Open Access Article
Open Access ArticleSARS-CoV-2 Mpro inhibition by a zinc ion: structural features and hints for drug design†
Deborah
Grifagni
 a,
Vito
Calderone
a,
Vito
Calderone
 abc,
Stefano
Giuntini
a,
Francesca
Cantini
abc,
Stefano
Giuntini
a,
Francesca
Cantini
 *abc,
Marco
Fragai
*abc,
Marco
Fragai
 *abc and
Lucia
Banci
*abc and
Lucia
Banci
 *abc
*abc
aMagnetic Resonance Center (CERM), University of Florence, via Sacconi 6, Sesto Fiorentino, 50019, Italy. E-mail: francesca.cantini@unifi.it; marco.fragai@unifi.it; lucia.banci@unifi.it
bDepartment of Chemistry “Ugo Schiff”, University of Florence, via della Lastruccia 3, Sesto Fiorentino, 50019, Italy
cConsorzio Interuniversitario Risonanze Magnetiche MetalloProteine (CIRMMP), via Sacconi 6, Sesto Fiorentino, 50019, Italy
First published on 16th July 2021
Abstract
Structural data on the SARS-CoV-2 main protease in complex with a zinc-containing organic inhibitor are already present in the literature and gave hints on the presence of a zinc binding site involving the catalytically relevant cysteine and histidine residues. In this paper, the structural basis of ionic zinc binding to the SARS-CoV-2 main protease has been elucidated by X-ray crystallography. The zinc binding affinity and its ability to inhibit the SARS-CoV-2 main protease have been investigated. These findings provide solid ground for the design of potent and selective metal-conjugated inhibitors of the SARS-CoV-2 main protease.
The SARS-CoV-2 main protease (SARS-CoV-2 Mpro), also referred to as 3-chymotrypsin-like protease (SARS-CoV-2 3CLpro) or nsp5 is a cysteine protease that hydrolyses viral polyproteins at several sites with a preference for the Leu-Gln (Ser, Ala, Gly) sequences.1 The protein exhibits 96% sequence identity and a very high structural similarity with the SARS-CoV Mpro protein.2,3 The enzyme represents one of the main drug-target candidates for covid-19 infection because it features a large and deep pocket at the active site and is crucial for viral replication.3–5 Presently more than 280 crystal structures of SARS-CoV-2 Mpro with covalently or non-covalently bound inhibitors or chemical fragments have been deposited into the protein data bank (PDB, https://www.rcsb.org/). The active form of the enzyme is a homodimer (67.6 kDa) where the N-terminal region of each monomer interacts with the Glu166 of the other, thus promoting and stabilizing binding of the substrate.3,4 The active sites in each monomer face away from one another and are formed by a Cys-His dyad (Cys 145 and His 41). The hydrolysis of the substrate takes place in two main steps named acylation and deacylation. During the first step, an acyl–enzyme complex is formed, via a covalent bond with the Sγ atom of Cys 145, with the release of a fragment of the substrate. During deacylation, the acyl–enzyme complex is released by the action of a water molecule, thus recovering the protease to its active state for a new catalytic cycle.6,7 Several inhibitors of SARS-CoV and SARS-CoV-2 Mpro have been designed that interfere with the formation of the acyl–enzyme complex due to the promotion of a covalent bond between the thiol group of the catalytic cysteine and the inihibitor.3,4,8
A similar approach has been exploited using zinc-coordinating inhibitors,9,10 where the zinc ion plays a role in stabilizing the protein–inhibitor complex through a tetrahedral coordination shared by the active site cysteine and histidine with two donor atoms of the inhibitor. The solved experimental structures9–11 however leave open interesting questions concerning the zinc binding ability of the protease in the cellular environment, even in the absence of a coordinating ligand, and the possible role of zinc ions in modulating its biological activity. Moreover, a recently published bioinformatics study12 stimulated our interest on the possible role of zinc in covid-19 infection and therapy.13
Zinc is second to iron as the most abundant transition metal ion in living organisms.14 In eukaryotes, 9–10% of proteins are zinc-binding proteins that depend on this metal ion to carry out their biological function. Zinc is involved in proliferation, cell signaling, differentiation, oxidative stress and immune response and many other cellular processes.15,16 It is well known that extracellular and intracellular zinc levels are tightly regulated in such a way that free zinc ions (Zn2+) represent a minimal fraction of total cellular zinc (∼0.0001%).17–20 Furthermore, zinc ions are not homogeneously distributed within the cells, with large differences among cellular compartments and organelles.21,22
Zinc has a key role in the signalling pathways of the innate and adaptive immune reactions.23,24 It has been reported that zinc is able to participate in many processes of defense against infections.25 Experimental and clinical evidence exists that assesses zinc as a direct antiviral, as well as a stimulant of antiviral immunity. For example, in vitro replication of the influenza virus is significantly decreased by the addition of the zinc ionophore pyrrolidine dithiocarbamate.26 Likewise, severe acute respiratory syndrome (SARS) coronavirus RdRp template binding and elongation was inhibited by zinc in Vero-E6 cells.27 Furthermore, zinc salts were shown to inhibit respiratory syncytial virus.28 Therefore, the supplementation of zinc has been proposed to treat viral infections. In particular, the role of zinc as an antiviral could be dual: on one side, zinc supplementation could improve the antiviral response and systemic immunity in patients with zinc deficiency,29 and, on the other side, zinc treatment could inhibit viral replication or infection-related symptoms.30
In this respect, advances in understanding the role of zinc in covid-19 pathogenesis require the characterization of the viral zincosome, and particularly how the different SARS-CoV-2 proteins interact with this metal ion.
Here, we provide an important piece of the puzzle by reporting the X-ray structure of SARS-CoV-2 Mpro both in the apo form and in complex with an isolated zinc ion, and an extensive biophysical analysis of the metal–protein interaction properties in solution.
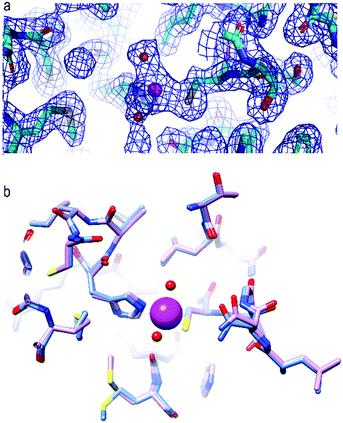
Crystals of apo SARS-CoV-2 Mpro were soaked for two days with 1.5 mM of the zinc ion solution (see the ESI† and Fig. S1 for experimental details). The apo protein structure discussed in this paper is totally superimposable with the others already deposited in the PDB along the entire sequence. The structure of Zn-bound SARS-CoV-2 Mpro (ESI,† Fig. S2) resembles that of the apo protein with average backbone and heavy atom RMSD values of 0.19 Å and 0.64 Å, respectively. Only the local loop regions showed backbone RMSD values higher than average (ESI,† Fig. S3). The electron density for the zinc ion is very well defined and the metal is present at full occupancy (Fig. 1a).
The zinc ion is coordinated by the sulfur atom of Cys145, the Nε atom of the imidazole ring of His41, a well-defined water molecule and a more labile one shuttling between two positions, thus completing a tetrahedral geometry. This is the first structure with a zinc ion bound to coronavirus Mpro (or homologous proteins in other viruses) as a cation, rather than as part of a coordination compound. Comparison of the present structure with that of apo SARS-CoV-2 Mpro shows that the residues involved in zinc binding do not undergo a significant structural rearrangement, supporting the idea that the binding site is ready to accommodate the metal (Fig. 1b and ESI,† Fig. S4).
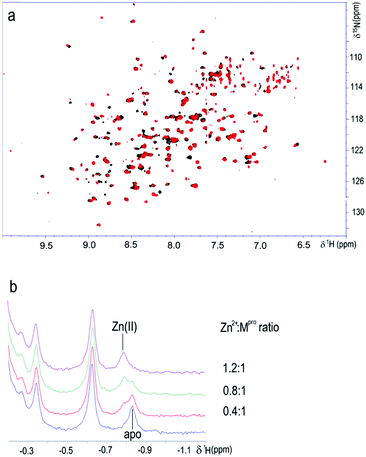
The interaction of SARS-CoV-2 Mpro with the zinc ion has been also investigated in solution using NMR. 15N isotopically enriched protein samples (at a concentration of 130 μM as the monomer) were titrated with the zinc ion and monitored using 1D 1H and 2D 1H–15N TROSY HSQC NMR spectroscopy (Fig. 2). Spectral changes were observed that indicate an intermediate-to-slow regime on the NMR time scale between the free and bound forms. Fig. 2b shows the methyl signal for both the apo- and metal-bound forms in a slow exchange regime. This observation indicates that the zinc ion binds SARS-CoV-2 Mpro in solution.
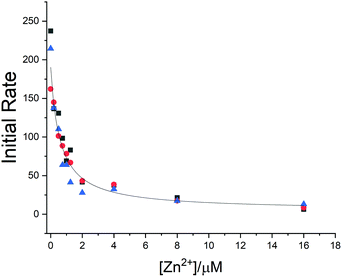
To estimate the affinity of the zinc ion for the protein and to investigate its effect on the proteolytic activity of the enzyme, a fluorimetric assay was carried out by monitoring the fluorescence increase due to the hydrolysis of the peptide substrate (Mca–AVLQ ↓ SGFR-K(Dnp)K).4 The addition of zinc ions inhibits the proteolytic activity of the enzyme, which is consistent with the interaction of zinc with the protein active site in solution. The fit of the kinetic data provided a Ki value of 0.58 ± 0.19 μM (see ESI,† Fig. S5 and Fig. 3).
The binding capability of SARS-CoV-2 Mpro towards the zinc ion is not completely unexpected as homologous proteases are reported to bind a zinc ion, not in its ionic form but as a metal-conjugated ligand. A comparison of the present structure with those of homologous proteases expressed by other viruses closely related to SARS-CoV-2 such as SARS-CoV (2Z9J, 2Z9K, 2Z9L, 2Z9G and 2Z94), Coxsackievirus B3 (2ZTX), HCoV-229E (2ZU2) and with the only report in the literature of a SARS-CoV-2 Mpro structure bound with zinc pyrithione (7B83) showed that, overall, the zinc binding site is well maintained with non-significant deviations from one structure to another (Fig. 4).
 | ||
| Fig. 4 Superposition of the zinc binding site in zinc bound SARS-COV-2 Mpro (light blue), 2Z94 (red), 2ZU2 (pink), 7B83 (green) and 2ZTX (gray). | ||
The zinc binding site has been also compared with that of other non-viral cysteine-proteases reported to bind zinc, such as cathepsin S both in the metal-bound and metal-free state (2HH5 and 2HHN), or enzymes such as dimethylarginine dimethylaminohydrolase I (2CI7). The structural features of the zinc binding site in these systems are very similar to those of SARS-CoV-2 Mpro.
The structural analysis and the inhibition activity of zinc towards SARS-CoV-2 Mpro suggest interesting considerations on how the metal ion and the viral protease could interplay in the cells. The affinity of the protein for zinc, although high, appears to be not sufficient to inactivate SARS-CoV-2 Mpro as almost all the intracellular zinc ion is bound to other proteins with similar or higher affinity. This is confirmed by the efficient viral replication and spreading of covid-19 infection and by the observation that proteins bearing the same metal binding site such as cathepsin S are highly active inside cells. For the same reasons, the potential use of zinc supplementation for SARS-CoV-2 Mpro inhibition does not seem viable. In particular, the presence of many zinc-binding proteins with similar or higher affinity would make a selective inhibition of SARS-CoV-2 Mpro by zinc supplementation unlikely. More promising for SARS-CoV-2 Mpro inhibition would be the design of suitable metallodrugs that incorporate zinc. A zinc-containing organic or peptidomimetic molecule, by simultaneously interacting with the catalytically relevant Cys-His dyad and with neighbouring additional sites or allosteric sites,11 could be the basis for the design of inhibitors with low nanomolar affinity for SARS-CoV-2 Mpro.31,32 This is not the case for zinc-pyrithione where the organic fragments stick-out from the protein surface without establishing any significant interaction with the protein (Fig. S6, ESI†). In contrast, the combination of the inhibitory effect of zinc with that of a molecule interacting with residues surrounding the catalytic dyad can be a strategy for the design of promising inhibitors. A similar approach has been exploited to develop inhibitors of zinc-dependent matrix metalloproteinases.33,34 In addition, our results suggest that drugs based on alternative metal ions, such as platinum or gold, could be investigated as potential SARS-CoV-2 Mpro inhibitors considering they are well established binders of cysteine thiolates.35,36
In summary, the present study highlights that a zinc ion inhibits SARS-CoV-2 Mpro by binding at the active site in a similar way to that observed for other cysteine proteases. The affinity of the protein for the metal ion has been experimentally determined and it does not seem high enough for any real therapeutic application of zinc supplementation, at least in its ionic state. However, our results suggest that a zinc ion coordinated to suitable ligands capable of interacting with additional sites on the protein surface could provide a significant increase in binding affinity, thus allowing the design of potent and more selective inhibitors of SARS-CoV-2 Mpro.
Instruct-ERIC, a landmark ESFRI project, and specifically the CERM/CIRMMP Italy centre are acknowledged. Moreover, the authors acknowledge H2020 – INFRAIA iNEXT-Discovery – Structural Biology Research Infrastructures for Translational Research and Discovery (contract no. 871037) and Fondazione Cassa di Risparmio di Firenze, project title “Dalla Struttura tridimensionale di antigeni proteici all’ottimizzazione di potenziali candidati vaccini”.
Conflicts of interest
There are no conflicts to declare.Notes and references
- W. Rut, K. Groborz, L. Zhang, X. Sun, M. Zmudzinski, B. Pawlik, X. Wang, D. Jochmans, J. Neyts, W. Mlynarski, R. Hilgenfeld and M. Drag, Nat. Chem. Biol., 2021, 17, 222–228 CrossRef CAS PubMed.
- K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Science, 2003, 300, 1763–1767 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao and H. Yang, Nature, 2020, 582, 289–293 CrossRef CAS PubMed.
- A. Douangamath, D. Fearon, P. Gehrtz, T. Krojer, P. Lukacik, C. D. Owen, E. Resnick, C. Strain-Damerell, A. Aimon, P. Abranyi-Balogh, J. Brandao-Neto, A. Carbery, G. Davison, A. Dias, T. D. Downes, L. Dunnett, M. Fairhead, J. D. Firth, S. P. Jones, A. Keeley, G. M. Keseru, H. F. Klein, M. P. Martin, M. E. M. Noble, P. O'Brien, A. Powell, R. N. Reddi, R. Skyner, M. Snee, M. J. Waring, C. Wild, N. London, F. von Delft and M. A. Walsh, Nat. Commun., 2020, 11, 5047 CrossRef CAS PubMed.
- J. Lee, L. J. Worrall, M. Vuckovic, F. I. Rosell, F. Gentile, A. T. Ton, N. A. Caveney, F. Ban, A. Cherkasov, M. Paetzel and N. C. J. Strynadka, Nat. Commun., 2020, 11, 5877 CrossRef CAS PubMed.
- J. Solowiej, J. A. Thomson, K. Ryan, C. Luo, M. He, J. Lou and B. W. Murray, Biochemistry, 2008, 47, 2617–2630 CrossRef CAS PubMed.
- C. A. Ramos-Guzman, J. J. Ruiz-Pernia and I. Tunon, ACS Catal., 2021, 11, 4157–4168 CrossRef CAS PubMed.
- C. C. Lee, C. J. Kuo, M. F. Hsu, P. H. Liang, J. M. Fang, J. J. Shie and A. H. Wang, FEBS Lett., 2007, 581, 5454–5458 CrossRef CAS PubMed.
- C. C. Lee, C. J. Kuo, T. P. Ko, M. F. Hsu, Y. C. Tsui, S. C. Chang, S. Yang, S. J. Chen, H. C. Chen, M. C. Hsu, S. R. Shih, P. H. Liang and A. H. Wang, J. Biol. Chem., 2009, 284, 7646–7655 CrossRef CAS PubMed.
- S. Günther, P. Y. A. Reinke, Y. Fernández-García, J. Lieske, T. J. Lane, H. M. Ginn, F. H. M. Koua, C. Ehrt, W. Ewert, D. Oberthuer, O. Yefanov, S. Meier, K. Lorenzen, B. Krichel, J. D. Kopicki, L. Gelisio, W. Brehm, I. Dunkel, B. Seychell, H. Gieseler, B. Norton-Baker, B. Escudero-Pérez, M. Domaracky, S. Saouane, A. Tolstikova, T. A. White, A. Hänle, M. Groessler, H. Fleckenstein, F. Trost, M. Galchenkova, Y. Gevorkov, C. Li, S. Awel, A. Peck, M. Barthelmess, F. Schlünzen, P. Lourdu Xavier, N. Werner, H. Andaleeb, N. Ullah, S. Falke, V. Srinivasan, B. A. França, M. Schwinzer, H. Brognaro, C. Rogers, D. Melo, J. J. Zaitseva-Doyle, J. Knoska, G. E. Peña-Murillo, A. R. Mashhour, V. Hennicke, P. Fischer, J. Hakanpää, J. Meyer, P. Gribbon, B. Ellinger, M. Kuzikov, M. Wolf, A. R. Beccari, G. Bourenkov, D. von Stetten, G. Pompidor, I. Bento, S. Panneerselvam, I. Karpics, T. R. Schneider, M. M. Garcia-Alai, S. Niebling, C. Günther, C. Schmidt, R. Schubert, H. Han, J. Boger, D. C. F. Monteiro, L. Zhang, X. Sun, J. Pletzer-Zelgert, J. Wollenhaupt, C. G. Feiler, M. S. Weiss, E. C. Schulz, P. Mehrabi, K. Karničar, A. Usenik, J. Loboda, H. Tidow, A. Chari, R. Hilgenfeld, C. Uetrecht, R. Cox, A. Zaliani, T. Beck, M. Rarey, S. Günther, D. Turk, W. Hinrichs, H. N. Chapman, A. R. Pearson, C. Betzel and A. Meents, Science, 2021, 372, 642–646 CrossRef PubMed.
- A. Pormohammad, N. K. Monych and R. J. Turner, Int. J. Mol. Med., 2021, 47, 326–334 CrossRef CAS PubMed.
- I. Wessels, B. Rolles and L. Rink, Front. Immunol., 2020, 11, 1712 CrossRef CAS PubMed.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, J. Proteome Res., 2006, 5, 3173–3178 CrossRef CAS PubMed.
- D. Beyersmann and H. Haase, Biometals, 2001, 14, 331–341 CrossRef CAS PubMed.
- D. D. Marreiro, K. J. Cruz, J. B. Morais, J. B. Beserra, J. S. Severo and A. R. de Oliveira, Antioxidants, 2017, 6 CrossRef CAS PubMed.
- R. A. Bozym, R. B. Thompson, A. K. Stoddard and C. A. Fierke, ACS Chem. Biol., 2006, 1, 103–111 CrossRef CAS PubMed.
- M. Malavolta, L. Costarelli, R. Giacconi, E. Muti, G. Bernardini, S. Tesei, C. Cipriano and E. Mocchegiani, Cytometry A, 2006, 69, 1043–1053 CrossRef PubMed.
- J. L. Vinkenborg, T. J. Nicolson, E. A. Bellomo, M. S. Koay, G. A. Rutter and M. Merkx, Nat. Methods, 2009, 6, 737–740 CrossRef CAS PubMed.
- D. J. Eide, Biochim. Biophys. Acta, 2006, 1763, 711–722 CrossRef CAS PubMed.
- D. Osman, M. A. Martini, A. W. Foster, J. Chen, A. J. P. Scott, R. J. Morton, J. W. Steed, E. Lurie-Luke, T. G. Huggins, A. D. Lawrence, E. Deery, M. J. Warren, P. T. Chivers and N. J. Robinson, Nat. Chem. Biol., 2019, 15, 241–249 CrossRef CAS PubMed.
- T. R. Young, M. A. Martini, A. W. Foster, A. Glasfeld, D. Osman, R. J. Morton, E. Deery, M. J. Warren and N. J. Robinson, Nat. Commun., 2021, 12, 1195 CrossRef CAS PubMed.
- M. Maywald, I. Wessels and L. Rink, Int. J. Mol. Sci., 2017, 18 CAS.
- J. R. de Jesus and T. de Araújo Andrade, Metallomics, 2020, 12, 1912–1930 CrossRef CAS PubMed.
- S. R. Hennigar and J. P. McClung, Am. J. Lifestyle Med., 2016, 10, 170–173 CrossRef PubMed.
- U. Noboru, O. Kunio, B. Toshio, Y. Bo and Y. Toshio, Antiviral Res., 2002, 56, 207–217 CrossRef.
- A. J. W. te Velthuis, S. H. E. van den Worm, A. C. Sims, R. S. Baric, E. J. Snijder and M. J. van Hemert, PLoS Pathog., 2010, 6, e1001176 CrossRef PubMed.
- R. O. Suara and J. E. Crowe, Antimicrob. Agents Chemother., 2004, 48, 783–790 CrossRef CAS PubMed.
- A. S. Prasad, J. Am. Coll. Nutr., 2009, 28, 257–265 CrossRef CAS PubMed.
- S. A. Read, S. Obeid, C. Ahlenstiel and G. Ahlenstiel, Adv. Nutr., 2019, 10, 696–710 CrossRef PubMed.
- S. Johnson, E. Barile, B. Farina, A. Purves, J. Wei, L.-H. Chen, S. Shiryaev, Z. Zhang, I. Rodionova, A. Agrawal, S. M. Cohen, A. Osterman, A. Strongin and M. Pellecchia, Chem. Biol. Drug Des., 2011, 78, 211–223 CrossRef CAS PubMed.
- A. Agrawal, S. L. Johnson, J. A. Jacobsen, M. T. Miller, L.-H. Chen, M. Pellecchia and S. M. Cohen, ChemMedChem, 2010, 5, 195–199 CrossRef CAS PubMed.
- I. Bertini, V. Calderone, M. Cosenza, M. Fragai, Y. M. Lee, C. Luchinat, S. Mangani, B. Terni and P. Turano, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5334–5339 CrossRef CAS PubMed.
- L. A. Alcaraz, L. Banci, I. Bertini, F. Cantini, A. Donaire and L. Gonnelli, J. Biol. Inorg. Chem., 2007, 12, 1197–1206 CrossRef CAS PubMed.
- L. Banci, I. Bertini, O. Blazevits, V. Calderone, F. Cantini, J. Mao, A. Trapananti, M. Vieru, I. Amori, M. Cozzolino and M. T. Carri, J. Am. Chem. Soc., 2012, 134, 7009–7014 CrossRef CAS PubMed.
- C. Zoppi, L. Messori and A. Pratesi, Dalton Trans., 2020, 49, 5906–5913 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: SARS-CoV-2 Mpro expression and purification, crystallographic data, NMR spectroscopy and enzymatic activity data. Apo and zinc bound SARS-CoV-2 Mpro structures have been deposited at the PDB (PDB ID codes: 7NXH and 7NWX, respectively). See DOI: 10.1039/d1cc02956h |
| This journal is © The Royal Society of Chemistry 2021 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: