Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic technologies for the synthesis and manipulation of biomimetic membranous nano-assemblies
Colin P.
Pilkington
 ab,
John M.
Seddon
ab,
John M.
Seddon
 a and
Yuval
Elani
a and
Yuval
Elani
 *b
*b
aDepartment of Chemistry, Molecular Science Research Hub, Imperial College London, 82 Wood Lane, London, W12 0BZ, UK
bDepartment of Chemical Engineering, Exhibition Road, Imperial College London, London, SW7 2AZ, UK. E-mail: y.elani@imperial.ac.uk
First published on 27th January 2021
Abstract
Microfluidics has been proposed as an attractive alternative to conventional bulk methods used in the generation of self-assembled biomimetic structures, particularly where there is a desire for more scalable production. The approach also allows for greater control over the self-assembly process, and parameters such as particle architecture, size, and composition can be finely tuned. Microfluidic techniques used in the generation of microscale assemblies (giant vesicles and higher-order multi-compartment assemblies) are fairly well established. These tend to rely on microdroplet templation, and the resulting structures have found use as comparmentalised motifs in artificial cells. Challenges in generating sub-micron droplets have meant that reconfiguring this approach to form nano-scale structures is not straightforward. This is beginning to change however, and recent technological advances have instigated the manufacture and manipulation of an increasingly diverse repertoire of biomimetic nano-assemblies, including liposomes, polymersomes, hybrid particles, multi-lamellar structures, cubosomes, hexosomes, nanodiscs, and virus-like particles. The following review will discuss these higher-order self-assembled nanostructures, including their biochemical and industrial applications, and techniques used in their production and analysis. We suggest ways in which existing technologies could be repurposed for the enhanced design, manufacture, and exploitation of these structures and discuss potential challenges and future research directions. By compiling recent advances in this area, it is hoped we will inspire future efforts toward establishing scalable microfluidic platforms for the generation of biomimetic nanoparticles of enhanced architectural and functional complexity.
Introduction
Bio-inspired membranous structures have a wide range of potential applications, most notably as simplified chassis for the study of cellular features and as functional soft-matter particles that can act as carriers for therapeutic molecules. They are also of interest to those working in the field of bottom-up synthetic biology, concerned with the design, synthesis, and manipulation of cell-like objects sometimes termed artificial cells. Membranous structures of varied size and phase behaviour (referring to the intrinsic order and morphology of said structures) have been produced, mimicking the basic architecture of living cells.1–3 Drawing inspiration from biological sources, many cell-like behaviours have been successfully modelled, including motility and intercellular communication.4–6 These biomimetic, synthetically derived structures have also been used to better understand cell fusion events and small molecule/membrane interactions.7,8 Their modular design and capacity to incorporate functional biomolecular componentry has allowed researchers to expand their use to specialised drug delivery,9,10 biosensing,11 controlled content release,12 protein synthesis,13,14 energy production,15 and most recently as carriers of immunotherapeutic mRNA strands with potential to aid the global effort against COVID-19.16,17 Controllable generation of these structures is key to their success, something that has been addressed via the integration of microfluidic methods. However, in contrast to microscale membrane assemblies,18 existing examples of nano-scale cell-like objects generated by microfluidics tend to show limited compositional and architectural diversity, which has restricted their technological potential. We know from biology that architectural complexity gives rise to functional and behavioural complexity. Indeed, this is one of the key drivers behind bottom-up synthetic biology. Widening the applicability of membranous particles is therefore reliant on the generation of structures more elaborate than simple uni-compartment bilayer capsules. In this review we highlight some of the effort that has been made toward establishing microfluidic methods capable of accessing and leveraging higher-order nano-scale biomimetic particles. To encourage future work, we also suggest ways in which other platforms may be adapted to better include these structures. The nano-assemblies we cover (summarised in Fig. 1) were chosen on the basis of their (i) biological relevance and (ii) deviation from the current structural and compositional norms of bottom-up synthetic biology.Self-assembly
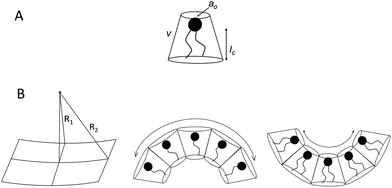
Before discussing the main characteristics and generation methods associated with higher-order biomimetic nanostructures, it is important to first outline the underlying principles behind their self-assembly. Here, the process can be considered a generalised grouping together of smaller components (molecules or protein subunits) forming larger structures with varying degrees of order. For the purposes of this paper, these components are naturally derived or at least bio-mimetic in some way, distinguishing their nano-assemblies from inorganic nanoparticles that have been covered extensively elsewhere (gold particles, silica colloids etc.).19 Nanostructures composed of amphiphilic small molecules (molecules with both hydrophilic and hydrophobic portions – either lipids or block copolymers) are predominantly stabilised by hydrophobic interactions in a polar environment. Amphiphiles are thought of in terms of their molecular shape, and grouped according to their geometric packing parameter P (see Fig. 2A):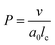 | (1) |
| K = c1c2 | (2) |
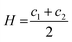 | (3) |
There is an energetic cost associated with packing hydrophobic portions of amphiphiles into self-assembled aggregates (gp), a process that is entropically driven by the expulsion of free water molecules. The surfaces generated also feature stored elastic free energy gc given by the Helfrich equation below:20–22
| gc = 2κ(H − H0)2 + κGK | (4) |
| gt = gc + gp + ginter | (5) |
A consideration of gt provides a thermodynamic rationale for how environmental conditions such as temperature, pressure and solvent type affect the phase behaviour of amphiphilic assemblies. As conditions change, so too do the preferred geometries of amphiphiles, and the resultant curvatures and phase behaviours of their aggregates. These phases are generally grouped according to their relative values of H and K, each with distinct topologies and varying degrees of liquid crystalline periodicity. Polymer assemblies are governed by the same thermodynamic principles, though their morphologies can differ significantly when compared to lipid systems.20–22 Both the physical properties and potential applications of a bio-inspired nano-assembly will vary from phase to phase, highlighting the need to maintain considerable control over the experimental parameters governing phase behaviour.
Where protein subunits are concerned (as is the case for virus-like particles) other forces such as H-bonding and covalent bonds control their self-assembly into nanostructures. Their formation is actively mediated by the cell-machinery of genetically engineered organisms, the thermodynamic principles of which will not be covered in this paper.24,25 The result are biomimetic particles composed of ordered arrays of repeating subunits that can be analysed in a similar fashion to amphiphilic assemblies (dynamic light scattering, X-ray diffraction techniques etc.).26
Microfluidics: advantages & current limiting factors
Microfluidics has revolutionised experimental design across many scientific disciplines. Through the manipulation of small volumes of liquids at low Reynolds numbers, entire laboratories have been miniaturised to fit on millimetre-sized chips, paving the way toward higher throughput and less wasteful scientific research.27–29 For bottom-up synthetic biology in particular, the use of microfluidics has shown great promise, whether in finely tuning the self-assembly of biologically inspired objects or in the subsequent application of those objects as “smart materials”.30–32 Microfluidic platforms have enabled researchers to achieve enhanced control over experimental parameters such as monomer composition, kinetics of self-assembly and particle morphology.32 However, microfluidics has tremendous untapped potential for the design, synthesis and manipulation of more complex bio-inspired materials, as most existing work has dealt with structures on the micrometre size range,33–35 rendering them unsuitable for a variety of industrial and medical applications.31,36 Existing “bulk” formation methods have worked well where nano-systems remain relatively simple. Most often used is the rehydration of a dried amphiphile film in a suitable polar medium, followed by agitation to promote self-assembly. The structures formed are extruded through filters of defined pore-size to achieve relatively monodisperse nano-sized particles.37 High-powered probe sonication and high-shear mixing achieve a similar result to extrusion, though both are often deemed unsuitable for use with sensitive biological material.38 In accessing more complex, higher-order structures, precise control over the self-assembly process is required, beyond the capability of these bulk methods. Though it is possible to produce particles of a narrow size distribution en masse, this can only be achieved either through secondary preparatory steps or by high energy input. As such, microfluidics has presented a superior alternative, bestowing:• high generation and experimental throughput.
• excellent size control.
• low energy costs, more efficient use of reagents.
• and, most importantly for higher-order supramolecular structure formation, an enhanced control over reaction conditions, such as temperature and solute concentration.
