 Open Access Article
Open Access ArticlePhotoswitches for controllable RNA binding: a future approach in the RNA-targeting therapy
Daria V.
Berdnikova

Universität Siegen, Organische Chemie II, Adolf-Reichwein-Str. 2, 57076 Siegen, Germany. E-mail: berdnikova@chemie-bio.uni-siegen.de
First published on 23rd September 2021
Abstract
RNA is an emerging drug target that opens new perspectives in the treatment of viral and bacterial infections, cancer and a range of so far incurable genetic diseases. Among the various strategies towards the design and development of selective and efficient ligands for targeting and detection of therapeutically relevant RNA, photoswitchable RNA binders represent a very promising approach due to the possibility to control the ligand–RNA and protein–RNA interactions by light with high spatiotemporal resolution. However, the field of photoswitchable RNA binders still remains underexplored due to challenging design of lead structures that should combine high RNA binding selectivity with efficient photochemical performance. The aim of this highlight article is to describe the development of photoswitchable noncovalent RNA binders and to outline the current situation and perspectives of this emerging interdisciplinary field.
1. Introduction
The ribonucleic acid (RNA) is the most diverse and multifunctional biomolecule in Nature that not only encodes genetic information, but also catalyses chemical reactions in the cell and provides intracellular recognition and transport.1–3 Due to the multiple and important roles of RNA in living processes it has as a huge and essential impact on human health. Structural changes and failure of the function of human RNAs often have significant health-related consequences. For example, more than 40 incurable neurodegenerative diseases, e.g. Huntington disease, are caused by formation of mutant RNAs in humans due to expansions of RNA sequence repeats.4,5 Moreover, the involvement of microRNAs (miRNA) in oncogenesis and in the development of metabolic, autoimmune and other disorders was recently discovered.6–8 Exogenous RNAs can also pose a serious threat to human health. The majority of human and animal pathogenic viruses are RNA viruses whose genetic information is stored in the form of RNA. These viruses cause a range of diseases such as influenza, measles, polio, rabies, Ebola fever, HIV infection, hepatitis C (HCV), COVID-19 and others.9–11 Before this background, RNA obviously represents a promising therapeutical target for treatment of a wide range of diseases, from genetically caused disorders and cancer to life-threating infections. RNA-targeting therapy could be applied also to bacterial infections because interaction of drugs with ribosomal RNA of bacteria can be used to fight pathogenic microorganisms.12 Along these lines, RNA-targeting therapy represents significant expansion, alternative or supplement to the majority of the existing therapeutical approaches that typically target proteins.13 The rapidly increasing amount of publications on RNA-targeting molecules points out the emerging importance of this interdisciplinary research area.14–19 Similar to DNA and proteins, RNA possesses a range of important features as a drug target. Like DNA, the chemical building units of RNA are fewer and less complex in comparison to those of proteins. RNA can also adopt a regular A-form double-helix, which, however, is often disrupted by regions of unpaired or mismatched bases that allow RNA to fold into more complex three-dimensional structures, analogous to those observed in proteins. The plethora of RNA structural elements and folding patterns20 that result in the formation of binding sites of unique size, shape, and electrostatic potential distribution is a prerequisite for highly selective interactions of RNA with small molecules. The targetability of RNA arises also from the absence of cellular RNA repair mechanisms and its accessibility in ribonucleoprotein complexes.21 Nevertheless, the rational design of selective and efficient RNA binders still represents a challenging task due to the absence of clear and universal design strategies arising from high variability of RNA structures.One of the major challenges in the development of RNA-targeting drugs is to establish complete spatial and temporal control of the drug activity, specifically the rate of drug delivery, its effective concentration in the proximity of a target, the actual drug–target interaction, as well as the final drug deactivation process. The latter is especially important to suppress potential side effects of the therapy. In this context, the introduction of a switchable functionality into an RNA binder that allows manipulation of its properties by an external stimulus is highly attractive. Among potential external stimuli for drug activation, light is particularly attractive as a non-invasive, bio-orthogonal tool possessing a high spatiotemporal resolution of action. In this context, two main approaches of the photochemical control of the structure and functions of nucleic acids can be applied.22 (i) With the first approach, the photoregulation is accomplished by a covalent integration of photoswitches into the nucleic acids.22–25 This method is very effective, but suffers from the disturbance of the native structure of nucleic acids, necessity of synthetically challenging modifications of nucleotides and potential difficulties of application in real systems, e.g. due to degradation by nucleases. (ii) The second approach considers the application of photoswitchable ligands as noncovalent nucleic acids binders, whose binding affinity can be regulated by light. Along these lines, a range of noncovalent photocontrollable DNA binders has been described that can be activated for binding interaction with DNA in a reversible26 or irreversible27 manner. At the same time, the field of photocontrollable noncovalent RNA binders has not been explored to the same extent, most likely, due to the ambiguity of the design strategies. Nevertheless, the accumulated data already allow to perform the first systematization of the noncovalent photoswitchable RNA binders, make some conclusions and suggest new ideas. This highlight outlines the achievements, challenges and future perspectives of the development of noncovalent light-controllable RNA binders with the aim to bring this emerging interdisciplinary area to the attention of chemists and biologists.
