Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exotic nuclear spin behavior in dendritic macromolecules†
Philip
Saul
 ab,
Shengjun
Yang‡
ab,
Salvatore
Mamone
ab,
Felipe
Opazo
ab,
Shengjun
Yang‡
ab,
Salvatore
Mamone
ab,
Felipe
Opazo
 bc,
Andreas
Meyer
d,
Silvio O.
Rizzoli
c and
Stefan
Glöggler
bc,
Andreas
Meyer
d,
Silvio O.
Rizzoli
c and
Stefan
Glöggler
 *ab
*ab
aResearch Group for NMR Signal Enhancement, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany. E-mail: stefan.gloeggler@mpibpc.mpg.de; Tel: +49 551 3961108
bCenter for Biostructural Imaging of Neurodegeneration, Von-Siebold-Straße 3A, 37075 Göttingen, Germany
cInstitute for Neuro- and Sensory Physiology, University Medical Center Göttingen, Humboldtallee 23, 37073 Göttingen, Germany
dResearch Group Electron Paramagnetic Resonance, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany
First published on 12th November 2021
Abstract
Dendrimers are a class of branched, highly symmetric macromolecules that have been shown to be useful for a vast number of different applications. Potential uses as fluorescence sensors, in catalysis and perhaps most importantly in medical applications as drug delivery systems or cytotoxica have been proposed. Herein we report on an exotic behaviour of the nuclear spins in a dendritic macromolecule in the presence of different paramagnetic ions. We show that the stability of the long lived nuclear singlet state, is affected by the presence of Cu(II), whereas other ions did not have any influence at all. This effect could not be observed in the case of a simple tripeptide, in which the nuclear singlet stability was influenced by all investigated paramagnetic ions, a potentially useful effect in the development of Cu(II) selective probes. By adding a fluorescent marker to our molecule we could show that the nuclear singlet multimer (NUSIMER) is taken up by living cells. Furthermore we were able to show that nuclear singlet state NMR can be used to investigate the NUSIMER in the presence of living cells, showing that an application in in vivo NMR can be feasible.
Introduction
Dendrimers are large, centrosymmetric, highly branched macromolecules that can be easily modified at the end of each branch by different means of chemical synthesis.1 Since the first report on dendritic structures,2 dendrimers quickly raised the interest of researchers in different fields and a large number of different possible applications have been proposed, ranging from the use as effective filtering systems3 over the use as chemical sensors4,5 to chemical catalysis.6,7 In the latter two cases, the high number of terminal functional groups in dendrimers drastically increases the effectiveness in the respective applications, since they allow for a large number of active residues to be condensed in one molecule and thus in the same area. Apart from the aforementioned possibilities, dendrimers have been investigated especially in the field of medical research. Cavities in the dendritic structures allow for the use as drug delivery systems by encapsulating different compounds.8,9 Additionally, dendrimer based cytotoxica have been introduced by attaching small cytotoxic compounds to dendrimers, again accumulating a high number of active compounds in the same place.10–12A fairly recent proposal for the use of dendrimers are nuclear singlet multimers (NUSIMERs).13 This approach makes use of the accumulation of up to 128 equivalent residues at the branches of a poly(amidoamide) dendrimer of the 5th generation (G5-PAMAM) in combination with nuclear singlet state NMR. A nuclear spin singlet state requires two spin-12 nuclei coupling with each other. While the triplet state with a total spin of 1, can be observed in regular NMR experiments, the singlet state with a total spin of 0 is NMR silent.14 Nuclear singlet states have been shown to have lifetimes often exceeding the spin–lattice relaxation T1. This particular property allows for the transfer of magnetization from the triplet state to the singlet state, in which the spin order can be stored and, at a later point, retrieved again.14–44 A combination of the transfer of magnetization to singlet state order and back with a singlet filter allows for selective observation of just the proton pair in which a singlet state has been populated, thus eliminating all background signal, including the solvent signal.13,44,45 For a better understanding of the behaviour of the nuclear spins, this phenomenon has been thoroughly investigated in perfect environments, such as low viscosity, perdeuterated solvents34 or under the exclusion of oxygen in order to minimize singlet leakage and thus extending the singlet state lifetime.15,21,33 The fact that the equilibration time between the NMR silent nuclear singlet state and the NMR active triplet state (Ts) oftentimes exceeds T120–23 leads to the possibility to analyze dynamics and has already been used to probe structures of proteins and to investigate drug binding as well as diffusion phenomena41–44,46,47 or even the selective detection of different amino acids in the same protein in vitro.48 However, most of those experiments have been carried out in deuterated solvents, degassed samples or with partially deuterated compounds as well. In few studies it could be shown that long-lived nuclear singlet states can be used in vitro in non-deuterated solvents and in near physiological conditions,13,44 or even in vivo in magnetic resonance spectroscopy to filter metabolic signals.49 In the latter example it could be shown that singlet filtering is possible in MRI experiments. Especially in this context, the inherently low sensitivity of NMR can be somewhat overcome by the accumulation of a large number of chemically equivalent proton pairs in the same molecule, for which the use of dendrimers is exceptionally well suited. The fact that dendritic structures have been shown to be a valid concept in the development of probes, lead us to investigate the possibilities to use dendritic structures to probe for paramagnetic ions and furthermore explore possibilities to use NUSIMERs in in-cell experiments.
General concept
A nuclear singlet multimer (NUSIMER) G5-PAMAM-GGA-NH2, is a poly(amidoamide) dendrimer of the 5th generation (G5-PAMAM) which has been modified by attaching the glycin–glycin–alanin tripeptide (GGA) (1) to it to obtain a macromolecule (2) that contains up to 128 proton pairs per molecule with the same chemical shift dispersion in which a nuclear singlet state can be populated13 (Fig. 1). Due to the high density of chemically equivalent proton pairs in which a singlet state can be populated, the use of NUSIMERs can greatly increase the sensitivity and NUSIMERs can be detected at low concentrations in reasonably quick experiments. The presence of amide and amine functions within the structure as well as at the end of each branch, makes them excellent candidates for the interaction with metal ions and thus the potential use as probes for metal ions.In a first set of experiments we investigated the effect of paramagnetic metal ions on T1−1 and Ts−1 in (1) and (2) to assess the possibility to use NUSIMERs to sense certain ions. In an investigation of the effect of some paramagnetic ions on T1 and Ts in a simple glycine–alanine dipeptide, it has been shown that the singlet state is less sensitive to paramagnetic relaxation than the conventional nuclear magnetization.40 This property can be useful in increasing the specificity compared to the standard NMR experiment. Metal ions play a pivotal role in many biological processes. Especially Cu(II) has gained some attention, since high Cu(II) concentrations are associated with the formation of Aβ-plaques which can be found in Alzheimer's disease patients.50 Furthermore, we investigated the effect of Fe(III), Gd(III), Mn(II) and Zn(II) on T1−1 and Ts−1.
To further explore the possibility to use NUSIMERs for the detection of said ions in a biological environment, we decided to investigate, if NUSIMERs can be viable for in vivo experiments in cells. In order to be able to trace the NUSIMER, compound (2) has been modified by attaching a fluorescent tag (Atto488) to it to obtain G5-PAMAM-GGA-Atto488 (3) (Fig. 1).
Results and discussion
Influence of paramagnetic metal ions on T1−1 and Ts−1
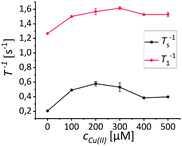
In order to simulate the environment most likely to be relevant in potential in vivo applications, samples of (2) have been prepared in aqueous HEPES buffered solutions (10 mM) and have been titrated with Gd(III), Mn(II), Fe(III), Cu(II) and Zn(II) respectively (experiments conducted at 600 MHz). Zn(II) is diamagnetic but also an important disease marker and was hence included in our investigation. After each addition of the respective ions T1−1 and Ts−1 have been determined (T1−1 plots are provided in the ESI†). We performed the same experiments with (1) to correlate the obtained data (Fig. 2).For (1) we observed changes in T1−1 and Ts−1 in a similar relation in the presence of paramagnetic ions as has been shown before.40 Notably, in the case of (1) in combination with Fe(III) we observed an effect on T1−1 and Ts−1 which can be explained by the slow complexation of Fe(III) by the HEPES buffer. The complexation of Fe(III) by different Goods buffers has been reported previously.51 We investigated this complexation more thoroughly by determining T1 and Ts in a sample of (1) in HEPES buffer at different times after the addition of Fe(III) (150 μM). We could observe a slow increase in T1 and Ts respectively (Fig. 2f). To show that this is an effect of the buffer as well as exclude any effect of the buffer in all cases, we investigated the observed phenomena in unbuffered D2O solutions of the paramagnetic ions, whereby the pD remained unchanged and independent of the ion concentration. These investigations have been conducted at a lower magnetic field (300 MHz) to exclude field dependency effects. We found that T1−1 and Ts−1 of the peptides and dendrimer investigated display a similar behavior as in the buffer, with Fe(III) following the expected behavior as it does not form a complex (Fig. 3a). In case of (2) we could not observe any effect on either Ts−1 or T1−1 except in the case of Cu(II) (Fig. 2d). This effect of Cu(II) is especially apparent at concentrations between 200 μM and 400 μM, which are the concentrations reported to be found in Aβ aggregates as well.50 If one closely examines the effect of copper on Ts−1, an increase in the singlet stability rate is experienced at first which reaches a minimum at 150 μM copper concentration. If the copper concentration is further increased, Ts−1 decreases as well. For copper we investigated if we can exclude effects of the buffer or if we, similarily to iron, see an interaction of buffer with the ions. Upon addition of copper to the dendrimer we observe in D2O that Ts−1 starts subceeding the initial value found in the dendrimer, when no ions had been added. We did not observe this behavior if T1−1 is investigated. T1−1 increases with copper concentration and reaches a plateau and did not go down again within the error over the investigated range.
We tried to confirm this behaviour at a higher magnetic field and observed a similar effect in D2O at 600 MHz, although Ts−1 did not subceed the initial value. The same effect could be observed for T1−1 (Fig. 4).
For an explanation of the effect of Cu(II) on Ts−1 we propose a direct influence of the ions on the constitution of (2). In dendrimers, typically three possible mechanisms are considered how ions can influence the constitution. They can interact with the chains, be incorporated into the center of the dendrimer and may be influenced by electrostatic repulsion of charged parts of the dendrimer and ions.52–55 Once copper is added, we observe complexation of Cu(II) to the amide functions at first. This interaction of Cu(II) with the amide functions is confirmed in UV/vis experiments (see ESI†). At a certain copper concentration, a saturation of the amide functions at the outer sphere seems to occur. With a further increase in copper concentration we hypothesize that the chain mobility further increases whereby no further copper ions have an effect on the singlet state equilibration. As a result, Ts−1 would become shorter due to an ongoing decrease in correlation time. A similar behaviour has been described for the interaction of the inner branches of a poly(amidoamide) dendrimer of the 4th generation with Cu(II).56
In order to verify this hypothesis, we performed diffusion-ordered spectroscopy (DOSY) measurements to analyze the change of chain mobility at increasing copper concentrations and how T1−1 and Ts−1 are influenced. A first result that we found is that the diffusion coefficient (D) of the glycine protons in the dendrimer increases linearly with the copper concentration (Fig. 5a). At the same time Ts−1 in the dendrimers increases sharply and then starts to decrease again. This effect, can be observed at 150 μM Cu(II) in Ts−1 whereby a plateau is reached for T1−1 at similar concentrations within the error. The increase in Cu(II) concentration did not show any effect on the diffusion coefficient of the water signal (data shown in the ESI†), showing that no change in viscosity takes place. Thus a contribution of a possible viscosity change can be excluded, as can be an effect of the buffer or the solvent, due to the effect not being observed in the case of the tripeptide GGA in buffered and unbuffered solutions (see ESI†).
To further investigate the kind of interaction between Cu(II) and (2), we recorded electron paramagnetic resonance (EPR) spectra of Cu(II) (500 μM), GGA in the presence of Cu(II) (500 μM), G5-PAMAM with 500 μM Cu(II) as well as (2) with 500 μM Cu(II) (Fig. 5). Additionally, simulations of the experimental spectra have been performed.
In a first experiment the EPR spectrum of Cu(II), or rather hexaaqua-d12 Cu(II), in D2O was recorded as a reference spectrum. This spectrum showed to be identical to the obtained spectrum of Cu(II) in D2O in the presence of GGA, indicating that no complexation of Cu(II) by GGA takes place (Fig. 5b). This observation has also been confirmed by UV/VIS measurements (see ESI†). The measurements for Cu(II) in the presence of G5-PAMAM and (2) (Fig. 5c and d) display different spectra not only to the one of hexaaqua Cu(II), with a gz of 2.4, but also to one another. Performed simulations indicated a ligation by the nitrogen atoms of the dendrimer and the Cu(II) with a gz at around 2.25 which is significantly different than the one of the hexaaqua complex of Cu(II) and characteristic for Cu(II)–N complexes.57–60 In both cases the shape of the EPR spectra were independent of the Cu(II) concentration. The simulations point towards a tetragonal symmetry around the Cu(II) in the presence of G5-PAMAM, with equal gx and gy, whereas in the presence of (2) a rhombic symmetry is obtained from the simulation where gx, gy and gz are all different from one another. It is, however, likely that the experimental spectra contain contributions from different Cu(II) complexes as has been shown to be the case in the presence of G4-PAMAM.56 The parameters obtained from the simulations are shown in the ESI.†
We herein propose a model in which, upon the first addition of Cu(II) to the dendrimer, a coordination of Cu(II) to the terminal amino functions as well as a coordination to the amide functions of the tripeptide residues occurs. This leads to an increase in the diffusion coefficient of the glycine residues and thus to an increased chain mobility. However upon further addition of Cu(II) the chain mobility of the tripeptide residues further increases and a decrease in Ts−1 takes place. The first increase in Ts−1 is likely induced by the binding of copper to the amide functions of the tripeptide resdiues and the thus increased proximity to the glycine protons. The subsequent decrease in Ts−1 then can be attributed to the further increasing mobility of the protons. The decrease in Ts−1 is likely due to a difference in the distance dependency of T1−1 and Ts−1 of the dipole–dipole relaxation (r6 for T1−1 and r8 for Ts−1)61 which might indicate a saturation of the outer sphere of the dendrimer with Cu(II) which then does only allow further Cu(II) to have an effect on T1−1 since it cannot get any closer to the relevant protons. Any further Cu(II) is likely incorporated into the dendritic core, an effect that has been made use of e.g. in dendrimer entrapped nanoparticles.52,62
At the point of the incorporation of Cu(II) into the dendritic core, a decrease in hydrodynamic radius of the dendrimers is likely to occur. Due to the difference in chain mobility a direct correlation of the diffusion coefficient determined by DOSY NMR and the hydrodynamic radius is challenging.63 For further insight into a possible change in size of the dendrimers, dynamic light scattering (DLS) measurements have been performed (see ESI†). The size of G5-PAMAM-GGA was thus determined to be around 3.5 nm which is in the order of magnitude of the determined size of G5-PAMAM.55 However no significant change in particle size upon the addition of Cu(II) could be determined. This indicates that Cu(II) at low concentrations, mainly interacts with the outer branches of the dendrimer, influencing their mobility, while the impact on the particle size seems to be minimal, if at all present.
The binding hypothesis is supported by the observation of a change in color upon the addition of (2) to a solution of Cu(II)Cl2 from a nearly colorless light blue to a considerably darker blue (see Fig. S24 in the ESI†). As dendrimers represent a complex system, the proposed mechanism above may serve as another step to support ongoing work on solving the structures of dendritic molecules. As already described above, this change is observed by UV/vis spectroscopy showing a change in absorption bands typically associated with amide bonds in proteins upon addition of Cu(II) to a solution of (2). We did not observe this change in the case of Gd(III), Mn(II) and Fe(III) (See Fig. S12–S23 in the ESI†).
Assessment of NUSIMERs as potential bioprobes
The observed selective effect of Cu(II) on T1−1 and Ts−1 gives rise to a possible application of the NUSIMERs as Cu(II) probes, especially, but not limited to, the detection of Cu(II) in cells or tissue. This is especially interesting due to the central role of Cu(II) in the formation of Aβ plaques. We would like to note that changes in Ts are already sensed below 100 μM copper concentrations which is well in the region in what is found in extracellular Aβ plaques. In order to test the viability of future tissue experiments, we performed additional measurements of a solution of (2) in an agarose gel which simulates the consistency of brain matter.64 In this case, two experiments have been performed: In both cases, the dendrimers were measured at a concentration of 10 μM in gel which consisted of a HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffered solution. One of the samples additionally contained 50 μM of Cu(II). Relaxation rate experiments gave a T1−1 of 0.016 s−1 and a Ts−1 of 2.42 ± 0.06 s for the sample without Cu(II) and T1−1 of 2.32 s−1 and a Ts−1 of 0.69 s−1 for the sample with Cu(II). These findings indicate that the same effect thoroughly studied in 100 μM concentrations in D2O can be observed in lower concentrations in a high viscosity environment and in the presence of buffered H2O solutions. Ts−1 appears thereby to be less sensitive to the presence of copper than T1−1. Those findings strongly indicate that detection of Cu(II) using NUSIMERs is possible in tissue.In order to evaluate the possibility to use NUSIMERs in cell experiments, we introduced a fluorescence tag (Atto488) to (2) to obtain the fluorescently labelled NURSIMER (3). We found an average of 5 fluorescence tags per molecule. At first, we have studied the molecular parameters T1−1 and Ts−1 of (3) in D2O and protonated phosphate saline buffer (PBS) to assess the viability for cell experiments (Table 1). We have discovered that (3) does not display a change in Ts−1 in protonated buffer with respect to the parent structure (2). In all cases, the equilibration rate Ts−1 is shorter than T1−1 and therefore represents an excellent starting point not just to filter signals but also to probe molecular dynamics in biological systems. Table 1 summarizes the results.
| Compound | Solvent | T 1 −1 [s−1] | T s −1 [s−1] |
|---|---|---|---|
| G5-PAMAM-GGA-NH2 (2) | D2O | 2.04 ± 0.04 | 0.14 ± 0.01 |
| G5-PAMAM-GGA-NH2 (2) | PBS | 2.85 ± 0.09 | 0.6 ± 0.06 |
| G5-PAMAM-GGA-Atto488 (3) | D2O | 1.4 ± 0.04 | 0.32 ± 0.03 |
| G5-PAMAM-GGA-Atto488 (3) | PBS | 1.54 ± 0.09 | 0.55 ± 0.01 |
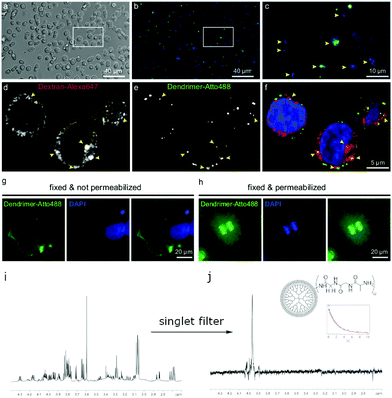
The observation that a long-lived state can be populated even in aqueous buffered solutions encouraged us to further investigate this behavior in the presence of cells by using fluorescence microscopy and the detection of long-lived states. At first, we have successfully tested the uptake of (3) into living human RAMOS B-cell line, in aldehyde-fixed COS-7 fibroblast cells, and in primary rat hippocampal neurons (see ESI†).65 Those first results included the population of a nuclear singlet state in the presence of lysed RAMOS cells in PBS buffered solutions. In another experiment RAMOS B-cells and COS-7 fibroblasts have been co-incubated with (3) as well as Dextran coupled to Alexa647 to determine if (3) are endocytosed in the same fashion as this classical endocytosis marker (Fig. 6).66
From the obtained images the internalization of (3) into the cells as well as the localization in the nucleus and the cell membranes could be shown. For subsequent NMR studies, we have focused on B-cells since they are cells growing in suspension. We supplied (3) to a pellet of cells and measured Ts. The performed NMR experiments on the B-cells after addition of (3) showed close to no change in Ts which indicates that (3) is stable in the presence of cells. Upon cell lysis, more macromolecules tend to bind to the cell membranes and nuclei. We confirmed this behavior by investigating COS-7 fibroblasts. An observed broadening of the NMR-signal of (3) supports the binding hypothesis. Despite the broadening, the signal observation of (3) in the presence of lysed cells was possible using a singlet NMR sequence45 with an additional singlet filter.67 This filtering experiment allows for direct observation of the glycine protons in (3) while suppressing other proton signals from cell membranes and water with a limit of detection of 6 μM concentrations of (3) in a single scan at 900 MHz. After extensive washing of the lysed cells, we have still observed (3) due to the possibility of populating singlet states. Additionally, we confirmed the binding by observing an orange coloring of the cells and solid state NMR experiments (see ESI†). Further experiments on cytosol of B-cells spiked with (3) after lysis and removal of cell membranes showed the stability: we observed no change of the lineshape of (3) and in the measured Ts over a period of several hours. As in the previous experiments, the glycine signal can solely be obtained while suppressing other signals (see Fig. S6, ESI†).
Conclusion
In conclusion we observed a unique behaviour of the molecular parameters T1 and Ts in NUSIMERs in the presence of different paramagnetic metal ions. Unexpectedly, the presence of Fe(III), Mn(II) and even Gd(III) did not have any effect on neither T1 nor Ts, which is a behaviour that, to the best of our knowledge, has not been observed so far. Furthermore we were able to observe a unique effect of the presence of Cu(II) on T1 and Ts, displaying a sharp decrease in both parameters at low Cu(II) concentrations followed by an increase at around 150 μM Cu(II). We propose an interaction between the terminal tripeptides of the NUSIMERs and Cu(II) to be responsible for this observation. The effect of the paramagnetic relaxation competes with a change in the mobility of the tripeptides at the outer sphere of the dendrimers which occurs upon addition of Cu(II). Based on this observed effect we explored the possibility to use NUSIMERs in an biological environment and evaluate the viability to use NUSIMERs as Cu(II) probes. We succeeded in attaching a fluorescence tag to the NUSIMERs and observing the uptake in different cells by fluorescence spectroscopy. Using the NUSIMER G5-PAMAM-GGA-Atto488, we were also able to eliminate any background signal using singlet NMR spectroscopy in the presence of cells. This is especially promising, because it opens up the possibility to branch out into Cu(II) detection in cells or even in vivo, which could in the future lead to a non-invasive technique for early diagnostics of neurodegenerative diseases. This prospect seems especially possible since it has recently be shown, that singlet filtering can be applied in MRI.49 Our described experiments could in general serve as an approach to design new molecular probes using nuclear spin singlet states.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge generous funding from the Max Planck Society and the Max Planck Institute for Biophysical Chemistry. We thank Prof. M. Bennati for enlightening discussions and access to her EPR instrumentation. K. Overkamp is acknowledged for her support in analyzing the synthesized compounds. We furthermore thank Dr V. Belov and the Chemical Synthesis Facility for a scale-up of compound (2). The authors thank Dr S. Becker and Dr N. Rezaie-Ghaleh for helpful discussions. We thank H. Sebesse for support in making the figure. Dr L. Andreas is acknowledged for providing access to the solid state NMR instrumentation. Dr R. Dervisoglu is acknowledged for help in preparing the samples for solid state NMR. We are indebted to Prof. C. Griesinger for access to his facilities. Open Access funding provided by the Max Planck Society.Notes and references
- G. M. Dykes, J. Chem. Technol. Biotechnol., 2001, 76, 903–918 CrossRef CAS.
- E. Buhleier, W. Wehner and F. Vögtle, Synthesis, 1978, 155–158 CrossRef CAS.
- M. S. Diallo, S. Christie, P. Swaminathan, J. H. Johnson Jr. and W. A. Goddard, Environ. Sci. Technol., 2005, 39, 1366–1377 CrossRef CAS PubMed.
- V. Balzani, P. Ceroni, S. Gestermann, C. Kauffmann, M. Gorka and F. Vögtle, Chem. Commun., 2000, 853–854 RSC.
- T. D. James, H. Shinmori, M. Takeuchi and S. Shinkai, Chem. Commun., 1996, 705–706 RSC.
- S. B. Garber, J. S. Kingsbury, B. L. Gray and A. H. Hoveyda, J. Am. Chem. Soc., 2000, 122, 8168–8179 CrossRef CAS.
- J. W. J. Knapen, A. W. van der Made, J. C. de Wilde, P. W. N. M. van Leeuwen, P. Wijkens, D. M. Grove and G. van Koten, Nature, 1994, 372, 659–663 CrossRef CAS.
- Y. Vida, D. Collado, F. Najera, S. Claros, J. Becerra, J. A. Andrades and E. Perez-Inestrosa, RSC Adv., 2016, 6, 49839–49844 RSC.
- S. P. Kambhampati, M. K. Mishra, P. Mastorakos, Y. Oh, G. A. Lutty and R. M. Kannan, Eur. J. Pharm. Biopharm., 2015, 95, 239–249 CrossRef CAS PubMed.
- F.-H. Liu, C.-Y. Hou, D. Zhang, W.-J. Zhao, Y. Cong, Z.-Y. Duan, Z.-Y. Qiao and H. Wang, Biomater. Sci., 2018, 6, 604–613 RSC.
- A. K. Sharma, L. Gupta, H. Sahu, A. Qayum, S. K. Singh, K. T. Nakhate, Ajazuddin and U. Gupta, Pharm. Res., 2018, 35, 9 CrossRef PubMed.
- S. P. Kuruvilla, G. Tiruchinapally, A. C. Crouch, M. E. H. ElSayed and J. M. Greve, PLoS One, 2017, 12, e0181944 CrossRef PubMed.
- P. Saul, S. Mamone and S. Glöggler, Chem. Sci., 2019, 10, 413–417 RSC.
- M. H. Levitt, Annu. Rev. Phys. Chem., 2012, 63, 89–105 CrossRef CAS PubMed.
- M. Carravetta and M. H. Levitt, J. Am. Chem. Soc., 2004, 126, 6228–6229 CrossRef CAS PubMed.
- M. Carravetta, O. G. Johannessen and M. H. Levitt, Phys. Rev. Lett., 2004, 92, 153003 CrossRef PubMed.
- M. Carravetta and M. H. Levitt, J. Chem. Phys., 2005, 122, 214505 CrossRef PubMed.
- G. Pileio and M. H. Levitt, J. Chem. Phys., 2009, 130, 214501 CrossRef PubMed.
- G. Pileio, M. Carravetta and M. H. Levitt, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17135–17139 CrossRef CAS PubMed.
- G. Pileio, M. Carravetta, E. Hughes and M. H. Levitt, J. Am. Chem. Soc., 2008, 130, 12582–12583 CrossRef CAS PubMed.
- G. Stevanato, J. T. Hill-Cousins, P. Håkansson, S. S. Roy, L. J. Brown, R. C. D. Brown, G. Pileio and M. H. Levitt, Angew. Chem., Int. Ed., 2015, 54, 3740–3743 CrossRef CAS PubMed.
- T. Theis, G. X. Ortiz Jr., A. W. J. Logan, K. E. Claytor, Y. Feng, W. P. Huhn, V. Blum, S. J. Malcolmson, E. Y. Chekmenev, Q. Wang and W. S. Warren, Sci. Adv., 2016, 2, e1501438 CrossRef PubMed.
- Y. Zhang, P. C. Soon, A. Jerschow and J. W. Canary, Angew. Chem., Int. Ed., 2014, 53, 3396–3399 CrossRef CAS PubMed.
- M. C. D. Tayler and M. H. Levitt, Phys. Chem. Chem. Phys., 2011, 13, 5556–5560 RSC.
- G. Pileio, M. Concistrè, M. Carravetta and M. H. Levitt, J. Magn. Reson., 2006, 182, 353–357 CrossRef CAS PubMed.
- T. Theis, Y. Feng, T. Wu and W. S. Warren, J. Chem. Phys., 2014, 140, 014201 CrossRef CAS PubMed.
- S. J. Elliott, L. J. Brown, J.-N. Dumez and M. H. Levitt, Phys. Chem. Chem. Phys., 2016, 18, 17965–17972 RSC.
- R. Buratto, D. Mammoli, E. Chiarparin, G. Williams and G. Bodenhausen, Angew. Chem., Int. Ed., 2014, 53, 11376–11380 CrossRef CAS PubMed.
- R. Buratto, A. Bornet, J. Milani, D. Mammoli, B. Vuichoud, N. Salvi, M. Singh, A. Laguerre, S. Passemard, S. Gerber-Lemaire, S. Jannin and G. Bodenhausen, ChemMedChem, 2014, 9, 2509–2515 CrossRef CAS PubMed.
- Y. Feng, R. M. Davis and W. S. Warren, Nat. Phys., 2012, 8, 831–837 Search PubMed.
- E. Vinogradov and A. K. Grant, J. Magn. Res., 2008, 194, 46–57 CrossRef CAS PubMed.
- W. S. Warren, E. Jenista, R. T. Branca and X. Chen, Science, 2009, 323, 1711–1714 CrossRef CAS PubMed.
- G. Pileio, S. Bowen, C. Laustsen, M. C. D. Wayler, J. T. Hill-Cousins, L. J. Brown, R. C. D. Brown, J. H. Ardenskjær-Larsen and M. H. Levitt, J. Am. Chem. Soc., 2013, 135, 5084–5088 CrossRef CAS PubMed.
- Y. Feng, T. Theis, X. Liang, Q. Wang, P. Zhou and W. S. Warren, J. Am. Chem. Soc., 2013, 135, 9632–9635 CrossRef CAS PubMed.
- Y. Zhang, X. Duan, P. C. Soon, V. Sychrovský, J. W. Canary and A. Jerschow, ChemPhysChem, 2016, 17, 2967–2971 CrossRef CAS PubMed.
- S. Cavadini, J. Dittmer, S. Antonijevic and G. Bodenhausen, J. Am. Chem. Soc., 2005, 127, 15744–15748 CrossRef CAS PubMed.
- P. Ahuja, R. Sarkar, P. R. Vasos and G. Bodenhausen, J. Am. Chem. Soc., 2009, 131, 7498–7499 CrossRef CAS PubMed.
- S. J. DeVience, R. L. Walsworth and M. S. Rosen, NMR Biomed., 2013, 26, 1204–1212 CrossRef CAS PubMed.
- R. Buratto, D. Mammoli, E. Canet and G. Bodenhausen, J. Med. Chem., 2016, 59, 1960–1966 CrossRef CAS PubMed.
- M. C. D. Tayler and M. H. Levitt, Phys. Chem. Chem. Phys., 2011, 13, 9128–9130 RSC.
- R. Sarkar, P. R. Vasos and G. Bodenhausen, J. Am. Chem. Soc., 2007, 129, 328–334 CrossRef CAS PubMed.
- P. Ahuja, R. Sarkar, P. R. Vasos and G. Bodenhausen, J. Chem. Phys., 2007, 127, 134112 CrossRef PubMed.
- N. Salvi, R. Buratto, A. Bornet, S. Ulzega, I. R. Rebollo, A. Angelini, C. Heinis and G. Bodenhausen, J. Am. Chem. Soc., 2012, 134, 11076–11079 CrossRef CAS PubMed.
- A. S. Kiryutin, A. N. Pravdivtsev, A. V. Yurkovskaya, H.-M. Vieth and K. L. Ivanov, J. Phys. Chem., 2016, 120, 11978–11986 CrossRef CAS PubMed.
- S. J. DeVience, R. L. Walsworth and M. S. Rosen, Phys. Rev. Lett., 2013, 111, 173002 CrossRef PubMed.
- C. Stavarache, A. Hanganu, A. Paun, C. Paraschivescu, M. Matache and P. R. Vasos, J. Magn. Res., 2017, 284, 15–19 CrossRef CAS PubMed.
- A. Bornet, P. Ahuja, R. Sarkar, L. Fernandes, S. Hadji, S. Y. Lee, A. Haririnia, D. Fushman, G. Bodenhausen and P. R. Vasos, ChemPhysChem, 2011, 12, 2729–2734 CrossRef CAS PubMed.
- S. Mamone, N. Rezaei-Ghaleh, F. Opazo, C. Griesinger and S. Glöggler, Sci. Adv., 2020, 6, eaaz1955 CrossRef CAS PubMed.
- S. Mamone, A. B. Schmidt, N. Schwaderlapp, T. Lange, D. von Elverfeldt, J. Hennig and S. Glöggler, NMR Biomed., 2021, 34, e4400 CrossRef PubMed.
- M. A. Lovell, J. D. Robertson, W. J. Teesdale, J. L. Campbell and W. R. Markesbery, J. Neurol. Sci., 1998, 158, 47–52 CrossRef CAS PubMed.
- B. S. Gupta, M. Taha and M.-J. Lee, J. Solution Chem., 2013, 42, 2296–2309 CrossRef CAS.
- X. Wang, X. Cai, J. Hu, N. Shao, F. Wang, Q. Zhang, J. Xiao and Y. Cheng, J. Am. Chem. Soc., 2013, 135, 9805–9810 CrossRef CAS PubMed.
- P. Welch and M. Muthukumar, Macromolecules, 1998, 31, 5892–5897 CrossRef CAS.
- S. García-Gallego, M. Cangiotti, L. Fiorani, A. Fattori, M. Ángeles Muñoz Fernández, R. Gomez, M. F. Ottaviani and F. J. de la Mata, Dalton Trans., 2013, 42, 5874–5889 RSC.
- D. Lombardo, Biochem. Res. Int., 2014, 837651 Search PubMed.
- M. F. Ottaviani, S. Bossmann, N. J. Turro and D. A. Tomalia, J. Am. Chem. Soc., 1994, 116, 661–671 CrossRef CAS.
- H. Motschi, Colloids Surf., 1984, 9, 333–347 CrossRef CAS.
- K. J. de Almeida, Z. Rinkevicius, H. W. Hugosson, A. C. Ferreira and H. Ågren, Chem. Phys., 2007, 332, 176–187 CrossRef CAS.
- J. V. Folgado, W. Henke, R. Allmann, H. Stratemeier, D. Beltran-Porter, T. Rojo and D. Reinen, Inorg. Chem., 1990, 29, 2035–2042 CrossRef CAS.
- A. Meyer, G. Schnakenburg, R. Glaum and O. Schiemann, Inorg. Chem., 2015, 54, 8456–8464 CrossRef CAS PubMed.
- S. J. Elliott, C. Bengs, L. J. Brown, J. T. Hill-Cousins, D. J. O'Leary, G. Pileio and M. H. Levitt, J. Chem. Phys., 2019, 150, 064315 CrossRef CAS PubMed.
- J. C. Garcia-Martinez and R. M. Crooks, J. Am. Chem. Soc., 2004, 126, 16170–16178 CrossRef CAS PubMed.
- L. F. Pinto, J. Correa, M. Martin-Pastor, R. Riguera and E. Fernandez-Megia, J. Am. Chem. Soc., 2013, 135, 1972–1977 CrossRef CAS PubMed.
- Z.-J. Chen, G. T. Gillies, W. C. Broaddus, S. S. Prabhu, H. Fillmore, R. M. Mitchell, F. D. Corwin and P. P. Fatouros, J. Neurosurg., 2019, 101, 314–323 Search PubMed.
- M. Maidorn, A. Olichon, S. O. Rizzoli and F. Opazo, MABS, 2019, 11, 305–321 CrossRef CAS PubMed.
- M. A. Gomes de Castro, C. Höbartner and F. Opazo, PLoS One, 2017, 12, e0173050 CrossRef PubMed.
- S. Mamone and S. Glöggler, Phys. Chem. Chem. Phys., 2018, 20, 22463–22467 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cp04483d |
| ‡ Present address: Key Laboratory of Pesticide and Chemical Biology of Ministry of Education, Hubei International Scientific and Technological Cooperation Base of Pesticide and Green Synthesis, College of Chemistry, Central China Normal University, Wuhan, 430079, China. |
| This journal is © the Owner Societies 2021 |





![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: