Open Access Article
Open Access ArticleThe importance of the bacterial cell wall in uranium(VI) biosorption†
Joseph
Hufton
 *a,
John
Harding
*a,
John
Harding
 b,
Thomas
Smith
b,
Thomas
Smith
 c and
Maria E.
Romero-González
c and
Maria E.
Romero-González
 *ad
*ad
aDepartment of Geography, The University of Sheffield, Sheffield, S10 2TN, UK. E-mail: j.m.hufton@sheffield.ac.uk
bDepartment of Materials Science and Engineering, Sir Robert Hadfield Building, University of Sheffield, Mappin Street, Sheffield, S1 3JD, UK
cBiomolecular Sciences Centre, Sheffield Hallam University, City Campus, Sheffield S1 1WB, UK
dSchool of Engineering and Materials Science (SEMS), Queen Mary University of London, Mile End Road, London E1 4NS, UK. E-mail: m.romero-gonzalez@qmul.ac.uk
First published on 17th December 2020
Abstract
The bacterial cell envelope, in particular the cell wall, is considered the main controlling factor in the biosorption of aqueous uranium(VI) by microorganisms. However, the specific roles of the cell wall, associated biomolecules, and other components of the cell envelope are not well defined. Here we report findings on the biosorption of uranium by isolated cell envelope components and associated biomolecules, with P. putida 33015 and B. subtilis 168 investigated as representative strains for the differences in Gram-negative and Gram-positive cell envelope architecture, respectively. The cell wall and cell surface membrane were isolated from intact cells and characterised by X-ray Photoelectron Spectroscopy (XPS) and Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FT-IR) spectroscopy; revealing variations in the abundance of functional moieties and biomolecules associated with components of the cell envelope. Uranium biosorption was investigated as a function of cell envelope component and pH, comparing with intact cells. The isolated cell wall from both strains exhibited the greatest uranium biosorption capacity. Deprotonation of favourable functional groups on the biomass as the pH increased from 3 to 5.5 increased their uranium biosorption capacity by approximately 3 fold. The results from ATR-FT-IR indicated that uranium(VI) biosorption was mediated by phosphate and carboxyl groups associated with proteins and phosphorylated biopolymers of the cell envelope. This includes outer membrane phospholipids and LPS of Gram-negative bacteria and teichoic acids, surface proteins and peptidoglycan from Gram-positive bacteria. As a result, the biosorption process of uranium(VI) to microorganisms is controlled by surface interactions, resulting in higher accumulation of uranium in the cell envelope. This demonstrates the importance of bacterial cell wall as the key mediator of uranium biosorption with microorganisms.
1 Introduction
Increased anthropogenic activity and the use of uranium in the nuclear fuel industry have caused large quantities of dissolved uranium to be released into the environment.1,2 Biosorption3–5 and biomineralisation6–8 provide opportunities for the use of environmentally friendly processes that utilise the power of microorganisms to immobilise and remove uranium from waste solutions. The realisation of these processes on large scales requires careful control of pH, redox conditions and concentrations of uranium in the effluent, since these all affect its mobility in solution and, in turn, govern the radioactive material's interactions with microorganisms.2,9–11 Despite the amount of existing literature in this area, the biosorption and accumulation processes exhibited by microorganisms are still not fully understood at a mechanistic level, making it difficult to consider their use and application in remediation, extraction and reuse of radioactive materials.It is known that the physiochemical interactions of bacterial cells with uranium are governed by intermolecular forces between functional groups associated with those of the interacting surface, in particular those associated with the cell wall.12–15 This has been suggested in numerous spectroscopic and microscopy techniques. Many studies and strains, including Gram-positive Bacillus species,16–21Paenibacillus sp. JG-TB8,22 Gram-negative strains Pseudomonas fluorescens,23Cupriavidus metallidurans24 and yeasts such as Saccharomyces cerevisiae25 have highlighted the cell wall's importance in uranium biosorption at the cell surface, as either the sole mechanism of uranium bioremediation or a precursor step to cell surface and intracellular biomineralisation.
However, the experimental approaches used in these studies make it difficult to see whether this process happens only at the outermost cell wall or can involve other components of the cell envelope. Uranium deposition onto the cell surface of Gram-negative strains Sphingomonas sp. S15-S1,26Rhodopseudomonas palustris24 and Acidovorax facilis27 indicate that uranium interacts with the cell surface membrane as well as with the outer membrane of the Gram-negative cell wall. Similarly, uranium deposits were apparent at both cell wall and cell surface membrane of the yeast Rhodotorula mucilaginosa B11-R8.28
Despite the evidence for uranium sequestration at the cell surface, the contribution of each component of the cell envelope and associated biomolecules to the biosorption of uranium(VI) is largely unknown. The availability of functional groups associated with the cell envelope varies with microbial strain, depending in particular on whether their cell wall architecture is of Gram-positive or Gram-negative origin.5,29 Therefore, understanding the role of individual cell envelope components extends the ability to determine the localisation, interaction strength and immobilisation of uranium with microorganisms.
Many studies have investigated how intact cells sequester U(VI) from acidic environments, focusing solely on intact cells and purified polymers commercially available.30–33 To date, no studies have been performed on components extracted from the cell envelope.
This work aims to reveal the role of the different components of the bacterial cell envelope, in particular the cell wall, of Pseudomonas putida 33015 and Bacillus subtilis 168 in uranium biosorption from acidic environments. This was achieved by isolating and characterising components of the bacterial cell envelope and quantifying their capacity for uranium biosorption in comparison with intact cells. We hypothesised that the cell wall components would retain a greater amount of uranium from acidic environments due to an increase in abundance of cell surface functional moieties sensitive to the presence of uranium in comparison with those present in intact cells.
A range of spectroscopic techniques were used to study the biosorption process. X-ray photoelectron spectroscopy (XPS) is an experimental technique that provides elemental analysis of surfaces, and was used to characterise the components of the bacterial cell envelope following extraction and purification. It is therefore ideal for the characterisation of functional groups and the quantification of biomolecules within the cell wall, such as peptides, polysaccharides and lipid-like material.12,34–37 Attenuated Total Reflectance Fourier Transform-Infrared Spectroscopy (ATR-FT-IR) was used to identify the relative abundance of biomolecules and associated functional groups in components of the cell envelope. This spectroscopic technique was used to identify those biomolecules involved in uranium biosorption.38–42
The results from this study will give insight into how the difference in molecular composition and functional groups associated with specific components of the cell envelope and biomolecules affect the mechanistic processes involved in uranium biosorption. Furthermore, understanding these mechanistic processes is essential for designing an effective remediation process using bacterial biomass.