2. Types of the noncovalent photoswitch–RNA interactions
Generally, there are four types of manipulation of noncovalent ligand–RNA interactions by light (Scheme 1). The first type is based on the photochemical transformation between two ligand forms that possess significantly different affinity towards RNA. In an ideal scenario, the light-controllable switching between the binding and non-binding isomer yields an “ON/OFF binding” system (Scheme 1a). Hence, the geometrical match of only one of the isomers to the binding pocket of the target RNA is an essential feature of these systems. | ||
| Scheme 1 Existing and prospective types of systems involving noncovalent photoswitch–RNA interactions. | ||
In the other three types of the photoswitch–RNA interactions, both the initial and photoinduced forms of the ligand remain bound to the nucleic acid, but the photoswitching allows to manipulate certain binding parameters of the ligand or/and RNA. Thus, the photoswitching can be accompanied by pronounced changes of the ligand fluorescence (“ON/OFF fluorescence”) (Scheme 1b),28 resulting in a photoswitchable stain, which can be applied in RNA bioimaging.29 Other possible effects may be the light-induced change of the ligand binding mode (Scheme 1c) and the light-controllable reversible un- or refolding of RNA (Scheme 1d). The photoinduced switching of the binding mode has been recently described for quadruplex DNA.26n To the best of my knowledge, however, similar examples for RNA species have not been reported, so far. Nevertheless, such systems can be envisaged for the naturally occurring RNA motifs of sufficient complexity, for example, three-helix junctions, riboswitches and pseudoknots,13 as well as RNA G-quadruplexes.30
In the next sections, the photoswitchable RNA binders reported up to date are highlighted. The majority of the systems belong to the “ON/OFF binding” type. Two single examples represent the “ON/OFF fluorescence” and “RNA un/re-folding” systems, respectively. The article is divided into sections according to the structure and therapeutical importance of RNA species, namely from synthetic RNAs having low therapeutical relevance to HIV-1 genome RNA.
3. Photoswitchable RNA binders
3.1. Photoswitchable binders for synthetic RNA homoduplexes and cellular double-stranded RNAs
RNA homoduplexes are RNA double helixes having precisely complementary bases in the two strands. Synthetic RNA homoduplexes are usually composed of two complementary homopolymeric single strands, e.g. polyriboadenylic acid (polyA) with polyribouridylic acid (polyU) or polyriboguanilic acid (polyA) with polyribocytidylic acid (polyC). Given to their availability, these RNAs are widely used in fundamental and medicinal studies of nucleic acids. The effect of synthetic RNA homoduplexes to promote antitumoral immune response has been explored in cancer immunotherapy for decades.31 However, as a drug target mimetic, such RNAs represent only a very simplifying model, whose structure and properties cannot be directly related to naturally occurring RNAs possessing much more variable composition and folding patterns. Nevertheless, studies with synthetic RNA homoduplexes often become the first step in the evaluation of the RNA-binding properties of ligands. These RNAs possess a highly ordered helical structure with a very shallow minor groove and a very deep and narrow major groove. As a consequence, binding of small ligands to polymeric double-stranded RNA (dsRNA) occurs mainly in two modes, namely intercalation between RNA base pairs and groove binding, like in the case of the double-stranded DNA (dsDNA). For this reason, it is not surprising that most DNA-binding ligands also associate with synthetic RNA homoduplexes.The first example of the photoactivated noncovalent RNA binding was reported in 2005.27a Thus, E-methyl-3-(furan-2-yl)-2-phenylacrylate 1 that has no affinity towards nucleic acids was photochemically transformed into polycyclic compound 2 that intercalates between the dsDNA/RNA base pairs (Scheme 2a). The photoreaction was performed directly in the presence of the double-stranded calf thymus DNA (ct DNA) or synthetic RNA homoduplexes polyA–polyU and polyG–polyC. However, intercalator 2 showed no selectivity towards a certain type of nucleic acids. Additionally, the photoreaction was irreversible, i.e. subsequent photochemical or thermal deactivation of intercalator 2 was not possible.
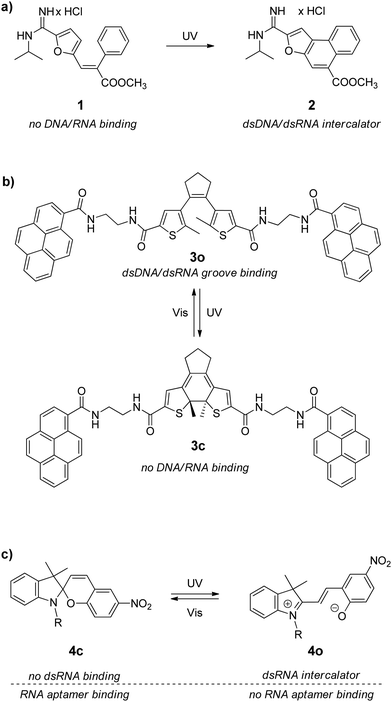 | ||
| Scheme 2 Photoswitchable binders for double-stranded RNAs (compound 4 also interacts with an RNA aptamer, see Section 3.2). | ||
Very recently, it was reported about the photoswitchable diarylethene–pyrene conjugate 3o (Scheme 2b and Table 1) that can be reversibly activated towards binding with polyA–polyU RNA and different types of double-stranded DNA (ct DNA, poly(dA:dT), poly(dG:dC)).32 The open form of 3o is accommodated in the dsDNA/RNA groove, whereas intramolecular stacking of two pyrene residues makes the closed form 3c too bulky for the groove binding resulting in the dissociation of the ligand–RNA complexes.
