Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Azulene-based fluorescent chemosensor for adenosine diphosphate†
Carlos M.
López-Alled
 ab,
Sang Jun
Park
c,
Dong Joon
Lee
c,
Lloyd C.
Murfin
ab,
Sang Jun
Park
c,
Dong Joon
Lee
c,
Lloyd C.
Murfin
 a,
Gabriele
Kociok-Köhn
a,
Gabriele
Kociok-Köhn
 d,
Jodie L.
Hann
d,
Jodie L.
Hann
 a,
Jannis
Wenk
a,
Jannis
Wenk
 *be,
Tony D.
James
*be,
Tony D.
James
 *abf,
Hwan Myung
Kim
*abf,
Hwan Myung
Kim
 *c and
Simon E.
Lewis
*c and
Simon E.
Lewis
 *ab
*ab
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: T.D.James@bath.ac.uk; S.E.Lewis@bath.ac.uk
bCentre for Sustainable Circular Technologies, University of Bath, Bath, BA2 7AY, UK. E-mail: J.H.Wenk@bath.ac.uk
cDepartment of Energy Systems Research, Ajou University, Suwon 443-749, South Korea. E-mail: kimhm@ajou.ac.kr
dMaterial and Chemical Characterisation Facility (MC2), University of Bath, Bath, BA2 7AY, UK
eDepartment of Chemical Engineering, University of Bath, Bath, BA2 7AY, UK
fSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
First published on 27th September 2021
Abstract
AzuFluor® 435-DPA-Zn, an azulene fluorophore bearing two zinc(II)-dipicolylamine receptor motifs, exhibits fluorescence enhancement in the presence of adenosine diphosphate. Selectivity for ADP over ATP, AMP and PPi results from appropriate positioning of the receptor motifs, since an isomeric sensor cannot discriminate between ADP and ATP.
As well as being the central constituent of DNA and RNA, the naturally occurring nucleotides (in various phosphorylation states) are crucial in numerous other biological contexts, for example as chemical messengers, as coenzymes, and in energy storage and transfer. Accordingly, there is sustained interest in methods for the selective detection and quantitation of the various nucleotides, to enable the elucidation of their diverse biological roles. Use of fluorescent chemosensors is an attractive technique for this purpose,1 since they typically have the advantages of high selectivity, sensitivity, rapid response, and reversibility, allowing nucleotide flux to be monitored over time. Adenosine triphosphate (ATP) is a nucleotide of particular importance for study owing to its role as Nature's universal energy carrier.2 The release of chemical energy through ATP hydrolysis entails formation of adenosine diphosphate (ADP), so co-occurrence of these two analytes is a common scenario, and hence the selectivity for one analyte over the other is an important parameter for any fluorescent probe.
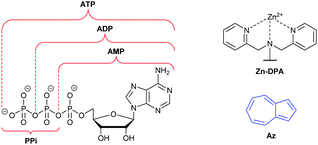
Amongst various receptor motifs for nucleotides that have been reported, Zn(II)–dipicolylamine (Zn-DPA) complexes have been employed in numerous sensors as they typically exhibit high selectivity for phosphates over other anions (Fig. 1).3 However, the selectivity profile for such sensors can vary significantly, depending on the nature of the fluorophore and scaffold to which the Zn-DPA recognition unit(s) are appended, as well as their position and orientation. Thus, for some Zn-DPA-based sensors, pyrophosphate anion (PPi) induces a greater fluorescence response than any nucleotides.4,5 Conversely, other Zn-DPA-based sensors exhibit their largest fluorescence response in the presence of a particular nucleotide, often ATP;6 for such sensors, discrimination between nucleotides can sometimes be effected on the basis of spectroscopic features such as change in ratiometric fluorescence intensities.7 However, it should be noted that even for the same sensor, a change in solvent composition can reverse the selectivity between nucleotides.8 Zn-DPA-based sensors with selectivity for Adenosine monophosphate (AMP)9 and for cyclic phosphates (cAMP, cGMP)10 have also been reported. Zn-DPA-based sensors for the various analytes discussed above whose readout is colorimetric (as opposed to fluorescent) have been described.11 To our knowledge, the work of Feng and co-workers constitutes the only Zn-DPA-based sensor to date having selectivity for ADP over ATP in terms of fluorescence enhancement.12 The desirability of methods for the effective discrimination of ATP and ADP specifically has also led to the development of numerous approaches based on sensor designs other than Zn-DPA receptors.13,14
Azulene (Az) is a non-benzenoid isomer of the more well-known naphthalene, from which its properties differ significantly.15 The smaller S0–S1 energy gap of azulene leads to absorption in the visible region and hence an intense blue colour. Furthermore, it has a large dipole moment of 1.08 D,16 and azulene and some derivatives are known to exhibit anomalous fluorescence, emitting primarily or entirely from higher excited states, in violation of Kasha's rule.17 Substituents on the azulene ring can profoundly alter the absorption and emission spectra,18 making an azulene ring a versatile reporter motif for chemosensors and chemodosimeters. Colorimetric azulene probes have been developed for a wide variety of analytes,19 whereas fluorescent azulene probes are fewer in number.20,21 In this communication we report the design, synthesis, and evaluation of a fluorescent sensor for nucleotides comprising Zn-DPA receptor motifs appended to an azulene reporter motif, which displays selectivity for ADP over ATP.
Our probe design involved connecting two Zn-DPA receptor motifs to the central azulene core (at the 1- and 3-positions) via a π-conjugated linker. Two isomeric linkers were employed (p-phenylene and m-phenylene), giving the two probes 1·2Zn and 2·2Zn (Fig. 2). Details of their synthesis are given in the ESI.† A key aspect of the probe design is that the amine groups of the DPA recognition units are directly conjugated to the fluorophore through the π-linker, in contrast to most previously reported Zn-DPA containing nucleotide sensors such as Feng's anthracene-containing ADP-selective system.12 For both probes, X-ray crystal structures of the unbound ligand forms were obtained and are shown in Fig. 3.
 | ||
| Fig. 3 ORTEP representation of the X-ray structures of 1 (left) and 2 (right). Ellipsoids are shown at 50% probability. Hydrogens are shown as spheres of arbitrary radius. CCDC† #2095982 and 2095983. | ||
Initially 1 and 2 were treated with Zn(NO3)2 and a selection of other metal salts, and absorption spectra were acquired (Fig. S1 and S2, ESI†). As expected, changes in the absorbance spectra were observed (more pronounced for 2), confirming that complexation of the amine lone pairs results in electronic perturbation of the azulene chromophore. The zinc complexes 1·2Zn and 2·2Zn were then exposed to a selection of anions and absorption spectra again acquired (Fig. S3 and S4, ESI†). The fluorescence response of the probes to the same anions was then studied (Fig. 4 and Fig. S5, ESI†). For both probes, a significant fluorescence enhancement is observed in response to ADP, whereas PPi induced a negligible response. Overall, 1·2Zn shows the better selectivity profile with respect to the potentially interfering anions that were evaluated. Since both probes possessed selectivity for ADP their responses to other phosphorylation states of this nucleotide were also evaluated (Fig. 5 and Fig. S6, ESI†).
Here the behaviour of the two probes differed markedly. 2·2Zn was unable to distinguish between ADP and ATP, with both analytes causing fluorescence enhancements of similar magnitude. In contrast, however, 1·2Zn displayed pronounced selectivity for ADP over ATP (and AMP), with the latter analytes inducing a near-negligible change in fluorescence over basal levels. We next considered the binding stoichiometry of the two probes with ADP (Fig. 6).
Job plot analysis indicated different stoichiometries of binding for the two probes: whereas 2·2Zn binds ADP in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, 1·2Zn seemingly binds ADP in a 1
1 ratio, 1·2Zn seemingly binds ADP in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio; fluorescence dose–response titrations (Fig. S7 and S8, ESI†) further support these stoichiometries. We noted that at concentrations above 1 mM 1·2Zn⊂2ADP began to precipitate from solution after standing for ≈5 min (Fig. S9, ESI†). Formation of a less-soluble complex upon analyte binding (and precipitation at high concentrations) has been reported for other Zn-DPA nucleotide sensors.5c,12a The response of 1·2Zn to other nucleobases was also studied (Fig. S10, ESI†). To rationalise the stoichiometry of binding and selectivity for ADP of 1·2Zn, we propose the binding mode shown in Fig. 7, namely a nucleobase–fluorophore–nucleobase π-stacked arrangement. Such an assembly rationalises the relative lack of response to PPi (lack of π-stacking) as well as to ATP (binding of triphosphate to Zn-DPA would lead to incorrect positioning of the purine ring for π-stacking). Analogous binding modes have been invoked for other Zn-DPA nucleotide sensors.12a
2 ratio; fluorescence dose–response titrations (Fig. S7 and S8, ESI†) further support these stoichiometries. We noted that at concentrations above 1 mM 1·2Zn⊂2ADP began to precipitate from solution after standing for ≈5 min (Fig. S9, ESI†). Formation of a less-soluble complex upon analyte binding (and precipitation at high concentrations) has been reported for other Zn-DPA nucleotide sensors.5c,12a The response of 1·2Zn to other nucleobases was also studied (Fig. S10, ESI†). To rationalise the stoichiometry of binding and selectivity for ADP of 1·2Zn, we propose the binding mode shown in Fig. 7, namely a nucleobase–fluorophore–nucleobase π-stacked arrangement. Such an assembly rationalises the relative lack of response to PPi (lack of π-stacking) as well as to ATP (binding of triphosphate to Zn-DPA would lead to incorrect positioning of the purine ring for π-stacking). Analogous binding modes have been invoked for other Zn-DPA nucleotide sensors.12a
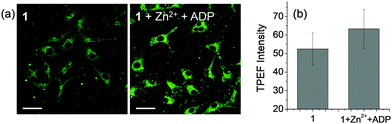
Two-Photon Microscopy (TPM) cell imaging studies were conducted to confirm the applicability of 1 in biological systems. First, to determine the appropriate two-photon excitation wavelength of the probe, two-photon excited fluorescence (TPEF) intensities were observed using HeLa cells stained with 1 (10 μM) (Fig. S11, ESI†). The TPEF intensity of 1 increased up to 740 nm excitation wavelength and gradually weakened with subsequent increases in wavelength, so we set the appropriate two-photon excitation wavelength of this probe at 740 nm. Next, we observed changes of TPEF intensity on treatment with Zn2+ (50 μM) and ADP (200 μM) in HeLa cells to confirm that 1 can detect ADP in living cells. As a result of treating HeLa cells with Zn2+ and ADP, the TPEF intensity of cells stained with 1 was observed to increase (Fig. 8). Cell viability assays (Fig. S12, ESI†) showed no significant cytotoxicity upon incubation of the cells with 1, Zn2+ and ADP at the concentrations specified above. These results indicate that 1·2Zn can directly detect the presence of ADP in biological systems.
In summary, we disclose the first azulene-based fluorescent chemosensor for nucleotides (1·2Zn), which we have named “AzuFluor® 435-DPA-Zn”, and which is selective for ADP over other phosphorylation states of adenosine. The receptor motif(s) employed (zinc(II)-dipicolylamine chelates) can exhibit differing selectivity profiles for various phosphate analytes, dependent on the structure of the probe in question. Thus, in the case of 1·2Zn we surmise that the selectivity for ADP is a consequence of the particular arrangement of the receptor motifs, which in turn arises from the use of the p-phenylene linker and the attachment at the azulene 1- and 3-positions. This conclusion is supported by the fact that changing the π-linker to m-phenylene (to give isomeric probe 2·2Zn) eradicates the system's ability to discriminate between ADP and ATP.
We thank EPSRC for IAA funding (EP/R51164X/1) and for DTG PhD studentships to L. C. M. and J. L. H. We are grateful for PhD funding to C. M. L.-A. from the EU Horizon 2020 research and innovation programme under grant agreement H2020-MSCA-CO-FUND, #665992. T. D. J. wishes to thank the Royal Society for a Wolfson Research Merit Award and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University for support (2020ZD01). The British-Spanish Society and Plastic Energy are thanked for a 2017 Scholarship to C.M.L.-A. The authors acknowledge the Material and Chemical Characterisation Facility (MC2) at University of Bath (doi:10.15125/mx6j-3r54). H. M. K. acknowledges the grants from the National Research Foundation of Korea (2019R1A2B5B03100278 and 2019R1A5A2026045).
Conflicts of interest
The trademark “AzuFluor®” is registered to the University of Bath, which may benefit financially from sales of AzuFluor® 435-DPA-Zn.References
- For reviews, see: (a) A. Nag and S. Das, Isr. J. Chem., 2021, 61, 169–184 CrossRef CAS; (b) P. Sun, H. Zhang, Y. Sun and J. Liu, Spectrochim. Acta, Part A, 2021, 245, 118919 CrossRef CAS PubMed; (c) W. Li, X. Gong, X. Fan, S. Yin, D. Su, X. Zhang and L. Yuan, Chin. Chem. Lett., 2019, 30, 1775–1790 CrossRef CAS; (d) A. M. Agafontsev, A. Ravi, T. A. Shumilova, A. S. Oshchepkov and E. A. Kataev, Chem. – Eur. J., 2019, 25, 2684–2694 CrossRef CAS PubMed; (e) Y. Zhou, Z. Xu and J. Yoon, Chem. Soc. Rev., 2011, 40, 2222–2235 RSC.
- For reviews, see: (a) Y. W. Jun, S. Sarkar, K. H. Kim and K. H. Ahn, ChemPhotoChem, 2019, 3, 214–219 CrossRef CAS; (b) Y. Wu, J. Wen, H. Li, S. Sun and Y. Xu, Chin. Chem. Lett., 2017, 28, 1916–1924 CrossRef CAS; (c) J. Dong and M. Zhao, TrAC, Trends Anal. Chem., 2016, 80, 190–203 CrossRef CAS.
- For a review, see: H. T. Ngo, X. Liu and K. A. Jolliffe, Chem. Soc. Rev., 2012, 41, 4928–4965 RSC.
- For reviews, see: (a) S. Lee, K. K. Y. Yuen, K. A. Jolliffe and J. Yoon, Chem. Soc. Rev., 2015, 44, 1749–1762 RSC; (b) S. Anbu, A. Paul, G. J. Stasiuk and A. J. L. Pombeiro, Coord. Chem. Rev., 2021, 431, 213744 CrossRef CAS.
- For selected examples, see: (a) S. Yang, W. Feng and G. Feng, Anal. Chim. Acta, 2018, 1034, 119–127 CrossRef CAS PubMed; (b) X. Liu, D. G. Smith and K. A. Jolliffe, Chem. Commun., 2016, 52, 8463–8466 RSC; (c) S.-Y. Jiao, K. Li, X. Wang, Z. Huang, L. Pu and X.-Q. Yu, Analyst, 2015, 140, 174–181 RSC; (d) S. Anbu, S. Kamalraj, A. Paul, C. Jayabaskaran and A. J. L. Pombeiro, Dalton Trans., 2015, 44, 3930–3933 RSC; (e) V. E. Zwicker, B. M. Long and K. A. Jolliffe, Org. Biomol. Chem., 2015, 13, 7822–7829 RSC; (f) L. Tang, Z. Zheng, Z. Huang, K. Zhong, Y. Bian and R. Nandhakumar, RSC Adv., 2015, 5, 10505–10511 RSC; (g) J. Wang, X. Liu and Y. Pang, J. Mater. Chem. B, 2014, 2, 6634–6638 RSC; (h) F. Huang and G. Feng, RSC Adv., 2014, 4, 484–487 RSC; (i) S.-i. Kondo, Y. Nakadai and M. Unno, RSC Adv., 2014, 4, 27140–27145 RSC; (j) Q.-C. Xu, X.-F. Wang, G.-W. Xing and Y. Zhang, RSC Adv., 2013, 3, 15834–15841 RSC; (k) X. Liu, H. T. Ngo, Z. Ge, S. J. Butler and K. A. Jolliffe, Chem. Sci., 2013, 4, 1680–1686 RSC; (l) L. G. Pathberiya, N. Barlow, T. Nguyen, B. Graham and K. L. Tuck, Tetrahedron, 2012, 68, 9435–9439 CrossRef CAS; (m) J. H. Lee, A. R. Jeong, J.-H. Jung, C.-M. Park and J.-I. Hong, J. Org. Chem., 2011, 76, 417–423 CrossRef CAS PubMed; (n) W.-H. Chen, Y. Xing and Y. Pang, Org. Lett., 2011, 13, 1362–1365 CrossRef CAS PubMed; (o) J. F. Zhang, S. Kim, J. H. Han, S.-J. Lee, T. Pradhan, Q. Y. Cao, S. J. Lee, C. Kang and J. S. Kim, Org. Lett., 2011, 13, 5294–5297 CrossRef CAS PubMed; (p) J. Veliscek-Carolan, S. J. Butler and K. A. Jolliffe, J. Org. Chem., 2009, 74, 2992–2996 CrossRef PubMed; (q) M. J. McDonough, A. J. Reynolds, W. Y. G. Lee and K. A. Jolliffe, Chem. Commun., 2006, 2971–2973 RSC; (r) H. N. Lee, Z. Xu, S. K. Kim, K. M. K. Swamy, Y. Kim, S.-J. Kim and J. Yoon, J. Am. Chem. Soc., 2007, 129, 3828–3829 CrossRef CAS PubMed; (s) D. H. Lee, S. Y. Kim and J.-I. Hong, Angew. Chem., Int. Ed., 2004, 43, 4777–4780 CrossRef CAS PubMed.
- (a) Y. Kurishita, T. Kohira, A. Ojida and I. Hamachi, J. Am. Chem. Soc., 2012, 134, 18779–18789 CrossRef CAS PubMed; (b) A. Ojida, I. Takashima, T. Kohira, H. Nonaka and I. Hamachi, J. Am. Chem. Soc., 2008, 130, 12095–12101 CrossRef CAS PubMed; (c) A. Ojida, Y. Miyahara, J. Wongkongkatep, S.-I. Tamaru, K. Sada and I. Hamachi, Chem. – Asian J., 2006, 1, 555–563 CrossRef CAS PubMed; (d) A. Ojida, H. Nonaka, Y. Miyahara, S.-I. Tamaru, K. Sada and I. Hamachi, Angew. Chem., Int. Ed., 2006, 45, 5518–5521 CrossRef CAS PubMed; (e) A. Ojida, S.-K. Park, Y. Mito-oka and I. Hamachi, Tetrahedron Lett., 2002, 43, 6193–6195 CrossRef CAS.
- (a) J. Kim, J. Oh and M. S. Han, RSC Adv., 2021, 11, 10375–10380 RSC; (b) Q.-C. Xu, H.-J. Lv, Z.-Q. Lv, M. Liu, Y.-J. Li, X.-F. Wang, Y. Zhang and G.-W. Xing, RSC Adv., 2014, 4, 47788–47792 RSC; (c) Z. Xu, D. R. Spring and J. Yoon, Chem. – Asian J., 2011, 6, 2114–2122 CrossRef CAS PubMed.
- S. Marbumrung, K. Wongravee, V. Ruangpornvisuti, G. Tumcharern, T. Tuntulani and B. Tomapatanaget, Sens. Actuators, B, 2012, 171–172, 969–975 CrossRef CAS.
- L. Reinke, M. Koch, C. Müller-Rennoc and S. Kubik, Org. Biomol. Chem., 2021, 19, 3893–3900 RSC.
- D. S. Philips, S. Ghosh, K. V. Sudheesh, C. H. Suresh and A. Ajayaghosh, Chem. – Eur. J., 2017, 23, 17973–17980 CrossRef CAS PubMed.
- (a) S. Minagawa, S. Fujiwara, T. Hashimoto and T. Hayashita, Int. J. Mol. Sci., 2021, 22, 4683 CrossRef CAS PubMed; (b) S. Yang, G. Feng and N. H. Williams, Org. Biomol. Chem., 2012, 10, 5606–5612 RSC; (c) F. Huang, C. Cheng and G. Feng, J. Org. Chem., 2012, 77, 11405–11408 CrossRef CAS PubMed; (d) K. M. Kim, D. J. Oh and K. H. Ahn, Chem. – Asian J., 2011, 6, 122–127 CrossRef CAS PubMed; (e) D. H. Lee, J. H. Im, S. U. Son, Y. K. Chung and J.-I. Hong, J. Am. Chem. Soc., 2003, 125, 7752–7753 CrossRef CAS PubMed.
- (a) F. Huang, G. Hao, F. Wu and G. Feng, Analyst, 2015, 140, 5873–5876 RSC; (b) P. Hu, S. Yang and G. Feng, Org. Biomol. Chem., 2014, 12, 3701–3706 RSC; (c) L. Shi, P. Hu, Y. Ren and G. Feng, Chem. Commun., 2013, 49, 11704–11706 RSC.
- For a review, see: S. J. Butler and K. A. Jolliffe, ChemPlusChem, 2021, 86, 59–70 CrossRef CAS PubMed.
- For selected examples, see: (a) S. Nakano, M. Shimizu, H. Dinh and T. Morii, Chem. Commun., 2019, 55, 1611–1614 RSC; (b) A. Kumar, P. Prasher and P. Singh, Org. Biomol. Chem., 2014, 12, 3071–3079 RSC; (c) K. Ghosh and I. Saha, Org. Biomol. Chem., 2012, 10, 9383–9392 RSC; (d) S. Kunzelmann and M. R. Webb, ACS Chem. Biol., 2010, 5, 415–425 CrossRef CAS PubMed; (e) D. Wang, X. Zhang, C. He and C. Duan, Org. Biomol. Chem., 2010, 8, 2923–2925 RSC; (f) S. Kunzelmann and M. R. Webb, J. Biol. Chem., 2009, 284, 33130–33138 CrossRef CAS PubMed; (g) K. M. Kleman-Leyer, T. A. Klink, A. L. Kopp, T. A. Westermeyer, M. D. Koeff, B. R. Larson, T. J. Worzella, C. A. Pinchard, S. A. T. van de Kar, G. J. R. Zaman, J. J. Hornberg and R. G. Lowery, Assay Drug Dev. Technol., 2009, 7, 56–67 CrossRef CAS PubMed; (h) L. Vial and P. Dumy, J. Am. Chem. Soc., 2007, 129, 4884–4885 CrossRef CAS PubMed; (i) S. Litvinchuk, N. Sordé and S. Matile, J. Am. Chem. Soc., 2005, 127, 9316–9317 CrossRef CAS PubMed; (j) J. Srinivasan, S. T. Cload, N. Hamaguchi, J. Kurz, S. Keene, M. Kurz, R. M. Boomer, J. Blanchard, D. Epstein, C. Wilson and J. L. Diener, Chem. Biol., 2004, 11, 499–508 CrossRef CAS PubMed; (k) M. Brune, J. E. T. Corrie and M. R. Webb, Biochemistry, 2001, 40, 5087–5094 CrossRef CAS PubMed.
- R. S. H. Liu, J. Chem. Educ., 2002, 79, 183 CrossRef CAS.
- A. G. Anderson Jr. and B. M. Steckler, J. Am. Chem. Soc., 1959, 81, 4941 CrossRef.
- (a) M. Beer and H. C. Longuet-Higgins, J. Chem. Phys., 1955, 23, 1390 CrossRef CAS; (b) B. D. Wagner, D. Tittelbach-Helmrich and R. P. Steer, J. Phys. Chem., 1992, 96, 7904 CrossRef CAS; (c) K. Veys and D. Escudero, J. Phys. Chem. A, 2020, 124, 7228 CrossRef CAS PubMed.
- R. S. H. Liu, R. S. Muthyala, X.-S. Wang and A. E. Asato, Org. Lett., 2000, 2, 269–271 CrossRef CAS PubMed.
- For selected examples, see: (a) Y. Jin, K. Akagawa, T. Mutai, I. Yoshikawa and K. Kudo, Tetrahedron, 2021, 88, 132146 CrossRef CAS; (b) H. Xin, J. Li, X. Yang and X. Gao, J. Org. Chem., 2020, 85, 70 CrossRef CAS PubMed; (c) L. C. Murfin, C. M. López-Alled, A. C. Segwick, J. Wenk, T. D. James and S. E. Lewis, Front. Chem. Sci. Eng., 2020, 14, 90 CrossRef CAS; (d) C. M. López-Alled, L. C. Murfin, G. Kociok-Köhn, T. D. James, J. Wenk and S. E. Lewis, Analyst, 2020, 145, 6262 RSC; (e) L. C. Murfin, K. Chiang, G. T. Williams, C. L. Lyall, A. T. A. Jenkins, J. Wenk, T. D. James and S. E. Lewis, Front. Chem., 2020, 8, 10 CrossRef CAS PubMed; (f) H. Xin, J. Li, X. Yang and X. Gao, J. Org. Chem., 2019, 85, 70 CrossRef PubMed; (g) D. Lichosyt, S. Wasiłek, P. Dydio and J. Jurczak, Chem. – Eur. J., 2018, 24, 11683 CrossRef CAS PubMed; (h) G.-O. Buica, I.-G. Lazar, L. Birzan, C. Lete, M. Prodana, M. Enachescu, V. Tecuceanu, A. B. Stoian and E.-M. Ungureanu, Electrochim. Acta, 2018, 263, 382 CrossRef CAS; (i) C. M. López-Alled, A. Sanchez-Fernandez, K. J. Edler, A. C. Sedgwick, S. D. Bull, C. L. McMullin, G. Kociok-Köhn, T. D. James, J. Wenk and S. E. Lewis, Chem. Commun., 2017, 53, 12580 RSC; (j) L. Birzan, M. Cristea, C. C. Draghici, V. Tecuceanu, M. Maganu, A. Hanganu, G.-L. Arnold, E.-M. Ungureanu and A. C. Razus, Tetrahedron, 2016, 72, 2316 CrossRef CAS; (k) S. Wakabayashi, M. Uchida, R. Tanaka, Y. Habata and M. Shimizu, Asian J. Org. Chem., 2013, 2, 786 CrossRef CAS.
- For a review, see: L. C. Murfin and S. E. Lewis, Molecules, 2021, 26, 353 CrossRef CAS PubMed.
- For selected examples, see: (a) L. C. Murfin, M. Weber, S. J. Park, W. T. Kim, C. M. Lopez-Alled, C. L. McMullin, F. Pradaux-Caggiano, C. L. Lyall, G. Kociok-Köhn, J. Wenk, S. D. Bull, J. Yoon, H. M. Kim, T. D. James and S. E. Lewis, J. Am. Chem. Soc., 2019, 141, 19389 CrossRef CAS PubMed; (b) Y. Zhou, G. Baryshnikov, X. Li, M. Zhu, H. Ågren and L. Zhu, Chem. Mater., 2018, 30, 8008 CrossRef CAS; (c) P. M. Gosavi, Y. S. Moroz and I. V. Korendovych, Chem. Commun., 2015, 51, 5347 RSC; (d) Y. S. Moroz, W. Binder, P. Nygren, G. A. Caputo and I. V. Korendovych, Chem. Commun., 2013, 49, 490 RSC; (e) H. Salman, Y. Abraham, S. Tal, S. Meltzman, M. Kapon, N. Tessler, S. Speiser and Y. Eichen, Eur. J. Org. Chem., 2005, 2207 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic details, fluorescence data, NMR spectra. All data generated in this project are available in the ESI. CCDC 2095982 and 2095983. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1cc04122c |
| This journal is © The Royal Society of Chemistry 2021 |