2 Experimental procedures
2.1 Bacteria and growth conditions
Two bacteria were used in this study; Pseudomonas putida 33015 (ATCC 33015) and Bacillus subtilis 168 (ATCC 23857) were obtained from LGC standards. P. putida 33015 was grown in sodium benzoate media (3.0 g L−1 (NH4)2HPO4, 1.2 g/LKH2PO4, 5.0 g L−1 NaCl, 0.2 g L−1 MgSO4·H2O, 0.5 g L−1 yeast extract, 3.0 g L−1 sodium benzoate) and B. subtilis 168 in nutrient broth (Sigma: 70122) in 1 L cultures until cells reached late exponential phase in their growth cycle, while shaking at 170 rpm (30 °C).2.2 Cell fractionation
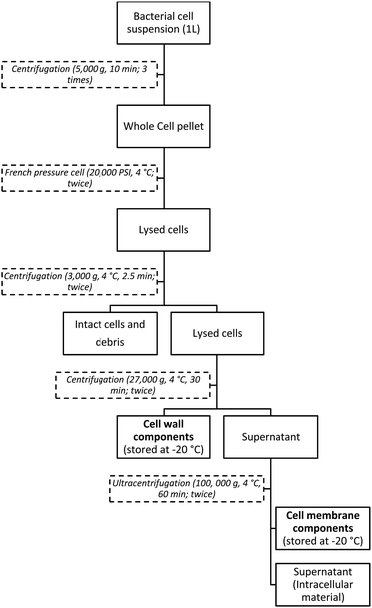
A cell fractionation method using a French pressure cell and ultracentrifuge was adapted and used43 to isolate cell wall and membrane components for spectroscopic characterisation and subsequent uranium biosorption experiments. This method of fractionation is widely reported for isolating pure cell wall and membrane components from multiple bacterial strains, without the need for assessing purity,44–46 and has been successfully adapted using different disruptive apparatus.47–49 A schematic of the isolation cell envelope components is shown in Fig. 1. Two main products are obtained through this process: cell wall components and cell membrane components. For Gram-negative bacteria, the cell wall components include peptidoglycan, capsular polysaccharides, lipopolysaccharides and porins. For Gram-positive bacteria, the cell wall components are mainly peptidoglycan, surface proteins, cell wall associated proteins, teichoic and lipoteichoic acids. The cell membrane components are common for both Gram-positive and Gram-negative bacteria and their composition is characterised by the presence of phospholipids, membrane associated proteins and membrane-derived oligosaccharides (MDO).The cells from a 1 L culture of P. putida 33015 and B. subtilis 168 were harvested by centrifugation at 5000g (10 min), washed 3 times in 0.1 M NaCl and re-suspended in 10 mL ice-cold 18.2 MΩ ultrapure water. H2O was used, rather than a buffered solution, as previously prepared,43 to prevent contamination from excess buffer constituents and to ensure the purity of extracted cell envelope components for uranium biosorption experiments. The concentrated cell suspension was passed through a French pressure cell (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PSI, 4 °C) three times to break open the cells to isolate cell wall and membrane components. The lysed cells were centrifuged (3000g, 4 °C, 2.5 min, twice) to remove any intact cells and any other debris from the cell disruption.
000 PSI, 4 °C) three times to break open the cells to isolate cell wall and membrane components. The lysed cells were centrifuged (3000g, 4 °C, 2.5 min, twice) to remove any intact cells and any other debris from the cell disruption.
To isolate cell wall components, the cell lysate was centrifuged (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 30 min), washed 18.2 MΩ ultrapure water multiple times, to ensure purity, and the pellet stored at −20 °C. The remaining supernatant was subject to the same centrifugation parameters and pellets pooled to give cell wall components. To isolate cell membrane components, the remaining supernatant was subject to ultracentrifugation (100
000g, 4 °C, 30 min), washed 18.2 MΩ ultrapure water multiple times, to ensure purity, and the pellet stored at −20 °C. The remaining supernatant was subject to the same centrifugation parameters and pellets pooled to give cell wall components. To isolate cell membrane components, the remaining supernatant was subject to ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 60 min). The pellet was washed as before and stored at −20 °C. The remaining supernatant was spun again with the pellet washed and pooled to give cell membrane components. The final supernatant was subsequently characterised as cell cytoplasm and associated intracellular components.
000g, 4 °C, 60 min). The pellet was washed as before and stored at −20 °C. The remaining supernatant was spun again with the pellet washed and pooled to give cell membrane components. The final supernatant was subsequently characterised as cell cytoplasm and associated intracellular components.
The isolated cell wall, membrane components and cytoplasm were lyophilized (Alpha 1-2 LD Plus freeze dryer) to determine dry weights and to investigate their uranium biosorption capabilities.
No chemical or enzymatic lysis techniques were applied to break open the cells. This was to ensure that all the uranium was interacting with the biomass and not with any chemicals or enzymes from the fractionation process.
2.3 Molecular characterisation of cell envelope components using XPS
The percentage abundance of peptide, polysaccharide and hydrocarbon-like material (i.e., lipids) in the isolated components of the cell envelope was characterised by XPS. XPS analysis was conducted using a KRATOS AXIS 165 Ultra Photoelectron spectrometer with an Al Kα X-ray source (1486.6 eV). Each sample was mounted on indium foil and analysed by a wide survey scan (pass energy 160 eV, 1.0 eV step size) and a high-resolution scan (pass energy 20 eV, 0.1 eV step size) for carbon, oxygen, nitrogen and phosphorus. Binding energies were determined using the C 1s component set at 285.0 eV, attributed to carbon bound only to carbon and hydrogen, using the CasaXPS software (Version 2.3.16). The full width half maximum was kept constant for subsequent peak calibration.2.4 Uranium biosorption
To determine the uranium biosorption capacity of each cell component, 1 mg mL−1 of biomass was incubated with increasing concentrations of uranium (0.125 mM to 4 mM) at pH 3, 4.25 and 5.5, and at 25 °C. U(VI) solutions were prepared using 0.1 M uranyl nitrate hexahydrate (UO2(NO3)2·6H2O) in 1% HNO3 (Fluka Analytical: 94270). The pH was adjusted by the addition of HCl or NaOH while the ionic strength was controlled by the addition of 0.1 M NaCl. Following uranium biosorption, the biomass was removed from solution (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 10 min) and the remaining uranium left within the supernatant was acidified using 1% HNO3 and analysed using a Spectro-Ciros-Vision ICP - OES. Instrument parameters were set at 1400 watts with a coolant flow of 12.00 L min−1, auxiliary flow at 1.00 L min−1, nebuliser flow at 0.85 L min−1 and a pump speed during analysis set at 1 mL min−1. Biomass that had retained uranium was recovered by centrifugation (10
000g, 4 °C, 10 min) and the remaining uranium left within the supernatant was acidified using 1% HNO3 and analysed using a Spectro-Ciros-Vision ICP - OES. Instrument parameters were set at 1400 watts with a coolant flow of 12.00 L min−1, auxiliary flow at 1.00 L min−1, nebuliser flow at 0.85 L min−1 and a pump speed during analysis set at 1 mL min−1. Biomass that had retained uranium was recovered by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 10 min), washed three times using 0.1 M NaCl and lyophilised (Alpha 1-2 LD Plus freeze dryer).
000g, 4 °C, 10 min), washed three times using 0.1 M NaCl and lyophilised (Alpha 1-2 LD Plus freeze dryer).
2.5 ATR-FT-IR
Measurements were performed using Attenuated Total Reflectance (ATR) – FT-IR to identify the functional groups of each cell surface component which interact with uranium. These were obtained using a Silver Gate Evolution ATR accessory, consisting of a germanium crystal, coupled to a PerkinElmer Spectrum One FT-IR spectrometer. A total of 30 scans were performed on each sample within the scanning range of 4000–800 cm−1. An average spectrum was obtained from 3 replicates, baseline corrected and normalised to 1.5 absorbance (arbitrary units) using the ∼1650 cm−1 amide I peak within the control sample (containing no uranium). Data acquisition and processing were performed using PerkinElmer Spectrum version 3.3 to obtain peak positions and relative peak intensities.3 Results and discussion
3.1 Characterisation of components of the cell envelope using XPS
In order to identify the role of the bacterial cell envelope in uranium biosorption, the key components of the cell envelope were first isolated, purified, and characterised by XPS. XPS was used to identify the surface functional groups characteristic of the intact bacteria and their cell envelope components since the probing X-rays penetrate only the top 2 to 5 nm of the sample, eliminating interference from intracellular components.36 XPS survey spectra and high-resolution C 1s, O 1s, N 1s and P 2p spectra of intact P. putida 33015 and B. subtilis 168 cells and cell envelope components are shown in Fig. S1 and S2 (ESI†). A detailed interpretation of high-resolution XPS spectra is given in Supplementary Information 1 (ESI†).Secondly, XPS was used to quantify the percentage elemental abundance of C, O, N and P within the outermost layer of each sample. Four biological replicates of each cell envelope component were used to calculate an average and standard deviation. From this, the abundances of polysaccharides, peptides and hydrocarbon-like compounds were determined.12,34,36 This approach compared the measured concentration ratios O/C and N/C, at the carbon concentration in the isolated components of the cell envelope, with the atomic concentration ratios O/C and N/C for model compounds representative of the three classes of cell envelope constituents, Table 1.
| Cell surface constituent | Model carbon compound | [Atomic] ratio (atom/atom) | [Carbon] (mmol g−1) | |
|---|---|---|---|---|
| O/C | N/C | |||
| a Outer membrane protein (OMP) amino acid analysis for peptide atomic concentration ratios and relative protein abundance. | ||||
| Polysaccharides | Glucan (C6H10O5) | 0.833 | 0.000 | 37.0 |
| Peptides | P. fluorescens OE 28.3 OMPa | 0.325 | 0.279 | 43.5 |
| Lipid | Hydrocarbon (CH2)n | 0.000 | 0.000 | 71.4 |
Eqn (1)–(3) and the carbon concentration of each model carbon compound (Table 1) were used to determine the percentage of polysaccharides, peptides and hydrocarbon-like compounds in the cell envelope, where CPEP, CPS, and CLIP are the atomic carbon concentrations of the peptides, polysaccharides and lipids within the bacterial surface biomass.12
| O/C = 0.325 (CPEP/[C]) + 0.833 (CPS/[C]) | (1) |
| N/C = 0.279 (CPEP/[C]) | (2) |
| 1 = (CPEP/[C]) + (CPS/[C]) + (CLIP/[C]) | (3) |
| Component | Concentration (% ± SD) | |||||
|---|---|---|---|---|---|---|
| P. putida 33015 | B. subtilis 168 | |||||
| Intact cells | Cell wall | Cell membrane | Intact cells | Cell wall | Cell membrane | |
| a Atomic concentration ratios with respect to total carbon, multiplied by 100. | ||||||
| Total C | 69.46 ± 1.84 | 71.37 ± 1.07 | 73.26 ± 0.68 | 65.96 ± 0.74 | 62.87 ± 1.87 | 73.61 ± 0.95 |
| Total N | 5.44 ± 0.38 | 7.66 ± 0.52 | 8.27 ± 0.24 | 6.03 ± 0.3 | 6.63 ± 0.56 | 6.89 ± 0.6 |
| Total O | 23.07 ± 1.67 | 19.49 ± 0.59 | 17.08 ± 0.34 | 25.46 ± 0.54 | 27.77 ± 1.41 | 18.31 ± 0.48 |
| Total P | 2.02 ± 0.04 | 1.48 ± 0.14 | 1.37 ± 0.12 | 2.56 ± 0.15 | 2.74 ± 0.11 | 1.19 ± 0.15 |
| O/Ca | 33.2 ± 3.2 | 27.3 ± 1.2 | 23.3 ± 0.7 | 38.6 ± 1.2 | 0.442 ± 3.35 | 24.9 ± 0.9 |
| N/Ca | 7.8 ± 0.7 | 10.7 ± 0.9 | 11.3 ± 0.4 | 9.1 ± 0.5 | 10.5 ± 1.2 | 9.4 ± 0.9 |
| P/Ca | 2.9 ± 0.1 | 2.1 ± 0.2 | 1.9 ± 0.2 | 3.9 ± 0.2 | 4.4 ± 0.2 | 1.6 ± 0.2 |
| Peptide | 33.97 ± 2.35 | 46.58 ± 3.15 | 49.79 ± 1.5 | 38.02 ± 1.89 | 42.08 ± 3.59 | 41.8 ± 3.55 |
| Polysaccharides | 34.31 ± 3.39 | 21.11 ± 0.52 | 14.71 ± 0.24 | 38.15 ± 1.03 | 41.71 ± 2.25 | 20.48 ± 1.5 |
| Hydrocarbons | 31.72 ± 4.36 | 32.31 ± 3.01 | 35.5 ± 1.61 | 23.83 ± 1.97 | 16.21 ± 4.92 | 37.72 ± 2.64 |
There was an almost equal abundance of peptide, polysaccharides and lipid compounds near the surface of intact P. putida cells. However, in the cell wall components there was an increase in the proportion of both peptides and a decrease in the proportion of polysaccharides compared with intact cells. A greater proportional decrease of polysaccharides in the cell membrane was attributed to a lack of polysaccharide-like compounds, such as peptidoglycan, present in small quantities in the Gram-negative cell wall. The increase in percentage abundance of peptides following cell fractionation was attributed to proteins present within the outer and cell surface membrane. No significant difference in hydrocarbon like material, associated with outer membrane phospholipids, were observed between intact cells and isolated cell wall components.
The surface of the intact B. subtilis cells was composed of almost equal amounts of peptides and polysaccharides while hydrocarbon-like compounds comprised 23.8 ± 2.0%. It has been suggested that the hydrocarbon-like material found in the surface of B. subtilis should be ascribed to lipoteichoic acids51 (rather than ruptures of the cell wall and X-ray penetration of cell surface membrane phospholipids) as it has been shown that freeze-drying intact bacteria does not rupture the cell walls.52 The proportion of peptides and polysaccharides to lipid material increased within isolated cell wall components from B. subtilis following the fractionation of intact cells. Previous XPS studies had found the cell surface to be rich in phosphate groups as well as protonated nitrogen, associated with protonated amino acids and alanine or (lipo) teichoic acids.50 A decrease in the relative abundance of lipids in the B. subtilis cell wall components was ascribed to a lack of lipoteichoic acid content due to that compound being covalently bound to the cell surface membrane.
Elevated phosphate concentrations were attributed to phosphorylated proteins and polysaccharides within the Gram-positive cell wall. The phosphate-related compounds create the surface negative charge at low pH with a point of zero charge at pH 2.2.53,54 Furthermore, the molecular composition of the B. subtilis cell wall can change as a function of pH and also in the presence of adsorbed heavy metal species. An increase in lipoteichoic acid content and a decrease in polysaccharide and peptide content was observed in the cell surface of intact B. subtilis cells as the pH of the solution that the cells were suspended in was increased.51
Hydrocarbon-like compounds were attributed to the outer membrane of the P. putida cell wall and phospholipids and other lipid material in the cell surface membrane in both bacterial strains. The low abundance of polysaccharides in cell membrane isolates could be attributed to bound polysaccharides such as lipoteichoic acids (in the Gram-positive B. subtilis).12,34–37
The characteristics of the fractions as obtained using XPS correspond to the typical morphological distribution of macromolecules in each of the different cell compartments for both P. putida and B. subtilis. As expected, there was a higher proportion of polysaccharides in the cell wall fraction compared to the cell membrane, for example, suggesting that this type of components preferred to be located mainly in the cell wall. This finding suggests that despite the limitations of the extraction method used, it provides a good representation of the molecular composition of the cell components per fraction.
3.2 Uranium biosorption
The ability of intact cells and isolated components of the cell envelope to retain uranium through biosorption from solution was studied by mixing 1 mg mL−1 biomass with solutions of increasing uranium concentration (0.125–4 mM) at pH 3, 4.25 and 5.5 for 48 hours at 30 °C. The biosorption capacity (qe) was calculated, eqn (4), in which Ce and C are the uranium concentrations (mol L−1) in solution before and after biomass interaction, respectively. V is the reaction volume (mL) and m is the biomass weight (mg).| qe = ((Ce − C) × V)/m | (4) |
Fig. 2 shows the biosorption isotherms for intact cells from both bacterial strains between pH 3–5.5 and for their cell wall and membrane isolates at pH 5.5, calculated from 3 biological replicates. Measurements were also performed in triplicate when analysed by ICP-OES. Biosorption isotherms for cell wall and membrane isolates of both strains in the pH range studied are displayed in Fig. S3 (ESI†).
For both strains and their respective cell envelope components studied, greater removal of uranium from solution was observed as the solution pH increased from 3 to 5.5. The isotherms obtained for all samples follow a typical curve where a plateau is observed towards higher concentrations of U(VI) in solution. For both strains in this study, there is a marked difference between the sorption capacity of intact cells at pH 3 and 4.25 compared to the process studied at pH 5.5.
The biosorption process is affected by pH since both the surface charge of the cell envelope components and the speciation of U(VI) are determined by the solution pH. The estimated speciation of U(VI) in solution, in the presence of 0.1 M NaCl, changed as a function of pH (Visual MINTEQ), Table S1 (ESI†). At pH 3, the dominant U(VI) species in solution was the highly mobile UO22+ ion (∼93%) within the concentration range studied. At pH 4.25, UO22+ was still the dominant species, however its abundance decreased as the uranium concentration in solution increased. Other species formed included UO2OH+ and (UO2)2(OH)22+, with the latter increasing as the uranium concentration increased. At pH 5.5, the dominant uranium species were (UO2)3(OH)5+ and (UO2)4(OH)7+, with the former decreasing and latter increasing as the uranium concentration in solution increased.
For intact B. subtilis cells the amount of uranium biosorbed was 0.73, 1.17 and 2.28 mol U(VI) kg−1 biomass from 4 mM U(VI) solution at pH 3, 4.25 and 5.5, respectively. This was substantially larger than that for intact P. putida cells which retained 0.66, 0.85 and 2.00 mol U(VI) kg−1 biomass at pH 3, 4.25 and 5.5, respectively.
The biosorption process is driven by interactions between aqueous U(VI) species and functional groups with a negative surface charge due to the influence of solution pH. The point of zero charge (pzc), at which the bacterial cell surfaces of intact P. putida and B. subtilis cells exhibit a net neutral charge, was calculated previously at pH 2.8 and 2.2 respectively.53–55 Therefore, the net negative charge of the bacterial cell surface of B. subtilis is greater than that of P. putida. This would result in a greater number of electrostatic interactions between deprotonated uranium-favourable functional groups and positively charged U(VI) species in the former case. Furthermore, a low pzc explains an increase in the biosorption capacity of both strains, approximately 3 fold, as the pH increased from 3 to 5.5.
Differences in the composition of the bacterial cell envelope and differences in the number of functional groups could be attributed to differences in uranium retention as a function of species. Uranium biosorption has been described in terms of interactions with phosphate, hydroxyl, carboxyl and amino groups.9 The bacterial cell envelope of B. subtilis is composed predominantly of polysaccharides, lipoteichoic acids and peptides, contributing to the increased number of phosphate hydroxyl, carboxyl and amino groups.50 The cell envelope of P. putida contains large quantities of phospholipids and peptides, with the former increasing the number of esters and phosphates. Esters have not been shown to readily interact with uranium from solution in previous studies. Phosphates, however, readily interact with uranium.23,24,26
An increase in the biosorption capacity of cell wall isolates compared to intact cells highlighted the importance of the functional groups associated with the bacterial cell wall in uranium biosorption. Cell wall components from B. subtilis biosorbed a larger amount of U(VI), 2.93 mol U(VI) kg−1 biomass, from 4 mM solution, at pH 5.5 than P. putida cell wall components, 2.22 mol U(VI) kg−1 biomass.
Cell wall components from both bacterial strains exhibited a higher uranium retention capability than that of intact cells, possibly due to the increased number of uranium-favourable functional groups associated with the cell walls of both strains. Cell wall isolation increases the surface area exposure for interactions with U(VI) species in comparison with intact cells.
Cell membrane components from both bacterial strains exhibited a lower U(VI) biosorption capacity to that observed for intact cells and cell wall isolates. B. subtilis cell membrane components retained 1.72 mol U(VI) kg−1 biomass following suspension in 4 mM uranium solution at pH 5.5. By comparison, P. putida cell membrane components retained 1.81 mol U(VI) kg−1 biomass. This decrease was attributed to the proportional increase in the number of uranium-unfavourable and non-reactive functional groups, such as those associated with the lipid bilayers. A decrease in the concentration of uranium-favourable groups from other cell surface biomolecules, such as polysaccharides, that were present within intact cell and cell wall components, would decrease the retention capacity of cell membrane components in comparison.
These results demonstrate that accounting for the removal of U(VI) from solution using intact cell dry weight underestimates the capacity of these organisms to sequester uranium using their cell wall. Intracellular components of intact cells contribute to their dry weight but may not contribute to the retention of uranium, and hence result in a lower uranium biosorption capacity per kg of biomass. Using the cell fractionation approach provides a better approximation of the retention capacity from intact cells, based on the direct contribution from key components of the cell envelope deemed responsible for uranium biosorption.
The Freundlich isotherm model was used to estimate the sorption parameters of the process studied here. This absorption model assumes a heterogeneous adsorbent surface with multiple adsorption sites and different binding affinities for the multiple U(VI) species. The bacterial cell surface can be considered as a multiple adsorption site system, rich in functional moieties that exhibit a deprotonation behaviour within the pH range studied.9
The Freundlich constants were calculated by a plot of triplicate data of the logarithms of Cevs. qe. The corresponding linear regression can be applied to the following equation (eqn (5)) from which ni and the Freundlich constant (KF) can be inferred.
log(qe) = ni![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(Ce) + log log(Ce) + log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF KF | (5) |
 | (6) |
ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD KD | (7) |
| pH | K F, mol kg−1 | K D, L g−1 | ΔG°, kJ mol−1 | |
|---|---|---|---|---|
| Pseudomonas putida 33015 | ||||
| Intact cells | 3 | 9.59 ± 0.45 | 199.75 ± 9.30 | −13.12 ± 0.11 |
| 4.25 | 20.89 ± 0.98 | 260.34 ± 2.24 | −13.78 ± 0.02 | |
| 5.5 | 273.02 ± 31.09 | 621.00 ± 12.99 | −15.94 ± 0.05 | |
| Cell wall | 3 | 14.68 ± 1.24 | 234.08 ± 6.17 | −13.52 ± 0.07 |
| 4.25 | 36.15 ± 1.89 | 322.50 ± 7.71 | −14.32 ± 0.06 | |
| 5.5 | 329.82 ± 19.44 | 685.12 ± 7.77 | −16.19 ± 0.03 | |
| Cell membrane | 3 | 6.78 ± 0.47 | 174.52 ± 4.63 | −12.80 ± 0.07 |
| 4.25 | 20.21 ± 2.04 | 255.82 ± 5.97 | −13.74 ± 0.06 | |
| 5.5 | 187.36 ± 9.01 | 565.42 ± 7.86 | −15.71 ± 0.03 | |
| Bacillus subtilis 168 | ||||
| Intact cells | 3 | 13.00 ± 0.50 | 218.00 ± 3.54 | −13.35 ± 0.04 |
| 4.25 | 47.54 ± 0.72 | 353.46 ± 2.37 | −14.55 ± 0.02 | |
| 5.5 | 326.44 ± 7.40 | 688.18 ± 7.70 | −16.20 ± 0.02 | |
| Cell wall | 3 | 24.69 ± 1.01 | 239.95 ± 12.57 | −13.58 ± 0.13 |
| 4.25 | 89.68 ± 1.60 | 366.83 ± 2.70 | −14.64 ± 0.02 | |
| 5.5 | 796.34 ± 66.23 | 796.62 ± 12.10 | −16.56 ± 0.04 | |
| Cell membrane | 3 | 3.48 ± 0.8 | 129.20 ± 4.64 | −12.05 ± 0.09 |
| 4.25 | 8.73 ± 0.22 | 189.21 ± 2.19 | −13.00 ± 0.03 | |
| 5.5 | 170.76 ± 32.28 | 539.93 ± 32.80 | −15.59 ± 0.15 | |
The Langmuir isotherm model was also used to examine the data, however, the quality of fits obtained was not adequate and hence it was deemed not appropriate to report here (but see Table S2 and Fig. S5, ESI†).
3.3 ATR-FT-IR
ATR-FT-IR was used to identify the functional groups for each component of the cell envelope. Using this technique, any changes in adsorption band shape and position in a spectrum provides identification of the functional groups of chemical molecules involved in uranium biosorption. The main infrared bands and functional groups identified for intact cells, cell wall and membrane components of P. putida and B. subtilis are summarised in Table 4.5,12–14,38–41,58–63 FT-IR spectra of the intact cells, cell wall and cell surface membrane of P. putida and B. subtilis, before and after U(VI) biosorption at pH 5.5 are shown in Fig. 3. Detailed interpretation of control spectra is given in Supplementary Information 2 (ESI†). Adsorption spectra were normalised with respect to the amide I absorption band, the adsorption band characteristic of functional groups associated with bacterial proteins,41 present in all isolates. Differences in adsorption band intensity, in comparison to intact cells, corresponded to changes in the relative abundance of biomolecules following the extraction, isolation and purification of the cell wall and cell membrane from both bacterial strains. Futhermore, this eliminated interference from IR radiation interacting with functional groups associated with intracellular components of intact cells.38,39,41 The spectra for the intact cell control, the cell wall and cell membrane (control) shows the signal characteristic of the components obtained through the XPS characterisation: peptides, polysaccharides and hydrocarbons. The spectra shows that the extraction process caused little changes to the structure at macromolecular level, the shape and position of the peaks in the spectra corresponds to previously reported FT-IR for bacteria cell wall.41,42 The intensity of the band in the fingerprint region for the polysaccharides (1080 cm−1) in the cell wall control spectrum is slightly higher than the cell membrane, since it is expected that there is more polysaccharides in this fraction since polysaccharides are the mainly located at the cell wall. This corroborates the results obtained from the calculated distribution of components using XPS.| Wavenumber (σ (cm−1)) | Functional group assignmenta |
|---|---|
| ∼3300 | ν(O–H) of hydroxyl groups |
| ∼3000–2850 | ν as(CH3), νas(CH2) and νs(CH2) from lipids |
| ∼1740–1720 |
ν
s(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) from esters in lipids, (lipo)teichoic acids, stretching of C O) from esters in lipids, (lipo)teichoic acids, stretching of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O from carboxylic acids O from carboxylic acids |
| ∼1650 |
ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) from proteins (Amide I) O) from proteins (Amide I) |
| ∼1540 | δ(N–H) from proteins (Amide II) |
| ∼1470–1450 | δ s(CH2) from lipids and fatty acids |
| ∼1420–1380 | ν s(COO−) from proteins |
| ∼1320 | ν(C–N) from proteins |
| ∼1240–1220 | ν as(PO2) from DNA, phospholipids and phosphorylated biopolymers |
| ∼1175 | ν s(PO2) from DNA, phospholipids and phosphorylated biopolymers |
| ∼1080–1040 | ν s(PO2, PO32−) from DNA, phospholipids and phosphorylated biopolymers; ν(C–OH, C—O–C, C–C) from polysaccharides, peptidoglycan |
| ∼976 | ν s(PO2) from DNA, phospholipids and phosphorylated biopolymers |
| ∼915–925 | ν(U–Oligand) from uranium following biomass accumulation. |
 | ||
| Fig. 3 FTIR spectra of control and uranium loaded biomass (pH 5.5) from P. putida 33015 [A] and B. subtilis 168 [B] within the region of 2000–800 cm−1. Insets in the region of 3000–2800 cm−1. | ||
The cell wall isolates from P. putida and B. subtilis exhibited different uranium biosorption mechanisms due to differences in cell wall architecture. Spectra suggested that biomolecules within the outer membrane of the Gram-negative cell wall of P. putida were responsible for uranium biosorption. A decrease in intensity of the νas(CH3), νas(CH2) and νs(CH2) absorption bands (∼3000–2850 cm−1), associated with lipid material, was seen following uranium binding. Broadening and changes in shape of protein adsorption bands (amide II and νs(COO−)) suggested uranium-favourable interactions with proteins in the cell wall. As for intact cells, broadening in absorption bands, the corresponding νas(PO2) of phosphorylated polymers and ν(C–O, C–O–C, C–C) of polysaccharides, suggested that phospholipids from the outer membrane and lipopolysaccharides (LPS) were involved in uranium biosorption by the Gram-negative bacterial cell wall.14,15,39,40,63 This was supported by previous studies suggesting that LPS sequestered uranium from solution using phosphate and carboxyl groups.30,32
There was no significant shift in absorption bands corresponding to the ν(C–O, C–O–C, C–C) of polysaccharides following uranium biosorption to cell wall components, except at high concentrations of uranium at pH 5.5, similar to the results observed for intact B. subtilis cells. This suggested that the biosorption of uranium with peptidoglycan within the Gram-positive cell wall was a pH dependent process and occurred to a greater extent when it was exposed to high uranium concentrations. The pKa values of purified peptidoglycan have been reported to be 4.55, 6.31 and 9.56, corresponding to the carboxyl groups of glutamic acid, diaminopimelic acid and a combination of hydroxyl/amino groups, respectively.31 Considering the experimental conditions used in this study, a deprotonation of the majority of functional groups associated with polysaccharides within the cell wall isolate at pH 5.5 was expected, enabling the biosorption of uranium by cell wall components. Shifts corresponding to changes in the νas(PO2) absorption band position indicated uranium biosorption with phosphate groups within the Gram-positive cell wall, such as those in (lipo)teichoic acids and proteins associated with the phosphorylated cell wall.41 The latter was further confirmed by changes in the shape of protein associated absorption bands following uranium exposure within the pH range studied. Broadening of the δ(N–H) and the νs(COO−) absorption bands indicate interactions with carboxyl groups associated with cell wall proteins.64,70 Conformational changes in protein structure following uranium binding would probably contribute to changes in δ(N–H) adsorption band shape rather than being due to uranium binding with amino groups. This is due to the pKa of amino groups being 9.0. Therefore, within the pH range studied, no binding interactions were expected since the positively charged amino groups prevent electrostatic binding with the positively charged U(VI) species. The small νs(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) band at 1734 cm−1 from (lipo)teichoic acids disappeared upon uranium biosorption suggested favourable interactions, with these cell wall constituents also anchored to the cell surface membrane.41
O) band at 1734 cm−1 from (lipo)teichoic acids disappeared upon uranium biosorption suggested favourable interactions, with these cell wall constituents also anchored to the cell surface membrane.41
The results from uranium biosorption to cell membrane isolates from P. putida and B. subtilis were comparable, probably due to similarities in macromolecular composition. A decrease in the intensity of the absorption bands for νas(CH3), νas(CH2) and νs(CH2) was observed following uranium biosorption. This was attributed to overall changes in membrane structure following uranium biosorption, rather than direct binding to unreactive non-polar lipid material.23,24,26 Similarly, the additional adsorption band at 1740 cm−1, attributed to the vibrational stretching of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O lipid bond and characteristic of cell membrane components following isolation from intact cells, disappeared following uranium biosorption. Shifts and broadening of νas(PO2) and νs(PO2) absorption bands from isolates of both bacterial strains suggested favourable U(VI)–phosphate interactions with phospholipids of the cell surface membrane. Finally, the broadening of the protein absorption bands associated with carboxyl groups in both membrane isolates indicated that there were interactions with membrane associated proteins.
O lipid bond and characteristic of cell membrane components following isolation from intact cells, disappeared following uranium biosorption. Shifts and broadening of νas(PO2) and νs(PO2) absorption bands from isolates of both bacterial strains suggested favourable U(VI)–phosphate interactions with phospholipids of the cell surface membrane. Finally, the broadening of the protein absorption bands associated with carboxyl groups in both membrane isolates indicated that there were interactions with membrane associated proteins.
Overall, the ATR-FT-IR data suggested that numerous functional groups are responsible for uranium biosorption at the cell surface and that this process is predominantly dependent on those groups associated with the cell wall. The level of biosorption varies due to solution pH and is influenced by the composition of bacterial cell envelope; whether it be of Gram-negative or Gram-positive origin. The major functional groups associated with uranium biosorption were phosphate and carboxyl, from phosphorylated proteins, phospholipids and polysaccharides. Differences in uranium biosorption mechanisms were observed between the intact cells and cell wall isolates of P. putida and B. subtilis. Cell membrane components exhibited a similar mechanism of uranium biosorption due to similarities in macromolecular composition, regardless of bacterial origin.
Based on these results, Fig. 4 shows the different interactions of the cell envelope of both Gram-negative and Gram-positive bacteria. U(VI) species will interact with negatively charged components of the outer membrane of the cell wall of Gram-negative bacteria, including lipopolysaccharides and capsular polysaccharides, membrane and surface proteins, and components of outer membrane phospholipids. Further interactions will occur with the small amounts of peptidoglycan below the outer membrane. In the case of Gram-positive bacteria, U(VI) species will readily bind to phosphorylated polysaccharides, including teichoic acids, and cell surface and wall associated proteins. U(VI) will also interact favourably with peptidogyclan towards circumneutral pH.
In the case of the cell membrane, both Gram-negative and Gram-positive bacteria display similar mechanisms of interaction with U(VI), mainly through phospholipids and membrane associated proteins. Additionally, U(VI) will interact with cell membrane bound lipoteichoic acids associated solely with Gram-positive bacteria. The sorption process is favoured by the ability of U(VI) to travel through the outer membrane layer of the Gram-negative cell wall and peptidoglycan layers of both cell envelopes The size of the uranyl ion is significantly smaller (approximately 0.242 nm71) compared to the pore size of peptidoglycan for B. subtilis (2.12–2.5 nm72–74), for example, suggesting that diffusion of uranyl that is not bound by the outer membrane layer can occur, facilitating the sorption process with cell membrane compounds.
4 Conclusions
The current study demonstrated that the bacterial cell wall is the vital component in the biosorption of U(VI). The capacity of cell wall components to retain uranium(VI) is larger than any other component of the cell envelope. The findings from this study indicate that Gram-positive bacteria and associated components of their cell envelope exhibited a greater biosorption capacity than components of the Gram-negative cell envelope. The preference for uranium biosorption with isolated cell envelope components and intact cells can be summarised as;| Cell wall > Intact cells > Cell membrane |
It was also found that carboxyl and phosphate functional moieties from proteins and phosphorylated biopolymers, associated with the cell wall, predominantly mediate U(VI) biosorption from aqueous solutions. This confirms that the biosorption mechanism is predominantly a surface process with the outermost components of the bacterial cell envelope. Furthermore, this study showed the cell membrane exhibits a lower capacity for U(VI) biosorption. Therefore uranium(VI) in solution will not be retained by cell membrane functional groups and accumulate at the cell wall since the chemical environment is more favourable for its retention. The question remains whether this is a diffusion or kinetically driven process, but this issue was beyond the scope of this study.
Overall, the findings from this study further aid with the design and use of bacteria for the remediation of uranium contaminated environments. A remediation process that uses Gram-positive bacteria would have a higher chance of success based on the superior ability of the components of their cell envelope to remove U(VI) from solution.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the EPSRC consortium “Hard-soft matter interfaces: from understanding to engineering” (EP/I001514/1) for financial support during the experimental study. Further thanks go to Dr Deborah Hammond and Dr Mathew Pringle (The University of Sheffield) for help with XPS data acquisition and interpretation.References
- T. Arnold, N. Baumann, E. Krawczyk-Baersch, S. Brockmann, U. Zimmermann, U. Jenk and S. Weiss, Geochim. Cosmochim. Acta, 2011, 75, 2200–2212 CrossRef CAS.
- J. R. Lloyd and J. C. Renshaw, in Metal Ions in Biological Systems, Vol 44, ed. A. Sigel, H. Sigel and R. K. O. Sigel, 2005, vol. 44, pp. 205–240 Search PubMed.
- G. M. Gadd, New Phytol., 1993, 124, 25–60 CrossRef CAS.
- N. Das, Hydrometallurgy, 2010, 103, 180–189 CrossRef CAS.
- J. Wang and C. Chen, Biotechnol. Adv., 2009, 27, 195–226 CrossRef CAS.
- L. E. Macaskie, R. M. Empson, A. K. Cheetham, C. P. Grey and A. J. Skarnulis, Science, 1992, 257, 782–784 CrossRef CAS.
- L. E. Macaskie, K. M. Bonthrone, P. Yong and D. T. Goddard, Microbiology, 2000, 146, 1855–1867 CrossRef CAS.
- S. Choudhary and P. Sar, Rev. Environ. Sci. Biotechnol., 2015, 14, 347–355 CrossRef CAS.
- L. Newsome, K. Morris and J. R. Lloyd, Chem. Geol., 2014, 363, 164–184 CrossRef CAS.
- M. Gavrilescu, L. V. Pavel and I. Cretescu, J. Hazard. Mater., 2009, 163, 475–510 CrossRef CAS.
- S. A. Cumberland, G. Douglas, K. Grice and J. W. Moreau, Earth-Sci. Rev., 2016, 159, 160–185 CrossRef CAS.
- J. J. Ojeda, M. E. Romero-Gonzalez, R. T. Bachmann, R. G. J. Edyvean and S. A. Banwart, Langmuir, 2008, 24, 4032–4040 CrossRef CAS.
- J. J. Ojeda, M. E. Romero-Gonzalez, H. M. Pouran and S. A. Banwart, Mineral. Mag., 2008, 72, 101–106 CrossRef CAS.
- J. J. Ojeda, M. E. Romero-Gonzalez and S. A. Banwart, Anal. Chem., 2009, 81, 6467–6473 CrossRef CAS.
- M. L. Merroun, G. Geipel, R. Nicolai, K. H. Heise and S. Selenska-Pobell, Biometals, 2003, 16, 331–339 CrossRef CAS.
- D. A. Fowle, J. B. Fein and A. M. Martin, Environ. Sci. Technol., 2000, 34, 3737–3741 CrossRef CAS.
- S. D. Kelly, K. M. Kemner, J. B. Fein, D. A. Fowle, M. I. Boyanov, B. A. Bunker and N. Yee, Geochim. Cosmochim. Acta, 2002, 66, 3855–3871 CrossRef CAS.
- D. Gorman-Lewis, P. E. Elias and J. B. Fein, Environ. Sci. Technol., 2005, 39, 4906–4912 CrossRef CAS.
- P. J. Panak, R. Knopp, C. H. Booth and H. Nitsche, Radiochim. Acta, 2002, 90, 779–783 CAS.
- M. L. Merroun, J. Raff, A. Rossberg, C. Hennig, T. Reich and S. Selenska-Pobell, Appl. Environ. Microbiol., 2005, 71, 5532–5543 CrossRef CAS.
- J. Zhang, H. Song, Z. Chen, S. Liu, Y. Wei, J. Huang, C. Guo, Z. Dang and Z. Lin, Chemosphere, 2018, 206, 682–692 CrossRef CAS.
- T. Reitz, A. Rossberg, A. Barkleit, S. Selenska-Pobell and M. L. Merroun, PLoS One, 2014, 9, e102447 CrossRef.
- L. Luetke, H. Moll and G. Bernhard, Dalton Trans., 2012, 41, 13370–13378 RSC.
- I. Llorens, G. Untereiner, D. Jaillard, B. Gouget, V. Chapon and M. Carriere, PLoS One, 2012, 7, e51783 CrossRef CAS.
- X.-Y. Zheng, X.-Y. Wang, Y.-H. Shen, X. Lu and T.-S. Wang, Chemosphere, 2017, 175, 161–169 CrossRef CAS.
- M. L. Merroun, M. Nedelkova, J. J. Ojeda, T. Reitz, M. Lopez Fernandez, J. M. Arias, M. Romero-Gonzalez and S. Selenska-Pobell, J. Hazard. Mater., 2011, 197, 1–10 CrossRef CAS.
- U. Gerber, I. Zirnstein, E. Krawczyk-Baersch, H. Luensdorf, T. Arnold and M. L. Merroun, J. Hazard. Mater., 2016, 317, 127–134 CrossRef CAS.
- M. Lopez-Fernandez, O. Fernandez-Sanfrancisco, A. Moreno-Garcia, I. Martin-Sanchez, I. Sanchez-Castro and M. Larbi Merroun, Appl. Geochem., 2014, 49, 77–86 CrossRef CAS.
- M. L. Merroun and S. Selenska-Pobell, J. Contam. Hydrol., 2008, 102, 285–295 CrossRef CAS.
- A. Barkleit, H. Moll and G. Bernhard, Dalton Trans., 2008, 2879–2886 RSC.
- A. Barkleit, H. Moll and G. Bernhard, Dalton Trans., 2009, 5379–5385, 10.1039/b818702a.
- A. Barkleit, H. Foerstendorf, B. Li, A. Rossberg, H. Moll and G. Bernhard, Dalton Trans., 2011, 40, 9868–9876 RSC.
- S. Xiang, X. Nie, F. Dong, W. Cheng, M. Liu, C. Ding, L. Bian, S. Sun and H. Li, J. Mol. Liq., 2020, 305, 112852 CrossRef CAS.
- P. G. Rouxhet, N. Mozes, P. B. Dengis, Y. F. Dufrene, P. A. Gerin and M. J. Genet, Colloids Surf., B, 1994, 2, 347–369 CrossRef CAS.
- Y. F. Dufrene and P. G. Rouxhet, Colloids Surf., B, 1996, 7, 271–279 CrossRef CAS.
- Y. F. Dufrene, A. VanderWal, W. Norde and P. G. Rouxhet, J. Bacteriol., 1997, 179, 1023–1028 CrossRef CAS.
- H. C. van der Mei, J. de Vries and H. J. Busscher, Surf. Sci. Rep., 2000, 39, 3–24 CrossRef.
- A. A. Kamnev, L. P. Antonyuk, L. Y. Matora, O. B. Serebrennikova, M. V. Sumaroka, M. Colina, M. F. Renou-Gonnord and V. V. Ignatov, J. Mol. Struct., 1999, 481, 387–393 CrossRef.
- L. Shakirova, M. Grube, R. Goodacre, M. Gavare, L. Auzina and P. Zikmanis, Vib. Spectrosc., 2013, 64, 51–57 CrossRef CAS.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sule-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS.
- W. Jiang, A. Saxena, B. Song, B. B. Ward, T. J. Beveridge and S. C. B. Myneni, Langmuir, 2004, 20, 11433–11442 CrossRef CAS.
- J. J. Ojeda and M. Dittrich, Methods Mol. Biol., 2012, 881, 187–211 CrossRef CAS.
- T. J. Smith and S. J. Foster, J. Bacteriol., 1995, 177, 3855–3862 CrossRef CAS.
- A. S. Eswayah, T. J. Smith, A. C. Scheinost, N. Hondow and P. H. Gardiner, Appl. Microbiol. Biotechnol., 2017, 101, 6713–6724 CrossRef CAS.
- T. J. Smith and S. J. Foster, FEMS Microbiol. Lett., 1997, 157, 141–147 CrossRef CAS.
- T. J. Smith, S. A. Blackman and S. J. Foster, Microbiology, 2000, 146, 249–262 CrossRef CAS.
- A. Kuroda and J. Sekiguchi, Microbiology, 1990, 136, 2209–2216 CrossRef CAS.
- J. E. Fein and H. J. Rogers, J. Bacteriol., 1976, 127, 1427–1442 CrossRef CAS.
- F. A. Nugroho, H. Yamamoto, Y. Kobayashi and J. Sekiguchi, J. Bacteriol., 1999, 181, 6230–6237 CrossRef CAS.
- F. Ahimou, C. J. P. Boonaert, Y. Adriaensen, P. Jacques, P. Thonart, M. Paquot and P. G. Rouxhet, J. Colloid Interface Sci., 2007, 309, 49–55 CrossRef CAS.
- M. Ramstedt, L. Leone, P. Persson and A. Shchukarev, Langmuir, 2014, 30, 4367–4374 CrossRef CAS.
- H. Moriwaki, R. Koide, R. Yoshikawa, Y. Warabino and H. Yamamoto, Appl. Microbiol. Biotechnol., 2013, 97, 3721–3728 CrossRef CAS.
- V. P. Harden and J. O. Harris, J. Bacteriol., 1953, 65, 198–202 CrossRef CAS.
- J. B. Fein, C. J. Daughney, N. Yee and T. A. Davis, Geochim. Cosmochim. Acta, 1997, 61, 3319–3328 CrossRef CAS.
- A. Borkowski, M. Szala and T. Clapa, Appl. Biochem. Biotechnol., 2015, 175, 1448–1459 CrossRef CAS.
- S. Xie, J. Yang, C. Chen, X. Zhang, Q. Wang and C. Zhang, J. Environ. Radioact., 2008, 99, 126–133 CrossRef CAS.
- M. Amini, H. Younesi and N. Bahramifar, J. Environ. Eng., 2013, 139, 410–421 CrossRef CAS.
- S. J. Parikh and J. Chorover, Langmuir, 2006, 22, 8492–8500 CrossRef CAS.
- D. S. Alessi, J. S. Lezama-Pacheco, J. E. Stubbs, M. Janousch, J. R. Bargar, P. Persson and R. Bernier-Latmani, Geochim. Cosmochim. Acta, 2014, 131, 115–127 CrossRef CAS.
- G. J. Lumetta, B. K. McNamara, B. M. Rapko and J. E. Hutchison, Inorg. Chim. Acta, 1999, 293, 195–205 CrossRef CAS.
- S. K. Kazy, S. F. D'Souza and P. Sar, J. Hazard. Mater., 2009, 163, 65–72 CrossRef CAS.
- S. Yagoubi, C. Renard, F. Abraham and S. Obbade, J. Solid State Chem., 2013, 200, 13–21 CrossRef CAS.
- S. K. Kazy, P. Sar and S. F. D'Souza, Biorem. J., 2008, 12, 47–57 CrossRef CAS.
- S. Choudhary and P. Sar, J. Hazard. Mater., 2011, 186, 336–343 CrossRef CAS.
- X. H. Pan, Z. Chen, F. B. Chen, Y. J. Cheng, Z. Lin and X. Guan, J. Hazard. Mater., 2015, 297, 313–319 CrossRef CAS.
- X. Lu, X.-J. Zhou and T.-S. Wang, J. Hazard. Mater., 2013, 262, 297–303 CrossRef CAS.
- N. Theodorakopoulos, V. Chapon, F. Coppin, M. Floriani, T. Vercouter, C. Sergeant, V. Camilleri, C. Berthomieu and L. Fevrier, J. Hazard. Mater., 2015, 285, 285–293 CrossRef CAS.
- N. Renninger, R. Knopp, H. Nitsche, D. S. Clark and J. D. Keasling, Appl. Environ. Microbiol., 2004, 70, 7404–7412 CrossRef CAS.
- F. Morcillo, M. T. Gonzalez-Munoz, T. Reitz, M. E. Romero-Gonzalez, J. M. Arias and M. L. Merroun, PLoS One, 2014, 9, e91305 CrossRef.
- F. Pagnanelli, M. P. Papini, L. Toro, M. Trifoni and F. Veglio, Environ. Sci. Technol., 2000, 34, 2773–2778 CrossRef CAS.
- Y. Oda and A. Aoshima, J. Nucl. Sci. Technol., 2002, 39, 647–654 CrossRef CAS.
- W. Vollmer, D. Blanot and M. A. De Pedro, FEMS Microbiol. Rev., 2008, 32, 149–167 CrossRef CAS.
- P. Demchick and A. L. Koch, J. Bacteriol., 1996, 178, 768–773 CrossRef CAS.
- R. Hughes, P. Thurman and E. Stokes, Z. Immunitaetsforsch., Exp. Klin. Immunol., 1975, 149, 126 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Characterisation of XPS spectra, uranium speciation, additional adsorption isotherms, Langmuir fittings, detailed FT-IR characterisation of cell envelope components and raw data. See DOI: 10.1039/d0cp04067c |
| This journal is © the Owner Societies 2021 |