Open Access Article
Open Access ArticleSynthesis and delivery of a stable phosphorylated ubiquitin probe to study ubiquitin conjugation in mitophagy†
Guy
Mann‡
 a,
Gandhesiri
Satish‡
a,
Prasad
Sulkshane‡
b,
Shaswati
Mandal
a,
Michael H.
Glickman
*b and
Ashraf
Brik
a,
Gandhesiri
Satish‡
a,
Prasad
Sulkshane‡
b,
Shaswati
Mandal
a,
Michael H.
Glickman
*b and
Ashraf
Brik
 *a
*a
aSchulich Faculty of Chemistry, Technion-Israel Institute of Technology Haifa, 3200008, Israel. E-mail: abrik@technion.ac.il
bFaculty of Biology, Technion-Israel Institute of Technology Haifa, 3200008, Israel. E-mail: glickman@tx.technion.ac.il
First published on 26th August 2021
Abstract
Protein post-translational modifications are involved in essentially all aspects of cellular signaling. Their dynamic nature and the difficulties in installing them using enzymatic approaches limits their direct study in human cells. Reported herein is the first synthesis, delivery and cellular study of a stable phosphoubiquitin probe. Our results compare Parkin's substrate preference during mitophagy via direct visualization of a phosphorylated ubiquitin probe in the cellular environment.
Studying the post-translational modifications (PTMs) of proteins is challenging due to the plethora of enzymes catalyzing the addition and removal of each modification.1 Probing the role of a given PTM in the cellular environment requires a system in which the modification state of the modified protein is predetermined and stable throughout the experiment.2
Chemical protein synthesis (CPS) is a powerful strategy for biochemical, structural and functional studies of PTMs.3 Chemical approaches enable not only the incorporation of naturally occurring PTMs at a desired site but also unnatural mimics that are stable in the cellular milieu.4–7 Despite the strength of CPS, utilizing synthetic proteins has been mostly limited to in vitro studies.8–12 Despite the current challenges, utilizing synthetic proteins to study PTMs in live cells has appealing advantages over conventional molecular biology approaches. The flexibility of functional design and freedom from extensive genetic manipulation are some of these advantages.
Clearance of damaged mitochondria, termed mitophagy,13 is triggered by ubiquitin (Ub) phosphorylation at serine 65 (pUb) by PTEN-induced kinase 1 (PINK1).14 PINK1 mediates the phosphorylation of Ub chains (conjugated to mitochondrial proteins) and E3 ligase Parkin at depolarized mitochondria leading to Parkin activation and amplification of a mitochondrial poly(p)Ub “eat me” signal.15 Subsequently, poly(p)Ub chains mediate the recruitment of autophagy components leading to degradation of tagged mitochondria in lysosomes.13 Plasmid mediated ectopic expression of Ub mutants (e.g. UbS65A, UbS65D and UbS65E) is the most direct approach to study how phosphorylation is involved in Ub conjugation during mitophagy. Notably, the reported Ub mutants (UbS65D and UbS65E) utilized in these studies as an overexpressed phosphoserine mimic,16 may not fully represent pUb function in the cellular context and can lead to contradicting results.17
To overcome these limitations, we envisioned an approach for the synthesis of a stable phosphorylated Ub analogue bearing a fluorescent label and a delivery unit, as a probe for studying the pathways involved in Ub phosphorylation. Therefore, 3-phosphonomethylenalanine, which is a stable phosphoserine (SPS) analogue,18 is a rational choice to study Ub phosphorylation. Due to its high structural similarity to phosphoserine, we believe that 3-phosphonomethylenalanine is superior to the commonly used UbS65D and UbS65E mutants for cellular studies.
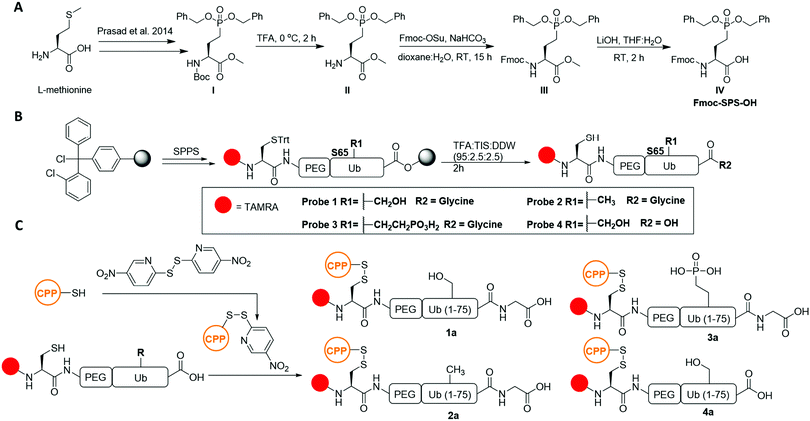
Current methods to prepare protected SPS for use in fluorenylmethyloxycarbonyl solid phase peptide synthesis (Fmoc-SPPS) produces only the mono-benzyl side chain protected form,19 which can be problematic in the synthesis of long polypeptides. Synthesis of SPS amino acids with Fmoc protected α-amine and di-benzyl side chain protection is therefore beneficial for synthesizing long polypeptides (e.g. Ub) via Fmoc-SPPS. We therefore designed a synthesis for Fmoc and di-benzyl protected SPS (Fmoc-SPS-OH) starting from tert-butyloxycarbonyl (BOC) building block I, which can be prepared from L-methionine in six steps.20 Deprotection of the BOC group in compound I afforded compound II. Protection of II with the Fmoc N-hydroxysuccinimide ester provided compound III. Methyl ester hydrolysis of III produced compound IV; the Fmoc-SPS-OH building block in 47% yield over three steps (Scheme 1A and Scheme S2, ESI†).
With IV in hand, we designed a synthetic strategy to prepare the Ub probe using Fmoc-SPPS. For cellular visualization, our design included (1) the N-terminal dye 5-carboxytetramethylrhodamin (TAMRA), (2) a Cys for transiently linking the cell penetrating peptide (CPP) cR10D,21 and (3) a flexible linker {2[2-(Fmoc-amino)ethoxy]ethoxy}acetic acid (PEG) as a spacer between Ub and the unnatural elements. Employing Fmoc-SPPS on a 2-chlorotrityl chloride (2-CTC) resin allowed the direct linear synthesis of Ub probes bearing the carboxylic acid functionality at the C-terminus (Scheme 1B). Probe 1 (wtUb) was prepared as we have reported previously in 19% yield (Fig. S2, ESI†).21 The nonphosphorylated S65A Ub mutant probe 2 (S65AUb) was similarly prepared in 19% yield (Fig. S3, ESI†). The stable phosphoserine 65 probe 3 (SPSUb) was prepared using Fmoc-SPS-OH, which was coupled at position 65 and isolated in 18% yield (Fig. S4, ESI†). As a negative control, we generated Ub probe 4 that lacked G76 (ΔG76) in 17% isolated yield (Fig. S2–S5, ESI†). Although probe 4 can exist in both the S65 phosphorylated or non-phosphorylated states, it cannot be conjugated by the ubiquitination machinery.
With 1–4 in hand, we proceeded to temporarily link them to cR10D through a reducible disulfide21 to generate the 1a–4a constructs, which were isolated in 50–65% yields (Scheme 1C and Fig. S6, ESI†). Notably, since probe 3 contains two additional negative charges, we failed in our initial attempts to deliver it using the previously reported cR10 as the CPP unit (Fig. S8, ESI†). In contrast, 3a was successfully delivered, in comparable efficiency as 2a, utilizing cR10D that was recently reported to be superior to the unmodified cR10 in Ub delivery (Fig. S8–S10, ESI†). Upon delivery, cytosolic GSH will generate the free probes 1–4 preventing cR10D from affecting their cellular localization.22 Using U2OS cells that stably express untagged human Parkin (+PRK) (Fig. S1, ESI†)23 and our previously reported conditions for delivery by cR10D, 1a–4a were successfully delivered after a one hour incubation followed by confocal laser scanning microscopy (CLSM) imaging and Z-stack reconstitution (Section 2.3, Fig. S9 and S10, ESI†).
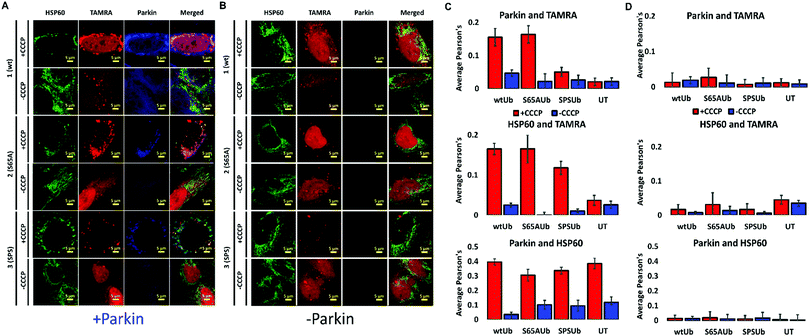
To measure the involvement of our probes in mitophagy, we delivered our probes to +PRK cells and treated the cells with the mitochondrial depolarization agent carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Fig. S12, ESI†).24 Mitophagy induction by a 4 h CCCP treatment, fixation and immunostaining of mitochondrial markers enabled the comparison of mitochondrial ubiquitination by our probes. We compared probes co-localized with Parkin, HSP60 and TOM20 and a mitochondrial outer membrane (MOM) protein reported to be polyubiquitinated and phospho-ubiquitinated during mitophagy.25 TOM20 is known to undergo proteasome-dependent degradation and to have a reduced stability during mitophagy when compared to other mitochondrial markers such as the mitochondrial matrix protein HSP60.26
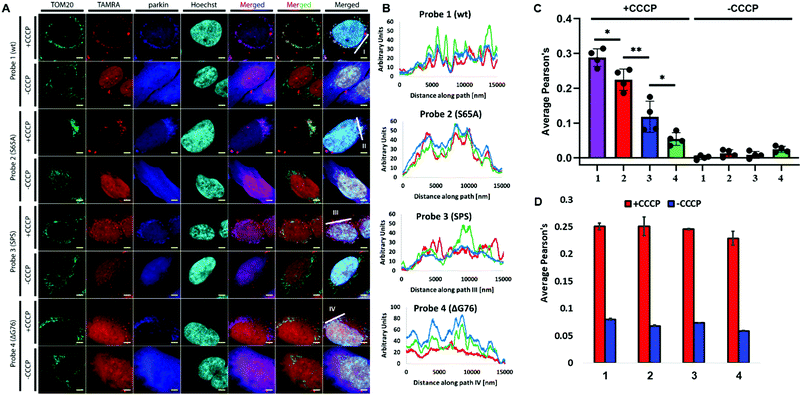
Probe 3 demonstrated diminished localization with TOM20, Parkin and HSP60 when compared to probes 1 and 2, which were massively recruited to these markers (Fig. 1 and 2). Notably, we could not detect any effect of our probes on the mitochondrial poly-ubiquitination pattern of endogenous Ub or co-localization of Parkin to the depolarized mitochondria, suggesting that they possibly cannot influence Parkin recruitment during mitophagy (Fig. S11 and S13, ESI†). Notably, in U2OS WT cells that do not express any endogenous Parkin (Fig. S1A, ESI†), depolarization induced substantial mitochondrial fragmentation without any co-localization between our probes and HSP60 due to the absence of mitophagy (Fig. 1B and Fig. S12D–F, ESI†).
To visualize Parkin's ability to incorporate our probes into poly-Ub chains, the localization of 1–3 was compared to that of 4. Immunostaining for TOM20 and Parkin revealed that probe 3 is marginally conjugated by Parkin. Conjugation of 3 occurred with substantially reduced efficiency compared to the unphosphorylated probes 1 and 2 (Fig. 2A–C). As expected, probe 4 did not exhibit any detectable incorporation by Parkin to the mitochondrial poly-Ub chains as evidenced by the lack of co-localization between 4 and Parkin or TOM20. Importantly, 1–4 did not have a visible effect on Parkin's recruitment as evidenced by the similar TOM20–Parkin co-localization in cells treated with these probes (Fig. 2D).
The ability to study post-translationally modified proteins in live cells, without drastic genetic manipulation, is a powerful approach to study a particular PTM in the cellular context. Although the preparation of modified proteins using chemical (semi)synthesis has reached advanced levels,27 tools for live cell delivery and preventing undesired enzymatic modifications are still needed.
Previous in vitro reports demonstrated that free pUb is not a preferred substrate for conjugation into poly(p)Ub chains by Parkin.28 This hypothesis is supported for the first time in the cellular environment by our results. Our results are also in line with the structural studies on the decreased preference of pUb monomers as a substrate for Parkin.29 Ub comes in two forms: extended and retracted. The overwhelming majority of free Ub exists in an extended form so that the RLRGG tail protrudes beyond the last beta strand in a flexible conformation. However, phosphorylation on position 65 stabilizes the retracted form so most of the Ub presents a much shorter C-terminus, which explains why free pUb is not conjugated efficiently by the ubiquitination enzymes (e.g. E1, E2 and Parkin) due to the limited accessibility of the Ub free C-terminus.30
In summary, we report for the first time, the preparation of a phosphatase stable and cell permeable pUb probe by combining the power of chemical protein synthesis and live cell delivery. We demonstrate the applicability of this probe to study how Ub phosphorylation at S65 affects its localization and conjugation by Parkin in human cells with minimal genetic manipulation. We envision that combining this approach with super resolution microscopy in neurodegenerative model systems (such as i-neurons31) without Parkin overexpression, could provide an additional understanding of Ub conjugation dynamics in mitophagy. In addition, stable phosphorylated Ub probes can find exciting applications in studying the role of additional phosphorylation sites, such as the recently reported one at Thr12 and its involvement in DNA damage.32
A. B. holds The Jordan and Irene Tark Academic Chair. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. [831783]) and is partially from grant 755/19 from the Israel Science Foundation (M. H. G.). P. S. received partial funding from a Technion Institute Cancer Center (TICC) fellowship. We are grateful to Dr Michael Lazarou of the Monash Biomedicine Discovery Institute for the pBMN Parkin plasmid.
Conflicts of interest
The authors declare that they have no known competing financial interests.Notes and references
- T. Hunter, Mol. Cell, 2007, 28, 730–738 CrossRef CAS PubMed.
- A. C. Conibear and Nat Rev, Chem, 2020, 4, 674–695 CAS.
- P. Siman and A. Brik, Org. Biomol. Chem., 2012, 10, 5684–5697 RSC.
- A. Hauser, M. Penkert and C. P. R. Hackenberger, Acc. Chem. Res., 2017, 50, 1883–1893 CrossRef CAS PubMed.
- P. Gopinath, S. Ohayon, M. Nawatha and A. Brik, Chem. Soc. Rev., 2016, 45, 4171–4198 RSC.
- N. D. Weikart and H. D. Mootz, ChemBioChem, 2010, 11, 774–777 CrossRef CAS.
- S. Eger, M. Scheffner, A. Marx and M. Rubini, J. Am. Chem. Soc., 2010, 132, 16337–16339 CrossRef CAS PubMed.
- G. Mann, G. Satish, R. Meledin, G. B. Vamisetti and A. Brik, Angew. Chem., Int. Ed., 2019, 58, 13540–13549 CrossRef CAS PubMed.
- Y. David, M. Vila-Perelló, S. Verma and T. W. Muir, Nat. Chem., 2015, 7, 394–402 CrossRef CAS PubMed.
- C. A. Ott, S. Shen, K. Yang, Z. Zhuang, W. Gui and J. S. Chung, J. Am. Chem. Soc., 2018, 140, 12424–12433 CrossRef PubMed.
- D. S. Hameed, A. Sapmaz, L. Gjonaj, R. Merkx and H. Ovaa, ChemBioChem, 2018, 19, 2553–2557 CrossRef CAS PubMed.
- A. E. Rabideau and B. L. Pentelute, ACS Cent. Sci., 2015, 1, 423–430 CrossRef CAS PubMed.
- D. Narendra, A. Tanaka, D. F. Suen and R. J. Youle, J. Cell Biol., 2008, 183, 795–803 CrossRef CAS PubMed.
- F. Koyano, K. Okatsu, H. Kosako, Y. Tamura, E. Go, M. Kimura, Y. Kimura, H. Tsuchiya, H. Yoshihara, T. Hirokawa, T. Endo, E. A. Fon, J. F. Trempe, Y. Saeki, K. Tanaka and N. Matsuda, Nature, 2014, 510, 162–166 CrossRef CAS PubMed.
- T. Wauer, M. Simicek, A. Schubert and D. Komander, Nature, 2015, 524, 370–374 CrossRef CAS PubMed.
- K. Okatsu, F. Koyano, M. Kimura, H. Kosako, Y. Saeki, K. Tanaka and N. Matsuda, J. Cell Biol., 2015, 209, 111–128 CrossRef CAS PubMed.
- K. E. Paleologou, A. W. Schmid, C. C. Rospigliosi, H. Y. Kim, G. R. Lamberto, R. A. Fredenburg, P. T. Lansbury, C. O. Fernandez, D. Eliezer, M. Zweckstetter and H. A. Lashuel, J. Biol. Chem., 2008, 283, 16895–16905 CrossRef CAS.
- R. Klingberg, J. O. Jost, M. Schümann, K. A. Gelato, W. Fischle, E. Krause and D. Schwarzer, ACS Chem. Biol., 2015, 10, 138–145 CrossRef CAS PubMed.
- J. Kang, H. X. Chen, S. Q. Huang, Y. L. Zhang, R. Chang, F. Y. Li, Y. M. Li and Y. X. Chen, Tetrahedron Lett., 2017, 58, 2551–2553 CrossRef CAS.
- V. P. Prasad, S. Wagner, P. Keul, S. Hermann, B. Levkau, M. Schäfers and G. Haufe, Bioorg. Med. Chem., 2014, 22, 5168–5181 CrossRef CAS PubMed.
- S. Mandal, G. Mann, G. Satish and A. Brik, Angew. Chem., Int. Ed., 2021, 60, 7333–7343 CrossRef CAS PubMed.
- A. F. L. Schneider, A. L. D. Wallabregue, L. Franz and C. P. R. Hackenberger, Bioconjugate Chem., 2019, 30, 400–404 CrossRef CAS.
- T. N. Nguyen, B. S. Padman, J. Usher, V. Oorschot, G. Ramm and M. Lazarou, J. Cell Biol., 2016, 215, 857–874 CrossRef CAS.
- N. Matsuda, S. Sato, K. Shiba, K. Okatsu, K. Saisho, C. A. Gautier, Y. S. Sou, S. Saiki, S. Kawajiri, F. Sato, M. Kimura, M. Komatsu, N. Hattori and K. Tanaka, J. Cell Biol., 2010, 189, 211–221 CrossRef CAS PubMed.
- A. Ordureau, J. A. Paulo, W. Zhang, T. Ahfeldt, J. Zhang, E. F. Cohn, Z. Hou, J. M. Heo, L. L. Rubin, S. S. Sidhu, S. P. Gygi and J. W. Harper, Mol. Cell, 2018, 70, 211–227 CrossRef CAS PubMed.
- N. C. Chan, A. M. Salazar, A. H. Pham, M. J. Sweredoski, N. J. Kolawa, R. L. J. Graham, S. Hess and D. C. Chan, Hum. Mol. Genet., 2011, 20, 1726–1737 CrossRef CAS.
- H. Sun and A. Brik, Acc. Chem. Res., 2019, 52, 3361–3371 CrossRef CAS PubMed.
- T. Wauer, K. N. Swatek, J. L. Wagstaff, C. Gladkova, J. N. Pruneda, M. A. Michel, M. Gersch, C. M. Johnson, S. M. Freund and D. Komander, EMBO J., 2015, 34, 307–325 CrossRef CAS PubMed.
- C. Gladkova, A. F. Schubert, J. L. Wagstaff, J. N. Pruneda, S. M. Freund and D. Komander, EMBO J., 2017, 36, 3555–3572 CrossRef CAS PubMed.
- C. Tang and W. P. Zhang, Biochemistry, 2020, 59, 26–33 CrossRef CAS PubMed.
- A. Ordureau, J. A. Paulo, J. Zhang, H. An, K. N. Swatek, J. R. Cannon, Q. Wan, D. Komander and J. W. Harper, Mol. Cell, 2020, 77, 1124–1142 CrossRef CAS PubMed.
- F. Walser, M. P. C. Mulder, B. Bragantini, S. Burger, T. Gubser, M. Gatti, M. V. Botuyan, A. Villa, M. Altmeyer, D. Neri, H. Ovaa, G. Mer and L. Penengo, Mol. Cell, 2020, 0, 1–14 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc04045f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |