR are commonly selected, initially answering the presence and loads of CoVs, through the detection of nucleic acid biomarkers. However, RT-PCR and RT-qPCR still have non-negligible drawbacks, such as relative quantification, unsatisfactory performance in the presence of inhibitors, and vulnerability to contamination. For example, samples with a known target concentration must be used to make a dilution series to obtain standard curves when qPCR is used for relative quantification. The accuracy of these curves is sensitive to PCR inhibitors and reaction conditions, resulting in data with high variability and low reproducibility, and even false negative data.28
Additionally, these PCR-based approaches normally require processes including nucleic acid extraction and PCR system preparation, leading to a contaminated laboratory environment and contaminated reagents, inevitably giving false-positive results. Furthermore, environmental aerosols contaminated with amplified nucleic acids are a bigger challenge for accurate and quick virus detection, particularly when many tests are performed during pandemic control. These drawbacks of RT-PCR and RT-qPCR together give poor comparability of quantitative data generated from different laboratories and experiment batches, thus posing a barrier to sharing and establishing a database.
Digital PCR (dPCR) is designed to precisely quantify RNA by providing absolute counts instead of relying on the standard curve. dPCR is developed based on limiting dilution, endpoint PCR, and Poisson statistics, with absolute quantification, which can be more immune to background noise than the conventional qPCR technique (shown in Fig. 4d).26 By randomly dividing the sample into enough separate reactions, such as thousands of droplets, theoretically either none or one target molecule will be present in each reaction chamber. Next, each chamber is used to perform an independent amplification to the endpoint, and the absolute quantification is estimated by counting PCR-positive reactions and using Poisson statistics. This core principle enables dPCR to provide a highly sensitive and precise absolute quantification, reduced false negative occurrence, and quantification of the target nucleic acids in a sample with a complex matrix effect. Given its ability to resist interference, dPCR can provide a more reliable result than a conventional PCR. Researchers employed an optimized droplet digital PCR (ddPCR) to detect SARS-CoV-2 RNA from 77 patients and compared them with RT-qPCR in diagnostic accuracy. Twenty-six patients with negative RT-qPCR but positive ddPCR reports were subsequently reported as positive by follow-up surveys, indicating the great potential of RT-ddPCR in screening patients with low viral loads.29 By further optimizing the detection method, Liu et al. applied RT-ddPCR to analyze SARS-CoV-2 in aerosols, giving a limit of detection of 2.18 and 0.42 copies per reaction (final volume of 20 μL) using two primers/probe sets targeting “ORF1ab” and “N” genes of SARS-CoV-2, respectively.9 The digital PCR technique significantly improves the detection sensitivity and accuracy of the virus, especially in the presence of high background interference.
Regarding environmental surveillance, ddPCR can provide high sensitivity and absolute quantitation, and has been used in the detection of SARS-CoV-2 in aerosols.9 Furthermore, high concordance of absolute quantification by measuring the endpoint signals rules out the potential influence of interference (which would lead to unstable amplification), enabling the comparison and data sharing of virus concentrations derived from different detection events. As ddPCR is a highly closed system, which helps to avoid contamination by aerosols during the amplification process, it is suitable for environmental monitoring. The characteristics of anti-interference and reliable quantification make the ddPCR technique a good choice for detecting CoVs from various environmental matrices.
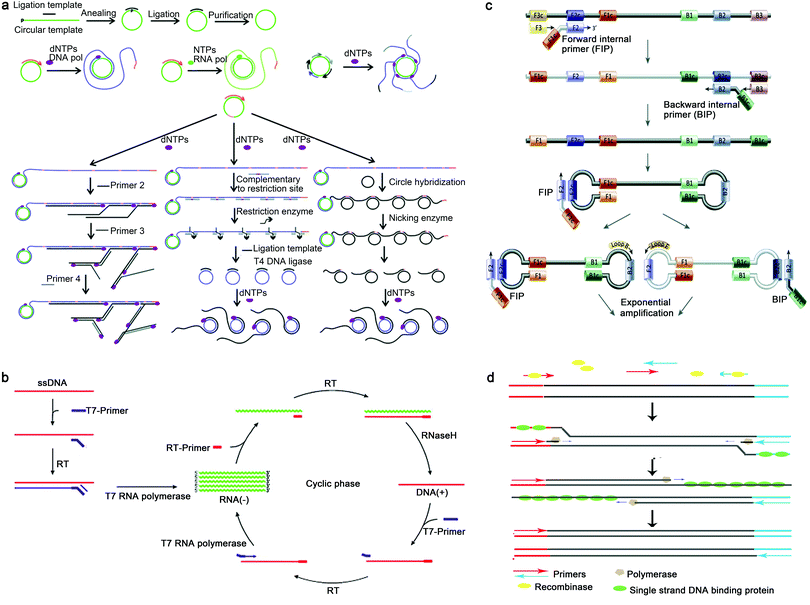 | ||
| Fig. 5 Principles of representative isothermal amplification techniques. (a) RCA: firstly, template-mediated enzymatic ligation leads to a DNA circle; secondly, following linear RCA reaction, long single-stranded deoxyribonucleic acid (ssDNA) with a pre-made circle and target nucleic acids was formed; thirdly, long RNA with a pre-made DNA circular template was generated; finally, after multi-primed RCA, multiple copies of the RCA products from a single circle were produced. Reproduced from ref. 33 with permission from the Royal Society of Chemistry, copyright 2014. (b) NASBA:NASBA is a sensitive transcription-based amplification system with multiple components, including avian myeloblastosis virus (AMV) reverse transcriptase (RT), RNase H, T7 DNA dependent RNA polymerase (DdRp) and two oligonucleotide primers specific to the analyte target. Reproduced from ref. 34 with permission from Elsevier B.V., copyright 2009. (c) LAMP: the LAMP method utilizes a single-stranded DNA shaped like a dumbbell with loops at both ends to initiate the reaction; DNA having an inverse structure relative to the dumbbell is produced. DNA products are amplified in reaction cycles that are attached to an inverted repeat structure at the amplified region. Amplified DNA products of various stem lengths are generated in the subsequent repeated elongation reactions. Reproduced from ref. 35 with permission from Elsevier Ltd, copyright 2015. (d) RPA: firstly, scanning of DNA with homologous sequences is performed by the complex of recombinase proteins and primers; secondly, the strand-displacement activity of the recombinase allows the primers to insert at the cognate site and single-stranded DNA binding proteins further stabilize the displaced ssDNA chain; thirdly, after recombinase disassembly, the 3′-end of the primers are accessible to a strand displacement DNA polymerase, which elongates the primer; finally, after cyclic repetition of this process, exponential amplification is achieved. | ||
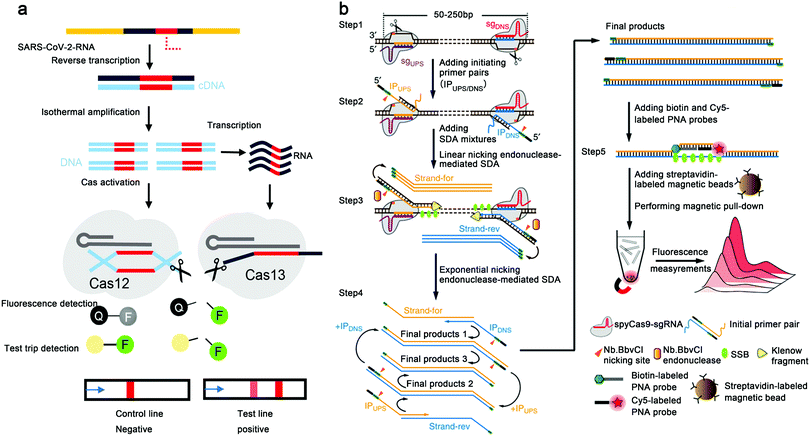 | ||
| Fig. 6 Mechanisms of CRISPR-based methods of nucleic acid detection. (a) Mechanisms of Cas12 and Cas13-based detection system. The purified SARS-CoV-2 RNA is reverse transcribed to cDNA and amplified through isothermal techniques, e.g., RT-RPA and RT-LAMP; Cas12 is activated by double-stranded deoxyribonucleic acid (dsDNA) with a CRISPR targeting sequence (in red) to cleave ssDNA reporters; Cas13 recognizes RNA containing CRISPR targeting sequences and cleaves its RNA reporters; in a fluorescence assay, the cleavage of the reporter generates fluorescence to provide naked eye color detection. Reproduced from ref. 37 with permission from the American Chemical Society, copyright 2020. (b) Schematic reaction mechanism of CRISDA. A pair of Cas9 ribonucleoproteins initiate the isothermal amplification and the amplicons are quantitatively determined by a PNA invasion-mediated endpoint measurement via magnetic pull-down and fluorescence measurements. Reproduced from ref. 32 with permission from the Springer Nature, copyright 2018. | ||
Different from the collateral cleavage of the CRISPR system discussed above, Zhou's group developed a method named CRISDA that employed Cas9-mediated isothermal amplification. Utilizing a peptide nucleic acid (PNA) invasion-mediated endpoint measurement, CRISDA achieves attomolar sensitivity and single-nucleotide specificity in the detection of a variety of DNA targets under a complex sample background (Fig. 6b).32 Besides, other groups also developed new detection systems based on the CRISPR technology (shown in Table S2, ESI†). The significant advantages of CRISPR-based techniques include simplicity, cost-effectiveness, rapid detection, absence of complex instrumentation, and high specificity, all of which make CRISPR a promising technique for onsite virus detection.
CRISPR is an emerging technique with great potential in environmental surveillance due to its high specificity and excellent sensitivity, allowing the accurate detection of target viruses under complex background disturbance in a short period. Using a combination of different sgRNAs targeting several sites simultaneously, the CRISPR method can identify any and multiple nucleic acid sequences, potentially enabling the multiplexed detection of target viruses during environmental surveillance.
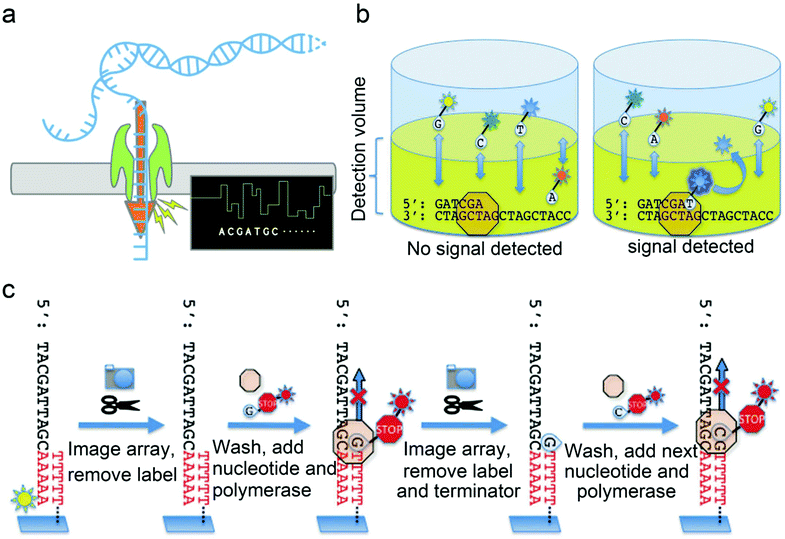 | ||
| Fig. 7 Mechanisms of next generation sequencing. (a) Nanopore sequencing utilizes nucleotides tagged with polymers in different sizes, helicase, DNA polymerase and hairpin adapter modified dsDNA. As the helicase “unzips” the dsDNA, ssDNA enters the pore causing current changes according to the specific nucleotide type. (b) Real-time single-molecule sequencing with polymerase. The zero-mode waveguide (ZMW) focuses laser energy to create an extremely small detection volume at the bottom of the ZMW where the polymerase and template molecule are localized. A mixture of all four nucleotides is added to the ZMW, each uniquely labeled with a different fluorescence moiety. As a nucleotide is incorporated, the fluorescent moiety is held within the detection volume long enough to be excited by the laser and give off a fluorescence signal which can be recorded vice versa. After the generation of the phosphodiester bond, the fluorophore is cleaved away and terminates the fluorescence signal. (c) Overview of the Helicos sequencing protocol. DNA templates are modified by fluorescently labeled 3′ poly-A tail and hybridized to poly-T oligonucleotides covalently linked to the surface of the flow cell which allows the equipment to record the initial location of each template. After the fluorescently labeled 3′ adenosine was cleaved and washed away, polymerase and a single fluorescently labeled nucleotide (A, T, G, or C) modified with a cleavable terminator residue which prevents multiple base incorporations are added to the flow cell every round of sequencing. Accurate DNA sequencing results are obtained based on the fluorescence signal. (b) and (c) were reproduced from ref. 39 with permission from MDPI, copyright 2010. | ||
Furthermore, nanopore sequencing is amplification- and fluorophore-free, which makes it a revolutionary technique with great potential in the application of onsite environmental surveillance. Shotgun metagenomic sequencing is a promising and disruptive technique in revealing unknown viruses from environmental samples. This technology analyzes all nucleic acids in a sample without bias, specifically allowing the microbial community to be detected at low abundance. Furthermore, non-target or non-selection sequencing via metagenomics also provides valuable genomic information for retrospective studies. When applying NGS methods for screening and identifying new viruses, the method of sequence-independent single primer amplification (SISPA) is normally adopted to amplify the unknown nucleotide sequence. NGS's main pitfall is the need for expensive equipment and reagents, the tedious sample preparation, and the lengthy turnaround time.
An important mission in environmental surveillance is to screen emerging viruses and track the evolutionary map of pathogenicity. It is of great significance for virology and origin tracking by knowing the viral mutation and evolution in gene sequence and expression, environmental tolerance, and viability. Analogous to discovering new chemicals in the environment, identifying emerging pathogenic viruses can be achieved through a strategy of suspect or non-target screening. The forward recognition of novel viruses is essentially a predictive prevention strategy for the potentially “unknown” epidemic. Newly recognized viruses should be further screened using criteria such as pathogenicity, infectivity, and transmissibility. Furthermore, the laboratory can research the detection method and the material in advance, aiming at the target viruses, which will accelerate the application and popularization of detection and medical technology in the event of an outbreak. The above objective can be accomplished through sequencing analysis of environmental samples, which is capable of providing comprehensive information about either a specific virus or a set of diverse viruses.
3.2 Protein based analysis
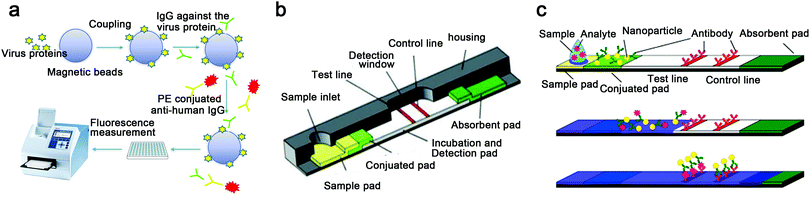 | ||
| Fig. 8 Protein based analysis. (a) The flowchart of MIA. (b) Commercial LFA and a schematic illustration of a typical test strip. (c) The detection principle of LFAs: firstly, loading aqueous sample including analytes to the sample pad; secondly, analytes binding to the conjugated tag; finally, analytes bind to the antibody of the test line to present results in color variation detectable by naked eye. (b) and (c) were reproduced from ref. 41 with permission from the Royal Society of Chemistry, copyright 2010. | ||
3.3 Virion detection
The basic requirement of the techniques used for virion characterization and detection is not to disrupt the viral integrity. The characterization of virions includes morphology, protein structure, and surface properties. In contrast, the detection of virions is normally achieved based on specific interactions between antibodies and spike proteins on the surface of CoVs.Atomic force microscopy (AFM) is a visualization technique that relies on a high-resolution scanning probe or functionalized probe to accomplish the analysis of viral properties, such as molecular interactions on the viral surface, viral isoelectric point, and viral chemistry variations under different conditions. The nanoscale characterization through AFM can be achieved in air and liquid environments, which enlightens the potential application of AFM in virion analysis in a different environmental matrix.
Aiming at the low-concentration virions in the environment, the main challenge of the above-visualized direct detection techniques and characterization is sensitivity. Further, complex sample preparation and high-cost facilities limit the practical application of this technique on a large scale. Although the direct detection of virions in environmental samples is challenging, the variation of the virus in physical and chemical properties under different environmental matrices or conditions can be investigated through the techniques described above, which is of great value in revealing the environmental behavior infection mechanism of pathogenic viruses.
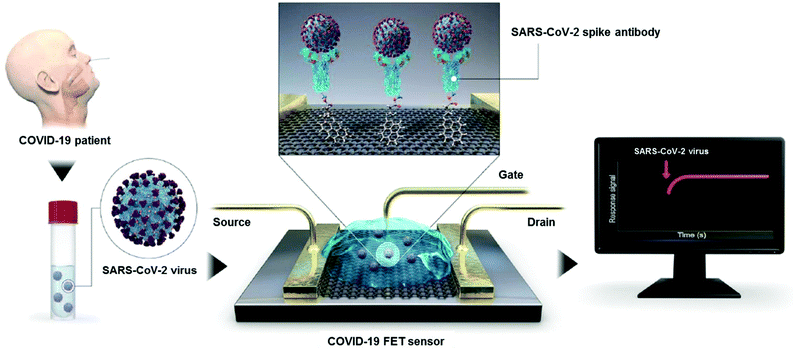 | ||
| Fig. 9 Procedure for SARS-CoV-2 detection on the FET sensor. Basically, the virions can be detected by the sensor while the device-immobilizing antibodies bind with the surface proteins of SARS-CoV-2. Reproduced from ref. 44 with permission from the American Chemical Society, copyright 2020. | ||
A surface-enhanced Raman scattering (SERS)-based portable device was reported for the onsite and in-time detection of SARS-CoV-2 in environmental water samples.45 This technique, named ACE2@SN-SERS, used the mechanism of SARS-CoV-2 entering human cells, which is a recognition and combination between the receptor-binding domain (RBD) on spike protein of SARS-CoV-2 and ACE2 of a human cell. Thus, researchers introduced the ACE2 on the fabricated silver-nanorod SERS array and analyzed the change in the Raman spectrum induced by a complex of ACE2 and SARS-CoV-2 spike protein; the device could detect SARS-CoV-2 directly in the sample. When applying ACE2@SN-SERS to the onsite detection of SARS-CoV-2 in real water samples, two positive results detected by the ACE2@SN-SERS assay turned to be negatively detected by RT-qPCR. Although there is room for further discussion and improvement, this technique has provided a potential strategy in the development of the onsite detection assay for environmental surveillance of the pathogenic virus.
The techniques of FET and SERS introduced above are capable of detecting SARS-CoV-2 particles through the combination of virus surface spike proteins and antibodies coated on sensors. However, if dissociative spike proteins are present in the detection matrix, they may generate detectable signals when bound in a sufficient number to the sensor's antibodies. Therefore, the precise quantification of virions through these techniques is currently challenging. However, these portable techniques do not require any sample pretreatment or preparation and can produce a readout in real-time. These advantages will encourage the development of fast and label-free detection techniques for the environmental surveillance of pathogens.
Quartz crystal microbalance (QCM) is a promising technique in analyzing virions. The principle of QCM is that the mass variation on the quartz crystal sensor will induce a change in the frequency of oscillation. By coating the functional substrate onto a quartz sensor, the QCM can be a promising fast technique to detect SARS-CoV-2 particles in an environmental matrix directly. Aiming at the hydrophobic and positively charged amino acid residues on spike proteins of SARS-CoV-2, Pandey discussed a potential detection strategy by functionalizing the quartz sensor's surface with hydrophobic and negatively charged groups to enable the adsorption of spike proteins of SARS-CoV-2 through electrostatic interaction.46
Flow cytometry techniques provide an alternative solution aiming at the quantitative detection of single virions. This technique can identify and analyze virions based on fluorescence signals and particle size. Yan's group has developed an analysis method targeting the single nanoparticles using a high-sensitivity flow cytometer (HSFCM). By improving the conventional flow cytometry approach, they proposed a single nanoparticle detection technique of HSFCM that enhanced the detection performance aiming at nanoparticles under 200 nm in diameter, enabling the characterization and detection of single extracellular vesicles as small as 40 nm at the detection speed of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles per minute.47 Although the HSFCM technique has yet to be applied in the analysis of single SARS-CoV-2 virions, it is potentially applicable considering the structural proteins anchored on SARS-CoV-2 particles' surface. Overall, in a biosafety laboratory with specific technicians, the quantitative detection of SARS-CoV-2 particles in either human specimens or an environmental matrix can be conducted through a reliable and high-throughput flow cytometry-based technique.
000 particles per minute.47 Although the HSFCM technique has yet to be applied in the analysis of single SARS-CoV-2 virions, it is potentially applicable considering the structural proteins anchored on SARS-CoV-2 particles' surface. Overall, in a biosafety laboratory with specific technicians, the quantitative detection of SARS-CoV-2 particles in either human specimens or an environmental matrix can be conducted through a reliable and high-throughput flow cytometry-based technique.
3.4 Viability and infectivity evaluation
Although cell culture is a traditional method, in the study of environmental CoVs, it is still the gold standard for evaluating viral viability. Using Vero E6 cells as a model host cell line, the viability of MERS-CoV, SARS-CoV-1, and SARS-CoV-2 in environmental samples has been evaluated (Table S1, ESI†). Among these studies, MERS-CoV and SARS-CoV-1 with viability were confirmed in aerosols and surface swabs from hospital ward and in the hospital wastewater, respectively (Table S1, ESI†). Additionally, the cell culture method can increase the feasibility of visualized analysis of virions through EM or AFM, as mentioned above. For instance, by inoculating aerosol samples into the cell culture medium, after days of cultivation, intact virions of MERS-CoV were identified by EM.16 When using a cell culture method for the virology study, the results could be highly affected by multiple factors, including suspending medium, initial inoculation amount, ambient temperature, and relative humidity, which suggested the requirement of standardized methodology and processes in future research.The cell culture method is time-consuming and labor-intensive, and it can be hard to optimize, which are considerable drawbacks. Techniques based on microscopy and cell culture are meaningful in virology research but are not suitable for routine large-scale environmental surveillance. The current data have confirmed the presence of SARS-CoV-2 RNA in environmental samples of aerosols, wastewater, river water, and inanimate surfaces (Table S1, ESI†). However, the viable SARS-CoV-2 determined through the cell culture method has yet been sufficiently reported in these environmental matrices. Multiple limitations resulted in the lack of viability data of environmental viruses. First of all, the analysis of viable CoVs requires a laboratory at the biosafety level of three or higher, and the availability of qualified laboratories is limited especially during the outbreak period of the epidemic. Secondly, techniques directly analyzing the viability of CoVs in original samples are not available, especially considering the low viral load in the environment compartments. Thirdly, efficient enrichment and purification of CoVs from environmental matrix while maintaining their viability are still technically challenging. Therefore, the load of viable CoVs in the actual environment has not yet been clearly profiled.
Due to the lack of viability and infectivity data, the transmission and infection of SARS-CoV-2 mediated by the environmental matrix are highly suspected rather than proved. For example, the intense discussion focused on the airborne transmission of SARS-CoV-2 is primarily caused by insufficient evidence on viral viability and infectivity in the actual air environment. Moreover, the confirmation of survivability and infectivity aimed at SARS-CoV-2 on inanimate surfaces is the basis for further study of the virion threshold and contact behavior that may cause an actual infection, and the transmission potential of the contaminated surface. Although the stability of SARS-CoV-2 in different environmental matrices was reported previously, most of the results were obtained through artificial experimental sets rather than using real environment samples, which were not sufficient for representing the realistic environmental conditions. The viability of SARS-CoV-2 is getting weak since it leaves the host, and the stability is sensitive to multiple factors, such as initial viral load, surface material, temperature, relative humidity, pH value, etc. Therefore, the attenuation of SARS-CoV-2 viability should be further studied and verified, especially in realistic environmental compartments, and standard protocols in evaluating viral viability should also be established. Overall, acquiring data about the viability and survivability of SARS-CoV-2 in various environmental conditions is an urgent objective, which may be highly achievable by developing detection techniques and evaluation methods suitable for environmental surveillance.
3.5 Integrated method for environmental surveillance
The integration of different detection techniques with outstanding performance provides opportunities for developing the applicable technology for environmental monitoring of CoVs. In particular, for the environmental surveillance, the sampling technique should be combined with the detection process. For instance, Yao's group have developed a real-time and online monitoring system aiming at the virus in aerosols, by integrating a bioaerosol-to-hydrosol sampling device with microfluidic a channel and a nanotechnology-based detection device.48 Most of techniques that have been applied to the clinical diagnosis of virus infection have potential in virus detection during environmental surveillance.Emerging in the early 1980s, microfluidics techniques can precisely manipulate and control the movements of tiny volume of fluids (normally 10−6 to 10−15 liters) in well-fabricated channels or chambers. Combining advanced biomarker detection methods (e.g., nucleic acid-based or immunology-based techniques) with microfluidics has exhibited great potential for realization of an integrated system for virus detection in POCT and environment monitoring. Microfluidics can aliquot samples into small droplets in chambers preloaded with reagents, and a parallel amplification or immunology-based analysis for different targets in their own chambers can be performed, achieving a multiplexed, automated, and high-throughput screening. Thus, microfluidics is regarded as a promising alternative to traditional methods in environmental monitoring with various advantages including, they are highly sensitive, fast, cost-effective, multiplexing and portable, and they have been applied in environment-related fields such as water quality testing, microbial identification, food spoilage detection, air screening, etc. Recently, researchers have established a portable microfluidic immunoassay system with high sensitivity and specifically for user-friendly, sensitive, quick (<15 min), multiple, and onsite detection of IgG/immunoglobulin M (IgM)/antigen of SARS-CoV-2 synchronously.49
However, in some microfluidic devices, sophisticated instrument or specialized consumables are required, which may impede their application potential in the analysis of a large number of samples or at resource-limited sites. Thus, recent advances in microfluidics are the implementation and integration of multiple components onto one single chip giving lab-on-a-chip (LOC) technologies for fully automated analysis. LOC systems combine a variety of devices with interconnected fluidic microchannel networks, valves, mixers, pumps, reaction chambers, and detectors based on different mechanisms, such as optical signals, electrochemical sensing and electronic properties. These highly integrated systems are capable of performing complex tasks in one chip such as reagent storage, fluid transport, fluid mixing, product detection, and possibly collection, without the need for any external equipment or human intervention. Thus, LOC techniques possess great advantages in fast and on-site pathogen screening (Table S2, ESI†). Researchers from the University of Arizona developed a microfluidic immunosensor system with detection limits of 1 and 10 pg mL−1 H1N1 antigens from real aerosol samples, using a spectrometer or a cell phone camera as an optical detector, respectively.50 Besides, lab-in-a-box is a highly integrated platform that combines all needed reagent synthesis and assay tools in a portable box for self-contained and mobile deployment of molecular detection, which is perfect for field virus diagnosis in developing countries or remote areas. The design of a lab-in-a-box system for virus detection has a biggest issue with integrating all parts and keeping all parts working excellently. Strictly speaking, there is no lab-in-a-box for viral diagnosis now, but researchers have put huge amounts of efforts to develop a well-performing and portable virus detection system. Microfluidic devices, and lab-on-a-chip and lab-in-a-box technology offer many advantages to on-site environmental analysis by reducing analysis time, improving detection limits, and allowing on-line, real time monitoring.
4. Perspectives and outlooks
4.1 Combination of environmental surveillance and clinical diagnosis in the prevention and control of a viral epidemic
An integrated approach combining environmental surveillance with clinical diagnosis could capture the emergence and evolution of an epidemic early on, thus predicting and revealing the development of viral epidemics.Compared with passive clinical diagnosis and screening without a specific symptom clue, the initiative to monitor pathogenic viruses in the environment under the guidance of a sampling and detection strategy is more precise and timely for preventing a viral epidemic. However, the occurrence of a pathogenic virus in the environment may not be enough to estimate the prevalence of an infectious disease, which needs to be further confirmed through clinical diagnosis and observation. Therefore, as a complementary tool, the environmental surveillance of CoVs will buy time for the preparation of clinical diagnosis and provide information to target the clinical symptoms. The combination of environmental surveillance and clinical diagnosis may be a viable approach to prevent and control an infectious disease due to the infection of CoVs before developing into an epidemic.
Although further verification is required, the correlation between a viral load in environmental matrices and the infected population may be a feasible direction in enhancing epidemiological prediction. In this perspective, environmental surveillance and clinical diagnosis should be integrated in terms of data sharing, analyzing, and modeling. The data collection should be conducted under the guidance of a unified methodology such as detecting CoVs in clinical specimens from different disease courses and the detection of CoVs in various environmental matrices from reasonable sites at multiple time points. The clear temporal relationship should be established among the infected individuals' clinical periods, CoVs in different clinical specimens, and CoVs in different environmental matrices. Based on sufficient and reliable data, epidemiological modeling can be conducted with confidence. It is possible to provide predictions aimed at the ongoing stage, change time-points, infected populations, diffusion scales, and future trends of an epidemic.
4.2 Challenges in virus detection and practical application
Due to the CoV-induced epidemics or pandemic in the first 20 years of the new century, virus detection techniques and products are being developed and emerging at an unprecedented speed. The function of environmental surveillance in viral monitoring and epidemic prevention is gaining more attention and is being discussed with great passion. How to apply the novel detection technology to environmental surveillance and promote the practical application of detection methods? This is a common challenge for multidisciplinary scientists and engineers. However, the existence of challenges likewise provides opportunities.Principles and methods based on chemistry have been involved in every aspect of environmental surveillance and environmental virology, including but not limited to material chemistry, interfacial chemistry, biological chemistry, and analytical chemistry. Therefore, to address current challenges in detection techniques, further researches towards multiple chemistry-related disciplines have to be strengthened. Here we propose critical guidelines in detection techniques, and their application potentials in environmental surveillance from the perspective of global surveillance and data sharing aimed at pathogenic CoVs: (1) the guarantee of sensitivity and reliability is always the priority of the detection method. The low abundance of CoVs in the environment and the complex matrix effect are the primary challenges for environmental detection. The false-positive or false-negative results in virus detection greatly hinders the control of an epidemic. Therefore, the acceptable detection limit and accuracy of the virus detection method should be achieved before its application in field monitoring. (2) Quantitative analysis of CoVs. The simple dichotomous data of positive and negative will not be enough for future research in environmental virology and epidemiology. Reliable quantitative data of specific viruses are essential for data comparison and sharing among different studies from global laboratories. When the CoVs loads in the environment are used to predict epidemiological trends, accurate quantitative data are essential. (3) The confirmation of viability. The presence of viral RNA in the environment is not always equivalent to the threat of pathogenicity and infectivity. Viability analysis would provide more information about the transmissibility and infectiousness of a virus. By confirming the viability of CoVs in the environment with the consideration of a potential transmission route, the probability and degree of the CoV-induced epidemic are predictable. Subsequently, the response level can be confirmed. (4) The integrated and automatic workflow for live detection and online data transfer is imperative in the future. Wide-ranging environmental surveillance of infectious CoVs in an adequate number of sites and during a sufficient period could provide the necessary data for in-depth research and accurate prediction in environmental virology and epidemiology. Therefore, an integrated technology that can automatically complete the onsite and in situ detection, in-time data analysis, and online data transfer will greatly improve big data collection, sharing, and computing. (5) The establishment of environmental sample banks and databases by standardizing sampling, pretreatment, analysis, and preservation of a specific target. The standard-agreed method and criteria-certified data will be of great help in establishing a global surveillance system aimed at pathogenic viruses. Once the novel CoV is identified in the future, a retrospective analysis is crucial to understand its origin, evolution, and pathogenicity. Further, along with the development of virus analysis technology and molecular biology, unexpected information can be obtained using new techniques and strategies to reanalyze the stored samples from sample banks. (6) Research collaboration and data sharing in different expertise fields and regions is imperative for future viral threats. The study of pathogenic CoVs, including pathogenesis, survivability, environmental transmission, variation, and mutation, among other features, is not independent of others. Furthermore, the geographical conditions, climatic factors, and public health foundations differ significantly in different regions. Global data sharing is especially desired for epidemiological research aimed at pathogenic viruses.
An ideal detection technique applied to environmental surveillance should balance sensitivity, accuracy, live detection, real-time data accessibility, portability, simplicity, and cost-effectiveness. Importantly, a certain compromise aimed at some specific property can be made depending on the different field surveillance purposes. Two examples illustrate this point: (1) when the goal of virus detection is the early warning of an epidemic, a sensitivity just below the alert threshold is acceptable, which is prudently set by simultaneously considering the outbreak potential and the unnecessary social panic based on epidemiological and virological data; and (2) when online monitoring and data transfer are available, the portability will not be the pre-requisite condition. In conclusion, the future of virus detection technology and its application in environmental surveillance rely on the collaboration of scientists and engineers as well as the integration of multidisciplinary solutions. This review is expected to inspire researchers interested in the topic by introducing the current situation profile and identifying the main challenges ahead.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21527901, 92043302, 92043000), the Science and Technology Innovation Commission of Shenzhen (JCYJ20180507182047316), and the Youth Innovation Promotion Association of Chinese Academy of Sciences.References
- A. Zumla, J. F. Chan, E. I. Azhar, D. S. Hui and K. Y. Yuen, Nat. Rev. Drug. Discovery, 2016, 15, 327–347 CrossRef CAS PubMed.
- A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng and T. Jiang, Cell Host Microbe, 2020, 27, 325–328 CrossRef CAS.
- L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, J. Xia, Q. Guo, H. L. Yen and J. Wu, N. Engl. J. Med., 2020, 382, 1177–1179 CrossRef PubMed.
- V. Stadnytskyi, C. E. Bax, A. Bax and P. Anfinrud, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11875–11877 CrossRef CAS PubMed.
- Y. Xu, X. Li, B. Zhu, H. Liang, C. Fang, Y. Gong, Q. Guo, X. Sun, D. Zhao, J. Shen, H. Zhang, H. Liu, H. Xia, J. Tang, K. Zhang and S. Gong, Nat. Med., 2020, 26, 502–505 CrossRef CAS PubMed.
- I. T. S. Yu, Y. G. Li, T. W. Wong, W. Tam, A. T. Chan, J. H. W. Lee, D. Y. C. Leung and T. Ho, N. Engl. J. Med., 2004, 350, 1731–1739 CrossRef CAS.
- N. van Doremalen, T. Bushmaker, D. H. Morris, M. G. Holbrook, A. Gamble, B. N. Williamson, A. Tamin, J. L. Harcourt, N. J. Thornburg, S. I. Gerber, J. O. Lloyd Smith, E. de Wit and V. J. Munster, N. Engl. J. Med., 2020, 382, 1564–1567 CrossRef PubMed.
- S. W. X. Ong, Y. K. Tan, P. Y. Chia, T. H. Lee, O. T. Ng, M. S. Y. Wong and K. Marimuthu, JAMA, 2020, 323, 1610–1612 CrossRef CAS PubMed.
- Y. Liu, Z. Ning, Y. Chen, M. Guo, Y. Liu, N. K. Gali, L. Sun, Y. Duan, J. Cai, D. Westerdahl, X. Liu, K. Xu, K. F. Ho, H. Kan, Q. Fu and K. Lan, Nature, 2020, 582, 557–560 CrossRef CAS PubMed.
- P. Y. Chia, K. K. Coleman, Y. K. Tan, S. W. X. Ong, M. Gum, S. K. Lau, X. F. Lim, A. S. Lim, S. Sutjipto, P. H. Lee, T. T. Son, B. E. Young, D. K. Milton, G. C. Gray, S. Schuster, T. Barkham, P. P. De, S. Vasoo, M. Chan, B. S. P. Ang, B. H. Tan, Y. S. Leo, O. T. Ng, M. S. Y. Wong, K. Marimuthu and Singapore Novel Coronavirus Outbreak Research Team, Nat. Commun., 2020, 11, 2800 CrossRef CAS PubMed.
- J. A. Lednicky, S. N. Shankar, M. A. Elbadry, J. C. Gibson, M. M. Alam, C. J. Stephenson, A. Eiguren-Fernandez, J. G. Morris, C. N. Mavian, M. Salemi, J. R. Clugston and C. Y. Wu, Aerosol Air Qual. Res., 2020, 20, 1167–1171 CrossRef CAS PubMed.
- L. Setti, F. Passarini, G. De Gennaro, P. Barbieri, M. G. Perrone, M. Borelli, J. Palmisani, A. Di Gilio, V. Torboli, F. Fontana, L. Clemente, A. Pallavicini, M. Ruscio, P. Piscitelli and A. Miani, Environ. Res., 2020, 188, 109754 CrossRef CAS PubMed.
- S. G. Rimoldi, F. Stefani, A. Gigantiello, S. Polesello, F. Comandatore, D. Mileto, M. Maresca, C. Longobardi, A. Mancon, F. Romeri, C. Pagani, F. Cappelli, C. Roscioli, L. Moja, M. R. Gismondo and F. Salerno, Sci. Total Environ., 2020, 744, 140911 CrossRef CAS PubMed.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Environ. Sci. Tech. Lett., 2020, 7, 511–516 CrossRef CAS.
- A. Bogler, A. Packman, A. Furman, A. Gross, A. Kushmaro, A. Ronen, C. Dagot, C. Hill, D. Vaizel-Ohayon, E. Morgenroth, E. Bertuzzo, G. Wells, H. R. Kiperwas, H. Horn, I. Negev, I. Zucker, I. Bar-Or, J. Moran-Gilad, J. L. Balcazar, K. Bibby, M. Elimelech, N. Weisbrod, O. Nir, O. Sued, O. Gillor, P. J. Alvarez, S. Crameri, S. Arnon, S. Walker, S. Yaron, T. H. Nguyen, Y. Berchenko, Y. Hu, Z. Ronen and E. Bar-Zeev, Nat. Sustain., 2020, 3, 981–990 CrossRef.
- S. H. Kim, S. Y. Chang, M. Sung, J. H. Park, H. Bin Kim, H. Lee, J. P. Choi, W. S. Choi and J. Y. Min, Clin. Infect. Dis., 2016, 63, 363–369 CrossRef PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins, J. W. O'Brien, P. M. Choi, M. Kitajima, S. L. Simpson, J. Li, B. Tscharke, R. Verhagen, W. J. M. Smith, J. Zaugg, L. Dierens, P. Hugenholtz, K. V. Thomas and J. F. Mueller, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS PubMed.
- X. W. Wang, J. S. Li, T. K. Guo, B. Zhen, Q. X. Kong, B. Yi, Z. Li, N. Song, M. Jin, X. M. Wu, W. J. Xiao, X. M. Zhu, C. Q. Gu, J. Yin, W. Wei, W. Yao, C. Liu, J. F. Li, G. R. Ou, M. N. Wang, T. Y. Fang, G. J. Wang, Y. H. Qiu, H. H. Wu, F. H. Chao and J. W. Li, World J. Gastroenterol., 2005, 11, 4390–4395 CrossRef CAS PubMed.
- W. Randazzo, P. Truchado, E. Cuevas-Ferrando, P. Simon, A. Allende and G. Sanchez, Water Res., 2020, 181, 115942 CrossRef CAS PubMed.
- B. A. Kocamemi, H. Kurt, S. Hacioglu, C. Yarali, A. M. Saatci and B. Pakdemirli, medRxiv, 2020 DOI:10.1101/2020.05.03.20089417.
- H. Green, M. Wilder, M. Collins, A. Fenty, K. Gentile, B. L. Kmush, T. Zeng, F. A. Middleton and D. A. Larsen, medRxiv, 2020 DOI:10.1101/2020.05.21.20109181.
- S. Faridi, S. Niazi, K. Sadeghi, K. Naddafi, J. Yavarian, M. Shamsipour, N. Z. S. Jandaghi, K. Sadeghniiat, R. Nabizadeh, M. Yunesian, F. Momeniha, A. Mokamel, M. S. Hassanvand and T. MokhtariAzad, Sci. Total Environ., 2020, 725, 138401 CrossRef CAS PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- P. Pokhrel, C. Hu and H. Mao, ACS Sens., 2020, 5, 2283–2296 CrossRef CAS PubMed.
- G. Adams, Biochemist, 2020, 42, 48–53 CrossRef CAS.
- B. Vogelstein and K. W. Kinzler, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS PubMed.
- S. P. Yip, S. S. T. To, P. H. M. Leung, T. S. Cheung, P. K. C. Cheng and W. W. L. Lim, Clin. Chem., 2005, 51, 1885–1888 CrossRef CAS PubMed.
- S. A. Bustin, V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl, G. L. Shipley, J. Vandesomepele and C. T. Wittwer, Clin. Chem., 2009, 55, 611–622 CrossRef CAS PubMed.
- T. Suo, X. Liu, J. Feng, M. Guo, W. Hu, D. Guo, H. Ullah, Y. Yang, Q. Zhang, X. Wang, M. Sajid, Z. Huang, L. Deng, T. Chen, F. Liu, K. Xu, Y. Liu, Q. Zhang, Y. Liu, Y. Xiong, G. Chen, K. Lan and Y. Chen, Emerg. Microbes Infect., 2020, 9, 1259–1268 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- F. Zhang, O. O. Abudayyeh and J. S. Gootenberg, A protocol for detection of COVID-19 using CRISPR diagnostics, 2020, 8 Search PubMed.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X. F. Yu, Nat. Commun., 2018, 9, 1–11 CrossRef PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D. K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- A. Böhmer, V. Schildgen, J. Lusebrink, S. Ziegler, R. L. Tillmann, M. Kleines and O. Schildgen, J. Virol. Methods, 2009, 158, 199–201 CrossRef PubMed.
- A. Alhassan, Z. Li, C. B. Poole and C. K. Carlow, Trends Parasitol., 2015, 31, 391–400 CrossRef PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- W. Feng, A. M. Newbigging, C. Le, B. Pang, H. Peng, Y. Cao, J. Wu, G. Abbas, J. Song, D. B. Wang, M. Cui, J. Tao, D. L. Tyrrell, X. E. Zhang, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 10196–10209 CrossRef CAS PubMed.
- L. Varadi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Chem. Soc. Rev., 2017, 46, 4818–4832 RSC.
- M. W. Anderson and I. Schrijver, Genes, 2010, 1, 38–69 CrossRef CAS PubMed.
- J. Du, E. Chu, D. Zhang, C. M. Lu, A. Zhang and Y. S. Michael, J. Virol. Methods, 2020, 287, 114009 CrossRef PubMed.
- D. Mark, S. Haeberle, G. Roth, F. von Stetten and R. Zengerle, Chem. Soc. Rev., 2010, 39, 1153–1182 RSC.
- F. M. Nachtigall, A. Pereira, O. S. Trofymchuk and L. S. Santos, Nat. Biotechnol., 2020, 38, 1168–1173 CrossRef CAS PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S. H. Baek, M. Choi, K. B. Ku, C. S. Lee, S. Jun, D. Park, H. G. Kim, S. J. Kim, J. O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- D. Zhang, X. Zhang, R. Ma, S. Deng, X. Wang, X. Zhang, X. Huang, Y. Liu, G. Li, J. Qu, Y. Zhu and J. Li, medRxiv, 2020 DOI:10.1101/2020.05.02.20086876.
- L. M. Pandey, Expert Rev. Proteomics, 2020, 17, 425–432 CrossRef CAS PubMed.
- Y. Tian, L. Ma, M. Gong, G. Su, S. Zhu, W. Zhang, S. Wang, Z. Li, C. Chen, L. Li, L. Wu and X. Yan, ACS Nano, 2018, 12, 671–680 CrossRef CAS.
- F. Shen, M. Tan, Z. Wang, M. Yao, Z. Xu, Y. Wu, J. Wang, X. Guo and T. Zhu, Environ. Sci. Technol., 2011, 45, 7473–7480 CrossRef CAS PubMed.
- Q. Lin, D. Wen, J. Wu, L. Liu, W. Wu, X. Fang and J. Kong, Anal. Chem., 2020, 92, 9454–9458 CrossRef CAS PubMed.
- H. J. Kwon, C. F. Fronczek, S. V. Angus, A. M. Nicolini and J. Y. Yoon, J. Lab. Autom., 2014, 19, 322–331 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cs00595a |
| ‡ These authors contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2021 |