Droplet microfluidics has been used extensively in micro-scale bottom-up synthetic biology, providing spherical templates around which the biomimetic structures are encouraged to assemble.18,39 This approach has been used to generate giant unilamellar vesicles (GUVs) and GUVs with multiple organelle-like sub-compartments called vesosomes.3,30–32,40 Though useful as bio-inspired synthetic vessels, in which enzymatic reactions and other biochemical pathways can be reproducibly mimicked, droplet-templated approaches have two key drawbacks:
(i) Size constraints: droplets often have diameters above 10 μm. The generation of droplets in the sub-micron regime can be difficult to achieve, requiring very high, impractical flow rates (in the case of cross-flow junction droplet generators) or nano-scale microfluidic chip features that are beyond the scope of existing lithographic fabrication techniques.41,42
(ii) Phases of lipidic/polymeric assemblies: biomimetic structures produced by droplet microfluidics are confined to lamellar phases only.18,31–33,39
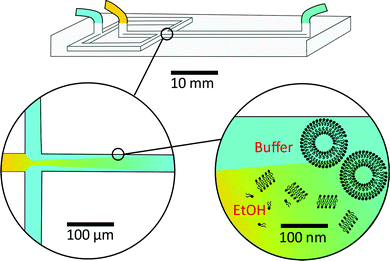
As a result, researchers have focused on continuous flow microfluidic methods to generate nano-assemblies (<150 nm), most notably microfluidic hydrodynamic focusing (MHF; see Fig. 3 for overview). Self-assembly in this case is mediated by diffusion-dominated mass transport.31 The technique has been used to generate nano-sized unilamellar lipid vesicles, though its potential for producing multi-compartment/architecturally and functionally diverse nano-assemblies has yet to be investigated in full. It presents an attractive alternative to existing bulk methods for nano-assembly preparation, particularly within an industrial setting, capable of generating structures of very low polydispersity indices (PDIs) at a high production rate. There are examples of it being used to produce polymersomes and to generate non-lamellar liposomal nanoparticles, as will be highlighted in later sections.23,43,44 An appropriate concentration of amphiphile dissolved in a water–miscible solvent is injected into a central microfluidic channel and the stream compressed into a continuous flow regime by two adjacent channels containing aqueous buffer. A diffusion gradient is established between the two solvents, along which amphiphiles reach their respective solubility thresholds and begin to nucleate and self-assemble. The process is governed by diffusion-dominated mass transport, and the average mixing time of the streams described by eqn (6).
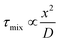 | (6) |
The method can be readily adapted to fit industrial requirements of scale and throughput. An impressive article by Carugo et al. demonstrated liposome production at concentrations suitable for most drug delivery applications (typically ranging between 5 and 15 mg ml−1) with total flow rates of up to 18 ml min−1, using chips with channel dimensions on the millimetre scale.31 Further efforts to scale-up MHF have led to the development of a technique termed “vertical flow focusing” (VFF), able to produce nano-sized liposomes two orders of magnitude faster than traditional MHF.47 The chip design extends channel dimensions along the z-axis only: channels are deep (5 mm) and narrow (50 μm). This minor change was observed to significantly increase the throughput of generated liposomes and further minimise the unwanted effects of a no-slip boundary condition on the process of self-assembly.47 Both examples successfully optimise the microfluidic method without jeopardising its main asset: the continuous and controllable generation of nano-assemblies. Future devices that incorporate parallelized MHF junctions are also expected to enhance throughput, taking inspiration from the mass production of emulsions via droplet microfluidics.48,49 This has been touched on by Carugo et al., who suggest the use of “off-the-shelf” components for parallel liposome generation, such as a four-way tubing splitter.31
MHF features heavily in the following sections, in many cases providing a likely route toward controllably generating atypical/higher-order biomimetic nanoassemblies. In addition, we believe it holds great promise for those assemblies where currently no microfluidic generation method yet exists.
Biomimetic nano-assemblies
Cubosomes
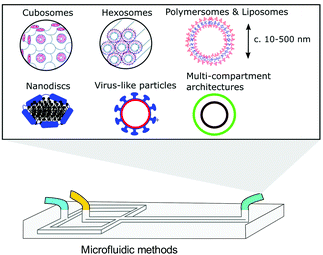
Bicontinuous cubosomes are a class of lyotropic liquid-crystalline nanostructure strongly implicated as part of “the next generation” of soft-matter particles.50,51 They consist of amphiphilic bilayers draped over minimal surfaces with zero mean curvature (H) and Gaussian curvature (K) < 0, featuring two interwoven but separate aqueous networks. They are usually subdivided into three forms depending on hydration levels, each with distinct crystallographic space groups: Primitive, Schwartz diamond or, less commonly, Gyroid (Im3m, Pn3m, and Ia3d respectively).21 They typically feature monoolein and/or phytantriol lipids, though modification of the permeability and lattice parameters of the heavily folded inner membranes has been achieved by incorporation of lipids with alternative packing parameters and other desired molecular properties.52–55 Although bicontinuous cubic phases can exist in bulk, these are not suitable for most biomedical applications. Instead, an amphiphilic polymer is often used to stabilise nano-scale particle dispersions of these liquid crystalline mesophases, incorporated either during lipid self-assembly or in a subsequent, post-assembly step. The inclusion of a stabilising polymer is crucial in preventing the aggregation of cubosomal particles and is expected to alter the dimensions of the interconnected aqueous channels, making a direct comparison with larger cubic counterparts difficult.50 The presence of a polymeric corona may also be advantageous, presenting a unique opportunity to introduce functionality into these systems, overlapping cubosome design with the rich field of synthetic polymer chemistry.50,56,57Biological membranes have been observed to adopt inverse bicontinuous cubic phases in vivo. They are thought to facilitate important biological processes such as cell fusion, division, endo/exocytosis and gas exchange in mammalian alveolar tissue.21 Synthetically mimicking these membrane structures to better understand their biological role falls in line with the overarching goals of bottom-up synthetic biology. Like their lamellar counterparts, engineered cubosomes are expected to find use in the pharmaceutical industry as delivery vehicles, their higher membrane surface area making them ideal structures for hydrophobic cargo transport and release.50,58–60 Some work has gone into cubosomal formulations with encapsulated gene-silencing RNA and other functional peptides, displaying exceptionally well-controlled diffusion rates.61–63 Cubosomes have been shown to provide a certain degree of in vivo protection to biomolecules that would otherwise be sensitive to enzymatic degradation, highly advantageous in the delivery of immunogenic therapeutics.62–64 Timely and impressive work from Sarkar et al. used cubosomal particles for the delivery of genetic material, attributing improved delivery efficiency to particle topology.65 There is potential for enhancing site specificity by leveraging the polymeric corona, and some preliminary work has gone into producing stimuli-responsive cubosomal particles for environmentally triggered cargo release.66–69 Sonication represents the most widely used technique in cubosome formation. A film of suitable lipid composition is generated, followed by hydration in a suitable buffer medium containing a stabilising polymer. The suspension is then homogenised via probe sonication.50,70,71 Though the technique has proved effective in reproducibly generating cubosomes of narrow size distributions, probe sonication is known to have a deleterious effect on biological material.72 As a result, alternative methods requiring lower energy input have also been explored. The most prominent of these has been solvent evaporation, whereby a volatile solvent with solubilised lipids is emulsified in a suitable aqueous buffer. Self-assembly into a cubosomal structure is initiated in a subsequent drying step.50,73 To the best of our knowledge, only one microfluidic method harnessing this emulsification approach exists, developed by Kim et al. Monoolein, DOTAP and a PEG-ylated stabiliser were dissolved in an ethanolic stream and emulsified via chaotic mixing in an adjacent buffer stream along a microfluidic channel with herringbone features (Fig. 4). After generating 50 nm sized ethanol-in-water droplets, an off-chip evaporation step promoted the gradual fusion of those droplets followed by self-assembly into stable cubosomes ranging from 75 nm to 200 nm (bulk methods typically produce particles between 200 nm and 500 nm with PDIs >0.1). They boast polydispersity indices as low as 0.04, and preliminary results indicate that size can be controlled by the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratios and flow rates used. They go on to demonstrate the successful loading of siRNA into the cubosomal membrane in a secondary off-chip step.63 Though a promising first attempt, the additional and necessary off-chip preparations eliminate one of the key advantages of microfluidic technologies: their potential for streamlining a process, requiring minimum effort on the part of the operator. The most widely used microfluidic method for generating small unilamellar vesicles (SUVs), microfluidic hydrodynamic focusing, has yet to be explored for its potential in controllably generating cubosomes. A careful consideration of other assembly routes may be necessary in designing a suitable microfluidic platform.
polymer ratios and flow rates used. They go on to demonstrate the successful loading of siRNA into the cubosomal membrane in a secondary off-chip step.63 Though a promising first attempt, the additional and necessary off-chip preparations eliminate one of the key advantages of microfluidic technologies: their potential for streamlining a process, requiring minimum effort on the part of the operator. The most widely used microfluidic method for generating small unilamellar vesicles (SUVs), microfluidic hydrodynamic focusing, has yet to be explored for its potential in controllably generating cubosomes. A careful consideration of other assembly routes may be necessary in designing a suitable microfluidic platform.
Hexosomes
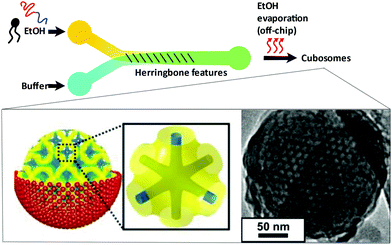
Hexosomes represent another class of non-lamellar liquid crystalline nanostructure with potential as functional soft-matter materials.74,75 Lipids capable of aggregating into surfaces with sufficiently negative mean curvature (H) are seen to spontaneously self-assemble into inverse hexagonal phases, generally visualised as seven or more discrete tubular structures.21 As with cubosomes, polymeric stabilisers are used to form a corona around the hexagonal morphology of these particles, necessary to prevent aggregation through Ostwald ripening. In fact, the lipid systems used for hexosome formation are often the same as those used for cubosomes, namely phytantriol and monoolein. Preferential formation of the H|| phase occurs where there is pronounced hydrophobic chain “splay”, usually seen at elevated temperatures, though transitions to H|| can be induced under ambient temperatures through careful choice of stabilising polymer and adjustment of other environmental conditions.21,22,76Biological examples of lipid assemblies adopting an inverse hexagonal phase tend to be more elusive, though they have been proposed as important intermediate structures in enzymatic activity pathways and cell fusion events.77,78 Colloidal dispersions of hexosomes have potential use as delivery vehicles for pharmaceutical agents and biomolecules. The discontinuous nature of the closed aqueous channels in hexosomal membranes is key, allowing the slow and sustained release of cargo.21,75,79,80 This property is particularly important for the delivery of hormonal and analgesic therapeutics. Triggered release mechanisms of hexosomes tend to involve controlled phase transitions, stimulated by temperature changes and pH.21,81,82 Techniques used in their formation mirror those used in cubosome assembly, including emulsification followed by solvent evaporation, and lipid film hydration with a secondary probe sonication or high shear homogenisation step.21,83 Interestingly, microfluidic hydrodynamic flow focusing (MHF) has been used to successfully generate hexosomes of relatively narrow size distribution, although only one example exists to date (Fig. 5). Yaghmur et al. developed an MHF chip capable of hexosome synthesis, featuring a long serpentine channel to aid the mixing of the various amphiphilic precursors (docosahexaenoic acid monoglyceride and Pluronic F127).84 Unfortunately, the system did not achieve the exceptional size control typical of an MHF regime, but this will no doubt be addressed in future iterations.84 The very fact that a complete “on-chip” synthesis was carried out with no additional preparatory steps is significant. Despite MHF having been predominantly used to generate SUVs, this work highlights its potential to include alternative architectures and phases.
Multi-lamellar vesicles
Rather surprisingly, nano-sized multi-lamellar vesicles (MLVs) have received limited attention for their potential use as soft matter machines, hydrophobic drug delivery agents and biomimetic artificial cells bearing architectural resemblance to various cellular organelles.85–87 For clarity, it is worth defining multi-lamellar vesicles as distinct from vesosomes, the latter referring to larger membrane structures with several smaller vesicular compartments encapsulated within.18 MLVs are typically observed as “stacked” lamellar bilayers, occasionally exhibiting sufficiently long-range order to give quasi-Bragg peaks in small-angle X-ray scattering analysis.88,89 Deliberate MLV generation is rare, researchers citing their undesirably low core volume and stability, and a processing step (extrusion etc.) is often included to remove these layered structures and ensure the unilamellarity of produced vesicles.88 Large values for spontaneous curvature (H0) and bending moduli, in addition to weak inter-bilayer repulsive forces, are thought to provide a thermodynamic explanation for preferential MLV formation at equilibrium, and as such their formation is highly dependent on the molecular properties of chosen amphiphiles.90,91 To the best of our knowledge, no examples exist for the controlled preparation of either nano-scale or micron-scale MLVs using bulk methods. This is presumably due to the absence of control one has over the exact number of bilayers present in generated vesicles, understandably eliminating the possibility of designing functional MLV particles and pursuing this architecture further.Microfluidic methods may address this shortfall, allowing enhanced control over the MLV self-assembly process, and some promising steps have already been made. Giant multi-lamellar vesicles with asymmetric bilayers were generated using a combination of droplet microfluidics and a Langmuir–Blodgett-type layering approach (Fig. 6). Lipid-stabilised water-in-oil droplets (∼50 μm) were generated using a flow focusing nozzle and captured in specialised traps on the same PDMS chip. When the traps were satisfactorily occupied, a second lipid-in-oil solution was introduced, followed by an aqueous buffer wash that resulted in the self-assembly of vesicles with asymmetric bilayers. The washing steps were repeated, producing vesicles with an additional bilayer.92 This work represents the first successfully controlled MLV generation, and the group go on to highlight the importance of this kind of assembly in mitochondrial protein function and other in vivo processes.92 Though this method is unprecedented, the scale at which they operate does limit its potential for generating drug delivery vehicles and functional nanomaterials. However, the stepwise introduction of subsequent bilayers could in theory be re-integrated into an alternative microfluidic platform. The various mixing regimes possible within modified MHF chips could be a likely route toward nano-MLV synthesis and have been utilised in a recent paper by Ghazal et al. The group produce nano-scale (∼160 nm) MLVs using a phytantriol glyceryl dioleate-PEG12, with low dispersity values, by including a long serpentine channel. They successfully couple this production with small-angle X-ray scattering analysis to gain an insight into the mechanism of formation.93 Though a promising first step, there is limited control over the size of MLVs produced (characteristic of MHF) and each bilayer is presumably composed of the same lipid/polymer mixture.
Nanodiscs
Nanodiscs, particles consisting of a small circular region of a lipid bilayer surrounded by an amphiphilic α-helical membrane scaffold protein (MSP), have been particularly useful in the study of membrane protein structure and function. They hold significant advantages over liposomal scaffolds in that the lipid environment they provide is often more biologically relevant. Normal protein function is conserved in these scaffolds and by changing the MSP length and lipid composition, these particles have been used to incorporate a wide range of membrane proteins with diverse functionality.94,95 Widening the lipid bilayer has allowed the study of multiple proteins immobilised on one nanodisc. These larger particles have significant implications for structural biology, as laid out in a recent review by Padmanabha Das et al.96 They are generated by mixing membrane proteins and lipids, both solubilised in detergent, with an MSP of defined length, followed by detergent removal using a suitable resin. Some work has gone into directly extracting membrane proteins from cells using amphiphilic copolymers (typically styrene maleic acid and derivatives). The polymer acts as an alternative to an MSP, and the native lipid environment is preserved. This is especially useful where scaffold proteins and/or solubilising detergents are thought to adversely affect the membrane protein of interest.97,98 In addition to this, the incorporation of a synthetic component into these assemblies opens the possibility for nanodisc functionalisation.Such research is often limited by the need for extensive optimisation of the conditions for self-assembly, necessary to best preserve protein structure and function but often time-consuming and expensive.94,99,100 This problem lends itself to adopting a microfluidic approach, as highlighted in recent work by Wade et al.(Fig. 7).100 They present a PDMS chip capable of nanodisc formation and purification. They benefit from the improved mass-transport and mixing characteristic of microfluidic flow regimes, and the inherent efficiency associated with the miniaturisation of a process. Lipids, MSPs and the membrane proteins of interest are fed into a chamber containing an immobilised detergent removal resin, then into a purification chamber with an Ni-NTA resin. They use UV-vis analysis to show the successful incorporation of Cytochrome P450 into nanodiscs as a proof of concept, acknowledging that much of the optimisation for this protein has already been accomplished. They propose the use of this platform in future optimisation studies, emphasising the potential for parallel nanodisc formation over a range of component concentrations.100
As far as we are aware this is the only published example of a microfluidic platform capable of nanodisc generation. However, some microfluidic methods have been used to manipulate pre-formed nanodiscs to study various biophysical phenomena. An interesting lipid grafting technique was developed by Goluch et al. for the study of the effect of lipid composition on protein incorporation into a bilayer. Nanodiscs of varied composition were layered onto a glass slide via PDMS channels, and a fluorescently labelled annexin protein was layered on top in a criss-cross pattern.101 The formation mechanism of copolymer stabilised nanodiscs was probed in a recent paper by Azouz et al., who reconfigure a microfluidic device used for the diffusional sizing of proteins.102 Their work provides an insight into the solubilisation process of lipid bilayers from large unilamellar vesicles by amphiphilic copolymers, which may be useful in the future design of such nano-assemblies.
 | ||
| Fig. 1 Graphical illustration of the various biomimetic nano-assemblies covered in this review, generated and/or manipulated using microfluidic methods. Some images adapted from Y. Huang and S. Gui, RSC Adv., 2018, 8, 6978 – Published by The Royal Society of Chemistry.79 | ||
 | ||
| Fig. 4 Graphical illustration of the ethanol-in-water emulsifying microfluidic chip, designed by Kim et al. Herringbone features stimulate chaotic mixing of the two streams. After a heating step at 65 °C, droplets fuse and the monoolein lipids self-assemble to monodisperse cubosomes. Complexation of fully formed particles with gene silencing RNA was achieved. Adapted with permission from Kim et al., ACS Nano, 2018, 12(9), 9196–9205. Copyright (2018) American Chemical Society.63 | ||
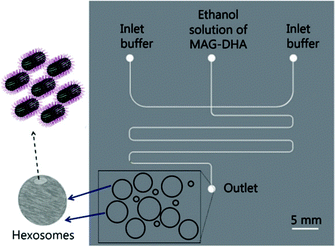 | ||
| Fig. 5 Graphical representation of the microfluidic hydrodynamic focusing chip used in hexosome generation developed by Yaghmur et al. The group also coupled this method with small angle X-ray scattering analysis at different flow rate ratios, confirming the presence of secondary micellar nanoobjects. Reproduced with permission from the PCCP Owner Societies.84 | ||
 | ||
| Fig. 6 Graphical representation of the microfluidic layer-by-layer approach used to generate double bilayer vesicles. (a–h) Represent the consecutive steps involved, where initial trapping of a lipid stabilised droplet is followed by several washing steps, controlling the self-assembly process. Image taken from Matosevic et al., 2013, Nature Chemistry.92 | ||
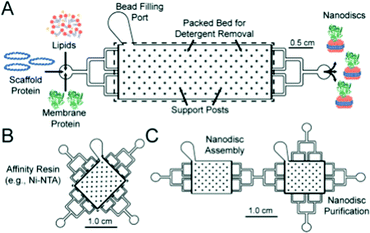 | ||
| Fig. 7 Graphical representation of the microfluidic chips used in the formation (A) and purification (B) of lipid nanodiscs. The group show the facile connection of both modules to generate a bifunctional chip. Reproduced with permission from The Royal Society of Chemistry.100 | ||
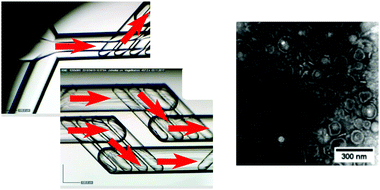 | ||
| Fig. 8 Optical microscopy images of the microfluidic chip, with a simple cross-flow junction and mixing chambers (left, red arrows indicating the direction of flow), used in the self-assembly of pH responsive polymersomes (right, TEM image), adapted with permission from Albuquerque et al., Langmuir, 2019, 35, 25, 8363–8372. Copyright (2019) American Chemical Society.103 | ||
Polymersomes
Polymersomes can be thought of as wholly synthetic alternatives to lipidic vesicles, formed by the self-assembly of amphiphilic block copolymers in a suitable aqueous medium.23 These structures have been used as bio-inspired nanomachines, where a loss of the inherent biocompatibility of lipid systems can be tolerated.23,104,105 Though widely varied, structures tend to have much larger membrane thicknesses (5–50 nm), bending rigidity values (κ values between 35–400 kBT) and an overall improved stability compared to their lipid counterparts.23 The synthetic origins of polymersomes also allow for greater functional diversity making them attractive targets in drug delivery research and nanoengineering. Aligning with advances in synthetic polymer chemistry (RAFT, ATRP etc.), polymersomes responsive to light, heat, magnetism and pH have been produced.23,106–109 Though spherical bilayers are the most common, other unusual polymer assemblies have been observed, including worm-like structures, “patchy” or “multiple surface-domain” vesicles and “raspberry”-like polymersomes.23,110,111 As with lipids, the molecular properties and resulting packing parameters of polymers in aqueous media determine the phase produced, in addition to the environmental conditions under which self-assembly takes place (temperature, pressure, salt concentration, solvent etc.).23 Current bulk assembly methods used to produce nano-scale polymersomes are like those seen for lipid SUV formation, including rehydration of dried polymer films followed by extrusion, shear mixing and probe sonication techniques.23,112–114 Some effort has gone into microfluidic polymersome generation at the μm scale, most notably droplet-templated methods that rely on an off-chip solvent evaporation step to induce self-assembly.115,116 However, as with lipid systems, droplet microfluidic production methods are not so easily transferred to the nano-scale. Continuous flow microfluidics have been used instead, specifically hydrodynamic focusing regimes. A standard MHF chip with micro-mixing features was used by Albuquerque et al. for the synthesis of pH-responsive nano polymersomes (Fig. 8), demonstrating the superb control (via flow rate ratio adjustment) over vesicle diameter characteristic of the method.103 In an interesting paper by Brown et al. the same tapered flow regime was manipulated to generate a controlled pH gradient in a microfluidic channel, triggering the self-assembly of the pH-sensitive polymer poly(2-(methacryloyloxy)ethyl phosphorylcholine)–poly(2-(diisopropylamino)ethyl methacrylate) (PMPC-b-PDPA) into polymersomes between 75 nm and 275 nm with PDI values of ∼0.1.117 The method relies on diffusive mixing of protons but differs from MHF in that there is poor size control and both streams are aqueous, avoiding the need for potentially bio-incompatible organic solvents (EtOH, MeCN etc.). Regardless, it is an interesting proof-of-concept, and future designs may address this reduced controllability.117 Relatively little attention (compared to liposomes) has been given to the specific design of microfluidic platforms for nano-scale polymersome generation. Based on existing examples however, it is reasonable to suggest that well established, lipid-focused designs may be successfully repurposed for polymer vesicles with some minor alterations, and due consideration of polymer size, degrees of molecular freedom and diffusive properties.Polymer/lipid hybrids are an emerging class of membranous nanostructure that seek to retain the advantages of both amphiphile class – the versatility and functionality of polymers coupled with the nature mimicking biocompatibility of lipids.23,118 Recent applications include viral tracking and specialised drug delivery.25,119,120 However, there are issues associated with their preparation; achieving reproducibility is often an onerous task and phase separation between polymeric and lipidic components can be difficult to both predict and prevent.23,118 It is therefore expected that adopting microfluidic approach to hybrid vesicle generation will propel the field considerably, though as far as we are aware no examples currently exist in the literature. As has been mentioned, MHF provides considerable control over amphiphile composition, concentration, and the self-assembly process itself, and future iterations may look to include these nano assemblies.
Virus-like particles
Virus-like particles (VLPs) consist of self-assembled, repeating protein subunits, often termed “coat proteins” or capsid proteins (CPs), that structurally mimic viruses, and are typically produced via modified gene expression in recombinant organisms.121,122 They have been shown to enhance and broaden immunological response when compared to singular protein/peptide immunotherapies; densely packed CPs can present multiple antigenic peptides not necessarily derived from the parent virus.123–125 This is particularly pertinent nowadays with the emergence of rapidly mutating viral strains.124,125 Their potential for use as “multi-valent vaccines” is highlighted in a recent publication from Garg et al., who present a VLP effective against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus.126 Beyond vaccinology, they are beginning to find use in point-of-care diagnostic research and biosensing,127,128 as novel delivery vehicles for small molecule drugs and gene therapies,129–131 and as biomimetic particles that could deepen our understanding of virus-cell interactions.132,133 Despite this range, research incorporating microfluidic techniques into the production and manipulation of VLPs is limited. This presents an exciting new opportunity given the benefits associated with the miniaturisation of a process, as regards cost-of-goods and reaction kinetics. One could envisage rapid, high throughput VLP separation, analysis, and screening on a single microfluidic platform. VLPs are successfully coupled with microfluidics in a device designed by Zang et al., where Tobacco Mosaic VLPs were immobilised on open microfluidic channel via gold–thiol (cysteine residue) interactions.128 VLPs were generated off-chip in recombinant E. coli, purified and fed into the device shown in Fig. 9 at low concentrations (0.2–4 mg ml−1). Evenly coated surfaces were achieved passively via a capillary flow and evaporation mechanism. Each CP was genetically modified to present a FLAG-tag receptor peptide, commonly used in antibody sensing, here used to detect anti-FLAG IgG. Changes in impedance over an interdigitated Au electrode were measured as a function of target molecule (antibody) concentration (ng ml−1), and a linear relationship was observed.128 They boast a surface functionalisation time of only 6 minutes compared to the 18 hours usually required to saturate an impedance sensor, attributing this to the local VLP concentration increase present after evaporation-capillary flow cycles.128 This is an impressive manipulation of microfluidic capillary action that displays the biosensing capability of VLPs, and will no doubt inspire future iterations of label-free biosensors.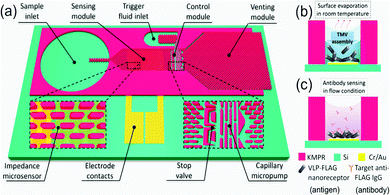 | ||
| Fig. 9 Graphical depiction of the (a) self-automated capillary-flow microfluidic chip, coupled with an interdigitated electrode for impedance measurements, (b) electrode deposition and functionalisation with Tobacco Mosaic VLPs via evaporation and (c) anti-FLAG IgG sensing. Reprinted with permission from Zang et al., ACS Appl. Mater. Interfaces, 2017, 9, 8471–8479. Copyright (2017) American Chemical Society.128 | ||
Future perspectives and concluding comments
Some interesting examples of microfluidic methods used in the controlled assembly of compositionally diverse/higher-order bio-inspired nanostructures have been presented, though the field is very much in its infancy. A growing appreciation for the potential applications of these structures as functional nanomaterials is expected to coincide with the emergence of new microfluidic methods.The use of these structures in biosensing has also been explored, and future optimisations and iterations could well see their use as novel, point-of-care diagnostic tools. The microfluidic assembly of solid/inorganic nanoparticles has been extensively reviewed elsewhere. Of note is a recent publication by Zhao et al., who outline existing platforms and give an exhaustive account of continuous flow microfluidic chip design.19 The process of particle assembly in these devices tends to follow similar principles to those of MHF. A route toward enhancing the complexity of biomimetic nano-assemblies may be to take inspiration from these existing designs. Chips capable of accurately tuning environmental conditions like temperature134 and salt concentration135,136 have already been optimised, and could well be re-engineered to controllably induce lipid/polymer-based nano-assembly phase changes, without any external preparatory steps.
As has been discussed, non-lamellar liquid crystalline nanoparticles (hexosomes and cubosomes) are likely to represent the next generation of functional drug delivery vehicles. Establishing robust microfluidic generation platforms could accelerate their incorporation into pharmaceutical formulations. The scalability of these microfluidic platforms will also determine their future in industry. Proof-of-concept research has benefited from photoresist soft lithography and is to date the most prolific microfabrication method. The process is time-consuming however, and requires significant operator training, making it an unlikely candidate for mass-scale production. The field may look to 3D printing techniques as an alternative. A highly automated fabrication process, 3D printing effectively eliminates human error and would assure the compliance of these devices with industrial standards. A review by Weisgrab et al. has excellently summarised recent advances in 3D microfabrication methods.137 Alongside scalability, microfluidic chip features capable of generating complex flow regimes, actuation and sensing can be more easily introduced.137,138
As discussed by Carugo et al., an extensive study of the encapsulation efficiency of nano-assemblies produced by MHF has yet to be carried out.31 This is an essential step for the comprehensive characterisation of the technique. The necessary separation of these particles from unencapsulated material will also require attention. Current methods including dialysis and size-exclusion chromatography take place off-chip, and detract from the streamlined, ease-of-operation so integral to microfluidic technologies. We expect future technologies will take inspiration from existing microfluidic methods for nanoparticle purification. In an interesting article by Hood et al., microfluidic channels capable of dialysis/solvent exchange were introduced downstream of a standard MHF junction, allowing “on-chip” purification of loaded liposomes.139
The problems facing the microfluidic assembly of biomimetic nano-assemblies are by no means insurmountable. The structures are rooted in a field of research that is inherently interdisciplinary and collaborative. Though current literature examples (summarised in Table 1) are limited, we expect these higher-order bioinspired particles to be at the centre of many future research efforts.
| Structure | Uses | Microfluidic method | Ref. no. |
|---|---|---|---|
| Cubosomes | Drug delivery, protein crystallography, nanomachines, biophysical studies | Micro-emulsification & solvent evaporation | 63 |
| Hexosomes | Biophysical studies, drug delivery, nanomachines, gene delivery | Microfluidic hydrodynamic focusing (MHF) with long serpentine channel | 84 |
| Multi-lamellar vesicles | Biophysical studies, (potentially) site-specific drug delivery and nanomachines | MHF (nano-scale); Langmuir–Blodgett type approach (microscale) | 92 and 93 |
| Nanodiscs | Membrane protein extraction and analysis | Micro-mixing | 100 |
| Polymersomes & hybrid vesicles | Functional and responsive nanomaterials, drug delivery | Microfluidic Hydrodynamic Focusing (MHF), microfluidic pH gradient | 103 and 117 |
| Virus-like particles | Vaccinology, immunotherapeutic delivery, antigen-directed drug delivery | No generation method; used as biosensor in microfluidic chip | 128 |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a UKRI Future Leaders Fellowship, grant reference number MR/S031537/1 awarded to Y. E. and a departmental scholarship (Imperial College) awarded to C. P. P.Notes and references
- M. A. J. Roberts, R. M. Cranenburgh, M. P. Stevens and P. C. F. Oyston, Synthetic biology: biology by design, Microbiology, 2013, 159(Pt 7), 1219–1220 CrossRef CAS.
- K. Göpfrich, I. Platzman and J. P. Spatz, Mastering Complexity: Towards Bottom-up Construction of Multifunctional Eukaryotic Synthetic Cells, Trends Biotechnol., 2018, 36(9), 938–951 CrossRef.
- A. Salehi-Reyhani, O. Ces and Y. Elani, Artificial cell mimics as simplified models for the study of cell biology, Exp. Biol. Med., 2017, 242(13), 1309–1317 CrossRef CAS.
- B. C. Buddingh’, J. Elzinga and J. C. M. van Hest, Intercellular communication between artificial cells by allosteric amplification of a molecular signal, Nat. Commun., 2020, 11(1), 1652 CrossRef.
- A. Joseph, C. Contini, D. Cecchin, S. Nyberg, L. Ruiz-Perez, J. Gaitzsch and G. Battaglia, Chemotactic synthetic vesicles: Design and applications in blood-brain barrier crossing, Sci. Adv., 2017, 3(8), e1700362 CrossRef.
- Y. Elani, Interfacing Living and Synthetic Cells as an Emerging Frontier in Synthetic Biology, Angew. Chem., Int. Ed., 2021 DOI:10.1002/anie.202006941.
- J. Yang, A. Bahreman, G. Daudey, J. Bussmann, R. C. L. Olsthoorn and A. Kros, Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes, ACS Cent. Sci., 2016, 2(9), 621–630 CrossRef CAS.
- C. Peetla, A. Stine and V. Labhasetwar, Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery, Mol. Pharmaceutics, 2009, 6(5), 1264–1276 CrossRef CAS.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu, A. K. Sood and S. Hua, Advances and Challenges of Liposome Assisted Drug Delivery, Front. Pharmacol., 2015, 6, 286 Search PubMed.
- M. Alavi, N. Karimi and M. Safaei, Application of Various Types of Liposomes in Drug Delivery Systems, Adv. Pharm. Bull., 2017, 7(1), 3–9 CrossRef CAS.
- T. Osaki and S. Takeuchi, Artificial Cell Membrane Systems for Biosensing Applications, Anal. Chem., 2017, 89(1), 216–231 CrossRef CAS.
- J. M. Thomas, M. S. Friddin, O. Ces and Y. Elani, Programming membrane permeability using integrated membrane pores and blockers as molecular regulators, Chem. Commun., 2017, 53(91), 12282–12285 RSC.
- E. D. Carlson, R. Gan, C. E. Hodgman and M. C. Jewett, Cell-free protein synthesis: applications come of age, Biotechnol. Adv., 2012, 30(5), 1185–1194 CrossRef CAS.
- S. N. Lai, X. Zhou, X. Ouyang, H. Zhou, Y. Liang, J. Xia and B. Zheng, Artificial Cells Capable of Long-Lived Protein Synthesis by Using Aptamer Grafted Polymer Hydrogel, ACS Synth. Biol., 2020, 9(1), 76–83 CrossRef CAS.
- J. P. Allen, Design of energy-transducing artificial cells, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(15), 3790–3791 CrossRef CAS.
- N. A. C. Jackson, K. E. Kester, D. Casimiro, S. Gurunathan and F. DeRosa, The promise of mRNA vaccines: a biotech and industrial perspective, npj Vaccines, 2020, 5(1), 11 CrossRef.
- A. M. Reichmuth, M. A. Oberli, A. Jaklenec, R. Langer and D. Blankschtein, mRNA vaccine delivery using lipid nanoparticles, Ther. Delivery, 2016, 7(5), 319–334 CrossRef CAS.
- T. Trantidou, M. S. Friddin, A. Salehi-Reyhani, O. Ces and Y. Elani, Droplet microfluidics for the construction of compartmentalised model membranes, Lab Chip, 2018, 18(17), 2488–2509 RSC.
- X. Zhao, F. Bian, L. Sun, L. Cai, L. Li and Y. Zhao, Microfluidic Generation of Nanomaterials for Biomedical Applications, Small, 2020, 16(9), 1901943 CrossRef CAS.
- G. C. Shearman, O. Ces, R. H. Templer and J. M. Seddon, Inverse lyotropic phases of lipids and membrane curvature, J. Phys.: Condens. Matter, 2006, 18(28), S1105–S1124 CrossRef CAS.
- R. Mezzenga, J. M. Seddon, C. J. Drummond, B. J. Boyd, G. E. Schröder-Turk and L. Sagalowicz, Nature-Inspired Design and Application of Lipidic Lyotropic Liquid Crystals, Adv. Mater., 2019, 31(35), 1900818 CrossRef.
- J. N. Israelachvili, Intermolecular and surface forces, Elsevier Science & Technology, 3rd edn, 2010 Search PubMed.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Liposomes and polymersomes: a comparative review towards cell mimicking, Chem. Soc. Rev., 2018, 47(23), 8572–8610 RSC.
- M. Schulz and W. H. Binder, Mixed Hybrid Lipid/Polymer Vesicles as a Novel Membrane Platform, Macromol. Rapid Commun., 2015, 36(23), 2031–2041 CrossRef CAS.
- H.-O. Kim, W. Na, M. Yeom, J.-W. Lim, E.-H. Bae, G. Park and D. Song, Dengue Virus–Polymersome Hybrid Nanovesicles for Advanced Drug Screening Using Real-Time Single Nanoparticle–Virus Tracking, ACS Appl. Mater. Interfaces, 2020, 12(6), 6876–6884 CrossRef CAS.
- D. Khaykelson and U. Raviv, Studying viruses using solution X-ray scattering, Biophys. Rev., 2020, 12(1), 41–48 CrossRef.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, The present and future role of microfluidics in biomedical research, Nature, 2014, 507(7491), 181–189 CrossRef CAS.
- N. Convery and N. Gadegaard, 30 years of microfluidics, Micro- Nanoeng., 2019, 2, 76–91 CrossRef.
- N.-T. Nguyen, M. Hejazian, C. H. Ooi and N. Kashaninejad, Recent Advances and Future Perspectives on Microfluidic Liquid Handling, Micromachines, 2017, 8(6), 186 CrossRef.
- D. van Swaay and A. deMello, Microfluidic methods for forming liposomes, Lab Chip, 2013, 13(5), 752–767 RSC.
- D. Carugo, E. Bottaro, J. Owen, E. Stride and C. Nastruzzi, Liposome production by microfluidics: potential and limiting factors, Sci. Rep., 2016, 6(1), 25876 CrossRef CAS.
- P. Supramaniam, O. Ces and A. Salehi-Reyhani, Microfluidics for Artificial Life: Techniques for Bottom-Up Synthetic Biology, Micromachines, 2019, 10(5), 299 CrossRef.
- S. Deshpande, Y. Caspi, A. E. C. Meijering and C. Dekker, Octanol-assisted liposome assembly on chip, Nat. Commun., 2016, 7(1), 10447 CrossRef CAS.
- O. Staufer, M. Schröter, I. Platzman and J. P. Spatz, Bottom-Up Assembly of Functional Intracellular Synthetic Organelles by Droplet-Based Microfluidics, Small, 2020, 1906424 CrossRef CAS.
- N.-N. Deng, M. Yelleswarapu, L. Zheng and W. T. S. Huck, Microfluidic Assembly of Monodisperse Vesosomes as Artificial Cell Models, J. Am. Chem. Soc., 2017, 139(2), 587–590 CrossRef CAS.
- P. Vega-Vásquez, N. S. Mosier and J. Irudayaraj, Nanoscale Drug Delivery Systems: From Medicine to Agriculture, Front. Bioeng. Biotechnol., 2020, 8, 79 CrossRef.
- S. G. M. Ong, M. Chitneni, K. S. Lee, L. C. Ming and K. H. Yuen, Evaluation of Extrusion Technique for Nanosizing Liposomes, Pharmaceutics, 2016, 8(4), 36 CrossRef.
- A. Wagner and K. Vorauer-Uhl, Liposome technology for industrial purposes, J. Drug Delivery, 2011, 2011, 591325 Search PubMed.
- Y. Elani, X. C. I. Solvas, J. B. Edel, R. V. Law and O. Ces, Microfluidic generation of encapsulated droplet interface bilayer networks (multisomes) and their use as cell-like reactors, Chem. Commun., 2016, 52(35), 5961–5964 RSC.
- T. Trantidou, L. Dekker, K. Polizzi, O. Ces and Y. Elani, Functionalizing cell-mimetic giant vesicles with encapsulated bacterial biosensors, Interface Focus, 2018, 8(5), 20180024 CrossRef.
- D. R. Link, S. L. Anna, D. A. Weitz and H. A. Stone, Geometrically mediated breakup of drops in microfluidic devices, Phys. Rev. Lett., 2004, 92(5), 054503 CrossRef CAS.
- A. M. Gañán-Calvo, R. González-Prieto, P. Riesco-Chueca, M. A. Herrada and M. Flores-Mosquera, Focusing capillary jets close to the continuum limit, Nat. Phys., 2007, 3(10), 737–742 Search PubMed.
- L. Brown, S. L. McArthur, P. C. Wright, A. Lewis and G. Battaglia, Polymersome production on a microfluidic platform using pH sensitive block copolymers, Lab Chip, 2010, 10(15), 1922–1928 RSC.
- M. Lu, A. Ozcelik, C. L. Grigsby, Y. Zhao, F. Guo, K. W. Leong and T. J. Huang, Microfluidic Hydrodynamic Focusing for Synthesis of Nanomaterials, Nano Today, 2016, 11(6), 778–792 CrossRef CAS.
- J. P. Golden, G. A. Justin, M. Nasir and F. S. Ligler, Hydrodynamic focusing – a versatile tool, Anal. Bioanal. Chem., 2012, 402(1), 325–335 CrossRef CAS.
- D. R. Mott, J. P. B. Howell, J. P. Golden, C. R. Kaplan, F. S. Ligler and E. S. Oran, Toolbox for the design of optimised microfluidic components, Lab Chip, 2006, 6(4), 540–549 RSC.
- R. R. Hood and D. L. DeVoe, High-Throughput Continuous Flow Production of Nanoscale Liposomes by Microfluidic Vertical Flow Focusing, Small, 2015, 11(43), 5790–5799 CrossRef CAS.
- D. Conchouso, D. Castro, S. A. Khan and I. G. Foulds, Three-dimensional parallelization of microfluidic droplet generators for a litre per hour volume production of single emulsions, Lab Chip, 2014, 14(16), 3011–3020 RSC.
- T. Nisisako and T. Torii, Microfluidic large-scale integration on a chip for mass production of monodisperse droplets and particles, Lab Chip, 2008, 8(2), 287–293 RSC.
- H. M. G. Barriga, M. N. Holme and M. M. Stevens, Cubosomes: The Next Generation of Smart Lipid Nanoparticles?, Angew. Chem., Int. Ed., 2019, 58(10), 2958–2978 CrossRef CAS.
- Z. A. Almsherqi, S. D. Kohlwein and Y. Deng, Cubic membranes: a legend beyond the Flatland* of cell membrane organisation, J. Cell Biol., 2006, 173(6), 839–844 CrossRef CAS.
- S. J. Fraser, X. Mulet, A. Hawley, F. Separovic and A. Polyzos, Controlling nanostructure and lattice parameter of the inverse bicontinuous cubic phases in functionalised phytantriol dispersions, J. Colloid Interface Sci., 2013, 408, 117–124 CrossRef CAS.
- W. Sun, J. J. Vallooran, A. Zabara and R. Mezzenga, Controlling enzymatic activity and kinetics in swollen mesophases by physical nano-confinement, Nanoscale, 2014, 6(12), 6853–6859 RSC.
- H. M. G. Barriga, O. Ces, R. V. Law, J. M. Seddon and N. J. Brooks, Engineering Swollen Cubosomes Using Cholesterol and Anionic Lipids, Langmuir, 2019, 35(50), 16521–16527 CrossRef CAS.
- A. Zabara, J. T. Y. Chong, I. Martiel, L. Stark, B. A. Cromer, C. Speziale and R. Mezzenga, Design of ultra-swollen lipidic mesophases for the crystallisation of membrane proteins with large extracellular domains, Nat. Commun., 2018, 9(1), 544 CrossRef.
- J. Y. T. Chong, X. Mulet, B. J. Boyd and C. J. Drummond, Chapter Five – Steric Stabilisers for Cubic Phase Lyotropic Liquid Crystal Nanodispersions (Cubosomes), in Advances in Planar Lipid Bilayers and Liposomes, ed. A. Iglič, C. V. Kulkarni and M. Rappolt, Academic Press, 2015, vol. 21, pp. 131–187 Search PubMed.
- J. Zhai, R. Suryadinata, B. Luan, N. Tran, T. M. Hinton, J. Ratcliffe and C. J. Drummond, Amphiphilic brush polymers produced using the RAFT polymerisation method stabilise and reduce the cell cytotoxicity of lipid lyotropic liquid crystalline nanoparticles, Faraday Discuss., 2016, 191(0), 545–563 RSC.
- C. Caltagirone, A. M. Falchi, S. Lampis, V. Lippolis, V. Meli, M. Monduzzi and S. Murgia, Cancer-Cell-Targeted Theranostic Cubosomes, Langmuir, 2014, 30(21), 6228–6236 CrossRef CAS.
- I. D. Azmi, S. M. Moghimi and A. Yaghmur, Cubosomes and hexosomes as versatile platforms for drug delivery, Ther. Delivery, 2015, 6(12), 1347–1364 CrossRef CAS.
- N. M. Lakshmi, P. R. Yalavarthi, H. C. Vadlamudi, J. Thanniru, G. Yaga and H. Kanishka, Cubosomes as targeted drug delivery systems – a biopharmaceutical approach, Curr. Drug Discovery Technol., 2014, 11(3), 181–188 CrossRef CAS.
- L. Boge, H. Bysell, L. Ringstad, D. Wennman, A. Umerska, V. Cassisa and M. Andersson, Lipid-Based Liquid Crystals As Carriers for Antimicrobial Peptides: Phase Behavior and Antimicrobial Effect, Langmuir, 2016, 32(17), 4217–4228 CrossRef CAS.
- S. B. Rizwan, W. T. McBurney, K. Young, T. Hanley, B. J. Boyd, T. Rades and S. Hook, Cubosomes containing the adjuvants imiquimod and monophosphoryl lipid A stimulate robust cellular and humoral immune responses, J. Controlled Release, 2013, 165(1), 16–21 CrossRef CAS.
- H. Kim, J. Sung, Y. Chang, A. Alfeche and C. Leal, Microfluidics Synthesis of Gene Silencing Cubosomes, ACS Nano, 2018, 12(9), 9196–9205 CrossRef CAS.
- L. Boge, K. L. Browning, R. Nordström, M. Campana, L. S. E. Damgaard, J. Seth Caous and M. Andersson, Peptide-Loaded Cubosomes Functioning as an Antimicrobial Unit against Escherichia coli, ACS Appl. Mater. Interfaces, 2019, 11(24), 21314–21322 CrossRef CAS.
- S. Sarkar, N. Tran, S. K. Soni, Z. Nasa, C. J. Drummond and C. E. Conn, Cuboplex-Mediated Nonviral Delivery of Functional siRNA to Chinese Hamster Ovary (CHO) Cells, ACS Appl. Mater. Interfaces, 2021, 13(2), 2336–2345 CrossRef CAS.
- N. R. Rarokar, S. D. Saoji, N. A. Raut, J. B. Taksande, P. B. Khedekar and V. S. Dave, Nanostructured Cubosomes in a Thermoresponsive Depot System: An Alternative Approach for the Controlled Delivery of Docetaxel, AAPS PharmSciTech, 2016, 17(2), 436–445 CrossRef CAS.
- E. Bukusoglu, X. Wang, J. A. Martinez-Gonzalez, J. J. de Pablo and N. L. Abbott, Stimuli-Responsive Cubosomes Formed from Blue Phase Liquid Crystals, Adv. Mater., 2015, 27(43), 6892–6898 CrossRef CAS.
- S. Murgia, S. Bonacchi, A. M. Falchi, S. Lampis, V. Lippolis, V. Meli and C. Caltagirone, Drug-Loaded Fluorescent Cubosomes: Versatile Nanoparticles for Potential Theranostic Applications, Langmuir, 2013, 29(22), 6673–6679 CrossRef CAS.
- O. Mertins, P. D. Mathews and A. Angelova, Advances in the Design of pH-Sensitive Cubosome Liquid Crystalline Nanocarriers for Drug Delivery Applications, Nanomaterials, 2020, 10(5), 963 CrossRef CAS.
- Z. Yang, X. Peng, Y. Tan, M. Chen, X. Zhu, M. Feng and C. Wu, Optimisation of the Preparation Process for an Oral Phytantriol-Based Amphotericin B Cubosomes, J. Nanomater., 2011, 2011, 308016 Search PubMed.
- M. A. Ali, S. Noguchi, Y. Iwao, T. Oka and S. Itai, Preparation and Characterisation of SN-38-Encapsulated Phytantriol Cubosomes Containing α-Monoglyceride Additives, Chem. Pharm. Bull., 2016, 64, 577–584 CrossRef CAS.
- P. B. Stathopulos, G. A. Scholz, Y.-M. Hwang, J. A. O. Rumfeldt, J. R. Lepock and E. M. Meiering, Sonication of proteins causes formation of aggregates that resemble amyloid, Protein Sci., 2004, 13(11), 3017–3027 CrossRef CAS.
- D.-H. Kim, S. Lim, J. Shim, J. E. Song, J. S. Chang, K. S. Jin and E. C. Cho, A Simple Evaporation Method for Large-Scale Production of Liquid Crystalline Lipid Nanoparticles with Various Internal Structures, ACS Appl. Mater. Interfaces, 2015, 7(36), 20438–20446 CrossRef CAS.
- S. Revathi and M. D. Dhanaraju, Hexosomes as a novel drug delivery system: A review, Int. J. Pharmacol. Pharm. Sci., 2014, 6, 58–63 Search PubMed.
- I. D. Azmi, S. M. Moghimi and A. Yaghmur, Cubosomes and hexosomes as versatile platforms for drug delivery, Ther. Delivery, 2015, 6(12), 1347–1364 CrossRef CAS.
- I. Amar-Yuli, E. Wachtel, E. B. Shoshan, D. Danino, A. Aserin and N. Garti, Hexosome and Hexagonal Phases Mediated by Hydration and Polymeric Stabilizer, Langmuir, 2007, 23(7), 3637–3645 CrossRef CAS.
- J. Jouhet, Importance of the hexagonal lipid phase in biological membrane organisation, Front. Plant Sci., 2013, 4, 494 Search PubMed.
- S.-J. Marrink and A. E. Mark, Molecular View of Hexagonal Phase Formation in Phospholipid Membranes, Biophys. J., 2004, 87(6), 3894–3900 CrossRef CAS.
- Y. Huang and S. Gui, Factors affecting the structure of lyotropic liquid crystals and the correlation between structure and drug diffusion, RSC Adv., 2018, 8(13), 6978–6987 RSC.
- I. Martiel, N. Baumann, J. J. Vallooran, J. Bergfreund, L. Sagalowicz and R. Mezzenga, Oil and drug control the release rate from lyotropic liquid crystals, J. Controlled Release, 2015, 204, 78–84 CrossRef CAS.
- M. A. Shetab Boushehri, D. Dietrich and A. Lamprecht, Nanotechnology as a Platform for the Development of Injectable Parenteral Formulations: A Comprehensive Review of the Know-Hows and State of the Art, Pharmaceutics, 2020, 12(6), 510 CrossRef CAS.
- Y. Li, A. Angelova, F. Hu, V. M. Garamus, C. Peng, N. Li and A. Zou, pH Responsiveness of Hexosomes and Cubosomes for Combined Delivery of Brucea javanica Oil and Doxorubicin, Langmuir, 2019, 35(45), 14532–14542 CrossRef CAS.
- Y. Chen, P. Ma and S. Gui, Cubic and Hexagonal Liquid Crystals as Drug Delivery Systems, BioMed Res. Int., 2014, 2014, 815981 Search PubMed.
- A. Yaghmur, A. Ghazal, R. Ghazal, M. Dimaki and W. E. Svendsen, A hydrodynamic flow focusing microfluidic device for the continuous production of hexosomes based on docosahexaenoic acid monoglyceride, Phys. Chem. Chem. Phys., 2019, 21(24), 13005–13013 RSC.
- D. Vorselen, M. Marchetti, C. López-Iglesias, P. J. Peters, W. H. Roos and G. J. L. Wuite, Multi-lamellar nanovesicles show distinct mechanical properties depending on their degree of lamellarity, Nanoscale, 2018, 10(11), 5318–5324 RSC.
- P. Lajoie, G. Guay, J. W. Dennis and I. R. Nabi, The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies, J. Cell Sci., 2005, 118(9), 1991 CrossRef CAS.
- V. E. Paquet, R. Lessire, F. Domergue, L. Fouillen, G. Filion, A. Sedighi and S. J. Charette, Lipid Composition of Multi-lamellar Bodies Secreted by Reveals Their Amoebal Origin, Eukaryotic Cell, 2013, 12(10), 1326 CrossRef CAS.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour and K. Nejati-Koshki, Liposome: classification, preparation, and applications, Nanoscale Res. Lett., 2013, 8(1), 102 CrossRef.
- J. A. Bouwstra, G. S. Gooris, W. Bras and H. Talsma, Small angle X-ray scattering: possibilities and limitations in characterisation of vesicles, Chem. Phys. Lipids, 1993, 64(1–3), 83–98 CrossRef CAS.
- J. Peters, J. Marion, F. J. Becher, M. Trapp, T. Gutberlet, D. J. Bicout and T. Heimburg, Thermodynamics of lipid multi-lamellar vesicles in presence of sterols at high hydrostatic pressure, Sci. Rep., 2017, 7(1), 15339 CrossRef CAS.
- D. D. Lasic, The mechanism of vesicle formation, Biochem. J., 1988, 256(1), 1–11 CrossRef CAS.
- S. Matosevic and B. M. Paegel, Layer-by-layer cell membrane assembly, Nat. Chem., 2013, 5(11), 958–963 CrossRef CAS.
- A. Ghazal, M. Gontsarik, J. P. Kutter, J. P. Lafleur, D. Ahmadvand, A. Labrador and A. Yaghmur, Microfluidic Platform for the Continuous Production and Characterisation of Multi-lamellar Vesicles: A Synchrotron Small-Angle X-ray Scattering (SAXS) Study, J. Phys. Chem. Lett., 2017, 8(1), 73–79 CrossRef CAS.
- I. G. Denisov and S. G. Sligar, Nanodiscs in Membrane Biochemistry and Biophysics, Chem. Rev., 2017, 117(6), 4669–4713 CrossRef CAS.
- I. G. Denisov and S. G. Sligar, Nanodiscs for structural and functional studies of membrane proteins, Nat. Struct. Mol. Biol., 2016, 23(6), 481–486 CrossRef CAS.
- K. M. Padmanabha Das, W. M. Shih, G. Wagner and M. L. Nasr, Large Nanodiscs: A Potential Game Changer in Structural Biology of Membrane Protein Complexes and Virus Entry, Front. Bioeng. Biotechnol., 2020, 8, 539 CrossRef.
- M. C. Fiori, W. Zheng, E. Kamilar, G. Simiyu, G. A. Altenberg and H. Liang, Extraction and reconstitution of membrane proteins into lipid nanodiscs encased by zwitterionic styrene-maleic amide copolymers, Sci. Rep., 2020, 10(1), 9940 CrossRef CAS.
- M. C. Fiori, Y. Jiang, W. Zheng, M. Anzaldua, M. J. Borgnia, G. A. Altenberg and H. Liang, Polymer Nanodiscs: Discoidal Amphiphilic Block Copolymer Membranes as a New Platform for Membrane Proteins, Sci. Rep., 2017, 7(1), 15227 CrossRef.
- R. Puthenveetil, K. Nguyen and O. Vinogradova, Nanodiscs and Solution NMR: preparation, application and challenges, Nanotechnol. Rev., 2017, 6(1), 111–126 CAS.
- J. H. Wade, J. D. Jones, I. L. Lenov, C. M. Riordan, S. G. Sligar and R. C. Bailey, Microfluidic platform for efficient Nanodisc assembly, membrane protein incorporation, and purification, Lab Chip, 2017, 17(17), 2951–2959 RSC.
- E. D. Goluch, A. W. Shaw, S. G. Sligar and C. Liu, Microfluidic patterning of nanodisc lipid bilayers and multiplexed analysis of protein interaction, Lab Chip, 2008, 8(10), 1723–1728 RSC.
- M. Azouz, M. Gonin, S. Fiedler, J. Faherty, M. Decossas, C. Cullin and A. Ciaccafava, Microfluidic diffusional sizing probes lipid nanodiscs formation, Biochim. Biophys. Acta, Biomembr., 2020, 1862(6), 183215 CrossRef CAS.
- L. J. C. Albuquerque, V. Sincari, A. Jäger, R. Konefa, J. I. Panek, P. Ĉernoch and E. Jäger, Microfluidic-Assisted Engineering of Quasi-Monodisperse pH-Responsive Polymersomes toward Advanced Platforms for the Intracellular Delivery of Hydrophilic Therapeutics, Langmuir, 2019, 35(25), 8363–8372 CAS.
- L. D. Blackman, S. Varlas, M. C. Arno, A. Fayter, M. I. Gibson and R. K. O’Reilly, Permeable Protein-Loaded Polymersome Cascade Nanoreactors by Polymerization-Induced Self-Assembly, ACS Macro Lett., 2017, 6(11), 1263–1267 CrossRef CAS.
- J. Kim and K. T. Kim, Polymersome-Based Modular Nanoreactors with Size-Selective Transmembrane Permeability, ACS Appl. Mater. Interfaces, 2020, 12(20), 23502–23513 CrossRef CAS.
- M. Kanamala, B. D. Palmer, S. M. F. Jamieson, W. R. Wilson and Z. Wu, Dual pH-sensitive liposomes with low pH-triggered sheddable PEG for enhanced tumor-targeted drug delivery, Nanomedicine, 2019, 14(15), 1971–1989 CrossRef CAS.
- H. Che and J. C. M. van Hest, Stimuli-responsive polymersomes and nanoreactors, J. Mater. Chem., 2016, 4(27), 4632–4647 RSC.
- O. Rifaie-Graham, S. Ulrich, N. F. B. Galensowske, S. Balog, M. Chami, D. Rentsch and N. Bruns, Wavelength-Selective Light-Responsive DASA-Functionalized Polymersome Nanoreactors, J. Am. Chem. Soc., 2018, 140(25), 8027–8036 CrossRef CAS.
- O. Bixner, S. Kurzhals, M. Virk and E. Reimhult, Triggered Release from Thermoresponsive Polymersomes with Superparamagnetic Membranes, Materials, 2016, 9(1), 29 CrossRef.
- J. Gaitzsch, L. Messager, E. Morecroft and W. Meier, Vesicles in Multiple Shapes: Fine-Tuning Polymersomes' Shape and Stability by Setting Membrane Hydrophobicity, Polymers, 2017, 9(10), 483 CrossRef.
- R. Salva, J.-F. Le Meins, O. Sandre, A. Brûlet, M. Schmutz, P. Guenoun and S. Lecommandoux, Polymersome Shape Transformation at the Nanoscale, ACS Nano, 2013, 7(10), 9298–9311 CrossRef CAS.
- Y. Men, F. Peng, Y. Tu, J. C. M. van Hest and D. A. Wilson, Methods for production of uniform small-sized polymersome with rigid membrane, Polym. Chem., 2016, 7(24), 3977–3982 RSC.
- J. Leong, J. Y. Teo, V. K. Aakalu, Y. Y. Yang and H. Kong, Engineering Polymersomes for Diagnostics and Therapy, Adv. Healthcare Mater., 2018, 7(8), 1701276 CrossRef.
- S. T. Poschenrieder, S. K. Schiebel and K. Castiglione, Polymersomes for biotechnological applications: Large-scale production of nano-scale vesicles, Eng. Life Sci., 2017, 17(1), 58–70 CrossRef CAS.
- Y. Hu and J. Pérez-Mercader, Microfluidic fabrication of polymersomes enclosing an active Belousov-Zhabotinsky (BZ) reaction: Effect on their stability of solute concentrations in the external media, Colloids Surf., B, 2016, 146, 406–414 CrossRef CAS.
- J. Petit, I. Polenz, J.-C. Baret, S. Herminghaus and O. Bäumchen, Vesicles-on-a-chip: A universal microfluidic platform for the assembly of liposomes and polymersomes, Eur. Phys. J. E: Soft Matter Biol. Phys., 2016, 39(6), 59 CrossRef.
- L. Brown, S. L. McArthur, P. C. Wright, A. Lewis and G. Battaglia, Polymersome production on a microfluidic platform using pH sensitive block copolymers, Lab Chip, 2010, 10(15), 1922–1928 RSC.
- M. Schulz and W. H. Binder, Mixed Hybrid Lipid/Polymer Vesicles as a Novel Membrane Platform, Macromol. Rapid Commun., 2015, 36(23), 2031–2041 CrossRef CAS.
- J. F. Le Meins, C. Schatz, S. Lecommandoux and O. Sandre, Hybrid polymer/lipid vesicles: state of the art and future perspectives, Mater. Today, 2013, 16(10), 397–402 CrossRef CAS.
- H. Zhang, W. Cui, X. Qu, H. Wu, L. Qu, X. Zhang and D. A. Weitz, Photothermal-responsive nano-sized hybrid polymersome as versatile therapeutics codelivery nanovehicle for effective tumor suppression, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(16), 7744–7749 CrossRef CAS.
- Z. Liu, J. Qiao, Z. Niu and Q. Wang, Natural supramolecular building blocks: from virus coat proteins to viral nanoparticles, Chem. Soc. Rev., 2012, 41(18), 6178–6194 RSC.
- A. Zeltins, Construction and Characterisation of Virus-Like Particles: A Review, Mol. Biotechnol., 2013, 53(1), 92–107 CrossRef CAS.
- H. K. Charlton Hume, J. Vidigal, M. J. T. Carrondo, A. P. J. Middelberg, A. Roldão and L. H. L. Lua, Synthetic biology for bioengineering virus-like particle vaccines, Biotechnol. Bioeng., 2019, 116(4), 919–935 CrossRef CAS.
- A. Roldão, M. C. Mellado, L. R. Castilho, M. J. Carrondo and P. M. Alves, Virus-like particles in vaccine development, Expert Rev. Vaccines, 2010, 9(10), 1149–1176 CrossRef.
- M. O. Mohsen, L. Zha, G. Cabral-Miranda and M. F. Bachmann, Major findings and recent advances in virus-like particle (VLP)-based vaccines, Semin. Immunol., 2017, 34, 123–132 CrossRef CAS.
- H. Garg, T. Mehmetoglu-Gurbuz and A. Joshi, Virus Like Particles (VLP) as multivalent vaccine candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus, Sci. Rep., 2020, 10(1), 4017 CrossRef CAS.
- K. M. Ricks, C. J. Shoemaker, L. C. Dupuy, O. Flusin, M. A. Voorhees, A. N. Fulmer and R. J. Schoepp, Development of a bead-based immunoassay using virus-like particles for detection of alphaviral humoral response, J. Virol. Methods, 2019, 270, 12–17 CrossRef CAS.
- F. Zang, K. Gerasopoulos, A. D. Brown, J. N. Culver and R. Ghodssi, Capillary Microfluidics-Assembled Virus-like Particle Bionanoreceptor Interfaces for Label-Free Biosensing, ACS Appl. Mater. Interfaces, 2017, 9(10), 8471–8479 CrossRef CAS.
- M. Zdanowicz and J. Chroboczek, Virus-like particles as drug delivery vectors, Acta Biochim. Pol., 2016, 63(3), 469–473 CrossRef CAS.
- H. Petry, C. Goldmann, O. Ast and W. Lüke, The use of virus-like particles for gene transfer, Curr. Opin. Mol. Ther., 2003, 5(5), 524–528 CAS.
- E. B. Lee, J.-H. Kim, W. Hur, J. E. Choi, S. M. Kim, D. J. Park and S. K. Yoon, Liver-specific Gene Delivery Using Engineered Virus-Like Particles of Hepatitis E Virus, Sci. Rep., 2019, 9(1), 1616 CrossRef.
- N. F. Steinmetz, S. Lim and F. Sainsbury, Protein cages and virus-like particles: from fundamental insight to biomimetic therapeutics, Biomater. Sci., 2020, 8(10), 2771–2777 RSC.
- J. Zepeda-Cervantes, J. O. Ramírez-Jarquín and L. Vaca, Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs, Front. Immunol., 2020, 11, 1100 CrossRef CAS.
- E. M. Chan, A. P. Alivisatos and R. A. Mathies, High-Temperature Microfluidic Synthesis of CdSe Nanocrystals in Nanoliter Droplets, J. Am. Chem. Soc., 2005, 127(40), 13854–13861 CrossRef CAS.
- M. J. Kennedy, H. D. Ladouceur, T. Moeller, D. Kirui and C. A. Batt, Analysis of a laminar-flow diffusional mixer for directed self-assembly of liposomes, Biomicrofluidics, 2012, 6(4), 44119 CrossRef.
- B. W. Muir, G. Zhen, P. Gunatillake and P. G. Hartley, Salt Induced Lamellar to Bicontinuous Cubic Phase Transitions in Cationic Nanoparticles, J. Phys. Chem. B, 2012, 116(11), 3551–3556 CrossRef CAS.
- G. Weisgrab, A. Ovsianikov and P. F. Costa, Functional 3D Printing for Microfluidic Chips, Adv. Mater. Technol., 2019, 4(10), 1900275 CrossRef CAS.
- A. Waldbaur, H. Rapp, K. Länge and B. E. Rapp, Let there be chip—towards rapid prototyping of microfluidic devices: one-step manufacturing processes, Anal. Methods, 2011, 3(12), 2681–2716 RSC.
- R. R. Hood, W. N. Vreeland and D. L. DeVoe, Microfluidic remote loading for rapid single-step liposomal drug preparation, Lab Chip, 2014, 14(17), 3359–3367 RSC.
| This journal is © the Owner Societies 2021 |