Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhancing the crystallisation of insulin using amino acids as soft-templates to control nucleation†
Frederik J.
Link
 a and
Jerry Y. Y.
Heng
a and
Jerry Y. Y.
Heng
 *ab
*ab
aDepartment of Chemical Engineering, Imperial College London, South Kensington Campus, London SW7 2AZ, UK. E-mail: jerry.heng@imperial.ac.uk
bInstitute for Molecular Science and Engineering, Imperial College London, South Kensington Campus, London SW7 2AZ, UK
First published on 31st March 2021
Abstract
Amino acids have been widely used in protein formulations to increase the protein's stability. In this study, amino acids have been introduced as soft-templates to control the nucleation of proteins. L-Arginine, L-glycine or L-leucine with concentrations between 0.01 M and 0.1 M were used in a hanging drop vapor diffusion set-up to investigate their role in the crystallisation of human insulin as a model protein at low supersaturation. All amino acids were in a dissolved state. Here we show that L-arginine and L-leucine clearly enhance the nucleation significantly. On the other hand, L-glycine does not enhance nucleation of human insulin at low supersaturation. We hypothesize that it is the intermolecular interaction between the protein's residues and the amino acid's residues that results in an enhancement in the formation of the initial protein nuclei. To prove that the enhanced nucleation is of a kinetic nature, the solubility of insulin in amino acid rich solution was also investigated. The solubility results show that amino acids increase insulin solubility. From that we derive that the enhancement in nucleation is not due to a change in the thermodynamic equilibrium between the crystalline and bulk-liquid phase. As this approach of using amino acids to enhance nucleation is based on a dissolved state in solution, we introduce here the concept of protein crystallisation by soft-templating.
1. Introduction
Since the first isolation and therapeutic use of the protein insulin for treating diabetes in the 1920s, the global polypeptide and protein drug market has grown significantly in size and economic value over the past decades, having a market value of $21.5 billion USD in 2016 and expecting a growth rate of 9.4% per annum for the coming 5 years until 2025.1 Stemming from the rapid growth in demand, significant improvements in upstream processing, such as more efficient fermentation and cell culture technologies, led to an increase in titre production from an average 0.2 g l−1 in 1985 to 2.56 g l−1 in 2014. This shifted the bottleneck in manufacturing towards the downstream processing which heavily relies on costly multi-step packed-bed chromatography2 and accounts for a total of up to 70% of the total production costs.3Crystallisation as a purification technique is already widely adapted in the manufacturing of small molecules. Crystallisation is seen as an efficient technique since no mass transfer limitation exists, no costly equipment and consumables are needed, and less buffer solutions are required, while still achieving a high maximum product purity.4,5 Not only is crystallisation a more economical and efficient technology compared to chromatography, but the crystalline form also offers pharmacokinetic advantages, such as better release control and higher bioavailability, and lower impurity loading.6 However, a protein's high number of residues and high structural flexibility paired with a lack in understanding of the underlying physico-chemical mechanism of protein crystallisation makes proteins notoriously difficult to crystallise. Additionally, it is very difficult to control the crystallisation process and challenging to produce protein crystals with desired crystal qualities.
It is believed that hydrogen bonding, electrostatic and hydrophobic interaction, which all are governing the protein–protein interaction, are most crucial in protein crystallisation.5 The stochastic event of nucleation is the governing and rate-determining step in crystallisation due to simultaneous occurrence of favourable protein–protein interaction and a local increase of protein concentration to enable the formation of the critical nucleus. Hence, nucleation is an extremely complex process which can be affected by both thermodynamic and kinetic factors.7,8 The complex interplay of attractive and repulsive short-range protein–protein interaction and the necessity of a threshold number of protein molecules to overcome the energy barrier and form the critical nucleus makes protein nucleation notoriously slow and uncontrollable. Hence, a major challenge in applying crystallisation is the lack of controllability of the nucleation mechanism.
The most commonly used route to achieve a controlled crystallisation is by controlling the level of supersaturation by either slowly decreasing the temperature (cooling crystallisation) or slowly increasing the content of precipitation-additives (salting-out).9 However, those approaches are rather time-consuming and often have limited success. Hence, major efforts have been done to facilitate and control nucleation with external stimuli, such as utilising an electric field10 or magnetic field,11 ultrasonic environment,12 microgravity,13 additional additives such as ionic liquids14 or heterogeneous nucleants.15,16
Heterogeneous nucleants have been demonstrated to be relatively successful in facilitating nucleation by stabilising the protein in the surface vicinity and lowering the interfacial energy needed to form a critical nucleus. The interplay of these effects results in a significantly faster crystallisation and reduces the necessary bulk-supersaturation needed to form nuclei.15,17,18 Heterogeneous nucleants include mineral surfaces,19 membranes,20 polymers,21 DNA-origami,22 nano- and mesoporous particles,23,24 porous surfaces,25,26 and surfaces with selected functional groups.4,17,25,27 Selective nucleants that have an engineering surface topography and morphology, such as molecular imprinted silica surfaces or polymers, have the ability to selectively crystallise proteins due to favourable geometrical and physico-chemical interaction between the protein and the nucleant's surface.4,15,18,28,29 We term those selective nucleants as hard-templates since the nucleant is rigid and does not dissolve in the crystallising solution.
In this study, we introduce the concept of soft-templating, which describes the facilitation and control of nucleation and crystallisation with soft organic molecules. In comparison to hard-templates, soft-templates aim to match and enhance favourable protein–protein interaction and block unfavourable protein–protein interaction resulting in an enhancement of crystallisation by facilitating nucleation (Fig. 1). Soft-templates can be sub-grouped as either additives or heterogeneous nucleants. Additives are soft organic molecules that fully dissolve in solution whereas heterogeneous nucleants are soft organic molecules which provide a soft heterogeneous surface for nucleation (e.g. pores of a DNA origami tube). Thus far, dissolved organic polymers30,31 and 3-dimensional DNA foldamers22 have been shown to facilitate nucleation.
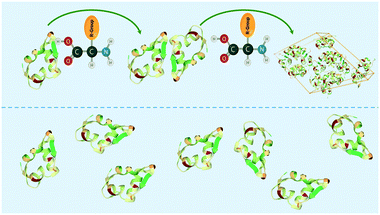 | ||
| Fig. 1 Schematic showing amino acid enhancing the interaction between protein molecules (e.g. insulin) in the soft-templating concept resulting in an enhancement of nucleation and crystallisation. | ||
Here, we would like to introduce natural amino acids as novel soft-templates. Besides being the building block for proteins, amino acids are widely utilised as excipients in freeze- and spray-drying,32 as stabilising additives,32–34 as osmolytes35 in liquid protein-based drug formulations or as stabilisers in crystalline formulations.36 In liquid protein formulations, hydrophobic and acidic hydrophilic amino acids have been used to thermally stabilise lysozyme37 and pig heart mitochondrial malate dehydrogenase32 respectively, while methionine and histidine increases the stability against oxidative stress to enhance the long-term storage capability.32,38,39 Furthermore, amino acids, especially arginine, have been used to prevent protein denaturation.32,34,40 Arginine,32,41,42 histidine,43 glutamic acid,42 isoleucine,42 glycine44 and combination of arginine and glycine44 have been found to stabilise and protect the stability of proteins, such as catalase, lysozyme and haemagglutinin proteins during freeze-drying.
We believe that amino acids as soft-templates are remarkable in controlling nucleation due to their similarities in surface chemistry to the protein surface. Additionally, amino acids are small in size compared to the protein surface which enables targeted interaction with a specific surface patch of the protein. Hence, we hypothesize that amino acids have the potential to 1) specifically enhance protein–protein interaction leading to the formation of the initial nucleus or 2) block unfavourable interaction to increase the likelihood of the formation of the initial nucleus.
To demonstrate the soft-templating approach, amino acids with specific residues and human insulin, which comprises of two unique chains, A and B, with a combined 51 amino acids was utilised. Insulin was utilised as a model protein since it comprises of amino acids with a wide variety of physico-chemical residues.
2. Material and methods
2.1. Materials
All materials were used without further purification or treatment, as received from the supplier, unless otherwise stated. Human insulin stock solution, zinc-sulphate (>99%), citric acid buffer components, amino acids (L-form, 99.9% purity) and methylene blue solution were purchased from Sigma-Aldrich (Gillingham, UK). 24-Well VDXTM plates with sealant were purchased from Hampton Research (California, USA). HPLC-grade acetonitrile, HPLC-grade methanol and potassium di-hydrate monobasic (>99%) (all from Sigma Aldrich, Gillingham, UK) as well as a Phenomenex Luna C-18 (average particle size 5 μm, pore size 100 A) column (150 × 4.6 mm) were utilised for the HPLC analysis.2.2. Solution preparation
Citrate buffer (0.48 M) was prepared at pH 7 and filtered with a 0.22 μm pore filter before further usage. The pH was determined using a SevenExcellence with InLab Expert Pro-ISM pH meter (Mettler Toledo, Switzerland), and conductivity was measured with a 4330 glass bodied conductivity probe (Jenway, UK). Zinc-sulphate and amino acids were separately dissolved in buffer solution. All solutions were filtrated with a 0.22 μm Millex-GS syringe filter units (Millipore) before further usage.2.3. Solubility determination of insulin in aqueous amino acid solutions
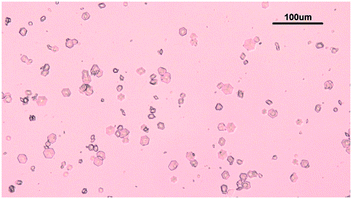
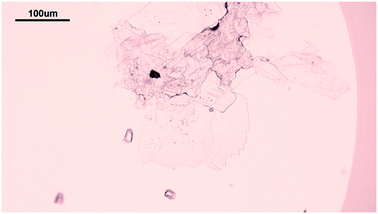
The solubility of insulin in aqueous amino acid and zinc-sulphate solution was determined by adding a small amount of insulin crystals to the amino acid, zinc-sulphate and buffer solution. The total volume of each solubility samples was 5 ml. The solution temperature was kept constant by placing the vials in a water bath, set at a temperature of 24 °C and the temperature was monitored with a digital thermometer (Hanna Instruments, UK). The vials in solution were tightly sealed to prevent solvent evaporation and it was ensured that the sample in the vials were submerged in the water bath. The insulin crystals were obtained via batch crystallisation under the same conditions as the solubility of insulin was probed. Fig. 2 shows the insulin crystals obtained from the batch crystallisation experiments that were added to the sample solutions.The insulin concentration in solution was determined by high performance liquid chromatography (HPLC) after 6 days. Full description of the chromatographic condition can be found in the ESI.†
2.4. Crystallisation of human insulin
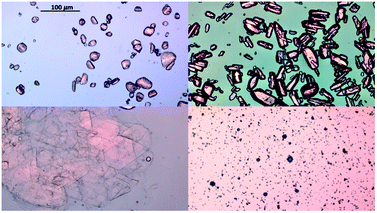
Hanging drop vapor diffusion experiments were carried out to investigate the impact of amino acids on human insulin crystallisation. Insulin crystallisation conditions were adapted from the literature and further optimized (e.g. type and pH of buffer and zinc containing salt).45–48 The buffer solution, the zinc-sulphate solution and the amino acids solution were pre-mixed in an Eppendorf tube for every experiment to achieve homogeneity in all droplets. No crystallisation occurred in the pre-mix process. From the Eppendorf tube a 2 μl droplet was placed on a hydrophobic borosilicate cover glass and then carefully inversed and sealed onto the well filled with reservoir solution. The zinc-sulphate concentration in the reservoir was twice as high as in the drops and the amino acid concentration in the reservoir was equal to the amino acid concentration in the drop. The 24-well plates were incubated at a constant temperature of 24 °C and monitored with an optical microscope (CX 41, Olympus, Japan) after 1, 2, 3, 4, 7, 10 and 14 days. A minimum of 72 drops from a minimum of 6 different plates were evaluated. Methylene blue dye was utilised to distinguish protein crystals from salt or amino acid crystals. It was demonstrated that pure amino acid crystals (arginine, glycine or leucine) are not stained by the methylene blue dye (Fig. 3). The amino acid crystals were obtained via cooling crystallisation in pure deionised water and in small batches.For simplification, only images of hanging drop vapor diffusion experiments in which crystallisation occurred are shown. The impact of amino acids on enhancing crystallisation was evaluated by calculating the occurrence of crystals (eqn (1)) after 4 and 7 days.
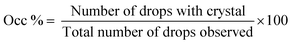 | (1) |
3. Results
3.1. Insulin crystallisation at low supersaturation
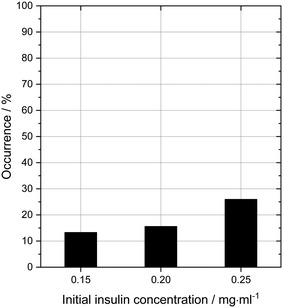
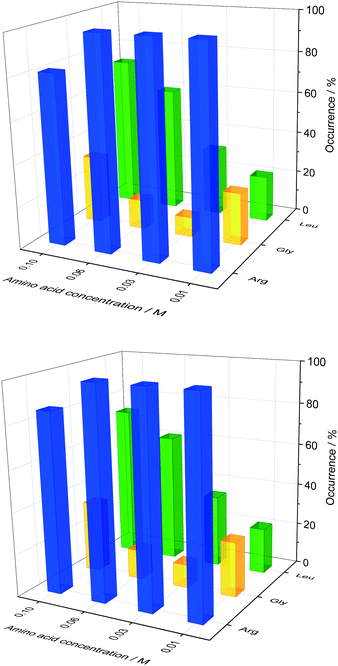
In order to probe the impact of amino acids as soft templates, an initial insulin concentration was chosen in which insulin was found to be difficult to crystallise due to the low supersaturation. As expected, the occurrence of insulin crystals decreases with decreasing insulin concentration at constant zinc-sulphate concentration and pH. Fig. 4 shows the insulin crystal occurrence at 3 distinct low initial insulin concentrations (ct=0). The standard deviation for the occurrence shown in Fig. 4 is ∼12%. The solubility of insulin (ce) at a zinc-sulphate concentration of 0.7 mM in a 0.48 M buffer (pH 7) is 0.011 mg ml−1 (see section 3.2). An initial insulin concentration of 0.2 mg ml−1 (supersaturation ratio S = ct=0/ce = 18.2) was selected because lower concentrations would increase the impact of external effects such as impurities, sample inhomogeneities or even inconstant drop size.49 On the other hand, a concentration higher than 0.2 mg ml−1 will increase the crystallisation of insulin within the control conditions and hence the impact of the amino acids on crystallisation will not be determinable.Glycine was chosen as the simplest amino acid with only one hydrogen as a residue to probe whether the amine or acidic functional groups interact with the amino acid residues of insulin. Leucine is often described as the most hydrophobic amino acid50 while arginine is a basic amino acid commonly used in solid protein formulations as a stabiliser and also increases the solubility of proteins. Fig. 5 presents the crystal occurrence of insulin crystals with the addition of different amino acids types and concentrations. The standard deviation for the crystal occurrence is between 0 to 20% for insulin concentration of 0.2 mg ml−1 (Fig. 5) after 4 days. The standard deviation of the control case (∼12%) is similar to the deviation for glycine (∼15%), but slightly higher for leucine (∼20%). For arginine as a soft-template, the standard deviation is ∼0% for 0.01, 0.03, 0.06 M and 17% for 0.1 M. The highest concentration used for each amino acid was 0.1 M, since the maximum concentration of leucine which can be dissolved in the utilised system was just slightly above 0.1 M. Fig. 5 shows that arginine and leucine increase the crystal occurrence significantly from 15.6% (control) to 100% (arginine) and 73% (leucine), whereas the crystal occurrence in presence of glycine was between 10 and 33% for all concentrations probed. The exact values of the crystal occurrence in addition of arginine, glycine or leucine can be found in the ESI.† At an arginine concentration higher than 0.1 M, no crystallisation was observed (data shown in the ESI†). This agrees with studies from Shiraki et al., who showed that arginine concentration higher than 0.2 M prevented completely (100%) the aggregation of the basic protein lysozyme.34 The increase in crystallisation rate (increase of crystal occurrence within a set period of time, e.g. 4 or 7 days) when arginine is added can also be attributed to the pH increase triggered by the dissolved arginine molecules in the solution. Measuring the pH in the Eppendorf-tube before setting up the drops revealed that even 0.03 M of arginine increases the pH from 7 (control) to 9.5 due to the exceeding of the buffer capacity. It has been shown that a pH opposite of the protein's isoelectric point (pI), e.g. acidic pH for a protein with a basic pI (e.g. lysozyme) favours the protein's crystallisation significantly, resulting in an improved crystallisation due to alteration of the solubility and super-solubility.49,51 Therefore, a more basic pH might increase the nucleation rate for insulin (pI = 5.4).
In the presence of leucine, the rate of nucleation increases (more drops with crystals are obtained in a set period of time, e.g. 4 or 7 days) with increased leucine concentration. While the crystal occurrence with leucine at concentration of 0.01 to 0.03 M is not statistically different to the control, leucine at concentrations of 0.06 and 0.1 M has a significant effect on promoting nucleation (see ESI† for standard deviation). At a leucine concentration of 0.1 M, which is just below the solubility of leucine in the initial crystallisation cocktail, leucine starts to crystallise out after ∼56 days. Fig. 6 shows sheet-like leucine crystals in the presence of rhombohedral insulin crystals. This could also be an indication that leucine is not incorporated in the insulin crystals, but instead only promotes insulin nucleation. It also shows that amino acids and insulin can be individually and selectively crystallised out from the same solution. While arginine and leucine are able to enhance the crystallisation of insulin, adding glycine did not result in an enhanced insulin crystallisation. This results in the consideration that the amine and acidic group of amino acids are not significantly involved in the nucleation-promoting interaction between insulin molecules which are important in the nuclei formation. Additionally, glycine does not promote nucleation which contradicts previous findings in which glycine is described as a “structure breaker”52 and could have acted as a Hoffmeister salt leading to an enhancement in crystallisation due to salting-out.
Since the supersaturation is very low, the overall insulin crystals are significantly smaller than reported in literature.53 When no amino acids were added, insulin crystallised out by forming the typical rhombohedral crystal shape which has been reported in literature.53 Adding glycine or leucine does not alter the crystal shape even at concentration of up to 0.1 M. On the other hand, adding arginine results in insulin crystals of rhombic dodecahedral crystal shape. The rhombic dodecahedral crystal shape was obtained for all arginine concentration probed (see Fig. 7 top row). With increasing arginine concentration, significantly larger insulin crystals were observed. Crystals obtained in the presence of arginine below a concentration of 0.1 M are significantly smaller in size than crystals obtained with glycine, leucine or even without amino acids, which indicates a rapid nucleation and a very limited growth.
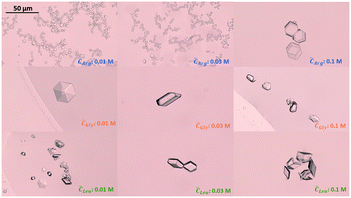 | ||
| Fig. 7 Insulin crystals obtained in the presence of arginine (top), glycine (middle) and leucine (bottom) for amino acid concentration of 0.01, 0.03 and 0.1 M. Images were taken at day 4. | ||
3.2. Solubility of insulin in aqueous amino acid solutions
The impact of amino acid concentration and type on the solubility of human insulin was studied to investigate the mechanism of the soft-templated crystallisation. Fig. 8 shows that the solubility of insulin increases if arginine, leucine or glycine is added with concentration between 0.001 and 0.1 M. While leucine or glycine increase the solubility of insulin of up to 3.2 folds, arginine increases the solubility significantly for concentration above 0.01 M to a maximum of 18.3 folds at arginine concentration of 0.06 M and 0.1 M. These findings agree with reported studies on the increasing effect of amino acids on peptide and protein solubility.48 The solubility also reveals that the effect of arginine and leucine is not due to a change in the thermodynamic equilibrium (solubility) but rather of kinetic nature. Hence, we hypothesize that both, arginine and leucine, promote favourable protein–protein interactions between insulin molecules which increases the likelihood of nucleation.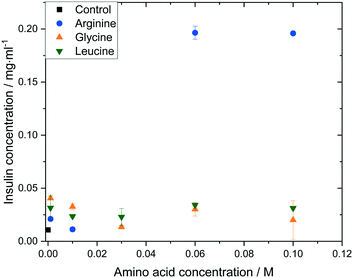 | ||
| Fig. 8 Insulin solubility (ce) in 0.48 M citric acid buffer (pH 7) with 0.7 mM zinc sulphate at different amino acid types and concentrations. | ||
4. Discussion: mechanism
4.1. General aspects
This study represents the first report of amino acids enhancing the nucleation of insulin at low supersaturation. Hanging drop vapor diffusion experiments were carried out and the initial concentration of amino acid within the drops was equal to the amino acid concentration in the reservoirs to exclude the impact of the amino acid on the partial pressure and the vapor diffusion rate.While leucine and arginine have shown crystallisation-promoting effects, glycine did not improve the crystallisation. To evaluate the impact of amino acids on the crystallisation of insulin, the concentration of amino acid in the drop was equal to the concentration of the amino acid in the reservoir. Since the buffer salt concentration and amino acid concentrations are significantly higher than the zinc-sulphate concentration (0.7 mM), it can be assumed that the water diffusion ceased after a short period of time (<4 days) since the occurrence does not change after 4 days. After 4 days, the drops behave as small (2 μl) hanging batch crystallisers. It has been shown that the air–liquid interface can impact nucleation significantly. Therefore, it is important to mention that dissolved amino acids change the surface tension of the crystallisation–solution–air interface50 and also alter the interfacial surface properties between the borosilicate cover glass and the crystallisation–solution. When arginine or leucine were added, the majority of crystals were observed in the centre of the drop, while for the control or glycine the majority of crystals were observed near the 3-phase boundary (air, cover-glass and crystallisation solution). Crystals which nucleate near the phase boundary do not sediment towards the centre of the drop but stay at the location where nucleation occurred (see Fig. 9). Since the majority of nucleation with addition of arginine or leucine occurred in the centre of the drop (Fig. 7 top and middle row), the enhancement of the nucleation rate is not due to the air–liquid interface. Although adding glycine changes the surface tension of the water–air interface,50 the majority of crystals in the control and glycine experiment occurred at the air–liquid–glass interface, indicating that the change in surface tension due to amino acids does not promote the nucleation near this interface. Since the nucleation rate for the control or addition of glycine is lower than for arginine or leucine, the interface induced nucleation is less impactful compared to the amino acid induced nucleation in the bulk solution.
We also investigated the fundamental mechanism of amino acid induced protein crystallisation. According to the classical nucleation theory, the nucleation rate can be expressed with the following equation
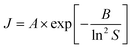 | (2) |
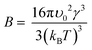 | (3) |
Our results on the solubility of insulin in amino acid rich solution is confirmed by previous studies which report that amino acids, especially arginine, increase protein solubility.32 If the supersaturation is not increased (i.e. solubility has increased by the addition of the amino acids), as observed in this study, the mechanism of amino acid enhanced nucleation must be due to either a reduction of the activation barrier of nucleation (parameter B) or an increase in the molecular attachment kinetic (parameter A).
4.2. Arginine
Although a pH of 7 is slightly higher than the pKa for citric acid (∼6.3 pH), citrate buffer at a pH of 7 is widely utilised as buffer for insulin crystallisation.48,55 However, adding arginine (>0.01 M) to the solution led to a change in the pH of the crystallisation cocktail (from 7 to 9.5 pH) since the amount added exceeds the buffer capacity. The effect of a change in buffer pH on protein crystallisation, as observed, has been widely studied in literature, as the pH is one of the most important parameters that governs crystallisation of proteins. By evaluating the relationship between solution pH and the protein's pI for hundreds of proteins, it was found that for proteins with a basic (acidic) isoelectric point a buffer pH below (above) the proteins isoelectric point enhances the crystallisation.56,57 Although this study finds that most proteins successfully crystalise if the difference between solution pH and protein's pI is between 1 to 3 pH units, it is also known that not every protein follows this trend.Lysozyme (pI = 10.7) and human insulin (pI = 5.4) are known to crystalise successfully at a buffer pH of 4 to 5 and 6 to 10 for lysozyme and human insulin respectively. Shifting the pH further away from the protein's isoelectric point has been shown to increase nucleation rate and enhances the formation of well-built crystals instead of amorphous precipitates.51,55,58 Additionally, an increasing pH could also increase insulin diffusivity resulting from increased preferential interaction, as it has been shown for lysozyme.59,60
Increasing the pH towards a more basic pH also increases the net negative charge on the insulin surface resulting in higher electrostatic repulsion. Hence a reduction in self-association and nucleation would be expected. The guanidinium group of arginine (pKa = 12.5) has a positive charge at the pH at which crystallisation takes place (pH 9.5). At this pH, arginine binds to the negatively charged residues on the insulin surface, such as glutamine or histidine, and neutralises repulsive electrostatic interaction, leading to an enhanced insulin aggregation and crystallisation. Enhanced insulin aggregation due to the addition of arginine at pH around 7.5 have already been reported in other studies.61 At higher arginine concentration (>0.06 M), the crystal occurrence starts to decrease (e.g. occurrence at 0.1 M arginine is 81% after 4 days compared to 100% at 0.06 M arginine, see Fig. 5 top). This could be due to 1) the increase in solubility as seen in Fig. 8 and/or 2) the prevention of insulin aggregation and crystallisation at higher arginine concentration (>0.1 M) which is widely reported in literature.34,60,62
We hypothesize that the combined effect of increased pH and addition of arginine reduces unfavourable (e.g. repulsive) interaction and enhances the attachment of insulin molecules during the formation of the critical nucleus in a controlled manner which depends on the arginine concentration. This can be expressed in eqn (2) by an increase in parameter A and hence an increase in the nucleation rate.
Besides promoting nucleation, the addition of arginine and the increase in pH also leads to a smaller mean crystal size, as it can be seen in Fig. 7. The effect of a smaller crystal size by changing the pH is already reported in literature. However, our findings contradict recent research which proposed that a pH further away from the protein's pI leads to the formation of bigger crystals as we obtained very small crystals for arginine concentrations between 0.01–0.06 M at 9.5 pH (see Fig. 7 top row).58 The increase in crystal size obtained with an arginine concentration of 0.1 M could be due to the suppression of insulin aggregation, resulting in a low nucleation rate and subsequent large insulin crystals (see Fig. 7).
Adding arginine leads to the formation of the rhombic dodecahedral crystal shape over the rhombohedral crystal shape, which we would have expected due to the addition of zinc. The formation of the rhombic dodecahedral shape has been shown to take place in a zinc free crystallisation environment in which a dimer is one growth unit.63 This change in crystal shape with the addition of arginine could originate from 1) arginine enhances the specific interaction that favours the formation of rhombic dodecahedra insulin crystals of which small molecules are capable.64 The specific interaction in this case refers to the interaction between the amino acid, acting as soft templates, with the insulin molecule. In order to crystalise within the rhombohedral crystal shape, three insulin dimers form a hexamer where the His10 of each dimer interacts with 2 Zn2+ to form a hexamer. However, arginine at higher ionic strengths and concentration also interacts with the dimer at the His10 position which could lead to a replacement of the Zn2+ ions with the guanidinium group of the arginine residues,65 leading to the zinc free rhombic dodecahedral crystal shape. Furthermore, it has been shown that the surface free energy of insulin crystals with the rhombic dodecahedral shape is lower than insulin crystals with a cubic shape,61 which could indicate a change in the interfacial energy between the critical nucleus and the bulk solution with the addition of arginine. Or 2) the increase in pH stemming from the protonated arginine molecules in solution promotes the formation of rhombic dodecahedral insulin crystals. It has been shown that protein crystal shapes are strongly dependent on the pH of the solution and a change in the pH can promote or inhibit a certain crystal shape (e.g. altering the aspect ratio or crystal form).58,66
4.3. Leucine
In comparison to arginine, leucine does not change the pH of the solution since leucine has a hydrophobic and not an electrically charged residue. According to Bull and Breese,50 increasing leucine concentration lowers the surface tension of the bulk solution. This indicates that the interfacial free energy at the crystal cluster and bulk–solution could be reduced and hence the energetic barrier of nucleation could be lowered (decrease of the activation barrier). Recent studies showed that amino acids can impact and alter the secondary structure of the native protein67 which also indicates a change in the interaction intensity between the protein molecules and bulk liquid phase. Additionally, the promoting effect of hydrophobic additives has already been shown in literature.68–70 Since leucine is a hydrophobic amino acid and reduces the interactions between the water molecules, the addition of leucine might lead to an increase in the hydrophobic interactions. The hydrophobic interaction has been reported to be a dominant interaction within a dense liquid cluster prior to crystallisation.69 On the other hand, leucine could suppress unfavourable hexamer contacts that do not lead to the formation of ordered hexamer arrangements. Leucine acts by binding to hydrophobic residues not buried in the interior of the insulin hexamer. Unfavourable contacts could be the interaction of a hydrophobic surface patch and a hydrophilic one which does not lead to crystallisation.70 This could be mathematically expressed with an increase in the kinetic constant (parameter A in eqn (2)) resulting in an increased nucleation rate.4.4. Glycine
Although glycine, and every other amino acid, is present in its zwitter-ion state in aqueous solution at neutral pH, and hence provides two opposite electrically charged groups, it was found that it does not promote crystallisation. Although the crystal occurrence of insulin in addition of glycine is higher than for the control, the difference is too small to derive an enhancing effect of glycine. Compared to arginine and similar to leucine, glycine does not change the pH nor the ionic strength of the solution. Hence, it can be hypothesized that the amine and acidic group of amino acids play a minor role in facilitating crystallisation.5. Conclusion
We have shown that amino acids have the propensity to control protein nucleation even at low supersaturation. Both arginine and leucine were found to enhance the nucleation, whilst glycine did not facilitate the nucleation of insulin. This enhancement in nucleation is not due to an increase in supersaturation as the addition of arginine resulted in an increase in the solubility of insulin, while leucine and glycine did not alter the solubility. The facilitation in nucleation is attributed to the promotion of attractive interactions and the suppression of unfavourable insulin hexamer contacts. Here, we demonstrated that the type and concentration of amino acid plays a significant role in the promoting effect.Conflicts of interest
There are no conflicts to declare.Acknowledgements
FL gratefully acknowledges the Department of Chemical Engineering at Imperial College London for a PhD Departmental Scholarship. This study is funded by the UK's EPSRC (EP/N015916/1).References
- S. Rastogi, S. Shukla, M. Kalaivani and G. S. Singh, Peptide-based therapeutics: quality specifications, regulatory considerations, and prospects, Drug Discovery Today, 2019, 24(1), 148–162 CrossRef CAS PubMed.
- A. C. A. Roque, A. S. Pina, A. M. Azevedo, R. Aires-Barros, A. Jungbauer, G. Di Profio, J. Y. Y. Heng, J. Haigh and M. Ottens, Anything but Conventional Chromatography Approaches in Bioseparation, Biotechnol. J., 2020, 15(8), 1900274–1900285 CrossRef CAS PubMed.
- C. Guillemot-Potelle, M. D. Costioli, H. Broly and C. Mitchell-Logean, Cost of Goods Modeling and Quality by Design for Developing Cost-Effective Processes, BioPharm Int., 2010, 23(6), 26–35 Search PubMed.
- T. Delmas, M. M. Roberts and J. Y. Y. Heng, Nucleation and Crystallization of Lysozyme: Role of Substrate Surface Chemistry and Topography, J. Adhes. Sci. Technol., 2012, 25(4–5), 357–366 Search PubMed.
- D. Hekmat, Large-scale crystallization of proteins for purification and formulation, Bioprocess Biosyst. Eng., 2015, 38(7), 1209–1231 CrossRef CAS PubMed.
- S. Pechenov, B. Shenoy, M. X. Yang, S. K. Basu and A. L. Margolin, Injectable controlled release formulations incorporating protein crystals, J. Controlled Release, 2004, 96(1), 149–158 CrossRef CAS PubMed.
- J. F. Lutsko, How crystals form: A theory of nucleation pathways, Sci. Adv., 2019, 5(4), eaav7399 CrossRef CAS PubMed.
- P. G. Vekilov, Nucleation, Cryst. Growth Des., 2010, 10(12), 5007–5019 CrossRef CAS PubMed.
- A. McPherson and J. A. Gavira, Introduction to protein crystallization, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2014, 70(Pt 1), 2–20 CrossRef CAS PubMed.
- E. Rubin, C. Owen and V. Stojanoff, Crystallization under an External Electric Field: A Case Study of Glucose Isomerase, Crystals, 2017, 7(7), 206–217 CrossRef.
- D.-C. Yin, Protein crystallization in a magnetic field, Prog. Cryst. Growth Charact. Mater., 2015, 61(1), 1–26 CrossRef CAS.
- H. Kim and K. Suslick, The Effects of Ultrasound on Crystals: Sonocrystallization and Sonofragmentation, Crystals, 2018, 8(7), 280–299 CrossRef.
- H. Yang, L. Huang, F. Zhang, V. Karde, Z. Yang and J. Y. Y. Heng, Gravity on Crystallization of Lysozyme: Slower or Faster?, Cryst. Growth Des., 2019, 19(12), 7402–7410 CrossRef CAS.
- Z. Wang, Q. Wang and L. Dang, Ionic liquids as selectors for controlling the crystallization nucleation of hen egg white lysozyme, Biotechnol. Bioprocess Eng., 2012, 17(5), 1025–1030 CrossRef CAS.
- R.-B. Zhou, H.-L. Cao, C.-Y. Zhang and D.-C. Yin, A review on recent advances for nucleants and nucleation in protein crystallization, CrystEngComm, 2017, 19(8), 1143–1155 RSC.
- U. V. Shah, C. Amberg, Y. Diao, Z. Yang and J. Y. Y. Heng, Heterogeneous nucleants for crystallogenesis and bioseparation, Curr. Opin. Chem. Eng., 2015, 8(1), 69–75 CrossRef.
- G. Tosi, S. Fermani, G. Falini, J. A. Gavira Gallardo and J. M. Garcia Ruiz, Crystallization of proteins on functionalized surfaces, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64(Pt 10), 1054–1061 CrossRef CAS PubMed.
- U. V. Shah, M. C. Allenby, D. R. Williams and J. Y. Y. Heng, Crystallization of Proteins at Ultralow Supersaturations Using Novel Three-Dimensional Nanotemplates, Cryst. Growth Des., 2012, 12(4), 1772–1777 CrossRef CAS.
- A. McPherson and P. Shlichta, Heterogeneous and Epitaxial Nucleation of Protein Crystals on Mineral Surfaces, Science, 1988, 239(4838), 385–387 CrossRef CAS PubMed.
- M. Polino, C. A. M. Portugal, G. Di Profio, I. M. Coelhoso and J. G. Crespo, Protein Crystallization by Membrane-Assisted Technology, Cryst. Growth Des., 2019, 19(8), 4871–4883 CrossRef CAS.
- E. Curcio, V. López-Mejías, G. Di Profio, E. Fontananova, E. Drioli, B. L. Trout and A. S. Myerson, Regulating Nucleation Kinetics through Molecular Interactions at the Polymer–Solute Interface, Cryst. Growth Des., 2014, 14(2), 678–686 CrossRef CAS.
- B. Zhang, A. R. Mei, M. A. Isbell, D. Wang, Y. Wang, S. F. Tan, X. L. Teo, L. Xu, Z. Yang and J. Y. Y. Heng, DNA Origami as Seeds for Promoting Protein Crystallization, ACS Appl. Mater. Interfaces, 2018, 10(51), 44240–44246 CrossRef CAS PubMed.
- W. Chen, S. J. Park, F. Kong, X. Li, H. Yang and J. Y. Y. Heng, High Protein-Loading Silica Template for Heterogeneous Protein Crystallization, Cryst. Growth Des., 2019, 20(2), 866–873 CrossRef.
- D. Ribeiro, A. Kulakova, P. Quaresma, E. Pereira, C. Bonifácio and M. J. Romão, Use of Gold Nanoparticles as Additives in Protein Crystallization, Cryst. Growth Des., 2013, 14(1), 222–227 CrossRef.
- U. V. Shah, D. R. Williams and J. Y. Y. Heng, Selective Crystallization of Proteins Using Engineered Nanonucleants, Cryst. Growth Des., 2012, 12(3), 1362–1369 CrossRef CAS.
- N. E. Chayen, E. Saridakis, R. El-Bahar and Y. Nemirovsky, Porous silicon: An effective nucleation-inducing material for protein crystallization, J. Mol. Biol., 2001, 312(4), 591–595 CrossRef CAS PubMed.
- K. Ino, I. Udagawa, K. Iwabata, Y. Takakusagi, M. Kubota, K. Kurosaka, K. Arai, Y. Seki, M. Nogawa, T. Tsunoda, F. Mizukami, H. Taguchi and K. Sakaguchi, Heterogeneous nucleation of protein crystals on fluorinated layered silicate, PLoS One, 2011, 6(7), e22582 CrossRef CAS PubMed.
- F. Artusio and R. Pisano, Surface-induced crystallization of pharmaceuticals and biopharmaceuticals: A review, Int. J. Pharm., 2018, 547(1–2), 190–208 CrossRef CAS PubMed.
- E. Saridakis, S. Khurshid, L. Govada, Q. Phan, D. Hawkins and G. V. Crichlow, Protein crystallization facilitated by molecularly imprinted polymers, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(27), 11081–11086 CrossRef CAS PubMed.
- X. Li, H. Liu, X. Tong, S. Dai, J. Zhang and W. Li, Charged polymeric additives affect the nucleation of lysozyme crystals, CrystEngComm, 2019, 21(12), 1992–2001 RSC.
- O. Galkin and P. G. Vekilov, Control of protein crystal nucleation around the metastable liquid–liquid phase boundary, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(12), 6277–6281 CrossRef CAS PubMed.
- T. Arakawa, K. Tsumoto, Y. Kita, B. Chang and D. Ejima, Biotechnology applications of amino acids in protein purification and formulations, Amino Acids, 2007, 33(4), 587–605 CrossRef CAS PubMed.
- T. Arakawa, D. Ejima, K. Tsumoto, N. Obeyama, Y. Tanaka, Y. Kita and S. N. Timasheff, Suppression of protein interactions by arginine: a proposed mechanism of the arginine effects, Biophys. Chem., 2007, 127(1–2), 1–8 CrossRef CAS PubMed.
- K. Shiraki, M. Kudou, S. Fujiwars, T. Imanaka and M. Takagi, Biophysical Effect of Amino acids on the Prevention of protein Aggregation, J. Biol. Chem., 2002, 132(4), 591–595 CAS.
- S. R. Wlodarczyk, D. Custodio, A. Pessoa Jr. and G. Monteiro, Influence and effect of osmolytes in biopharmaceutical formulations, Eur. J. Pharm. Biopharm., 2018, 131(1), 92–98 CrossRef CAS PubMed.
- M. Norrman, F. Hubálek and G. Schluckebier, Structural characterization of insulin NPH formulations, Eur. J. Pharm. Sci., 2007, 30(5), 414–423 CrossRef CAS PubMed.
- T. J. Kamerzell, R. Esfandiary, S. B. Joshi, C. R. Middaugh and D. B. Volkin, Protein-excipient interactions: mechanisms and biophysical characterization applied to protein formulation development, Adv. Drug Delivery Rev., 2011, 63(13), 1118–1159 CrossRef CAS PubMed.
- R. J. Falconer, Advances in liquid formulations of parenteral therapeutic proteins, Biotechnol. Adv., 2019, 37(7), 107412–107420 CrossRef CAS PubMed.
- J. Y. Lim, N. A. Kim, D. G. Lim, K. H. Kim, S. Hada and S. H. Jeong, Evaluation of etanercept degradation under oxidative stress and potential protective effects of various amino acids, Int. J. Pharm., 2015, 492(1–2), 127–136 CrossRef CAS PubMed.
- J. L. Cleland and D. I. C. Wang, Cosolvent Assisted Protein Refolding, Nat. Biotechnol., 1990, 8(1), 1274–1278 CrossRef CAS PubMed.
- P. Stärtzel, Arginine as an Excipient for Protein Freeze-Drying: A Mini Review, J. Pharm. Sci., 2018, 107(4), 960–967 CrossRef PubMed.
- S. H. Paik, Y. J. Kim, S. K. Han, J. M. Kim, J. W. Huh and Y. I. Park, Mixture of three amino acids as stabilizers replacing albumin in lyophilization of new third generation recombinant factor VIII GreenGene F, Biotechnol. Prog., 2012, 28(6), 1517–1525 CrossRef CAS PubMed.
- A. Al-Hussein and H. Gieseler, Investigation of histidine stabilizing effects on LDH during freeze-drying, J. Pharm. Sci., 2013, 102(3), 813–826 CrossRef CAS PubMed.
- A. Ajmera and R. Scherliess, Stabilisation of proteins via mixtures of amino acids during spray drying, Int. J. Pharm., 2014, 463(1), 98–107 CrossRef CAS PubMed.
- L. Bergeron, L. F. Filobelo, O. Galkin and P. G. Vekilov, Thermodynamics of the Hydrophobicity in Crystallization of Insulin, Biophys. J., 2003, 85(6), 3935–3942 CrossRef CAS PubMed.
- J. V. Parambil, M. Schaepertoens, D. R. Williams and J. Y. Y. Heng, Effects of Oscillatory Flow on the Nucleation and Crystallization of Insulin, Cryst. Growth Des., 2011, 11(10), 4353–4359 CrossRef CAS.
- M. M. Harding, D. C. Hodgkin, A. F. Kennedy, A. O'Connor and P. D. J. Weitzmann, The Crystal Structure of Insulin 2. An Investigation of Rhombohedral Zinc Insulin Crystals and a Report of Other Crystalline Forms, J. Mol. Biol., 1966, 16(1), 212–226 CrossRef CAS PubMed.
- C. N. Nanev, F. V. Hodzhaoglu and I. L. Dimitrov, Kinetics of Insulin Crystal Nucleation, Energy Barrier, and Nucleus Size, Cryst. Growth Des., 2011, 11(1), 196–202 CrossRef CAS.
- J. Newman, J. Xu and M. C. Willis, Initial evaluations of the reproducibility of vapor-diffusion crystallization, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2007, 63(Pt 7), 826–832 CrossRef CAS PubMed.
- H. B. Bull and K. Breese, Surface tension of amino acid solutions A hydrophobicity scale of the amino acid residues, Arch. Biochem. Biophys., 1974, 161(2), 665–670 CrossRef CAS.
- C. Y. Zhang, Z. Q. Wu, D. C. Yin, B. R. Zhou, Y. Z. Guo, H. M. Lu and R. B. Zhou, A strategy for selecting the pH of protein solutions to enhance crystallization, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2013, 69(Pt 7), 821–826 CrossRef CAS PubMed.
- N. Samanta, D. Das Mahanta, S. Choudhury, A. Barman and R. Kumar Mitra, Collective hydration dynamics in some amino acid solutions: A combined GHz-THz spectroscopic study, J. Chem. Phys., 2017, 146(12), 125101 CrossRef PubMed.
- C. N. Nanev, I. L. Dimitrov and F. V. Hodzhaoglu, Growth of rhombohedral insulin crystals and in vitro modeling of their dissolution in the blood stream, Cryst. Res. Technol., 2011, 46(2), 119–126 CrossRef CAS.
- D. Kashchiev and G. M. van Rosmalen, Review: Nucleation in solutions revisited, Cryst. Res. Technol., 2003, 38(78), 555–574 CrossRef CAS.
- F. V. Hodzhaoglu, M. Conejero-Muriel, I. L. Dimitrov and J. A. Gavira, Optimization of the classical method for nucleation and growth of rhombohedral insulin crystals by pH titration and screening, Bulg. Chem. Commun., 2016, 48(A), 29–37 Search PubMed.
- K. A. Kantardjieff and B. Rupp, Protein isoelectric point as a predictor for increased crystallization screening efficiency, Bioinformatics, 2004, 20(14), 2162–2168 CrossRef CAS PubMed.
- M. Charles, S. Veesler and F. Bonnete, MPCD: a new interactive on-line crystallization data bank for screening strategies, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62(Pt 11), 1311–1318 CrossRef PubMed.
- M. Liang, F. Jin, R. Liu, Y. Yu, R. Su, L. Wang, W. Qi and Z. He, Shape evolution and thermal stability of lysozyme crystals: effect of pH and temperature, Bioprocess Biosyst. Eng., 2013, 36(1), 91–99 CrossRef CAS PubMed.
- D. E. Kuehner, C. Heyer, C. Rämsch, U. M. Fornefeld, H. W. Blanch and J. M. Prausnitz, Interactions of Lysozyme in Concentrated Electrolyte Solutions from Dynamic Light-Scattering Measurements, Biophys. J., 1997, 73(6), 3211–3224 CrossRef CAS PubMed.
- Y. Kita, T. Arakawa, T.-Y. Lin and S. N. Timasheff, Contribution of the Surface Free Energy Perturbation to Protein—Solvent Interactions, Biochemistry, 1994, 33(50), 15178–15189 CrossRef CAS PubMed.
- M. M. Nuhu and R. Curtis, Arginine dipeptides affect insulin aggregation in a pH- and ionic strength-dependent manner, Biotechnol. J., 2015, 10(3), 404–416 CrossRef CAS PubMed.
- S. Haghighi-Poodeh, B. Kurganov, L. Navidpour, P. Yaghmaei and A. Ebrahim-Habibi, Characterization of arginine preventive effect on heat-induced aggregation of insulin, Int. J. Biol. Macromol., 2020, 145(2), 1039–1048 CrossRef CAS PubMed.
- M. Ootaki, S. Endo, Y. Sugawara and T. Takahashi, Crystal habits of cubic insulin from porcine pancreas and evaluation of intermolecular interactions by macrobond and EET analyses, J. Cryst. Growth, 2009, 311(17), 4226–4234 CrossRef CAS.
- A. McPherson, C. Nguyen, R. Cudney and S. B. Larson, The Role of Small Molecule Additives and Chemical Modification in Protein Crystallization, Cryst. Growth Des., 2011, 11(5), 1469–1474 CrossRef CAS.
- K. Brezina, E. Duboue-Dijon, V. Palivec, J. Jiracek, T. Krizek, C. M. Viola, T. R. Ganderton, A. M. Brzozowski and P. Jungwirth, Can Arginine Inhibit Insulin Aggregation? A Combined Protein Crystallography, Capillary Electrophoresis, and Molecular Simulation Study, J. Phys. Chem. B, 2018, 122(44), 10069–10076 CrossRef CAS PubMed.
- M. Mohamed Abubakkar, K. Saraboji and M. N. Ponnuswamy, Purification, crystallization and preliminary crystallographic studies of haemoglobin from mongoose (Helogale parvula) in two different crystal forms induced by pH variation, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2013, 69(Pt 2), 126–129 CrossRef CAS PubMed.
- S. Dai, H. Liu, W. Li and J. Zhang, Effects of Small Biomolecules on Lysozyme Crystallization, Trans. Tianjin Univ., 2020 DOI:10.1007/s12209-020-00251-x.
- C. N. Nanev, E. Saridakis, L. Govada, S. C. Kassen, H. V. Solomon and N. E. Chayen, Hydrophobic Interface-Assisted Protein Crystallization: Theory and Experiment, ACS Appl. Mater. Interfaces, 2019, 11(13), 12931–12940 CrossRef CAS PubMed.
- L. Ito, K. Shiraki and H. Yamaguchi, Comparative analysis of amino acids and amino-acid derivatives in protein crystallization, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2010, 66(6), 744–749 CrossRef CAS PubMed.
- S. Tanaka, M. Ataka, T. Kubota, T. Soga, K. Homma, W. C. Lee and M. Tanokura, The effect of amphiphilic additives on the growth and morphology of Aspergillus niger acid proteinase A crystals, J. Cryst. Growth, 2002, 234(1), 247–254 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Section S1. Summary of insulin crystal occurrence after 4 and 7 days. Section S2. Chromatographic conditions for HPLC analysis. See DOI: 10.1039/d1ce00026h |
| This journal is © The Royal Society of Chemistry 2021 |