| Type of photoreaction | Photoswitch class | Species | λ fwd/λbkwd/nmb |
φ
fwd/φbkwd/10−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c c |
α
fwd/αbkwd/(% c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) o) or (%E o) or (%E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z)d Z)d |
τ 25, °Ce | RNA target | RNA-binding isomer |
K
A/105 M−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f f |
Application | Exp. condition | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a All data are obtained in aqueous buffer systems of different compositions. b Irradiation wavelengths for the forward and backward photoswitching. c Photoisomerization quantum yields of the forward and backward reactions. d Maximum conversion of the photoswitches in the forward and backward reactions determined without RNA in the corresponding photostationary states (PSS). e Half-lifes of the photoinduced forms. f Association constants with RNA; if nothing is additionally indicated in brackets, the constant values are calculated for the RNA-binding form of the photoswitch; in the case of several RNA targets, the RNA and the corresponding binding constant are placed in one row in the table. g The photoreaction of compound 1 is irreversible. h The original data are given in the form of dissociation constants. The data in this table are recalculated and presented in the form of association constants for uniformity and ease of comparison. i Although a clear evidence for the association of the E-isomer of 10 with TAR and RRE RNA was provided, the binding constants for the corresponding complexes were not determined. | ||||||||||||
| Electrocyclization | 1,2-Arenyl ethene g | 1 | >300/n.d.g | n.d. | n.d.g | n.d.g | • polyA–polyU dsRNA | 2 | 0.200 | Photoactivatable dsRNA intercalator | In vitro | 27a |
| • polyC–polyG dsRNA | 0.794 | |||||||||||
| Dithenylethene | 3 | 254–315/400–700 | n.d. | n.d. | n.d. | polyA–polyU dsRNA | 3o | 12.6h | dsRNA photoswitchable groove binder | In vitro | 32 | |
| Spiropyran | 4 | 254/420 | 1.5/1.4 | PSS254 (63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37)/n.d. 37)/n.d. |
n.d. | • polyA–polyU dsRNA | 4o | 0.074–0.111 | Detection of synthetic and cellular dsRNAs | In vitro | 33 | |
| • polyC–polyG dsRNA | 0.039–0.070 | |||||||||||
| • endogenous dsRNAs from HeLa cells | — | |||||||||||
| 365/>490 | n.d. | PSS365 (<5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) >95)/PSS490 (>95 >95)/PSS490 (>95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) <5) <5) |
<1 h | Synthetic RNA aptamer | 4c | 0.526h | Selective photoswitchable binder for an RNA aptamer | In vitro | 42 | |||
| Dihydropyrene | 5 | 280–375/>400 | n.d. | n.d. | n.d. | Synthetic RNA aptamer (alone and as a part of an allosteric hammerhead ribozyme) | 5c | 3.70 (with the aptamer alone)h | Reversible photocontrol of a hammerhead ribozyme | In vitro | 43 | |
| 0.833 (with the same aptamer implemented in the ribozyme)h | ||||||||||||
| E–Z isomerization | Azobenzene | 6 | 365/420 | n.d. | PSS365 (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80)/PSS420 (>95 80)/PSS420 (>95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :<5) :<5) |
27 d | Synthetic RNA aptamer | Z-6 | 18.3h | Selective photoswitchable binder for an RNA aptamer | In vitro | 44 |
| 8 | 360/430 | n.d. | PSS360 (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95)/PSS430 (79 95)/PSS430 (79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21) 21) |
10 h | Several synthetic RNA aptamers | E-8 | 4.17–14.1h | Reversible photocontrol of peptide–RNA interaction in solution and on the surface | In vitro | 52 | ||
| 9 | 365/>400 | n.d. | n.d. | n.d. | • 164-mRNA-EGFP | E-9 | n.d. | Reversible photocontrol of gene expression | In vitro | 57 | ||
| • 156-mRNA-DsRedM | ||||||||||||
| Stiff-stilbene | 7 | 342/372 | n.d. | n.d. | n.d. | Synthetic RNA aptamer as a ligand-binding domain of a modified bacterial riboswitch | Z-7 | 0.009 (Z-7)h | Reversible photocontrol of gene expression | In vitro | 45 | |
| 0.00009 (E-7)h | In vivo | |||||||||||
| Hemi-indigo | 10 | 470/590 | 2.7/0.20 (no RNA) | PSS470 (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80)/PSS590 (97 80)/PSS590 (97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
10.8 h (no RNA) | • HIV-1 TAR RNA | Both | 1.1 (Z-10–TAR)i | RNA stain with photoswitchable fluorescence | In vitro | 28 | |
| 2.1/0.06 (with TAR) | 51.0 h (with RRE) | • HIV-1 RRE-IIB RNA | 1.7 (Z-10–RRE)i | |||||||||
| 1.2/0.02 (with RRE) | ||||||||||||
Another recent example of the photocontrollable interaction with dsRNA is spiropyran derivative 4 (Scheme 2c and Table 1).33 The closed form 4c did not bind to dsRNA whereas the open form 4o intercalated between the base pairs of the RNA A-helix. Studies with synthetic RNA homoduplexes polyA–polyU and polyG–polyC revealed that 4o had a slight preference towards interaction with GC-pairs (Table 1). Moreover, it was shown that the open form 4o associated with cellular dsRNAs obtained from HeLa cells. The long endogenous dsRNAs were isolated by the treatment of the total HeLa RNA samples with RNase T1 followed by phenol extraction that allowed to remove single-stranded RNAs and hairpin RNAs. The binding of 4o to the cellular dsRNAs was detected photometrically. However, the identification of the particular structure of the dsRNA species involved in this interaction has not been performed. Additionally, the open form of spiropyran 4o was applied for the optical detection of the increased dsRNA expression in the HCT116 colorectal cancer cells after treatment with anticancer drug decitabine.33 The analysis was performed on the isolated dsRNAs samples.
Overall, the association of small ligands with dsRNA homopolymers and long cellular dsRNAs does not significantly differ from the interaction with polymeric dsDNA. For this reason, photoswitchable ligands 1, 3 and 4 lack selectivity in discrimination of dsRNA against dsDNA.
3.2. Photoswitchable binders for RNA aptamers
The next approximation towards native RNA structures was realized by photocontrollable interactions with RNA aptamers, i.e. short single-stranded RNAs that can selectively associate with a specific target.34–36 The high affinity and selectivity of the aptamer–target binding is provided by the unique tertiary structure of aptamers arising from their flexibility and shape-forming properties.37 To date, a wide range of aptamers has been obtained for various targets, from small organic and inorganic molecules to protein complexes and even cells.38 In medicine, aptamers designed for certain therapeutic targets are used as analogues of antibodies for diagnostic and treatment purposes.39–41 RNA aptamers represent a more realistic model of native RNAs in comparison to the synthetic RNA homoduplexes. However, the majority of RNA aptamers obtained up to now does not correspond to the native RNA sequences, although they can be artificially implemented, for example, into bacterial plasmids.The peculiarities of the aptamer–ligand interactions allowed to obtain systems with highly efficient, light-controllable switching of the ligand binding state (“ON/OFF binding”). Thus, four examples of photoswitchable binders for RNA aptamers were reported that are based on the reversible photoinduced transformations of spiropyran 4,42 benzodihydropyrene 5,43 azobenzene 6,44 and stiff-stilbene 745 (Schemes 2c, 3 and Table 1). The RNA aptamers that bind specifically only to one of the two forms of the ligands 4–6 were selected in vitro from RNA aptamer libraries by the Systematic Evolution of Ligands by EXperimental enrichment (SELEX) procedure46i.e. by screening the geometrical fit of the ligands to the aptamer binding pockets. In the case of stiff-stilbene 7, the selection was performed using an RNA pool derived from a bacterial SAM-1 riboswitch by replacing its ligand-binding domain with a random region of 45 nucleotides and partially randomizing the terminator and anti-terminator hairpins.45 As a result, the selected RNA aptamer was isolated as a fused construct together with a riboswitch expression platform. The reversible photoswitching between the isomers of ligands 4–7 gave the systems an advantage of accomplishing repeated binding/dissociation cycles with RNA. Whereas the studies on spiropyran 4 and azobenzene 6 derivatives provided a proof-of-principle for the selective light-controllable RNA binding, compounds 5 and 7 were used in more complex systems for the manipulation of the biological functions. Thus, introduction of the corresponding RNA aptamer into a hammerhead ribozyme yielded a two-state system with a >900-fold difference in catalytic activity that was governed by the photoswitchable interaction with dihydropyrene derivative 5.43 In the second example, a synthetic riboswitch comprising the stiff-stilbene 7-binding aptamer has been successfully implemented into a bacterial plasmid that allowed to perform reversible photocontrol of the Fluc gene expression in the modified E. coli in vivo.45
Nevertheless, all the studied aptamer models were selected from the randomized RNA libraries and had, therefore, no therapeutic relevance, thus reducing the applicability of these systems for real human disease models.
3.3. Special case: photoswitchable peptide–RNA interactions
The investigation of photocontrollable protein–RNA interactions is especially important for a therapeutic perspective because aberrant protein–RNA interactions are involved in pathogenic mechanisms of various diseases. These interactions involve abnormal protein binding to endogenous mutant RNAs, which happens, for example, in RNA repeat expansion diseases.16 In the case of viral infections, association of certain proteins to viral genome RNA plays an important role in the living cycle of viruses. For example, binding of the Tat protein to the transactivation response element (TAR) of the human immunodeficiency virus (HIV) RNA triggers the transcription of HIV in infected cells.47 In this context, targeted disruption of disease-related protein–RNA complexes is critical for achieving a therapeutic effect in the treatment of RNA-associated diseases. Along these lines, reversible regulation of protein–RNA interactions by light may act not only as a therapeutical approach but also as a powerful model for the understanding of molecular mechanisms that underlie corresponding pathogenic processes.Although the range of light-responsive peptides has been developed for peptide–DNA-interactions,48–51 the photoswitchable RNA-targeting peptides have been, again, surprisingly underexplored. The first and, so far, the only fully light-controllable peptide−RNA interaction was accomplished by the introduction of the photoswitchable azobenzene moiety into a short peptide.52 Several RNA aptamers that selectively bind to the photoresponsive peptide 8 in its E-form (Scheme 4 and Table 1) were obtained by in vitro selection from 70 random nucleotides sequence pool of RNA. The E–Z photoisomerization of the azobenzene chromophore upon UV irradiation resulted in significant changes in the structure of peptide Z-8 leading to the dissociation of the peptide–RNA complex. The backwards Z–E isomerization and re-activation of the peptide E-8 towards RNA binding was performed upon irradiation with visible light.
Overall, the studies with synthetic RNA homoduplexes and aptamers provided a clear proof-of-principle for the “ON/OFF binding” concept to control ligand–RNA interactions by light. Nevertheless, these studies were mainly performed on non-native RNA sequences or undefined natural RNAs that may not be appropriate models for the therapeutically relevant human or viral RNAs. Application of the UV light for the photoswitching in model systems is, in principle, justified because RNA is more UV-resistant in comparison with DNA.53 However, in real systems the necessity to use UV light for the activation of the binders may significantly hamper their potential application due to the damage of biomolecules and low UV penetration depth of the tissues.54
3.4. Photoswitchable binders for messenger RNAs (mRNAs)
A messenger RNA (mRNA) is a single-stranded ribonucleic acid that is complementary to one of the DNA strands of a gene.2 The genetic information on the protein structure is copied from DNA into an mRNA molecule during the transcription process. Once it is complete, the mRNA moves to the cytoplasm where ribosomes decode it and synthesize the corresponding protein during the translation process. The importance of mRNA studies has recently drawn public attention in connection with the application of mRNA-based vaccines for the immunization against COVID-19 that started in December 2020.55,56Despite the large amount of publications on mRNA, there is only one example of the interaction of an organic photoswitch with mRNA, so far.57 Thus, a positively charged azobenzene derivative E-9 (Scheme 5 and Table 1) that acts as an unspecific electrostatic binder induced aggregation of 759-base mRNA coding the Enhanced Green Fluorescent Protein (EGFP)58 in the in vitro experiments.57a The system represents the “RNA un/re-folding” principle (Scheme 1d). Since the protein translation directly depends on the condensation state of the nucleic acid, mRNA aggregation yielded the inhibition of the EGFP synthesis. The E–Z photoisomerization of E-9 upon irradiation with 365 nm light triggered the dissociation of the mRNA aggregates and re-activated the translation process. The inhibition and re-activation of the translation was followed by the detection of the fluorescence intensity of EGPF. In a follow-up study,57b the E-9-induced condensation of mRNAs was accomplished in the presence of gene-silencing small RNAs. Two mRNAs called 164-mRNA-EGFP and 156-mRNA-DsRedM, which code EGFP and DsRedM59 fluorescent proteins and additionally contain antisense sequences of two small RNAs (miR164 and miR156), respectively, were used in this experiment. In this case, the E–Z isomerization of E-9 led to the selective photoactivation of genes that were not silenced by small RNAs.
3.5. Photoswitchable binders for natural therapeutically relevant RNAs
The first example of a noncovalent photoactive ligand for natural therapeutically relevant RNA was reported in 2019.28 In this study, a photoswitchable hemi-indigo binder Z-10 for human immunodeficiency virus RNA was developed. It was shown that compound Z-10 (Scheme 6 and Table 1) associated with the transactivation response element (TAR) and the stem IIB region of the Rev response element (RRE-IIB) of HIV-1 RNA. In contrast to the previously reported systems, both forward and backward photoisomerization of 10 could be activated by visible light and did not require the use of UV. However, both the initial Z-10 and photoinduced E-10 forms showed significant affinity towards RNA, thus hampering the photoregulation of binding. Nevertheless, photoswitching of the RNA-bound Z-10 resulted in significant changes in the fluorescent properties of the ligand which allowed to turn the fluorescent staining of RNA on (Z-10-RNA) and off (E-10-RNA) by repeated irradiation with blue and amber light (“ON/OFF fluorescence”). On/off switching of the fluorescence could be performed only upon binding of 10 to RNA because the ligand showed no emission in bulk solution. In this context, binding of both isomers of 10 to RNA is advantageous because the ligand can be used as a tunable fluorescent stain to study dynamic processes of the RNA.4. Summary and outlook
To sum up, the approaches towards development of photocontrollable interactions with RNA are far from being fully explored and realized. This is not surprising because the design of photoswitchable RNA binders is particularly challenging due to the limited possibility of the ligand modification. Thus, structural modifications for adjustment of the RNA binding properties can be mainly performed on the binder periphery keeping the photoswitchable core intact such that the photochemical properties are not disrupted. Additionally, for the realization of the “ON/OFF binding”, only one of the two isomers of the photoswitch should possess a strong affinity towards RNA to ensure the photocontrol of the binding. For this reason, in the majority of cases, it was the RNA structure that was selected from the pool to bind selectively to a photoswitch with known structure. The only attempt to target pre-defined therapeutically relevant sequences (TAR and RRE-IIB HIV RNA) with a photoswitchable ligand yielded the RNA binder 10 of “ON/OFF fluorescence” type. However, the selectivity towards a certain RNA structure as well as the photocontol of binding to naturally occurring RNAs still was not achieved.A significant potential of photoswitchable ligands for controllable binding and manipulation of the spatial structure of therapeutically relevant RNAs, including RNA repeat expansions is foreseen. Light-controllable manipulation of the RNA folding will, in turn, open the opportunity of reversible regulation of the RNA–protein interactions for investigation and treatment purposes. Additionally, selective RNA markers with photoswitchable fluorescence are perspective for the real-time monitoring of RNA dynamics by novel bioimaging approaches.29 Interestingly, all the examples highlighted in this article were developed experimentally without the use of any computational methods and modelling. Along these lines, future application of computational approaches that facilitate the design of photoswitchable RNA binders of different types (Scheme 1) is envisaged.60 As have been shown by collected examples, different classes of photoswitches are compatible with RNA binding (Table 1). However, for real applications, special attention should be paid to the shifting of the excitation energy to longer wavelength that, in an ideal case, should fit to the biooptical transparency window (650–900 nm).54 In this context, all-visible indigoid photoswitches, such as hemi-indigo,61 hemithioindigo61 and N,N′-substituted indigo,62 represent promising candidates for the further development of the photoswitchable binders for disease-related RNA species. Overall, I hope that this highlight will provide a basis for the development of various photoswitchable RNA binders for future therapeutical applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Generous financial support from Deutsche Forschungsgemeinschaft (project BE 7202/1-1) is gratefully acknowledged. I thank Prof. Dr Heiko Ihmels (Universität Siegen, Germany), Dr Anna Berdnikova (Moscow State University of Medicine and Dentistry, Russia) and Dr Ekaterina Chernikova (INEOS RAS, Russia) for fruitful discussions and valuable advices.Notes and references
- RNA Worlds: From Life's Origins to Diversity in Gene Regulation, ed. P. V. Peplow, B. Martinez, G. A. Calin and A. Esquela-Kerscher, Cold Spring Harbor Laboratory Press, New York, 5th edn, 2019 Search PubMed.
- Nucleic Acids in Chemistry and Biology, ed. G. M. Blackburn, RSC Publishing, Cambridge, 3rd edn, 2006 Search PubMed.
- P. A. Sharp, Cell, 2009, 136, 577 CrossRef CAS PubMed.
- H. Paulson, Handb. Clin. Neurol., 2018, 147, 105 Search PubMed.
- V. Bernat and M. D. Disney, Neuron, 2015, 87, 28 CrossRef CAS PubMed.
- MicroRNAs in Diseases and Disorders: Emerging Therapeutic Targets, ed. J. F. Atkins, R. F. Gesteland and T. R. Cech, RSC Publishing, Cambridge, 2019 Search PubMed.
- S. M. Hammond, Curr. Opin. Genet. Dev., 2006, 16, 4 CrossRef CAS PubMed.
- T. A. Farazi, J. I. Spitzer, P. Morozov and T. Tuschl, J. Pathol., 2011, 223, 102 CrossRef CAS PubMed.
- E. J. Lefkowitz, D. M. Dempsey, R. C. Hendrickson, R. J. Orton, S. G. Siddell and D. B. Smith, Nucleic Acids Res., 2018, 46, D708 CrossRef CAS PubMed.
- E. D. Holmes, The Evolution and Emergence of RNA Viruses, Oxford University Press, New York, 2009 Search PubMed.
- S. J. Russell, Cancer Gene Ther., 2002, 9, 961 CrossRef CAS PubMed.
- D. N. Wilson, Nat. Rev. Microbiol., 2014, 12, 35 CrossRef CAS PubMed.
- K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discovery, 2018, 17, 547 CrossRef CAS PubMed.
- J. P. Falese, A. Donlic and A. E. Hargrove, Chem. Soc. Rev., 2021, 50, 2224 RSC.
- S. M. Meyer, C. C. Williams, Y. Akahori, T. Tanaka, H. Aikawa, Y. Tong, J. L. Childs-Disney and M. D. Disney, Chem. Soc. Rev., 2020, 49, 7167 RSC.
- A. Ursu, J. L. Childs-Disney, R. J. Andrews, C. A. O’Leary, S. M. Meyer, A. J. Angelbello, W. N. Moss and M. D. Disney, Chem. Soc. Rev., 2020, 49, 7252 RSC.
- M. D. Disney, J. Am. Chem. Soc., 2019, 141, 6776 CrossRef CAS PubMed.
- RNA Therapeutics, ed. A. L. Garner, Springer, Cham, 2018 Search PubMed.
- J. R. Thomas and P. J. Hergenrother, Chem. Rev., 2008, 108, 1171 CrossRef CAS PubMed.
- J. Nowakowski and I. Tinoco Jr, Semin. Virol., 1997, 8, 153 CrossRef CAS.
- A. R. Ferré-D’Amaré and J. A. Doudna, Annu. Rev. Biophys. Biomol. Struct., 1999, 28, 57 CrossRef PubMed.
- (a) A. S. Lubbe, W. Szymanski and B. L. Feringa, Chem. Soc. Rev., 2017, 46, 1052 RSC; (b) W. Szymanski, J. M. Beierle, H. A. V. Kistemaker, W. A. Velema and B. L. Feringa, Chem. Rev., 2013, 113, 6114 CrossRef CAS PubMed.
- Z. L. Pianowski, Chem. – Eur. J., 2019, 25, 5128 CrossRef CAS PubMed.
- M. You and S. R. Jaffrey, Ann. N. Y. Acad. Sci., 2015, 1352, 13 CrossRef CAS PubMed.
- (a) K. Murayama, Y. Yamano and H. Asanuma, J. Am. Chem. Soc., 2019, 141, 9485 CrossRef CAS PubMed; (b) Y. Kamiya, Y. Arimura, H. Ooi, K. Kato, X.-G. Liang and H. Asanuma, ChemBioChem, 2018, 19, 1305 CrossRef CAS PubMed; (c) T. Doi, H. Kashida and H. Asanuma, Org. Biomol. Chem., 2015, 13, 4430 RSC; (d) Y. Kamiya and H. Asanuma, Acc. Chem. Res., 2014, 47, 1663 CrossRef CAS PubMed.
- (a) C. Dohno, S. Uno and K. Nakatani, J. Am. Chem. Soc., 2007, 129, 11898 CrossRef CAS PubMed; (b) J. Andersson, S. Li, P. Lincoln and J. Andréasson, J. Am. Chem. Soc., 2008, 130, 11836 CrossRef CAS PubMed; (c) C. Dohno, T. Yamamoto and K. Nakatani, Eur. J. Org. Chem., 2009, 4051 CrossRef CAS; (d) X. Wang, J. Huang, Y. Zhou, S. Yan, X. Weng, X. Wu, M. Deng and X. Zhou, Angew. Chem., Int. Ed., 2010, 49, 5305 CrossRef CAS PubMed; (e) X. Xing, X. Wang, L. Xu, Y. Tai, L. Dai, X. Zheng, W. Mao, X. Xu and X. Zhou, Org. Biomol. Chem., 2011, 9, 6639 RSC; (f) A. Mammana, G. T. Carroll, J. Areephong and B. L. Feringa, J. Phys. Chem. B, 2011, 115, 11581 CrossRef CAS PubMed; (g) S. V. Paramonov, V. Lokshin, H. Ihmels and O. A. Fedorova, Photochem. Photobiol. Sci., 2011, 10, 1279 CrossRef CAS PubMed; (h) H. Ihmels, J. Mattay, F. May and L. Thomas, Org. Biomol. Chem., 2013, 11, 5184 RSC; (i) M. Hammarson, J. R. Nilsson, S. Li, P. Lincoln and J. Andréasson, Chem. – Eur. J., 2014, 20, 15855 CrossRef CAS PubMed; (j) M. P. O'Hagan, S. Haldar, M. Duchi, T. A. A. Oliver, A. J. Mulholland, J. C. Morales and M. C. Galan, Angew. Chem., Int. Ed., 2019, 58, 4334 CrossRef PubMed; (k) B. Heinrich, K. Bouazoune, M. Wojcik, U. Bakowsky and O. Vázquez, Org. Biomol. Chem., 2019, 17, 1827 RSC; (l) S. Müller, J. Paulus, J. Mattay, H. Ihmels, V. I. Dodero and N. Sewald, Beilstein J. Org. Chem., 2020, 16, 60 CrossRef PubMed; (m) S. Kölsch, H. Ihmels, J. Mattay, N. Sewald and B. O. Patrick, Beilstein J. Org. Chem., 2020, 16, 111 CrossRef PubMed; (n) M. P. O’Hagan, J. Ramos-Soriano, S. Haldar, S. Sheikh, J. C. Morales, A. J. Mulholland and M. C. Galan, Chem. Commun., 2020, 56, 5186 RSC.
- (a) K. Starčević, G. Karminski-Zamola, I. Piantanida, M. Žinić, L. Šuman and M. Kralj, J. Am. Chem. Soc., 2005, 127, 1074 CrossRef PubMed; (b) M. L. Di Pietro, F. Puntoriero, F. Tuyéras, P. Ochsenbein, P. P. Lainé and S. Campagna, Chem. Commun., 2010, 46, 5169 RSC; (c) M. I. Sánchez, O. Vázquez, M. E. Vázquez and J. L. Mascareñas, Chem. Commun., 2011, 47, 11107 RSC; (d) D. Berdnikova, O. Fedorova, E. Gulakova and H. Ihmels, Chem. Commun., 2012, 48, 4603 RSC; (e) D. V. Berdnikova, T. M. Aliyeu, T. Paululat, Y. V. Fedorov, O. A. Fedorova and H. Ihmels, Chem. Commun., 2015, 51, 4906 RSC; (f) D. V. Berdnikova, J. Heider, H. Ihmels, N. Sewald and P. M. Pithan, ChemPhotoChem, 2020, 4, 520 CrossRef CAS.
- (a) D. V. Berdnikova, Chem. Commun., 2019, 55, 8402 RSC; (b) D. V. Berdnikova, Beilstein J. Org. Chem., 2019, 15, 2822 CrossRef CAS PubMed.
- (a) Y. Zhang, S. Tang, E. R. Thapaliya, L. Sansalone and F. M. Raymo, Chem. Commun., 2018, 54, 8799 RSC; (b) Y. Zhang, S. Tang, L. Sansalone, J. D. Baker and F. M. Raymo, Chem. – Eur. J., 2016, 22, 15027 CrossRef CAS PubMed; (c) Y. Xiong, P. Rivera-Fuentes, E. Sezgin, A. V. Jentzsch, C. Eggeling and H. L. Anderson, Org. Lett., 2016, 18, 3666 CrossRef CAS PubMed; (d) M. N. Tran and D. M. Chenoweth, Angew. Chem., Int. Ed., 2015, 54, 6442 CrossRef CAS PubMed.
- M. M. Fay, S. M. Lyons and P. Ivanov, J. Mol. Biol., 2017, 429, 2127 CrossRef CAS PubMed.
- For example, see: (a) J. Lacour, A. Spira, J.-Y. Petit, D. Sarrazin, F. Lacour, M. Michelson, G. Delage, G. Contesso and J. Viguier, Lancet, 1980, 316, 161 CrossRef; (b) G. Gatti, N. G. Nuñez, D. A. Nocera, L. Dejager, C. Libert, C. Giraudo and M. Maccioni, Eur. J. Immunol., 2013, 43, 1849 CrossRef CAS PubMed.
- I. Orehovec, M. Matković, I. Pehar, D. Majhen and I. Piantanida, Int. J. Mol. Sci., 2021, 22, 4916 CrossRef CAS PubMed.
- A. A. Ali, M. Kang, R. Kharbash and Y. Kim, BMC Biomed. Eng., 2019, 1, 6 CrossRef PubMed.
- M. R. Dunn, R. M. Jimenez and J. C. Chaput, Nat. Rev. Chem., 2017, 1, 0076 CrossRef CAS.
- K. N. Kang and Y. S. Lee, RNA Aptamers: A Review of Recent Trends and Applications, in Future Trends in Biotechnology. Advances in Biochemical Engineering/Biotechnology, ed. J. J. Zhong, Springer, Berlin, Heidelberg, 2012, vol. 131 Search PubMed.
- P. Röthlisberger and M. Hollenstein, Adv. Drug Delivery Rev., 2018, 134, 3 CrossRef PubMed.
- T. Adachi and Y. Nakamura, Molecules, 2019, 24, 4229 CrossRef CAS PubMed.
- D. Bunka and P. Stockley, Nat. Rev. Microbiol., 2006, 4, 588 CrossRef CAS PubMed.
- Y. S. Kim, N. H. A. Raston and M. B. Gu, Biosens. Bioelectron., 2016, 76, 2 CrossRef PubMed.
- J. Zhou, M. L. Bobbin, J. C. Burnett and J. J. Rossi, Front. Genet., 2012, 3, 234 CAS.
- A. B. Iliuk, L. Hu and W. A. Tao, Anal. Chem., 2011, 83, 4440 CrossRef CAS PubMed.
- (a) D. D. Young and A. Deiters, ChemBioChem, 2008, 9, 1225 CrossRef CAS PubMed; (b) X. Zhang, J. Zhang, Y.-L. Ying, H. Tian and Y.-T. Long, Chem. Sci., 2014, 5, 2642 RSC.
- H. W. Lee, S. G. Robinson, S. Bandyopadhyay, R. H. Mitchel and D. Sen, J. Mol. Biol., 2007, 371, 1163 CrossRef CAS PubMed.
- T. S. Lotz, T. Halbritter, C. Kaiser, M. M. Rudolph, L. Kraus, F. Groher, S. Steinwand, J. Wachtveitl, A. Heckel and B. Suess, Nucleic Acids Res., 2019, 47, 2029 CrossRef CAS PubMed.
- K. A. Rotstan, M. M. Abdelsayed, L. F. M. Passalacqua, F. Chizzolini, K. Sudarshan, A. R. Chamberlin, J. Míšek and A. Luptak, eLife, 2020, 9, e51737 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, Biomol. Eng., 2007, 24, 381 CrossRef CAS PubMed.
- J. Karn, J. Mol. Biol., 1999, 293, 235 CrossRef CAS PubMed.
- L. Albert and O. Vázquez, Chem. Commun., 2019, 55, 10192 RSC.
- J. Rodriguez, J. Mosquera, S. Learte-Aymamí, M. E. Vázquez and J. L. Mascareñas, Acc. Chem. Res., 2020, 53, 2286 CrossRef CAS PubMed.
- A. A. Beharry and G. A. Woolley, Chem. Soc. Rev., 2011, 40, 4422 RSC.
- V. Peddie and A. D. Abell, J. Photochem. Photobiol., C, 2019, 40, 1 CrossRef CAS.
- (a) G. Hayashi, M. Hagihara, C. Dohno and K. Nakatani, J. Am. Chem. Soc., 2007, 129, 8678 CrossRef CAS PubMed; (b) G. Hayashi, M. Hagihara and K. Nakatani, Chem. – Eur. J., 2009, 15, 424 CrossRef CAS PubMed; (c) G. Hayashi and K. Nakatani. Development of Photoswitchable RNA Aptamer–Ligand Complexes, in Artificial Riboswitches. Methods in Molecular Biology (Methods and Protocols), ed. A. Ogawa, Humana Press, Totowa, NJ, 2014, vol. 1111 Search PubMed.
- L. M. Kundu, U. Linne, M. Marahiel and T. Carell, Chem. – Eur. J., 2004, 10, 5697 CrossRef CAS PubMed.
- R. Weissleder, Nat. Biotechnol., 2001, 19, 316 CrossRef CAS PubMed.
- K. S. Park, X. Sun, M. E. Aikins and J. J. Moon, Adv. Drug Delivery Rev., 2021, 169, 137 CrossRef CAS PubMed.
- D. Pushparajah, S. Jimenez, S. Wong, H. Alattas, N. Nafissi and R. A. Slavcev, Adv. Drug Delivery Rev., 2021, 170, 113 CrossRef CAS PubMed.
- (a) A. Estévez-Torres, C. Crozatier, A. Diguet, T. Hara, H. Saito, K. Yoshikawa and D. Baigl, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12219 CrossRef PubMed; (b) S. Rudiuk, H. Saito, T. Hara, T. Inoue, K. Yoshikawa and D. Baigl, Biomacromolecules, 2011, 12, 3945 CrossRef CAS PubMed.
- B. P. Cormack, R. H. Valdivia and S. Falkow, Gene, 1996, 173, 33 CrossRef CAS PubMed.
- D. E. Strongin, B. Bevis, N. Khuong, M. E. Downing, R. L. Strack, K. Sundaram, B. S. Glick and R. J. Keenan, Protein Eng., Des. Sel., 2007, 20, 525 CrossRef CAS PubMed.
- D. V. Berdnikova, P. Carloni, S. Krauß and G. Rossetti, Molecules, 2021, 26, 3384 CrossRef CAS PubMed.
- C. Petermayer and H. Dube, Acc. Chem. Res., 2018, 51, 1153 CrossRef CAS PubMed.
- C.-Y. Huang, A. Bonasera, L. Hristov, Y. Garmshausen, B. M. Schmidt, D. Jacquemin and S. Hecht, J. Am. Chem. Soc., 2017, 139, 15205 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |





